Abstract
Methods for the preparation of DNA duplexes containing interstrand covalent cross-links may facilitate research in the fields of biochemistry, molecular biology, nanotechnology, and materials science. Here we report methods for the synthesis and isolation of DNA duplexes containing a site-specific, chemically-stable, reduced covalent interstrand cross-link between a guanine residue and an abasic site. The method uses experimental techniques and equipment that are common in most biochemical laboratories and inexpensive, commercially available oligonucleotides and reagents.
Keywords: Interstrand DNA cross-link, cross-link synthesis, abasic site, gel electrophoresis, reductive amination, post-synthetic oligonucleotide modification
1. Introduction
Covalent interstrand cross-links in duplex DNA have significant biological effects because they prevent the strand separation that is required for read-out of the genetic information stored in the nucleotide sequence of the double helix (1, 2). Interstrand cross-links can arise from environmental chemicals, anticancer drugs, and unavoidable endogenous processes (1, 3–9). The repair of interstrand cross-links is important in human health and disease, but the complex processes required for cellular recognition and repair of cross-links is not yet well understood (2, 10–13). In the fields of materials science, nanotechnology, and diagnostic medicine, interstrand cross-linking can be employed for the preparation of novel materials and for the sensitive, selective detection of particular DNA sequences (14–20).
Studies that employ cross-linked DNA in biology, medicine, biochemistry, and materials science will be facilitated by the development of practical methods for the preparation of duplexes that contain site specific, chemically-defined cross-links. We have recently characterized interstrand cross-links derived from abasic (Ap) sites in duplex DNA (9, 21–25). These cross-links may be biologically significant and, more relevant to this methods report, also may provide easy access to synthetic cross-linked DNA duplexes (26). Here we describe preparative methods for the synthesis and isolation of DNA duplexes containing site-specific, chemically-stable covalent cross-links resulting from a reductive amination reaction between a guanine residue and an Ap site. The process involves enzymatic generation of DNA duplexes containing a single Ap site at a defined location. This is accomplished by treatment of the corresponding 2’-deoxyuridine-containing duplex with uracil DNA glycosylase (UDG, Fig. 1) (27). Incubation of the Ap-containing duplex in buffered solution (pH 5) containing the reducing agent sodium cyanoborohydride generates the reduced dG-Ap cross-link (dG-Apred, Fig. 2) via a reductive amination reaction (22, 28). The chemical structure and location of the cross-link has been established in our previous studies (22). The cross-link attachment is chemically stable under physiologically-relevant conditions (8, 22, 25). The preparative method described here employs simple benchtop procedures and inexpensive commercially available oligonucleotides and chemical reagents. The method enables preparation of cross-linked duplexes of mixed sequence. Proximity enforced by the DNA duplex constrains cross-link formation to a single, defined guanine residue in the DNA sequence (Fig. 3). If desired, cross-linked duplexes can be readily separated from uncross-linked DNA using gel electrophoretic methods that are common in most biochemical laboratories. The separation exploits the markedly decreased migration of cross-linked duplexes relative to the component single-stranded oligonucleotides in denaturing gels (22, 29). The method can be used to generate nmol quantities of cross-linked duplexes for use in biochemical and structural applications.
Fig. 1.
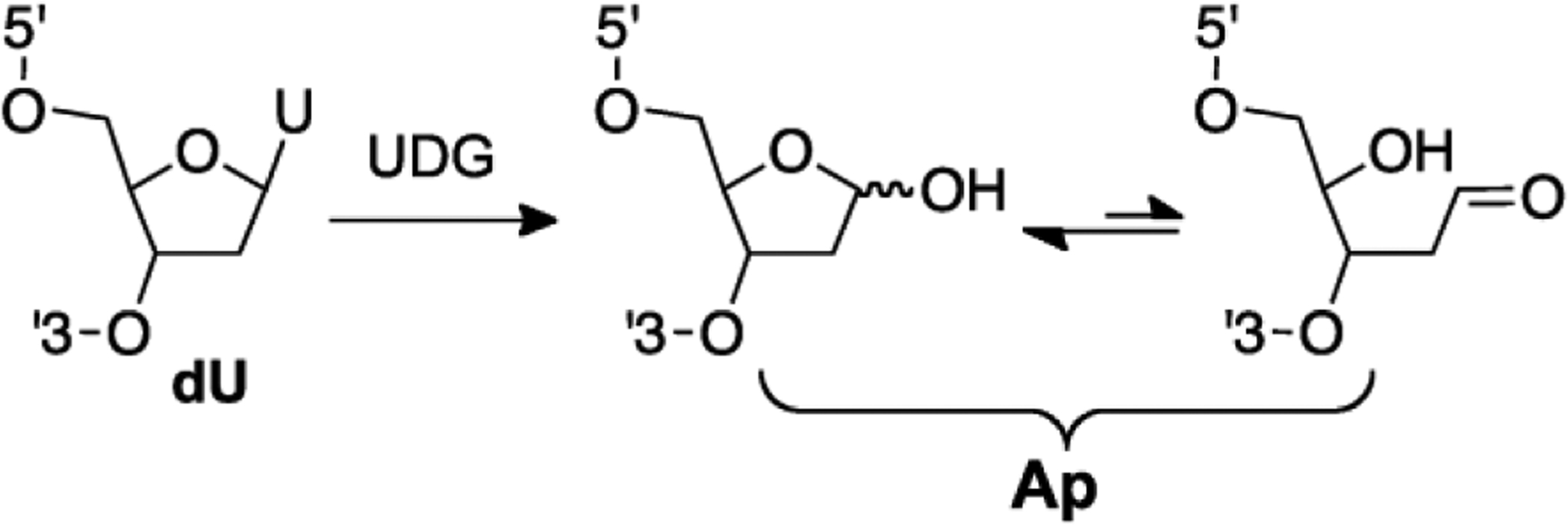
An abasic (Ap) site can be generated site specifically in duplex DNA by treatment of the corresponding 2’-deoxyuridine-containing duplex with the enzyme uracil DNA glycosylase (UDG).
Fig. 2.
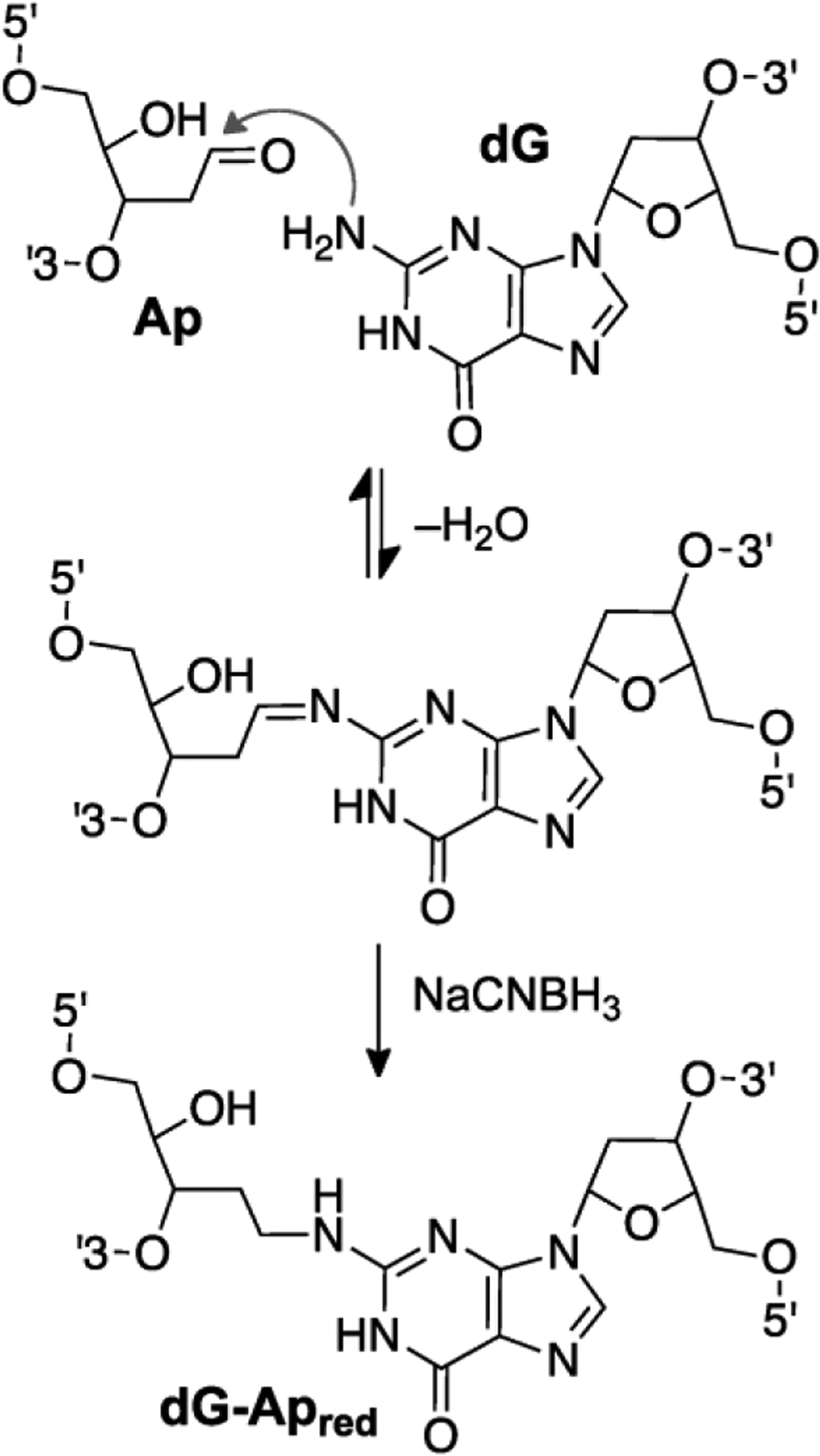
A reductive amination reaction generates a site-specific, chemically-stable, reduced interstrand cross-link between a guanine residue and an abasic (Ap) site in duplex DNA.
Fig. 3.
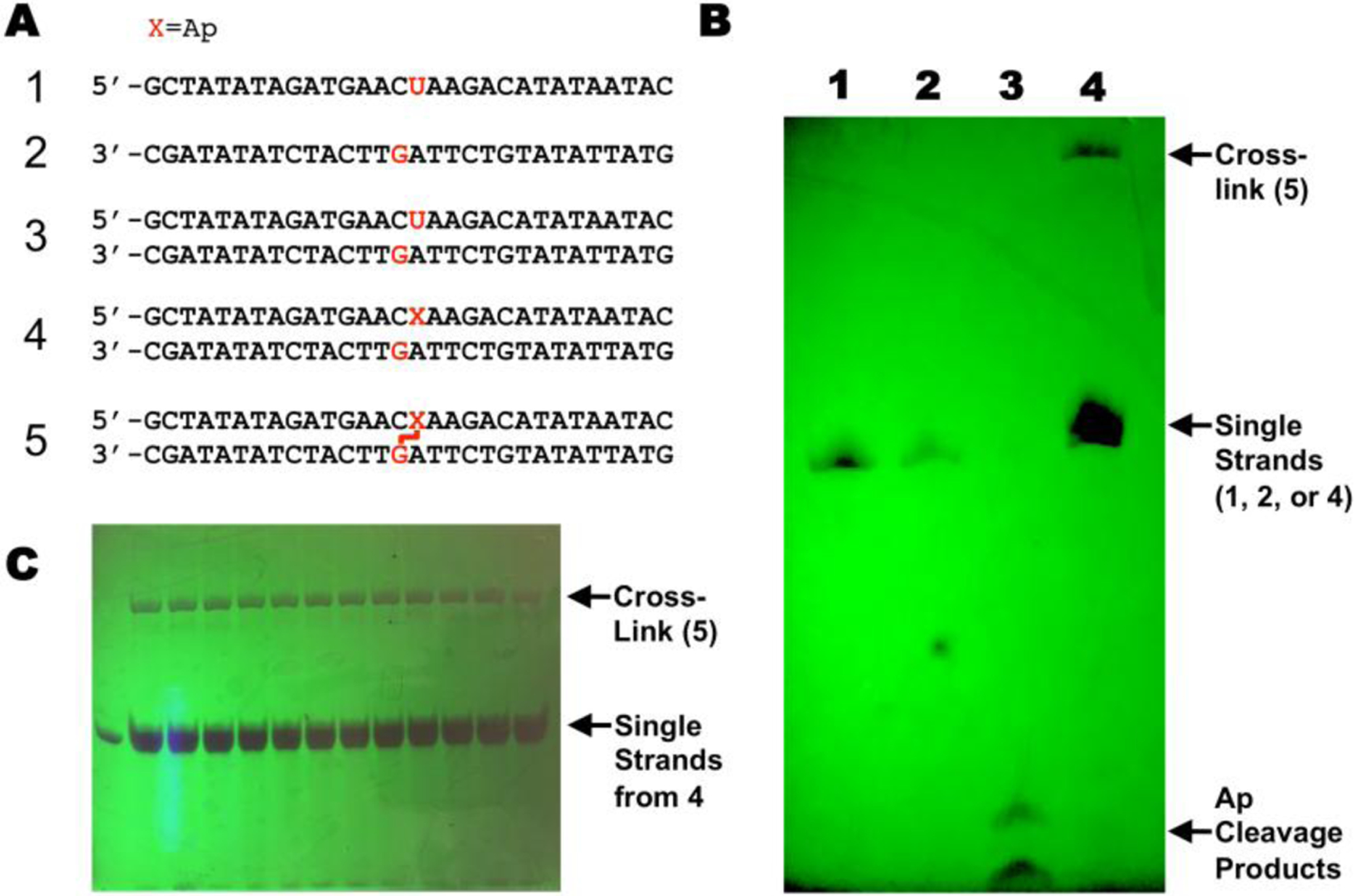
Generation of a covalent cross-link between a guanine residue and a reduced abasic site in duplex DNA. Panel A: DNA sequences used in this study (X = Ap site). Panel B: Photograph of UV-shadowed preparative denaturing polyacrylamide gel: Lane 1. Single strand 1; lane 2. Single strand 2; lane 3. Single strand 2 treated with piperidine (0.1 M, 90 °C, 30 min) to induce strand cleavage; lane 4. Cross-linking reaction mixture containing a mixture of duplexes 4 and 5. Panel C: Photograph of UV-shadowed preparative gel from which the cross-linked DNA was isolated.
2. Materials
2.1. Analytical detection of cross-link formation in 32P-labeled 2’-deoxyoligonucleotide duplexes
2.2. Materials for Preparative Cross-linking reaction
1 mM Oligodeoxynucleotide 1 dissolved in HPLC grade water
1 mM Oligodeoxynucleotide 2 in water
Uracil-DNA glycosylase “10×” reaction buffer (UDG buffer; New England Biolabs)
Uracil-DNA glycosylase enzyme (UDG, New England Biolabs)
25:24:1 Phenol-chloroform-isoamyl alcohol mixture
500 mM 3-(N-Morpholino)propanesulfonic acid, pH 7 (MOPS)
1 M Sodium chloride
3 M Sodium acetate, pH 5.2
2.5 M Sodium cyanoborohydride, freshly prepared
1.5 mL microcentrifuge tubes
Vortex-mixer
Thermostat-controlled oven-incubator set at 37 °C
Benchtop centrifuge
Benchtop centrifuge in 4 °C cold room
Speed-vac Concentrator
Thermostat-controlled heating block
2.3. Materials for Purification and Isolation of a Cross-linked Duplex Using Denaturing Polyacrylamide Gel Electrophoresis
20% Polyacrylamide solution (19:1 acrylamide/bis-acrylamide, 8 M urea)
N,N,N’,N’-Tetramethylethylenediamine (TEMED)
10% (w/v) Aqueous ammonium persulfate (for molecular biology, for electrophoresis)
Tris-borate-EDTA buffer (TBE, 89 mM tris-borate, 2 mM EDTA, pH 8.3) prepared by dilution of a 10× stock solution.
Formamide loading buffer composed of 17.75 M formamide (deionized), 0.01 M EDTA, and bromophenol blue dye (ACS reagent) in water (30).
Elution buffer (0.2 M NaCl, 0.001 M EDTA, pH 8)
Crushed dry ice
Water (HPLC grade)
Glass gel plates (16 × 19.7 cm) with 2 mm thick spacers and 12-well comb
1.5 and 2 mL microcentrifuge tubes
100 mL beaker and magnetic stir bar
20 mL disposable syringe and 18 gauge, 1.5 in needle
Electrophoresis power source
Vortex-mixer
All-Purpose Laboratory Wrap (clear, Saran-type wrap)
UV lamp and silica gel TLC plate impregnated with UV-254 fluorophore
Disposable, single edge razor blade
Glass rod (round tip, 5 mm × 15 cm)
Poly-Prep Chromatography column (spin column, Bio-Rad Laboratories)
Clinical centrifuge (swinging bucket type)
C-18 Sep-Pak cartridges (1 mL, 100 mg, Waters, cat. no.WAT023590)
Speed-vac Concentrator
Syringe-tip filter, Titan3 PES (polyethersulfone, ThermoFisher Scientific)
Vertical slab gel electrophoresis stand
3 M Sodium acetate, pH 5.2
Ethanol, 200 proof
Micro-90 concentrated cleaning solution
Precision wipes
Medium size binder clips (heavy duty office-type paper clips)
3. Methods
All manipulations were carried out at room temperature unless otherwise noted.
3.1. Preparation of the dU-Containing Oligonucleotide Duplex 3
Concentrations of single-strand oligonucleotides were determined using UV-vis absorbance and the extinction coefficient provided by the manufacturer (or calculated using the IDT oligo analyzer web app).
To a 1.5 mL microcentrifuge tube add 5 μL of oligonucleotide 1, 5 μL of oligonucleotide 2, 2.5 μL of 500 mM pH 7 MOPS buffer, 5 μL of 1 M NaCl and 32.5 μL water (final concentrations of 25 mM MOPS, and 100 mM NaCl and 5 nmols of DNA). In the method described here, twelve reactions each containing 5 nmols of DNA were run in parallel.
Mix for 5 s using a vortex-mixer.
Incubate in a thermostat-controlled aluminum heating block for 5 min at 95 °C.
Remove the aluminum block containing the reaction tube from the heating source and cool to room temperature overnight to allow annealing of the duplex.
3.2. Preparation of the Ap-Containing Oligonucleotide Duplex 4
Mix the solution containing the annealed dU-duplex for 30 s using a vortex-mixer.
Add 10 μL “10x” UDG buffer, 33 μL water and 7 μL UDG enzyme (35 U).
Mix for 10 s using a vortex-mixer.
Incubate for 1 h at 37 °C.
Add 100 μL of phenol:chloroform:isoamyl alcohol and vortex for a few seconds until the solution gets cloudy.
Centrifuge for 3 min at 15000 rpm using a benchtop microcentrifuge at 24 °C.
Remove the top aqueous layer containing the DNA using a micropipetter set to 100 μL.
Precipitate the DNA by addition of 10 μL of 3 M sodium acetate, pH 5.2 to the aqueous solution, followed by vortex mixing for 10 s.
Add 550 μL of cold absolute ethanol (–20 °C) and mix the solution for 10 s.
Place the sample on crushed dry ice for 1 h.
Centrifuge the sample for 1 h at 13300 rpm at 4 °C.
Remove the supernatant, being careful not to disturb the DNA pellet at the bottom of the tube.
Wash the pellet by addition of 90 μL of 8:2 ethanol:water and centrifuge for 20 min at 13300 rpm at 4 °C.
Remove the supernatant, being careful not to disturb the DNA pellet at the bottom of the tube.
Dry the Ap-containing oligonucleotide duplex 4 under vacuum in a Speed-vac concentrator for 2 to 3 min at room temperature (24 °C, see Notes 2 and 3).
3.3. Preparation of the Cross-linked Duplex 5
Add 33.5 μL water to the freshly prepared Ap-containing duplex 4.
Add to the microcentrifuge tube containing the Ap-duplex 4, 12.5 μL of 3 M sodium acetate, pH 5.2 and 5 μL of 2.5 M NaCNBH3 in water and vortex for 10 s (final concentrations: NaCNBH3 250 mM and sodium acetate 750 mM). See Notes 4 and 5.
Incubate the sample for 24 h at 37 °C.
Ethanol precipitate the DNA as described above in steps 8–15 above and store the sample dry at −20 °C until gel purification.
3.4. Gel Electrophoresis Set-Up
CAUTION: Gel electrophoresis involves the use of a high voltage power source and the toxic chemicals, acylamide and bis-acrylamide. Use a gel apparatus with proper safety designs after receiving proper safety training and take appropriate precautions when dispensing and working with acrylamide and bis-acrylamide.
Wash the glass gel plates and spacers with Micro-90 concentrated cleaning solution and rinse them well with DI water. Hold the plates on the edges and rinse them with absolute ethanol.
Dry the plates with Precision Wipes. Assemble the plates separated by spacers (a small amount of grease on the top and bottom of the spacers will keep them from moving during plate assembly), clamp the assembly together with binder clips, and add some grease to the corners to prevent leaking when the gel is poured.
In a 100 mL beaker equipped with a magnetic stir bar, pour 40 mL of 20% polyacrylamide solution containing 8 M urea (see Note 6).
Add TEMED (neat, 20 to 25 μL) to the stirred 100 mL beaker from step 3.
Add 200 μL of aqueous ammonium persulfate to the stirred 100 mL beaker from step 4 and stir for 1–2 min.
Holding the gel assembly at an incline, with the top of the assembly approximately 5–8 cm above the top, pour the mixture into the gel plate assembly.
Insert a 12-well comb between the plates assembly. Well size 5 mm wide × 9 mm high × 2 mm thickness.
Allow gel polymerization to occur (1.5 to 2 h at room temperature).
After polymerization is complete, remove the comb and bottom spacer slowly and flush the wells with 10 mL of 1×TBE in a 20 mL disposable syringe equipped with a disposable needle.
Mount the gel-plate assembly onto the gel stand according to manufacturer’s instructions (see Note 7). Fill the top and bottom buffer trays of the gel stand with 1× TBE.
Before loading the samples on the gel, electrophorese the gel for 30 min at 300 V.
3.5. Separation of Cross-linked Duplex 5 from Uncross-linked DNA Using Denaturing Gel Electrophoresis
This protocol describes the purification and isolation of approximately 5 nmol of cross-linked duplex 5 via denaturing polyacrylamide gel electrophoresis.
Add 20 μL formamide loading buffer to the dry cross-linked duplex 5 sample and vortex mix for 5 s.
Spin the sample to the bottom of the tube using a brief (3 s) spin in a benchtop microcentrifuge.
Before loading on the gel, heat the samples at 95 °C for 2 min to denature uncross-linked material.
Turn off the power supply and completely disconnect the gel apparatus from the power supply. Load the first sample into a well of the gel. Each reaction is loaded into a separate lane of the gel.
Flush the wells of the gel once again with 10 to 15 mL 1×TBE dispensed from a 20 mL disposable syringe equipped with a 18 gauge needle. Flushing the wells removes urea that diffuses from the gel. If the wells are not flushed, the samples in loading buffer will not sink properly to the bottom of the wells when loaded. Samples should be loaded soon after flushing the wells to ensure proper layering of the sample on the bottom of the well.
Load the sample into a well of the gel.
Rinse the sample tube with 10 μL formamide loading buffer and spin the sample to the bottom of the tube using a brief (3 s) spin in a benchtop microcentrifuge and load the rinse into the same lane as in step 5.
Repeat steps 6 and 7 to load each reaction into a separate well of the gel.
Carefully reconnect the gel apparatus to the power supply. Turn the power supply on and electrophorese the gel at 300 V.
Turn off the power supply and disconnect the gel from the power supply once the dye has traveled approximately 12 to 14 cm from the well (approximately 4 h).
Remove the plate assembly from the stand and separate the plates carefully and remove the gel from both plates.
3.6. Isolation of Cross-linked Duplex 5 from the Gel
Wrap the gel with plastic wrap and place on top of a large (20 × 20 cm) silica gel TLC plate containing UV-254 fluorophore.
Illuminate the gel from above with a handheld UV-254 lamp to “UV-shadow” the DNA bands in the gel (30). The DNA will appear as purple bands against the light green fluorescence background of the TLC plate. The cross-linked DNA will be located approximately 5 to 7 cm from the wells for the oligonucleotides used here (Fig. 3C, see Notes 8 and 9). Minimize the amount of time the DNA is exposed to the handheld UV light. UV light can cause DNA damage. Using a permanent marker, outline the location of the band on the plastic wrap covering the gel.
Cut the band from the gel with a disposable razor blade. Peel the plastic wrap from the gel slice and place the gel slice into a 2 mL microcentrifuge tube.
Crush the gel slice with the tip of a glass rod and add 0.3 mL elution buffer.
Agitate the sample utilizing a vortex mixer for at least 1 h.
Spin the sample through a Poly-prep column according to manufacturer’s instructions.
Ethanol precipitate the cross-linked duplex 5 as described above.
Resuspend the sample in 300 μL water and filter it through a Titan3 PES syringe filter to remove residual polyacrylamide.
Rinse the filter with 300 μL water.
Freeze the solution on crushed dry ice for 2 to 3 min.
Lyophilize the sample using a Speed-vac concentrator at room temperature (approximately 3 h).
Estimate the concentration of the cross-linked duplex when dissolved in aqueous solution using the extinction coefficient of the uncross-linked duplex 3 calculated by literature methods (31). The procedures described here, involving the isolation of DNA from twelve separate 5 nmol reactions, purified in twelve separate lanes of a preparative gel, yields approximately 9 nmols of cross-linked duplex (15% overall yield). See Notes 10–12.
4. Notes
The sequence of purchased oligonucleotides can be confirmed using ESI-TOF-MS (32) and Maxam-Gilbert sequencing reactions (33).
Following ethanol precipitation, care should be taken to dry oligonucleotides for only 2–3 min under vacuum at room temperature in the Speed-vac Concentrator. Excessive drying can dehydrate the DNA making it difficult (or impossible) to resuspend.
Heating the Ap-containing duplex during Speed-vac evaporation causes unwanted strand cleavage via β-elimination at the Ap site (34, 35).
Read the material safety data sheet (MSDS) for the reagent NaCNBH3 and take appropriate precautions. For example, avoid mixing NaCNBH3 with strong acid.
Cross-links can form at adenine residues in the absence of NaCNBH3 in the sequence 5’-ApT/AA (24). Our unpublished data indicates that incubation of duplexes containing the sequence 5’CApT/AAG, in the presence of NaCNBH3, generates a mixture of cross-linked duplexes (separable on a denaturing polyacrylamide sequencing gel) presumably including both the dA-Ap and dG-Apred cross-links. Such sequences should be avoided if generation of a single cross-linked species is desired.
Polyacrylamide is neurotoxic. Read the MSDS sheet and use appropriate safety precautions.
Gel electrophoresis involves the use of a high voltage power source. Use a gel apparatus with proper safety designs after receiving proper safety training.
It may be useful to load a sample of single-stranded oligonucleotide in one lane of the gel to provide a size marker (Fig. 3).
It may be useful to carry out a piperidine work-up on a small sample of the single-stranded Ap-oligonucleotide treated with UDG to ensure that the enzyme is active in the conversion of dU to Ap (Fig. 3B, lane 3). The cleavage products will migrate faster than full-length oligonucleotides.
Differences in flanking sequence can affect cross-link yields. Analytical gel measurements using 32P-labeled oligonucleotides indicate that the cross-link yields for duplexes that contain various bases flanking the 5’-CAp/AG cross-link site produce cross-linked DNA in yield ranging from 10–40%. The sequence employed here typically gives approximately 20% yield of cross-linked DNA using 32P-labeled oligonucleotides using NaCNBH3 under the conditions described here (22).
The dG-Apred cross-linkage can be generated in duplexes of about 20 base pairs and larger. Shorter Ap-containing duplexes may not be fully hybridized at room temperature and therefore may provide poor cross-link yields.
The cross-linked duplex obtained by this protocol displays temperature-dependent hyperchromicity consistent with melting of the helical regions flanking the cross-link. When the cross-linked duplex is annealed with two slow heat-cool (95 °C to room temp) cycles the melting curve such as that shown in Fig. 4 will be obtained. As expected (23, 36), the melting temperature of the cross-linked duplex is higher than the corresponding uncross-linked dU-duplex 3 or the Ap-duplex 4. Finally, consistent with the presence of double-helical character in the DNA flanking the cross-link, we have shown that a 21-base pair duplex insert containing a centrally-located dG-Apred cross-link can be enzymatically ligated into a linearized plasmid (25).
Vendors are identified for products where we believe that there could be significant differences between suppliers or proprietary technology makes it difficult to identify comparable products.
Fig. 4.
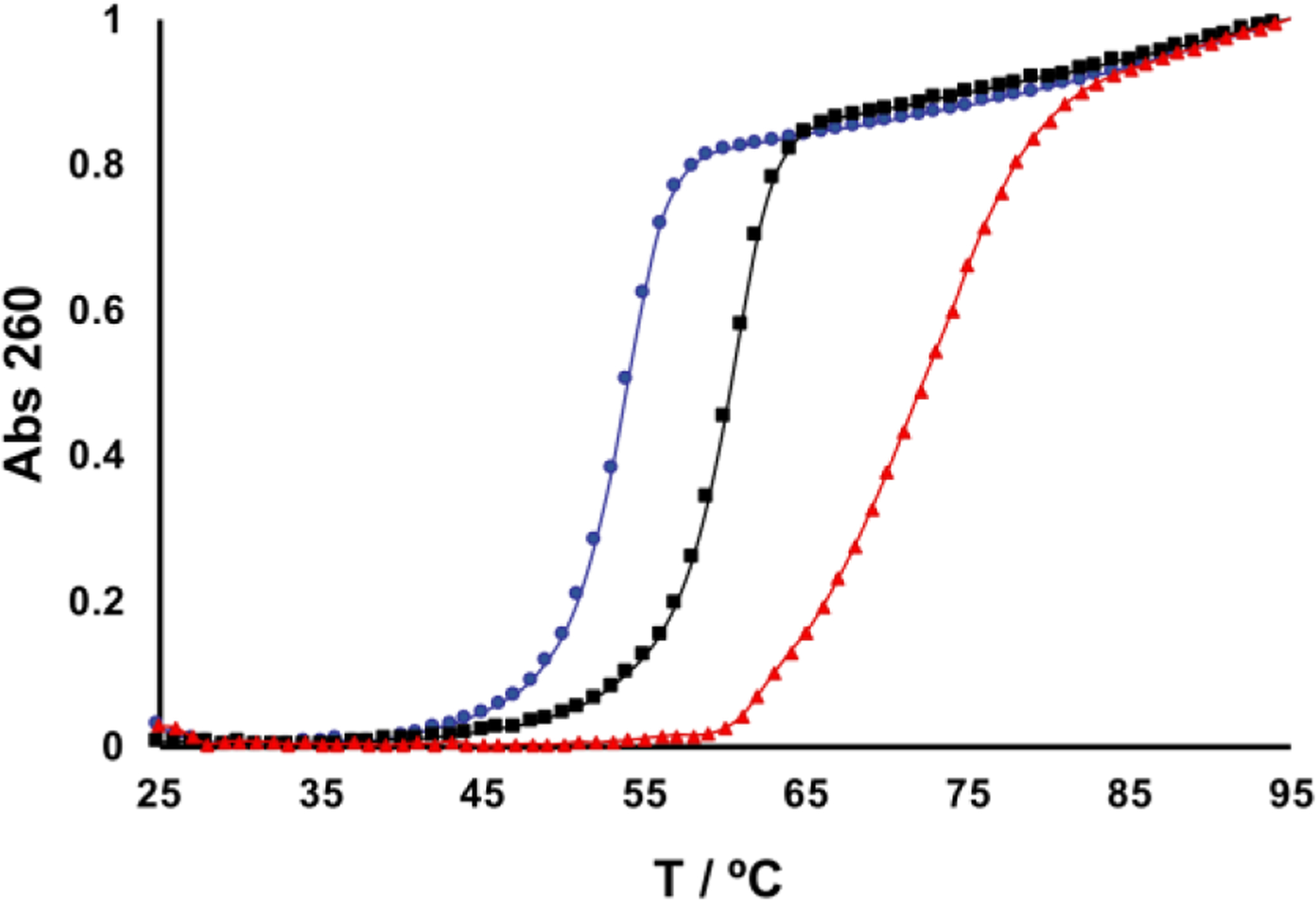
The cross-linked duplex 5 displays temperature-dependent hyperchromicity and melts at a substantially higher temperature than uncross-linked duplexes 3 and 4. Duplexes (1.4 μM) were dissolved in 50 mM HEPES (pH 7) containing 100 mM NaCl and the absorbance was measured while heating at 0.5 °C/min. The melting curves were normalized to set the starting absorbance to 0 and the final absorbance to 1. The total change in absorbance upon melting was similar for duplexes 3-5, at approximately 0.2 units. From left to right, the curves show melting of the Ap-duplex (blue, 4), the dU-duplex (black, 3), and the cross-linked duplex (red, 5).
Acknowledgement
We are grateful to the National Institutes of Health for supporting this work (ES021007)
References
- (1).Schärer OD (2005) DNA interstrand crosslinks: natural and drug-induced DNA adducts that induce unique cellular responses. ChemBioChem 6, 27–32. [DOI] [PubMed] [Google Scholar]
- (2).Clauson C, Schärer OD and Niedernhofer LJ (2013) Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harbor Perspectives in Biology 5, a012732/012731–a012732/012725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Imani-Nejad M, Johnson KM, Price NE and Gates KS (2016) A new cross-link for an old cross-linking drug: the nitrogen mustard anticancer agent mechlorethamine generates cross-links derived from abasic sites in addition to the expected drug-bridged cross-links. Biochemistry 55, 7033–7041. [DOI] [PubMed] [Google Scholar]
- (4).Stone MP, Cho YJ, Huang H, Kim HY, Kozekov ID, Kozekova A, Wang H, Minko IG, Lloyd RS, Harris TM and Rizzo CJ (2008) Interstrand cross-links induced by α,β-unsaturated aldehydes derived from lipid peroxidation and environmental sources. Acc. Chem. Res 41, 793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Greenberg MM (2014) Abasic and oxidized abasic site reactivity in DNA: enzyme inhibition, cross-linking, and nucleosome-catalyzed reactions. Acc. Chem. Res 47, 646–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Rajski SR and Williams RM (1998) DNA cross-linking agents as antitumor drugs. Chem. Rev 98, 2723–2795. [DOI] [PubMed] [Google Scholar]
- (7).Yang Z, Price NE, Johnson KM, Wang Y and Gates KS (2017) Interstrand cross-links arising from strand breaks at true abasic sites in duplex DNA. Nucleic Acids Res 45, 6275–6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Catalano MJ, Liu S, Andersen N, Yang Z, Johnson KM, Price NA, Wang Y and Gates KS (2015) Chemical structure and properties of the interstrand cross-link formed by the reaction of guanine residues with abasic sites in duplex DNA. J. Am. Chem. Soc 137, 3933–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Price NE, Catalano MJ, Liu S, Wang Y and Gates KS (2015) Chemical and structural characterization of interstrand cross-links formed between abasic sites and adenine residue in duplex DNA. Nucleic Acids Res 43, 3434–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kato N, Kawasoe Y, Williams HL, Coates E, Roy U, Shi Y, Beese LS, Schärer OD, Yan H, Gottesman ME, Takahashi TS and Gautier J (2017) Sensing and processing of DNA interstrand crosslinks by the mismatch repair pathway. Cell Rep 21, 1375–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Yang Z, Nejad MI, Gamboa Varela J, Price NE, Wang Y and Gates KS (2017) A role for the base excision repair enzyme NEIL3 in replication-dependent repair of interstrand cross-links derived from psoralen and abasic sites. DNA Repair 52, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Semlow DR, Zhang J, Budzowska M, Drohat AC and Walter JC (2016) Replication-dependent unhooking of DNA interstrand cross-links by the NEIL3 glycosylase. Cell 167, 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Huang J-C, Liu S, Bellani MA, Thazhathveetil AK, Ling C, de Winter JP, Wang Y, Wang W and Seidman MM (2013) The DNA translocase FANCM/MHF promotes replication traverse of DNA interstrand cross-links. Mol. Cell 52, 434–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Tomás-Gamasa M, Serdjukow S, Su M, Müller M and Carell T (2014) “Post-it” type connected DNA created with a reversible covalent cross-link. Angew. Chem. Int. Ed. Eng 53, 796–800. [DOI] [PubMed] [Google Scholar]
- (15).Imani-Nejad M, Shi R, Zhang X, Gu L-Q and Gates KS (2017) Sequence-specific covalent capture coupled with high-contrast nanopore detection of a disease-derived nucleic acid sequence. ChemBioChem 18, 1383–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Vieregg JR, Nelson HM, Stoltz BM and Pierce NA (2013) Selective nucleic acid capture with shielded covalent probes. J. Am. Chem. Soc 135, 9691–9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Peng X and Greenberg MM (2008) Facile SNP detection using bifunctional cross-linking oligonucleotide probes. Nucleic Acids Res 36, e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Fujimoto K, Yamada A, Yoshimura Y, Tsukaguchi T and Sakamoto T (2013) Details of the ultrafast DNA photo-cross-linking reaction of 3-cyanovinylcarbazole nucleoside: cis-trans isomeric effect and the application for SNP-based genotyping. J. Am. Chem. Soc 135, 16161–16167. [DOI] [PubMed] [Google Scholar]
- (19).Rajendran A, Endo M, Katsuda Y, Hidaka K and Sugiyama H (2011) Photo-cross-linking-assisted thermal stability of DNA origami structures and its application for higher-temperature self assembly. J. Am. Chem. Soc 133, 14488–14491. [DOI] [PubMed] [Google Scholar]
- (20).Chen W and Schuster GB (2013) Structural stabilization of DNA-templated nanostructures: cross-linking with 2,5-bis(2-thienyl)-pyrrole monomers. Org. Biomol. Chem 11, 35–40. [DOI] [PubMed] [Google Scholar]
- (21).Dutta S, Chowdhury G and Gates KS (2007) Interstrand crosslinks generated by abasic sites in duplex DNA. J. Am. Chem. Soc 129, 1852–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Johnson KM, Price NE, Wang J, Fekry MI, Dutta S, Seiner DR, Wang Y and Gates KS (2013) On the Formation and Properties of Interstrand DNA-DNA Cross-links Forged by Reaction of an Abasic Site With the Opposing Guanine Residue of 5’-CAp Sequences in Duplex DNA. J. Am. Chem. Soc 135, 1015–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Gamboa Varela J and Gates KS (2015) A Simple, High-Yield Synthesis of DNA Duplexes Containing a Covalent, Thermally-Reversible Interstrand Cross-link At a Defined Location Angew. Chem. Int. Ed. Eng 54, 7666–7669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Price NE, Johnson KM, Wang J, Fekry MI, Wang Y and Gates KS (2014) Interstrand DNA−DNA Cross-Link Formation Between Adenine Residues and Abasic Sites in Duplex DNA. J. Am. Chem. Soc 136, 3483–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Price NE, Li L, Gates KS and Wang Y (2017) Replication and repair of a reduced 2’-deoxyguanosine-abasic site cross-link in human cells. Nucleic Acids Res 45, 6486–6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Gamboa Varela J and Gates KS (2016) Simple, high-yield syntheses of DNA duplexes containing interstrand DNA-DNA cross-links between an N4-aminocytidine residue and an abasic site. Curr. Protoc. Nucleic Acid Chem 65, 5.16.11–15.16.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Varshney U and van de Sande JH (1991) Specificities and kinetics of uracil excision from uracil-containing DNA oligomers by Escherichia coli uracil DNA glycosylase. Biochemistry 30, 4055–4061. [DOI] [PubMed] [Google Scholar]
- (28).Borch RF and Hassid AI (1972) A new method for the methylation of amines. J. Org. Chem 37, 1673–1674. [Google Scholar]
- (29).Romero RM, Rojsittisak P and Haworth IS (2013) Electrophoretic mobility of duplex DNA cross-linked by mechlorethamine at a cytosine-cytosine mismatch pair. Electrophoresis 34, 917–924. [DOI] [PubMed] [Google Scholar]
- (30).Sambrook J, Fritsch EF and Maniatis T (1989) Molecular Cloning: A Lab Manual, Cold Spring Harbor Press, Cold Spring Harbor, NY. [Google Scholar]
- (31).Tataurov AV, You Y and Owczarzy R (2008) Predicting ultraviolet spectrum of single stranded and double stranded doexyribonucleic acids. Biophys. Chem 133, 66–70. [DOI] [PubMed] [Google Scholar]
- (32).Melton D, Lewis CD, Price NE and Gates KS (2014) Covalent adduct formation between the antihypertensive drug hydralazine and abasic sites in double- and single-stranded DNA. Chem. Res. Toxicol 27, 2113–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Maxam AM and Gilbert W (1980) Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol 65, 499–560. [DOI] [PubMed] [Google Scholar]
- (34).Gates KS (2009) An overview of chemical processes that damage cellular DNA: spontaneous hydrolysis, alkylation, and reactions with radicals. Chem. Res. Toxicol 22, 1747–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Gates KS, Nooner T and Dutta S (2004) Biologically relevant chemical reactions of N7-alkyl-2’-deoxyguanosine adducts in DNA. Chem. Res. Toxicol 17, 839–856. [DOI] [PubMed] [Google Scholar]
- (36).Shi Y-B and Hearst JE (1986) Thermostability of double-stranded deoxyribonucleic acids: effects of covalent additions of a psoralen. Biochemistry 25, 5895–5902. [DOI] [PubMed] [Google Scholar]


