Abstract
The transfer of ermA and ermC genes, the two most common resistance determinants of erythromycin resistance, was studied with Luria-Bertani broth in the absence of additional Ca2+ or Mg2+ ions. Fifteen human and five poultry isolates of Staphylococcus aureus, which were resistant to erythromycin but carried different genetic markers for erythromycin resistance, were used for conjugation. Since both the donors (Amps-Tetr) and recipients (Ampr-Tets) were resistant to erythromycin, the transconjugants were initially picked up as ampicillin- and tetracycline-resistant colonies. The resistance transfer mechanisms of the chromosomally located erythromycin rRNA methylase gene ermA and the plasmid-borne ermC gene were monitored by a multiplex PCR and gene-specific internal probing assay. Four groups of transconjugants, based upon the transfer of the ermA and/or ermC gene, were distinguished from each other by the use of this method. Selective antibiotic screening revealed only one type of transconjugant that was resistant to ampicillin and tetracycline. A high frequency of transfer (4.5 × 10−3) was observed in all of the 23 transconjugants obtained, and the direction of tetracycline and erythromycin resistance marker transfer was determined to be from poultry to clinical isolates. The transfers of the ermA and ermC genes were via transposition and transformation, respectively.
Staphylococcus aureus is an important cause of nosocomial as well as community-based infections. The use of antibiotics in humans, to treat infections, and in animals, to promote growth and prevent colonization by pathogenic bacteria, has led to an increased resistance among bacteria (2, 21, 25). The resistance often is transferable at interspecies and intergeneric levels (3, 18, 28). The relative ease with which bacteria become resistant to currently used antimicrobial agents is of concern to public health officials (8, 9, 31). The spread of resistance to antimicrobial agents in S. aureus is largely due to the acquisition of plasmids and/or transposons (19). Although transfer of resistance between staphylococcal strains in the laboratory has been shown to occur via transformation, transduction, and conjugation (6, 14, 15, 17, 35), only conjugative transfer appears to be significant in vivo (17, 35). In staphylococci, the conjugative transfer of resistance determinants is usually mediated by conjugative plasmids (5, 20, 38, 39) but has also been shown to occur in the absence of detectable conjugative plasmids (4). Conjugative plasmids, usually 35 to 50 kb (7), spread resistance determinants between species and genera (3, 17, 18, 28, 35). Besides transferring the resistance determinants, they can mobilize nonconjugative plasmids (5, 17), recombine with nonconjugative plasmids to form new plasmids (37), or acquire and transfer resistance transposons (36).
Studies with human staphylococcal strains indicate that Staphylococcus epidermidis is a reservoir of antibiotic resistance genes that can be transferred to S. aureus under in vitro and in vivo conditions (5, 10, 19, 20). Studies of drug resistance transfer between staphylococcal strains have been done mostly on human isolates; studies of transfer between animal and human staphylococcal strains are rare (16, 24), and little or no information is available about the transfer of drug resistance between avian and human staphylococci. In the context of the prevalent use of antibiotics in the poultry industry and the limited data about the role of poultry staphylococcal isolates in drug resistance transfer, it is pertinent to ask whether poultry isolates also contribute to the spread of antibiotic resistance in human staphylococcal strains. The objective of this study was to investigate the role of poultry S. aureus isolates in the dissemination of antibiotic resistance to human clinical S. aureus isolates in vitro and the development of a method to study the resistance transfer between bacterial strains resistant to the same antibiotic. In this study, a PCR-based method is reported that was used in combination with the selective antibiotic screening method to study the direction and mechanism of resistance transfer between poultry and human staphylococcal isolates, both of which were resistant to erythromycin but carried different genetic markers.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Erythromycin-resistant S. aureus strains were isolated from the hock joints and internal organs of diseased chickens. These were identified using the Automicrobic System (BioMerieux Vitek, Inc., Hazelwood, Mo.) and maintained as in-house stocks (27). The clinical S. aureus strains were either from the University of Arkansas for Medical Sciences, Little Rock, or from the Center for Food Safety and Applied Nutrition, Food and Drug Administration, Washington, D.C. All the isolates, which were highly resistant to erythromycin (MIC of >256 μg/ml), were stored in Luria-Bertani (LB) broth containing 20% glycerol at −70°C. Organisms were grown overnight at 37°C in LB broth or on tryptic soy agar plates supplemented with 5% sheep's blood (Remel, Lenexa, Kans.). The plasmids pEM9698 and pE194, both maintained in Bacillus subtilis, were used as controls for the detection of ermA and ermC genes, respectively.
Antibiotic resistance profile of bacterial strains.
The antibiotic resistance profiles of various S. aureus strains obtained from poultry and human sources were determined by the disk-diffusion assay method (1). The diameter of the inhibition zone was measured in triplicate and interpreted according to standards set by the National Committee for Clinical Laboratory Standards (26). Ampicillin, penicillin, streptomycin, tetracycline, erythromycin, lincomycin, azithromycin, and ciprofloxacin were used for determining the sensitivity of the above strains.
Transfer of antibiotic resistance in mixed liquid cultures.
Bacterial cultures were grown for 12 h in LB broth at 30°C (∼3 × 109 CFU/ml). Equal volumes (200 μl) of the poultry strains (resistant to tetracycline) and clinical S. aureus cultures (resistant to ampicillin and ciprofloxacin but sensitive to tetracycline and streptomycin) were mixed in an Eppendorf tube. The cultures were supplemented with 200 μl of LB broth and incubated at 37°C without shaking. Aliquots of 50 μl each were withdrawn after 6 h and plated in triplicate on tetracycline (30 μg/ml)- and ampicillin (100 μg/ml)-tetracycline (30 μg/ml)-containing hard agar plates after appropriate dilution. After 24 h, colonies resistant to ampicillin and tetracycline were picked up as transconjugants. The choice of these antibiotics was purposefully made so that the resistant colonies might represent the transconjugants where the transfer of ampicillin resistance from clinical to poultry strains or the transfer of tetracycline resistance from poultry to clinical strains had taken place. In either case, the transconjugants would be resistant to ampicillin and tetracycline. Although the transconjugants were initially picked up on these plates, the transfer mechanisms of the erythromycin resistance markers, ermA and ermC genes, were studied.
Isolation of DNA and gel electrophoresis.
The total DNA from the overnight-grown cultures was isolated by the method of Thakker-Varia et al. (33), and the plasmid DNA was isolated by other methods (13, 30). The electrophoresis of plasmid DNA, total DNA (chromosomal and plasmid DNA), or their EcoRI digestion products was carried out on a 1.0% agarose gel. The gels were run at 40 mA for 4 h, stained with ethidium bromide, and photographed.
Detection of ermA and ermC genes by PCR and gene-specific probing.
The ermA and ermC genes from the donors, recipients, and transconjugants were detected by multiplex PCR analysis using the gene-specific PCR primers (12). The transfer of these genes was studied by in-gel probing of the EcoRI-digested chromosomal DNA for the detection of ermA gene inserts or Southern blotting and hybridization of the total DNA for detection of the ermC gene by using gene-specific probes as described earlier (11, 12).
Frequency of transfer.
The transfer frequency of tetracycline resistance was calculated as the ratio of ampicillin- and tetracycline-resistant cells (average of three platings) to the total number of tetracycline-resistant cells (average of three platings).
RESULTS
Antibiotic resistance profiles of poultry, clinical, and transconjugant strains.
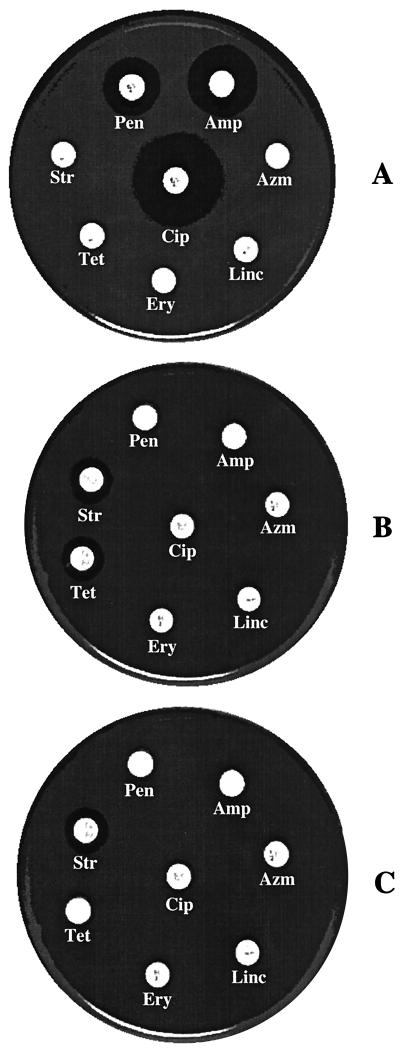
The poultry isolates were sensitive to ampicillin, penicillin, and ciprofloxacin but resistant to streptomycin, tetracycline, erythromycin, lincomycin, and azithromycin (Fig. 1A). The clinical strains were sensitive to streptomycin and tetracycline but resistant to the other six antibiotics (Fig. 1B). After mixed culture transfer experiments and plating on ampicillin (100 μg/ml)-tetracycline (30 μg/ml)-containing plates, 23 transconjugants were obtained. The resistance profiles of the transconjugants (Fig. 1C) were similar to those of the clinical strains (Fig. 1B), except that the transconjugants were resistant to tetracycline.
FIG. 1.
Antibiotic resistance and sensitivity profile of the donors, recipients, and transconjugants. The strains were tested against eight different antibiotics. The antibiotic resistance profiles of the representative strains are shown for poultry isolates (A), clinical isolates (B), and transconjugants (C). The antibiotics and concentrations were as follows: ampicillin (Amp; 10 μg/ml), penicillin (Pen; 10 U/ml), streptomycin (Str; 10 μg/ml), tetracycline (Tet; 30 μg/ml), erythromycin (Ery; 15 μg/ml), lincomycin (Linc; 2 μg/ml), azithromycin (Azm; 15 μg/ml), and ciprofloxacin (Cip; 5 μg/ml).
DNA profiles of the donors, recipients, and transconjugants.
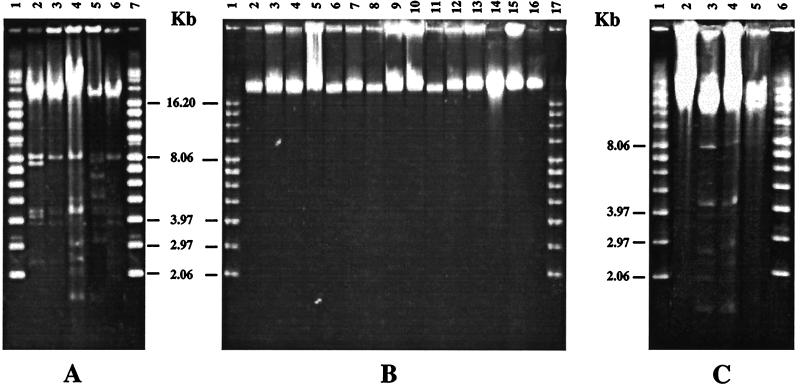
All of the poultry strains contained plasmids ranging from 1.6 to 8.0 kb in size (Fig. 2A). The clinical strains, on the other hand, contained no plasmids (Fig. 2B). DNA analysis of the transconjugants revealed that only 8 of the 23 transconjugants contained plasmids (Fig. 2C) and the other 15 did not (data shown for only two transconjugants). This observation suggested that there were at least two types of transconjugants, ones that had acquired the plasmid DNA and others that had not.
FIG. 2.
Total DNA profile of the poultry and clinical S. aureus isolates. Approximately 1 μg (5 μl) of total DNA was loaded on 1.0% agarose gels and electrophoresed. The gels were stained with ethidium bromide and photographed. The letters p and c followed by numbers represent poultry and clinical strains, respectively. (A) DNA from poultry isolates. Lanes 1 and 7, supercoiled DNA ladder; lane 2, p45; lane 3, p46; lane 4, p58; lane 5, p62; lane 6, p63. (B) DNA from clinical isolates. Lanes 1 and 17, supercoiled DNA ladder; lane 2, c16; lane 3, c18; lane 4, c29; lane 5, c656; lane 6, c657; lane 7, c660; lane 8, c661; lane 9, c712; lane 10; c714; lane 11, c716; lane 12, c720; lane 13, c722; lane 14, c772; lane 15, c796; lane 16, c803. (C) DNA from transconjugants. Lanes 1 and 6, supercoiled DNA ladder; lane 2, c716; lane 3, p58; lane 4, p58-c716; lane 5, p58-c803.
Multiplex PCR and the direction of resistance transfer.
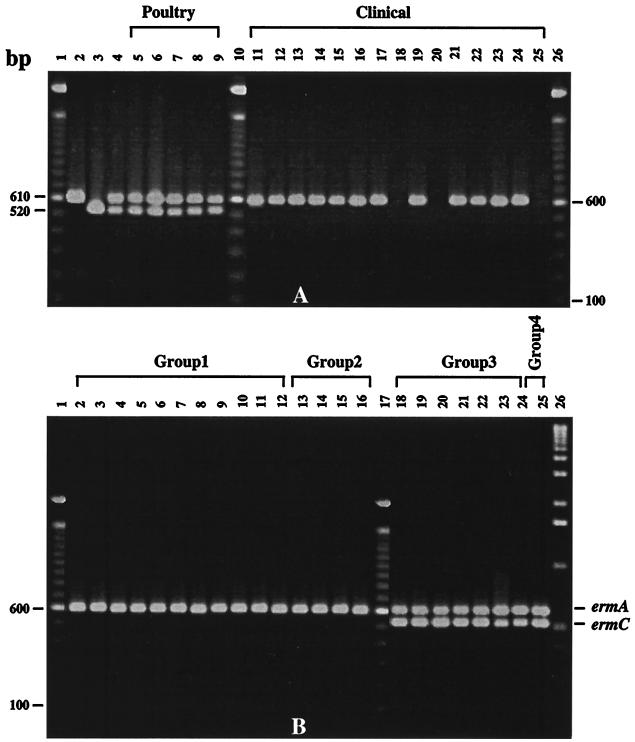
The antibiotic resistance and the DNA profiles of the transconjugants were helpful in screening and differentiating the transconjugants, but they did not reveal any information about the transfer of either ermA or the ermC gene. To determine whether the transfer of the chromosomally located ermA and the plasmid-borne ermC gene had taken place or not, the bacterial lysates from parents and transconjugants were subjected to multiplex PCR analysis to determine the presence of the two genes. The PCR revealed that the poultry strains had both the ermA and the ermC genes (Fig. 3A, lanes 5 to 9). The clinical strains, on the other hand, had only the ermA gene, except for strains 712, 716, and 803, which had neither ermA nor the ermC gene (Fig. 3A, lanes 18, 20, and 25). The transconjugants that had acquired the plasmid DNA had also acquired the ermC gene (Fig. 3B, lanes 18 to 25). The transconjugants that did not acquire the plasmid DNA were also missing the ermC gene (Fig. 3B, lanes 2 to 16). All the transconjugants, however, possessed the ermA gene (Fig. 3B, lanes 2 to 16). The above observations suggested that the transfer of these markers occurred from poultry to clinical isolates.
FIG. 3.
Multiplex PCR analysis of the donors, recipients, and transconjugants. PCR samples (5 μl) from different bacterial strains were loaded on a 1.0% agarose gel, and the gels were photographed after staining with ethidium bromide. (A) Lanes 1, 10, and 26, 100-bp DNA ladder; lane 2, ermA control from plasmid pEM9698; lane 3, ermC control from plasmid pE194; lane 4, ermA-ermC control; lane 5, p45; lane 6, p46; lane 7, p58; lane 8, p62; lane 9, p63; lane 11, c16; lane 12, c18; lane 13, c29; lane 14, c656; lane 15, c657; lane 16, c660; lane 17, c661; lane 18, c712; lane 19, c714; lane 20, c716; lane 21, c720; lane 22, c722; lane 23, c772; lane 24, c796; lane 25, c803. (B) Lanes 1, 17, and 26, 100-bp DNA ladder; lane 2, p45-c16; lane 3, p45-c18; lane 4, p45-c772; lane 5, p58-c657; lane 6, p62-c656; lane 7, p62-c657; lane 8, p62-c660; lane 9, p62-c661; lane 10, p62-c720; lane 11, p62-c722; lane 12, p63-c18; lane 13, p46-c716; lane 14, p58-c803; lane 15, p62-c712; lane 16, p63-c716; lane 18, p45-c29; lane 19, p45-c722; lane 20, p46-c714; lane 21, p46-c796; lane 22, p58-c18; lane 23, p58-c714; lane 24, p63-c29; lane 25, p58-c716. The transconjugants are identified as donor-recipient pairs. The transconjugants belonging to different groups are indicated. Designations beginning with “p” are for poultry strains; those beginning with “c” are for clinical strains.
Based upon the presence or absence of the ermA and/or ermC gene, the transconjugants were classified into four groups (Table 1). The transconjugants that belonged to group 1 possessed the ermA gene (Fig. 3B, lanes 2 to 12). The group 2 transconjugants also had the ermA gene (Fig. 3B, lanes 13 to 16), but the DNA-recipient parent strains did not have the ermA gene (Fig. 3A, lanes 18, 20, and 25). The third group of transconjugants showed the presence of both the ermA and the ermC genes (Fig. 3B, lanes 18 to 24). The only transconjugant strain that belonged to group 4 also had the ermA and the ermC genes (Fig. 3B, lane 25), but like group 2 transconjugants, the recipient parent did not have either of the two genes (Fig. 3A, lane 20).
TABLE 1.
Transfer frequency of tetracycline resistance and the presence of ermA and ermC genes in transconjugants
| Transconjugant | Transfer frequencya (10−3) | ermA | ermC |
|---|---|---|---|
| Group 1b | |||
| p45-c16 | 4.12 | + | − |
| p45-c18 | 4.56 | + | − |
| p45-c772 | 4.32 | + | − |
| p58-c657 | 4.51 | + | − |
| p62-c656 | 4.78 | + | − |
| p62-c657 | 4.66 | + | − |
| p62-c660 | 4.58 | + | − |
| p62-c661 | 4.90 | + | − |
| p62-c720 | 4.05 | + | − |
| p62-c722 | 4.02 | + | − |
| p63-c18 | 4.00 | + | − |
| Group 2c | |||
| p46-c716 | 4.22 | + | − |
| p58-c803 | 4.72 | + | − |
| p62-c712 | 4.85 | + | − |
| p63-c716 | 4.11 | + | − |
| Group 3d | |||
| p45-c29 | 4.32 | + | + |
| p45-c722 | 4.50 | + | + |
| p46-c714 | 4.35 | + | + |
| p46-c796 | 4.12 | + | + |
| p58-c18 | 4.22 | + | + |
| p58-c714 | 4.81 | + | + |
| p63-c29 | 4.10 | + | + |
| Group 4e | |||
| p58-c716 | 4.22 | + | + |
The transfer frequency represents the average of three plating experiments.
The recipients had multiple inserts of the ermA gene but acquired an extra copy of the gene.
The recipients had neither the ermA nor the ermC gene but acquired the ermA gene.
The recipients had ermA but acquired the ermC gene and an extra copy of the ermA gene.
The recipient acquired both ermA and ermC genes.
In-gel and Southern hybridization.
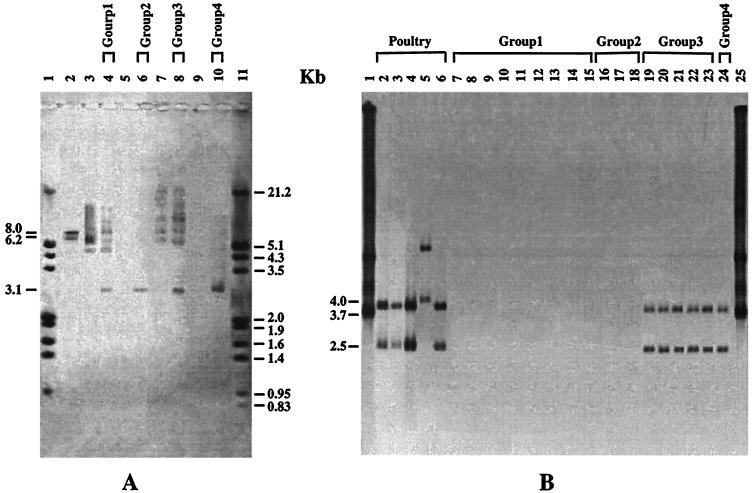
To determine the locations of the ermA and/or ermC gene, each group of transconjugants was probed with gene-specific internal probes for ermA and ermC genes. To determine the copy number of the ermA gene, EcoRI-digested chromosomal DNA molecules from the donors, recipients, and transconjugants were separated on agarose gels and subjected to in-gel probing with an ermA gene-specific internal oligonucleotide probe. The avian isolates (donors) had only two inserts, of 8.0 and 6.2 kb each (data shown for p58 only), which hybridized with the ermA gene-specific probe (Fig. 4A, lane 2), whereas the clinical isolates (recipients) had three to four copies (Fig. 4A, lanes 3 and 7). The recipients that did not test positive for the ermA gene by PCR also did not show any hybridization signal with the ermA gene-specific probe (Fig. 4A, lanes 5 and 9). All the transconjugants, however, had acquired an extra copy of the ermA gene (Fig. 4A, lanes 4, 6, 8, and 10) as 3.1-kb chromosomal DNA inserts (data shown for only one representative from each group of transconjugants).
FIG. 4.
In-gel and Southern hybridization. (A) EcoRI-digested DNAs of the donors, recipients, and transconjugants were probed with an ermA gene-specific internal oligonucleotide probe. Lanes 1 and 11, digoxigenin-labeled DNA molecular size marker III (Boehringer Mannheim); lane 2, p58; lane 3, c657; lane 4, p58-c657; lane 5, c803; lane 6, p58-c803; lane 7, c18; lane 8, p58-c18; lane 9, c716; lane 10, p58-c716. (B) Southern hybridization of the donor and transconjugant total DNA with an ermC gene-specific internal oligonucleotide probe. Lanes 1 and 25, control plasmid pE194 DNA; lane 2, p45; lane 3, p46; lane 4, p58; lane 5, p62; lane 6, p63; lane 7, p45-c16; lane 8, p45-c772; lane 9, p58-c657; lane 10, p62-c656; lane 11, p62-c660; lane 12, p62-c661; lane 13, p62-c720; lane 14, p62-c722; lane 15, p63-c18; lane 16, p46-c716; lane 17, p58-c803; lane 18, p62-c712; lane 19, p45-c29; lane 20, p45-c722; lane 21, p46-c796; lane 22, p58-c714; lane 23, p63-c29; lane 24, p58-c716. Designations are explained in the legend to Fig. 3.
To determine which plasmid harbored the ermC gene, the DNA gel was blotted onto a nylon membrane and probed with an ermC gene-specific internal oligonucleotide probe. The probe hybridized with a 2.5-kb plasmid in group 3 and 4 transconjugants (Fig. 4B, lanes 19 to 24), suggesting that the ermC gene was present on this plasmid only. The transconjugants belonging to groups 1 (Fig. 4B, lanes 7 to 15) and 2 (Fig. 4B, lanes 16 to 18) showed no hybridization signal with the ermC gene-specific internal probe. No hybridization signal was detected with the chromosomal DNA in any group of transconjugants (Fig. 4B, lanes 2 to 24).
Frequency and efficiency of transfer of genetic markers.
Based upon the antibiotic resistance phenotype and the PCR data, the transfer of tetracycline resistance and the two major determinants of erythromycin resistance, namely, ermA and ermC, occurred from poultry to clinical isolates. The efficiency of transfer was, however, different for different genetic markers. The transfer frequency of tetracycline resistance was calculated to be 4.5 × 10−3. Since all the 23 transconjugants that were resistant to tetracycline also acquired an extra copy of the ermA gene, it was assumed that the transfer frequency of the ermA gene was at least equal to that of tetracycline resistance. The ermC gene was present in only 8 of the 23 transconjugants, and therefore, its transfer frequency appears to be three times lower than that for ermA or tet.
DISCUSSION
The ampicillin and tetracycline resistance phenotype of the transconjugants could arise by the transfer either of ampicillin resistance from clinical to poultry strains or of tetracycline resistance from poultry to clinical isolates. Upon comparison, the antibiotic resistance and sensitivity profile of the transconjugants was found to be similar to that of the clinical strains, except that the transconjugants were resistant to tetracycline. The sensitivity of transconjugants to streptomycin and their resistance to ciprofloxacin suggested that they had more in common with the clinical isolates than with the poultry isolates. The clinical strains appeared to have acquired the tetracycline resistance from the poultry isolates. In the event of transfer from clinical to poultry isolates, the ampicillin, penicillin, and ciprofloxacin resistance has to have been transferred in order to explain the resistance and sensitivity profile of the transconjugants. Further, the sensitivity of the transconjugants to streptomycin could not be explained if the transfer of resistance(s) occurred from clinical to poultry strains. The direction of tetracycline resistance transfer was, therefore, determined to be from poultry to clinical S. aureus isolates.
The use of ampicillin and tetracycline as selective antibiotics was helpful in determining the direction of tetracycline resistance transfer but it was of no use in determining if the transfer of erythromycin resistance markers, ermA and ermC, between poultry and clinical strains had also occurred. Both the poultry and the clinical strains were resistant to high concentrations of erythromycin (MIC of >256 μg/ml). Based upon the total DNA analysis of the transconjugants, only two types of transconjugants, the ones that acquired the plasmids and the ones that did not, were obtained. The PCR amplification of the ermA and ermC genes, however, indicated four types of transconjugants. The hybridization of a 3.1-kb insert with the ermA gene-specific probe in all the transconjugants and the presence of the ermC gene bearing plasmids in group 3 and 4 transconjugants also confirmed the PCR data. The use of a selective antibiotic in determining the resistance transfer between the donors and the recipients that show resistance to the same antibiotic is of little or no use. The use of PCR and gene-specific probing along with the selective antibiotic screening for the transconjugants is not only useful in determining the direction of resistance transfer but also helpful in determining the mechanism of resistance transfer.
The transfer of drug resistance in mixed liquid cultures was initially thought to require phage mediation and Ca2+ or Mg2+ ions (34), but it was shown later on that it can also occur in the absence of the phage (6, 22). The absence of externally added metal ions and transducing phages in our experiments, however, excluded the possibility of phage-mediated conjugation and transduction and pointed toward a phage-independent transfer mechanism(s). The transfer of the chromosomally located ermA gene, which is known to be associated with transposon Tn554 (29), suggested the possibility of transposon-mediated drug resistance transfer. The presence of an extra 3.1-kb chromosomal DNA insert in all the transconjugants confirmed that the transposition of the ermA gene to a different site on the recipient's chromosome had taken place. Our results are similar to those of an earlier study (32) in which the transposition of chromosomal gene markers was observed, but unlike our observations, the transfer was achieved during filter mating only and not in mixed culture transfer experiments.
Although the transfer of the ermA gene was shown to occur via transposition, the transfer of the plasmid-based ermC gene could not be explained by this mechanism. If transposition was responsible for the mobilization of smaller plasmids, all the transconjugants would have acquired the ermC gene. Since the transposition of the ermA gene took place in all the transconjugants and only 8 of the 23 transconjugants acquired the ermC gene, transposition does not seem to be responsible for the transfer of ermC. The involvement of conjugative plasmids that are known to mobilize the smaller plasmids was also ruled out because of our repeated failure to isolate larger conjugative plasmids by known techniques (13, 30). In the absence of detectable conjugative plasmids and phage-mediated conjugation, transformation seems to be the most likely mechanism for the transfer of the ermC gene. In fact, in an earlier mixed liquid culture transfer study (23), the transfer of smaller plasmids bearing penicillin resistance was observed in the absence of Ca2+ and transformation was believed to be the mechanism of transfer. Since our results were also similar to those of the above study, the transfer of smaller plasmids is believed to take place via transformation.
The demonstration that the poultry S. aureus strains can transfer the resistance to human S. aureus strains indicates that the strains belonging to different ecosystems can contribute to the spread of antibiotic resistance genes. Up until now, the transfer of resistance was studied between donors and recipients that were resistant to different antibiotics. The data presented in this study clearly demonstrate the usefulness of PCR and gene-specific probing along with the conventional selective antibiotic screening method to study the transfer of drug resistance between organisms that are resistant to the same antibiotic but carry different genetic markers. The method presented here offers a unique approach to analyze the transconjugants and could be extended to other such systems. By the use of this procedure, the direction of resistance transfer was clearly established to be from avian to human isolates of S. aureus. The transfer of resistance was found to be via transposition and transformation.
ACKNOWLEDGMENTS
We gratefully acknowledge the receipt of plasmids pEM9698 and pE194 provided by E. Murphy, Department of Public Health, Research Institute of the City of New York, and B. Weisblum, University of Wisconsin, Madison. We also acknowledge the receipt of erythromycin-resistant clinical strains from F. Khambaty, Center for Food Safety and Applied Nutrition (CFSAN), Food and Drug Administration, Washington, D.C.
The work was supported by an appointment of S. Khan to the Postgraduate Research Program at the National Center for Toxicological Research and administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the U.S. Food and Drug Administration.
REFERENCES
- 1.Bauer A W, Kirby W M M, Sherris J C, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493–496. [PubMed] [Google Scholar]
- 2.Davies J. Inactivation of antibiotics and the dissemination of resistance genes. Science. 1994;264:375–382. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- 3.Dutka-Malen S, Leclercq R, Coulant V, Duval J, Courvalin P. Phenotypic and genotypic heterogeneity of glycopeptide resistance determinants in gram-positive bacteria. Antimicrob Agents Chemother. 1990;34:1875–1879. doi: 10.1128/aac.34.10.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El Solh N, Allignet J, Bismuth R, Buret B, Fouace J M. Conjugative transfer of staphylococcal antibiotic resistance markers in the absence of detectable plasmid DNA. Antimicrob Agents Chemother. 1986;30:161–169. doi: 10.1128/aac.30.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forbes B A, Schaberg D R. Transfer of resistance plasmids from Staphylococcus epidermidis to Staphylococcus aureus: evidence of conjugative exchange of resistance. J Bacteriol. 1983;153:627–634. doi: 10.1128/jb.153.2.627-634.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fouace J. Mixed cultures of Staphylococcus aureus: some observations concerning transfer of antibiotic resistance. Ann Inst Pasteur (Paris) 1981;132:375–386. [PubMed] [Google Scholar]
- 7.Gillespi M T, May J W, Skurray R A. Antibiotic susceptibilities and plasmid profiles of nosocomial methicillin-resistant Staphylococcus aureus: a retrospective study. J Med Microbiol. 1984;17:295–310. doi: 10.1099/00222615-17-3-295. [DOI] [PubMed] [Google Scholar]
- 8.Grabo W O K, Middendorf I G, Prozesky O W. Survival in maturation ponds of coliform bacteria with transferable drug resistance. Water Res. 1973;7:1589–1597. [Google Scholar]
- 9.Harada K, Mitsuhashi S. Physiology of R factors. In: Mitsuhashi S, editor. R factor, drug resistance plasmid. Baltimore, Md: University Park Press; 1977. pp. 135–160. [Google Scholar]
- 10.Jaffe H W, Sweeney H M, Nathan C, Weinstein R A, Kabins S A, Cohen S. Identity and interspecific transfer of gentamicin resistance plasmids in Staphylococcus aureus and Staphylococcus epidermidis. J Infect Dis. 1980;141:738–747. doi: 10.1093/infdis/141.6.738. [DOI] [PubMed] [Google Scholar]
- 11.Khan S A, Nawaz M S, Khan A A, Cerniglia C E. Direct in-gel hybridization of digoxigenin-labelled non-radioactive probes. Mol Cell Probes. 1999;13:233–237. doi: 10.1006/mcpr.1999.0241. [DOI] [PubMed] [Google Scholar]
- 12.Khan S A, Nawaz M S, Khan A A, Cerniglia C E. Simultaneous detection of erythromycin-resistant methylase genes ermA and ermC by multiplex-PCR in Staphylococcus sp. Mol Cell Probes. 1999;13:381–387. doi: 10.1006/mcpr.1999.0265. [DOI] [PubMed] [Google Scholar]
- 13.Labigne-Roussel A, Gerbaud G, Courvalin P. Translocation of sequences encoding antibiotic resistance from the chromosome to a receptor plasmid in Salmonella ordonez. Mol Gen Genet. 1981;182:390–408. doi: 10.1007/BF00293927. [DOI] [PubMed] [Google Scholar]
- 14.Lacey R W. Antibiotic resistance plasmids of Staphylococcus aureus and their clinical importance. Bacteriol Rev. 1975;39:1–32. doi: 10.1128/br.39.1.1-32.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lacey R W. Evidence for two mechanisms of plasmid transfer in mixed cultures of Staphylococcus aureus. J Gen Microbiol. 1980;119:423–435. doi: 10.1099/00221287-119-2-423. [DOI] [PubMed] [Google Scholar]
- 16.Lacey R W. Rarity of gene transfer between animal and human isolates of Staphylococcus aureus in vitro. J Gen Microbiol. 1980;119:437–442. doi: 10.1099/00221287-119-2-437. [DOI] [PubMed] [Google Scholar]
- 17.Lacey R W. Antibiotic resistance in Staphylococcus aureus and streptococci. Br Med Bull. 1984;40:77–83. doi: 10.1093/oxfordjournals.bmb.a071951. [DOI] [PubMed] [Google Scholar]
- 18.Leclercq R, Derlot E, Weber M, Duval J, Courvalin P. Transferable vancomycin and teicoplanin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1989;33:10–15. doi: 10.1128/aac.33.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyon B R, Skurray R A. Antimicrobial resistance of Staphylococcus aureus: genetic basis. Microbiol Rev. 1987;51:88–134. doi: 10.1128/mr.51.1.88-134.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDonnell R W, Sweeney H M, Cohen S. Conjugal transfer of gentamicin resistance plasmids intra- and interspecifically in Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 1983;23:151–160. doi: 10.1128/aac.23.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGowan J E., Jr Abrupt changes in antibiotic resistance. J Hosp Infect. 1991;18:202–210. doi: 10.1016/0195-6701(91)90025-4. [DOI] [PubMed] [Google Scholar]
- 22.Meijers J A, Winkler K C, Stobberingh E E. Resistance transfer in mixed cultures of Staphylococcus aureus. J Med Microbiol. 1981;14:21–39. doi: 10.1099/00222615-14-1-21. [DOI] [PubMed] [Google Scholar]
- 23.Mitra M, Mukherjee M, Gupta M S, Chakrabarty A N. Phage-mediated conjugation in staphylococci as a means of transfer of drug-resistance. Indian J Exp Biol. 1995;33:505–508. [PubMed] [Google Scholar]
- 24.Muhammad G, Hoblet K H, Jackwood D J, Nielsen S B, Smith K L. Interspecific conjugal transfer of antibiotic resistance among staphylococci isolated from the bovine mammary gland. Am J Vet Res. 1993;54:1432–1440. [PubMed] [Google Scholar]
- 25.Munoz P, Diaz M D, Rodriguez-Creixems M, Cercenado E, Pelaez T, Bouza E. Antimicrobial resistance of Salmonella isolates in a Spanish hospital. Antimicrob Agents Chemother. 1993;36:1200–1202. doi: 10.1128/aac.37.5.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. 6th ed. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 27.Nawaz M S, Khan A A, Khan S A, Paine D D, Pothuluri J V, Cerniglia C E. Biochemical and molecular characterization of erythromycin resistant Staphylococcus spp. isolated from diseased chicken. Poult Sci. 1999;78:1191–1197. doi: 10.1093/ps/78.8.1191. [DOI] [PubMed] [Google Scholar]
- 28.Noble W C, Virani Z, Cree R G. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol Lett. 1992;72:195–198. doi: 10.1016/0378-1097(92)90528-v. [DOI] [PubMed] [Google Scholar]
- 29.Philipps S, Novick R P. Tn554—a site specific repressor-controlled transposon in Staphylococcus aureus. Nature. 1979;278:476–478. doi: 10.1038/278476a0. [DOI] [PubMed] [Google Scholar]
- 30.Portnoy D A, Moseley S L, Falkow S. Characterization of plasmids and plasmid-associated determinants of Yersinia enterocolitica pathogenesis. Infect Immun. 1981;31:775–782. doi: 10.1128/iai.31.2.775-782.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaw D R, Cabelli V J. R plasmid transfer frequencies from environmental isolates of Escherichia coli to laboratory and fecal strains. Appl Environ Microbiol. 1980;40:756–764. doi: 10.1128/aem.40.4.756-764.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stout V G, Iandolo J J. Chromosomal gene transfer during conjugation by Staphylococcus aureus is mediated by transposon-facilitated mobilization. J Bacteriol. 1990;172:6148–6150. doi: 10.1128/jb.172.10.6148-6150.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thakker-Varia S, Jenssen W D, Moon-McDermott L, Weinstein M P, Dubin D T. Molecular epidemiology of macrolide-lincosamide-streptogramin B resistance in Staphylococcus aureus and coagulase-negative staphylococci. Antimicrob Agents Chemother. 1987;31:735–743. doi: 10.1128/aac.31.5.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Townsend D E, Bolton S, Ashdown N, Taheri S, Grubb W B. Comparison of phage-mediated and conjugative transfer of staphylococcal plasmids in vitro and in vivo. J Med Microbiol. 1986;22:107–114. doi: 10.1099/00222615-22-2-107. [DOI] [PubMed] [Google Scholar]
- 35.Townsend D E, Hollander D L, Bolton S, Grubb W B. Clinical isolates of staphylococci conjugate on contact with dry absorbent surfaces. Med J Aust. 1986;144:166. doi: 10.5694/j.1326-5377.1986.tb112260.x. [DOI] [PubMed] [Google Scholar]
- 36.Udo E E, Grubb W B. Excision of a conjugative plasmid from the Staphylococcus chromosome. J Med Microbiol. 1990;31:207–212. doi: 10.1099/00222615-31-3-207. [DOI] [PubMed] [Google Scholar]
- 37.Udo E E, Grubb W B. Transfer of resistance determinants from a multi-resistant Staphylococcus aureus isolate. J Med Microbiol. 1991;35:72–79. doi: 10.1099/00222615-35-2-72. [DOI] [PubMed] [Google Scholar]
- 38.Udo E E, Grubb W B. Transfer of plasmid-borne resistance from a multiply resistant Staphylococcus aureus isolate, WBG1022. Curr Microbiol. 1995;31:71–76. doi: 10.1007/BF00294278. [DOI] [PubMed] [Google Scholar]
- 39.Yu L, Baldwin J N. Intraspecific transduction in Staphylococcus epidermidis and interspecific transduction between Staphylococcus aureus and Staphylococcus epidermidis. Can J Microbiol. 1971;17:767–773. doi: 10.1139/m71-122. [DOI] [PubMed] [Google Scholar]