Abstract
Mechanisms that directly control mammalian ovarian primordial follicle (PF) growth activation and the selection of individual follicles for survival are largely unknown. Follicle cells produce factors that can act as potent inducers of cellular stress during normal function. Consistent with this, we show here that normal, untreated ovarian cells, including pre-granulosa cells of dormant PFs, express phenotype and protein markers of the activated integrated stress response (ISR), including stress-specific protein translation (phospho-Serine 51 eukaryotic initiation factor 2; P-EIF2), active DNA damage checkpoints, and cell-cycle arrest. We further demonstrate that mRNAs upregulated in primary (growing) follicles versus arrested PFs mostly include stress-responsive upstream open reading frames (uORFs). Treatment of a granulosa cell (GC) line with the PF growth trigger tumor necrosis factor alpha results in the upregulation of a ‘stress-dependent’ translation profile. This includes further elevated P-eIF2 and a shift of uORF-containing mRNAs to polysomes. Because the active ISR corresponds to slow follicle growth and PF arrest, we propose that repair and abrogation of ISR checkpoints (e.g. checkpoint recovery) drives the GC cell cycle and PF growth activation (PFGA). If cellular stress is elevated beyond a threshold(s) or, if damage occurs that cannot be repaired, cell and follicle death ensue, consistent with physiological atresia. These data suggest an intrinsic quality control mechanism for immature and growing follicles, where PFGA and subsequent follicle growth and survival depend causally upon ISR resolution, including DNA repair and thus the proof of genomic integrity.
Keywords: aging, eukaryotic initiation Factor 2 (EIF2), follicle, integrated stress response, oocyte, ovary, menopause, translational control
Introduction
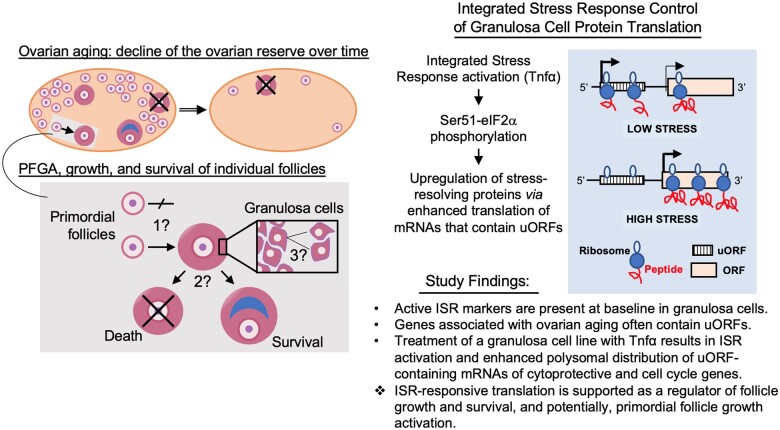
Mammalian ovarian aging consists of a regular commitment of primordial follicles (PFs) to a growth phase. This is referred to as PF growth activation (PFGA) (Kallen et al., 2018). Once committed, most growing follicles die in a process termed atresia, and only a small remaining fraction survives to ovulate. In women, this is usually one egg per menstrual cycle. Human menopause occurs at a median age of 51 years when the number of arrested PFs drops below a threshold of hundreds to a few thousand per ovary (Santoro and Johnson, 2019; Ford et al., 2020). Genetic model systems and large-scale genome-wide association studies (GWAS) have revealed pathways and individual genes that influence the rate of ovarian aging and the age of natural menopause (ANM). GWAS dataset(s) reported by Stolk et al. (2012) revealed the importance of DNA repair pathways and inflammatory signaling in ovarian aging, and this was confirmed and extended by Day et al. (2015). Mechanisms that select individual PFs for growth activation and how growing follicles are selected for survival versus death are largely unknown (see ‘knowledge gaps’, Fig. 1) (Reddy et al., 2010; Zhang and Liu, 2015; Ernst et al., 2017; Kallen et al., 2018). Factors that regulate the overall rate of PFGA are known, however. For example, anti-Müllerian hormone (Amh) negatively regulates the PFGA rate (Durlinger et al., 1999; Pankhurst, 2017). Amh knockout mice display an acceleration of the loss of the PF reserve, and a corresponding increase in growing follicles (Durlinger et al., 1999). Conversely, in vivo elevation of Amh slows PFGA and ovarian aging (Kano et al., 2017). Phosphatase and tensin homolog deleted on chromosome ten (Pten) also negatively regulates PFGA (Reddy et al., 2008; Cheng et al., 2015), acting upon downstream mammalian RAC-alpha serine/threonine-protein kinase (Akt)/Target of Rapamycin (mTOR) signaling to regulate protein synthesis and the cell cycle (Yu et al., 2011; Cheng et al., 2015). Pten inhibition accelerates PFGA in mouse models in vivo and human PFs in experimental clinical interventions (Kawamura et al., 2013; Novella-Maestre et al., 2015), and manipulation of mTOR activity has been used to regulate follicle growth and/or survival (McLaughlin et al., 2011; Goldman et al., 2017). Instead, deficiencies for either tumor necrosis factor alpha (Tnf) (Cui et al., 2011), or, tumor necrosis factor Receptor 2 (Tnfr2) (Greenfeld et al., 2007) significantly slow the PFGA rate and extend ovarian function. While Tnf protein production is limited to oocytes in the mouse ovary (Chen et al., 1993), follicular somatic cells also produce the protein in women (Kondo et al., 1995).
Figure 1.
Knowledge gap(s), concepts, and study findings. Ovals represent aging ovaries that contain declining numbers of the reserve of primordial follicles (small circles) over time (double arrow). Primordial follicle growth activation (PFGA) is the commitment of primordial follicles to a growth phase (gray boxes). Growing follicles most often die in a process termed atresia (follicles crossed by ‘X’), and only a small fraction survive to ovulation. The lower grey box shows two adjacent primordial follicles, one that stays growth-arrested, and the other that undergoes PFGA. Mechanisms that control whether individual primordial follicles undergo PFGA or stay arrested, for what can be decades, are mostly unknown (1?). Similarly, mechanisms that select individual growing follicles for death or survival are unclear (2?). Finally, follicle growth is characterized by the slow proliferation of granulosa cells (3?), such that early growth stages are thought to take months (Gougeon, 1986). Factors that enforce this slow growth rate are also mostly unknown. The hypothesis of this study and study findings are summarized at right. We hypothesized that regulation of protein translation by the integrated stress response (ISR; pathway diagram in Supplementary Fig. S1) controls granulosa cell growth and survival. In support of this hypothesis, genes previously identified in large scale studies as associated with human ovarian aging were shown to preferentially contain stress-responsive upstream open reading frames (uORFs), whose action is summarized in the diagram within the blue box. Under conditions of low stress, ribosomes occupy upstream reading frames and translation of the depicted mRNA is inefficient and low. Under conditions of elevated stress, ribosomes ‘skip’ the uORF and more efficiently translate the mRNA’s encoded protein. Activating transcription factor 4 (ATF4) is one well-studied example of translational control by stress and uORFs (Vattem and Wek, 2004). Overall, we found that markers of the activated ISR are present under normal baseline conditions in mouse and human granulosa cells and the pregranulosa cells of primordial follicles. In addition, analysis of genes shown to be involved in PFGA and ovarian aging revealed that uORFs are present and have higher ‘quality scores’ (McGillivray et al., 2018) than found in randomly selected genes. Last, we found that treatment of a mouse granulosa cell line with PFGA activator (Cui et al., 2011; Greenfeld et al., 2007) tumor necrosis factor alpha (Tnf resulted in the redistribution of transcripts that contain uORFs to the actively translating polysome subcellular fraction. The data were interpreted as supporting the ISR as a regulator of follicle growth and survival, and potentially PFGA.
Stress and oxidative damage have long been associated with ovarian aging (Oktay et al., 2020). Indeed, exogenous DNA-damaging agents, including chemotherapeutic drugs, can accelerate the process. There is evidence that PFGA occurs prior to overt measures of follicle death after chemotherapeutic treatment, a phenomenon referred to as follicular ‘burnout’ (Kalich-Philosoph et al., 2013; Roness et al., 2013). In other settings, the endogenous PFGA activator TNF has been shown to act as a cell damage agent and ‘potent endogenous mutagen’ (Yan et al., 2006, 2009). TNF exposure can result in increased reactive oxygen species, and in the oxidative DNA lesion 8-oxo-7,8-dihydroguanine (8-oxoG) (Wu et al., 2004; Menoni et al., 2012; Ba et al., 2014). Other endogenous factors produced within the ovary are also known to induce specific types of cellular damage, e.g. acetaldehyde (Kawai et al., 2012). Compared to exogenous agents (e.g. chemotherapies), links between physiologically induced cellular damage and PFGA (and, thus, ovarian aging) are understudied.
Different classes of cellular stress activate the four integrated stress response (ISR) eukaryotic initiation Factor 2 (eIF2) activator kinases (EIF2AK1-4), which in turn can upregulate cytoprotective/DNA repair and cell-cycle machinery (Supplementary Fig. S1) (Pakos-Zebrucka et al., 2016; Costa-Mattioli and Walter, 2020). DNA damage response (DDR) activation of protein kinase, DNA-activated, catalytic subunit/Ku (DNA-PKcs/PRKDC; Ku/XRCC5) overlaps with the ISR in its phosphorylation of EIF2AK4/GCN2 (Kondrashov et al., 2009; Powley et al., 2009; Clementi et al., 2020) and ataxia telangiectasia and Rad3-related (ATR) kinase, while activation of ataxia telangiectasia mutated (ATM) kinase can activate the ISR kinase EIF2AK3/PERK (Kondrashov et al., 2009; Powley et al., 2009; Clementi et al., 2020). ATR (Dai and Grant, 2010; Zhang and Hunter, 2014) and PERK (via ATM) (Malzer et al., 2010) phosphorylate cell-cycle checkpoint kinases Chk1 and Chk2 (Bartek and Lukas, 2007; Dai and Grant, 2010) and induce cell-cycle arrest (Supplementary Fig. S1) (Levitus et al., 2006). DDR activation of ISR proteins, thus, induces cell-cycle arrest until DNA damage lesions are repaired (Marini et al., 2006).
Activated ISR kinases also phosphorylate the core regulator eukaryotic initiation Factor 2 subunit alpha (eIF2/EIF2S1) on Serine 51. Under ‘non-stressed’ conditions, low eIF2 phosphorylation allows its activation by eIF2B guanine nucleotide exchange (eIF2-GTP) and initiation of 5’ cap-dependent mRNA translation. P-eIF2 (P-eIF2-GDP, in complex with eIF2B) does not initiate protein translation and a reduction in active eIF2-containing ternary complexes (TCs) results in the use of alternative initiation mechanisms. These can include a switch to eIF3-dependent initiation (Guan et al., 2017) and the use of alternative eIFs (eIF2A, eIF5B, and eIF2D/MCT-1/DENR) (Komar and Merrick, 2020). Genetic variants in protein translation factors have previously been associated with accelerated human ovarian aging. Variants in eIF2B (Fogli et al., 2004), eIF3M (Day et al., 2015) and eIF4E (Ernst et al., 2017), and mutations in the eIF4E nucleocytoplasmic shuttle EIF4ENIF1 show strong associations with accelerated human ovarian aging (Kasippillai et al., 2013; Zhao et al., 2019). In a prior study of galactosemic mice, our group detected P-eIF2 in mutant and wild-type (WT) ovarian total protein lysates (Balakrishnan et al., 2019).
Upstream open reading frames (uORFs) are ‘sequences defined by an initiation codon in frame with a termination codon located upstream or downstream of the main AUG’ (Barbosa et al., 2013). uORFs often occur within the 5’ untranslated region (UTR) of mRNAs and, as defined, can overlap the coding region of the gene product itself (Vattem and Wek, 2004; Pakos-Zebrucka et al., 2016; Costa-Mattioli and Walter, 2020). These motifs are distinct from upstream in-frame alternative start codons that, if used, result in the production of longer proteins (Orr et al., 2020). Ribosomal occupancy of uORFs, which sometimes includes the translation of uORF-encoding peptides, usually comes at the expense of the efficient translation of an mRNA’s encoded protein. Thus, a switch from uORF occupancy to translation of the mRNA-encoded peptide can be used to regulate protein translation (Fig. 1, right panel). Relevant to our work here, ‘genes with [upstream open reading frame (uORF) motifs] in their transcripts are good candidates for upregulated translation in response to eIF2 phosphorylation’ (Barbosa et al., 2013). Enhanced translation of mRNAs that harbor uORFs occurs as follows (Takahashi et al., 2020). Under low-stress conditions, eIF2-GTP is available to initiate translation in the TC with Met-tRNAi. Here, there are sufficient eIF2-GTP levels so that uORF translation initiation is favored, at the expense of initiation at the gene’s translational start. Under conditions of stress, limited eIF2-GTP levels limit active TC availability to participate in initiation events. Continued 40S ribosomal subunit scanning when TCs are limited in this way favors uORF bypass, and initiation at the gene product’s bona fide ORF translation start site (Vattem and Wek, 2004; Barbosa et al., 2013).
Female individuals with the condition classic galactosemia often exhibit a severe primary ovarian insufficiency (POI) phenotype, reflective of an accelerated loss of PFs (see Hagen-Lillevik et al., in press, for a review). We have shown (Balakrishnan et al., 2017, 2019) that maintenance of eIF2 Ser51 phosphorylation using the pharmacological agent Salubrinal (Boyce et al., 2005) rescues PF numbers and normalizes normal ovarian function, fertility, and fecundity long-term in a mouse model of classic galactosemia (galactose-1-phosphate uridylyltransferase knockouts [GalTKO]) that demonstrates a severe POI-like phenotype when untreated (Balakrishnan et al., 2017, 2019). However, it was not clear in those studies whether ISR activity at the level of P-eIF2 might be related to PFGA and ovarian aging under normal circumstances. The correspondence between the maintenance of the active ISR with Salubrinal, and PF arrest and overall improved follicle survival in galactosemic conditions, suggested that the ISR might be active under physiological conditions in the WT ovary. While perhaps unanticipated, baseline ISR activation could indeed correspond to cytoprotection and cell-cycle arrest within dormant and slowly growing ovarian follicles.
The above information led us to the hypothesis that constitutive elevation of the ISR occurs in WT granulosa cells (GC), and that resolution of physiological stress/DNA damage results in P-S51 eIF2 dephosphorylation and a checkpoint resolution-dependent ‘switch’ to an active cell-cycle (Kaufmann and Paules, 1996; Marini et al., 2006; Bartek and Lukas, 2007; Shaltiel et al., 2015; Smits and Gillespie, 2015). If correct, transcripts that contain uORFs might be over-represented in cells of immature ovarian follicles, at least relative to their expected overall incidence of approximately 49% in all human genes (Barbosa et al., 2013). We addressed this using publicly available datasets. The first dataset, produced by Zhang et al., reports oocyte and GC transcriptomes from cells isolated at different stages of follicle development (Zhang et al., 2018). Because DDR pathways have been implicated in ovarian aging (Stolk et al., 2012; Day et al., 2015), including specifically at the level of PFGA (Roness et al., 2013; Lin et al., 2017; Oktay et al., 2020), ISR regulation of cytoprotective and cell-cycle genes was our focus. Separately, McGillivray et al. generated a searchable whole human genome catalog of uORFs (http://github.gersteinlab.org/uORFs/; McGillivray et al., 2018). uORF functionality prediction involved the calculation of ‘uORF [quality] scores’, that classify uORFs as ‘high quality’ (HiQ) and thus likely functional, and those with the ‘top 10%’ (Top10) highest probability of being functional.
After determining the pattern of P-eIF2 and other markers of cellular stress in human and mouse ovarian follicles, we catalogued uORF content in transcripts expressed by GCs of immature follicles. We then tested whether TNF treatment leads to ISR activation in a way that, first, impacted GC proliferation and, second, involved a stress-activated redistribution of uORF-containing mRNAs to polysomal fractions, indicating ISR influence upon protein translation.
Materials and methods
Bioinformatic analyses
Publicly available human granulosa cell trancriptome (Zhang et al., 2018) and uORF (McGillivray et al., 2018) data were downloaded and analyzed using R (R Core Team, 2018). Statistical tests used are indicated in legends and P < 0.05 was considered significant.
Tissue collection and processing
Mice were maintained under an approved protocol at the University of Utah School of Medicine (Lai); GalTKO and WT control mice were sacrificed and ovaries were collected at postnatal Day 36. Approval for the use of paraffin-embedded human ovary samples was granted by the Colorado Multiple Institutional Review Board as de-identified, discarded tissue (COMIRB Protocol #17-1428). Paraffin embedding was performed using a standard protocol by the University of Colorado Department of Pathology.
Immunohistochemical staining
For fluorescence immunohistochemical (IHC) staining of paraffin sections, deparaffinization and rehydration were followed by antigen retrieval (10 mM citrate buffer, pH 6 at 120°C for 30 min). Peroxidase blocking was performed using 3% hydrogen peroxide (Labchem, Pittsburgh, PA, USA, #LC154301). Sections were washed in water, followed by PBS containing 0.05% Tween 20 (0.05% TBST). Blocking was performed using 1% (wt/vol) bovine serum albumin, 10% normal goat serum at room temperature for 90 min. Sections were then incubated overnight at 4 with rabbit anti P-Ser51 eIF2 (Cell Signaling Technology [CST], Danvers, MA, USA, #3398).
After washes, Alexa Fluor 647 fluorophore-conjugated anti-rabbit second antibody (Abcam, Cat. No. ab150075) was applied.
Colorimetric IHC was used for P-Chk1 (AbCam, Cambridge, UK, #Ab58567) and P-Rpa2 (AbCam, #Ab10359). After three washes 0.05% TBST, sections were incubated for 30 min. in biotinylated goat anti-rabbit antibody (Abcam, #6721). Signal was detected using an ABC Kit (Vector Laboratories, Burlingame, CA, USA, Vectastain #PK6100) with DAB (Vector Labs, #SK-4100). Harris hematoxylin was used for counterstaining. Slides were then dehydrated and sections were mounted in Permafluor (Thermo Fisher Scientific, Waltham, MA, USA #SP15-500). Negative controls were prepared identically, except that first antibody was omitted. Microscopy was performed with Olympus microscope (#CKX41SF) with 4×, 10× and 20× objectives.
Protein lysate preparation and western blotting
OV3121 cells (Yanagihara et al., 1995) were plated in T75 flasks using Advanced DMEM/F12 (Invitrogen Gibco, Waltham, MA, USA, #12634), 2% l-glutamine 200 mM (Invitrogen Gibco, #25030-081), 2% fetal bovine serum (FBS: Invitrogen Gibco, #26140-079), and 1% Pen Strep (Invitrogen Gibco, #15140-122). Once at 50% confluency, 10 ng/ml TNF (R&D Systems, Minneapolis, MN, USA, #210-TA-100) treatment or PBS (Gibco, Ref 20012-043) vehicle were added as indicated. Treatments were performed in triplicate unless noted. For protein isolation, cells were washed with cold PBS followed by RIPA lysis buffer (Thermo Fisher Scientific, #78410) including Halt Protease Inhibitor (Thermo Fisher Scientific, #78410). Flasks containing treated OV3121 cells were scraped, and cells were disrupted by pulling through a 20G syringe 20×. Homogenates were centrifuged at 14 000g for 5 min and BCA protein quantification was performed (Thermo Fisher Scientific, #23225), reading at 562 nm.
Protein (10 μg or 20 μg) was combined with 4X NuPAGE Sample Loading Buffer (Invitrogen, #NP0007) and boiled at 95°C for 10 min (VWR Scientific, Radnor, PA, USA, #13259-005). Samples were run on 4–15% gradient Mini-Protean TGX gels (Bio-Rad, Hercules, CA, USA, #456-1083) for 1 h at 120 V and transferred to Immuno-Blot PVDF membranes (Bio-Rad, #162-0182) overnight at 35 V at 4°C. Membranes were washed 3× with TBST then blocked by incubation with TBST plus 10% nonfat milk for 1 h at room temperature. Membranes were then incubated with primary antibodies diluted in 2% milk overnight at 4. These were anti-P-eIF2 (CST, #3398) or anti-p65 (CST, #8242). After TBST washes, membranes were incubated with horse-radish peroxidase-linked goat secondary antibody (CST, #7074S) diluted 1:3000 for 1 h at room temperature and again washed. Signal was detected using the SuperSignal West Dura chemiluminescence substrate detection kit (Thermo Fisher Scientific, #34076). Densitometry was performed by using a GBOX Chemi XT4 gel imaging system and Image J analysis software. Amido Black staining was used for loading control normalization.
Polysome profiling and Nanostring transcript partition analysis
OV3121 cells were grown as above, here in T175 flasks. At 70% confluency, TNF (R&D Systems, #210-TA-100) was added at 10 ng/ml for 3 h and 12 h; vehicle (VEH) controls were collected at 3 h. For collection, media was removed and 500 l lysis buffer was added (20 mM HEPES, pH 7.4, 15 mM MgCl2, 200 mM NaCl, 1% Triton X-100, 0.1 mg/ml cycloheximide, 2 mM DTT, and 100 U/ml ribonuclease inhibitor). Homogenates were collected into prechilled 1.5 ml Eppendorf tubes.
Homogenates were centrifuged for 5 min at 13 000g to pellet debris, then 500 l clarified lysate was loaded onto 10–60% sucrose gradients in SW41 tubes, in lysis buffer lacking Triton X-100. Gradients were prepared using a BioComp system and chilled to 4 before use. Samples were ultracentrifuged in a SW41 rotor at 160 000g for 3 h and 10 min, at 4, then samples were fractionated using the BioComp system, monitoring absorbance at 260 nm while collecting fractions of approximately 0.4 ml. Fractions were stored at −80°C until use.
For Nanostring analysis, RNA was isolated using Qiazol (Qiagen, Inc., Germantown, MD, USA, #79306). Individual fractions were combined into ‘SUB’-polysomal and POLYsomal pooled samples (Heyer and Moore, 2016). RNA purity and concentrations were determined using a NanoDrop 2000 Spectrophotometer (Thermo Scientific, #D467). Samples were normalized to 15 ng/l by diluting in DEPC-treated water. Hybridization reaction master mix was prepared by adding 70 l hybridization buffer to the reporter code set: 8 l of master mix was added to 5 l of sample for 13 l total volume. Then 2 l of capture probe set was added, tubes were mixed by inverting and spun down briefly. Samples were incubated at 65°C using a Verti thermal cycler (Applied Biosystems LLC, Norwalk, CT, USA) for 20 h before adding 15 l of hybridization buffer. A total of 30 l volumes were then loaded into each well of the nCounter mouse PanCancer Pathways IO360 cartridge (Nanostring Technologies Inc., Seattle, WA, USA, #LBL-10545-01). The cartridge run was performed using a Nanostring Sprint Profiler (Nanostring Technologies Inc.) at the VA Medical Center Research Core Facility (Aurora, CO, USA). Data quality control analysis was performed using nSolver software, and ROSALIND (Rosalind, Inc., San Diego, USA) and R were used for transcript count determination.
Results
The ISR is active in cells of arrested primordial and growing follicles
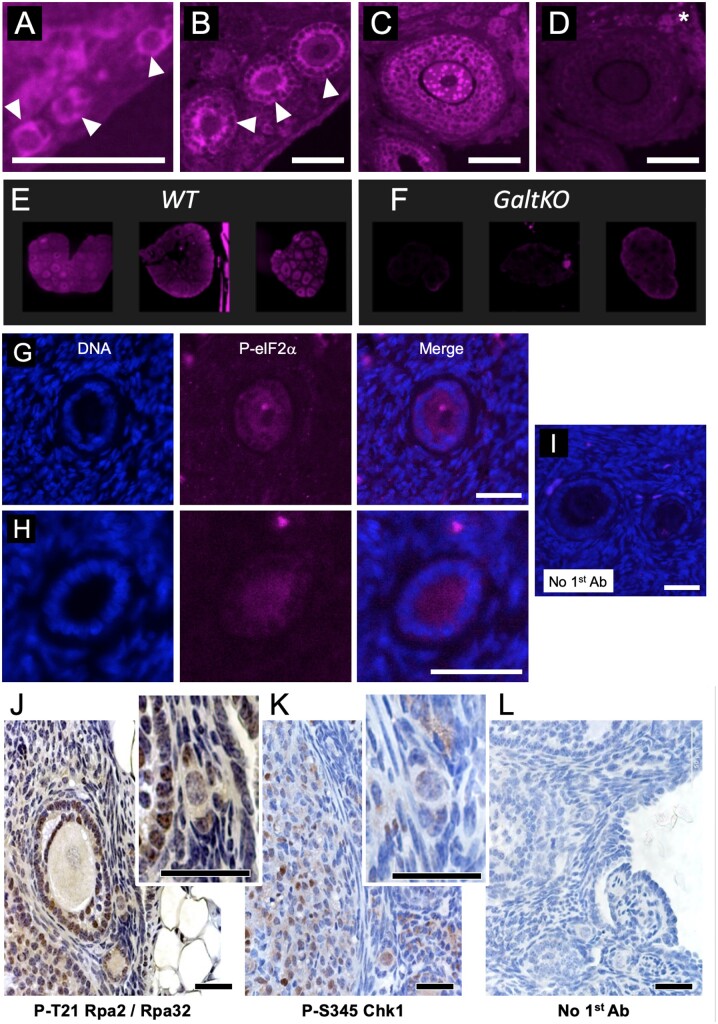
We first evaluated eIF2 Ser 51 phosphorylation in the WT mouse ovary (Fig. 2A–E) by immunofluorescence, comparing signal with that in the GalTKO ovary (Fig. 2F). The phosphoprotein is detectable in WT ovaries in both GCs and oocytes within a range of follicle sizes, and direct fluorescence comparison (Fig. 2E vs F) supports the presence of higher P-eIF2 in WT than GalTKO ovarian follicles (background staining, Fig. 2D). P-eIF2 is also detectable in immature follicles in the adult human ovary (representative images from two unique specimens, Fig. 2G and H). Overlap between DDR and ISR downstream machinery (Supplementary Fig. S1) led us to next evaluate two biomarkers of ongoing DNA damage, phospho-threonine 21 replication protein A2 (P-Rpa2/RPA34), indicative of single-stranded DNA repair intermediates (Iftode et al., 1999; Block et al., 2004; Overmeer et al., 2011), and Phosphoserine 345 checkpoint kinase 1 (Chk/Chek1) (Dai and Grant, 2010; Patil et al., 2013; Zhang and Hunter, 2014). Both DDR markers are detectable (Fig. 2J and K) in multiple ovarian cell types, including PF pregranulosa cells and oocytes (Fig. 2J and K, insets). These DDR marks are consistent with an active ISR, and also, active cell-cycle checkpoints.
Figure 2.
Active ISR marker detection in immature follicles of the mouse and human ovary. (A) Phosphoserine 51-eukaryotic Initiation Factor 2 subunit alpha (P-eIF2) is detectable via immunofluorescence (purple color is positive signal) in histological preparations of the mouse ovary. PFs are shown in A (arrowheads), primary follicles in (B) (arrowheads), and a small pre-antral follicle is shown in (C). Note that in all cases, both the oocyte and granulosa cells (GCs) exhibit signal; intense punctate staining is visible in some growing oocytes (example in C). A control image where the first antibody to was omitted is shown in (D) (asterisk indicates autofluorescent background) and is the adjacent tissue section to the image shown in C. Three panels are provided in (E) that show wild-type (WT) levels of P-eIF2 for direct comparison with galactose-1-phosphate uridylyltransferase knockout mouse (GalTKO) ovaries in F, images collected using identical settings. (G) and (H) show representative staining for P-eIF2 in immature human follicles from separate patient biopsies (DNA/DAPI, P-eIF2 channels and merged image labeled). (I) is a merged image of DAPI and background staining when first antibody to P-eIF2 is omitted, identical settings as H and I. Panels (J) and (K) show colorimetric staining of mouse ovaries for two DNA damage marks, P-T21 replication protein A2 (Rpa2) and P-S345 checkpoint kinase 1 (Chk1), respectively. Insets show electronically magnified areas that include PFs. Panel (L) is a no 1st antibody control. Scale bars for Panels A–D = 100, G–I = 20 and J–L = 75 m, respectively.
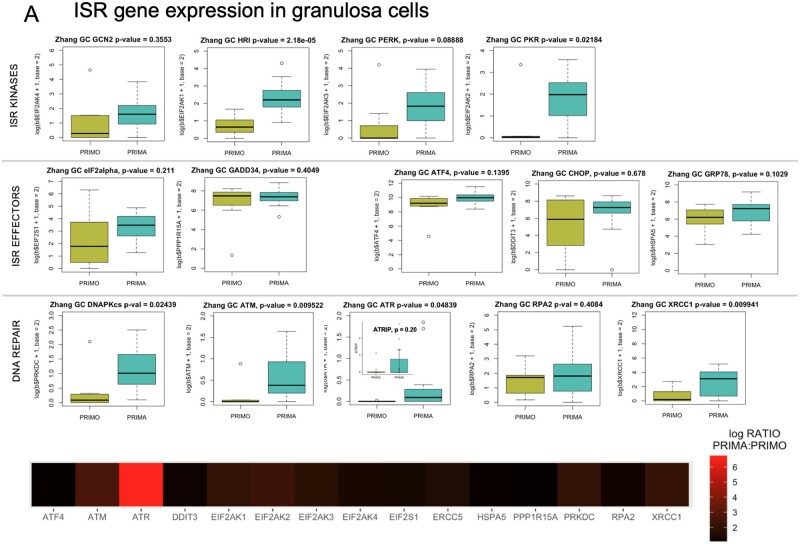
We then evaluated ISR core kinase, key effector, and DDR expression in the Zhang et al. (2018) granulosa cell datasets of human primordial and primary follicle transcripts. GCN2, HRI, PERK, PKR, and DNA-PKcs were all detected, with HRI, PKR, and DNA-PKcs at significantly higher levels at the primary than the PF stage (Fig. 3A, plots, matched heatmap). eIF2 (EIF2S1) expression was confirmed, as was that of the regulatory phosphatase GADD34, known to de-phosphorylate its Ser51. ISR-responsive ATF4 was also reported at both stages, along with two factors known to be upregulated by ATF4, CHOP/DDIT3, and HSPA5/GRP78. DDR genes ATM, ATR (and cofactor ATRIP), RPA2, and XRCC1 were all present at both stages, and all of these excepting ATRIP and RPA2 were significantly higher in primary than PF GCs. An active ISR would also mean that ATF4 (itself translated in response to ISR activation) transcriptional targets should be transcribed. ATF4 response-element containing genes ASNS, TRIB3, NLRP1, SLC7A1, SLC38A2, ATF3, VEGFA, and 4E-BP/EIF4EBP1, are all expressed in PF and primary follicle GCs (Supplementary Fig. S2) (Kilberg et al., 2009).
Figure 3.
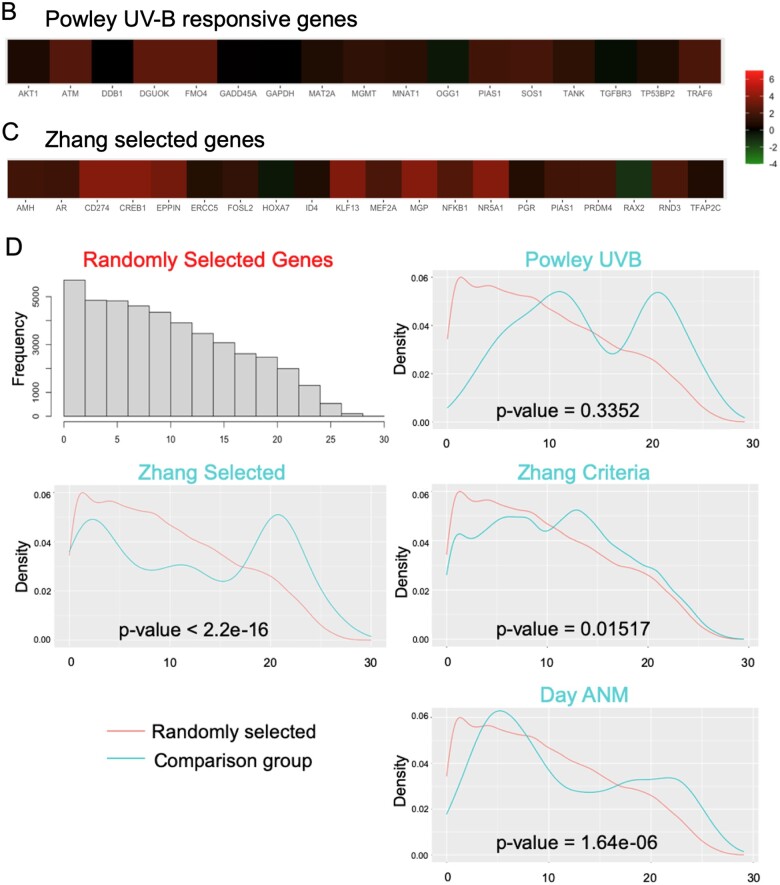
Gene expression and uORF analysis of control cell and GC gene sets. Zhang et al. (2018) gene expression data were explored and comparative expression of ISR machinery and DNA repair genes between PF and primary follicle GCs was determined (A). Heme-regulated inhibitor (HRI/EIF2AK1), protein kinase RNA-activated (PKR/EIF2AK2), protein kinase, DNA-activated, catalytic subunit (DNAPk-cs/PRKDC), ataxia telangiectasia mutated (ATM), ataxia telangiectasia and rad3 related (ATR), and X-ray repair cross complementing 1 (XRCC1) were all significantly higher in primary GCs. Data in panel A graphs are summarized as a heatmap immediately below, with the log (Base 2) ratio of expression in primary GCs to PF GCs depicted. (B) shows a heatmap generated as in (A) of the Powley UVB-responsive gene set, and Panel (C) is a heatmap of the same expression ratio for the Zhang selected GC gene set. (D) is a comparison of Gerstein uORFs quality scores between the selected gene sets and randomly selected genes. Histograms of uORF score frequencies were prepared for a set of randomly selected genes (top left) and each of the previously evaluated gene sets (Powley, Zhang selected, Zhang criteria, and Day ANM: age of natural menopause). Density plots reveal that in each case, a rightward ‘shift’ in uORF scores is present (blue) compared to randomly selected genes (red). In the cases of Powley UVB, Zhang selected GC, and Day ANM gene sets, a high score ‘bump’ where uORF scores within a particular range are over-represented is detectable compared to randomly selected genes. These shifts correspond directly to increased mean HiQ uORF scores for each gene set (Table II). P values shown comparing uORF densities were calculated using the Kolmogorov–Smirnov test.
Figure 3.
Continued.
Because cell-cycle regulators are directly regulated by the ISR (Blais and Bell, 2006; Lee et al., 2009; Malzer et al., 2010), and loss of function studies show that at least one of the G1 cyclins (D- or E-type) is required for mammalian cell proliferation (Liu et al., 2017), we compared their expression in non-growing PF pregranulosa to expression in growing GCs of primary follicles. G1 cyclin mRNA levels did not significantly differ between GCs at the two follicle stages (Supplementary Fig. S3). We found that all five G1 cyclin genes contain uORFs, four of the five have HiQ uORFs, and Cyclins D1 and D2 have Top10 uORFs. We reasoned that post-transcriptional ISR activity might control G1 cyclin translation and, thus, cell-cycle progression.
ISR and stress-responsive gene expression in the GC of immature human ovarian follicles: a correspondence with uORF content
The above data supported possible stress-dependent protein translation, including the regulatory involvement of uORFs (Vattem and Wek, 2004; Pakos-Zebrucka et al., 2016; Costa-Mattioli and Walter, 2020). We, therefore, predicted that uORFs would be over-represented in the genes whose expression levels change in GC during PFGA, relative to their approximately 49% presence in all human genes (Barbosa et al., 2013). We tested this as follows. Powley et al. (2009) assessed UVB induced stress-responsive translation in HeLa cells, and found that activation of DNA-PKcs corresponded to increased downstream P-eIF2 and an altered mRNA translation profile. As a ‘test set’ for our study, we used their reported list of genes whose mRNAs shifted from sub-polysomal fractions to polysomal fractions after UVB exposure for uORF analysis. All 17 genes (100%) whose mRNAs exhibited a UVB stress-responsive shift are expressed by human PF and primary GCs (Fig. 4B), with none expressed at lower levels in primary GCs than PF pregranulosa cells. mRNAs of all 17 genes contain uORFs (Table I, ‘Powley UVB’); 15 of 17 contain HiQ uORFs and six of those, Top10 uORFs.
Figure 4.
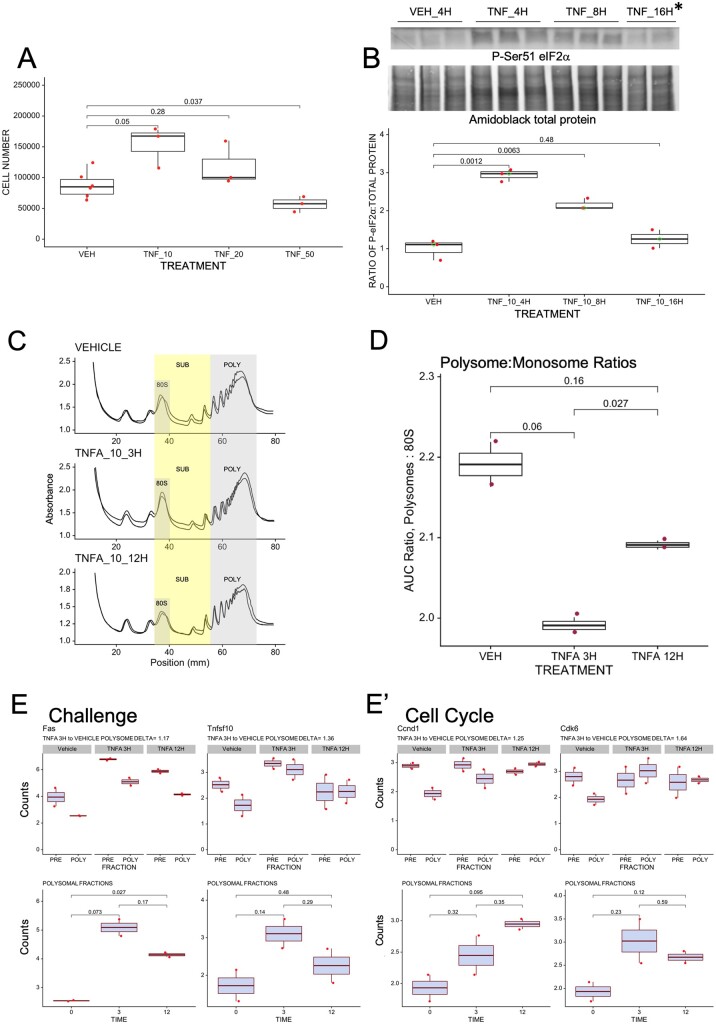
ISR activation in a murine granulosa cell line using Tnf results in stimulated growth and an altered translational profile that corresponds to uORF content. To establish a model of GC ISR activation, we treated OV3121 cells with a dose curve of Tnf (Tnfα at 10 ng/ml is labeled TNF_10; 20 ng/ml, TNF_20; and 50 ng/ml, TNF_50) or vehicle (VEH) and evaluated cell growth 24H post-treatment
Table I.
Incidence of upstream open reading frames in gene sets of interest.
| Gene Set | Top10 uORFs | HiQ uORFs | Other uORFs | No uORF |
|---|---|---|---|---|
| Powley UVB | AKT1, DDB1, GADD45A, PIAS1, TGFBR3, TP53B2 | ATM, DGUOK, FMO4, GAPDH, MGMT, MNAT1, OGG1, TANK, TRAF69 | MAT2A, SOS1 | |
| Zhang Selected GC | CREB1, FOSL2, KLF13, NFKB1, PIAS1, PRDM4 | HOXA7, MEF2A, TFAP2C, AR, PGR, ID4, MGP, CD274 | RAX2, NR5A1, RND3, EPPIN | AMH |
| Day ANM | APEX1, C16orf72, DIDO1, EIF4B, FBXO18, GSPT1, GTF3C2, INO80, PAPD7, POLG, POLR2E, REV3L, SH3PXD2B, SPPL3, STX6 | ABAT, APTX, BRE, CDK12, CHD7, FAM175A, FANCI, HELQ, KNTC1, MCM8, MSH5, MYCBP, PARP2, PGAP3, PITPNM2, POLR2H, PNP, PRIM1, STAR, STARD3, UIMC1 | ASCL1, BCAR4, BRCA1, BRSK1, CENPU, CHEK2, DDX17, DMC1, EIF3M, EXO1, FSHB*, GAD1, HELB, KISS1R*, MAK, MSH6, NLRP11, PARL, PIWIL1, RAD51, RAD54L, RHBDL2, RPAIN, SLCO4A1, SRSF9, SYCP2L, TAC3, TDRD3, TLK1, U2AF2, UBE2MP1, ZNF729 |
Not detectable in granulosa cells.
ANM, age at natural menopause; GC, granulosa cells; uORF, upstream Open Reading Frame; UVB, ultraviolet B light.
Figure 4.
Continued by direct cell counts (A) and with a fluorescent assay (Supplementary Fig. S4A). In each case, 10 ng/ml Tnf stimulated cell growth 24 H post-treatment, 20 ng/ml resulted in no net change in cell number (Panel A, Supplementary Fig. S4A), and 50 ng/ml resulted in a significant reduction in cell number (A). Panel (B) shows that 10 ng/ml Tnf significantly increases Ser51 P-eIF2 at 4 and 8 h (H) compared to cells collected 4H post-VEH (asterisk * indicates that two replicates were performed for 16H time point instead of three as in other cases). Panels (C) and (D) show normalized ribosomal fractionation traces and corresponding Polysome: Monosome ratios for two OV3121 cell replicates treated with VEH or 10 ng/ml Tnf for 3 or 12 H. Areas under the curve for POLY(some) and 80S regions indicated in C were used to calculate ratios in D. Plots in (E) are representative examples of Nanostring quantification of mRNAs in SUB (polysomal) versus POLY fractions in (C). POLY: SUB deltas were determined by calculating the ratio between the POLY:SUB ratio for 3 h Tnf treatment and VEH-treated cells. Sub-panels in Fig. 4E denote functional gene groups, and sub-panel 4E”’ is a data summary where mRNAs are organized by function and threshold ISR activity is indicated by the dashed vertical line. Panel (F) shows a summary density plot of Gerstein uORF quality scores for human mRNAs whose mouse orthologues are enriched (UP) in OV3121 polysome fractions 3H post-Tnf compared to those that are diminished (DOWN) in polysomes. P values for A, B, D and E were determined using Welch’s t test, for F, the Kolmogorov–Smirnov test.
We next evaluated uORFs in the mRNAs of genes that Zhang et al. (2018) identified as reflective of PFGA due to altered expression between PF and primary stages (‘Zhang selected GC’ gene set) (Zhang et al., 2018). One of the Zhang selected GC genes, protein inhibitor of activated stat-1 (PIAS1) (Sudharsan and Azuma, 2012), was included independently in the Powley et al. UV-B responsive gene set. A heatmap of the GC expression ratio for this gene set is provided for validation purposes and for comparison to other gene sets in Fig. 3C. As seen in the Powley UVB set, most Zhang selected GC mRNAs upregulated after PFGA were found to have uORFs (Table I). A total of 18 of 19 Zhang selected GC genes have any uORF (excepting AMH; 95% incidence), 14 have HiQ uORFs, and 6 have Top10 uORFs. All but two genes in this gene set (HOXA7 and RAX2) are upregulated in primary follicle GCs compared to pregranulosa cells, and most of those are significantly upregulated (P ≤ 0.05; Fig. 4C). Qualitative analysis of the gene set revealed that 12/13 Zhang selected GC genes with HiQ uORFs are transcriptional regulators, and nine of those gene products upregulate DNA repair genes after cellular insult.
Because uORF incidence and quality in the selected gene sets positively correlated with genes whose expression changes between arrested PF pregranulosa cells and growing primary follicle GCs, we expanded our analysis of the Zhang et al. dataset. We selected all human genes that are expressed in primary GCs above a threshold of 1.5 (raw mean value), with a ratio of expression between primary: PF GCs greater than or equal to two. This ‘Zhang all criteria’ gene set included 2656 total genes, 2251 of which had 168 933 uORFs (85% had uORFs, 75.0 uORFs/gene).
We also interrogated uORFs in 68 genes implicated in the human ANM (Day et al., 2015) (Day ANM gene set). We first probed Zhang et al. (2018) expression data for the Day ANM genes and removed two that lacked expression in either PF pregranulosa cells or primary follicle GCs (FSH subunit B (FSHB) and kisspeptin receptor 1 receptor (KISS1R)). All of the genes selected in this way contain uORFs (100% incidence, Table I) and exhibit a higher incidence of Top10 and HiQ uORFs than would be expected given the incidence of those uORFs in all transcripts. Day et al. genes with Top10 uORFs include DNA repair factors (APEX1, FBXO18, PAPD7, REV3L) and transcription and translation machinery (EIF4B, GSPT1, GTF3C2, INO80, POL2RE).
We then compared uORF content and characteristics between gene sets and randomly selected genes (Table II and Fig. 3D). In all cases, the experimental group gene sets exhibited very highly significantly elevated mean HiQ uORF scores compared to those selected randomly (Table II). Zhang all criteria and Day ANM gene sets also had mean Top10 category uORF scores that were significantly higher than randomly selected genes. Density plots revealed that individual HiQ uORF quality scores exhibit distinct patterns from randomly selected genes, with HiQ uORF over-representation visible as a high score ‘bump’ within particular ranges (Fig. 3D). At this point, we added assessment of the gene set of a high-throughput analysis panel (Nanostring pan cancer, see Materials and methods).
Table II.
Mean uORF score by gene set.
| HiQ |
Top10 |
|||
|---|---|---|---|---|
| Random comparison, mean ± SEM (n) | 9.78 ± 0.03 (43 799) |
22.95 ± 0.03 (2698) |
||
| Mean ± SEM (n) | P-value | Mean ± SEM (n) | P-value | |
| Powley selected UV-B | 13.99 ± 0.39 (298) | <2.2E−16 | 23.07 ± 0.20 (58) | 0.55 |
| Zhang selected PFGA | 11.97 ± 0.43 (382) | 4.53E−07 | 22.78 ± 0.22 (66) | 0.45 |
| Zhang all criteria | 10.99 ± 0.04 (28 454) | <2.2E−16 | 23.07 ± 0.03 (2227) | <0.005 |
| Day menopause related | 11.53 ± 0.25 (887) | 8.60E−12 | 23.26 ± 0.12 (138) | 0.01 |
| Nanostring pan cancer | 11.57 ± 0.07 (8981) | <2.2E−16 | 22.94 ± 0.05 (910) | 0.86 |
P-values calculated using Welch's t-test.
PFGA, primordial follicle growth activation.
Differences in mean uORF lengths and uORF start codon usage were also detected between gene sets and randomly selected genes (not shown). A trend toward the over-representation of alternative start codons CTG, GTG, and TTG (relative to ATG) was observed in GC and ANM gene sets. Overall, uORFs were overrepresented in terms of their incidence in genes associated with PFGA, GC function and ovarian aging, and differences in uORF ‘quality’ and features such as length and start codon usage suggested that they might have a role in controlling GC protein translation downstream of the activated ISR.
Tnf treatment induces transfer of mRNAs consistent with the ISR to polysomes
We next addressed the question of whether the presence of uORFs corresponded to increased transit from sub-polysome to polysomal fractions in a mouse GC model, namely OV3121 cells (Yanagihara et al., 1995). Such transit to polysome fractions is indicative of the active translation of the corresponding mRNAs. In initial studies, we tested the hypothesis that the PFGA activator Tnf would stimulate growth of the cell line as seen in the stimulation of PFGA (Greenfeld et al., 2007). Treatment of OV3121 cells with 10 ng/ml Tnf significantly stimulated OV3121 proliferation above that of VEH-treated cells, with a significant near-doubling of cell number at 24 h (Fig. 4A and Supplementary Fig. S4A). No difference in cell number was seen with 20 ng/ml, and cell number instead declined with 50 ng/ml Tnf (Fig. 4A). Tnf bioactivity at 10 ng/ml was validated by detection of transit of the NFB factor p65 to OV3121 nuclear fractions 12 h post-treatment (Supplementary Fig. S4B) (Martin and Fresno, 2000, Ding et al., 2003).
We then evaluated the impact of Tnf treatment upon Ser 51 eIF2 phosphorylation. P-eIF2 significantly increased 4 and 8 h post-treatment, returning to VEH levels at 16 h (Fig. 4B). Because Tnf upregulation of P-eIF2 corresponds to ISR activation, we evaluated potential changes in overall protein synthesis. After treating OV3121 cells with VEH or 10 ng/ml Tnf for 3 or 12 h, we visualized and fractionated the polysome populations using sucrose gradient centrifugation. Polysomal profiles (Fig. 4C) showed that Tnf induces a distribution change, where polysomal mRNAs (actively translating) were significantly reduced compared to VEH-treated cells (Fig. 4C, polysome: monosome ratios in Fig. 4D). We tested whether Tnf alters the stress-responsive translation of uORF-containing genes as follows.
Individual ribosomal fractions were pooled into ‘SUB’-polysomal (80S through the two ribosome peak) and POLYsomal samples (see highlighted regions of polysomal profiles in Fig. 4C). Quantification of a panel of 700 genes was then performed upon the pooled fractions (Nanostring Pan-Cancer, uORF incidence in Tables II and III). From the dataset produced, we first determined the Nanostring counts for the 700 individual panel mRNAs in the POLYsomal fractions and the SUB-polysomal fractions, and calculated the mean ratio between counts for each gene (mean ratio of Nanostring counts per gene, plus-minus [pm] SEM). The mean POLY: SUB ratio for VEH-treated cells was 0.978 pm 0.037, and for Tnf0.879 pm 0.034 (P value 0.051, Welch’s t test). Thus, as seen in the polysomal profiles, mRNAs for individual genes considered in aggregate were reduced in actively translating polysomal fractions 3 h after treatment with Tnfα. Determination of mRNA copy levels of individual genes in the ribosomal fractions followed.
Table III.
uORF incidence in human orthologues of genes identified in the OV3121 granulosa cell line experiment.
| OV3121 gene set | Total genes (n) | Any uORF (%) | HiQ uORFs (%) | Top10 uORFs (%) |
|---|---|---|---|---|
| Polysome upregulated | 401 | 385 (96.0%) | 287 (71.6%) | 83 (20.1%) |
| Polysome downregulated | 349 | 333 (95.4%) | 223 (63.9%) | 54 (15.5% |
| Top 25% polysome redistribution | 188 | 180 (95.7%) | 113 (60.1%) | 25 (13.3%) |
| Bottom 25% polysome redistribution | 188 | 177 (94.1%) | 107 (56.9%) | 16 (8.5%) |
As above, we determined mRNA copy number in SUB and POLY fractions (Fig. 4E–E” ’, Supplementary Fig. S4C and F) and determined the ratio of polysomal mRNA copies to sub-polysomal mRNA copies (POLY: SUB ratio) for cells treated with Tnf for 3 h and separately, for VEH-treated cells. Plots were produced for panel mRNAs implicated in the ISR response (cell cycle or cytoprotection/DDR) and we compare both POLY and SUB levels (top plot panels Fig. 4E–E”’), and POLY mRNA counts alone (bottom plot panels). We also calculated a ‘delta’ for the change between the POLY: SUB ratios by dividing the POLY: SUB ratio for Tnf 3 h cells by the VEH-treated ratio. This delta reflects flux to or from the polysomal fraction resulting from Tnfα treatment (values included in plot subtitles). Genes with a positive POLY: SUB delta, or increased polysomal copies in response to Tnf are candidates for uORF regulation (Barbosa et al., 2013, Fig. 1).
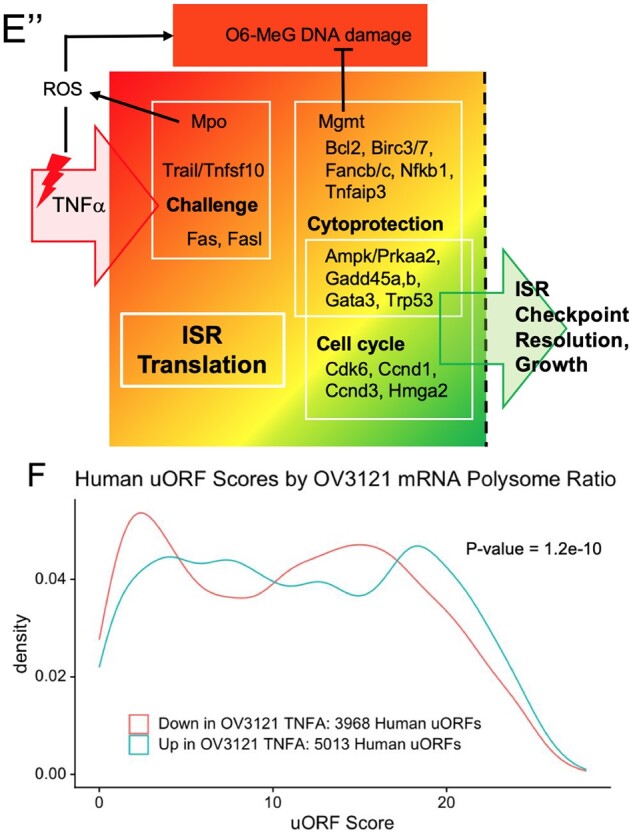
Panel mRNAs that transmit cellular damage signals were classified as cellular ‘Challenge’ genes. As examples, both Fas death receptor and Tnfsf10/Trail mRNAs exhibit elevated POLY: SUB deltas and elevated polysomal levels 3 h post Tnf (Fig. 4E, ‘Challenge’), and this was matched by their corresponding ligand and receptor, Fasl and Tnfrsf10b (Supplementary Fig. S4C, ‘Challenge’). ‘Cell Cycle’ regulators Cdk6, Ccnd1 (Fig. 4E’), Ccnd2, and Hmga2 (Vignali and Marracci, 2020) (Supplementary Fig. S4D) all exhibited deltas greater than 1, and elevated polysomal levels. Several cytoprotective/DDR factors that also impact cell-cycle progression exhibited elevated mRNA polysomal partitioning in the same way. These were ‘Cell Cycle/Cytoprotection’ genes Growth Arrest and DNA Damage Inducible Beta (Gadd45b; Schäfer, 2013), O-6-Methylguanine-DNA Methyltransferase (Mgmt; Mooney and Sahingur, 2021), TNF Alpha Induced Protein 3 (A20/Tnfaip3; Mooney and Sahingur, 2021) and Tumor Protein P53 (Trp53) (not shown, see also Supplementary Fig. S4E). Concurrent upregulation of cytoprotective and DDR factors, as well as ‘Challenge’ factors (e.g., Tnfsf10/Trail) that can induce cellular stress and DNA damage (Lovric and Hawkins, 2010, Zhang et al., 2017) is suggestive of a balance between physiologically induced damage and repair. A graphical summary of key genes whose mRNAs partition in this way, with genes grouped according to their function is included in Fig. 4E”. mRNAs for two factors that negatively regulate PFGA, Amh and Pten, did not differ in ribosomal partitioning between groups at either time point (not shown).
As a first step in determining if human GC translation is regulated in the same way, we tested whether human orthologues of genes that redistribute to polysomal fractions in the mouse GC line have more or higher quality uORFs compared to those that do not. We evaluated human uORF incidence and quality in two different ways. Delta measurements were used to rank genes from those with the highest flux into the polysome fraction at 3 h post-treatment with those with the lowest flux. The lowest delta values also included genes that changed their distribution such that their mRNAs were at lower levels in POLY than SUB fractions after Tnf treatment. uORF incidence and quality of human orthologues of polysome upregulated (401/750 genes) versus polysome downregulated (349/750) genes, and also the top 25th percentile of polysome-upregulated genes versus the bottom 25th percentile are summarized in Tables III and IV.
Table IV.
uORF quality in human orthologues of genes identified in OV3121 granulosa cell line ISR induction experiment.
| OV3121 gene set (n genes) | HiQ uORFs (n) | uORFs/gene | Mean uORFs Quality Score | SD | P-value (up vs down) |
|---|---|---|---|---|---|
| Polysome upregulated (287) | 5013 | 17.5 | 11.9 | 7.1 | P < 1E−7 |
| Polysome downregulated (223) | 3968 | 17.8 | 11.1 | 7 | |
| Top 25% polysome redistribution (113) | 1696 | 15.0 | 11.7 | 7.3 | P < 1E−9 |
| Bottom 25% polysome redistribution (107) | 1758 | 16.4 | 10.2 | 6.7 |
P-values calculated using Welch's t-test.
While the incidence of total, HiQ, and Top10 uORFs was similar between groups (Table III), mean human orthologue uORF quality scores significantly differed between genes that became more prevalent in polysomes versus those that did not (Table IV). As seen with the control and ovarian aging gene sets (Fig. 3D), a density plot comparing human uORF scores between groups (Fig. 4F) showed that the orthologous human genes exhibited a rightward ‘shift’ of higher uORF quality. These data support uORF content and quality as a conserved regulator of protein translation in response to cellular stress in GCs.
uORFs are overrepresented in the genes of the human pregranulosa cell transcriptome
While all of the above data support a role for the ISR and uORFs in GC translational regulation during follicle development, the question of high-level control of the transcriptome remained open. We performed a final analysis of genes whose mRNAs are produced in PF pregranulosa cells (Zhang et al., 2018) and determined uORF content and characteristics.
Remarkably, having one or more Top10 or HiQ uORFs in a gene is highly predictive of that gene being transcribed in pregranulosa cells (Table V). Of more than 25 000 unique genes in the McGillivray et al. database (McGillivray et al., 2018) with any uORF, approximately 3000 have Top10 uORFs. All but 16 of those genes are transcribed in pregranulosa cells. In addition, 11 000 genes have HiQ uORFs and, of those, all but 638 are transcribed. In contrast, only 30% of genes that lack any uORF are transcribed in pregranulosa cells. How and why this transcriptional control occurs, with potentially important consequences for gene expression regulation in this cell lineage, is worthy of further study.
Table V.
Pregranulosa cell genes and their transcription in the presence or absence of uORFs.
| Gene list/uORF category | Number of genes | Number of genes with detectable* mRNA in pregranulosa cells (%) |
|---|---|---|
| All genes with NO uORFs | 8569 | 2539 (29.6%) |
| All genes with uORFs | 19 075 | 15 381 (80.6%) |
| Genes with HiQ uORFs | 10 925 | 10 287 (94.2%) |
| Genes with Top10 uORFs | 2979 | 2963 (99.5%) |
Detection threshold = 0.005 (raw), log base 2 = 0.0072.
Discussion
Measures of an activated ISR and DDR (Supplementary Fig. S1) are detectable in normal ovarian follicles under baseline physiological conditions. This can be seen in high baseline levels of P-Ser 51 eIF2, P-Chk1, and P-Rpa2 in follicular GCs and oocytes of WT mice, and levels of P-eIF2 in human non-growing follicles, above those in surrounding stroma. It is informative that levels of P-eIF2 in WT ovaries exceed those seen in ovaries collected from GalTKO mice (Balakrishnan et al., 2017). This is because when the functional role of P-eIF2 in the response to elevated stress is considered, the activated ISR in the WT state can be interpreted as ‘normal’ while lower levels in the mutant that exhibits accelerated ovarian follicle loss instead likely reflects compromised signaling. Further, our analysis of a publicly available transcriptomic database revealed that stress-translated ATF4 and its transcriptional targets (Kilberg et al., 2009) are all transcribed at similar levels in non-growing pregranulosa cells and growing GCs in the human (Zhang et al., 2018). Similarly, G1 cyclin mRNA expression does not significantly differ between pregranulosa cells and growing GCs. In combination, these data suggested that a stress-responsive mechanism might play a role in driving ATF4 and G1 cyclin translation in these cells. ATF4 translational control by uORFs is well understood (Kilberg et al., 2009), and because an active ISR can favor the translation of mRNAs that contain uORFs (Barbosa et al., 2013) more generally, we hypothesized that an active ISR controls GC translation and, thus, follicle growth in an uORF-dependent manner.
Using a tool that provided unbiased prediction of uORF quality (McGillivray et al., 2018), we went on to find that mRNAs associated with GC function (Zhang gene sets) and also the ANM in women (Day menopause gene set) often contain uORFs with HiQ scores. HiQ scores in GC gene sets were significantly higher than matched groups of randomly selected genes in all cases, and density plots show that specific high uORF score ranges are over-represented in mRNAs expressed by GCs. It is possible that some uORFs detected and quantified using the bioinformatic tool represent proteins of extended length owing to alternative in-frame translational starts (Orr et al., 2020), and this is worthy of additional investigation. Unexpectedly, uORFs were not only over-represented in ovarian function and ovarian aging gene sets but also when all pregranulosa cell transcripts were evaluated, uORF presence and quality were very highly predictive of their transcription. HiQ uORFs corresponded to a nearly 95% likelihood of transcription, and Top10 uORFs, greater than 99%. A relation between uORF content and transcription has been detected during yeast meiosis (Hinnebusch et al., 2016) but how the correspondence between uORF content and the mammalian GC transcriptome arises is currently unknown.
A mouse GC line model was used to assess ISR activation and translational control (e.g. mRNA redistribution between SUB- and POLYsomal fractions) in response to the follicle growth activator, Tnf. In the longer term, treatment with 10 ng/ml Tnf results in significantly accelerated OV3121 cell growth (24 h). In the shorter term, the same treatment results in elevated P-eIF2 (3 h post-treatment). This corresponded to decreased protein translation (3and 12 h post-treatment) as seen in POLY: SUB ratios. Tnf stimulated a flux of a subset of mRNAs to actively translating POLY fractions. These genes were categorized into three groups: cell-cycle control, cytoprotection, and those that induce cell damage (‘challenge’ genes). Also in accordance with the expected impact upon protein translation, uORF content was significantly higher in the human orthologues of genes that shifted to polysomes in the mouse GC cell line model. These data support ISR control of translation of genes whose mRNAs include stress-responsive uORF motifs as an evolutionarily conserved mechanism that regulates the GC cell cycle and also GC survival.
We do not interpret faster GC growth after activation of the ISR as paradoxical. Instead, we interpret these data as an initial upregulation of both cellular damage and repair protein translation that eventually results in a net reduction of damage (Fig. 5). This is concurrent with temporarily elevated cell-cycle machinery translation (e.g. Cdk6, Ccnd1) that can support cell-cycle progression. In this paradigm, elevated stress increases the probability of overcoming ISR, DDR, and cell-cycle checkpoints if ‘complete’ repair beyond a threshold is achieved. This is only one interpretation, however, and further information is needed about the precise ISR influence upon GC cytoprotection and cell-cycle control, and, thus, the overall survival and growth of ovarian follicles.
Figure 5.
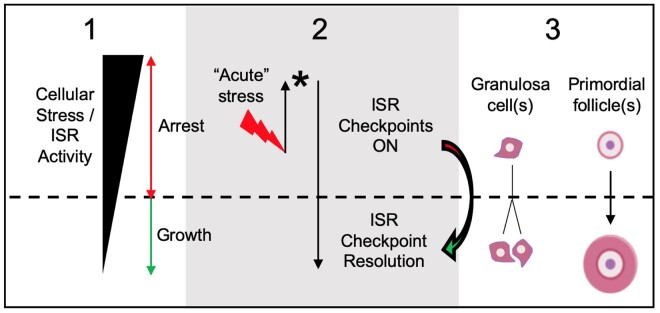
Integrated stress response checkpoint resolution as a trigger of the GC cell cycle and PF growth activation. Box 1 depicts fluctuating ISR activity over time, as controlled by levels physiological stressors. As long as the ISR is active/high (red double-headed arrow), GC stay growth-arrested. Box 2 depicts the action of an acute stressor (red lightning bolt; e.g. by local elevation of an ISR activating factor such as Tnf(Chen et al., 1993)) that initially elevates ISR activity. In response, the cell upregulates the expression of stress-resolving damage repair genes at the level of ISR translational control (asterisk; Fig. 4, Supplementary Fig. S4C–E). Cellular repair activity then increases, and net ISR activity declines. Increased translation of positive cell cycle regulators including G1 cyclins and CDK6 also occurs at this time. If ISR activity declines beneath a threshold (horizontal dashed line), ISR checkpoint resolution can occur and the cell can enter the cell cycle Box 3. In pregranulosa cells of primordial follicles, ISR checkpoint resolution would correspond to PFGA. How oocyte: granulosa cell interactions contribute to ISR action in this model is an open question.
Our data support a situation where ovarian follicles exist within an ‘ISR ON’ stress state, favoring cytoprotective protein translation and cell-cycle arrest. An active ISR is also consistent with the slow GC cell cycle during follicle growth. Growth of individual human ovarian follicles from the time of recruitment/PFGA to periovulatory size can take months, with GC cell-cycle lengths as high as 240 h in immature follicles (Gougeon, 1986). Active ISR checkpoints would allow growth only when checkpoint recovery occurs. PFs may utilize active ISR checkpoints similarly to remain arrested within a protected state (Fig. 5), which in the human would reflect the dormant/non-growing ovarian reserve. A low probability of pregranulosa cell-cycle checkpoint recovery would correspond to the probability that individual PFs begin to grow and, thus, the rate that the reserve becomes depleted (bioRxiv preprint, Johnson et al., 2016).
A model that includes an active ISR as the default for ovarian follicles implies an internal ‘quality control’ mechanism. This is because cell-cycle progression depends upon checkpoint resolution, which may only take place if cell stress is resolved and DNA is of high integrity (overlap of ISR and DDR machinery, Supplementary Fig. S1). Because overlapping signals control the cell cycle and cell survival in somatic GC, it is interesting to speculate about oocyte quality control. If GCs are intact (and optionally, growing), at least their ability to resolve damage has been assured. Poor-quality oocytes can predominate in infertile individuals whose follicles demonstrate normal survival, growth, and development. Oocytes have the capacity to repair at least double-stranded DNA breaks with high efficiency (Stringer et al., 2020), and, thus, the capacity to ensure that most eggs have high-to-maximal DNA integrity. An important question, then, is whether oocyte quality control is being impacted in a fashion as appears to be regulated by the ISR in somatic GCs. An additional question is how aging-related degeneration (e.g. fibrosis) of non-follicular ovarian tissue like the ovarian stroma (Amargant et al., 2020) is influenced by, or influences, ISR action within GCs and follicles. Information provided here about GC ISR function can be used as a platform for the investigation of the impact of stressors upon different cell types and tissues in the ovary.
A counterintuitive hypothesis is raised where mammalian ovaries may use physiological activation of the ISR to control GC survival and the rates at which they divide, including cell-cycle resumption of pregranulosa cells at the time of PFGA. This same mechanism would influence which follicles survive and which die, and, thus, the overall rate of female reproductive aging. The ovary is often considered a ‘cradle’ that functions to protect the precious oocytes within. This model suggests that the ovary may instead function as a ‘gauntlet’, where physiological factors that activate the ISR and favor DNA damage act ultimately to select follicles for survival that are capable of resolving that stress and damage.
Supplementary data
Supplementary data are available at Molecular Human Reproduction online.
Data availability
Publicly available human GC trancriptome (Zhang et al., 2018) and uORF (McGillivray et al., 2018) data were downloaded from their respective sources and analyzed using R (R Core Team, 2018). Raw Nanostring panel data are available in a publicly available repository at https://doi.org/10.6084/m9.figshare.14251652.
Supplementary Material
Acknowledgements
Mary Zelinski, PhD and Brian Kennedy, PhD are thanked for valuable comments provided during the development of this manuscript. Jeremy Rahkola of the Denver Veterans Affairs Medical Center, Aurora, CO, USA is acknowledged for performance of Nanostring analysis.
Authors’ roles
E.L.C., S.H-L., A.G. and J.J. performed experiments and helped revise the manuscript, A.K., A.G., K.L., N.S., L.A., A.P., M.P. and B.B. provided critical feedback during the project and contributed to the manuscript, J.J., K.L. and J.K. designed experiments; J.J. drafted and completed the manuscript.
Funding
Studies reported here were supported by the University of Colorado School of Medicine (Anschutz Medical Campus), University of Colorado School of Medicine Department of Obstetrics and Gynecology Research Funds, McPherson Family Funds, and a Florence Crozier Cobb Endowment Undergraduate Student Research Award to A.G.
Conflict of interest
The authors have declared no competing interest.
References
- Amargant F, Manuel SL, Tu Q, Parkes WS, Rivas F, Zhou LT, Rowley JE, Villanueva CE, Hornick JE, Shekhawat GS. et al. Ovarian stiffness increases with age in the mammalian ovary and depends on collagen and hyaluronan matrices. Aging Cell 2020;19:e13259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ba X, Bacsi A, Luo J, Aguilera-Aguirre L, Zeng X, Radak Z, Brasier AR, Boldogh I. 8-Oxoguanine DNA glycosylase-1 augments proinflammatory gene expression by facilitating the recruitment of site-specific transcription factors. J Immunol 2014;192:2384–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan B, Nicholas C, Siddiqi A, Chen W, Bales E, Feng M, Johnson J, Lai K. Reversal of aberrant PI3K/Akt signaling by Salubrinal in a GalT-deficient mouse model. Biochim Biophys Acta Mol Basis Dis 2017;1863:3286–3293. [DOI] [PubMed] [Google Scholar]
- Balakrishnan B, Siddiqi A, Mella J, Lupo A, Li E, Hollien J, Johnson J, Lai K. Salubrinal enhances eIF2alpha phosphorylation and improves fertility in a mouse model of Classic Galactosemia. Biochim Biophys Acta Mol Basis Dis 2019;1865:165516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa C, Peixeiro I, Romão L. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet 2013;9:e1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol 2007;19:238–245. [DOI] [PubMed] [Google Scholar]
- Blais J, Bell JC. Novel therapeutic target: the PERKs of inhibiting the integrated stress response. Cell Cycle 2006;5:2874–2877. [DOI] [PubMed] [Google Scholar]
- Block WD, Yu Y, Lees-Miller SP. Phosphatidyl inositol 3-kinase-like serine/threonine protein kinases (PIKKs) are required for DNA damage-induced phosphorylation of the 32 kDa subunit of replication protein A at threonine 21. Nucleic Acids Res 2004;32:997–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, Kaufman RJ, Ma D, Coen DM, Ron D. et al. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science 2005;307:935–939. [DOI] [PubMed] [Google Scholar]
- Chen HL, Marcinkiewicz JL, Sancho-Tello M, Hunt JS, Terranova PF. Tumor necrosis factor-alpha gene expression in mouse oocytes and follicular cells. Biol Reprod 1993;48:707–714. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Kim J, Li XX, Hsueh AJ. Promotion of ovarian follicle growth following mTOR activation: synergistic effects of AKT stimulators. PLoS One 2015;10:e0117769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementi E, Inglin L, Beebe E, Gsell C, Garajova Z, Markkanen E. Persistent DNA damage triggers activation of the integrated stress response to promote cell survival under nutrient restriction. BMC Biol 2020;18:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Walter P. The integrated stress response: from mechanism to disease. Science 2020;368:eaat5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui LL, Yang G, Pan J, Zhang C. Tumor necrosis factor alpha knockout increases fertility of mice. Theriogenology 2011;75:867–876. [DOI] [PubMed] [Google Scholar]
- Dai Y, Grant S. New insights into checkpoint kinase 1 in the DNA damage response signaling network. Clin Cancer Res 2010;16:376–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day FR, Ruth KS, Thompson DJ, Lunetta KL, Pervjakova N, Chasman DI, Stolk L, Finucane HK, Sulem P, Bulik-Sullivan B et al. Large-scale genomic analyses link reproductive aging to hypothalamic signaling, breast cancer susceptibility and BRCA1-mediated DNA repair. Nat Genet 2015;47:1294–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding GR, Honda N, Nakahara T, Tian F, Yoshida M, Hirose H, Miyakoshi J. Radiosensitization by inhibition of IkappaB-alpha phosphorylation in human glioma cells. Radiat Res 2003;160:232–237. [DOI] [PubMed] [Google Scholar]
- Durlinger AL, Kramer P, Karels B, Jong FH, de Uilenbroek JT, Grootegoed JA, Themmen AP. Control of primordial follicle recruitment by anti-Müllerian hormone in the mouse ovary. Endocrinology 1999;140:5789–5796. [DOI] [PubMed] [Google Scholar]
- Ernst EH, Grøndahl ML, Grund S, Hardy K, Heuck A, Sunde L, Franks S, Andersen CY, Villesen P, Lykke-Hartmann K. Dormancy and activation of human oocytes from primordial and primary follicles: molecular clues to oocyte regulation. Hum Reprod 2017;32:1684–1700. [DOI] [PubMed] [Google Scholar]
- Fogli A, Gauthier-Barichard F, Schiffmann R, Vanderhoof VH, Bakalov VK, Nelson LM, Boespflug-Tanguy O. Screening for known mutations in EIF2B genes in a large panel of patients with premature ovarian failure. BMC Womens Health 2004;4:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford EA, Beckett EL, Roman SD, McLaughlin EA, Sutherland JM. Advances in human primordial follicle activation and premature ovarian insufficiency. Reproduction 2020;159:R15–R29. [DOI] [PubMed] [Google Scholar]
- Goldman KN, Chenette D, Arju R, Duncan FE, Keefe DL, Grifo JA, Schneider RJ. mTORC1/2 inhibition preserves ovarian function and fertility during genotoxic chemotherapy. Proc Natl Acad Sci U S A 2017;114:3186–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gougeon A. Dynamics of follicular growth in the human: a model from preliminary results. Hum Reprod 1986;1:81–87. [DOI] [PubMed] [Google Scholar]
- Greenfeld CR, Roby KF, Pepling ME, Babus JK, Terranova PF, Flaws JA. Tumor necrosis factor (TNF) receptor type 2 is an important mediator of TNF alpha function in the mouse ovary. Biol Reprod 2007;76:224–231. [DOI] [PubMed] [Google Scholar]
- Guan BJ, Hoef V, van Jobava R, Elroy-Stein O, Valasek LS, Cargnello M, Gao XH, Krokowski D, Merrick WC, Kimball SR. et al. A unique ISR program determines cellular responses to chronic stress. Mol Cell 2017;68:885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen-Lillevik S, Rushing J, Appiah L, Longo N, Andrews A, Lai K, Johnson J. (in press). Pathophysology and management of galactosemic primary ovarian insufficiency. Reprod Fertil 2021;2:R67–R84. Doi:10.1530/RAF-21-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer EE, Moore MJ. Redefining the translational status of 80S monosomes. Cell 2016;164:757–769. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5’-untranslated regions of eukaryotic mRNAs. Science 2016;352:1413–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iftode C, Daniely Y, Borowiec JA. Replication protein A (RPA): the eukaryotic SSB. Crit Rev Biochem Mol Biol 1999;34:141–180. [DOI] [PubMed] [Google Scholar]
- Johnson J, Chen X, Xu X, Emerson J. (2016). ŌvSim: A simulation of the population dynamics of mammalian ovarian follicles (Vienna, Austria: BioRXiv, The Preprint Server for Biology). Doi: 10.1101/034249. [DOI]
- Kalich-Philosoph L, Roness H, Carmely A, Fishel-Bartal M, Ligumsky H, Paglin S, Wolf I, Kanety H, Sredni B, Meirow D. Cyclophosphamide triggers follicle activation and “burnout”; AS101 prevents follicle loss and preserves fertility. Sci Transl Med 2013;5:185ra62. [DOI] [PubMed] [Google Scholar]
- Kallen A, Polotsky AJ, Johnson J. Untapped reserves: controlling primordial follicle growth activation. Trends Mol Med 2018;24:319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano M, Sosulski AE, Zhang L, Saatcioglu HD, Wang D, Nagykery N, Sabatini ME, Gao G, Donahoe PK, Pépin D. AMH/MIS as a contraceptive that protects the ovarian reserve during chemotherapy. Proc Natl Acad Sci USA 2017;114:E1688–E1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasippillai T, MacArthur DG, Kirby A, Thomas B, Lambalk CB, Daly MJ, Welt CK. Mutations in eIF4ENIF1 are associated with primary ovarian insufficiency. J Clin Endocrinol Metab 2013;98:E1534–E1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann WK, Paules RS. DNA damage and cell cycle checkpoints. FASEB J 1996;10:238–247. [DOI] [PubMed] [Google Scholar]
- Kawai T, Mihara T, Kawashima I, Fujita Y, Ikeda C, Negishi H, Richards JS, Shimada M. Endogenous acetaldehyde toxicity during antral follicular development in the mouse ovary. Reprod Toxicol 2012;33:322–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura K, Cheng Y, Suzuki N, Deguchi M, Sato Y, Takae S, Ho CH, Kawamura N, Tamura M, Hashimoto S. et al. Hippo signaling disruption and Akt stimulation of ovarian follicles for infertility treatment. Proc Natl Acad Sci USA 2013;110:17474–17479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilberg MS, Shan J, Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol Metab 2009;20:436–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar AA, Merrick WC. A retrospective on eIF2A-and not the alpha subunit of eIF2. Int J Mol Sci 2020;21: 2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo H, Maruo T, Mochizuki M. Immunohistochemical evidence for the presence of tumor necrosis factor-alpha in the infant and adult human ovary. Endocr J 1995;42:771–780. [DOI] [PubMed] [Google Scholar]
- Kondrashov AV, Spriggs KA, Bushell M, Willis AE. Co-ordinated regulation of translation following DNA damage. Cell Cycle 2009;8:3067–3068. [DOI] [PubMed] [Google Scholar]
- Lee YY, Cevallos RC, Jan E. An upstream open reading frame regulates translation of GADD34 during cellular stresses that induce eIF2alpha phosphorylation. J Biol Chem 2009;284:6661–6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitus M, Joenje H, de Winter JP. The Fanconi anemia pathway of genomic maintenance. Cell Oncol 2006;28:3–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Titus S, Moy F, Ginsburg ES, Oktay K. Ovarian aging in women with BRCA germline mutations. J Clin Endocrinol Metab 2017;102:3839–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Michowski W, Inuzuka H, Shimizu K, Nihira NT, Chick JM, Li N, Geng Y, Meng AY, Ordureau A. et al. G1 cyclins link proliferation, pluripotency and differentiation of embryonic stem cells. Nat Cell Biol 2017;19:177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovric MM, Hawkins CJ. TRAIL treatment provokes mutations in surviving cells. Oncogene 2010;29:5048–5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malzer E, Daly ML, Moloney A, Sendall TJ, Thomas SE, Ryder E, Ryoo HD, Crowther DC, Lomas DA, Marciniak SJ. Impaired tissue growth is mediated by checkpoint kinase 1 (CHK1) in the integrated stress response. J Cell Sci 2010;123:2892–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini F, Nardo T, Giannattasio M, Minuzzo M, Stefanini M, Plevani P, Muzi Falconi M. DNA nucleotide excision repair-dependent signaling to checkpoint activation. Proc Natl Acad Sci U S A 2006;103:17325–17330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AG, Fresno M. Tumor necrosis factor-alpha activation of NF-kappa B requires the phosphorylation of Ser-471 in the transactivation domain of c-Rel. J Biol Chem 2000;275:24383–24391. [DOI] [PubMed] [Google Scholar]
- McGillivray P, Ault R, Pawashe M, Kitchen R, Balasubramanian S, Gerstein M. A comprehensive catalog of predicted functional upstream open reading frames in humans. Nucleic Acids Res 2018;46:3326–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin M, Patrizio P, Kayisli U, Luk J, Thomson TC, Anderson RA, Telfer EE, Johnson J. mTOR kinase inhibition results in oocyte loss characterized by empty follicles in human ovarian cortical strips cultured in vitro. Fertil Steril 2011;96:1154–1159. [DOI] [PubMed] [Google Scholar]
- Menoni H, Hoeijmakers JH, Vermeulen W. Nucleotide excision repair-initiating proteins bind to oxidative DNA lesions in vivo. J Cell Biol 2012;199:1037–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney EC, Sahingur SE. The ubiquitin system and A20: implications in health and disease. J Dent Res 2021;100:10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novella-Maestre E, Herraiz S, Rodr?Guez-Iglesias B, D?Az-Garc?A C, Pellicer A. Short-term PTEN inhibition improves in vitro activation of primordial follicles, preserves follicular viability, and restores AMH levels in cryopreserved ovarian tissue from cancer patients. PLoS One 2015;10:e0127786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oktay KH, Bedoschi G, Goldfarb SB, Taylan E, Titus S, Palomaki GE, Cigler T, Robson M, Dickler MN. Increased chemotherapy-induced ovarian reserve loss in women with germline BRCA mutations due to oocyte deoxyribonucleic acid double strand break repair deficiency. Fertil Steril 2020;113:1251–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr MW, Mao Y, Storz G, Qian SB. Alternative ORFs and small ORFs: shedding light on the dark proteome. Nucleic Acids Res 2020;48:1029–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmeer RM, Moser J, Volker M, Kool H, Tomkinson AE, Zeeland AA, van Mullenders LH, Fousteri M. Replication protein A safeguards genome integrity by controlling NER incision events. J Cell Biol 2011;192:401–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep 2016;17:1374–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankhurst MW. A putative role for anti-Müllerian hormone (AMH) in optimising ovarian reserve expenditure. J Endocrinol 2017;233:R1–R13. [DOI] [PubMed] [Google Scholar]
- Patil M, Pabla N, Dong Z. Checkpoint kinase 1 in DNA damage response and cell cycle regulation. Cell Mol Life Sci 2013;70:4009–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powley IR, Kondrashov A, Young LA, Dobbyn HC, Hill K, Cannell IG, Stoneley M, Kong YW, Cotes JA, Smith GC. et al. Translational reprogramming following UVB irradiation is mediated by DNA-PKcs and allows selective recruitment to the polysomes of mRNAs encoding DNA repair enzymes. Genes Dev 2009;23:1207–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2018. [Google Scholar]
- Reddy P, Liu L, Adhikari D, Jagarlamudi K, Rajareddy S, Shen Y, Du C, Tang W, Hamalainen T, Peng SL. et al. Oocyte-specific deletion of Pten causes premature activation of the primordial follicle pool. Science 2008;319:611–613. [DOI] [PubMed] [Google Scholar]
- Reddy P, Zheng W, Liu K. Mechanisms maintaining the dormancy and survival of mammalian primordial follicles. Trends Endocrinol Metab 2010;21:96–103. [DOI] [PubMed] [Google Scholar]
- Roness H, Gavish Z, Cohen Y, Meirow D. Ovarian follicle burnout: a universal phenomenon? Cell Cycle 2013;12:3245–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro N, Johnson J. Diagnosing the onset of menopause. JAMA 2019;322:775. [DOI] [PubMed] [Google Scholar]
- Schäfer A. Gadd45 proteins: key players of repair-mediated DNA demethylation. Adv Exp Med Biol 2013;793:35–50. [DOI] [PubMed] [Google Scholar]
- Shaltiel IA, Krenning L, Bruinsma W, Medema RH. The same, only different – DNA damage checkpoints and their reversal throughout the cell cycle. J Cell Sci 2015;128:607–620. [DOI] [PubMed] [Google Scholar]
- Smits VA, Gillespie DA. DNA damage control: regulation and functions of checkpoint kinase 1. FEBS J 2015;282:3681–3692. [DOI] [PubMed] [Google Scholar]
- Stolk L, Perry JR, Chasman DI, He C, Mangino M, Sulem P, Barbalic M, Broer L, Byrne EM, Ernst F. et al. Meta-analyses identify 13 loci associated with age at menopause and highlight DNA repair and immune pathways. Nat Genet 2012;44:260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer JM, Winship A, Zerafa N, Wakefield M, Hutt K. Oocytes can efficiently repair DNA double-strand breaks to restore genetic integrity and protect offspring health. Proc Natl Acad Sci USA 2020;117:11513–11522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudharsan R, Azuma Y. The SUMO ligase PIAS1 regulates UV-induced apoptosis by recruiting Daxx to SUMOylated foci. J Cell Sci 2012;125:5819–5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Miyaki S, Onouchi H, Motomura T, Idesako N, Takahashi A, Murase M, Fukuyoshi S, Endo T, Satou K. et al. Exhaustive identification of conserved upstream open reading frames with potential translational regulatory functions from animal genomes. Sci Rep 2020;10:16289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci USA 2004;101:11269–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignali R, Marracci S. HMGA genes and proteins in development and evolution. Int J Mol Sci 2020;21:654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LL, Chiou CC, Chang PY, Wu JT. Urinary 8-OHdG: a marker of oxidative stress to DNA and a risk factor for cancer, atherosclerosis and diabetics. Clin Chim Acta 2004;339:1–9. [DOI] [PubMed] [Google Scholar]
- Yan B, Wang H, Rabbani ZN, Zhao Y, Li W, Yuan Y, Li F, Dewhirst MW, Li CY. Tumor necrosis factor-alpha is a potent endogenous mutagen that promotes cellular transformation. Cancer Res 2006;66:11565–11570. [DOI] [PubMed] [Google Scholar]
- Yan B, Peng Y, Li CY. Molecular analysis of genetic instability caused by chronic inflammation. Methods Mol Biol 2009;512:15–28. [DOI] [PubMed] [Google Scholar]
- Yanagihara K, Nii M, Tsumuraya M, Numoto M, Seito T, Seyama T. A radiation-induced murine ovarian granulosa cell tumor line: introduction of v-ras gene potentiates a high metastatic ability. Jpn J Cancer Res 1995;86:347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Yaba A, Kasiman C, Thomson T, Johnson J. mTOR controls ovarian follicle growth by regulating granulosa cell proliferation. PLoS One 2011;6:e21415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu K. Cellular and molecular regulation of the activation of mammalian primordial follicles: somatic cells initiate follicle activation in adulthood. Hum Reprod Update 2015;21:779–786. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hunter T. Roles of Chk1 in cell biology and cancer therapy. Int J Cancer 2014;134:1013–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Tang H, Jiang Y, Mao Z. The transcription factor GATA3 is required for homologous recombination repair by regulating CtIP expression. Oncogene 2017;36:5168–5176. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yan Z, Qin Q, Nisenblat V, Chang HM, Yu Y, Wang T, Lu C, Yang M, Yang S. et al. Transcriptome landscape of human folliculogenesis reveals oocyte and granulosa cell interactions. Mol Cell 2018;72:1021–1034. [DOI] [PubMed] [Google Scholar]
- Zhao M, Feng F, Chu C, Yue W, Li L. A novel EIF4ENIF1 mutation associated with a diminished ovarian reserve and premature ovarian insufficiency identified by whole-exome sequencing. J Ovarian Res 2019;12:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available human GC trancriptome (Zhang et al., 2018) and uORF (McGillivray et al., 2018) data were downloaded from their respective sources and analyzed using R (R Core Team, 2018). Raw Nanostring panel data are available in a publicly available repository at https://doi.org/10.6084/m9.figshare.14251652.