Abstract
Human cytomegalovirus (HCMV) infection was monitored retrospectively by qualitative determination of immediate-early (IE) mRNA by nucleic acid sequence-based amplification (NASBA) in a series of 51 bone marrow transplant (BMT) recipients. The qualitative results for IE mRNA obtained by NASBA were compared with those obtained by prospective quantitation of HCMV viremia and antigenemia and retrospective quantitation of DNA in blood (DNAemia) by PCR as well as by qualitative determination of late pp67 mRNA by NASBA. On the whole, of the 39 HCMV-positive patients (all asymptomatic), HCMV was detected in 14 (35.9%) by quantitation of viremia, 15 (38.5%) by detection of pp67 mRNA by NASBA, 32 (82.1%) by quantitation of DNAemia, and 33 (84.6%) by quantitation of antigenemia, while HCMV was detected in 38 (97.4%) patients by detection of IE mRNA by NASBA. In the immunocompetent host, IE mRNA was not detected by NASBA in 100 blood donors or during reactivated infections in 30 breast-feeding mothers. Likewise, NASBA did not detect IE mRNA in 56 solid-organ transplant recipients in the first 21 days after transplantation. By using NASBA for detection of IE mRNA as the reference standard for detection of HCMV infection in blood samples, the diagnostic sensitivities were 67.7% for quantitation of DNAemia, 59.0% for quantitation of antigenemia, 18.3% for detection of pp67 mRNA by NASBA, and 16.0% for quantitation of viremia. Specificities and negative and positive predictive values were >90.0, >70.0, and >80.0%, respectively, for all four assays. The mean times to first HCMV detection after bone marrow transplantation were 37.7 ± 15.4 days for detection of IE mRNA by NASBA, 39.6 ± 15.6 days for quantitation of antigenemia, 40.9 ± 15.2 days for quantitation of DNAemia, and 43.7 ± 16.3 or 43.7 ± 17.5 days for quantitation of viremia and detection of pp67 mRNA by NASBA, respectively. On the whole, 31 BMT recipients received preemptive therapy by using confirmed antigenemia positivity as a cutoff, while 35 patients could have been treated by using NASBA positivity as a cutoff and 31 could have been treated by using quantitation of DNAemia as a cutoff. In single patients, IE mRNA was detected in every episode of active HCMV infection, mostly preceding and sometimes accompanying antigenemia and DNAemia, whereas pp67 mRNA was detected only concomitantly with the highest peaks of infection. HCMV IE mRNA detection may represent a useful parameter for initiation of preemptive therapy in BMT recipients.
Among different assays developed and introduced in the last decade in diagnostic virology laboratories for detection and monitoring of human cytomegalovirus (HCMV) infections in different patient populations who have received transplants, qualitative and quantitative determinations of HCMV antigenemia (34, 40) and DNA in blood (DNAemia) (7, 10, 13, 15, 16, 38) in peripheral blood leukocytes (PBLs) have been widely used for preemptive (presymptomatic) therapy (21, 27, 31, 36). However, since viral products detected in PBLs by different diagnostic assays are taken up in vivo by leukocytes from HCMV-infected cells (26, 33), neither quantitation of antigenemia nor quantitation of DNAemia appears to correlate consistently with the actual viral replication in vivo (21). In this respect, detection of viral transcripts is currently considered a more direct marker of HCMV replication in vivo, and in particular, it seems that the detection of late transcripts may better reflect active HCMV replication, dissemination, and disease (24, 25, 29, 32).
However, reverse transcription-PCR, the method currently most used for viral RNA detection, bears the risk of false-positive results because of problems in differentiating between RNA- and DNA-derived PCR products. To overcome this problem, the recently introduced nucleic acid sequence-based amplification (NASBA) assays, which allow specific amplification of unspliced mRNA in a DNA background, appear to be very promising, as shown by the first published reports (1, 3, 14). In a recent retrospective study, we showed that detection of pp67 late HCMV transcript by NASBA in PBLs from solid-organ transplant recipients preceded the antigenemia threshold of 100 pp65-positive PBLs/2 × 105 PBLs (the currently used cutoff value in our hospital for initiation of preemptive therapy) by a mean time of 3.5 ± 2.6 days. On the other hand, late transcripts were detected by NASBA a mean time of 2.0 ± 5.1 days after initial positivity for antigenemia in a group of bone marrow transplant (BMT) recipients and solid-organ transplant recipients with primary infection (14). Thus, this delay appeared to place patients at risk if pp67 mRNA detection was used as a cutoff for treatment of BMT patients, in whom early treatment is requested, given the risk of early HCMV interstitial pneumonia in the posttransplantation period (12). In fact, in BMT recipients the severe impairment of anti-HCMV-specific immunity forces the clinician to rely on antiviral therapy as the sole tool to control HCMV infection, thus requiring an ultrasensitive diagnostic approach.
In the present study, we investigated retrospectively whether detection of immediate-early (IE) mRNA by NASBA could be a better cutoff than detection of antigenemia for preemptive treatment in BMT recipients. This approach is based on recent preliminary reports that have shown that IE mRNA detection by NASBA is the most sensitive assay for early detection of HCMV infection (2, 4). Results show that detection of IE mRNA may be a new useful parameter for early intervention strategies in BMT recipients.
MATERIALS AND METHODS
Patients and control subjects.
From July 1997 to January 1999 a total of 51 allogeneic BMT recipients, including 31 children (median age, 8 years; age range, 2 to 20 years) and 20 adults (median age, 42.5 years; age range, 21 to 55 years) were enrolled in the study. Their underlying hematologic diseases that required bone marrow transplantation were the following: chronic myeloid leukemia (n = 12), acute myeloid leukemia (n = 12), acute lymphoblastic leukemia (n = 11), myelodysplastic syndrome (n = 8), aplastic anemia (n = 2), thalassemia major (n = 3), Wiskott-Aldrich syndrome (n = 1), infantile malignant osteopetrosis (n = 1), and chronic granulomatous disease (n = 1). BMT recipients received an allogeneic marrow transplant at the Department of Pediatrics and at the Division of Hematology, University Hospital Istituto di Ricovero e Cura a Carattere Scientifico Policlinico San Matteo, Pavia, Italy. All patients were monitored for HCMV infection for a 3-month period, which was prolonged, when required, until HCMV disappearance from blood or until 20 weeks after transplantation. During follow-up, 544 heparinized or EDTA-treated blood samples were collected weekly or more frequently, according to the kinetics of HCMV infection. The serological statuses of the donors and recipients were determined by enzyme-linked immunosorbent assay prior to transplantation (19).
Prophylaxis for graft-versus-host disease (GVHD) consisted of cyclosporine for patients who received transplants from a compatible sibling, whereas patients given BMT from an HLA partially matched family donor received in vitro T-cell-depleted marrow, and patients who received the allograft from a matched but unrelated donor received, in addition to cyclosporine, short-term treatments with methotrexate, steroids, and the monoclonal antibody Campath-1G. Patients with acute GVHD were treated with steroids as first-line therapy, and resistant patients were treated with horse anti-lymphocyte globulin (16, 31). All BMT patients were given multiple transfusions of leukocyte-depleted red blood cells, platelets, and frozen plasma from unscreened blood donors.
Preliminarily, the clinical significance of IE mRNA detection by NASBA was assessed by testing three groups of control subjects: (i) 83 HCMV-seropositive and 17 HCMV-seronegative blood donors, (ii) 48 HCMV-seropositive and 8 HCMV-seronegative BMT and solid-organ transplant recipients tested within the first 20 days after transplantation, and (iii) 30 breast-feeding mothers with reactivated HCMV infection, as shown by HCMV isolation from their milk.
Antiviral treatment.
A preemptive therapy approach was adopted for this study. Ganciclovir (GCV) was administered intravenously at a standard dosage of 10 mg/kg of body weight/day for 14 days or until pp65 antigenemia clearance (i.e., three consecutive samples were negative for antigenemia). Alternatively, foscarnet (PFA) was administered intravenously at a dosage of 180 mg/kg/day for 14 days (unless otherwise clinically indicated) or until antigenemia clearance (i.e., three subsequent samples were negative for antigenemia). In 32 of 51 patients in this study, therapy was started either immediately when two or more pp65-positive PBLs were detected or when detection of one pp65-positive PBL was confirmed in the following 2 to 3 days by similar or higher levels of pp65-positive PBLs. Secondary courses of either GCV or PFA treatment were administered to 10 of 32 patients who presented with a single recurrence episode of HCMV infection and to 4 of 32 patients who presented with two or three secondary episodes of infection. When considered clinically necessary on the basis of delayed viral clearance, some patients were treated with a combination of the two antiviral drugs. Patients who had active GVHD and who were experiencing HCMV reactivation were consistently treated for 1 more week with either GCV or PFA after viral clearance (16, 31).
Virologic follow-up.
All patients were virologically monitored for HCMV infection by prospective quantitation of pp65 antigenemia and viremia, whereas quantitation of DNAemia and qualitative determination of IE and pp67 (late) RNAemia were performed retrospectively. Quantitation of pp65 antigenemia was achieved under a fluorescence microscope by counting the number of PBLs positive for pp65/2 × 105 PBLs examined on cytospin preparations stained with a pool of three pp65-specific monoclonal antibodies, according to a previously reported (18) and a more recently standardized (17) procedure. The quantitation of viremia was achieved by inoculating 2 × 105 PBLs onto human embryonic lung fibroblast cell cultures by the shell vial technique and then by counting the number of fibroblast nuclei positive for HCMV IE antigen p72 at 16 to 24 h after inoculation (20). DNAemia was quantitated by PCR following DNA extraction from whole blood as reported below for NASBA. The previously described PCR method for HCMV DNA quantification (15) was modified by using a new internal control (pAC2) as well as new external standards (pCM2) (21), which were constructed according to the same principle described in the original report (41). Briefly, samples were amplified in the presence of 100 copies of plasmid pAC2 and in parallel with semilogarithmic dilutions (104 to 101 copies) of plasmid pCM2 containing the same HCMV IE1 region currently amplified from clinical samples. HCMV DNA quantification was then achieved by plotting the pCM2/pAC2 ratio values from the external standards following densitometric analysis of gel signals and then by interpolating the HCMV/pAC2 ratio values from clinical samples on the standard curve that was generated (15, 21). This method allowed reproducible HCMV DNA quantification in the range of 101 to 104 genome equivalents/10 μl of DNA from whole blood when the single-step quantitative PCR was used.
NASBA.
The Nuclisens HCMV pp67 NASBA was carried out according to the manufacturer's instructions (Organon Teknika B.V., Boxtel, The Netherlands), as reported previously (3, 14). The NASBA procedure for detection of IE mRNA was carried out essentially as described for the NASBA for detection of pp67 (2). Briefly, nucleic acids from 100 μl of whole blood were isolated by the method of Boom et al. (8). System control (SC) RNA (∼8,000 cRNA copies) was added to the samples prior to nucleic acid isolation, thus serving as a positive control for isolation, amplification, and detection. The SC RNA included part of the IE1 mRNA corresponding to nucleotides 171,797 to 172,050 of the HCMV (AD169) genome (9) and could be distinguished from wild-type (wt) RNA by insertion of a fragment of 134 nucleotides, as reported previously (2). wt and SC IE mRNAs were amplified with a primer that contained a T7 promoter and a reverse primer. Amplification products were detected by electrochemiluminescence with capture probes coupled to magnetic beads and wt- and SC-specific ruthenium-labeled oligonucleotide detection probes (2). The analytical sensitivity of the NASBA for detection of IE mRNA was about 70 copies/10 μl of whole blood. The same containment measures as those used for PCR protocols (28) were adopted for the performance of NASBA (the use of three separate rooms as well as the use of separate reagents, micropipettes, and aerosol-resistant filter tips). In addition, negative controls were included in each test run.
Statistical analysis.
Differences in the means of parametric data (by the Lilliefors test for normal distributions) were tested by using the t test, while the Kolmogorov-Smirnov test for unpaired data was used for nonparametric data. Differences in distributions were analyzed by using the Pearson chi-square test.
RESULTS
Detection of IE mRNA by NASBA in control groups.
To define the clinical significance of IE mRNA detection by NASBA, we preliminarily tested the blood of 100 healthy blood donors, of whom 17 were HCMV seronegative, and 30 breast-feeding women excreting HCMV in their breast milk. IE mRNA was not detected in any subject. In addition, 56 transplant recipients, of whom 8 were HCMV seronegative, showed no IE mRNA in their blood when they were tested in the first 3 weeks after transplantation. Eleven additional control transplant patients found to be IE mRNA positive in the first 21 days after transplantation were shown to be in the initial phase of an HCMV infection, as subsequently confirmed by the positive results of the other assays. In conclusion, on the basis of these results we considered the NASBA for detection of IE mRNA the reference standard method for the diagnosis of active HCMV infection.
Incidence of HCMV infection in BMT recipient population.
The serological statuses of the donors and recipients for the indicated numbers of patients with HCMV infection among the numbers of patients tested were as follows: for 25 of 30 patients (12 of 17 pediatric patients), donor positive and recipient positive; for 1 of 3 patients (all pediatric patients), donor positive and recipient negative; for 12 of 17 patients (7 of 11 pediatric patients), donor negative and recipient positive; for 1 of 1 (adult) patient, donor negative and recipient negative. Thus, of the 51 BMT recipients examined, 39 (76.5%) developed HCMV infection. No patient developed overt HCMV disease due to the preemptive therapy approach. Twelve patients remained HCMV negative during the entire follow-up period. Of the 20 HCMV-positive (64.5%) pediatric patients, 19 (95.0%) were positive by detection of IE mRNA by NASBA and 14 (70.0%) were treated according to the antigenemia-guided preemptive therapy protocol. In parallel, of the 19 HCMV-positive (95.0%) adult patients, all 19 (100%) were IE mRNA positive and 17 (89.5%) were treated. On the whole, 31 of 39 (79.5%) patients who developed HCMV infection were treated. The single HCMV-seronegative adult patient, given BMT from a seronegative donor and who developed HCMV infection after transplantation, had been transfused with several units of platelets, erythrocytes, and plasma.
Of the 39 of 51 HCMV-positive patients identified when considering results provided by any virologic assay, HCMV was detected in 14 by quantitation of viremia, 15 by NASBA for detection of pp67 mRNA, 32 by quantitation of DNAemia, and 33 by quantitation of antigenemia, while HCMV was detected in 38 patients by NASBA for detection of IE mRNA (Table 1). By using IE mRNA detection by NASBA as the reference standard for detection of active HCMV infection in BMT patients, the highest sensitivities were given by the combination of any assay for (100%) followed by assays for DNAemia and antigenemia (84.2%), and this was also true for the negative predictive value (NPV). On the other hand, specificities and positive predictive values (PPVs) were very high (>90%) for all assays.
TABLE 1.
Detection of HCMV infection by different assays and diagnostic value of each assay for 51 BMT recipients by using NASBA for detection of IE mRNA as the reference standard
| Assay for detection of: | No. (%) of patients with diagnosis of HCMV infection in blood
|
Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |
|---|---|---|---|---|---|---|
| Positive | Negative | |||||
| Antigenemia | 33 (64.7) | 18 (35.3) | 84.2 | 92.3 | 97.0 | 66.7 |
| Viremia | 14 (27.5) | 37 (72.5) | 36.8 | 100 | 100 | 35.1 |
| DNAemia | 32 (62.7) | 19 (37.3) | 84.2 | 100 | 100 | 68.4 |
| pp67 mRNA | 15 (29.4) | 36 (70.6) | 39.5 | 100 | 100 | 36.1 |
| IE mRNAa | 38 (74.6) | 13 (25.4) | NAb | NA | NA | NA |
| Any | 39 (76.5) | 12 (23.5) | 100 | 92.3 | 97.4 | 100 |
Reference assay.
NA, not applicable.
In more detail, when considering patient positivity for HCMV infection in blood by different assay combinations, 9 of 51 patients (17.6%) were positive by all five assays, 11 (21.6%) were positive by four assays (6 patients were positive by assays for antigenemia, DNAemia, pp67 mRNA, and IE mRNA, and 5 patients were positive by assays for antigenemia, viremia, DNAemia, and IE mRNA), 10 (19.6%) were positive by three assays (assays for antigenemia, DNAemia, and IE mRNA), four (7.8%) were positive by two assays (2 patients were positive by assays for antigenemia and IE mRNA, and 2 patients were positive by assays for DNAemia and IE mRNA), and 5 (9.8%) were positive by one assay (1 patient was positive by the assay for antigenemia and 4 patients were positive by the assay for IE mRNA). The single patient found to be HCMV positive only by the assay for antigenemia was a true positive (and was treated), since the assay for antigenemia was found to be repeatedly positive at a very low level (1 pp65-positive PBL/2 × 105 PBLs examined), whereas the NASBA for detection of IE mRNA and the other assays were repeatedly negative (tests performed in replicate with sequential blood samples). Of the four patients (two were donor positive and recipient positive, one was donor positive and recipient negative, and one was donor negative and recipient positive) positive only by the assay for IE mRNA, a single blood sample from three patients was positive and two subsequent blood samples from one patient were positive. All these patients received transfusions of several units of leukocyte-depleted blood components.
Detection of HCMV infection.
As reported in Table 2, of the 137 DNAemia-positive samples, as many as 113 (82.5%) were IE mRNA positive, whereas of the 383 DNAemia-negative samples, as many as 329 (85.9%) were IE mRNA negative. Similarly, of the 122 antigenemia-positive samples, 98 (80.3%) were IE mRNA positive, whereas of the 396 antigenemia-negative samples, 328 (82.8%) were IE mRNA negative. On the other hand, as far as assays for viremia and pp67 mRNA are concerned, the level of positive results concordant with positivity for IE mRNA was in the range of 93.8 to 100%, and that of concordant negative results was in the range of 71.5 to 72.0%. The difference in the distribution of concordant and discordant results, according to comparisons of the results between different pairs of assays, is reported in Table 2. Statistical analysis showed that the proportion of concordant results was significantly greater than that of discordant results for each pair of comparisons.
TABLE 2.
Comparison of results of NASBA for IE mRNA, assays for antigenemia, DNAemia, and viremia, and NASBA for pp67 mRNA
| HCMV assay results | No. (%) of blood samples
|
Pa | ||
|---|---|---|---|---|
| IE mRNA positive | IE mRNA negative | Total | ||
| Antigenemia assay positive | 98 (18.9) | 24 (4.6) | 122 (23.5) | |
| Antigenemia assay negative | 68 (13.1) | 328 (63.4) | 396 (76.5) | <0.0001 |
| Total | 166 (32.0) | 352 (68.0) | 518 | |
| DNAemia assay positive | 113 (21.7) | 24 (4.6) | 137 (26.3) | |
| DNAemia assay negative | 54 (10.4) | 329 (63.3) | 383 (73.7) | <0.0001 |
| Total | 167 (32.1) | 353 (67.9) | 520 | |
| Viremia assay positive | 25 (5.2) | 0 | 25 (5.2) | |
| Viremia assay negative | 131 (27.0) | 329 (67.8) | 460 (94.8) | <0.0001 |
| Total | 156 (32.2) | 329 (67.8) | 485 | |
| NASBA for pp67 mRNA positive | 30 (5.9) | 2 (0.4) | 32 (6.3) | |
| NASBA for pp67 mRNA negative | 134 (26.2) | 345 (67.5) | 479 (93.7) | <0.0001 |
| Total | 164 (32.1) | 347 (67.9) | 511 | |
Concordant versus discordant results (pearson chi-square test).
By using the NASBA for detection of IE mRNA as the reference standard for detection of HCMV infection in single blood samples, the diagnostic sensitivities of assays for DNAemia and antigenemia were relatively low but were largely superior to those of the assay for viremia and the NASBA for detection of pp67 mRNA (Table 3). Similarly, NPVs for the assays for antigenemia and DNAemia were higher than those for the assay for viremia and the NASBA for detection of pp67 mRNA. On the contrary, specificities and PPVs were high (80 to 100%) for all four assays.
TABLE 3.
Diagnostic value of assays for antigenemia, viremia, and DNAemia and NASBA for detection of pp67 mRNA for detection of HCMV infection in 544 blood samples from 51 BMT recipients by using the NASBA for detection of IE mRNA as the “gold standard”
| Assay for detection of: | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|
| Antigenemia | 59.0 | 93.2 | 80.3 | 82.8 |
| Viremia | 16.0 | 100 | 100 | 71.5 |
| DNAemia | 67.7 | 93.2 | 82.5 | 85.9 |
| pp67 mRNA | 18.3 | 99.4 | 93.7 | 72.0 |
Time to detection of HCMV infection.
The mean time to the first detection of HCMV infection following transplantation in the 39 BMT recipients is reported in Table 4. The first assay that detected HCMV infection was the NASBA for detection of IE mRNA, followed by assays for antigenemia, DNAemia, and viremia and the NASBA for detection of pp67 mRNA. These differences were statistically significant only for the assay for viremia and the NASBA for pp67 mRNA versus the NASBA for IE mRNA. However, when considering the mean times (in days) of delay (with respect to the time to detection by the NASBA for IE mRNA) for different assays to become positive during both first and relapse episodes, it was found that such delays were in the range of 4.2 ± 7.4 (assay for DNAemia) to 8.4 ± 8.8 (assay for viremia) days. Thus, on average, the NASBA for IE mRNA preceded the other four assays in detecting HCMV in blood for the overall number of episodes (first and secondary) of HCMV infection per patient by about 1 week (Table 4).
TABLE 4.
Times to first detection of HCMV in blood following transplantation and to first disappearance of HCMV from blood following onset of antiviral treatment according to different viral assays
| Assay for detection of: | Time to first HCMV detection
|
Time lag by which IE mRNA detection precedes positivity by other assaysa
|
Time to first HCMV disappearance during therapy
|
|||
|---|---|---|---|---|---|---|
| Mean ± SD days | Pb | Mean ± SD days | Pc | Mean ± SD days | Pd | |
| IE mRNA | 37.7 ± 15.4 | NAe | NA | NA | 7.7 ± 9.8 | NA |
| Antigenemia | 39.6 ± 15.6 | NSf | 5.7 ± 7.7 | NS | 8.9 ± 10.5 | NS |
| DNAemia | 40.9 ± 15.2 | NS | 4.2 ± 7.4 | NS | 8.9 ± 10.1 | NS |
| Viremia | 43.7 ± 16.3 | <0.0005 | 8.4 ± 8.8 | NS | 1.6 ± 2.5 | <0.01 |
| pp67 mRNA | 43.7 ± 17.5 | <0.005 | 6.7 ± 5.6 | NS | 4.9 ± 8.9 | NS |
Determined upon first HCMV appearance as well as relapse episodes.
t test. A significant difference was also found for the assay for DNAemia versus the assay for viremia (P < 0.005) and the NASBA for pp67 mRNA (P < 0.05).
Kolmogorov-Smirnov test.
Kolmogorov-Smirnov test. A significant difference was also found for the assay for DNAemia versus the assay for viremia (P < 0.0001) and the NASBA for pp67 mRNA (P < 0.005) and for the assay for antigenemia versus the assay for viremia (P < 0.01).
NA, not applicable.
NS, not significant.
Finally, the mean time to first HCMV negativity (disappearance) for blood following initiation of antiviral treatment was significantly lower (1.6 ± 2.5 days) for the assay for viremia than for the NASBA for detection of IE mRNA, whereas it was comparable to that for the NASBA for detection of IE mRNA for the other three assays (Table 4). The longer duration of treatment for the latter assays was due to the time required to confirm negativity by the assay for antigenemia in three sequential blood samples.
NASBA for IE mRNA and antiviral treatment.
On the whole, 31 of 39 (79.5%) BMT recipients received antigenemia assay-guided preemptive therapy. Among the eight untreated patients, four were positive only by the NASBA for IE mRNA, two were positive only by the NASBA for IE mRNA and the assay for DNAemia, and two were positive only by the NASBA for IE mRNA and the assay for antigenemia, yet for the last two patients a single pp65-positive PBL was detected in a single blood sample.
As shown in Table 5, if confirmed positivity of qualitative IE mRNA determination by NASBA had been used as a virologic parameter for the initiation of preemptive therapy instead of the assay for antigenemia, 35 of 39 (89.7%) HCMV-positive BMT patients would have been treated (with the exclusion of the 3 patients positive by the NASBA for IE mRNA in a single blood sample and the single patient positive only by the assay for antigenemia). On the other hand, if confirmed positivity by the assay for DNAemia had been used for the initiation of preemptive therapy, the same absolute number of patients (n = 31) whose HCMV infection was detected by the assay for antigenemia could have been treated. Finally, if first positivity for pp67 mRNA had been chosen as a criterion for the initiation of therapy, only 15 of 39 (38.5%) patients would have been treated. However, pp67 mRNA expression could be directly influenced by early initiation of antiviral therapy in these patients.
TABLE 5.
Predicted treatment under guidance of NASBAs for IE mRNA and pp67 mRNA and assay for DNAemia versus the actual treatment under guidance of assay for antigenemia
| Patient characteristic | Actual treatment under the guidance of assay for antigenemia
|
Predicted treatmenta under the guidance of:
|
||||||
|---|---|---|---|---|---|---|---|---|
| NASBA for IE mRNA
|
NASBA for pp67 mRNA
|
Assay for DNAemia
|
||||||
| No. of treated patients | No. of untreated patients | No. of treated patients | No. of untreated patients | No. of treated patients | No. of untreated patients | No. of treated patients | No. of untreated patients | |
| Antigenemia positive | 31 | 2b | 32b | 1 | 15 | 18b | 30b | 3b |
| Antigenemia negative | NAc | 18 | 3 | 15 | 0 | 18 | 1 | 17 |
| IE mRNA positive | 30 | 8 | 35 | 3d | 15 | 23 | 31 | 7 |
| IE mRNA negative | 1e | 12 | NA | 13 | 0 | 13 | 0 | 13 |
| pp67 mRNA positive | 15 | 0 | 15 | 0 | 15 | 0 | 15 | 0 |
| pp67 mRNA negative | 16 | 20 | 20 | 16 | NA | 36 | 16 | 20 |
| DNAemia positive | 29 | 3 | 32 | 0 | 15 | 17 | 31 | 1f |
| DNAemia negative | 2e | 17 | 3 | 16 | 0 | 19 | NA | 19 |
| Total | 31 | 20 | 35 | 16 | 15 | 36 | 31 | 20 |
Predicted starting treatment upon first confirmed positive result by NASBA for IE mRNA and assay for DNAemia and upon first positive result by NASBA for pp67 mRNA.
Although antigenemia positive, two patients (patients 19 and 32) were not treated due to detection of a single pp65-positive polymorphonuclear leukocyte not confirmed subsequently (they were both IE mRNA positive and one was also DNAemia positive).
NA, not applicable.
Three patients (patients 36, 37, and 38) had only a single IE-mRNA-positive sample during the follow-up period.
One patient (patient 51), treated upon the first confirmed detection of one pp65-positive polymorphonuclear leukocyte on day 17 after transplantation, did not show the presence of IE mRNA or HCMV DNA in blood.
One of the 32 DNAemia-positive patients (patient 33) had only a single (not subsequently confirmed) positive sample (the patient was also antigenemia negative).
Kinetics of HCMV infection in treated BMT recipients by different assays.
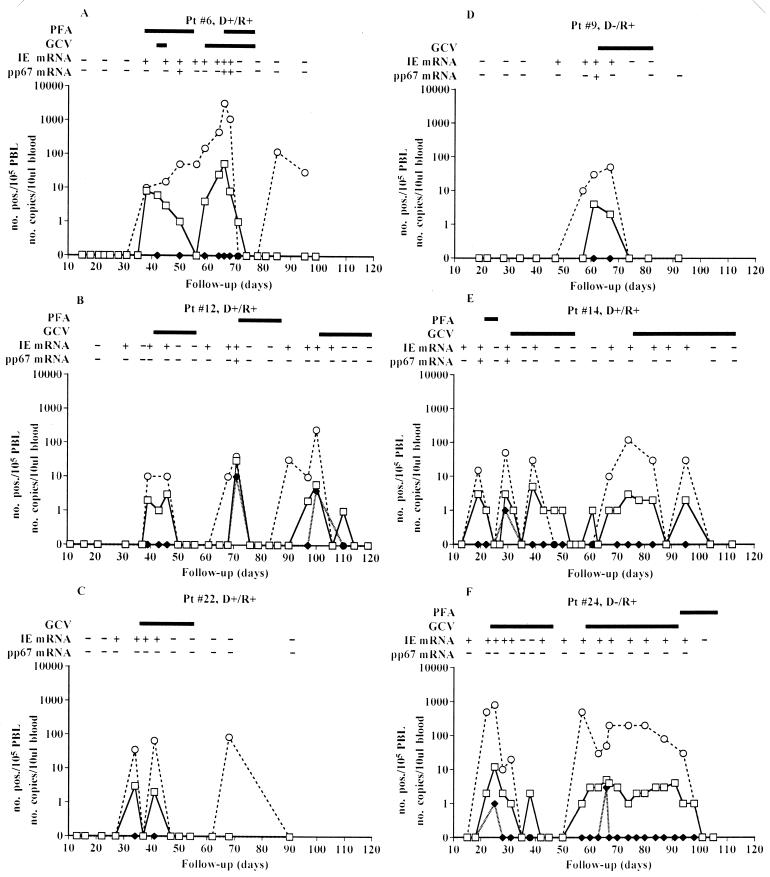
The kinetics of HCMV infection as detected by different viral parameters in six representative treated BMT patients is reported in Fig. 1. In patients 22 and 9 (Fig. 1C and D, respectively), a single course of antiviral treatment was able to block HCMV reactivation, whereas the other four patients whose data are reported in Fig. 1 needed multiple treatment courses to overcome the subsequent episodes of HCMV reactivation. The appearance of pp67 mRNA in the blood of these patients was restricted to the highest peak of HCMV infection (patients 6, 9, and 12), which was reached sometimes after the onset of treatment (patient 6), or to the first episode of reactivation (patient 14), while antiviral treatment induced negativity for this viral parameter. In some other patients (patients 22 and 24), pp67 mRNA was never detected, possibly due to the very early therapy. On the other hand, IE mRNA was detected in nearly every episode of HCMV active infection, mostly (patients 9, 12, 14, 22, and 24) preceding by some days or sometimes (patient 6) appearing concomitantly with pp65 antigenemia and DNAemia. During antiviral treatment, the NASBA for IE mRNA required approximately the same time as the assays for pp65 antigenemia and DNAemia to become negative. However, in patient 6 (Fig. 1A), while the assay for pp65 antigenemia became negative after a first course of combined antiviral treatment, the NASBA for IE mRNA remained positive (and the level of DNAemia was increasing). Thus, a second course of combined treatment was needed in this patient to control HCMV infection after pp65 antigenemia reappearance; later, HCMV DNA alone was again detected in blood (as in patient 22), but neither pp65 antigenemia nor IE mRNA was further detected.
FIG. 1.
Kinetics of HCMV infection in six preemptively treated BMT recipients. Pt, patient; D, donor; R, recipient; −, HCMV negative; +, HCMV positive; □, assay for antigenemia; ⧫, assay for viremia; ○, assay for DNAemia.
DISCUSSION
HCMV infection still represents one of the major causes of morbidity and mortality in BMT recipients, mainly as HCMV-related interstitial pneumonia. This life-threatening disease is difficult to treat successfully once it is established, with prevention representing the most effective approach. Early detection of HCMV infection is particularly relevant in BMT recipients, as the severe impairment of their specific antiviral immunity makes the timely initiation of antiviral therapy mandatory for prevention of HCMV disease. This consideration is in contrast to the situation for solid-organ recipients, in whom modulation of immunosuppressive therapy is an additional option, whether or not it is combined with antiviral agents. The availability of highly active HCMV-specific antiviral drugs such as GCV and PFA has led to prompt treatment of the HCMV infection before the appearance of clinical symptoms. Demonstration of infectious virus in blood (viremia) or in bronchoalveolar lavage specimens has reduced the number of patients who develop interstitial pneumonia through preemptive treatment of infection.
However, in 10 to 15% of patients interstitial pneumonia developed contemporaneously with the identification of infectious virus either in blood or in bronchoalveolar lavage specimens, and in these patients treatment was ineffective (23, 37). More recently, more sensitive assays for the early detection of virus in blood have been developed, and either antigenemia or DNAemia positivity has been used as a virologic indicator for the initiation of antiviral treatment in BMT recipients (5, 6, 11, 22, 30, 31, 39, 42), with satisfactory results in terms of disease prevention. However, because viral transcripts may represent a more direct marker of viral biologic activity in patients compared to antigenemia and DNAemia, the presence of both late and IE mRNAs has been proposed as a better virologic parameter to be relied on for the planning of preemptive therapy strategies in patients who receive transplants. In this respect, the development of the NASBA technology has represented a recent major advance, as it overcomes the problems (usually encountered with reverse transcription-PCR technology) inherent to amplification of RNA in a background of DNA. In a recent retrospective study, in which an antigenemia cutoff of 100 was used for preemptive therapy of reactivated HCMV infections in solid-organ transplant (heart, lung) recipients, it was shown that NASBA detection of the pp67 viral transcript in blood could usefully be used as a virologic marker for initiation of preemptive therapy in this patient population (14). On the other hand, in the same study, use of pp67 mRNA detection in BMT recipients appeared to be potentially too risky, as the NASBA for pp67 mRNA became positive a mean time of 2.0 ± 5 days later than assays for antigenemia. Actually, in that study it was found that the sensitivity of the NASBA for pp67 mRNA detection for detection of HCMV infection in transplant recipients was intermediate (58%) between those of assays for antigenemia (78%) and viremia (54%). In view of the need for the very early diagnosis of HCMV infection in severely immunocompromised BMT patients, the NASBA for detection of pp67 mRNA may not be a suitable method for determination of the need for initiation of antiviral therapy in this clinical setting. However, it should be considered that the results for pp67 mRNA in the current retrospective study may have been influenced by the early start of preemptive therapy. A prospective study that compares preemptive therapy based on either the results of the assay for pp65 antigenemia or the NASBA for pp67 mRNA has recently been started with heart and lung transplant recipients, and it may provide more conclusive data on this issue for BMT recipients as well.
On this basis, the present study was aimed at investigating the following issues: (i) the level of sensitivity of the NASBA for detection of IE mRNA for detection HCMV infection in the blood of BMT recipients compared to those of the other assays tested, including the NASBA for pp67 mRNA; (ii) the effectiveness of the very early detection of HCMV infection in blood by the NASBA for IE mRNA compared to that by the other four assays; and (iii) the (predicted) number of BMT patients with HCMV infection to be treated by use of the results of the NASBA for IE mRNA as a virologic marker compared to use of the results of the assays for antigenemia and DNAemia (the most commonly used assays) and to the NASBA for pp67 mRNA.
Prior to discussing the clinical sensitivity and specificity of the NASBA for IE mRNA, we must consider the results obtained by performing the NASBA for IE mRNA with the three groups of controls mentioned above. IE mRNA were not detected on repeat testing in any control subject or patient. These results allow us to reliably conclude that detection of IE mRNA by NASBA in patients reveals an active HCMV infection in the immunocompromised host. IE mRNA was not detected in blood by NASBA during reactivated infections in breast-feeding mothers, yet it was detected for a short period of a few months during the convalescent phase of a primary HCMV infection (M. G. Revello, D. Lilleri, and G. Gerna, unpublished data) or during congenital HCMV infections (Revello et al., unpublished data). In immunocompetent subjects, the lack of IE mRNA in blood during HCMV reactivations corresponds to the lack of viral DNA or antigenemia (35).
The sensitivity of the NASBA for IE mRNA in detecting HCMV infection in individual BMT patients was shown in this study to be slightly superior (38 HCMV-positive patients detected) to those of the assays for antigenemia (33 patients) and DNAemia (32 patients) and largely superior to those of the NASBA for pp67 mRNA (15 patients) and the assay for viremia (14 patients). By using the NASBA for IE mRNA as the reference test, the assays for antigenemia and DNAemia had sensitivities of greater than 80% and NPVs of greater than 60% for the detection of active HCMV infection, whereas values of both parameters were lower than 40% for the NASBA for pp67 mRNA and the assay for viremia. On the other hand, specificities and PPVs were greater than 90% for all five assays. In addition, of the 39 HCMV-positive BMT patients, HCMV infection was detected in as many as 34 (87.2%) patients by at least two assays. Of the five patients in whom HCMV infection was detected by only one assay, infection was detected in four patients by the NASBA for IE mRNA and in one patient by the assay for antigenemia. Of the four patients positive by the NASBA for IE mRNA, one was repeatedly positive, whereas only a single blood sample from each of the three remaining patients was positive. Thus, the last three patients would not have been treated because of the requested confirmation of positivity. As mentioned above, sequential blood samples from the single patient who was positive only by the assay for antigenemia were found to be positive by detection of a single pp65-positive PBL and thus the patient was treated.
The greater sensitivity of the NASBA for IE mRNA was also shown by the very early detection of the HCMV infection in blood compared to the times to detection for the other assays. Earlier HCMV detection by the NASBA for IE mRNA was in the range of 4 to 8 days as compared to the other assays. These data allow us to predict that, by using positivity by the NASBA for IE mRNA as a virologic parameter for the initiation of preemptive therapy, not only a greater number of patients would be treated but the patients would also be treated some days earlier. In addition, they would be treated for a slightly shorter period of time.
Treatment guided by the assay for antigenemia required confirmation of positivity within 2 days when it was limited to a single pp65-positive PBL. By using this criterion, 31 BMT patients were treated in this study, and no HCMV disease or HCMV-related clinical complications were observed. Treatment predicted by use of confirmed positivity by the assay for DNAemia as a virologic indicator would have involved an identical absolute number of patients. By use of first positivity for pp67 mRNA as an indicator for treatment, less than 50% of the patients (15 instead of 31) would have been treated. However, we must consider that in some patients the appearance of pp67 mRNA might have been prevented by early therapy guided by the results of the assay for antigenemia. If first confirmed positivity by the NASBA for IE mRNA had been used to predict the need for treatment of the BMT patients in this study, as many as 35, i.e., 4 more than the number of patients predicted by the assays for antigenemia or DNAemia, would have been treated. However, the single patient positive only by the assay for antigenemia would not have been treated.
The clinical finding that these additional patients, who were not actually treated on the basis of guidance from the results of the assay for antigenemia, did not present any clinical manifestation related to HCMV disease would suggest that these patients did not need treatment for the HCMV infection, which actually resolved spontaneously. On the other hand, in contrast to other reports (4), in our department we never observed cases of HCMV disease in patients preemptively treated on the basis of the results of the assay for antigenemia. This finding justifies trials based on even more sensitive tests than the assay for antigenemia, such as detection of IE mRNA by NASBA. In fact, the risk of the early onset of interstitial HCMV pneumonia together with the easy performance of NASBA, as well as the need for more extensive knowledge of the effect of an earlier treatment of HCMV infection in a BMT population, suggest the need to perform comparative controlled trials based on treatment schedules guided by the results of assays for antigenemia (or DNAemia) and the NASBA for IE mRNA detection. A better understanding of the clinical impact of earlier treatment of HCMV infections in BMT patients would improve management of HCMV infections in this important transplant patient population.
Finally, the occurrence of late HCMV disease following prolonged prophylactic GCV treatment as a result of a lack of an immune response due to early abrogation of antigenic stimuli (5) was never observed in our department when the preemptive therapy protocol guided by the results of the assay for antigenemia was used. It is reasonable to speculate that our patients could have developed specific antiviral immunity due to both the limited treatment duration and the consideration that HCMV infection is detected in PBLs of transplant patients when virus replication in endothelial cells of blood vessels has already occurred, as recently suggested by the in vitro model developed in our laboratory for the study of the interactions between PBLs and endothelial cells (33).
ACKNOWLEDGMENTS
We thank Linda D'Arrigo for revision of the English. We are also indebted to Luca Dossena and Sebastiano Scandurra for excellent technical assistance and to Franca Bordoni for typing the manuscript.
This work was partially supported by the Ministero della Sanità, Ricerca Corrente, Istituto di Ricovero e Cura a Carattere Scientifico Policlinico San Matteo, Pavia, Italy (grant 820 RCR 96/01), and by the Ministero della Sanità, Istituto Superiore di Sanità, II Programma di Ricerca sull'AIDS (grant 50B.21).
REFERENCES
- 1.Aono T, Kondo K, Miyoshi H, Tanaka-Taya K, Kondo M, Osugi Y, Hara J, Okada S, Yamanishi K. Monitoring of human cytomegalovirus infections in pediatric bone marrow transplant recipients by nucleic acid sequence-based amplification. J Infect Dis. 1998;178:1244–1249. doi: 10.1086/314449. [DOI] [PubMed] [Google Scholar]
- 2.Blok M J, Christiaans M H L, Goossens V J, van Hooff J P, Sillekens P, Middeldorp J M, Bruggeman C A. Early detection of human cytomegalovirus infection after kidney transplantation by nucleic acid sequence-based amplification. Transplantation. 1999;67:1274–1277. doi: 10.1097/00007890-199905150-00013. [DOI] [PubMed] [Google Scholar]
- 3.Blok M J, Goossens V J, Vanherle S J V, Top B, Tacken N, Middeldorp J M, Christiaans M H L, van Hooff J P, Bruggeman C A. Diagnostic value of monitoring human cytomegalovirus late pp67 mRNA expression in renal-allograft recipients by nucleic acid sequence-based amplification. J Clin Microbiol. 1998;36:1341–1346. doi: 10.1128/jcm.36.5.1341-1346.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blok M J, Lautenschlager I, Christiaans M H L, van Hooff J P, Goossens V J, Middeldorp J M, Sillekens P, Ramon A, Höckerstedt K, Bruggeman C A. Nucleic acid sequence-based amplification: a new technique for monitoring cytomegalovirus infection in transplant recipients. Transplant Proc. 1999;31:308–309. doi: 10.1016/s0041-1345(98)01639-x. [DOI] [PubMed] [Google Scholar]
- 5.Boeckh M, Gooley T A, Myerson D, Cunningham T, Schoch G, Bowden R A. Cytomegalovirus pp65 antigenemia-guided early treatment with ganciclovir versus ganciclovir at engraftment after allogeneic marrow transplantation: a randomized double-blind study. Blood. 1996;88:4063–4071. [PubMed] [Google Scholar]
- 6.Boeckh M, Stevens-Ayers T, Bowden R A. Cytomegalovirus pp65 antigenemia after autologous marrow and peripheral blood stem cell transplantation. J Infect Dis. 1996;174:907–912. doi: 10.1093/infdis/174.5.907. [DOI] [PubMed] [Google Scholar]
- 7.Boivin G, Olson C A, Quirk M R, St-Cyr S M, Jordan M C. Quantitation of cytomegalovirus glycoprotein H gene in cells using competitive PCR and a rapid fluorescent-based detection system. J Virol Methods. 1995;51:329–342. doi: 10.1016/0166-0934(94)00128-4. [DOI] [PubMed] [Google Scholar]
- 8.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, Van der Nordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Crny R, Horsnell T, Hutchinson III C A, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 10.Einsele H, Ehninger G, Hebart H, Wittkowski K M, Schule U, Jahn G, Mackes P, Herter M, Klingebiel T, Loeffler J, Wagner S, Muller C. Polymerase chain reaction monitoring reduces the incidence of cytomegalovirus disease and the duration and side effects of antiviral therapy after bone marrow transplantation. Blood. 1995;86:2815–2820. [PubMed] [Google Scholar]
- 11.Einsele H, Ehninger G, Steidle M, Vallbracht A, Muller M, Schmidt H, Saal J G, Waller H D, Muller C. Polymerase chain reaction to evaluate antiviral therapy for cytomegalovirus disease. Lancet. 1991;338:1170–1172. doi: 10.1016/0140-6736(91)92032-w. [DOI] [PubMed] [Google Scholar]
- 12.Enright H, Haake R, Weisdorf D, Ramsay N, McGleave P, Kersey J, Thomas W, McKenzie D, Miller W. Cytomegalovirus pneumonia after bone marrow transplantation: risk factors and response to therapy. Transplantation. 1993;55:1339–1346. doi: 10.1097/00007890-199306000-00024. [DOI] [PubMed] [Google Scholar]
- 13.Fox J C, Griffiths P D, Emery V C. Quantitation of human cytomegalovirus DNA using the polymerase chain reaction. J Gen Virol. 1992;73:2405–2408. doi: 10.1099/0022-1317-73-9-2405. [DOI] [PubMed] [Google Scholar]
- 14.Gerna G, Baldanti F, Middeldorp J M, Furione M, Zavattoni M, Lilleri D, Revello M G. Clinical significance of expression of human cytomegalovirus pp67 late transcript in heart, lung, and bone marrow transplant recipients as determined by nucleic acid sequence-based amplification. J Clin Microbiol. 1999;37:902–911. doi: 10.1128/jcm.37.4.902-911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerna G, Baldanti F, Sarasini A, Furione M, Percivalle E, Revello M G, Zipeto D, Zella D the Italian Foscarnet Study Group. Effect of foscarnet induction treatment on quantitation of human cytomegalovirus (HCMV) DNA in peripheral blood polymorphonuclear leukocytes and aqueous humor of AIDS patients with HCMV retinitis. Antimicrob Agents Chemother. 1994;38:38–44. doi: 10.1128/aac.38.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerna G, Furione M, Baldanti F, Percivalle E, Comoli P, Locatelli F. Quantitation of human cytomegalovirus DNA in bone marrow transplant recipients. Br J Hematol. 1995;91:674–683. doi: 10.1111/j.1365-2141.1995.tb05368.x. [DOI] [PubMed] [Google Scholar]
- 17.Gerna G, Percivalle E, Torsellini M, Revello M G. Standardization of the human cytomegalovirus antigenemia assay by means of in vitro-generated pp65-positive peripheral blood polymorphonuclear leukocytes. J Clin Microbiol. 1998;36:3585–3589. doi: 10.1128/jcm.36.12.3585-3589.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerna G, Revello M G, Percivalle E, Morini F. Comparison of different immunostaining techniques and monoclonal antibodies to the lower matrix phosphoprotein (pp65) for optimal quantitation of human cytomegalovirus antigenemia. J Clin Microbiol. 1992;30:1232–1237. doi: 10.1128/jcm.30.5.1232-1237.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerna G, Revello M G, Percivalle E, Torsellini M. A 6-hour microneutralization assay for human cytomegalovirus antibody by using monoclonal antibodies. Serodiagn Immunother Infect Dis. 1990;4:243–247. [Google Scholar]
- 20.Gerna G, Revello M G, Percivalle E, Zavattoni M, Parea M, Battaglia M. Quantification of human cytomegalovirus viremia by using monoclonal antibodies to different viral proteins. J Clin Microbiol. 1990;28:2681–2688. doi: 10.1128/jcm.28.12.2681-2688.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerna G, Zavattoni M, Baldanti F, Sarasini A, Chezzi L, Grossi P, Revello M G. Human cytomegalovirus (HCMV) leukoDNAemia correlates more closely with clinical symptoms than antigenemia and viremia in heart and heart-lung transplant recipients with primary HCMV infection. Transplantation. 1998;65:1378–1385. doi: 10.1097/00007890-199805270-00016. [DOI] [PubMed] [Google Scholar]
- 22.Gerna G, Zipeto D, Parea M, Revello M G, Silini E, Percivalle E, Zavattoni M, Grossi P, Milanesi G. Monitoring of human cytomegalovirus infections and ganciclovir treatment in heart transplant recipients by determination of viremia, antigenemia and DNAemia. J Infect Dis. 1991;164:488–498. doi: 10.1093/infdis/164.3.488. [DOI] [PubMed] [Google Scholar]
- 23.Goodrich J M, Mori M, Gleaves C A, Mond C O, Cays M, Ebeling D F, Buhles W C, De Armond B, Meyers J D. Early treatment with ganciclovir to prevent cytomegalovirus disease after allogeneic bone marrow transplantation. N Engl J Med. 1991;325:1601–1607. doi: 10.1056/NEJM199112053252303. [DOI] [PubMed] [Google Scholar]
- 24.Gozlan J, Salord J M, Chouaïd C, Duvivier C, Picard O, Mejohas M C, Petit J C. Human cytomegalovirus (HCMV) late mRNA detection in peripheral blood of AIDS patients: diagnostic value for HCMV disease compared with those of viral culture and HCMV DNA detection. J Clin Microbiol. 1993;31:1943–1945. doi: 10.1128/jcm.31.7.1943-1945.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gozlan J, Laporte J P, Lesage S, Labopin M, Najman A, Gorin N C, Petit J C. Monitoring of cytomegalovirus infection and disease in bone marrow recipients by reverse-transcription-PCR and blood and urine culture. J Clin Microbiol. 1996;34:2085–2088. doi: 10.1128/jcm.34.9.2085-2088.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grefte A, Harmsen M C, van der Giessen M, Knollema S, van Son W J, The T H. Presence of human cytomegalovirus (HCMV) immediate early mRNA but not ppUL83 (lower matrix protein pp65) mRNA in polymorphonuclear and mononuclear leukocytes during active HCMV infection. J Gen Virol. 1994;75:1989–1998. doi: 10.1099/0022-1317-75-8-1989. [DOI] [PubMed] [Google Scholar]
- 27.Grossi P, Minoli L, Percivalle E, Irish W, Viganò M, Gerna G. Clinical and virological monitoring of human cytomegalovirus infection in 294 heart transplant recipients. Transplantation. 1995;59:847–851. [PubMed] [Google Scholar]
- 28.Kwok S. Procedures to minimize PCR product carry-over. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. A guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 142–145. [Google Scholar]
- 29.Lam K M C, Oldenburg N, Khan M A, Gaylore V, Mikhail G W, Strouhal P D, Middeldorp J M, Banner N, Yacoub M. Significance of reverse transcription polymerase chain reaction in the detection of human cytomegalovirus gene transcripts in thoracic organ transplant recipients. J Heart Lung Transplant. 1998;17:555–565. [PubMed] [Google Scholar]
- 30.Ljungman P, Lore K, Aschan J, Klaesson S, Lewensohn-Fuchs I, Lonnqvist B, Ringden O, Winiarski J, Ehrnst A. Use of a semiquantitative PCR for cytomegalovirus DNA as a basis for preemptive antiviral therapy in allogeneic bone marrow transplant patients. Bone Marrow Transplant. 1996;17:583–587. [PubMed] [Google Scholar]
- 31.Locatelli F, Percivalle E, Comoli P, Maccario R, Zecca M, Giorgiani G, De Stefano P, Gerna G. Human cytomegalovirus (HCMV) infection in pediatric patients given allogeneic bone marrow transplantation: role of early antiviral treatment for HCMV antigenemia on patients' outcome. Br J Hematol. 1994;88:64–71. doi: 10.1111/j.1365-2141.1994.tb04978.x. [DOI] [PubMed] [Google Scholar]
- 32.Nelson P N, Kawal B K, Boriskin Y S, Mathers K E, Powels R L, Steel H M, Tryhorn Y S, Butcher P D, Booth J C. A polymerase chain reaction to detect a spliced late transcript of human cytomegalovirus in the blood of bone marrow transplant recipients. J Virol Methods. 1996;56:139–148. doi: 10.1016/0166-0934(95)01900-6. [DOI] [PubMed] [Google Scholar]
- 33.Revello M G, Percivalle E, Arbustini E, Pardi R, Sozzani S, Gerna G. In vitro generation of human cytomegalovirus pp65 antigenemia, viremia and leukoDNAemia. J Clin Invest. 1998;101:2686–2692. doi: 10.1172/JCI1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Revello M G, Percivalle E, Zavattoni M, Parea M, Grossi P, Gerna G. Detection of human cytomegalovirus immediate-early antigen in leukocytes as a marker of viremia in immunocompromised patients. J Med Virol. 1989;29:88–93. doi: 10.1002/jmv.1890290204. [DOI] [PubMed] [Google Scholar]
- 35.Revello M G, Zavattoni M, Sarasini A, Percivalle E, Simoncini L, Gerna G. Human cytomegalovirus in blood of immunocompetent persons during primary infection: prognostic implications for pregnancy. J Infect Dis. 1998;177:1170–1175. doi: 10.1086/515277. [DOI] [PubMed] [Google Scholar]
- 36.Rubin R H. Preemptive therapy in immunocompromised hosts. N Engl J Med. 1991;324:1057–1059. doi: 10.1056/NEJM199104113241509. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt G M, Horak D A, Niland J C, Duncan S R, Forman S J, Zaia J A. A randomized controlled trial of prophylactic ganciclovir for cytomegalovirus pulmonary infection in recipients of allogeneic bone marrow transplants. The City of Hope-Stanford-Syntex CMV Study Group. N Engl J Med. 1991;324:1005–1011. doi: 10.1056/NEJM199104113241501. [DOI] [PubMed] [Google Scholar]
- 38.Shinkai M, Spector S A. Quantitation of human cytomegalovirus (HCMV) in cerebrospinal fluid by competitive PCR in AIDS patients with different HCMV central nervous system diseases. Scand J Infect Dis. 1995;27:559–561. doi: 10.3109/00365549509047067. [DOI] [PubMed] [Google Scholar]
- 39.Van den Berg A P, Tegzess A M, Scholten-Sampson A, Schirm J, van der Giessen M, The T H, Van Son W J. Monitoring antigenemia is useful in guiding treatment of severe cytomegalovirus disease after organ transplantation. Transplant Int. 1992;5:101–106. doi: 10.1007/BF00339224. [DOI] [PubMed] [Google Scholar]
- 40.Van Der Bij W, Schirm J, Torensma R, Van Son W J, Tegzess A M, The T H. Comparison between viremia and antigenemia for detection of cytomegalovirus in blood. J Clin Microbiol. 1998;26:2531–2535. doi: 10.1128/jcm.26.12.2531-2535.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zipeto D, Baldanti F, Zella D, Furione M, Cavicchini A, Milanesi G, Gerna G. Quantification of human cytomegalovirus DNA in peripheral blood leukocytes of immunocompromised patients by the polymerase chain reaction. J Virol Methods. 1993;44:45–56. doi: 10.1016/0166-0934(93)90006-d. [DOI] [PubMed] [Google Scholar]
- 42.Zipeto D, Revello M G, Silini E, Parea M, Percivalle E, Zavattoni M, Milanesi G, Gerna G. Development and clinical significance of a diagnostic assay based on the polymerase chain reaction for detection of human cytomegalovirus DNA in blood samples from immunocompromised patients. J Clin Microbiol. 1992;30:527–530. doi: 10.1128/jcm.30.2.527-530.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]