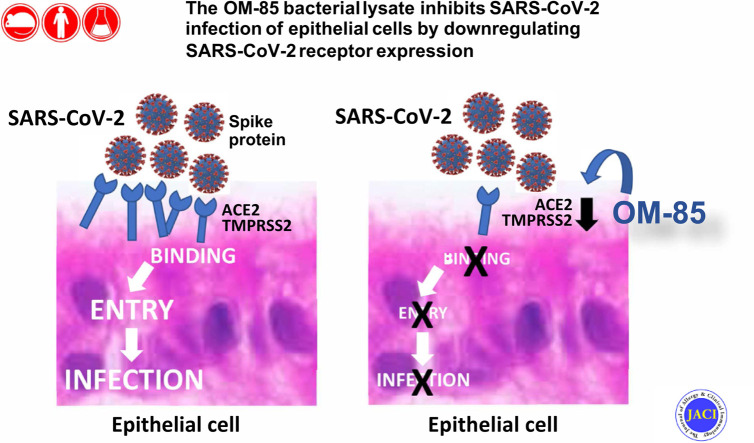
Abstract
Background
Treatments for coronavirus disease 2019, which is caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), are urgently needed but remain limited. SARS-CoV-2 infects cells through interactions of its spike (S) protein with angiotensin-converting enzyme 2 (ACE2) and transmembrane protease serine 2 (TMPRSS2) on host cells. Multiple cells and organs are targeted, particularly airway epithelial cells. OM-85, a standardized lysate of human airway bacteria with strong immunomodulating properties and an impeccable safety profile, is widely used to prevent recurrent respiratory infections. We found that airway OM-85 administration inhibits Ace2 and Tmprss2 transcription in the mouse lung, suggesting that OM-85 might hinder SARS-CoV-2/host cell interactions.
Objectives
We sought to investigate whether and how OM-85 treatment protects nonhuman primate and human epithelial cells against SARS-CoV-2.
Methods
ACE2 and TMPRSS2 mRNA and protein expression, cell binding of SARS-CoV-2 S1 protein, cell entry of SARS-CoV-2 S protein–pseudotyped lentiviral particles, and SARS-CoV-2 cell infection were measured in kidney, lung, and intestinal epithelial cell lines, primary human bronchial epithelial cells, and ACE2-transfected HEK293T cells treated with OM-85 in vitro.
Results
OM-85 significantly downregulated ACE2 and TMPRSS2 transcription and surface ACE2 protein expression in epithelial cell lines and primary bronchial epithelial cells. OM-85 also strongly inhibited SARS-CoV-2 S1 protein binding to, SARS-CoV-2 S protein–pseudotyped lentivirus entry into, and SARS-CoV-2 infection of epithelial cells. These effects of OM-85 appeared to depend on SARS-CoV-2 receptor downregulation.
Conclusions
OM-85 inhibits SARS-CoV-2 epithelial cell infection in vitro by downregulating SARS-CoV-2 receptor expression. Further studies are warranted to assess whether OM-85 may prevent and/or reduce the severity of coronavirus disease 2019.
Key words: COVID-19, SARS-CoV-2, bacterial lysate, OM-85, epithelial cells, ACE2, TMPRSS2
Abbreviations used: ACE2, Angiotensin-converting enzyme 2; COVID-19, Coronavirus disease 2019; GFP, Green fluorescent protein; pfu, Plaque-forming units; SARS-CoV-2, Severe acute respiratory syndrome coronavirus-2; S, Spike; TMPRSS2, Transmembrane protease serine 2; TU, Transducing units; VSV, Vesicular stomatitis virus
Graphical abstract
The emergence of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) in December 2019 triggered a global pandemic marked by a broad spectrum of clinical manifestations. These range from a mild, self-limiting flu-like respiratory illness to life-threatening multiorgan failure and are collectively referred to as coronavirus disease 2019 (COVID-19).1 , 2 Angiotensin-converting enzyme 2 (ACE2) on host cells serves as the main receptor for SARS-CoV-2 spike (S) protein attachment,3, 4, 5 whereas the endogenous transmembrane protease serine 2 (TMPRSS2) cleaves the S protein, thus allowing fusion of viral and cellular membranes.3 These events promote efficient viral entry and productive infection of target cells. ACE2 and TMPRSS2 are expressed in several tissues and in multiple airway epithelial cell types, particularly nasal6 and alveolar type II cells, goblet cells, and ciliated cells.7 Although the receptor expression pattern and aerosol mode of transmission8 of SARS-CoV-2 render the airways a primary viral target, kidney and intestinal epithelial cells also express ACE2 and TMPRSS2 and can become infected in patients.9 , 10
Despite rapid progress in our understanding of COVID-19 pathogenesis, treatment options for this disease remain limited. Although several vaccines are being deployed, inoculating the world population will require much time, and the emergence of viral mutants with decreased sensitivity to vaccines remains a distinct possibility.11 Novel, safe, and accessible strategies to reduce the frequency and/or severity of SARS-CoV-2 infection are therefore highly desirable. Oral administration of OM-85 (Broncho-Vaxom), a standardized lysate of 21 bacterial strains often found in the human airways,12 is widely used empirically in Europe, South America, and Asia for the prophylaxis of upper airway recurrent infections in adults12 and children,13 with an excellent safety profile.14 A National Institutes of Health–sponsored trial (NCT02148796) is currently ongoing in the United States, where OM-85 is not yet approved. The trial is testing whether oral administration of the lysate prevents wheezing lower respiratory illnesses or asthma-like symptoms in young high-risk children. The mechanisms underlying OM-85–mediated protection from respiratory infections are complex15 , 16 and remain incompletely understood. We recently found that OM-85 boosts human airway epithelial cell barrier function in vitro and regulates multiple airway barrier-related transcriptional networks in the lung following intranasal administration to mice.17 These findings prompted us to investigate whether OM-85 also affects the expression of genes involved in SARS-CoV-2 infection of epithelial cells.
Methods
Cell lines and primary airway epithelial cells
The Vero E6 African green monkey (Chlorocebus sabaeus) kidney-derived cell line was kindly provided by Dr M. Kraft (University of Arizona). Calu-3 human lung cells and HEK293T/17 cells (HTB55 and CRL-11268, respectively) were purchased from ATCC (Manassas, Va). Caco-2 human colon cells were a kind gift from Dr J. Wilson (University of Arizona). HEK293T cells stably expressing human ACE2 were purchased from GeneCopoeia (Rockville, Md). All cell lines were propagated in Dulbecco modified Eagle medium (Gibco, Thermo Fisher Scientific, Waltham, Mass) supplemented with FBS (10%, Sigma, St Louis, Mo), GlutaMax (2 mM), penicillin (50 units/mL), and streptomycin (50 μg/mL, all from Gibco). Vero cells for SARS-CoV-2 infection experiments were obtained from ATCC (CCL-81).
To obtain human primary airway cells, participants were recruited from the population in Tucson, Arizona, and the surrounding areas. Before undergoing any procedure, informed consent according to an institutional review board–approved protocol was obtained from each participant. Healthy participants had no evidence of airway obstruction and no history of pulmonary disease; atopy (as determined by clinical history, allergen skin testing, and blood eosinophil levels) was not an exclusion criterion. Participants underwent bronchoscopy with endobronchial-protected brushing, as previously described.18 To obtain bronchial epithelial cells, brushing of the proximal airways was performed using a separate protected cytologic brush for each pass, for a total of 10 passes. Participants were discharged after their FEV1 reached 90% of their prebronchoscopy postalbuterol value.
Freshly isolated airway bronchial epithelial cells from endobronchial brushing were cultured with PneumaCult-EX Plus Medium (StemCell Technologies, Vancouver, British Columbia, Canada). Once confluent, cells were trypsinized and seeded onto collagen-coated polyester 12-mm-diameter Transwell insert membranes (Corning, Waltham, Mass) at 4 × 104 cells/well. To allow for differentiation, cells were cultured at air-liquid interface for 2 weeks with PneumaCult-Air-Liquid Interface Medium (StemCell Technologies).
Plasmids
The lentiviral packaging plasmid, psPAX2, and the vesicular stomatitis virus (VSV)-G envelope expressing plasmid, pMD2.G (Addgene plasmid no. 12260; http://n2t.net/addgene:1226; RRID:Addgene_12260, and Addgene plasmid no. 12259; http://n2t.net/addgene:12259; RRID:Addgene_12259), were gifts from Didier Trono (École Polytechnique Fédérale de Lausanne, Switzerland). The lentiviral green fluorescent protein (GFP)-expressing reporter plasmid pCIB-GFP was kindly provided by Dr Michael Johnson (University of Arizona). The SARS-CoV-2 S protein expressing plasmid HDM-SARS2-Spike-delta21 (Addgene plasmid no. 155130; http://n2t.net/addgene:155130; RRID:Addgene_155130)19 was a gift from Jesse Bloom (University of Washington, Seattle, Wash).
Functional S1 protein binding assay
Cells were grown in 6- or 12-well plates until confluent and treated with OM-85 (0.12, 0.24, 0.48, 0.96, or 1.92 mg/mL) or PBS. After 72 hours, cells were harvested, washed, and resuspended in FACSWash buffer (PBS with 1% BSA and 0.1% NaN3) at 0.5 to 1 × 107 cells/mL. To block nonspecific staining, cells were incubated on ice for 10 minutes in the same buffer containing normal rat serum (2%) followed by a 30-minute incubation with recombinant His-tagged S1 protein (5.625 μg/mL: Sino Biological US, Wayne, Pa). After an additional 30-minute incubation with an anti–His tag-phycoerythrin antibody (Cat# 362603, Biolegend, San Diego, Calif), cells were washed with FACSWash buffer and analyzed by flow cytometry. At least 20,000 events were acquired on an LSR II or a FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ) and analyzed with FlowJo software (Version 10.6.0, Becton Dickinson, 2019, Ashland, Ore). S1 binding was expressed as the percentage of S1-binding cells in each sample relative to the average of PBS-treated samples.
Production of pseudotyped lentiviral particles
HEK 293T/17 cells were transfected overnight in 100-mm dishes with packaging plasmid psPAX2 (15 μg) plus envelope plasmids (either HDM-SARS2-Spike-delta21 or pMD2.G: 15 μg) and HIV reporter plasmid expressing GFP (pCIB-GFP: 15 μg) using Lipofectamine 3000 (ThermoFisher Scientific, Waltham, Mass) according to the manufacturer’s instructions. The next day, the transfection medium was replaced with fresh complete Dulbecco modified Eagle medium. Virus-containing supernatants were collected 48 hours later and kept frozen at −80°C. The concentration of viral particles was estimated using an HIV p24 ELISA (Abcam, Cambridge, Mass). The functional titer of VSV-G–pseudotyped lentivirus (positive control), that is, the number of virions capable of productively integrating into cells per milliliter of viral preparation, was determined by flow cytometry quantification of GFP+ Vero E6 cells as described20 , 21 and was estimated to be 2.34 × 106 transducing units (TU)/mL. The titer of SARS-CoV-2-S protein–pseudotyped virus was estimated to be 1 × 104 TU/mL through side-by-side comparison with serial dilutions of VSV-G–pseudotyped lentivirus in Vero E6 cells.
Cell transduction with pseudotyped viral particles
Cells were cultured in 24- or 96-well plates until confluent and then treated with OM-85 (0.48 mg/mL). After 48 hours, the OM-85–containing medium was replaced with medium that was supplemented with polybrene (6 μg/mL) and contained either SARS-CoV-2 S protein– (2 × 103 TU) or VSV-G protein– (2.34 × 104 TU) pseudotyped lentivirus. Plates were centrifuged at 2000g for 1 hour at room temperature and cultured overnight at 37°C in a 5% CO2 atmosphere. The GFP fluorescence generated on transcription and translation of the reporter gene by transduced cells was assessed 72 hours posttransduction using an Axio Vert microscope (Carl Zeiss Microscopy, White Plains, NY) under 10× magnification. At least 20 (SARS-CoV-2–pseudotyped virus) or 10 (VSV-G–pseudotyped virus) frames/well were recorded using an Axiocam 305 camera (Carl Zeiss Microscopy) and quantified by counting fluorescent pixels with the Slide Analyzer software (https://github.com/dpivniouk/slide_analyzer).
SARS-CoV-2 infection
SARS-CoV-2, isolate USA-WA1/2020, was deposited by Dr Natalie J. Thornburg at the Centers for Disease Control and Prevention and obtained from the World Reference Center for Emerging Viruses and Arboviruses. Stocks of SARS-CoV-2 were generated as a single passage from received stock vial on mycoplasma-negative Vero cells (ATCC CCL-81).
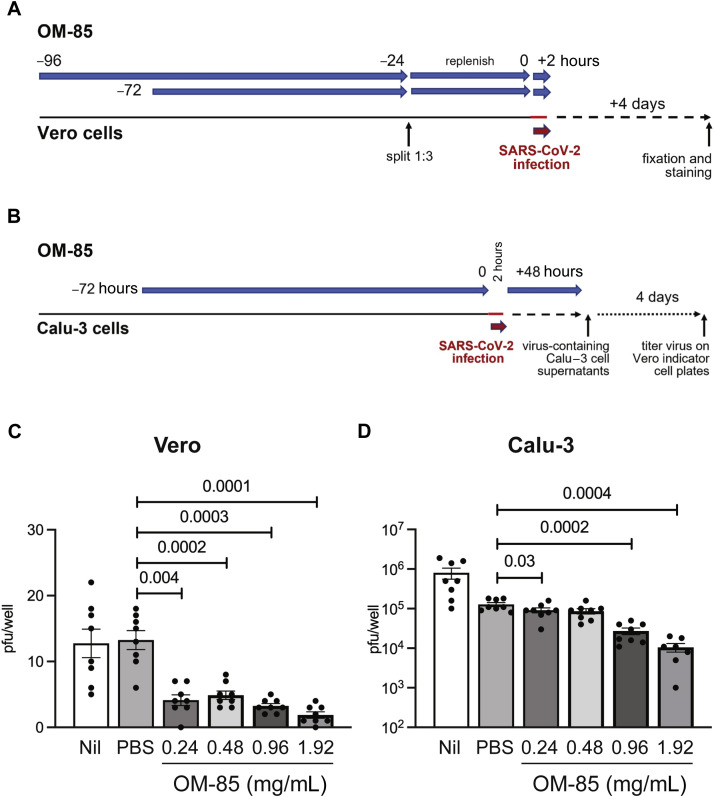
For the virus inhibition assay, preliminary flow cytometry experiments established that OM-85 stimulation decreased S1 protein binding to Vero cells (ATCC CCL-81) to levels comparable to those seen in Vero E6 cells (not shown). Vero cells (1 × 106 per 96-well, flat-bottom plate) were pretreated with PBS or increasing OM-85 concentrations (0.24-1.92 mg/mL) for a total of 72 or 96 hours. Twenty-four hours before adding SARS-CoV-2, cells were dissociated with 0.25% trypsin, washed once with media in the original plate, and replated from one to three 96-well plates; OM-85 was replenished at the original concentrations. At the 72- or 96-hour treatment time point, SARS-CoV-2 (15 plaque-forming units, pfu/well) was added in the presence or absence of OM-85. The virus was incubated for 2 hours to allow infection of the monolayer and then dumped off. Cells were then overlayed with 1% methylcellulose in 5% Dulbecco modified Eagle medium without OM-85. Four days later, plates were fixed in 10% neutral buffered formalin for 30 minutes and stained with 1% crystal violet. Plaques were imaged using an ImmunoSpot Versa plate reader (Cellular Technology Limited, Cleveland, Ohio) and counted. Results were expressed as average numbers of pfu/well detected in quadruplicate wells for each condition.
Calu-3 cells were plated at 0.5 × 106 per 96-well plate for 24 hours and then treated with PBS or OM-85 (0.24-1.92 mg/mL). After 72 hours, the medium was removed and SARS-CoV-2 (WA/2020, ∼100 pfu) was added to all wells. After 2 hours, the virus was removed and replaced with fresh medium containing the original OM-85 concentrations; cells were then cultured for 48 additional hours. Plates were then frozen at −80°C to release virus and centrifuged to pellet debris. To determine viral titers in Calu-3 cultures, virus-containing supernatants from each row of a Calu-3 plate were titrated by 10-fold serial dilutions on 96-well indicator Vero plates and incubated for 2 hours. Indicator Vero plates were then overlayed with 1% methylcellulose in fresh medium and incubated for 4 days to allow plaques to develop. Plates were fixed and stained as above, and plaques were counted. The number of pfu/well of Calu-3 plates was calculated by dividing the number of plaques on indicator Vero plates by the dilution factor. Results were expressed as average numbers of pfu/well detected in 8 replicates from the original Calu-3 plate.
OM-85 preparations and all other experimental procedures and statistical methods are detailed in this article’s Online Repository at www.jacionline.org.
Results
OM-85 inhibits Ace2 and Tmprss2 transcription in the mouse lung through the Myd88/Trif pathway
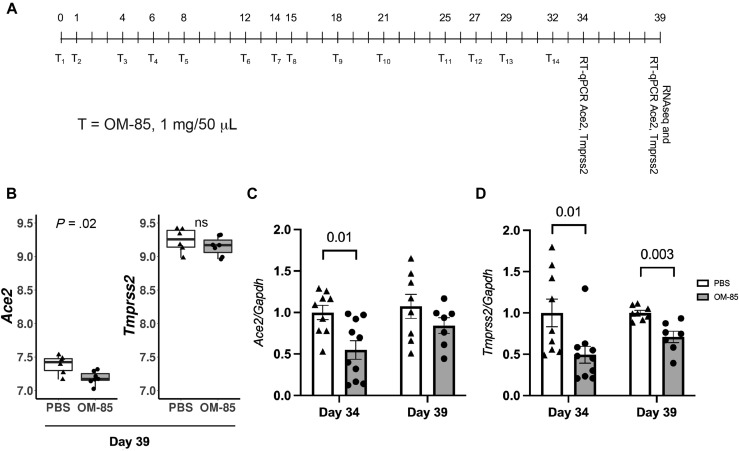
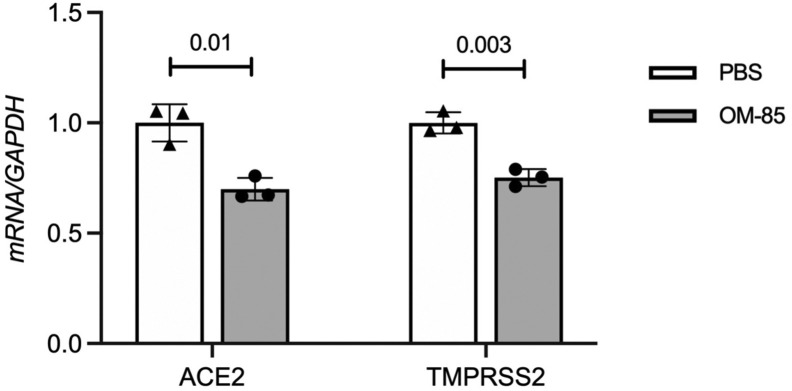
Our recent studies assessing how OM-85 administration to the airway compartment affects immune responses in the mouse lung17 provided the first clue that OM-85 might affect SARS-CoV-2/host interactions. RNA-sequencing profiling of the lung transcriptome in Balb/c mice treated intranasally with OM-85 according to an optimized protocol (1 mg/treatment × 14 treatments over 32 days, with terminal assessments at day 39: Fig 1 , A) identified multiple genetic networks regulated by the lysate. Especially notable was the upregulation of signature genes related to tight junctions and epithelial barrier function.17 Interestingly, a preliminary analysis focused on the expression of the SARS-CoV-2 receptors Ace2 and Tmprss2 revealed a nominally significant (P = .02) inhibition of Ace2 expression in OM-85–treated mice compared with PBS-treated controls. Tmprss2 expression was also decreased, albeit not significantly (Fig 1, B). RT-quantitative PCR with Ace2- and Tmprss2-specific primers was then used to measure lung expression of these genes and compare their mRNA levels at day 34 and day 39 of our protocol (ie, 2 and 7 days after the last OM-85 treatment: Fig 1, A). We chose the 7-day time point because it was optimal for inhibition of experimental allergic asthma and was used to generate the RNA-sequencing data presented here.17 However, the 2-day time point allowed us to explore shorter-term effects. Relative to PBS-treated mice, Tmprss2 was significantly downregulated at both time points (P = .01 and P = .003, respectively: Fig 1, D), whereas Ace2 was significantly (P = .01) inhibited only at the earlier time point (Fig 1, C). These experiments showed that delivery of OM-85 to the airways affected the expression of the major SARS-CoV-2 receptor components in the lung, with distinct time-dependent effects.
Fig 1.
OM-85 inhibits Ace2 and Tmprss2 transcription in the mouse lung. A, Balb/c mice were treated intranasally with OM-85 (1 mg in 50 μL) 14 times as indicated in the chart (T1-T14). The last OM-85 treatment was administered at day 32. Lungs for transcriptional analyses were collected 2 (day 34) or 7 (day 39) days later. B,Ace2 and Tmprss2 mRNA levels in the lungs of mice treated intranasally with OM-85 (1 mg/treatment × 14 treatments) or PBS (n = 6/group) over 32 days and sacrificed at day 39. Data were extracted from DESeq2-normalized RNA-sequencing counts after adjusting for latent factors. P values were calculated from differential expression analysis (DESeq2 Wald test). C and D,Ace2 and Tmprss2 expression in the lungs of Balb/c mice treated intranasally with OM-85 or PBS as in Fig 1, A, was measured by RT-qPCR at day 34 or 39 (ie, 2 or 7 days after the last OM-85 treatment, respectively; n = 9-10 mice/group at day 2, and n = 7-8 mice/group at day 7). mRNA levels normalized by Gapdh are shown relative to PBS. A Wilcoxon 2-sample test was used for statistical analysis after testing for normality of sample distribution. GAPDH, Glyceraldehyde 3-phosphate dehydrogenase; ns, nonsignificant; RT-qPCR, RT-quantitative PCR.
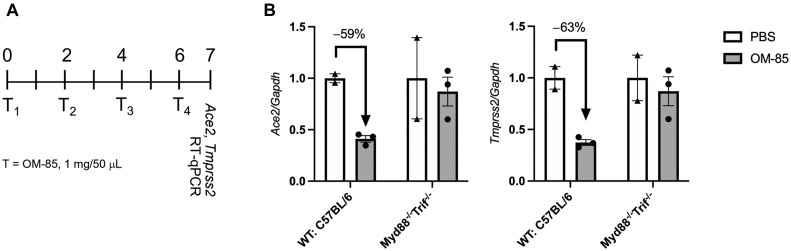
Because OM-85 is a bacterial lysate whose ability to protect from asthma in vivo depends on Myd88/Trif innate immune signaling,17 we then assessed whether OM-85–induced inhibition of Ace2 and Tmprss2 transcription in the mouse lung also required this pathway. These experiments, which were performed in C57BL/6 mice, revealed that as few as 4 intranasal treatments with OM-85 (1 mg/treatment: see Fig E1, A, in this article’s Online Repository at www.jacionline.org) were sufficient to reduce Ace2 and Tmprss2 expression by 59% and 63%, respectively, in lung cells isolated from wild-type mice. In contrast, negligible changes in the expression of these genes were detected in Myd88 −/− Trif −/− mice (Fig E1, B). These findings indicated that OM-85–induced downregulation of Ace2 and Tmprss2 transcription in mouse lung tissue is Myd88/Trif-dependent and strain-independent.
Fig E1.
OM-85–induced downregulation of Ace2 and Tmprss2 in the mouse lung is Myd88/Trif-dependent. A, WT and Myd88−/−Trif−/− C57BL/6 mice were treated intranasally with OM-85 (1 mg in 50 μL) or PBS every 2 days for 4 times as indicated in the chart (T1-T4). Lung cells for transcriptional analyses were collected at day 7. B,Ace2 and Tmprss2 mRNA levels in the lungs of WT and Myd88−/−Trif−/− C57BL/6 mice treated intranasally with OM-85 or PBS were measured by RT-qPCR and normalized by Gapdh (n = 2 mice each/PBS group, and n = 3 mice each/OM-85 group). GAPDH, Glyceraldehyde 3-phosphate dehydrogenase; RT-qPCR, RT-quantitative PCR; WT, wild-type.
OM-85 inhibits Ace2 and Tmprss2 expression in nonhuman primate and human epithelial cells in vitro
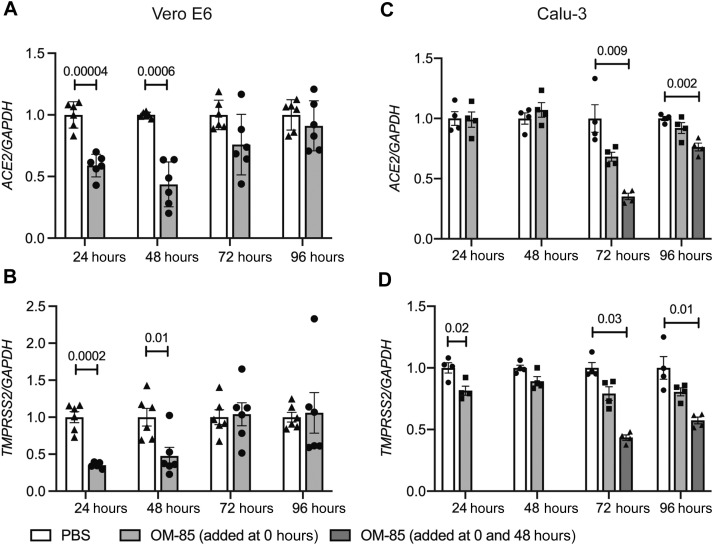
Our initial results illustrated the in vivo effects of OM-85 in the airways, a main target of SARS-CoV-2. However, further mechanistic dissections in these models were problematic because lung Ace2 and Tmprss2 expression likely derived from multiple cell types.7 , 22 Moreover, mice are not naturally susceptible to infection by most SARS-CoV-2 strains.23 Therefore, subsequent experiments relied on RT-quantitative PCR to assess the effects of OM-85 on ACE2 and TMPRSS2 expression in nonhuman primate Vero E6 as well as human Calu-3 and Caco-2 epithelial cells. These cells derive from SARS-CoV-2 target organs (kidney, lung, and colon, respectively) of naturally susceptible species23 and thus provide ideal models to characterize the interactions of epithelial cells with, and their infection by, SARS-CoV-2.3 , 4 , 24, 25, 26 OM-85 (0.48 mg/mL) significantly and strongly inhibited ACE2 expression in kidney Vero E6 cells after 24 and 48 hours of stimulation (P = .00004 and P = .0006, respectively: Fig 2 , A). TMPRSS2 was also significantly decreased at the same time points (P = .0002 and P = .01, respectively: Fig 2, B). In contrast, ACE2 expression by lung Calu-3 cells was significantly inhibited only after 72 and 96 hours, and only when fresh OM-85 was replaced after 48 hours of culture (P = .009 and P = .002, respectively: Fig 2, C). TMPRSS2 expression was inhibited at the 24-hour time point (P = .02) as well as at 72 and 96 hours, but only when at 48 hours spent medium was replaced with fresh medium containing the original concentration of OM-85 (P = .03 and P = .01, respectively: Fig 2, D). Therefore, OM-85 inhibited the expression of both ACE2 and TMPRSS2 in Vero E6 and Calu-3 epithelial cells, but overall downregulation occurred more rapidly in the former than in the latter. A 48-hour stimulation with OM-85 (0.48 mg/mL) also significantly reduced ACE2 and TMPRSS2 mRNA levels in intestinal Caco-2 cells (P = .01 and P = .003, respectively: see Fig E2 in this article’s Online Repository at www.jacionline.org).
Fig 2.
OM-85 inhibits ACE2 and TMPRSS2 transcription in NHP and human epithelial cell lines from kidney and lung. ACE2 (A and C) and TMPRSS2 (B and D) mRNA levels were measured by RT-qPCR in Vero E6 (Fig 2, A and B) and Calu-3 (Fig 2, C and D) epithelial cells treated with OM-85 (0.48 mg/mL) or PBS for 24 to 96 hours. mRNA levels at each time point were normalized by GAPDH and are shown relative to PBS. Data are from 2 pooled experiments (Fig 2, A and C) and 1 representative experiment of 2 (Fig 2, B and D) (n = 3-4 wells/condition, each run in triplicate). An unpaired, 2-tailed t test (Fig 2, A-C) or a Wilcoxon 2-sample test (Fig 2, D) was used for statistical analysis after testing for normality of sample distribution. GAPDH, Glyceraldehyde 3-phosphate dehydrogenase; NHP, nonhuman primate; RT-qPCR, RT-quantitative PCR.
Fig E2.
OM-85 inhibits ACE2 and TMPRSS2 transcription in human colon Caco-2 cells. ACE2 and TMPRSS2 expression was measured by RT-qPCR in cells treated with OM-85 (0.48 mg/mL) or PBS for 48 hours. mRNA levels were normalized by GAPDH and are shown relative to PBS. Data are from 1 representative experiment (n = 3-4 wells/condition, each run in triplicate) of 2. An unpaired, 2-tailed t test was used for statistical analysis after testing for normality of sample distribution. GAPDH, Glyceraldehyde 3-phosphate dehydrogenase; RT-qPCR, RT-quantitative PCR.
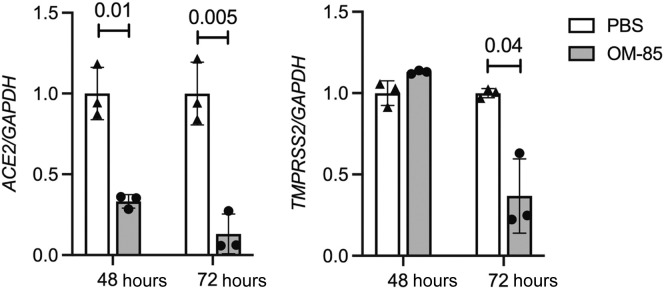
To further explore the significance of our findings, we turned to human primary airway cells, a preeminent natural SARS-CoV-2 target. Bronchial epithelial cells isolated from a healthy individual were differentiated at the air-liquid interface for 2 weeks. ACE2 and TMPRSS2 mRNA levels were then measured by RT-quantitative PCR after a 48- or 72-hour incubation with PBS or OM-85 (1.92 mg/mL). Fig 3 shows strong, significant inhibition of ACE2 at 48 and 72 hours (P = .01 and P = .005, respectively) and a more modest, but still significant inhibition of TMPRSS2 at 72 hours (P = .04). These results from OM-85–treated human normal primary epithelial cells overall validated those generated in epithelial cell lines.
Fig 3.
OM-5 inhibits ACE2 and TMPRSS2 transcription in primary normal human bronchial epithelial cells. Bronchial epithelial cells freshly isolated from a healthy donor were cultured to confluence and differentiated onto collagen-coated polyester 12-mm Transwell insert membranes for 2 weeks at air-liquid interface. Cells were then stimulated apically with PBS or OM-85 (1.92 mg/mL) for 48 or 72 hours. ACE2 and TMPRSS2 expression was determined by RT-qPCR (n = 3 wells/condition, each run in triplicate). mRNA levels at each time point were normalized by GAPDH and are shown relative to PBS. An unpaired, 2-tailed t test was used for statistical analysis after assessing the normality of sample distribution. RT-qPCR, RT-quantitative PCR.
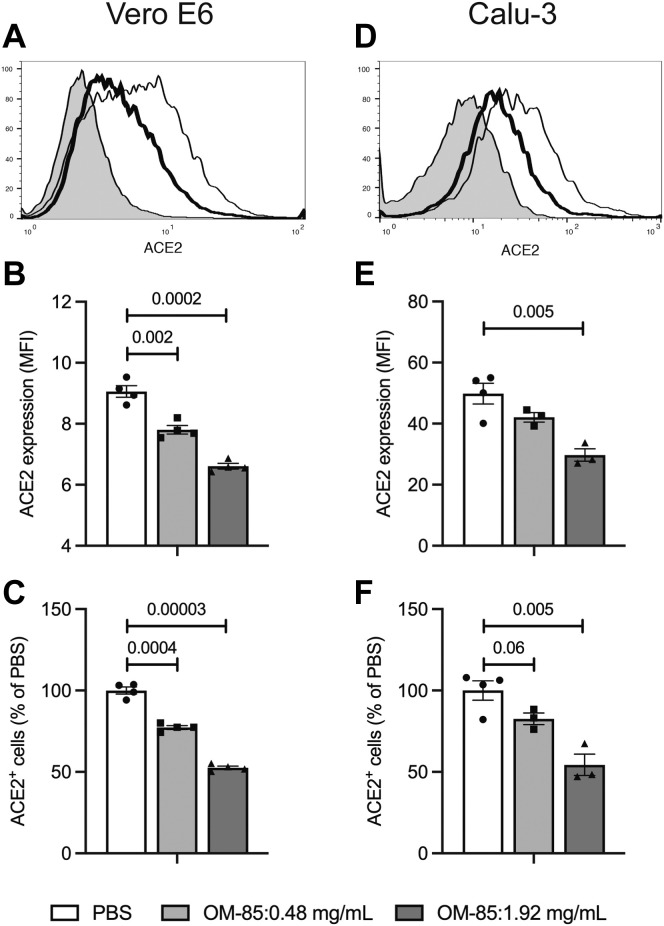
We next assessed whether decreased SARS-CoV-2 receptor transcription was associated with reduced surface levels of receptor protein. To this end, Vero E6 and Calu-3 cells were incubated with PBS or OM-85 (0.48 and 1.92 mg/mL) for 72 hours and examined by flow cytometry with ACE2-specific mAbs. OM-85 treatment significantly and dose-dependently decreased both ACE2 mean fluorescence intensity (P = .002 and P = .0002, respectively) and the percentage of ACE2-positive cells in Vero E6 cells (Fig 4 , A-C) and Calu-3 cells (P = .005 for ACE2 mean fluorescence intensity and ACE2-positive cell percentages at 1.92 mg/mL: Fig 4, D-F). In combination, these results demonstrate that in vitro stimulation with the OM-85 bacterial lysate dampens SARS-CoV-2 receptor expression on epithelial cells from distinct organs.
Fig 4.
OM-5 downregulates surface ACE2 protein expression in epithelial cells. Vero E6 (A-C) and Calu-3 (D-F) cells were treated with OM-85 (0.48 or 1.92 mg/mL) for 72 hours and assessed for ACE2 protein expression by flow cytometry with ACE2-specific mAbs and relevant isotype controls. A and D, Representative ACE2 flow cytometry plots (shaded area: isotype control; thin line: PBS-treated cells; thick line: cells treated with OM-85, 1.92 mg/mL); B and E, ACE2 MFI and percentages of ACE2+ cells in PBS- and OM-85–treated cells (Fig 4, C and F). Data are from 1 experiment with 4 samples/condition. An unpaired, 2-tailed t test was used for statistical analysis after testing for normality of sample distribution. MFI, Mean fluorescence intensity.
OM-85 reduces S1 protein–mediated attachment to epithelial cells
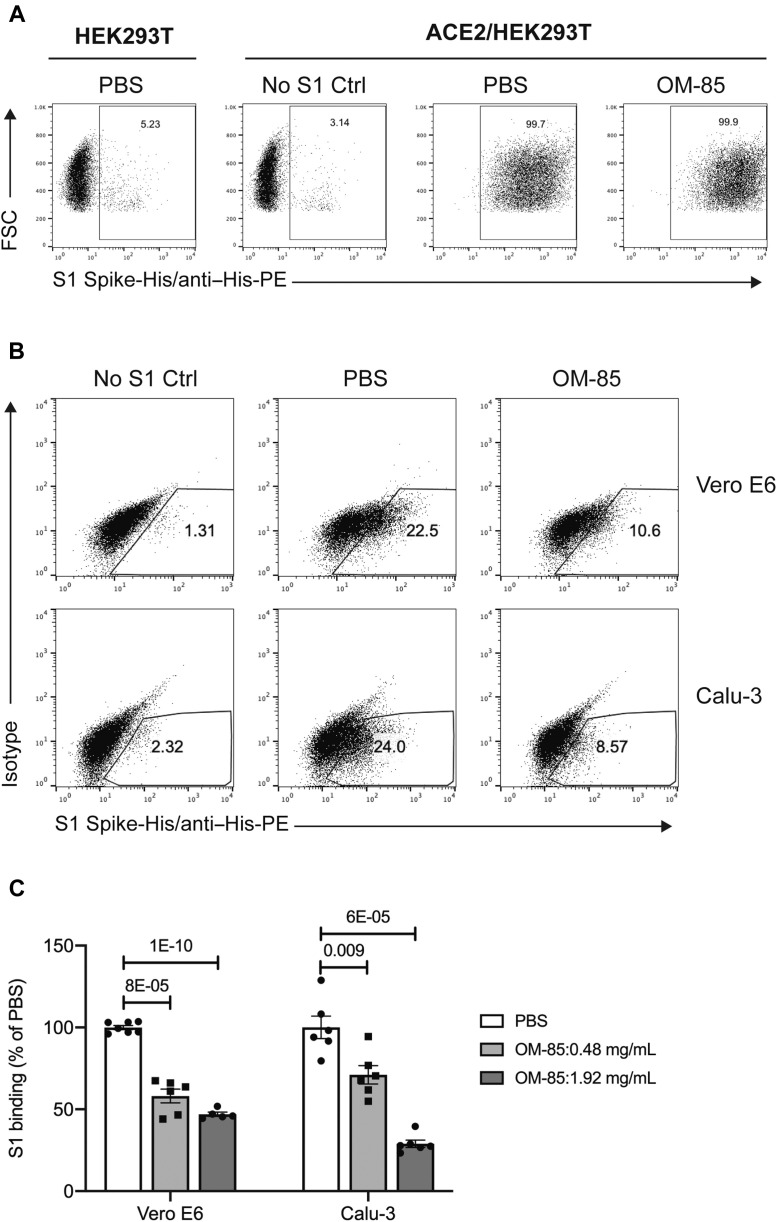
Because ACE2 and TMPRSS2 enable SARS-CoV-2 infection of epithelial cell by mediating S protein attachment to, and viral entry into, these cells, we next investigated whether OM-85–induced downregulation of ACE2 and TMPRSS2 interferes with these processes. To assess the effects of OM-85 on SARS-CoV-2 S protein attachment, we developed an S1 protein binding assay. Cells were incubated with a recombinant His-tagged S1 subunit comprising the SARS-CoV-2 receptor binding domain,3 followed by a phycoerythrin-conjugated anti-His antibody. S1 protein binding to cells was assessed by flow cytometry. We validated the ability of this assay to specifically detect ACE2-mediated S1 protein cellular binding using HEK293T cells, untransfected or stably transfected with human ACE2 (ACE2/HEK293T). S1 binding was detected in only 5% of untransfected HEK293T cells and ACE2/HEK293T cells incubated with anti–His-phycoerythrin antibody without S1 protein, whereas more than 99% of PBS-treated ACE2/HEK293T cells bound S1 protein (Fig 5 , A).
Fig 5.
OM-85 reduces S1 protein–mediated attachment to epithelial cells. Representative flow cytometry plots showing SARS-CoV-2 S1 protein binding to parental HEK293T cells and HEK293T cells stably transfected with human ACE2 (ACE2/HEK293T) (A) and Vero E6 and Calu-3 (B) epithelial cells. Cells were treated with PBS or OM-85 (1.92 mg/mL) for 72 hours and then incubated with or without recombinant His-tagged S1 protein, followed by an anti–His-PE antibody. S1 binding was assessed by flow cytometry. C, Effects of OM-85 on S1 protein binding to Vero E6 or Calu-3 cells. Data are shown as percentages of S1 protein–binding cells in OM-85–treated vs PBS-treated cultures (n = 5-6 wells/group from 3 [Vero E6 cells] or 2 [Calu-3 cells] independent experiments). An unpaired, 2-tailed t test was used for statistical analysis after testing for normality of sample distribution. FSC, Forward scatter; PE, phycoerythrin.
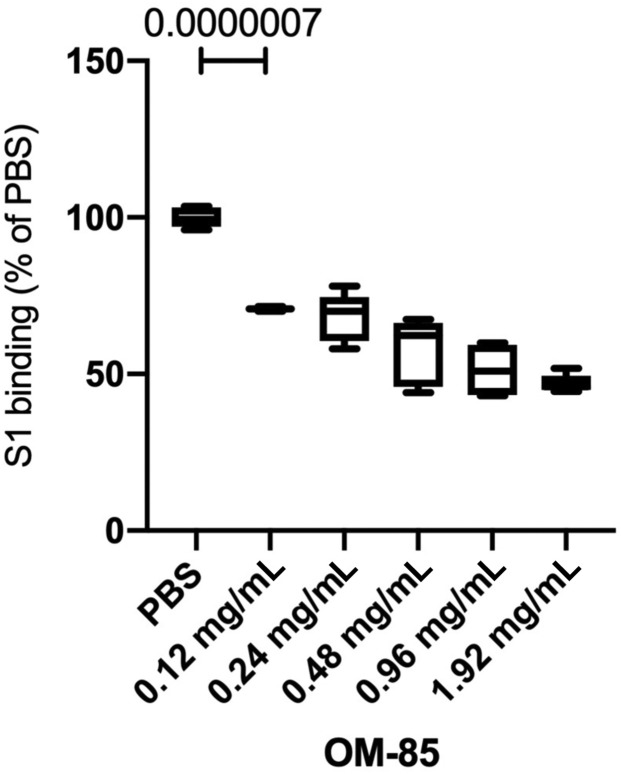
Using this S1 protein binding assay, we then found that a substantial proportion of Vero E6 (22% ± 0.5%) and Calu-3 (24% ± 1%) epithelial cells bound SARS-CoV-2 S1 protein (Fig 5, B). Preliminary dose-response curves with a broad range of OM-85 concentrations (0.12-1.92 mg/mL) showed dose-dependent inhibition of S1 binding to Vero E6 cells, with significant effects (P = 7.00 × 10−07) even at the lowest concentration (0.12 mg/mL) and robust inhibition at concentrations greater than or equal to 0.48 mg/mL (see Fig E3 in this article’s Online Repository at www.jacionline.org). A 72-hour incubation with OM-85 (0.48 or 1.92 mg/mL) dose-dependently reduced S1 binding by 42% ± 4% (P = 8 × 10−05) and 53% ± 1.3% (P = 1 × 10−10) for Vero E6 cells, and 29% ± 5.6% (P = .009) and 71% ± 2.3% (P = 6 × 10−05) for Calu-3 cells (Fig 5, C).
Fig E3.
OM-85 dose-dependently inhibits S1 protein binding to Vero E6 cells. Cells were treated with PBS or increasing OM-85 concentrations for 72 hours and then incubated with or without recombinant His-tagged S1 protein, followed by an anti–His-PE antibody. S1 binding was assessed by flow cytometry. Data are shown as percentages of S1 protein–binding cells in OM-85-treated vs PBS-treated cultures (n = 5-6 wells/group). An unpaired, 2-tailed t test was used for statistical analysis after testing for normality of sample distribution. PE, Phycoerythrin.
These results demonstrated that OM-85 efficiently inhibited S1 protein attachment to epithelial cells derived from natural SARS-CoV-2 target organs. Mechanistically, our findings strongly suggested that this inhibition reflected OM-85–dependent interference with the physiologic regulation of ACE2 expression. In support of this notion, even a maximal concentration of OM-85 (1.92 mg/mL) failed to inhibit ACE2 protein expression on (see Table E1 in this article’s Online Repository at www.jacionline.org), and S1 protein binding to (Fig 5, A, right), ACE2/HEK293T cells in which human ACE2 transcription is driven by a heterologous, OM-85–unresponsive cytomegalovirus promoter.
OM-85 inhibits the entry of S protein–pseudotyped lentiviral particles into epithelial cells
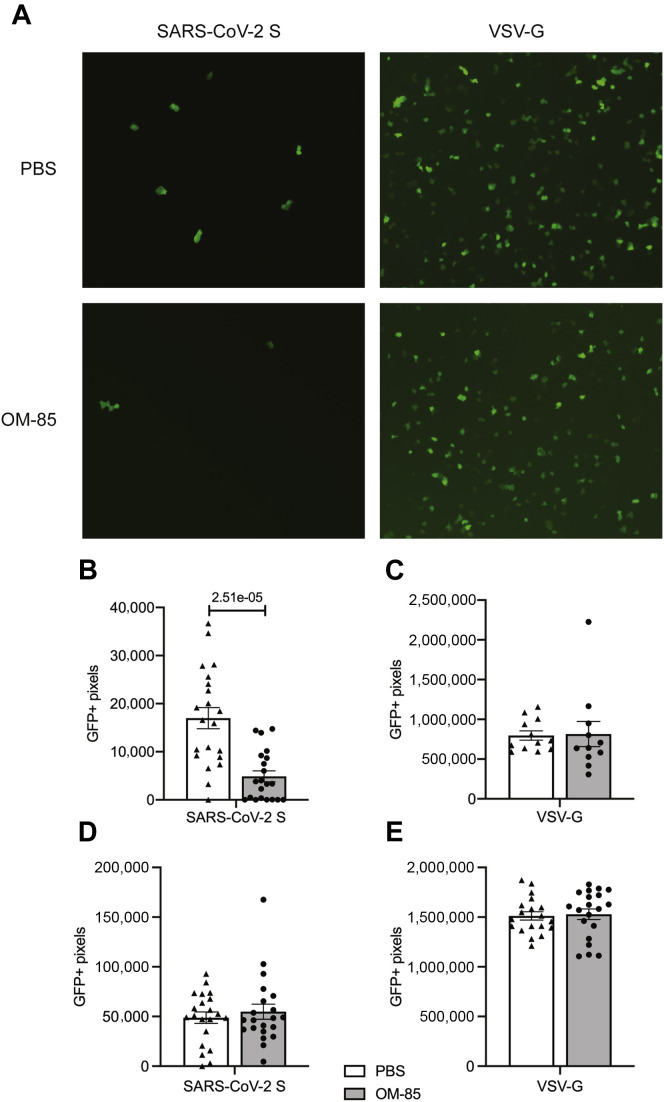
To investigate whether OM-85–dependent inhibition of SARS-CoV-2 receptor expression in, and SARS-CoV-2 S1 protein attachment to, epithelial cells also reduces S protein–mediated SARS-CoV-2 entry into these cells, we directly measured the entry of replication-deficient, SARS-CoV-2 S protein–pseudotyped lentiviral particles into epithelial cells preincubated with OM-85 or PBS. Our lentiviral particles carried a GFP reporter gene that is transcribed and translated by transduced cells, and were pseudotyped with SARS-CoV-2 S protein or the G glycoprotein of the pantropic VSV (positive transduction control).3 As expected, VSV-G–pseudotyped particles efficiently transduced Vero E6 cells, with functional titers reaching 2.3 × 106 TU/mL (Fig 6 , A, top right). Vero E6 transduction by SARS-CoV-2–pseudotyped particles was less efficient but still robust and consistent (Fig 6, A, top left), with titers reaching 1 × 104 TU/mL.
Fig 6.
OM-85 inhibits entry of S protein–pseudotyped lentiviral particles into Vero E6 cells. A, Vero E6 cells were treated with OM-85 (0.48 mg/mL) or PBS for 48 hours, washed, and transduced with lentiviral particles that were pseudotyped with SARS-CoV-2 S protein (0.2 × 104 TU) or VSV-G protein (2 × 104 TU) and carried a GFP reporter. Shown are frames representative of GFP immunofluorescence under various experimental conditions. B-E, GFP fluorescence was assessed under a microscope (10× magnification) 72 hours posttransduction and quantified using the Slide Analyzer software in Vero E6 (Fig 6, B and C) and ACE2/HEK293T (Fig 6, D and E) cells transduced with either SARS-CoV-2-S–pseudotyped virus (Fig 6, B and D) or VSV-G–pseudotyped virus (Fig 6, C and E). Results represent the average ± SEM of 20 (SARS-CoV-2–S-pseudotyped virus) or 10 (VSV-G–pseudotyped virus) random frames from 1 representative experiment of 2 independent experiments. A Wilcoxon 2-sample test was used for statistical analysis after assessing the normality of sample distribution.
OM-85 pretreatment (0.48 mg/mL) strongly (P = 2.5 × 10−05) inhibited SARS-CoV-2 S protein–mediated Vero E6 cell transduction (Fig 6, A, left, and Fig 6, B). This effect was specific because transduction by VSV-G–pseudotyped particles remained unaffected (Fig 6, A, right, and Fig 6, C). The effects of OM-85 on viral entry into Calu-3 cells could not be reliably estimated because these cells were poorly transduced under our experimental conditions. Indeed, both percentages of GFP+ cells and GFP mean fluorescence intensity in these cells were consistently low following transduction with either SARS-CoV-2 S protein– or VSV-G– (positive control) pseudotyped particles (not shown). Notably, ACE2/HEK293T cells were readily transduced by both SARS-CoV-2 S– and VSV-G–pseudotyped viral particles, but OM-85 failed to inhibit viral entry into these cells (Fig 6, D and E). Because ACE2/HEK293T cells do not downregulate ACE2 on OM-85 stimulation (Table E1), these findings were consistent with the notion that OM-85–induced suppression of events leading to SARS-CoV-2 infection involves a reduction of ACE2 expression.
OM-85 inhibits SARS-CoV-2 infection of epithelial cells from distinct tissues
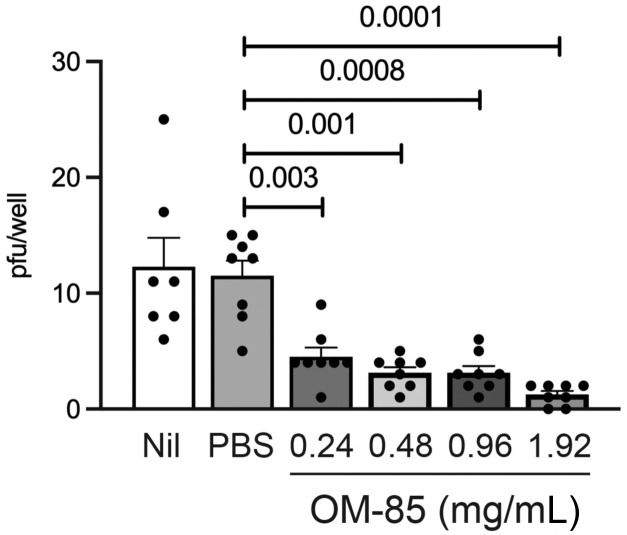
The data discussed above had shown that OM-85 effectively inhibits both SARS-CoV-2 S protein attachment and S protein–mediated pseudotyped virus entry into epithelial cells. Therefore, the last set of experiments assessed whether OM-85 treatment also suppressed epithelial cell infection with live SARS-CoV-2. To this end, kidney-derived Vero cells were pretreated with PBS or OM-85 (0.24-1.92 mg/mL) for 72 or 96 hours and then incubated for 2 hours with SARS-CoV-2 (isolate USA-WA1/2020). Numbers of pfu/well were counted 4 days later (Fig 7 , A). SARS-CoV-2 infection was strongly and significantly inhibited in cultures pretreated with OM-85 for 72 hours (Fig 7, C) or 96 hours (see Fig E4 in this article’s Online Repository at www.jacionline.org) but not in PBS-pretreated cultures. Inhibition was evident even at the lowest OM-85 concentration and reflected effects of OM-85 pretreatment on epithelial cells rather than SARS-CoV-2 itself because infection was comparably reduced in cultures that did or did not receive OM-85 during the 2-hour infection period (not shown). SARS-CoV-2 infection of lung-derived Calu-3 cells was also significantly inhibited by a 72-hour pretreatment with OM-85, especially at the highest concentrations of bacterial lysate (Fig 7, B and D). These results clearly demonstrate that OM-85 inhibits in vitro SARS-CoV-2 infection of epithelial cells sourced from distinct tissues.
Fig 7.
OM-5 inhibits epithelial cell infection by SARS-CoV-2. A, Vero cells were pretreated with PBS or OM-85 (0.24-1.92 mg/mL) for a total of 72 hours and then infected with SARS-CoV-2 (isolate USA-WA1/2020, 15 pfu/well) for 2 hours. Plaques were counted as described in Methods. B, Calu-3 cells treated with PBS or OM-85 (0.24-1.92 mg/mL) for 72 hours were infected with SARS-CoV-2 (WA/2020, ∼100 pfu) for 2 hours. Cells were then cultured for 48 hours in medium containing the original OM-85 concentrations. Viral titers were determined by dispensing 10-fold serial dilutions of Calu-3 culture supernatants on 96-well indicator Vero plates. Plaques were counted as described in Methods. The number of pfu/well of Calu-3 plates was calculated by dividing the number of plaques on indicator Vero plates by the dilution factor. Shown are average numbers ± SEM of pfu detected in 4 (C) or 8 (D) SARS-CoV-2–infected replicate wells pretreated with PBS or increasing OM-85 concentrations. Data were pooled from 2 independent experiments (Fig 7, A and C) and 1 representative experiment (Fig 7, B and D). A 2-tailed t test was used for statistical analysis after assessing the normality of sample distribution.
Fig E4.
A 96-hour pretreatment with OM-85 inhibits Vero cell infection by SARS-CoV-2. Vero cells were pretreated with PBS or OM-85 (0.24-1.92 mg/mL) for 96 hours and then infected with SARS-CoV-2 (isolate USA-WA1/2020, 15 pfu/well) for 2 hours. Plaques were counted as described in Methods. Shown are average numbers ± SEM of pfu detected in 8 SARS-CoV-2–infected replicate wells pretreated with PBS or increasing OM-85 concentrations. Data were pooled from 2 independent experiments. A 2-tailed t test was used for statistical analysis after assessing the normality of sample distribution.
Discussion
Bacterial lysates are receiving increasing attention for their ability to act as potent response modulators in immune disorders.27 , 28 Our results demonstrate that OM-85, a standardized lysate of human airway–derived bacterial strains, efficiently inhibits live SARS-CoV-2 infection of epithelial cells derived from distinct tissues. OM-85 interfered with multiple steps in the chain of events leading to SARS-CoV-2 epithelial cell infection: it suppressed SARS-CoV-2 receptor (ACE2 and TMPRSS2) expression, SARS-CoV-2 S1 protein–mediated cell attachment, and SARS-CoV-2 S protein–mediated cell entry. Remarkably, OM-85–dependent inhibition occurred only when the lysate downregulated ACE2, which initiates SARS-CoV-2 infection by mediating SARS-CoV-2 S protein attachment to target cells. In combination, these data strongly suggest that decreased transcription of SARS-CoV-2 receptor components, primarily ACE2, is an essential mechanism for the inhibition of SARS-CoV-2 epithelial cell infection by OM-85. Further studies are needed to identify the molecular pathways underlying ACE2 downregulation by OM-85. However, OM-85–dependent inhibition of SARS-CoV-2 infection in vitro in isolated epithelial cells points to epithelial-intrinsic effects of the lysate.
Our findings have translational implications because the COVID-19 pandemic is not abating and the therapeutic arsenal against COVID-19 is expanding but remains limited. In addition to corticosteroids,29 , 30 biologics that interfere with selected proinflammatory pathways (eg, IL-6, IL-1, and Janus kinase inhibitors),31, 32, 33 and anti–SARS-CoV-2 antibodies aimed at disrupting progression of COVID-19 infection,34 , 35 current COVID-19 vaccines seek to induce humoral and cellular immune responses against the SARS-CoV-2 S protein so as to neutralize its ability to latch onto cellular receptors and mediate infection.36 , 37 Despite their remarkable overall efficacy, though, vaccines remain cumbersome to distribute and administer. Moreover, viral variants of concern may emerge that resist or even escape the immunity generated by the current vaccines.11 , 37 Antivirals that could be used against SARS-CoV-2 and future emerging viruses are therefore under intense investigation. Remdesivir, an RNA-dependent RNA polymerase inhibitor, was initially reported to shorten COVID-19 hospitalization times38 but failed in a large clinical trial examining hospitalized patients.39 Other interesting drugs with potential anti–SARS-CoV-2 properties have been identified through drug-repurposing screens,40, 41, 42, 43 an approach that is becoming increasingly attractive because it involves the use of derisked compounds, potentially lower development costs, and shorter development timelines.44
In this context, our current results indicate that OM-85 also deserves active consideration. Indeed, the capacity of OM-85 to suppress multiple steps of SARS-CoV-2 cell infection by downregulating the receptor machinery in the epithelium, a primary viral target, may be leveraged to prevent infection and/or decrease its severity by limiting the infection/reinfection cycle. Further studies are needed to better understand the impact of OM-85 on ACE2- and TMPRSS2-expressing primary cells from distinct portions of the airways and from other SARS-CoV-2 target organs. The administration route resulting in optimal inhibition of SARS-CoV-2 infection in vivo also remains to be established. However, it is noteworthy that OM-85–induced SARS-CoV-2 receptor downregulation would be expected to protect against multiple SARS-CoV-2 variants and even against other coronaviruses that rely on ACE2 for host cell infection. The impeccable safety profile of OM-85 demonstrated by decades of clinical use as an immunomodulator,14 , 45 the lack of reported side effects on ACE2 physiologic functions,14 and the ease of administration of this agent suggest that this standardized bacterial extract may eventually complement the current COVID-19 therapeutic toolkit.
Key messages.
-
•
The OM-85 bacterial lysate downregulates the SARS-CoV-2 receptor ACE2 and TMPRSS2 in epithelial cells and strongly inhibits SARS-CoV-2 S-1 protein binding to, SARS-CoV-2 S protein–pseudotyped lentivirus entry into, and SARS-CoV-2 infection of these cells.
-
•
The ability of OM-85 to inhibit SARS-CoV-2 infection of epithelial cells in vitro and its excellent safety profile warrant further studies of its effects against COVID-19.
Footnotes
This work was funded in part by a research grant provided by OM Pharma SA to the University of Arizona. Support was also provided by postdoctoral NIH fellowships from T32 ES007091 and the BIO5 Institute (to A.D.V.), a predoctoral T32 HL007249 fellowship (to S.R.V.L.), and P01AI148104, R21AI144722, and R25HL126140 (to D.V.).
Disclosure of potential conflict of interest: D. Vercelli and V. Pivniouk are inventors in PCT/EP2019/074562, “Method of Treating and/or Preventing Asthma, Asthma Exacerbations, Allergic Asthma and/or Associated Conditions with Microbiota Related to Respiratory Disorders.” M. Kraft has received grants from the National Institutes of Health, the American Lung Association, Chiesi, Sanofi, and Astra-Zeneca, speaker fees from Chiesi outside this work, and consulting fees from Astra-Zeneca and Sanofi, outside the submitted work. J. Nikolich-Zugich is cochair of the Scientific Advisory Board of and receives research funding from Young Blood Institute, Inc. The rest of the authors declare that they have no relevant conflicts of interest.
Methods
Mice
OM-85 (1 mg in 50 μL, 25 μL/nostril) was instilled intranasally every 2 to 3 days (14 times total) beginning at day 0 (Fig 1, A) into adult (6-7-week-old) male Balb/c mice (Charles Rivers Laboratories, Wilmington, Mass) maintained on a standard hypoallergenic diet under specific pathogen-free conditions. In selected experiments, OM-85 (1 mg in 50 μL) was instilled intranasally 4 times into 8-week-old wild-type C57BL/6 mice (Charles Rivers Laboratories) or Myd88 −/− Trif −/− C57BL/6 mice (kindly provided by Dominik Schenten, University of Arizona). Lungs were collected and preserved in RNA later (Qiagen, Germantown, Md). All animal procedures conform to the principles set forth by the Animal Welfare Act and the National Institutes of Health guidelines for the care and use of laboratory animals in biomedical research and were approved by the University of Arizona Institutional Animal Care and Use Committee.
OM-85
OM-85 concentrate was provided by OM Pharma and is the soluble supernatant obtained after bacterial lysis. It represents the drug substance before its lyophilization and final manufacturing as Broncho-Vaxom. OM-85 lots number 1618006 (22.9 mg/mL of dry residue) and 1620074 (23.1 mg/mL of dry residue) were used in these experiments.
RNA sequencing from mouse lung tissue
Balb/c mice were treated with OM-85 or PBS as depicted in Fig 1, A. Unfractionated lung tissue was collected at day 39 and processed for RNA sequencing. Raw and normalized data for the complete data set were deposited in the GEO database (GSE167867), where detailed information on data processing and normalization can be found. Briefly, RNA-sequencing reads (>25 bp after trimming adapter sequences) were mapped to the BALB/cJ genome (version mm10) using STAR,E1 and genomic coordinates were shifted to the standard mm10 genome using MARGE (http://cistrome.org/MARGE/index. html). Uniquely aligning reads were used to generate gene counts relying on HOMER (http://homer.ucsd.edu/homer/index.html). Latent factors potentially introducing unwanted variation were removed using RUVSeq.E2 Data for Ace2 and Tmprss2 expression in OM-85– and PBS-treated mice were extracted from DESeq2-normalized counts,E3 and P values from differential expression analysis (DESeq2) were reported. P values less than .05 were considered significant.
RNA extraction, cDNA synthesis, and RT-quantitative PCR
RNA from cell lines or mouse lung tissue was extracted with the RNeasy kit (Qiagen). After extraction, total RNA (500 ng) was used as a template to synthesize cDNA with the QuantiTect Reverse Transcription Kit (Qiagen). RT-quantitative PCR was carried out using the QuantiTect SYBR Green PCR kit (Qiagen) on an ABI 7900 Applied Biosystems thermocycler (ThermoFisher Scientific). All genes except African green monkey (Chlorocebus sabaeus) TMPRSS2 were amplified using commercially available primers (QuantiTect Primer Assays, Qiagen). For green monkey TMPRSS2, the following primers were used: TGCATCAGCTCCTCTAACTG (forward) and GAGATGAGTACACCTGAAGG (reverse). Each sample was run in triplicate. The change in gene expression relative to PBS was normalized to glyceraldehyde 3-phosphate dehydrogenase and calculated using the 2−ΔΔct method.E4
Flow cytometry evaluation of ACE2 expression
Vero E6 and Calu-3 cells were grown in 12-well plates until confluent and treated with OM-85 (0.48 or 1.92 mg/mL) or PBS. After 72 hours, cells were harvested, and a single-cell suspension was prepared in FACSWash buffer (PBS with 1% BSA and 0.1% NaN3) at 0.5 to 1 × 107 cells/mL. To block nonspecific staining, cells were incubated on ice for 10 minutes in the same buffer containing normal mouse serum (5%) followed by a 30-minute incubation with either mouse anti-human ACE2-phycoerythrin (Sino Biological, clone 36,E5, E6 for Vero E6 cells) or mouse anti-human ACE2-AF647 (R&D Systems, Minneapolis, Minn: clone 535919,E7, E8, E9, E10, E11 for Calu-3 cells). We further validated the ACE2 specificity of these mAbs by showing that they detected more than 99% of ACE2-transfected HEK293T cells, but less than 1% of parental, untransfected HEK293T cells. A total of 20,000 to 30,000 events were acquired on a FACSCalibur flow cytometer (BD Biosciences) and analyzed using FlowJo software (Version 10.6.0, Becton Dickinson, 2019).
Statistical analyses
Statistical differences between treatment groups were assessed by an unpaired, 2-tailed t test or a Wilcoxon 2-sample test after assessing the normality of sample distribution using the Shapiro-Wilk test. P values less than .05 were considered statistically significant. Analyses were conducted in Stata (version 14.2, StataCorp LLC, College Station, Tex), R (version 3.5.0, R Core Team, Vienna, Austria), GraphPad Prism (version 9.1.1, GraphPad Software, San Diego, Calif), and Microsoft Excel (Microsoft Corporation, Redmond, Wash).
Table E1.
Effect of OM-85 on human ACE2 expression by ACE2/HEK293T cells
| PBS | OM-85 | |
|---|---|---|
| ACE2/HEK293T | 99.3 ± 0.3 | 99.4 ± 0.1 |
Cells were treated with PBS or OM-85 (1.92 mg/mL) for 72 hours, and human ACE2 expression was evaluated by flow cytometry with an AF647-conjugated antihuman ACE2 antibody or isotype control. Data are shown as mean percentages ± SE of ACE2-positive cells in PBS- or OM-85–treated cultures (n = 3/condition). Negligible proportions of positive cells were detected in the isotype control samples.
References
- 1.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;183:1735. doi: 10.1016/j.cell.2020.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kimura H., Francisco D., Conway M., Martinez F.D., Vercelli D., Polverino F., et al. Type 2 inflammation modulates ACE2 and TMPRSS2 in airway epithelial cells. J Allergy Clin Immunol. 2020;146:80–88.e8. doi: 10.1016/j.jaci.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sungnak W., Huang N., Becavin C., Berg M., Queen R., Litvinukova M., et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung K.S., Hung I.F.N., Chan P.P.Y., Lung K.C., Tso E., Liu R., et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020;159:81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puelles V.G., Lutgehetmann M., Lindenmeyer M.T., Sperhake J.P., Wong M.N., Allweiss L., et al. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krause P.R., Fleming T.R., Longini I.M., Peto R., Briand S., Heymann D.L., et al. SARS-CoV-2 variants and vaccines. N Engl J Med. 2021;385:179–186. doi: 10.1056/NEJMsr2105280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaad U.B., Mutterlein R., Goffin H., Group BV-Child Study Group Immunostimulation with OM-85 in children with recurrent infections of the upper respiratory tract: a double-blind, placebo-controlled multicenter study. Chest. 2002;122:2042–2049. doi: 10.1378/chest.122.6.2042. [DOI] [PubMed] [Google Scholar]
- 13.Gutierrez-Tarango M.D., Berber A. Safety and efficacy of two courses of OM-85 BV in the prevention of respiratory tract infections in children during 12 months. Chest. 2001;119:1742–1748. doi: 10.1378/chest.119.6.1742. [DOI] [PubMed] [Google Scholar]
- 14.Esposito S., Soto-Martinez M.E., Feleszko W., Jones M.H., Shen K.L., Schaad U.B. Nonspecific immunomodulators for recurrent respiratory tract infections, wheezing and asthma in children: a systematic review of mechanistic and clinical evidence. Curr Opin Allergy Clin Immunol. 2018;18:198–209. doi: 10.1097/ACI.0000000000000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasquali C., Salami O., Taneja M., Gollwitzer E.S., Trompette A., Pattaroni C., et al. Enhanced mucosal antibody production and protection against respiratory infections following an orally administered bacterial extract. Front Med (Lausanne) 2014;1:41. doi: 10.3389/fmed.2014.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roth M., Pasquali C., Stolz D., Tamm M. Broncho Vaxom (OM-85) modulates rhinovirus docking proteins on human airway epithelial cells via Erk1/2 mitogen activated protein kinase and cAMP. PLoS One. 2017;12 doi: 10.1371/journal.pone.0188010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pivniouk V, Gimenes JA, Ezeh P, Michael AN, Pivniouk O, Hahn S, et al. Airway administration of OM-85, a bacterial lysate, blocks experimental asthma by targeting dendritic cells and the epithelium/IL-33/ILC2 axis [Published online ahead of print September 22, 2021. J Allergy Clin Immunol. https://doi.org/10.1016/j.jaci.2021.09.013. [DOI] [PMC free article] [PubMed]
- 18.Kraft M., Adler K.B., Ingram J.L., Crews A.L., Atkinson T.P., Cairns C.B., et al. Mycoplasma pneumoniae induces airway epithelial cell expression of MUC5AC in asthma. Eur Respir J. 2008;31:43–46. doi: 10.1183/09031936.00103307. [DOI] [PubMed] [Google Scholar]
- 19.Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., et al. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12:513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sastry L., Johnson T., Hobson M.J., Smucker B., Cornetta K. Titering lentiviral vectors: comparison of DNA, RNA and marker expression methods. Gene Ther. 2002;9:1155–1162. doi: 10.1038/sj.gt.3301731. [DOI] [PubMed] [Google Scholar]
- 21.White S.M., Renda M., Nam N.Y., Klimatcheva E., Zhu Y., Fisk J., et al. Lentivirus vectors using human and simian immunodeficiency virus elements. J Virol. 1999;73:2832–2840. doi: 10.1128/jvi.73.4.2832-2840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansen M.D., Irving A., Montagutelli X., Tate M.D., Rudloff I., Nold M.F., et al. Animal and translational models of SARS-CoV-2 infection and COVID-19. Mucosal Immunol. 2020;13:877–891. doi: 10.1038/s41385-020-00340-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dittmar M., Lee J.S., Whig K., Segrist E., Li M., Kamalia B., et al. Drug repurposing screens reveal cell-type-specific entry pathways and FDA-approved drugs active against SARS-Cov-2. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.108959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Felgenhauer U., Schoen A., Gad H.H., Hartmann R., Schaubmar A.R., Failing K., et al. Inhibition of SARS-CoV-2 by type I and type III interferons. J Biol Chem. 2020;295:13958–13964. doi: 10.1074/jbc.AC120.013788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., III, et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larenas-Linnemann D., Rodriguez-Perez N., Arias-Cruz A., Blandon-Vijil M.V., Del Rio-Navarro B.E., Estrada-Cardona A., et al. Enhancing innate immunity against virus in times of COVID-19: trying to untangle facts from fictions. World Allergy Organ J. 2020;13 doi: 10.1016/j.waojou.2020.100476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suarez N., Ferrara F., Rial A., Dee V., Chabalgoity J.A. Bacterial lysates as immunotherapies for respiratory infections: methods of preparation. Front Bioeng Biotechnol. 2020;8:545. doi: 10.3389/fbioe.2020.00545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., Linsell L., et al. Dexamethasone in hospitalized patients with Covid-19. N Engl J Med. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramakrishnan S., Nicolau D.V., Jr., Langford B., Mahdi M., Jeffers H., Mwasuku C., et al. Inhaled budesonide in the treatment of early COVID-19 (STOIC): a phase 2, open-label, randomised controlled trial. Lancet Respir Med. 2021;9:763–772. doi: 10.1016/S2213-2600(21)00160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rubin E.J., Longo D.L., Baden L.R. Interleukin-6 receptor inhibition in Covid-19—cooling the inflammatory soup. N Engl J Med. 2021;384:1564–1565. doi: 10.1056/NEJMe2103108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pontali E., Volpi S., Signori A., Antonucci G., Castellaneta M., Buzzi D., et al. Efficacy of early anti-inflammatory treatment with high doses of intravenous anakinra with or without glucocorticoids in patients with severe COVID-19 pneumonia. J Allergy Clin Immunol. 2021;147:1217–1225. doi: 10.1016/j.jaci.2021.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guimaraes P.O., Quirk D., Furtado R.H., Maia L.N., Saraiva J.F., Antunes M.O., et al. Tofacitinib in patients hospitalized with Covid-19 pneumonia. N Engl J Med. 2021;385:406–415. doi: 10.1056/NEJMoa2101643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen M.S. Monoclonal antibodies to disrupt progression of early Covid-19 infection. N Engl J Med. 2021;384:289–291. doi: 10.1056/NEJMe2034495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dougan M., Nirula A., Azizad M., Mocherla B., Gottlieb R.L., Chen P., et al. Bamlanivimab plus etesevimab in mild or moderate Covid-19. N Engl J Med. 2021;385:1382–1392. doi: 10.1056/NEJMoa2102685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kyriakidis N.C., Lopez-Cortes A., Gonzalez E.V., Grimaldos A.B., Prado E.O. SARS-CoV-2 vaccines strategies: a comprehensive review of phase 3 candidates. NPJ Vaccines. 2021;6:28. doi: 10.1038/s41541-021-00292-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.COVID-19 Vaccine Resource Center. N Engl J Med. Available at: https://www-nejm-org.ezproxy2.library.arizona.edu/covid-vaccine. Accessed September 5, 2021.
- 38.Pardo J., Shukla A.M., Chamarthi G., Gupte A. The journey of remdesivir: from Ebola to COVID-19. Drugs Context. 2020;9 doi: 10.7573/dic.2020-4-14. 2020-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., et al. Remdesivir for the treatment of Covid-19—final report. N Engl J Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drayman N., DeMarco J.K., Jones K.A., Azizi S.A., Froggatt H.M., Tan K., et al. Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2. Science. 2021;373:931–936. doi: 10.1126/science.abg5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahoney M., Damalanka V.C., Tartell M.A., Chung D.H., Lourenco A.L., Pwee D., et al. A novel class of TMPRSS2 inhibitors potently block SARS-CoV-2 and MERS-CoV viral entry and protect human epithelial lung cells. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2108728118. e2108728118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A., et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18:41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 45.Cao C., Wang J., Li Y., Li Y., Ma L., Abdelrahim M.E.A., et al. Efficacy and safety of OM-85 in paediatric recurrent respiratory tract infections which could have a possible protective effect on COVID-19 pandemic: a meta-analysis. Int J Clin Pract. 2021;75 doi: 10.1111/ijcp.13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D., Ngai J., Speed T.P., Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol. 2014;32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Wang C., Wang S., Chen Y., Zhao J., Han S., Zhao G., et al. Membrane nanoparticles derived from ACE2-rich cells block SARS-CoV-2 infection. ACS Nano. 2021;15:6340–6351. doi: 10.1021/acsnano.0c06836. [DOI] [PubMed] [Google Scholar]
- Li J., Zou W., Yu K., Liu B., Liang W., Wang L., et al. Discovery of the natural product 3',4',7,8-tetrahydroxyflavone as a novel and potent selective BRD4 bromodomain 2 inhibitor. J Enzyme Inhib Med Chem. 2021;36:903–913. doi: 10.1080/14756366.2021.1906663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Wang A., Geng K., Honnen W., Wang X., Bruiners N., et al. Human immunodeficiency viruses pseudotyped with SARS-CoV-2 spike proteins infect a broad spectrum of human cell lines through multiple entry mechanisms. Viruses. 2021;13:953. doi: 10.3390/v13060953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neises J.Z., Hossain M.S., Sultana R., Wanniarachchi K.N., Wollman J.W., Nelson E., et al. Seroprevalence of SARS-CoV-2 antibodies among rural healthcare workers. J Med Virol. 2021;93:6611–6618. doi: 10.1002/jmv.27224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Liu J., Zhao S., Gomez Castro M.F., Laurent-Rolle M., Dong J., et al. SARS-CoV-2 exacerbates proinflammatory responses in myeloid cells through C-type lectin receptors and Tweety family member 2. Immunity. 2021;54:1304–1319.e9. doi: 10.1016/j.immuni.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Soh W.T., Kishikawa J.I., Hirose M., Nakayama E.E., Li S., et al. An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies. Cell. 2021;184:3452–3466.e18. doi: 10.1016/j.cell.2021.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]