Abstract
Objectives
To compare the performance of the PRECISE scoring system against several MRI-derived delta-radiomics models for predicting histopathological prostate cancer (PCa) progression in patients on active surveillance (AS).
Methods
The study included AS patients with biopsy-proven PCa with a minimum follow-up of 2 years and at least one repeat targeted biopsy. Histopathological progression was defined as grade group progression from diagnostic biopsy. The control group included patients with both radiologically and histopathologically stable disease. PRECISE scores were applied prospectively by four uro-radiologists with 5–16 years’ experience. T2WI- and ADC-derived delta-radiomics features were computed using baseline and latest available MRI scans, with the predictive modelling performed using the parenclitic networks (PN), least absolute shrinkage and selection operator (LASSO) logistic regression, and random forests (RF) algorithms. Standard measures of discrimination and areas under the ROC curve (AUCs) were calculated, with AUCs compared using DeLong’s test.
Results
The study included 64 patients (27 progressors and 37 non-progressors) with a median follow-up of 46 months. PRECISE scores had the highest specificity (94.7%) and positive predictive value (90.9%), whilst RF had the highest sensitivity (92.6%) and negative predictive value (92.6%) for predicting disease progression. The AUC for PRECISE (84.4%) was non-significantly higher than AUCs of 81.5%, 78.0%, and 80.9% for PN, LASSO regression, and RF, respectively (p = 0.64, 0.43, and 0.57, respectively). No significant differences were observed between AUCs of the three delta-radiomics models (p-value range 0.34–0.77).
Conclusions
PRECISE and delta-radiomics models achieved comparably good performance for predicting PCa progression in AS patients.
Key Points
• The observed high specificity and PPV of PRECISE are complemented by the high sensitivity and NPV of delta-radiomics, suggesting a possible synergy between the two image assessment approaches.
• The comparable performance of delta-radiomics to PRECISE scores applied by expert readers highlights the prospective use of the former as an objective and standardisable quantitative tool for MRI-guided AS follow-up.
• The marginally superior performance of parenclitic networks compared to conventional machine learning algorithms warrants its further use in radiomics research.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00330-021-08151-x.
Keywords: Prostate cancer, Magnetic resonance imaging, Active surveillance, PRECISE, Machine learning
Introduction
Prostate cancer (PCa) is the second commonest and the fifth deadliest male cancer worldwide [1]. In the USA and the UK, nearly half of men present with low- and intermediate-risk localised disease [2, 3], for which active surveillance (AS) is the recommended management option [4–6]. Simultaneously, a cumulative 5-year AS dropout rate due to disease progression is 27% [7], which is partially driven by the lack of consensus on AS protocols and the definition of disease progression across guidelines and individual centres [8–11]. Whilst tumour progression is the expected natural outcome of AS, there is a lack of objective non-invasive diagnostic tools enabling continuous re-evaluation of the risk of PCa progression. If proven accurate and standardisable, such tools could help clinicians make more informed decisions on the need for switching to radical treatment without repeat biopsies, thereby reducing the risk of associated complications and costs to healthcare systems.
At this stage, this unmet clinical need is partially addressed by the increasing reliance on magnetic resonance imaging (MRI) as an integral part of AS follow-up [12–15]. The PRECISE scoring system was developed in 2017 [16] in order to standardise reporting of serial MRI scans in patients on AS to detect clinically significant radiological changes sufficient to either trigger additional investigations or switch to immediate radical treatment. Since then, several groups [17–21] have demonstrated a high negative predictive value of PRECISE, suggesting that it can be used to avoid routine repeat biopsy if the disease is regarded as radiologically stable. However, the comparatively moderate positive predictive value of PRECISE for predicting histopathological disease progression emphasises the relatively subjective nature of the system, particularly when determining PRECISE category 4 lesions as demonstrating “clinically significant” radiological progression [12]. Furthermore, PRECISE has been validated exclusively by expert readers in academic centres, which may limit its generalisability and lead to a reduced performance when used by non-expert radiologists.
Quantitative imaging techniques may prove clinically useful by providing more objective and expertise-independent measures of the underlying biological changes occurring over the course of PCa natural history on AS. One such technique is delta-radiomics, which analyses changes in MRI-derived texture features obtained at two time points [22]. In the AS setting, delta-radiomic features may be compared between any two consecutive MRI scans obtained from the same patient to provide a quantitative readout of the underlying histological changes within a lesion of interest. Therefore, in this proof-of-concept study, we compared the diagnostic performance of PRECISE applied by expert readers against several MRI-derived delta-radiomics models for the purpose of predicting histopathological PCa progression in patients enrolled on AS.
Methods
Patient population
The local institutional review board (NRES Committee East of England, UK) waived the need for informed consent for retrospective data analysis obtained as part of a service evaluation of the prostate diagnostic pathway. The study included consecutive patients with biopsy-proven PCa enrolled on the local AS programme with a minimum follow-up of 2 years, with their first and last 3T MRI scans performed on the same magnet, and at least one repeat targeted biopsy performed within a year of the last MRI. The exclusion criteria were the absence of MR-visible lesions, prior or interim treatment for PCa or benign disease, and the presence of total hip replacement or other pelvic metalwork. A total of 281 patients enrolled on AS in our centre between November 2012 and November 2018 were screened, of whom 217 were excluded.
The remaining 64 patients were divided into two groups depending on their disease progression status. AS progression (n = 27) was defined as a switch to radical treatment prompted by confirmed histopathological progression on repeat targeted biopsy (grade group progression from diagnostic biopsy). The control group (n = 37) included patients whose disease remained both radiologically (PRECISE score 3 [16]) and histopathologically stable over the course of AS.
MRI acquisition parameters
Patients underwent prostate MRI on a 3-T MR750 scanner (GE Healthcare) using a 32-channel receiver coil. Unless clinically contraindicated, intravenous injection of hyoscine butylbromide (Buscopan, 20 mg/mL; Boehringer) was administered prior to imaging to reduce peristaltic movement [23]; no additional patient preparation measures were taken. Multiparametric MRI protocol included axial T1, multiplanar high-resolution T2-weighted 2D fast recovery FSE, spin-echo echo-planar imaging pulse DWI, and dynamic contrast enhancement imaging, with acquisition parameters summarised in Supplementary Table S1.
Biopsy technique
Depending on clinical recommendation, either transrectal (DynaCAD, InVivo Corp) or transperineal (Biopsee, Oncology Systems Limited) biopsies were performed by three urologists with 8–20 years’ experience using MRI/ultrasound fusion. Twelve systematic cores were taken as part of the transrectal approach, and 24 systematic cores were obtained during transperineal procedures, following the Ginsburg protocol [24]. In addition, 2–4 separate target cores corresponding to lesions outlined on MRI were sampled, as previously described [25]. Repeat targeted biopsies were performed at time points specified by the local protocol (12 and 36 months) if not triggered earlier by clinical suspicion of progression, encompassing either three consecutive elevated PSA levels above the pre-defined threshold or suspected radiological progression (PRECISE scores 4–5). At baseline, 13 and 51 biopsies were performed using transrectal and transperineal approaches, respectively. At follow-up, 18 and 46 biopsies were performed using transrectal and transperineal approaches, respectively.
Image segmentation and analysis
Tumour ROIs were drawn on anatomical T2WI (Fig. 1) and ADC maps by a fellowship-trained uro-radiologist (T.B.) with 13 years’ experience of reporting prostate MRI and an imaging research fellow (N.S.) with 4 years’ experience. The segmentation was performed in consensus using the open-source software ITK-SNAP [26], with all cases outlined jointly by the two readers. The reliability of image segmentation by readers was evaluated by applying ROI morphological perturbations using the “Scipy.ndimage.morphology” functions (binary_opening and binary_closing) in the SciPy version 1.3.2 multi-dimensional image processing package.
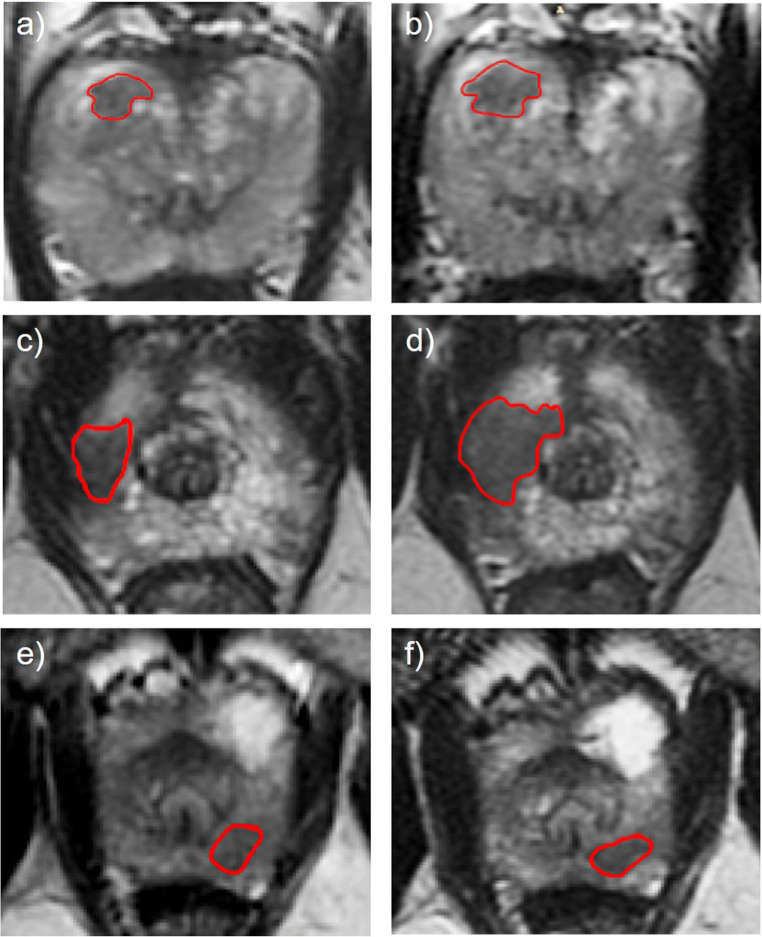
Fig. 1.
Comparison of T2-weighted images of the prostate obtained at baseline pre-biopsy (a, c, e) and follow-up (b, d, f) MRI scans from patients enrolled on active surveillance. Images (a, b) were obtained from a patient with stable 3 + 3 = 6 disease that showed neither radiological nor histopathological progression over a follow-up period of 3 years (PRECISE 3). Images (c, d) were obtained from a patient with both radiological (PRECISE 5) and histopathological (3 + 3 = 6 to 4 + 3 = 7) progression. Images (e, f) were obtained from a patient with confirmed histopathological progression (3 + 3 = 6 to 3 + 4 = 7) but radiologically stable disease (PRECISE 3). In all presented cases, the clinical outcome was successfully predicted by all three delta-radiomics models used
Follow-up MRI studies were scored on a 5-point PRECISE scale described in Supplementary Table S2 [16]. PRECISE scores were applied prospectively by four sub-specialist uro-radiologists with 5–16 years’ experience of reporting prostate MRI, with each having read > 2,000 cases and considered to be experts [27, 28]. At the time of reporting, the readers were not blinded to clinical information, including PSA and PSA density dynamics. For the purposes of predictive modelling, the PRECISE scores were dichotomised at a cutoff value of 4 since values 1–3 and 4–5 indicate resolution of previous features/stable disease and radiological disease progression, respectively.
Delta-radiomics analysis
The overall workflow of the radiomics pipeline utilised to develop and validate predictive models for PCa progression on AS is illustrated in Fig. 2. The T2WI- and ADC-derived texture features (summarised in Supplementary Table S3) were extracted using PyRadiomics version 2.0 and Python version 3.7.5 [29, 30]. 3D feature computation without any resampling was used to avoid interpolation artifacts. According to the Imaging Biomarker Standardisation Initiative (IBSI) [30], the use of the number of bins is favoured over the bin width in the case of arbitrary intensity units, such as MRI. Hence, no re-segmentation (i.e. the voxels outside a specified range being removed from the mask prior to texture feature calculation) was applied.
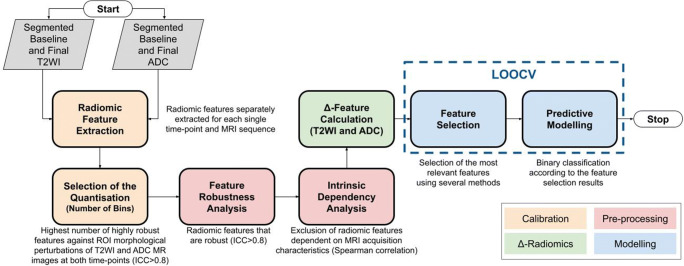
Fig. 2.
Flow diagram summarising the key stages of delta-radiomics analysis used in this study, including calibration, pre-processing, delta-radiomics feature calculation, and predictive modelling using the leave-one-out cross-validation (LOOCV) approach. ADC, apparent diffusion coefficient; ICC, intraclass correlation coefficient; MRI, magnetic resonance imaging; T2WI, T2-weighted imaging
Feature robustness was assessed by applying ROI morphological perturbations and evaluating the relationship between individual features and MRI acquisition parameters [31–33] as described in detail in the Calibration and pre-processing section of the Electronic Supplementary Material. This procedure also simulates the ROI variability by considering intra- and inter-reader dependence during manual image segmentation [34]. Only features considered robust at both time points were included in the delta-radiomics predictive modelling.
Delta-radiomics predictive modelling
The delta-radiomics features were computed as the arithmetic difference between the final and baseline features. For each radiomic feature (separately for T2WI and ADC), let fbase and ffinal be its value at the baseline and final scan, respectively, Δf was calculated as:
Predictive modelling was then carried out using three machine learning methods: parenclitic networks [35], least absolute shrinkage and selection operator (LASSO) logistic regression [36], and random forests [37]. For the latter, we used 500 trees with a maximum depth of 10 (for reproducibility, a random_state parameter of 42 was set). The binary outcome of PCa progression was used as an outcome, with all delta-radiomics features used as predictors. The predictive performance of each feature was assessed using a leave-one-out cross-validation (LOOCV) scheme. The prediction was made for each sample by excluding (withholding) it from the dataset, training the classifier on the remaining (independent) samples, and then generating predictions for the withheld samples using the trained model.
Statistical and computational analysis
Normal distribution of the data was assessed using the D’Agostino-Pearson test (threshold p ≥ 0.05). Intergroup comparison of patient age, PSA, gland volume, PSA density, and AS follow-up length was performed using the Mann-Whitney U test. The relationship between the radiomic feature and individual MRI acquisition parameters was assessed using Spearman’s correlation analysis.
Random forests and LASSO regression were implemented in Python 3.9.0 using the pandas version 1.2.4, scikit-learn version 0.24.2, glmnet_python version 1.0, and rpy2 version 3.4.2 modules. In parenclitic networks, a support vector machine (SVM) with radial basis functions was trained for each pair of features using the “e1071” R package with default settings. For each sample, a network was then built wherein vertices corresponded to features and the edge weight was the disease progression probability as predicted by the SVM classifier. The mean of the vertices degrees was calculated, and a generalised linear model (GLM) classification was then constructed using the “stats” R package with default settings. The performance of PRECISE-based and delta-radiomics-based predictive models was assessed with measures of discrimination. Specificity and sensitivity were derived using non-parametric stratified resampling with the percentile method (2,000 bootstrap replicates) [38]. In addition, areas under the ROC curve (AUCs) were calculated for each model, alongside 95% confidence intervals using DeLong’s asymptomatically exact method to evaluate the uncertainty of each AUC [39]. AUC values were compared between the models using DeLong’s test for correlated/paired AUCs [39]. Positive and negative predictive values (PPV and NPV) and their 95% confidence intervals were computed using the standard approach [40]. Statistical analysis was performed in R version 3.5.1 (R Foundation for Statistical Computing) using the “pROC” and “reportROC” packages.
Results
Patient characteristics
The study included 64 patients enrolled on the AS programme in our centre between May 2013 and May 2018. The key clinicopathological characteristics of the patient cohort are summarised in Table 1. Twenty-seven patients showed histopathological disease progression and 37 patients harboured stable disease. Non-progressors were followed up for a median time of 47 months (IQR, 44–60 months), and time to progression in progressors was significantly shorter at 43 months (IQR, 24–29 months; p = 0.009). PSA and PSA density were significantly higher in progressors compared to non-progressors (p = 0.01 and p = 0.002, respectively), whilst age, gland volume, and PI-RADS scores were similar and demonstrated no significant difference between the two groups (Table 1). At enrollment biopsy, 51/65 (79%) and 14/65 (21%) lesions represented ISUP grade group 1 and 2 tumours, respectively, with 46/65 (71%) and 19/65 (29%) target lesions located in the peripheral and transition zones, respectively.
Table 1.
Summary baseline clinicopathological characteristics of the study cohort. The p-values are presented for an intergroup comparison between progressors and non-progressors performed using the Mann-Whitney U test. AS, active surveillance; PSA, prostate-specific antigen
| Parameter | Total cohort (n = 64) | Progressors (n = 27) | Non-progressors (n = 37) | p-value |
|---|---|---|---|---|
| Age, years | 67 (60–69) | 66 (60–69) | 67 (61–69) | 0.9218 |
| PSA, ng/mL | 5.6 (3.6–7.7) | 7.0 (5.2–8.7) | 5.0 (3.17–6.9) | 0.0102 |
| Gland volume, mL | 45.0 (33.0–63.8) | 45.0 (29.0–52.0) | 45.0 (36.6–66.2) | 0.1851 |
| PSA density | 0.11 (0.08–0.19) | 0.17 (0.11–0.27) | 0.09 (0.06–0.16) | 0.0024 |
| AS follow-up, mo | 46 (35–52) | 43 (24–49) | 47 (44–60) | 0.0093 |
| Biopsy grade group 1 (3 + 3 = 6), n | 51 (78%) | 23 (85%) | 28 (74%) | - |
| Biopsy grade group 2 (3 + 4 = 7), n | 14 (22%) | 4 (15%) | 10 (26%) | |
| Target lesion in the peripheral zone, n | 46 (70%) | 17 (63%) | 29 (76%) | - |
| Target lesion in the transition zone, n | 19 (30%) | 10 (27%) | 9 (24%) |
Delta-radiomics analysis: calibration and pre-processing
The numbers of highly robust features per number of bins identified at the calibration and pre-processing stage are presented in Table 2. Spearman’s correlation analysis showed no significant relationship between any of the texture features and MRI acquisition parameters (p > 0.05 for all with no multiplicity correction applied). The selected number of bins was, therefore, 128, with 34 T2WI- and 53 ADC-derived texture features overlapping at both time points (Supplementary Table S4).
Table 2.
The number of features with high robustness (ICC > 0.8) by varying the number of bins in the quantisation step for radiomic feature extraction for T2-weighted and ADC MR images at both baseline and final time points. ADC, apparent diffusion coefficient; T2WI, T2-weighted imaging
| Number of bins | Number of features with high robustness | ||||
|---|---|---|---|---|---|
| Baseline | Final | Total | |||
| T2WI | ADC | T2WI | ADC | ||
| 8 | 38 | 47 | 67 | 47 | 199 |
| 16 | 34 | 49 | 67 | 51 | 201 |
| 32 | 32 | 52 | 63 | 54 | 201 |
| 64 | 33 | 55 | 64 | 54 | 206 |
| 128 | 34 | 57 | 69 | 54 | 214 |
| 256 | 33 | 56 | 70 | 55 | 214 |
PRECISE versus delta-radiomics for predicting disease progression
Table 3 presents summary performance characteristics of PRECISE and the three machine learning approaches: parenclitic networks, LASSO regression, and random forests. The analysis of sensitivity, specificity, PPV, and NPV did not allow to identify the best-performing approach: PRECISE score had the highest specificity (94.7%) and PPV (90.9%), whilst random forests had the highest sensitivity (92.6%) and NPV (92.6%). AUC for PRECISE (84.4%) was the highest of all, compared to 81.5%, 78.0%, and 80.9% for parenclitic networks, LASSO regression, and random forests, respectively (Fig. 3). This difference, however, was non-significant: p = 0.643 for PRECISE versus parenclitic networks, p = 0.342 for PRECISE versus LASSO regression, and p = 0.572 for PRECISE versus random forests. No significant differences were reported between AUCs of the three delta-radiomics models (p-value range 0.342–0.768).
Table 3.
Summary performance characteristics of PRECISE, alongside parenclitic networks, LASSO regression, and random forests delta-radiomics models for predicting histopathological progression of prostate cancer in patients on active surveillance. AUC, area under the receiver operator characteristic curve; LASSO, least absolute shrinkage and selection operator; NPV, negative predictive value; PPV, positive predictive value; PRECISE, Prostate Cancer Radiological Estimation of Change in Sequential Evaluation
| Method | Sensitivity | Specificity | PPV | NPV | AUC |
|---|---|---|---|---|---|
| PRECISE | 74.1 (57.5–90.6) | 94.7 (87.6–1) | 90.9 (78.9–1) | 83.7 (72.7–94.8) | 84.4 (72.6–96.2) |
| Parenclitic networks | 85.2 (71.8–98.6) | 73.7 (59.7–87.7) | 69.7 (54–85.4) | 87.5 (76–99) | 81.6 (70.6–92.5) |
| LASSO regression | 70.4 (53.1–87.6) | 84.2 (72.6–95.8) | 76.0 (59.3–92.7) | 80.0 (67.6–92.4) | 78.0 (65.8–90.1) |
| Random forests | 92.6 (82.7–1) | 65.8 (50.7–80.9) | 65.8 (50.7–80.9) | 92.6 (82.7–1) | 80.9 (70–91.9) |
Fig. 3.
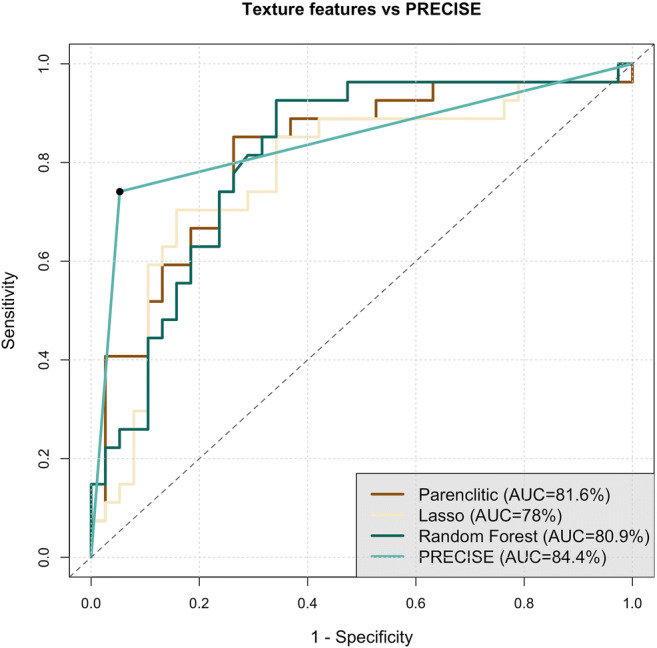
Receiver operating characteristic (ROC) curves for PRECISE, parenclitic networks, lasso regression, and random forest for predicting histopathological progression of prostate cancer in patients on active surveillance. The embedded legend denotes areas under ROC curves for each method
Discussion
This proof-of-concept study investigates the comparative performance of the MRI-derived PRECISE scoring system applied in routine clinical practice by expert uro-radiologists versus three delta-radiomics models developed using the parenclitic networks, LASSO regression, and random forests machine learning methods. The primary performance metric was the ability to predict histopathological progression of PCa in patients enrolled on AS. Although PRECISE performance characteristics appeared marginally superior, there was no significance difference between the AUCs achieved by all methods used in this study. These results pave the way for future investigations evaluating the role of MRI-derived delta-radiomics as an alternative quantitative means of MRI-guided follow-up of AS patients that could be standardisable and less dependent on reader experience.
In this study, PRECISE scoring demonstrated a high performance for predicting PCa histopathological progression with an AUC of 0.84, which is in line with AUCs of 0.83 and 0.82 reported by Dieffenbacher et al [17] and Caglic et al [21], respectively. It should be noted that, in addition to the readers’ expertise, the availability of clinical parameters may help supplement clinical decision-making and further improve performance, for example knowledge of PSA kinetics may help call PRECISE 4, when features of radiological progression may be otherwise equivocal. Among the machine learning techniques, random forests showed the best performance with the highest sensitivity (92.6%) and NPV (92.6%).
To our knowledge, this is the first study applying delta-radiomics in the setting of MRI follow-up in PCa AS. The comparable performance of standalone delta-radiomics models, which only account for quantitative image-derived texture features of the disease and were subject to rigorous cross-validation to reduce overfitting, offers potential for future studies attempting to introduce accurate and objective tools for evaluating PCa radiological progression on AS. Previous studies in PCa using MRI-derived delta-radiomics to predict tumour response to radiotherapy [41, 42] have also shown promising results. In other tumour types, delta-radiomics studies have likewise focused on predicting treatment response, including lung [22, 43], rectal [44, 45], and gastric [46] cancers, as well as bone and soft tissue malignancies [47, 48]. With the concept of AS being relatively unique to PCa, using delta-radiomics to track intratumoural morphological changes occurring naturally in the absence of any external interventions may offer insights into the biological significance of certain texture features that show strong correlation with the underlying cytoarchitectural patterns; this technique may additionally be applicable to other tumour types.
In this study, we were the first to apply the parenclitic networks for delta-radiomics predictive modelling. The particular strength of this novel machine learning approach is that it is deemed useful in a setting where the number of features is large whilst the sample size is limited [49], which is often the case with radiomics studies. Another strength of parenclitic networks is the ability to embed any multivariate data into the low-dimensional space of topological indices, which avoids the risk of overfitting. In this study, no difference in performance was observed between any of the machine learning algorithms used. The marginally better performance of the clinically applied PRECISE system may be explained by the use of stringent and unbiased strategies to reduce overfitting in the delta-radiomics modelling. These proof-of-concept results, therefore, offer promise for the development of improved delta-radiomics performance in larger cohorts.
Our study has several limitations. The sample size was relatively small, which was dictated by the stringent inclusion criteria requiring the presence of MR-visible lesions, which represents only around 50% of patients in AS cohorts [21] and mandating repeat targeted biopsies performed within a year of the most recent MRI, which was necessary for histopathological confirmation of progression. Gaining access to a larger cohort will also enable us to fit a single parenclitic networks model and fine-tune the hyperparameters for the SVM. Furthermore, the lesion-centred delta-radiomics approach was selected for several reasons. Firstly, the presence of MR-visible lesions is associated with an increased risk of progression on AS [21], thereby warranting closer follow-up of such patients. Secondly, as this is the first study to use delta-radiomics in this setting, focusing on the analysis of exclusively intratumoural rather than whole-gland texture features was deliberate in order to reduce the complexity of the analysis. The use of a whole-gland image segmentation approach [50, 51] coupled with MRI-derived habitat radiomics [52, 53] is an area for future work and would be applicable to all patients enrolled on AS. This will, however, require even more stringent pre-processing, calibration, modelling, and cross-validation steps to ensure that the resulting predictive models are not overfitted by features possibly arising from temporal changes in concurrent benign conditions (such as benign prostatic hyperplasia or prostatitis). Finally, as discussed previously, the reporting clinicians were not prospectively blinded to clinical information, which may have artificially increased the performance of the PRECISE scoring system. However, benchmarking alternative diagnostic modalities against the real-life performance of those already applied in routine clinical practice offers the fastest route to understanding the feasibility of their clinical translation.
In conclusion, MRI-derived delta-radiomics demonstrated comparable performance to expert prostate MRI readers and offer an objective and standardisable method for predicting histopathological progression of PCa in patients enrolled on AS programmes. These results pave the way for future multicentre studies investigating the diagnostic utility of more complex predictive models incorporating delta-radiomics features alongside standard-of-care clinicopathological predictors of PCa progression across different centres.
Supplementary Information
(DOCX 1238 kb)
Abbreviations
- ADC
Apparent diffusion coefficient
- AS
Active surveillance
- AUC
Area under the receiver operator curve
- LASSO
Least absolute shrinkage and selection operator
- LOOCV
Leave-one-out cross-validation
- MRI
Magnetic resonance imaging
- NPV
Negative predictive value
- PCa
Prostate cancer
- PN
Parenclitic networks
- PPV
Positive predictive value
- PRECISE
Prostate Cancer Radiological Estimation of Change in Sequential Evaluation
- PSA
Prostate-specific antigen
- RF
Random forests
- ROC
Receiver operator curve
- T2WI
T2-weighted imaging
Funding
The authors acknowledge support from National Institute of Health Research Cambridge Biomedical Research Centre, Cancer Research UK (Cambridge Imaging Centre grant number C197/A16465), the Engineering and Physical Sciences Research Council Imaging Centre in Cambridge and Manchester, and the Cambridge Experimental Cancer Medicine Centre. N. Sushentsev is supported by the Gates Cambridge Trust. T. Nazarenko is supported by a Medical Research Council grant (MR/R02524X/1). A. Suvorov is supported by the Ministry of Science and Higher Education of the Russian Federation within the programme developing World-Class Research Centres “Digital Biodesign and Personalized Healthcare” (075-15-2020-926).
Declarations
Guarantor
The scientific guarantor of this publication is Dr Tristan Barrett.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
Four of the authors have significant statistical expertise (OB, TN, AS, LR).
Informed consent
Written informed consent was waived by the Institutional Review Board (NRES Committee East of England, UK).
Ethical approval
Institutional Review Board approval was obtained.
Study subjects or cohorts overlap
Some study subjects or cohorts have been previously reported in:
Caglic I, Sushentsev N, Gnanapragasam V, Sala E, Shaida N, Koo B, et al. MRI-derived PRECISE scores for predicting pathologically-confirmed radiological progression in prostate cancer patients on active surveillance. Eur Radiol 2020. 2020; 1–10. doi:10.1007/s00330-020-07336-0.
Methodology
• Retrospective
• Observational
• Performed at one institution
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Nikita Sushentsev and Leonardo Rundo contributed equally to this work.
References
- 1.Sung H, Ferlay J, Siegel RL et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 10.3322/caac.21660 [DOI] [PubMed]
- 2.Negoita S, Feuer EJ, Mariotto A, et al. Annual Report to the Nation on the Status of Cancer, part II: recent changes in prostate cancer trends and disease characteristics. Cancer. 2018;124:2801–2814. doi: 10.1002/cncr.31549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Results of the NPCA (2020) Prospective Audit in England and Wales for men diagnosed from 1 National Prostate Cancer Audit Seventh Year Annual Report-Results of the NPCA Prospective Audit in England and Wales for men diagnosed 1
- 4.Mohler JL, Antonarakis ES, Armstrong AJ, et al. Prostate cancer, version 2.2019. JNCCN J Natl Compr Cancer Netw. 2019;17:479–505. doi: 10.6004/jnccn.2019.0023. [DOI] [PubMed] [Google Scholar]
- 5.Mottet N, Bellmunt J, Bolla M, et al. EAU-ESTRO-SIOG guidelines on prostate cancer. Part 1: screening, diagnosis, and local treatment with curative intent. Eur Urol. 2017;71:618–629. doi: 10.1016/j.eururo.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 6.NICE Guidance Prostate cancer: diagnosis and management. BJU Int. 2019;124:9–26. doi: 10.1111/bju.14809. [DOI] [PubMed] [Google Scholar]
- 7.Van Hemelrijck M, Ji X, Helleman J, et al. Reasons for discontinuing active surveillance: assessment of 21 centres in 12 countries in the Movember GAP3 Consortium. Eur Urol. 2019;75:523–531. doi: 10.1016/j.eururo.2018.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kinsella N, Helleman J, Bruinsma S, et al. Active surveillance for prostate cancer: a systematic review of contemporary worldwide practices. Transl Androl Urol. 2018;7:83–97. doi: 10.21037/tau.2017.12.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Washington SL, Jeong CW, Lonergan PE, et al. Regional variation in active surveillance for low-risk prostate cancer in the US. JAMA Netw Open. 2020;3:e2031349. doi: 10.1001/jamanetworkopen.2020.31349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Preisser F, Cooperberg MR, Crook J, et al. Intermediate-risk prostate cancer: stratification and management. Eur Urol Oncol. 2020;3:270–280. doi: 10.1016/j.euo.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Gnanapragasam VJ, Barrett T, Thankapannair V, et al. Using prognosis to guide inclusion criteria, define standardised endpoints and stratify follow-up in active surveillance for prostate cancer. BJU Int. 2019;124:758–767. doi: 10.1111/bju.14800. [DOI] [PubMed] [Google Scholar]
- 12.Barrett T, Haider MA (2017) The emerging role of MRI in prostate cancer active surveillance and ongoing challenges. AJR Am J Roentgenol 208:131–139. 10.2214/AJR.16.16355 [DOI] [PubMed]
- 13.Sanda MG, Cadeddu JA, Kirkby E, et al. Clinically localized prostate cancer: AUA/ASTRO/SUO guideline Part II: recommended approaches and details of specific care options. J Urol. 2018;199:990–997. doi: 10.1016/j.juro.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Sushentsev N, Caglic I, Sala E et al (2020) The effect of capped biparametric magnetic resonance imaging slots on weekly prostate cancer imaging workload. Br J Radiol:93. 10.1259/bjr.20190929 [DOI] [PMC free article] [PubMed]
- 15.Thurtle D, Barrett T, Thankappan-Nair V, et al. Progression and treatment rates using an active surveillance protocol incorporating image-guided baseline biopsies and multiparametric magnetic resonance imaging monitoring for men with favourable-risk prostate cancer. BJU Int. 2018;122:59–65. doi: 10.1111/bju.14166. [DOI] [PubMed] [Google Scholar]
- 16.Moore CM, Giganti F, Albertsen P, et al. Reporting magnetic resonance imaging in men on active surveillance for prostate cancer: the PRECISE recommendations—a report of a European School of Oncology Task Force. Eur Urol. 2017;71:648–655. doi: 10.1016/j.eururo.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 17.Dieffenbacher S, Nyarangi-Dix J, Giganti F et al (2019) Standardized magnetic resonance imaging reporting using the prostate cancer radiological estimation of change in sequential evaluation criteria and magnetic resonance imaging/transrectal ultrasound fusion with transperineal saturation biopsy to select men on active surveillance. Eur Urol Focus. 10.1016/j.euf.2019.03.001 [DOI] [PubMed]
- 18.Giganti F, Stabile A, Stavrinides V, et al. Natural history of prostate cancer on active surveillance: stratification by MRI using the PRECISE recommendations in a UK cohort. Eur Radiol. 2020;31:1644–1655. doi: 10.1007/s00330-020-07256-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ullrich T, Arsov C, Quentin M, et al. Multiparametric magnetic resonance imaging can exclude prostate cancer progression in patients on active surveillance: a retrospective cohort study. Eur Radiol. 2020;30:6042–6051. doi: 10.1007/s00330-020-06997-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Connor LP, Wang AZ, Yerram NK et al (2020) Changes in magnetic resonance imaging using the prostate cancer radiologic estimation of change in sequential evaluation criteria to detect prostate cancer progression for men on active surveillance. Eur Urol Oncol. 10.1016/j.euo.2020.09.004 [DOI] [PMC free article] [PubMed]
- 21.Caglic I, Sushentsev N, Gnanapragasam V, et al. MRI-derived PRECISE scores for predicting pathologically-confirmed radiological progression in prostate cancer patients on active surveillance. Eur Radiol. 2020;2020:1–10. doi: 10.1007/s00330-020-07336-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fave X, Zhang L, Yang J et al (2017) Delta-radiomics features for the prediction of patient outcomes in non-small cell lung cancer. Sci Rep 7. 10.1038/s41598-017-00665-z [DOI] [PMC free article] [PubMed]
- 23.Caglic I, Barrett T. Optimising prostate mpMRI: prepare for success. Clin Radiol. 2019;74:831–840. doi: 10.1016/j.crad.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Kuru TH, Wadhwa K, Chang RTM, et al. Definitions of terms, processes and a minimum dataset for transperineal prostate biopsies: a standardization approach of the Ginsburg Study Group for enhanced prostate diagnostics. BJU Int. 2013;112:568–577. doi: 10.1111/bju.12132. [DOI] [PubMed] [Google Scholar]
- 25.Barrett T, Slough R, Sushentsev N et al (2019) Three-year experience of a dedicated prostate mpMRI pre-biopsy programme and effect on timed cancer diagnostic pathways. Clin Radiol:74. 10.1016/j.crad.2019.06.004 [DOI] [PubMed]
- 26.Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 27.de Rooij M, Israël B, Tummers M et al (2020) ESUR/ESUI consensus statements on multi-parametric MRI for the detection of clinically significant prostate cancer: quality requirements for image acquisition, interpretation and radiologists’ training. Eur Radiol. 10.1007/s00330-020-06929-z* [DOI] [PMC free article] [PubMed]
- 28.Barrett T, Padhani AR, Patel A et al (2020) Certification in reporting multiparametric magnetic resonance imaging of the prostate: recommendations of a UK consensus meeting. BJU Int. 10.1111/bju.15285 [DOI] [PubMed]
- 29.van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zwanenburg A, Vallières M, Abdalah MA, et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295:328–338. doi: 10.1148/radiol.2020191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fornacon-Wood I, Mistry H, Ackermann CJ, et al. Reliability and prognostic value of radiomic features are highly dependent on choice of feature extraction platform. Eur Radiol. 2020;30:6241–6250. doi: 10.1007/s00330-020-06957-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.IBSI (2021) Image Biomarker Standardisation Initiative. https://theibsi.github.io/ibsi2/. Accessed 6 May
- 34.Zwanenburg A, Leger S, Agolli L, et al. Assessing robustness of radiomic features by image perturbation. Sci Rep. 2019;9:1–10. doi: 10.1038/s41598-018-36938-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitwell HJ, Blyuss O, Menon U, Timms JF, Zaikin A (2018) Parenclitic networks for predicting ovarian cancer. Oncotarget 9:22717–22726. 10.18632/oncotarget.25216 [DOI] [PMC free article] [PubMed]
- 36.Tibshirani R (1996) Regression shrinkage and selection via the lasso. J R Stat Soc Ser B 58. 10.1111/j.2517-6161.1996.tb02080.x
- 37.Ziegel ER (2003) The elements of statistical learning. Technometrics 45. 10.1198/tech.2003.s770
- 38.Carpenter J, Bithell J. Bootstrap confidence intervals: when, which, what? A practical guide for medical statisticians. Stat Med. 2000;19:1141–1164. doi: 10.1002/(SICI)1097-0258(20000515)19:9<1141::AID-SIM479>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 39.DeLong ER, DeLong DM, Clarke-Pearson DL (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics:44. 10.2307/2531595 [PubMed]
- 40.Altman DG, Bland JM. Statistics notes: diagnostic tests 2: predictive values. BMJ. 1994;309:102. doi: 10.1136/bmj.309.6947.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang F, Ford JC, Dogan N, et al. Magnetic resonance imaging (MRI)-based radiomics for prostate cancer radiotherapy. Transl Androl Urol. 2018;7:445–458. doi: 10.21037/tau.2018.06.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Escande A, Betrouni N, Tresch E, et al. Magnetic resonance imaging based delta radiomics as biomarker for intermediate risk prostate cancer treated by hypofractionated stereotactic boost: a phase II CKNO PRO Ancillary Study. Int J Radiat Oncol. 2019;105:E274–E275. doi: 10.1016/j.ijrobp.2019.06.1897. [DOI] [Google Scholar]
- 43.Shi L, Rong Y, Daly M, et al. Cone-beam computed tomography-based delta-radiomics for early response assessment in radiotherapy for locally advanced lung cancer. Phys Med Biol. 2020;65:15009–15019. doi: 10.1088/1361-6560/ab3247. [DOI] [PubMed] [Google Scholar]
- 44.Boldrini L, Cusumano D, Chiloiro G, et al. Delta radiomics for rectal cancer response prediction with hybrid 0.35 T magnetic resonance-guided radiotherapy (MRgRT): a hypothesis-generating study for an innovative personalized medicine approach. Radiol Med. 2019;124:145–153. doi: 10.1007/s11547-018-0951-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeon SH, Song C, Chie EK, et al. Delta-radiomics signature predicts treatment outcomes after preoperative chemoradiotherapy and surgery in rectal cancer. Radiat Oncol. 2019;14:43. doi: 10.1186/s13014-019-1246-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mazzei MA, Nardone V, Di Giacomo L, et al. The role of delta radiomics in gastric cancer. Quant Imaging Med Surg. 2018;8:719–721. doi: 10.21037/qims.2018.07.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin P, Yang PF, Chen S, et al. A Delta-radiomics model for preoperative evaluation of neoadjuvant chemotherapy response in high-grade osteosarcoma. Cancer Imaging. 2020;20:7. doi: 10.1186/s40644-019-0283-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crombé A, Périer C, Kind M, et al. T 2 -based MRI Delta-radiomics improve response prediction in soft-tissue sarcomas treated by neoadjuvant chemotherapy. J Magn Reson Imaging. 2019;50:497–510. doi: 10.1002/jmri.26589. [DOI] [PubMed] [Google Scholar]
- 49.Zanin M, Alcazar JM, Carbajosa JV, et al. Parenclitic networks: uncovering new functions in biological data. Sci Rep. 2014;4:1–6. doi: 10.1038/srep05112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rundo L, Militello C, Russo G et al (2017) Automated prostate gland segmentation based on an unsupervised fuzzy C-means clustering technique using multispectral T1w and T2w MR imaging. Inf. 10.3390/info8020049
- 51.Alkadi R, Taher F, El-baz A, Werghi N (2019) A deep learning-based approach for the detection and localization of prostate cancer in T2 magnetic resonance images. J Digit Imaging. 10.1007/s10278-018-0160-1 [DOI] [PMC free article] [PubMed]
- 52.Stoyanova R, Takhar M, Tschudi Y, et al. Prostate cancer radiomics and the promise of radiogenomics. Transl Cancer Res. 2016;5:432–447. doi: 10.21037/tcr.2016.06.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fehr D, Veeraraghavan H, Wibmer A, et al. Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proc Natl Acad Sci U S A. 2015;112:E6265–E6273. doi: 10.1073/pnas.1505935112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 1238 kb)