Abstract
cGAS, a DNA sensor in mammalian cells, catalyzes the generation of 2’-3’-cyclic AMP-GMP (cGAMP) once activated by the binding of free DNA. cGAMP can bind to STING, activating downstream TBK1-IRF-3 signaling to initiate the expression of type I interferons. Although cGAS has been considered a traditional DNA-binding protein, several lines of evidence suggest that cGAS is a potential RNA-binding protein (RBP), which is mainly supported by its interactions with RNAs, RBP partners, RNA/cGAS-phase-separations as well as its structural similarity with the dsRNA recognition receptor 2’-5’ oligoadenylate synthase. Moreover, two influential studies reported that the cGAS-like receptors (cGLRs) of fly Drosophila melanogaster sense RNA and control 3′-2′-cGAMP signaling. In this review, we summarize and discuss in depth recent studies that identified or implied cGAS as an RBP. We also comprehensively summarized current experimental methods and computational tools that can identify or predict RNAs that bind to cGAS. Based on these discussions, we appeal that the RNA-binding activity of cGAS cannot be ignored in the cGAS-mediated innate antiviral response. It will be important to identify RNAs that can bind and regulate the activity of cGAS in cells with or without virus infection. Our review provides novel insight into the regulation of cGAS by its RNA-binding activity and extends beyond its DNA-binding activity. Our review would be significant for understanding the precise modulation of cGAS activity, providing the foundation for the future development of drugs against cGAS-triggering autoimmune diseases such as Aicardi-Gourtières syndrome.
Keywords: cGAS, RNA-binding activity, RNA-binding protein, phase-separated condensates, innate antiviral response
Introduction
As highlighted by the current COVID-19 pandemic, viral infection poses a great threat to human health and economics (1). The elimination of viruses by the host largely depends on the immune system, including innate immunity and adaptive immunity. Innate immunity is the first line of defense against viral infection due to its timely response and broad-spectrum effect (2). The innate antiviral response mainly refers to the process of the elimination of the virus, which involves NF-κB and interferon signaling (3). Numerous pattern recognition receptors (PRRs) can recognize pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs), triggering the expression of innate antiviral factors and inflammatory factors (3). Viral PAMPs mainly include viral nucleic acids such as DNA and RNA (4). Currently, the common intracellular DNA sensors include endosomal Toll-like receptor 9 (TLR9), cytosolic absent in melanoma 2 (AIM2), interferon gamma inducible protein 16 (IFI16), DNA-dependent protein kinase (DNA-PK), RNA polymerase III, and cyclic GMP-AMP synthase (cGAS) (5, 6). Recently, two novel DNA receptors, heterogeneous nuclear ribonucleoprotein A2B1 (7) and the transmembrane protein CCDC25 (8), have been identified. Among these DNA sensors, cGAS has been well characterized and widely studied (9). Once free DNA, including viral DNA and host DNA, is recognized, cGAS synthesizes the crucial second messenger cGAMP to bind and activate the downstream stimulator of interferon genes (STING). Consequently, TANK1 binding kinase 1 (TBK1)-interferon regulatory factor-3 (IRF-3) signaling is activated to initiate the expression of type I interferons (I-IFNs), establishing the innate antiviral response (10). The absence of cGAS leads to a serious deficiency of immune defense against a series of viruses (11). Indeed, cGAS-STING is a conserved and primordial defense pathway (10, 12, 13). cGAS also plays a key role in the regulation of autoimmune diseases (14, 15) and tumor immunity (16).
Although cGAS is traditionally recognized as a DNA-binding protein (17), numerous studies have suggested the RNA-binding activity of cGAS. cGAS also plays a redundant or essential role in mediating the defense against infection with RNA viruses, such as West Nile virus (WNV) (18), dengue virus (DENV) (19), and murine norovirus (MNV) (20). Moreover, numerous studies have implied the potential of RNA-binding activity of cGAS and its involvement in cGAS-mediated innate antiviral response, which were supported by the direct binding of RNA to cGAS, the RNA-binding protein (RBP) partners of cGAS, and the structural similarity of cGAS with dsRNA recognition receptor 2’-5’ oligoadenylate synthase (OAS) (21–25). In addition, cGAS undergoes a variety of post-translational modifications in the process of performing its function (26), which partially depend on RBP modifiers. Furthermore, considering the role of noncoding RNAs and RNA-related metabolic enzymes in the DNA virus-activated innate antiviral response supported by clinical studies (23, 27, 28), we cannot ignore the RNA-binding potential of cGAS in the initiation of the innate antiviral response in addition to the traditionally recognized DNA-binding activity.
Herein, we highlighted recent studies supporting or implying the RNA-binding activity of cGAS. Simultaneously, we discussed the subcellular location of cGAS based on the reported membrane, cytosol, and nucleus localization. Moreover, we summarized current experimental and computational methods that can identify or predict the RNA interactors of cGAS. We used bioinformatics tools to analyze the interaction propensity between mouse- or human-derived cGAS and RNA. This review provides novel insight into the noncanonical regulatory manner of cGAS activity in the innate antiviral response.
cGAS-STING Defense Pathway
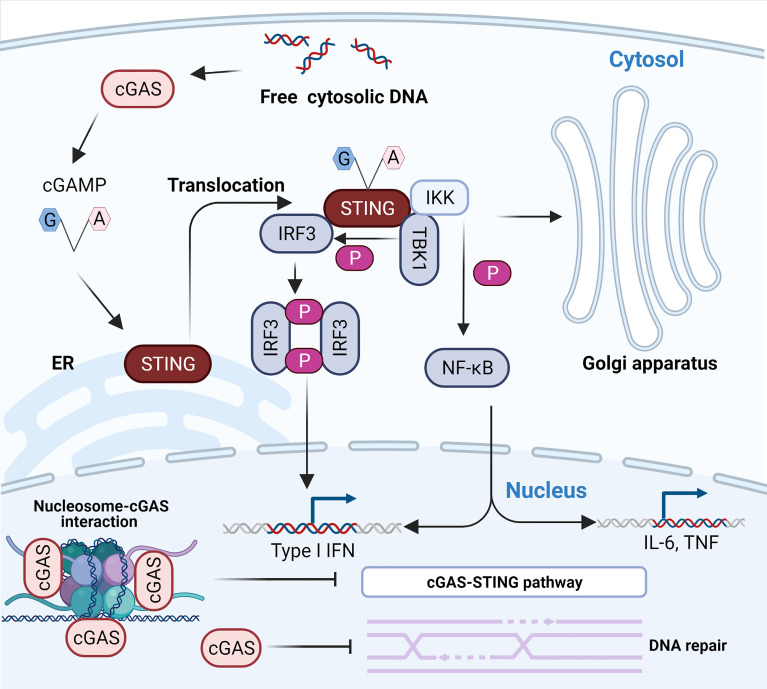
cGAS-STING signaling is the main pathway mediating immune defense against viruses containing DNA or producing DNA during their life cycle, such as DNA viruses and retroviruses (29, 30). Once it has recognized DNA, cGAS synthesizes 2’-3’-cGAMP using GTP and ATP as materials (10). 2’-3’-cGAMP acts as a second messenger to activate endoplasmic reticulum membrane (ER)-resident STING to form a dimer that is subsequently transported from the ER to the Golgi intermediate compartment and inter-Golgi (31) ( Figure 1 ). In this process, STING recruits and activates TBK1 via the carboxyl terminus to phosphorylate IRF-3 (32–34) ( Figure 1 ). Phosphorylated IRF-3 is dimerized and subsequently enters the nucleus to initiate the expression of IFN-β (35–37). STING also activates inhibitors of nuclear factor κB (IκB) kinase (IKK) that phosphorylate IκB, an inhibitor of NF-κB (32). The phosphorylated IκB protein is degraded by the ubiquitin–proteasome pathway, leading to the nuclear import of NF-κB (32, 38). Notably, IKKϵ allows for the redundant activation of NF-κB by STING (39). Nuclear transported NF-κB acts in concert with IRF-3 to drive the expression of IFN-β and inflammatory cytokines such as tumor necrosis factor (TNF), interleukin-1β (IL-1β), and interleukin-6 (IL-6) (32, 38) ( Figure 1 ). Binding of IFN-β to the type I IFN receptor (IFNAR), which is composed of subunits IFNAR1 and IFNAR2, induces the Janus kinase (JAK)-signal transcription and activator of transcription (STAT) signaling pathway in an autocrine and paracrine manner, leading to the downstream expression of hundreds of antiviral host effector proteins (ISGs) (40, 41). These proteins block the completeness of the virus life cycle and assist neighboring cells in establishing their resistance to infection by targeting viral proteins and nucleic acids (42). However, the binding of DNA to cGAS does not always activate cGAS because the activation of downstream immune signaling pathways requires the stabilization of activated cGAS-DNA complexes and the formation of cGAS dimers (43). For example, short-stranded DNA can bind to cGAS, but such binding induces weak dimerization of cGAS (44). For longer DNA, the clustering oligomerization and arrangement of DNA induced by cGAS forms stable active dimer structures (43) and facilitates the formation of large-scale liquid-liquid phase-separated condensates (45). In these condensates, cGAS is highly concentrated, stabilizing the active dimer state and promoting its catalytic activity (45). In general, dsDNA activates cGAS more efficiently than ssDNA (46), and cGAS is much more sensitive for longer DNA than shorter DNA (47, 48).
Figure 1.
The cytosolic DNA-activated cGAS-STING pathway. The recognition of free dsDNA in the cytoplasm by cGAS activates the production of 2’-3’-cGAMP, a natural ligand of ER-resident STING. The binding of 2’-3’-cGAMP to STING results in its translocation to the ER-Golgi intermediate compartment (ERGIC) and the Golgi apparatus. The relocated STING activates TBK1 and IKK. First, activated TBK1 phosphorylates STING, which consequently recruits and phosphorylates IRF3. The phosphorylated activated IRF3 dimerizes and enters the nucleus to initiate transcription of type I IFN. In addition, activated IKK results in the activation and nuclear transport of NF-κB to induce the expression of type I IFN and inflammatory cytokines such as TNF and IL-6. The interaction between cGAS and nucleosomes prevents the activation of cGAS. Notably, nuclear cGAS suppresses homologous recombination and promotes tumorigenesis.
Although cGAS has been well recognized in the process of defending against DNA viruses, many studies in recent years have shown that it also plays an instrumental role in the defense against RNA viruses (49). For instance, cGas−/− mice are more vulnerable to lethal WNV infection than wild-type mice (18). Consistently, another in vitro study showed that ablation of cGAS enhances MNV replication by regulating the transcription of ISGs (20). This may be because the lack of cGAS leads to decreased RIGI/MDA5 sensing of RNA (18). In addition, there is growing evidence that DNA leaking from mitochondria can engage cGAS in cells infected with RNA viruses, such as lymphocytic choriomeningitis virus (LCMV), dengue virus (DENV), encephalomyocarditis virus (EMCV), and influenza virus (19, 50–52). Specifically, influenza virus and EMCV stimulate mitochondrial DNA (mtDNA) release into the cytosol through their viroporin activity. Subsequently, cytosolic mtDNA promotes cGAS-STING-dependent innate antiviral immunity and confers resistance to RNA viruses (52). The resistance of cGAS to RNA viruses may not be due to viral RNA binding and activation of cGAS and most likely occurs in an indirect way, such as virus-induced leakage of mtDNA. Nevertheless, further research is urgently needed on the mechanism of action of cGAS against RNA virus infection.
The DNA Sensor cGAS: A Cytosolic, Nuclear, or Membrane Protein?
cGAS was first reported as a cytoplasmic protein that can prevent cGAS from interacting with deoxyribonucleic acid in the nucleus or mitochondria (53). However, copious studies have recently found that cGAS is also localized at the cell membrane (54) and nucleus (53, 55). In detail, Orzalli et al. (55) first observed nuclear accumulation of cGAS in human fibroblasts and keratinocytes transfected with plasmid DNA. Liu et al. found that cGAS can translocate into the nucleus in response to DNA damage in the immortalized human fibroblast HCA2-TERT cell line and two primary human skin fibroblast cell lines and localize at the site where DNA damage occurs (56). Subsequently, numerous studies have shown that cGAS is constitutively present in the nucleus and even predominantly distributed in the nucleus of immune cells (57–59). More specifically, endogenous cGAS is almost exclusively localized to the nucleus in HeLa cells, primary bone marrow-derived macrophages (BMMs) (57), bone marrow‐differentiating monocytes (BMDMos) (58), and primary human monocyte-derived dendritic cells (DCs) (60), except for exogenously expressed GFP‐tagged human cGAS (GFP‐hcGAS) in HEK 293T cells (58).
Although cGAS is abundant in the nucleus, several studies almost simultaneously found that cGAS does not interact with nucleosome DNA but interacts with histones 2A and 2B of chromosomes and tightly anchors them to the “acidic patch” (59, 61–65) ( Figure 1 ). This interaction masks one of the DNA-binding sites of cGAS and hinders the formation of active cGAS dimers, thereby preventing cGAS from interacting with its own genomic DNA (59, 61–65). Moreover, some extra regulatory mechanisms also limit cGAS activation, ensuring that cGAS is highly unresponsive to endogenous DNA (66, 67). For instance, barrier-to-autointegration factor 1 (BAF) dynamically inhibits the binding of cGAS to DNA and limits cGAS enzyme activity when cytoplasmic cGAS accumulates on chromatin around the nucleus during nuclear membrane rupture (66). Furthermore, the selective inhibition of cGAS activity during mitosis is attributed to two parallel mechanisms, including the hyperphosphorylation of cGAS at the N-terminus blocking the sensing of chromatin and inhibition of oligomerization of chromatin-bound cGAS (67). Intriguingly, Barnett et al. first pointed out that cGAS is not a cytoplasmic protein, as previously understood, but a membrane-localized protein that binds to plasma membrane lipids through its N-terminal domain (54). Membrane sequestration of the intracellular localization can keep cGAS away from endogenous DNA and help cGAS effectively detect viral DNA (54). Indeed, the N-terminal region of cGAS constitutes a major cytoplasmic retention signal that enables its detection and recognition in the cytosol (57) and fills an important role in regulating its activation (68). Different results regarding the localization of cGAS may be attributed to specific cells, advances in experimental techniques, and the specificity of cGAS antibodies (57). The future determination of cGAS localization requires specific cGAS antibodies that are screened by using Cgas -/- cells.
cGAS Could Bind RNA and Recognize DNA-RNA Hybrids
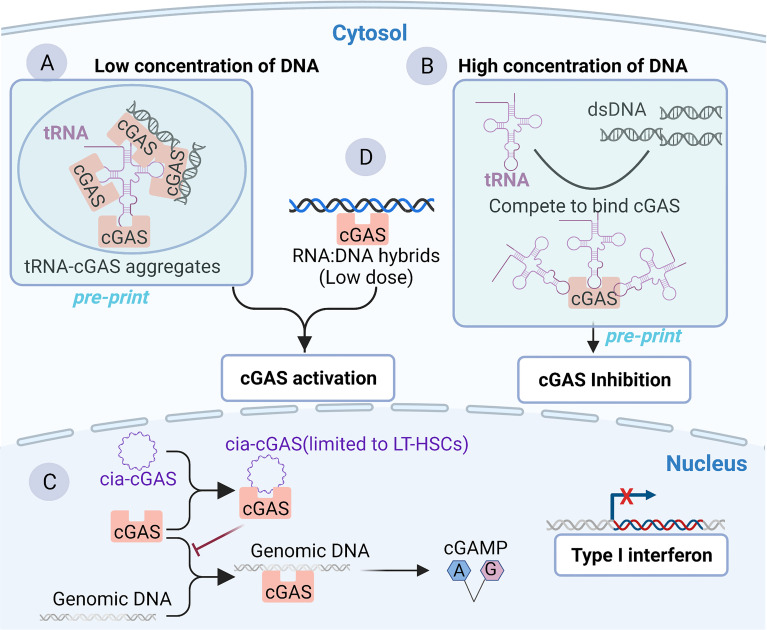
Although cGAS has been widely regarded as a classical DNA-binding protein, some recent studies have uncovered or implied RNA-binding activity of cGAS. Recent in vitro and in vivo research has shown that 2′-O-methyl (2′-OMe) gapmer antisense oligonucleotides (ASOs) exhibit sequence-dependent inhibition of sensing by the RNA sensor TLR7 (69) and two major DNA sensors, cGAS and TLR9 (70). The 2’-OMe-modified RNA motifs within synthetic oligonucleotides can inhibit the recognition of cGAS DNA in a sequence-dependent manner which is independent of cGAS mRNA targeting. The 2’-OMe-modified RNA motifs compete directly for DNA binding to cGAS. Interestingly, a few ASOs can enhance cGAS sensing when used at low concentrations. The binding of these ASOs to the third DNA binding domain within cGAS was speculated to facilitate the formation of the cGAS oligomer and thus increases its enzymatic activity (70, 71). These works suggest that RNA may compete for cGAS binding to dsDNA and inhibit cGAS activity, which has also been confirmed by a preprint study (25). The test tube assay showed that RNAs compete for binding to cGAS with dsDNA, promote the formation of phase separations, enhance cGAS activity when the dsDNA concentration is low ( Figure 2A ) and inhibit cGAS activity when the dsDNA concentration is high ( Figure 2B ) (25). Indeed, the concentration of RNA (both tRNA and mRNA) is much higher than that of DNA in the cytoplasm (72–74) under normal conditions, which is also established under the condition of virus infection (75, 76). This makes it reasonable that RNA binds to cGAS and regulates its activity in the cytosol. However, the preliminary nature of this work must be noted: this conclusion remains to be fully supported experimentally in cells because endogenous RNAs (such as tRNA) can modulate the sensing of DNA by cGAS through the modulation of phase separations. In the absence of dsDNA, human cGAS preferentially binds to intracellular tRNA to form phase-separated droplets in the cytoplasm via an unidentified mechanism, ensuring a nonactivated state of cGAS; when exposed to dsDNA, the tRNAs bound by cGAS in the phase-separated particles are gradually replaced by dsDNAs due to multivalent interactions between the DNA-binding domain within cGAS and DNA (25, 45).
Figure 2.
Binding of cGAS to RNA or RNA: DNA hybrids. (A, B) Binding of cGAS to tRNA as reported by a preprint server, which remains to be fully supported experimentally in cells. Cytoplasmic tRNA regulates cGAS activity by interfering with the formation of cGAS-containing aggregates. (A) In the context of low concentrations of DNA, cytoplasmic tRNA forms aggregates with cGAS, providing a platform for dsDNA-mediated cGAS activation. (B) In the context of a high concentration of DNA in the cytoplasm, DNA is sufficient to induce phase separation and activate cGAS. However, tRNA harbors a higher affinity than dsDNA for cGAS. Consequently, cGAS competes with dsDNA to bind cGAS and inhibit cGAS activity to avoid an excessive immune response. (C) Binding of cGAS to cia-cGAS. A circular RNA named cia-cGAS was highly expressed in the nucleus of LT-HSCs. Under homeostatic conditions, cia-cGAS binds cGAS in the nucleus to inhibit its binding to genomic DNA. As a consequence, cia-cGAS suppressed cGAS-mediated production of type I IFNs, thereby protecting dormant LT-HSCs from cGAS-mediated exhaustion. (D) RNA : DNA hybridization products can bind and activate cGAS.
Another study reported the RNA-binding activity of cGAS. Covalently closed single-stranded circular RNAs (circRNAs), a class of noncoding RNAs, are characterized by a covalent bond linking the 3’ and 5’ ends during the process of RNA splicing (77, 78). cia-cGAS, an exonic circular RNA containing a paired stem region, showed a strong affinity with the DNA binding domains within cGAS. In detail, in both human and murine long-term hematopoietic stem cells (LT-HSCs), cia-cGAS binds cGAS and inhibits the binding of self-DNA to cGAS, thereby reducing the production of I-IFNs to avoid the exhaustion of HSCs to maintain the resting state of HSCs ( Figure 2C ) (22). cia-cGAS is also expressed in several tissues. Importantly, the inhibitory effect of cia-cGAS on cGAS activity also manifested in tissues expressing cia-cGAS (22). Therefore, the cia-cGAS-mediated inhibition of cGAS activity may exist in cell types but is not limited to LT-HSCs (22). However, whether cia-cGAS can bind to cGAS under viral infection conditions and participate in the regulation of antiviral natural immune responses needs to be further investigated. Of note, it has been shown earlier that cGAS can also bind to dsRNA with an affinity comparable to that of binding dsDNA, while dsRNA does not activate cGAS (21). As mentioned earlier in the article, cGAS binds to DNA in a sequence-independent manner, which is mainly associated with the length and structures of DNA (21). However, for RNA, it is clear that short synthetic oligos can compete with DNA to bind cGAS in a sequence-dependent manner (70, 79), suggesting that it is the sequence rather than the length of the RNA that may be important for the DNA binding activity of cGAS. In summary, a series of RNAs, such as synthetic oligos (70), tRNA (25), and circRNA (22), which all belong to ssRNA, can bind to cGAS and regulate its activity, whereas dsRNA can bind cGAS but does not affect the activity of cGAS (21). These results indicate that ssRNA may be able to influence cGAS activity to a greater extent than dsRNA.
cGAS can also recognize synthetic RNA : DNA hybrids and activate downstream STING-TBK1-IRF-3 signaling to initiate the expression of I-IFNs in MAVS-knockout THP-1 cells ( Figure 2C ) (80). In vitro studies also showed that recombinant cGAS could produce cGAMP in the presence of RNA : DNA hybrids ( Figure 2D ) (80). Moreover, although synthetic heterodimers were used in the experiments, natural RNA : DNA heterodimers can be detected from the intracellular replication cycle of retroviruses and DNA viruses such as Herpesviridae (81, 82). Therefore, there may be aberrant recognition of endogenous nucleic acids by cGAS during the life cycle of several viruses. Further studies need to be carried out based on the improvement of experimental techniques such as purification and detection of RNA : DNA hybrids (82, 83). Notably, the dose of cytoplasmic RNA : DNA hybrids that can induce potent antiviral immune responses by binding and activating cGAS is 10-fold less than that of dsDNA (80), suggesting that RNA/DNA hybrids are potent activators of cGAS. Alternatively, there is a study implying that cGAS may bind to RNA. Briefly, following exogenous DNA and KSHV infection, cGAS binds to the ribonucleoprotein complex HDP-RNP (HEXIM1-DNA-PK-paraspeckle) containing lncRNA NEAT1, leading to the remodeling of HDP-RNP and subsequent release of the paraspeckle protein from this complex and the recruitment of STING (84). As a consequence, downstream TBK1-IRF-3 signaling is activated. This implied that lncRNA NEAT1 may bind to cGAS and serve a regulatory function in the cGAS-mediated innate antiviral response (84). However, it will be important to address in greater detail whether cGAS can interact with NEAT1 directly, as this study did not investigate the interaction between cGAS and NEAT1 (84). Furthermore, as mentioned above, the subcellular localization of cGAS may also prompt its interaction with RNA. The distribution of cGAS in the cytosol may be involved in RNA-related physiological functions in different membrane compartments, such as stress granules (SGs) (85), liquid-liquid phase, or separated aggregates (54). Given that RNAs are first produced in the nucleus and that an RNA-filling environment is also formed in the nucleus, there may be some nuclear RNAs that regulate the activity of cGAS in the nucleus and affect the immune response: of note is the example of cia-cGAS (22). Collectively, RNA may be a crucial medium regulating the activity of cGAS by competitively binding to dsDNA rather than directly activating cGAS, ensuring precise recognition of intracellular free dsDNA to avoid immune overload.
The RBP Interactors of cGAS
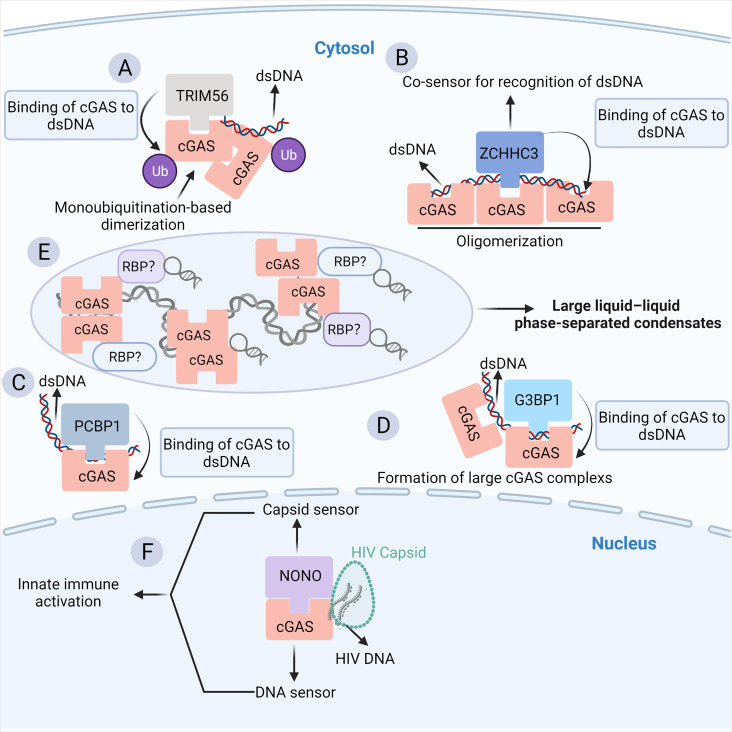
In addition to the cGAS-RNA interaction, convincing evidence indicates that the binding of some classical and emerging RBPs to cGAS prompts the pivotal role of the RNA-binding activity of cGAS in the innate antiviral response. In particular, cGAS is subjected to numerous post-translational modifications to maintain immune homeostasis (26). The post-translational modifications (PTMs) that cGAS undergoes include proteasomal breakdown, acetylation, glutamylation, ubiquitination, SUMOylation, and phosphorylation (26). Most of the factors involved are RBPs, implying a strong link between cGAS and RNA. Tripartite motif (TRIM) family proteins, as multifunctional ubiquitin E3 ligases, play a central role in host defense against viral infection, which is achieved partially by regulating the PTMs of cGAS (86). TRIM56, a recently identified RBP (87, 88), binds to ZIKV RNA in infected cells and acts as a restriction factor of ZIKV (89). The binding of TRIM56 to cGAS in the cytosol induces monoubiquitylation-based dimerization of cGAS, which is important for sensing dsDNA by cGAS (24) ( Figure 3A ). Human TRIM14 recruits USP14 to cleave K48-linked cGAS ubiquitination at the K414 site, disrupting cGAS-p62 interactions to stabilize cGAS (90, 91). In addition, other classical RBPs, such as zinc finger (ZnF) CCHC-type containing 3 (ZCCHC3) ( Figure 3B ) (92) and poly (C)-binding protein 1 (PCBP1) ( Figure 3C ) (93), can bind cGAS in the cytoplasm as a cofactor to enhance the binding of cGAS to DNA, enhancing downstream antiviral natural immune responses (92, 94, 95).
Figure 3.
The RBP partners of cGAS. (A) The binding of TRIM56 to cGAS induces the Lys335 monoubiquitylation of cGAS, thereby increasing the dimerization and DNA-binding activity of cGAS. (B) ZCCHC3 acts as a co-sensor for the recognition of dsDNA by cGAS. Briefly, ZCCHC3 binds to dsDNA and interacts with cGAS in the cytoplasm, enhancing the binding of cGAS to dsDNA and the formation of a large cGAS complex. (C) PCBP1 is a critical regulator of DNA recognition by cGAS. PCBP1 was recruited to cGAS in a viral infection-dependent manner. PCBP1 bound to DNA and enhanced cGAS binding to dsDNA. (D) G3BP1 physically interacts with and primes cGAS for efficient activation. G3BP1 enhanced the DNA binding of cGAS by promoting the formation of large cGAS complexes. (E) The binding of cytosolic dsDNA to cGAS induces a robust phase transition to liquid-like droplets, which are considered as microreactors with concentrated RNA and RBPs, suggesting a potential link between cGAS and RBPs. (F) NONO is an essential sensor of the HIV capsid in the nucleus. NONO forms a complex with cGAS in the nucleus. Detection of the nuclear viral capsid by NONO promotes the recognition of DNA by cGAS.
In particular, G3BP1, a core component of SGs, is a classical RBP (85). Additionally, the G3BP1-RNA interaction network is a key node in the formation of SG particles (96–98). G3BP1 can bind and promote the binding of cGAS to DNA, which in turn enhances downstream I-IFN signaling ( Figure 3D ) (99). The close relationship between G3BP1, SG, and cGAS implies that the abundant RNA and RBPs inside SG particles may be crucial regulators of cGAS activity ( Figure 3E ). Indeed, SGs are produced in response to stress, such as virus infection. In such a context, RNAs, a large number of RBPs, and other molecules are rapidly condensed into cytoplasmic SGs by phase separation (100). Moreover, it has been shown that cGAS occurs in this liquid-liquid phase separation structure (53), indicating that cGAS may interact with related components of SGs ( Figure 3E ). NONO is another multifunctional RBP implicated in transcription, splicing, DNA damage response, circadian rhythm, and neuronal development (101). NONO can recognize the HIV capsid and facilitate the binding of cGAS to HIV DNA, which is required for cGAS activation ( Figure 3F ) (101, 102). A recent study also showed that the disruption of the ribosome-associated protein quality control (RQC) pathway under stressful conditions such as viral infection, which detects and resolves ribosome collision during translation, results in cGAS-dependent ISG expression and causes relocalization of cGAS from the nucleus to the cytosol (103). Nevertheless, the RBP interactors of cGAS only suggest the potential RNA-binding activity of cGAS. In the future, it will be important to uncover the RNA interactors of cGAS in more detail by using RNA pull-down and RNA-immunoprecipitation experiments.
Structure-Based Analysis of the RNA-Binding Activity of cGAS
Typical RBPs are defined by the presence of RNA-binding domains (RBDs), such as hnRNP K homology domains, RNA recognition motifs, dsRNA-binding domains, and ZnF domains (104–106). However, many proteins lacking conventional RBDs or even DNA-binding proteins have been gradually recognized as factors with RNA-binding activity (23, 107–109). From the perspective of protein structure, cGAS consists of a disordered, positively charged N-terminal domain, a central nucleotidyltransferase (NTase) structural domain, a zinc-finger domain, and a C-terminal Mab-21 homologous structural domain (17, 110). The unique zinc-finger domain might be the decisive structural domain for its DNA binding activity. Interestingly, cGAS shares striking structural and functional similarities with the dsRNA recognition receptor 2′-5′-OAS (21, 111). In detail, both cGAS and 2′-5′-OAS are template-independent NTases. Moreover, cGAS and 2′-5′-OAS share overall folding structure, characterization of several active sites, the arrangement of blade 1 and blade 2, and even the 3D structural elements of the nucleic acid binding site (49, 112). Furthermore, the activation of both cGAS and 2′-5′-OAS is associated with the length of nucleic acids (111). From the perspective of function, cGAS and 2′-5′-OAS can be activated by similar double-stranded nucleic acid-inducible structural switches (21, 111). Once activated, cGAS and 2′-5′-OAS can generate a unique 2’-5’ phosphodiester-linked second messenger of nucleotides to initiate the relevant IFN-I immune pathway (21, 111). Based on evolutionary terms, the structures of cGAS are highly similar to those of 2′-5′-OAS (21, 46), especially the shared structure fold (21, 46), suggesting a common evolutionary ancestral origin as a structurally related but functionally distinct family of cytoplasmic nucleic acid sensors. Indeed, some scholars have merged cGAS and 2′-5′-OAS into a new family of catalytic OAS-like second messenger receptors (OLRs) (46). Notably, two recent studies have identified a class of cGAS-like receptors (cGLRs) in Drosophila that share a bi-lobed architecture, a caged nucleotidyltransferase core, a Gly-[Gly/Ser] activation loop, and a putative catalytic triad with cGAS (113). cGLRs recognize dsRNA and activate the production of the nucleotide product 3′-2′-cGAMP, and the signal is preferentially recognized by the Drosophila stimulator of interferon genes (dSTING), which exerts antiviral immunity in conjunction with the NF-κB pathway (114). It can be speculated that this conserved nucleic acid recognition structure is of great significance in the evolution of postnatal 2′-5′-OAS, cGAS, and cGLR animal proteins.
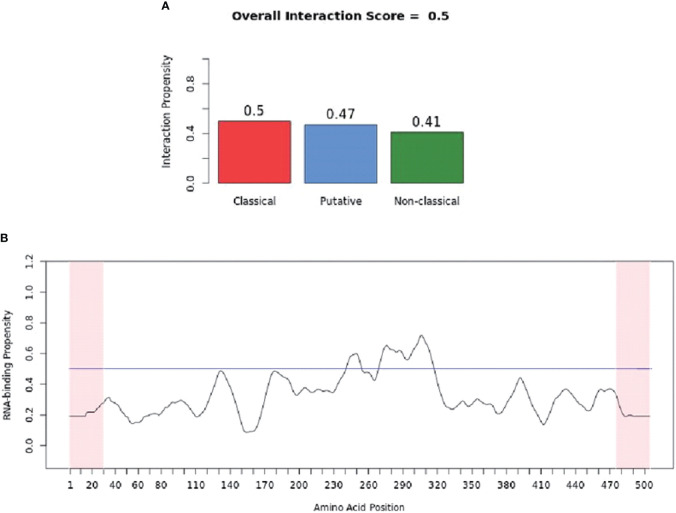
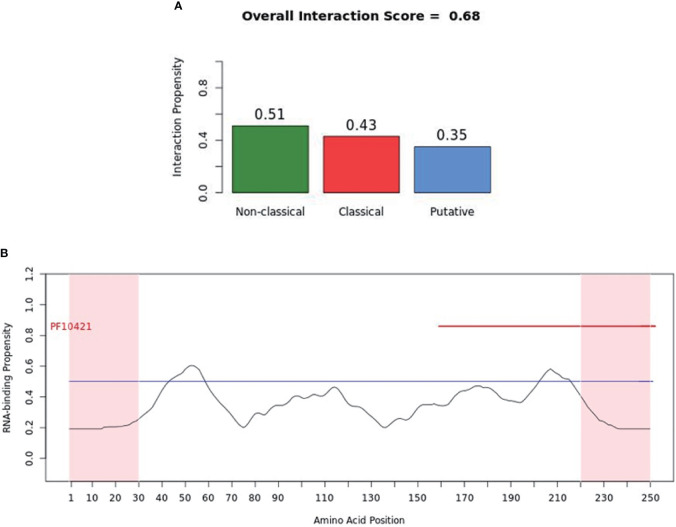
catRAPID (http://service.tartaglialab.com/page/catrapid_group) is an online algorithm predicting the propensity of protein-RNA interactions based on the respective contributions of secondary structure, hydrogen bonding, and Van Der Waals interactions (115). catRAPID has been widely used to study protein-RNA interactions with high accuracy (116–120). We first used the catRAPID signature (121) to predict the RNA-binding potential of murine cGAS. The overall prediction score of mcGAS was 0.5 ( Figure 4 ), reflecting potential RNA-binding activity of mcGAS. Further analysis indicated that the RNA-binding activity of cGAS was mainly attributed to amino acids 234-331 within the Mab-21 structural domain ( Figure 4 ). We also analyzed the RNA binding activity of 2’-5’-OAS1 as a well-characterized RBP using the catRAPID signature, and the results indicated a higher predicted RNA binding score of 0.68 compared to cGAS ( Figure 5 ). We next used catRAPID omics (123, 124) to discover the potential RNA interactors of murine cGAS ( Table 1 ) and human cGAS ( Table 2 ), which provides novel insight for future study of the RNA interactors of cGAS. However, the potential RNA interactors of cGAS should be determined by using relevant experimental methods. Nevertheless, these predicted results based on bioinformatic analysis further demonstrated the potential RNA-binding activity of cGAS.
Figure 4.
RNA-binding ability of murine-cGAS determined by catRAPID signature. (A) The propensity of murine cGAS for classical (0.5), putative (0.47), and nonclassical (0.41) RBPs. The overall interaction score (0.5) for murine cGAS as an RBP was also present. (B) The profile shows protein regions and their propensity to interact with RNA. All results were predicted by using catRAPID.
Figure 5.
RNA-binding ability of murine-2’-5’-OAS1 predicted by catRAPID signature. (A) The propensity of murine-2’-5’-OAS1 for the nonclassical (0.51), classical (0.43) and putative (0.35) RBPs. The overall interaction score (0.68) for murine-2’-5’-OAS1 as an RBP was also present. (B) The profile shows protein regions and their propensity to interact with RNA. The catRAPID signature correctly identifies the RNA binding domain of murine-2’-5’-OAS1, which carries the region of enzymatic activity between 320 and 344 at the extreme C-terminal end (122).
Table 1.
The RNA interactors of murine cGAS predicted by catRAPID omics (version 2.0).
| RNA ID | Gene Name | Interaction Propensity | Z-score | RBP Propensity | RNA-Binding Domains | RNA-Binding Motifs | Conserved Interactions | Ranking | |
|---|---|---|---|---|---|---|---|---|---|
| protein-coding RNAs | ENSMUST00000000080 | Kruppel-like Factor 6 (Klf6) | 88.83 | 1.73 | 0.5 | 1 | 0 | 0/0 | 0.405431 |
| ENSMUST00000000291 | max binding protein (Mnt) | 73.94 | 1.34 | 0.5 | 1 | 0 | 0/0 | 0.389061 | |
| ENSMUST00000000579 | sex determining region Y-box 9 (Sox9) | 56.82 | 0.89 | 0.5 | 1 | 0 | 0/0 | 0.37024 | |
| ENSMUST00000000619 | chloride channel, voltage-sensitive 4 (Clcn4) | 74.44 | 1.35 | 0.5 | 1 | 0 | 0/0 | 0.389611 | |
| ENSMUST00000000724 | K (lysine) acetyltransferase 2B (Kat2b) | 61.09 | 1 | 0.5 | 1 | 0 | 0/0 | 0.374934 | |
| long noncoding RNAs | ENSMUST00000047876 | Gm10710 | 45.58 | 0.59 | 0.5 | 1 | 0 | 0/0 | 0.357883 |
| ENSMUST00000052189 | B230317F23Rik | 49.89 | 0.7 | 0.5 | 1 | 0 | 0/0 | 0.362621 | |
| ENSMUST00000097612 | Gm10545 | 49.12 | 0.68 | 0.5 | 1 | 0 | 0/0 | 0.361774 | |
| ENSMUST00000098303 | Gm9934 | 42.98 | 0.52 | 0.5 | 1 | 0 | 0/0 | 0.355024 | |
| ENSMUST00000100713 | Gm10384 | 41.97 | 0.49 | 0.5 | 1 | 0 | 0/0 | 0.353914 | |
| small noncoding RNAs | ENSMUST00000082459 | Gm23627 | 25.98 | 0.07 | 0.49 | 1 | 0 | 0/0 | 0.333001 |
| ENSMUST00000082490 | Gm22749 | 28.99 | 0.15 | 0.49 | 1 | 0 | 0/0 | 0.33631 | |
| ENSMUST00000082508 | Gm26225 | 27.07 | 0.1 | 0.49 | 1 | 0 | 0/0 | 0.3342 | |
| ENSMUST00000082509 | Gm26226 | 36.2 | 0.34 | 0.49 | 1 | 0 | 0/0 | 0.344237 | |
| ENSMUST00000082534 | Gm25081 | 25.39 | 0.06 | 0.49 | 1 | 0 | 0/0 | 0.332353 |
Table summarization of the top 5 predicted coding-RNA interactors (RNA ID and Gene Name) of murine-cGAS by catRAPID omics (version 2.0). The table shows Z-scores (interaction propensity normalization relative to experimental cases), discriminative ability (relative to training sets), interaction strength (enrichment relative to random interactions), the presence of RNA-binding domains, and RNA motifs. RNAs were ranked by the score, which is the sum of three individual values: 1) catRAPID normalized propensity, 2) RBP propensity and 3) presence of known RNA-binding motifs. The full score is 1.
Table 2.
The RNA interactors of human cGAS predicted by catRAPID omics (version 2.0).
| RNA ID | Gene Name | Interaction Propensity | Z-score | RBP Propensity | RNA-Binding Domains | RNA-Binding Motifs | Conserved Interactions | Ranking | |
|---|---|---|---|---|---|---|---|---|---|
| protein-coding RNAs | ENST00000078429 | G protein subunit alpha 11 (GNA11) | 57.38 | 0.9 | 0.35 | 1 | 0 | 0/0 | 0.320855 |
| ENST00000169298 | ST6 beta-galactoside alpha-2,6-sialyltransferase 1 (ST6GAL1) | 56.82 | 0.89 | 0.35 | 1 | 0 | 0/0 | 0.32024 | |
| ENST00000170168 | RNA exonuclease 1 homolog (REXO1) | 59.47 | 0.96 | 0.35 | 1 | 0 | 0/0 | 0.323153 | |
| ENST00000174618 | MAX network transcriptional repressor (MNT) | 60.79 | 0.99 | 0.35 | 1 | 0 | 0/0 | 0.324604 | |
| ENST00000193322 | osteoclastogenesis associated transmembrane protein 1 (OSTM1) | 54.06 | 0.81 | 0.35 | 1 | 0 | 0/0 | 0.317205 | |
| long noncoding RNAs | ENST00000242109 | KIAA0087 | 24.68 | 0.04 | 0.35 | 1 | 0 | 0/0 | 0.284905 |
| ENST00000309874 | AC020659.1 | 19.83 | -0.09 | 0.35 | 1 | 0 | 0/0 | 0.279573 | |
| ENST00000316786 | C9orf106 | 21.42 | -0.05 | 0.35 | 1 | 0 | 0/0 | 0.281321 | |
| ENST00000325390 | AC018865.2 | 20.01 | -0.09 | 0.35 | 1 | 0 | 0/0 | 0.279771 | |
| ENST00000329618 | B3GALT5-AS1 | 20.99 | -0.06 | 0.35 | 1 | 0 | 0/0 | 0.280849 | |
| small noncoding RNAs | ENST00000347538 | MIR133A2 | 4.53 | -0.49 | 0.35 | 1 | 0 | 0/0 | 0.262753 |
| ENST00000362117 | MIR16-2 | 6.2 | -0.45 | 0.35 | 1 | 0 | 0/0 | 0.264589 | |
| ENST00000362317 | MIR365B | 7.6 | -0.41 | 0.35 | 1 | 0 | 0/0 | 0.266128 | |
| ENST00000362349 | RNU6-500P | 6.08 | -0.45 | 0.35 | 1 | 0 | 0/0 | 0.264457 | |
| ENST00000362356 | RNU6-50P | 5.19 | -0.48 | 0.35 | 1 | 0 | 0/0 | 0.263478 |
Table summarization of the top 5 predicted coding-RNA interactors (RNA ID and Gene Name) of murine-cGAS by catRAPID omics (version 2.0). The table shows Z-scores (interaction propensity normalization relative to experimental cases), discriminative ability (relative to training sets), interaction strength (enrichment relative to random interactions), the presence of RNA-binding domains, and RNA motifs. RNAs were ranked by the score, which is the sum of three individual values: 1) catRAPID normalized propensity, 2) RBP propensity and 3) presence of known RNA-binding motifs. The full score is 1.
Computational and Experimental Methods for Detecting or Predicting cGAS-RNA Interactions
Numerous experimental methods and computational methods have been devised to reveal RNA-protein interactions (125, 126). We comprehensively summarized the current experimental methods ( Table 3 ) and computational tools ( Table 4 ) for studying RNA-protein interactions as well as their respective advantages and disadvantages. These methods would be beneficial for us to identify RNAs that bind to cGAS and reveal the RNA-binding activity of cGAS and its regulatory function in the antiviral immune response. The experimental methods can be further classified according to cells and test tubes in terms of experimental conditions. The latter mainly include electrophoretic mobility shift assay (EMSA), RNA pull-down, systematic evolution of ligands by exponential enrichment (SELEX), RNAcompete, and RBNS ( Table 3 ). The in cells assays mainly include RNA binding protein immunoprecipitation (RIP) and crosslinking immunoprecipitation (CLIP) assays ( Table 3 ). In fact, the concentrations of RBPs and RNAs used in test tubes are much higher than those levels in a physiological context (144). Furthermore, the artificially created in vitro environment is pure and susceptible to technical influence (145). Therefore, the results of the test tube assays cannot represent the true physiological mechanisms of interactions in vivo.
Table 3.
Experimental methods for uncovering the interaction between proteins and RNAs.
| Classes | Methods | Advantages | Disadvantages | References |
|---|---|---|---|---|
| In the test tube | EMSA | Separation of numerous types of complexes, such as monomer and dimer; Works well with crude cell extracts. | Low-throughput; Failure of detecting binding sites. | (125, 127) |
| RNA pull-down | A simple protocol; Enrichment of low-abundance RBPs. | Failure of confirming direct or indirect interaction; Failure of forming RNA-protein interactions that only occur in vitro under non-physiological conditions. | (125, 128) | |
| SELEX | Direct interaction of the oligonucleotides with the target is closely application-oriented; Independence of in-depth knowledge regarding the respective target for aptamer selection. | No standardized aptamer selection protocol for target. | (129–132) | |
| RNAcompete | Accurate estimation of the relative preference for a large numbers of individual sequences; Querying preferences are available for structured RNA; Time-saving. | Limited size of the current RNA pool for the represented combinations of sequence and structure. | (125, 133) | |
| RBNS | Accurate estimation of dissociation constants of RBP-RNA complex; A more reliable prediction of RNA folding and a better identification of the binding structural determinants than RNAcompete. | N/A | (125, 134, 135) | |
| In cells | RIP Assay | A simple and standard protocol; Preserves the intracellular native complexes. | Failure of confirmation of direct or indirect interaction between RNA and protein; Failure of determination of the precise site within the RNA interacting protein; Poor resolution; Antibody-consuming. | (125, 136) |
| CLIP | A sensitive determination of binding sites. | Low abundance of RNA-ribonucleoprotein complexes; Potentially inefficient library preparation; Large amounts of raw material required. | (108, 125, 137) | |
| PIP-seq | A simultaneous view of the global landscapes of both RNA secondary structure and RNA-protein interactions. | High concentrations of structure-specific RNases. | (138) |
EMSA, Electrophoretic Mobility Shift Assay; SELEX, Systematic Evolution of Ligands by Exponential Enrichment; RBNS; RNA Bind-n-Seq; RIP, RNA Immunoprecipitation; CLIP, Cross-linked Immunoprecipitation; PIP-seq, Protein Interaction Profile Sequencing; N/A, not applicable.
Table 4.
Computational tools for the prediction of RNA-protein interactions.
| Methods | Advantages | Disadvantages | Links | References |
|---|---|---|---|---|
| catRAPID | Outperforms other algorithms in the identification of RBPs and detection of non-classical RNA-binding regions. | Spurious binding may occur. | http://s.tartaglialab.com/page/catrapid_group | (115, 121) |
| lncPro | Time-saving. | Limited ability of computational prediction of RNA secondary structure for lncRNAs. | bioinfo.bjmu.edu.cn/lncpro/ | (139) |
| RNAcontext | A more accurate elucidation of RBP-specific sequence and structural preferences. | Misleading results will be produced for those RBPs with non-trivial structural preferences. | http://www.cs.toronto.edu/~hilal/rnacontext/ | (140) |
| RPI-Pred | A comprehensive understanding of PRIs based on the sequence features and the high-order structures of both proteins and RNAs. | Limited to prediction of ncRNA-Protein interaction. | http://ctsb.is.wfubmc.edu/projects/rpi-pred. | (141) |
| PRIPU | Employs SVM for predicting PRIs using only positive and unlabeled examples based on proposing a new performance measure called EPR; Outperforms existing methods and predicts unknown PRIs. | N/A | http://admis.fudan.edu.cn/projects/pripu.htm | (142) |
| RPISeq | Reliable prediction based on sequence. | Requirement for larger and more diverse experimental datasets. | RNA-Protein Interaction Prediction (RPISeq) (iastate.edu) | (143) |
RBPs, RNA-binding proteins; SVM, biased-support vector machine; lncRNA, long noncoding RNA; PRIS, Protein-RNA interactions; EPR, explicit positive recall; N/A, not applicable.
Although experimental methods could provide convincing evidence supporting the RNA-protein interaction, most of the experimental methods are technically challenging and time-consuming ( Table 3 ). In contrast, the computational approaches are simple, fast, and time-saving and can be used as ideal amendments to the experimental methodology (125, 146–149) ( Table 4 ). The growing numbers of public experimental data also offer the possibility to train computational models for inferring new interactions. Most of these computational methods for predicting RNA-protein interactions (such as catRAPID, lncPro, and RNAcontext) calculate their propensity based on the physicochemical properties of peptides and nucleotide chains (e.g., propensity for hydrogen bonding, van der Waals interactions, and secondary structure) (125) ( Table 4 ). On the basis of these principles, there are also methods such as RPI-Pred and PRIPU that further introduced statistical and machine learning algorithms, including vector machines, in the prediction of RNA-protein interactions (141, 142) ( Table 4 ).
Conclusions and Future Perspectives
In this review, we highlighted the RNA-binding activity of cGAS based on the RNA interactors of cGAS, the numerous RBP partners of cGAS, and its structural similarity with 2′-5′-OAS. We also elucidated and discussed current experimental methods and computational tools that can be used for exploring and calculating cGAS-RNA interactions. These clues indicated that the RNA-binding activity of cGAS may play an important role in the innate antiviral response, which needs to be further confirmed in the future. Although cGAS showed potential RNA-binding activity, it should be noted that the RNA binding activity of cGAS is different from that of known RNA sensors, such as RIG-1 and TLR3, which mainly recognize viral RNA (23). In contrast, known dsRNA interactors cannot activate cGAS (21). Furthermore, given that the known interacting RNAs of cGAS are not derived from a virus (21, 22) and that cGAS plays a redundant role in recognizing RNA viruses (49, 150), the RNA-binding activity of cGAS may largely affect the DNA-binding activity of cGAS but does not mean that cGAS can recognize viral RNA or RNA interactors which directly activate cGAS. In fact, since the discovery of cGAS as a novel receptor for dsDNA, researchers have used a variety of physical and biochemical methods to explore the mechanism of action of dsDNA recognition by cGAS. However, numerous reports have also begun to prove the RNA-binding activity of cGAS. Of note, no studies have proven that binding RNA could act as a PAMP to directly activate cGAS. The RNA-binding ability of cGAS in the innate antiviral response represents a scientific question that urgently needs to be resolved. In summary, the RNA interactor of cGAS could largely modulate cGAS function rather than activate it. Nevertheless, we cannot ignore the role of the RNA-binding activity of cGAS in the innate antiviral response, although cGAS is recognized as a classical DNA-binding protein. Moreover, cGAS can be activated by self-DNA, which triggers the development of autoimmune diseases such as Aicardi-Gourtières syndrome (151). Therefore, it is important to address the RNA-binding activity of cGAS in the innate antiviral response in more detail to understand the precise modulation of cGAS activity. Our review provides a comprehensive understanding of the regulation of cGAS activity, which extends beyond the DNA-binding activity of cGAS. Our review is significant for understanding cGAS signaling.
Author Contributions
YM, XW, HS, YilW, WS, JX, and WL: conception and design, collection and/or assembly of references, data analysis and interpretation, and manuscript writing; HS, ZR, YifW, and YilW: final interpretation and helpful discussion of the manuscript. All authors read and approved the final manuscript and agreed to this publication. YM and XW contributed equally to this article.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81872908 and 82072274), Key Laboratory of Virology of Guangdong Province, Guangdong Provincial Natural Science Foundation Project (No. 2019A1515010046), Study on the mechanism of estrogen promoting the repair of urethral function in the treatment of female stress urinary incontinence with hUCMSCs (No. 2021A1515011221), and Guangdong Modern Agricultural Industrial Technology System Innovation Team (No. 2017YFC1701100).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Luo W, Huang L, Wang X, Ma Y, Xiao J, Song X, et al. SARS-CoV-2 Infection Activates a Subset of Intrinsic Pathways to Inhibit Type I Interferons In Vitro and In Vivo. Int J Med Sci (2021) 18(12):2561. doi: 10.7150/ijms.56630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ike AC, Onu CJ, Ononugbo CM, Reward EE, Muo SO. Immune Response to Herpes Simplex Virus Infection and Vaccine Development. Vaccines (Basel) (2020) 8(2):302. doi: 10.3390/vaccines8020302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ivashkiv LB, Donlin LT. Regulation of Type I Interferon Responses. Nat Rev Immunol (2014) 14(1):36–49. doi: 10.1038/nri3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Unterholzner L, Bowie AG. The Interplay Between Viruses and Innate Immune Signaling: Recent Insights and Therapeutic Opportunities. Biochem Pharmacol (2008) 75(3):589–602. doi: 10.1016/j.bcp.2007.07.043 [DOI] [PubMed] [Google Scholar]
- 5. Zhao J, Qin C, Liu Y, Rao Y, Feng P. Herpes Simplex Virus and Pattern Recognition Receptors: An Arms Race. Front Immunol (2020) 11:613799. doi: 10.3389/fimmu.2020.613799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hu MM, Shu HB. Innate Immune Response to Cytoplasmic DNA: Mechanisms and Diseases. Annu Rev Immunol (2020) 38:79–98. doi: 10.1146/annurev-immunol-070119-115052 [DOI] [PubMed] [Google Scholar]
- 7. Wang L, Wen M, Cao X. Nuclear Hnrnpa2b1 Initiates and Amplifies the Innate Immune Response to DNA Viruses. Science (2019) 365(6454):656–+. doi: 10.1126/science.aav0758 [DOI] [PubMed] [Google Scholar]
- 8. Yang L, Liu Q, Zhang X, Liu X, Zhou B, Chen J, et al. DNA of Neutrophil Extracellular Traps Promotes Cancer Metastasis via CCDC25. Nature (2020) 583(7814):133–8. doi: 10.1038/s41586-020-2394-6 [DOI] [PubMed] [Google Scholar]
- 9. Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal Roles of cGAS-cGAMP Signaling in Antiviral Defense and Immune Adjuvant Effects. Science (2013) 341(6152):1390–4. doi: 10.1126/science.1244040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ma Z, Damania B. The cGAS-STING Defense Pathway and Its Counteraction by Viruses. Cell Host Microbe (2016) 19(2):150–8. doi: 10.1016/j.chom.2016.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, et al. Pan-Viral Specificity of IFN-Induced Genes Reveals New Roles for cGAS in Innate Immunity. Nature (2015) 525(7567):144. doi: 10.1038/nature14555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gui X, Yang H, Li T, Tan X, Shi P, Li M, et al. Autophagy Induction via STING Trafficking Is a Primordial Function of the cGAS Pathway. Nature (2019) 567(7747):262–6. doi: 10.1038/s41586-019-1006-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morehouse BR, Govande AA, Millman A, Keszei AFA, Lowey B, Ofir G, et al. STING Cyclic Dinucleotide Sensing Originated in Bacteria. Nature (2020) 586(7829):429–33. doi: 10.1038/s41586-020-2719-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gao D, Li T, Li XD, Chen X, Li QZ, Wight-Carter M, et al. Activation of Cyclic GMP-AMP Synthase by Self-DNA Causes Autoimmune Diseases. Proc Natl Acad Sci U S A (2015) 112(42):E5699–705. doi: 10.1073/pnas.1516465112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. An J, Durcan L, Karr RM, Briggs TA, Rice GI, Teal TH, et al. Expression of Cyclic GMP-AMP Synthase in Patients With Systemic Lupus Erythematosus. Arthritis Rheumatol (2017) 69(4):800–7. doi: 10.1002/art.40002 [DOI] [PubMed] [Google Scholar]
- 16. Chen Q, Sun L, Chen ZJ. Regulation and Function of the cGAS-STING Pathway of Cytosolic DNA Sensing. Nat Immunol (2016) 17(10):1142–9. doi: 10.1038/ni.3558 [DOI] [PubMed] [Google Scholar]
- 17. Sun LJ, Wu JX, Du FH, Chen X, Chen ZJJ. Cyclic GMP-AMP Synthase Is a Cytosolic DNA Sensor That Activates the Type I Interferon Pathway. Science (2013) 339(6121):786–91. doi: 10.1126/science.1232458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, et al. Pan-Viral Specificity of IFN-Induced Genes Reveals New Roles for cGAS in Innate Immunity. Nature (2014) 505(7485):691–+. doi: 10.1038/nature12862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aguirre S, Luthra P, Sanchez-Aparicio MT, Maestre AM, Patel J, Lamothe F, et al. Dengue Virus NS2B Protein Targets cGAS for Degradation and Prevents Mitochondrial DNA Sensing During Infection. Nat Microbiol (2017) 2(5):17037. doi: 10.1038/nmicrobiol.2017.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu P, Miao Z, Li Y, Bansal R, Peppelenbosch MP, Pan Q. cGAS-STING Effectively Restricts Murine Norovirus Infection But Antagonizes the Antiviral Action of N-Terminus of RIG-I in Mouse Macrophages. Gut Microbes (2021) 13(1):1959839. doi: 10.1080/19490976.2021.1959839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Civril F, Deimling T, Mann CCD, Ablasser A, Moldt M, Witte G, et al. Structural Mechanism of Cytosolic DNA Sensing by cGAS. Nature (2013) 498(7454):332–+. doi: 10.1038/nature12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xia P, Wang S, Ye B, Du Y, Li C, Xiong Z, et al. A Circular RNA Protects Dormant Hematopoietic Stem Cells From DNA Sensor cGAS-Mediated Exhaustion. Immunity (2018) 48(4):688–701.e7. doi: 10.1016/j.immuni.2018.03.016 [DOI] [PubMed] [Google Scholar]
- 23. Wang Y, Wang Y, Luo W, Song X, Huang L, Xiao J, et al. Roles of Long Non-Coding RNAs and Emerging RNA-Binding Proteins in Innate Antiviral Responses. Theranostics (2020) 10(20):9407–24. doi: 10.7150/thno.48520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seo GJ, Kim C, Shin WJ, Sklan EH, Eoh H, Jung JU. TRIM56-Mediated Monoubiquitination of cGAS for Cytosolic DNA Sensing. Nat Commun (2018) 9(1):613. doi: 10.1038/s41467-018-02936-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen S, Rong M, Lv Y, Zhu D, Xiang Y. Regulation of cGAS Activity Through RNA-Mediated Phase Separation. bioRxiv (2020). doi: 10.1101/2020.09.27.316166 [DOI] [Google Scholar]
- 26. Baker PJ, De Nardo D, Moghaddas F, Tran LS, Bachem A, Nguyen T, et al. Posttranslational Modification as a Critical Determinant of Cytoplasmic Innate Immune Recognition. Physiol Rev (2017) 97(3):1165–209. doi: 10.1152/physrev.00026.2016 [DOI] [PubMed] [Google Scholar]
- 27. Zhang SY, Clark NE, Freije CA, Pauwels E, Taggart AJ, Okada S, et al. Inborn Errors of RNA Lariat Metabolism in Humans With Brainstem Viral Infection. Cell (2018) 172(5):952–65.e18. doi: 10.1016/j.cell.2018.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lafaille FG, Harschnitz O, Lee YS, Zhang P, Hasek ML, Kerner G, et al. Human SNORA31 Variations Impair Cortical Neuron-Intrinsic Immunity to HSV-1 and Underlie Herpes Simplex Encephalitis. Nat Med (2019) 25(12):1873–84. doi: 10.1038/s41591-019-0672-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Abe T, Marutani Y, Shoji I. Cytosolic DNA-Sensing Immune Response and Viral Infection. Microbiol Immunol (2019) 63(2):51–64. doi: 10.1111/1348-0421.12669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu F, Zhao S, Yu B, Chen Y, Wang W, Song Z-G, et al. A New Coronavirus Associated With Human Respiratory Disease in China. Nature (2020) 579(7798):1–8. doi: 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Motwani M, Pesiridis S, Fitzgerald KA. DNA Sensing by the cGAS-STING Pathway in Health and Disease. Nat Rev Genet (2019) 20(11):657–74. doi: 10.1038/s41576-019-0151-1 [DOI] [PubMed] [Google Scholar]
- 32. Ishikawa H, Barber GN. STING Is an Endoplasmic Reticulum Adaptor That Facilitates Innate Immune Signalling. Nature (2008) 455(7213):674–8. doi: 10.1038/nature07317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ishikawa H, Ma Z, Barber GN. STING Regulates Intracellular DNA-Mediated, Type I Interferon-Dependent Innate Immunity. Nature (2009) 461(7265):788–92. doi: 10.1038/nature08476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, et al. Phosphorylation of Innate Immune Adaptor Proteins MAVS, STING, and TRIF Induces IRF3 Activation. Science (2015) 347(6227):aaa2630. doi: 10.1126/science.aaa2630 [DOI] [PubMed] [Google Scholar]
- 35. Sato M, Tanaka N, Hata N, Oda E, Taniguchi T. Involvement of the IRF Family Transcription Factor IRF-3 in Virus-Induced Activation of the IFN-Beta Gene. FEBS Lett (1998) 425(1):112–6. doi: 10.1016/S0014-5793(98)00210-5 [DOI] [PubMed] [Google Scholar]
- 36. Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. Direct Triggering of the Type I Interferon System by Virus Infection: Activation of a Transcription Factor Complex Containing IRF-3 and CBP/P300. EMBO J (1998) 17(4):1087–95. doi: 10.1093/emboj/17.4.1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schwanke H, Stempel M, Brinkmann MM. Of Keeping and Tipping the Balance: Host Regulation and Viral Modulation of IRF3-Dependent IFNB1 Expression. Viruses (2020) 12(7):733. doi: 10.3390/v12070733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fang R, Wang C, Jiang Q, Lv M, Gao P, Yu X, et al. NEMO-IKKbeta Are Essential for IRF3 and NF-kappaB Activation in the cGAS-STING Pathway. J Immunol (2017) 199(9):3222–33. doi: 10.4049/jimmunol.1700699 [DOI] [PubMed] [Google Scholar]
- 39. Balka KR, Louis C, Saunders TL, Smith AM, Calleja DJ, D’Silva DB, et al. TBK1 and IKKepsilon Act Redundantly to Mediate STING-Induced NF-kappaB Responses in Myeloid Cells. Cell Rep (2020) 31(1):107492. doi: 10.1016/j.celrep.2020.03.056 [DOI] [PubMed] [Google Scholar]
- 40. Wang Y, Jia J, Wang Y, Li F, Song X, Qin S, et al. Roles of HSV-1 Infection-Induced Microglial Immune Responses in CNS Diseases: Friends or Foes? Crit Rev Microbiol (2019) 45(5-6):581–94. doi: 10.1080/1040841X.2019.1660615 [DOI] [PubMed] [Google Scholar]
- 41. Taniguchi T, Takaoka A. The Interferon-Alpha/Beta System in Antiviral Responses: A Multimodal Machinery of Gene Regulation by the IRF Family of Transcription Factors. Curr Opin Immunol (2002) 14(1):111–6. doi: 10.1016/S0952-7915(01)00305-3 [DOI] [PubMed] [Google Scholar]
- 42. Iwasaki A. A Virological View of Innate Immune Recognition. Annu Rev Microbiol (2012) 66:177–96. doi: 10.1146/annurev-micro-092611-150203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Andreeva L, Hiller B, Kostrewa D, Lässig C, de Oliveira Mann CC, Jan Drexler D, et al. cGAS Senses Long and HMGB/TFAM-Bound U-Turn DNA by Forming Protein-DNA Ladders. Nature (2017) 549(7672):394–8. doi: 10.1038/nature23890 [DOI] [PubMed] [Google Scholar]
- 44. Herzner AM, Hagmann CA, Goldeck M, Wolter S, Kübler K, Wittmann S, et al. Sequence-Specific Activation of the DNA Sensor cGAS by Y-Form DNA Structures as Found in Primary HIV-1 cDNA. Nat Immunol (2015) 16(10):1025–33. doi: 10.1038/ni.3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Du M, Chen ZJ. DNA-Induced Liquid Phase Condensation of cGAS Activates Innate Immune Signaling. Science (2018) 361(6403):704–9. doi: 10.1126/science.aat1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kranzusch PJ, Lee ASY, Berger JM, Doudna JA. Structure of Human cGAS Reveals a Conserved Family of Second-Messenger Enzymes in Innate Immunity. Cell Rep (2013) 3(5):1362–8. doi: 10.1016/j.celrep.2013.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Luecke S, Holleufer A, Christensen MH, Jonsson KL, Boni GA, Sorensen LK, et al. cGAS Is Activated by DNA in a Length-Dependent Manner. EMBO Rep (2017) 18(10):1707–15. doi: 10.15252/embr.201744017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gantier MP. Length Does Matter for cGAS. EMBO Rep (2017) 18(10):1675–6. doi: 10.15252/embr.201744773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ni G, Ma Z, Damania B. cGAS and STING: At the Intersection of DNA and RNA Virus-Sensing Networks. PLoS Pathog (2018) 14(8):e1007148. doi: 10.1371/journal.ppat.1007148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, et al. Mitochondrial DNA Stress Primes the Antiviral Innate Immune Response. Nature (2015) 520(7548):553. doi: 10.1038/nature14156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang H, Hu S, Chen X, Shi H, Chen C, Sun L, et al. cGAS Is Essential for the Antitumor Effect of Immune Checkpoint Blockade. Proc Natl Acad Sci (2017) 114(7):1637–42. doi: 10.1073/pnas.1621363114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Moriyama M, Koshiba T, Ichinohe T. Influenza A Virus M2 Protein Triggers Mitochondrial DNA-Mediated Antiviral Immune Responses. Nat Commun (2019) 10:4624. doi: 10.1038/s41467-019-12632-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hopfner KP, Hornung V. Molecular Mechanisms and Cellular Functions of cGAS-STING Signalling. Nat Rev Mol Cell Biol (2020) 21(9):501–21. doi: 10.1038/s41580-020-0244-x [DOI] [PubMed] [Google Scholar]
- 54. Barnett KC, Coronas-Serna JM, Zhou W, Ernandes MJ, Cao A, Kranzusch PJ, et al. Phosphoinositide Interactions Position cGAS at the Plasma Membrane to Ensure Efficient Distinction Between Self- and Viral DNA. Cell (2019) 176(6):1432–46.e11. doi: 10.1016/j.cell.2019.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Orzalli MH, Broekema NM, Diner BA, Hancks DC, Elde NC, Cristea IM, et al. cGAS-Mediated Stabilization of IFI16 Promotes Innate Signaling During Herpes Simplex Virus Infection. Proc Natl Acad Sci U S A (2015) 112(14):E1773–81. doi: 10.1073/pnas.1424637112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu H, Zhang H, Wu X, Ma D, Wu J, Wang L, et al. Nuclear cGAS Suppresses DNA Repair and Promotes Tumorigenesis. Nature (2018) 563(7729):131–6. doi: 10.1038/s41586-018-0629-6 [DOI] [PubMed] [Google Scholar]
- 57. Volkman HE, Cambier S, Gray EE, Stetson DB. Tight Nuclear Tethering of cGAS Is Essential for Preventing Autoreactivity. Elife (2019) 8:e47491. doi: 10.7554/eLife.47491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jiang H, Xue X, Panda S, Kawale A, Hooy RM, Liang F, et al. Chromatin-Bound cGAS Is an Inhibitor of DNA Repair and Hence Accelerates Genome Destabilization and Cell Death. EMBO J (2019) 38(21):e102718. doi: 10.15252/embj.2019102718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Michalski S, de Oliveira Mann CC, Stafford CA, Witte G, Bartho J, Lammens K, et al. Structural Basis for Sequestration and Autoinhibition of cGAS by Chromatin. Nature (2020) 587(7835):678–82. doi: 10.1038/s41586-020-2748-0 [DOI] [PubMed] [Google Scholar]
- 60. Gentili M, Lahaye X, Nadalin F, Nader GPF, Lombardi EP, Herve S, et al. The N-Terminal Domain of cGAS Determines Preferential Association With Centromeric DNA and Innate Immune Activation in the Nucleus (Vol 26, pg 2377, 2019). Cell Rep (2019) 26(13):3474–. doi: 10.1016/j.celrep.2019.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhao B, Xu P, Rowlett CM, Jing T, Shinde O, Lei Y, et al. The Molecular Basis of Tight Nuclear Tethering and Inactivation of cGAS. Nature (2020) 587(7835):673–7. doi: 10.1038/s41586-020-2749-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pathare GR, Decout A, Glück S, Cavadini S, Makasheva K, Hovius R, et al. Structural Mechanism of cGAS Inhibition by the Nucleosome. Nature (2020) 587(7835):668–72. doi: 10.1038/s41586-020-2750-6 [DOI] [PubMed] [Google Scholar]
- 63. Boyer JA, Spangler CJ, Strauss JD, Cesmat AP, Liu P, McGinty RK, et al. Structural Basis of Nucleosome-Dependent cGAS Inhibition. Science (2020) 370(6515):450–4. doi: 10.1126/science.abd0609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kujirai T, Zierhut C, Takizawa Y, Kim R, Negishi L, Uruma N, et al. Structural Basis for the Inhibition of cGAS by Nucleosomes. Science (2020) 370(6515):455–8. doi: 10.1126/science.abd0237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cao D, Han X, Fan X, Xu RM, Zhang X. Structural Basis for Nucleosome-Mediated Inhibition of cGAS Activity. Cell Res (2020) 30(12):1088–97. doi: 10.1038/s41422-020-00422-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Guey B, Wischnewski M, Decout A, Makasheva K, Kaynak M, Sakar MS, et al. BAF Restricts cGAS on Nuclear DNA to Prevent Innate Immune Activation. Science (2020) 369(6505):823–8. doi: 10.1126/science.aaw6421 [DOI] [PubMed] [Google Scholar]
- 67. Li T, Huang T, Du M, Chen X, Du F, Ren J, et al. Phosphorylation and Chromatin Tethering Prevent cGAS Activation During Mitosis. Science (2021) 371(6535):eabc5386. doi: 10.1126/science.abc5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gekara NO, Jiang H. The Innate Immune DNA Sensor cGAS: A Membrane, Cytosolic, or Nuclear Protein? Sci Signal (2019) 12(581):eaax3521. doi: 10.1126/scisignal.aax3521 [DOI] [PubMed] [Google Scholar]
- 69. Alharbi AS, Garcin AJ, Lennox KA, Pradeloux S, Wong C, Straub S, et al. Rational Design of Antisense Oligonucleotides Modulating the Activity of TLR7/8 Agonists. Nucleic Acids Res (2020) 48(13):7052–65. doi: 10.1093/nar/gkaa523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Valentin R, Wong C, Alharbi AS, Pradeloux S, Morros MP, Lennox KA, et al. Sequence-Dependent Inhibition of cGAS and TLR9 DNA Sensing by 2 ‘-O-Methyl Gapmer Oligonucleotides. Nucleic Acids Res (2021) 49(11):6082–99. doi: 10.1093/nar/gkab451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Xie W, Lama L, Adura C, Tomita D, Glickman JF, Tuschl T, et al. Human cGAS Catalytic Domain has an Additional DNA-Binding Interface That Enhances Enzymatic Activity and Liquid-Phase Condensation. P Natl Acad Sci U S A (2019) 116(24):11946–55. doi: 10.1073/pnas.1905013116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Feijo Delgado F, Cermak N, Hecht VC, Son S, Li Y, Knudsen SM, et al. Intracellular Water Exchange for Measuring the Dry Mass, Water Mass and Changes in Chemical Composition of Living Cells. PLoS One (2013) 8(7):e67590. doi: 10.1371/journal.pone.0067590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Palazzo AF, Lee ES. Non-Coding RNA: What Is Functional and What Is Junk? Front Genet (2015) 6:2. doi: 10.3389/fgene.2015.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Frenkel-Morgenstern M, Danon T, Christian T, Igarashi T, Cohen L, Hou YM, et al. Genes Adopt Non-Optimal Codon Usage to Generate Cell Cycle-Dependent Oscillations in Protein Levels. Mol Syst Biol (2012) 8:572. doi: 10.1038/msb.2012.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Russo J, Russo IH. Techniques and Methodological Approaches in Breast Cancer Research. Springer; (2014). pp. 287. [Google Scholar]
- 76. Karalyan ZA, Izmailyan RA, Abroyan LO, Avetisyan AS, Hakobyan LA, Zakaryan HS, et al. Evaluation of Viral Genome Copies Within Viral Factories on Different DNA Viruses. J Histochem Cytochem (2018) 66(5):359–65. doi: 10.1369/0022155417749490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chen LL. The Biogenesis and Emerging Roles of Circular RNAs. Nat Rev Mol Cell Bio (2016) 17(4):205–11. doi: 10.1038/nrm.2015.32 [DOI] [PubMed] [Google Scholar]
- 78. Shang QF, Yang Z, Jia RB, Ge SF. The Novel Roles of circRNAs in Human Cancer. Mol Cancer (2019) 18:6. doi: 10.1186/s12943-018-0934-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Steinhagen F, Zillinger T, Peukert K, Fox M, Thudium M, Barchet W, et al. Suppressive Oligodeoxynucleotides Containing TTAGGG Motifs Inhibit cGAS Activation in Human Monocytes. Eur J Immunol (2018) 48(4):605–11. doi: 10.1002/eji.201747338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Mankan AK, Schmidt T, Chauhan D, Goldeck M, Höning K, Gaidt M, et al. Cytosolic RNA : DNA Hybrids Activate the cGAS-STING Axis. EMBO J (2014) 33(24):2937–46. doi: 10.15252/embj.201488726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Prichard MN, Jairath S, Penfold ME, St Jeor S, Bohlman MC, Pari GS. Identification of Persistent RNA-DNA Hybrid Structures Within the Origin of Replication of Human Cytomegalovirus. J Virol (1998) 72(9):6997–7004. doi: 10.1128/JVI.72.9.6997-7004.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rennekamp AJ, Lieberman PM. Initiation of Epstein-Barr Virus Lytic Replication Requires Transcription and the Formation of a Stable RNA-DNA Hybrid Molecule at OriLyt. J Virol (2011) 85(6):2837–50. doi: 10.1128/JVI.02175-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Telesnitsky A, Goff SP. “Reverse Transcriptase and the Generation of Retroviral DNA”. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor (NY: Cold Spring Harbor Laboratory Press; (1997). [PubMed] [Google Scholar]
- 84. Morchikh M, Cribier A, Raffel R, Amraoui S, Cau J, Severac D, et al. HEXIM1 and NEAT1 Long Non-Coding RNA Form a Multi-Subunit Complex That Regulates DNA-Mediated Innate Immune Response. Mol Cell (2017) 67(3):387–99.e5. doi: 10.1016/j.molcel.2017.06.020 [DOI] [PubMed] [Google Scholar]
- 85. Sidibé H, Dubinski A, Vande Velde C. The Multi-Functional RNA-Binding Protein G3BP1 and Its Potential Implication in Neurodegenerative Disease. J Neurochem (2020) 157(4):944–62. doi: 10.1111/jnc.15280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. van Gent M, Sparrer KMJ, Gack MU. TRIM Proteins and Their Roles in Antiviral Host Defenses. Annu Rev Virol (2018) 5(1):385–405. doi: 10.1146/annurev-virology-092917-043323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Goyani S, Roy M, Singh R. TRIM-NHL as RNA Binding Ubiquitin E3 Ligase (RBUL): Implication in Development and Disease Pathogenesis. Bba-Mol Basis Dis (2021) 1687(7):166066. doi: 10.1016/j.bbadis.2020.166066 [DOI] [PubMed] [Google Scholar]
- 88. Thapa P, Shanmugam N, Pokrzywa W. Ubiquitin Signaling Regulates RNA Biogenesis, Processing, and Metabolism. Bioessays (2020) 42(1):e1900171. doi: 10.1002/bies.201900171 [DOI] [PubMed] [Google Scholar]
- 89. Yang D, Li NL, Wei D, Liu B, Guo F, Elbahesh H, et al. The E3 Ligase TRIM56 Is a Host Restriction Factor of Zika Virus and Depends on Its RNA-Binding Activity But Not miRNA Regulation, for Antiviral Function. PLoS Negl Trop Dis (2019) 13(6):e0007537. doi: 10.1371/journal.pntd.0007537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chen M, Meng Q, Qin Y, Liang P, Tan P, He L, et al. TRIM14 Inhibits cGAS Degradation Mediated by Selective Autophagy Receptor P62 to Promote Innate Immune Responses. Mol Cell (2016) 64(1):105–19. doi: 10.1016/j.molcel.2016.08.025 [DOI] [PubMed] [Google Scholar]
- 91. Williams FP, Haubrich K, Perez-Borrajero C, Hennig J. Emerging RNA-Binding Roles in the TRIM Family of Ubiquitin Ligases. Biol Chem (2019) 400(11):1443–64. doi: 10.1515/hsz-2019-0158 [DOI] [PubMed] [Google Scholar]
- 92. Lian H, Zang R, Wei J, Ye W, Hu MM, Chen YD, et al. The Zinc-Finger Protein ZCCHC3 Binds RNA and Facilitates Viral RNA Sensing and Activation of the RIG-I-Like Receptors. Immunity (2018) 49(3):438–48.e5. doi: 10.1016/j.immuni.2018.08.014 [DOI] [PubMed] [Google Scholar]
- 93. Liao CY, Lei CQ, Shu HB. PCBP1 Modulates the Innate Immune Response by Facilitating the Binding of cGAS to DNA. Cell Mol Immunol (2020) 18(10):2334–43. doi: 10.1038/s41423-020-0462-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lian H, Wei J, Zang R, Ye W, Yang Q, Zhang XN, et al. ZCCHC3 Is a Co-Sensor of cGAS for dsDNA Recognition in Innate Immune Response. Nat Commun (2018) 9(1):3349. doi: 10.1038/s41467-018-05559-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zang R, Lian H, Zhong X, Yang Q, Shu HB. ZCCHC3 Modulates TLR3-Mediated Signaling by Promoting Recruitment of TRIF to TLR3. J Mol Cell Biol (2020) 12(4):251–62. doi: 10.1093/jmcb/mjaa004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yang P, Mathieu C, Kolaitis RM, Zhang P, Messing J, Yurtsever U, et al. G3BP1 Is a Tunable Switch That Triggers Phase Separation to Assemble Stress Granules. Cell (2020) 181(2):325–45.e28. doi: 10.1016/j.cell.2020.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Sanders DW, Kedersha N, Lee DSW, Strom AR, Drake V, Riback JA, et al. Competing Protein-RNA Interaction Networks Control Multiphase Intracellular Organization. Cell (2020) 181(2):306–24.e28. doi: 10.1016/j.cell.2020.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Guillen-Boixet J, Kopach A, Holehouse AS, Wittmann S, Jahnel M, Schlussler R, et al. RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell (2020) 181(2):346–61.e17. doi: 10.1016/j.cell.2020.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Liu ZS, Cai H, Xue W, Wang M, Xia T, Li WJ, et al. G3BP1 Promotes DNA Binding and Activation of cGAS. Nat Immunol (2019) 20(1):18–28. doi: 10.1038/s41590-018-0262-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Eiermann N, Haneke K, Sun Z, Stoecklin G, Ruggieri A. Dance With the Devil: Stress Granules and Signaling in Antiviral Responses. Viruses (2020) 12(9):984. doi: 10.3390/v12090984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Knott GJ, Bond CS, Fox AH. The DBHS Proteins SFPQ, NONO and PSPC1: A Multipurpose Molecular Scaffold. Nucleic Acids Res (2016) 44(9):3989–4004. doi: 10.1093/nar/gkw271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Lahaye X, Gentili M, Silvin A, Conrad C, Picard L, Jouve M, et al. NONO Detects the Nuclear HIV Capsid to Promote cGAS-Mediated Innate Immune Activation. Cell (2018) 175(2):488–501.e22. doi: 10.1016/j.cell.2018.08.062 [DOI] [PubMed] [Google Scholar]
- 103. Wan L, Juszkiewicz S, Blears D, Bajpe PK, Han Z, Faull P, et al. Translation Stress and Collided Ribosomes Are Co-Activators of cGAS. Mol Cell (2021) 81(13):2808–22.e10. doi: 10.1016/j.molcel.2021.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Anantharaman V, Koonin EV, Aravind L. Comparative Genomics and Evolution of Proteins Involved in RNA Metabolism. Nucleic Acids Res (2002) 30(7):1427–64. doi: 10.1093/nar/30.7.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Gerstberger S, Hafner M, Tuschl T. A Census of Human RNA-Binding Proteins. Nat Rev Genet (2014) 15(12):829–45. doi: 10.1038/nrg3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Corley M, Burns MC, Yeo GW. How RNA-Binding Proteins Interact With RNA: Molecules and Mechanisms. Mol Cell (2020) 78(1):9–29. doi: 10.1016/j.molcel.2020.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Albihlal WS, Gerber AP. Unconventional RNA-Binding Proteins: An Uncharted Zone in RNA Biology. FEBS Lett (2018) 592(17):2917–31. doi: 10.1002/1873-3468.13161 [DOI] [PubMed] [Google Scholar]
- 108. Ramanathan M, Porter DF, Khavari PA. Methods to Study RNA-Protein Interactions. Nat Methods (2019) 16(3):225–34. doi: 10.1038/s41592-019-0330-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Hentze MW, Castello A, Schwarzl T, Preiss T. A Brave New World of RNA-Binding Proteins. Nat Rev Mol Cell Biol (2018) 19(5):327–41. doi: 10.1038/nrm.2017.130 [DOI] [PubMed] [Google Scholar]
- 110. Ablasser A, Chen ZJ. cGAS in Action: Expanding Roles in Immunity and Inflammation. Science (2019) 363(6431):1055–+. doi: 10.1126/science.aat8657 [DOI] [PubMed] [Google Scholar]
- 111. Hornung V, Hartmann R, Ablasser A, Hopfner KP. OAS Proteins and cGAS: Unifying Concepts in Sensing and Responding to Cytosolic Nucleic Acids. Nat Rev Immunol (2014) 14(8):521–8. doi: 10.1038/nri3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Cadena C, Hur S. Filament-Like Assemblies of Intracellular Nucleic Acid Sensors: Commonalities and Differences. Mol Cell (2019) 76(2):243–54. doi: 10.1016/j.molcel.2019.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Slavik KM, Morehouse BR, Ragucci AE, Zhou W, Ai XL, Chen YQ, et al. cGAS-Like Receptors Sense RNA and Control 3 ‘ 2 ‘-cGAMP Signalling in Drosophila. Nature (2021) 597(7874):109. doi: 10.1038/s41586-021-03743-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Holleufer A, Winther KG, Gad HH, Ai XL, Chen YQ, Li LH, et al. Two cGAS-Like Receptors Induce Antiviral Immunity in Drosophila. Nature (2021) 597(7874):114. doi: 10.1038/s41586-021-03800-z [DOI] [PubMed] [Google Scholar]
- 115. Bellucci M, Agostini F, Masin M, Tartaglia GG. Predicting Protein Associations With Long Noncoding RNAs. Nat Methods (2011) 8(6):444–5. doi: 10.1038/nmeth.1611 [DOI] [PubMed] [Google Scholar]
- 116. Lang B, Armaos A, Tartaglia GG. RNAct: Protein-RNA Interaction Predictions for Model Organisms With Supporting Experimental Data. Nucleic Acids Res (2019) 47(D1):D601–D6. doi: 10.1093/nar/gky967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Colantoni A, Rupert J, Vandelli A, Tartaglia GG, Zacco E. Zooming in on Protein-RNA Interactions: A Multi-Level Workflow to Identify Interaction Partners. Biochem Soc Trans (2020) 48(4):1529–43. doi: 10.1042/BST20191059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Yang W, Zhou J, Zhang K, Li L, Xu Y, Ma K, et al. Identification and Validation of the Clinical Roles of the VHL-Related LncRNAs in Clear Cell Renal Cell Carcinoma. J Cancer (2021) 12(9):2702–14. doi: 10.7150/jca.55113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Ribeiro DM, Zanzoni A, Cipriano A, Delli Ponti R, Spinelli L, Ballarino M, et al. Protein Complex Scaffolding Predicted as a Prevalent Function of Long Non-Coding RNAs. Nucleic Acids Res (2018) 46(2):917–28. doi: 10.1093/nar/gkx1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Wang Y, Luo W, Huang L, Xiao J, Song X, Li F, et al. A Novel lncRNA Linc-AhRA Negatively Regulates Innate Antiviral Response in Murine Microglia Upon Neurotropic Herpesvirus Infection. Theranostics (2021) 11(19):9623–51. doi: 10.7150/thno.64880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Livi CM, Klus P, Delli Ponti R, Tartaglia GG. catRAPID Signature: Identification of Ribonucleoproteins and RNA-Binding Regions. Bioinformatics (2016) 32(5):773–5. doi: 10.1093/bioinformatics/btv629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Hartmann R, Justesen J, Sarkar SN, Sen GC, Yee VC. Crystal Structure of the 2'-Specific and Double-Stranded RNA-Activated Interferon-Induced Antiviral Protein 2'-5'-Oligoadenylate Synthetase. Mol Cell (2003) 12(5):1173–85. [DOI] [PubMed] [Google Scholar]
- 123. Agostini F, Zanzoni A, Klus P, Marchese D, Cirillo D, Tartaglia GG. catRAPID Omics: A Web Server for Large-Scale Prediction of Protein-RNA Interactions. Bioinformatics (2013) 29(22):2928–30. doi: 10.1093/bioinformatics/btt495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Armaos A, Colantoni A, Proietti G, Rupert J, Tartaglia GG. catRAPID Omics V2.0: Going Deeper and Wider in the Prediction of Protein-RNA Interactions. Nucleic Acids Res (2021) 49(W1):W72–9. doi: 10.1093/nar/gkab393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Ferre F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA Interaction. Brief Bioinform (2016) 17(1):106–16. doi: 10.1093/bib/bbv031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Xu J, Wang Z, Jin X, Li L, Pan T. Methods for Identification of Protein-RNA Interaction. Adv Exp Med Biol (2018) 1094:117–26. doi: 10.1007/978-981-13-0719-5_12 [DOI] [PubMed] [Google Scholar]
- 127. Hellman LM, Fried MG. Electrophoretic Mobility Shift Assay (EMSA) for Detecting Protein-Nucleic Acid Interactions. Nat Protoc (2007) 2(8):1849–61. doi: 10.1038/nprot.2007.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Bai Q, Bai Z, Sun L. Detection of RNA-Binding Proteins by In Vitro RNA Pull-Down in Adipocyte Culture. J Vis Exp (2016) 113):e54207. doi: 10.3791/54207 [DOI] [PubMed] [Google Scholar]
- 129. Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M. HuR Regulates Cyclin A and Cyclin B1 mRNA Stability During Cell Proliferation. EMBO J (2000) 19(10):2340–50. doi: 10.1093/emboj/19.10.2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Tuerk C, Gold L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science (1990) 249(4968):505–10. doi: 10.1126/science.2200121 [DOI] [PubMed] [Google Scholar]
- 131. Lorenz C, von Pelchrzim F, Schroeder R. Genomic Systematic Evolution of Ligands by Exponential Enrichment (Genomic SELEX) for the Identification of Protein-Binding RNAs Independent of Their Expression Levels. Nat Protoc (2006) 1(5):2204–12. doi: 10.1038/nprot.2006.372 [DOI] [PubMed] [Google Scholar]
- 132. Stoltenburg R, Reinemann C, Strehlitz B. SELEX–a (R)Evolutionary Method to Generate High-Affinity Nucleic Acid Ligands. Biomol Eng (2007) 24(4):381–403. doi: 10.1016/j.bioeng.2007.06.001 [DOI] [PubMed] [Google Scholar]
- 133. Ray D, Kazan H, Chan ET, Pena Castillo L, Chaudhry S, Talukder S, et al. Rapid and Systematic Analysis of the RNA Recognition Specificities of RNA-Binding Proteins. Nat Biotechnol (2009) 27(7):667–70. doi: 10.1038/nbt.1550 [DOI] [PubMed] [Google Scholar]
- 134. Lambert N, Robertson A, Jangi M, McGeary S, Sharp PA, Burge CB. RNA Bind-N-Seq: Quantitative Assessment of the Sequence and Structural Binding Specificity of RNA Binding Proteins. Mol Cell (2014) 54(5):887–900. doi: 10.1016/j.molcel.2014.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Lambert NJ, Robertson AD, Burge CB. RNA Bind-N-Seq: Measuring the Binding Affinity Landscape of RNA-Binding Proteins. Methods Enzymol (2015) 558:465–93. doi: 10.1016/bs.mie.2015.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Gagliardi M, Matarazzo MR. RIP: RNA Immunoprecipitation. Methods Mol Biol (2016) 1480:73–86. doi: 10.1007/978-1-4939-6380-5_7 [DOI] [PubMed] [Google Scholar]
- 137. Ule J, Jensen K, Mele A, Darnell RB. CLIP: A Method for Identifying Protein-RNA Interaction Sites in Living Cells. Methods (2005) 37(4):376–86. doi: 10.1016/j.ymeth.2005.07.018 [DOI] [PubMed] [Google Scholar]
- 138. Foley SW, Gregory BD. Protein Interaction Profile Sequencing (PIP-Seq). Curr Protoc Mol Biol (2016) 116:27 5 1– 5 15. doi: 10.1002/cpmb.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, et al. A Long Noncoding RNA Mediates Both Activation and Repression of Immune Response Genes. Science (2013) 341(6147):789–92. doi: 10.1126/science.1240925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Kazan H, Ray D, Chan ET, Hughes TR, Morris Q. RNAcontext: A New Method for Learning the Sequence and Structure Binding Preferences of RNA-Binding Proteins. PLoS Comput Biol (2010) 6(7):e1000832. doi: 10.1371/journal.pcbi.1000832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Suresh V, Liu L, Adjeroh D, Zhou X. RPI-Pred: Predicting ncRNA-Protein Interaction Using Sequence and Structural Information. Nucleic Acids Res (2015) 43(3):1370–9. doi: 10.1093/nar/gkv020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Cheng ZZ, Zhou SG, Guan JH. Computationally Predicting Protein-RNA Interactions Using Only Positive and Unlabeled Examples. J Bioinf Comput Biol (2015) 13(3):1541005. doi: 10.1142/S021972001541005X [DOI] [PubMed] [Google Scholar]
- 143. Muppirala UK, Honavar VG, Dobbs D. Predicting RNA-Protein Interactions Using Only Sequence Information. BMC Bioinf (2011) 12:489. doi: 10.1186/1471-2105-12-489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Ankö ML, Neugebauer KM. RNA-Protein Interactions In Vivo: Global Gets Specific. Trends Biochem Sci (2012) 37(7):255–62. doi: 10.1016/j.tibs.2012.02.005 [DOI] [PubMed] [Google Scholar]
- 145. Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-Binding Proteins Interact With Functionally Related Sets of RNAs, Suggesting an Extensive Regulatory System. PLoS Biol (2008) 6(10):e255. doi: 10.1371/journal.pbio.0060255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Hu H, Zhu C, Ai H, Zhang L, Zhao J, Zhao Q, et al. LPI-ETSLP: lncRNA-Protein Interaction Prediction Using Eigenvalue Transformation-Based Semi-Supervised Link Prediction. Mol Biosyst (2017) 13(9):1781–7. doi: 10.1039/C7MB00290D [DOI] [PubMed] [Google Scholar]
- 147. Zhao Q, Zhang Y, Hu H, Ren G, Zhang W, Liu H. IRWNRLPI: Integrating Random Walk and Neighborhood Regularized Logistic Matrix Factorization for lncRNA-Protein Interaction Prediction. Front Genet (2018) 9:239. doi: 10.3389/fgene.2018.00239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Kloetgen A, Munch PC, Borkhardt A, Hoell JI, McHardy AC. Corrigendum to: Biochemical and Bioinformatic Methods for Elucidating the Role of RNA-Protein Interactions in Posttranscriptional Regulation. Brief Funct Genomics (2019) 18(6):433. doi: 10.1093/bfgp/elz014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Pan X, Yang Y, Xia CQ, Mirza AH, Shen HB. Recent Methodology Progress of Deep Learning for RNA-Protein Interaction Prediction. Wiley Interdiscip Rev RNA (2019) 10(6):e1544. doi: 10.1002/wrna.1544 [DOI] [PubMed] [Google Scholar]
- 150. Tan X, Sun L, Chen J, Chen ZJ. Detection of Microbial Infections Through Innate Immune Sensing of Nucleic Acids. Annu Rev Microbiol (2018) 72:447–78. doi: 10.1146/annurev-micro-102215-095605 [DOI] [PubMed] [Google Scholar]
- 151. Gray EE, Treuting PM, Woodward JJ, Stetson DB. Cutting Edge: cGAS Is Required for Lethal Autoimmune Disease in the Trex1-Deficient Mouse Model of Aicardi-Goutieres Syndrome. J Immunol (2015) 195(5):1939–43. doi: 10.4049/jimmunol.1500969 [DOI] [PMC free article] [PubMed] [Google Scholar]