Abstract
Aim/hypothesis
Five subgroups were described in European diabetes patients using a data driven machine learning approach on commonly measured variables. We aimed to test the applicability of this phenotyping in Indian individuals with young-onset type 2 diabetes.
Methods
We applied the European-derived centroids to Indian individuals with type 2 diabetes diagnosed before 45 years of age from the WellGen cohort (n = 1612). We also applied de novo k-means clustering to the WellGen cohort to validate the subgroups. We then compared clinical and metabolic-endocrine characteristics and the complication rates between the subgroups. We also compared characteristics of the WellGen subgroups with those of two young European cohorts, ANDIS (n = 962) and DIREVA (n = 420). Subgroups were also assessed in two other Indian cohorts, Ahmedabad (n = 187) and PHENOEINDY-2 (n = 205).
Results
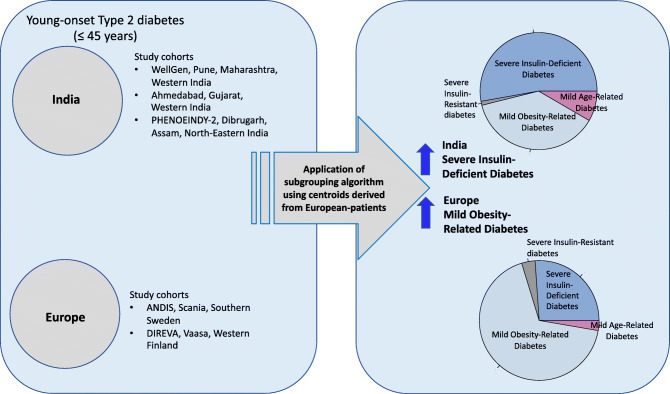
Both Indian and European young-onset type 2 diabetes patients were predominantly classified into severe insulin-deficient (SIDD) and mild obesity-related (MOD) subgroups, while the severe insulin-resistant (SIRD) and mild age-related (MARD) subgroups were rare. In WellGen, SIDD (53%) was more common than MOD (38%), contrary to findings in Europeans (Swedish 26% vs 68%, Finnish 24% vs 71%, respectively). A higher proportion of SIDD compared with MOD was also seen in Ahmedabad (57% vs 33%) and in PHENOEINDY-2 (67% vs 23%). Both in Indians and Europeans, the SIDD subgroup was characterised by insulin deficiency and hyperglycaemia, MOD by obesity, SIRD by severe insulin resistance and MARD by mild metabolic-endocrine disturbances. In WellGen, nephropathy and retinopathy were more prevalent in SIDD compared with MOD while the latter had higher prevalence of neuropathy.
Conclusions /interpretation
Our data identified insulin deficiency as the major driver of type 2 diabetes in young Indians, unlike in young European individuals in whom obesity and insulin resistance predominate. Our results provide useful clues to pathophysiological mechanisms and susceptibility to complications in type 2 diabetes in the young Indian population and suggest a need to review management strategies.
Graphical abstract
Supplementary Information
The online version contains peer-reviewed but unedited supplementary material available at 10.1007/s00125-021-05543-y.
Keywords: Europe, India, Insulin deficiency, Subgroups, Type 2 diabetes, Young-onset type 2 diabetes
Introduction
Type 2 diabetes has been traditionally considered as one disease characterised by both insulin resistance and insulin deficiency. Nevertheless, the disease is heterogeneous [1]. A formal description of five distinct subgroups was proposed in a large Swedish cohort [2] and replicated in other populations [2–5]: severe autoimmune diabetes (SAID) severe insulin-deficient diabetes (SIDD); severe insulin-resistant diabetes (SIRD); mild obesity-related diabetes (MOD); and mild age-related diabetes (MARD). The subgroups differed not only in their clinical characteristics at diagnosis but also in their pathophysiological mechanisms and susceptibility to complications.
India is referred to as one of the diabetes capitals of the world, and Indian individuals with type 2 diabetes differ from Europeans in that they develop diabetes at a younger age and are thinner [6, 7]. Indians also differ in body composition, having higher fat and lower lean proportions at the same BMI [8]. Given the role of adiposity in insulin resistance, it has therefore been assumed that type 2 diabetes in Indians is primarily driven by insulin resistance [9]. However, it is increasingly recognised that insulin deficiency may be a significant driver of diabetes in Indians [10]. Recent studies show that both the increase in the diabetes prevalence and the characteristics of the affected individuals vary in different parts of India [11, 12]. Lean type 2 diabetes is prevalent in India, especially in undernourished regions [13]. Recent studies have shown that subgroups of type 2 diabetes in Indians show partial concordance with those in Europeans [14, 15].
Type 2 diabetes is diagnosed at a younger age in India and its pathophysiology and heterogeneity warrants further investigation. Younger age at diagnosis has distinct implications for treatment, long-term complications and mortality as well as socioeconomic burden [16]. Therefore, early identification of subclasses may be vital for appropriate treatment to reduce adverse outcomes [17]. To address this, we implemented the Swedish algorithm [2] to identify subgroups of young Indians with type 2 diabetes diagnosed before 45 years of age from the WellGen cohort from Pune, India [18]. We then compared Indian and European type 2 diabetes subgroups to obtain information on the relative distributions and characteristics in the two populations. We also performed de novo clustering of individuals from the WellGen study to assess whether clusters obtained were similar in proportion and characteristics to those from the European-derived centroids. Finally, we investigated the subclassification of type 2 diabetes in cohorts from two other geographical regions across India.
Methods
Study population
WellGen (Pune, Maharashtra, western India)
The WellGen study includes patients visiting the Diabetes Unit, KEM Hospital, Pune and associated clinics for routine diabetes management between 2004 and 2006 [18]. Individuals diagnosed with diabetes below 45 years of age using the WHO guidelines were included [1]. Diagnosis of type 2 diabetes was based on clinical criteria: age at diagnosis >20 years; no history of ketoacidosis; central obesity (waist–hip ratio: WHR >0.80 in women and >0.90 in men); and response to treatment with oral glucose-lowering agents. Individuals with a clinical diagnosis of type 1 diabetes (diagnosis before 20 years of age, history of ketoacidosis, continuous insulin treatment since diagnosis), fibrocalculous pancreatic diabetes (FCPD) or fulfilling criteria for monogenic diabetes were excluded. In total, 1612 individuals were included (Table 1).
Table 1.
Clinical and biochemical characteristics of participants enrolled in WellGen and ANDIS study with age at diagnosis less than 45 years
| Characteristic | WellGen | ANDIS | WellGen vs ANDIS | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Male participants | Female participants | All | Male participants | Female participants | All | p value (male participants) | p value (female participants) | p value (all) | |
| No. of participants | 902 | 710 | 1612 | 567 | 395 | 962 | |||
| Age at diabetes diagnosis, years | 37.39 (5.58) | 36.47 (5.99) | 36.98 (5.78) | 39.13 (5.8) | 38.4 (6.16) | 38.83 (5.96) | <0.0001a | <0.0001a | <0.0001a |
| BMI, kg/m2 | 25.43 (3.74) | 27.14 (4.27) | 26.18 (4.07) | 32.47 (6.60) | 33.66 (7.30) | 32.96 (6.92) | <0.0001a | <0.0001a | <0.0001a |
| Fasting glucose, mmol/l | 9.08 (3.23) | 9.48 (3.45) | 9.26 (3.33) | 9.28 (3.36) | 8.34 (2.80) | 8.89 (3.18) | 0.23 | <0.0001a | 0.007a |
| HbA1c, mmol/mol | 73.23 (23.04) | 71.62 (22.19) | 72.52 (22.68) | 75.09 (25.55) | 60.93 (24.03) | 69.28 (25.88) | 0.16 | <0.0001a | 0.0008a |
| HbA1c, % | 8.8 (4.2) | 8.7 (4.2) | 9.1 (4.2) | 9.0 (4.5) | 7.7 (4.4) | 8.5 (4.5) | 0.16 | <0.0001a | 0.0008a |
| Fasting C-peptide, nmol/l | 0.76 (0.46) | 0.76 (0.45) | 0.76 (0.46) | 1.20 (0.57) | 1.97 (0.56) | 1.20 (0.56) | <0.0001a | <0.0001a | <0.0001a |
| HOMA2-B | 58.7 (40.28) | 56.39 (41.93) | 57.69 (41.02) | 81.09 (50.59) | 88.36 (45.48) | 84.11 (48.91) | <0.0001a | <0.0001a | <0.0001a |
| HOMA2-IR | 2.12 (1.38) | 2.15 (1.35) | 2.13 (1.37) | 3.27 (1.62) | 3.12 (1.51) | 3.21 (1.58) | <0.0001a | <0.0001a | <0.0001a |
Values are mean (SD)
aBonferroni corrected significant p values
p values were calculated using t test
Clinical information including age, sex, age at diabetes diagnosis, family history and socioeconomic status was obtained through a standardised questionnaire. Height, weight, waist and hip circumferences, and BP were measured using standardised methods [18, 19]. Fasting plasma glucose, total cholesterol, HDL-cholesterol, triacylglycerols and HbA1c were measured using standard laboratory assays as described previously [18, 19]. Fasting C-peptide was measured by ELISA (Diagnostic Biochem Canade, ON, Canada). Fasting glucose and C-peptide measurements were used to calculate Homeostatic model assessment 2 estimates of β-cell function (HOMA2-B) and insulin resistance (HOMA2-IR) values [20, 21]. Details of treatment (insulin, oral glucose-, BP- and lipid-lowering medication) were recorded.
Cardiovascular complications were defined by ICD-10 codes (http://apps.who.int/classifications/icd10/browse/2016/en): Coronary artery disease (CAD) I20–21, I24, I251 and I253–259; stroke I60–61 and I63–64. Nephropathy was diagnosed by urine strip albumin measurement (nil, trace, and +), and by eGFR calculation (ml min−1 [1.73 m body surface area]−2) by the Modification of Diet in Renal Disease (MDRD) formula (>90 ml min−1 [1.73 m]−2 normal; and 90–60 ml min−1 [1.73 m]−2 mild, 60–30 ml min−1 [1.73 m]−2 moderate and <30 ml min−1 [1.73 m]−2 severe chronic kidney disease [CKD]). Diagnosis of retinopathy was based on dilated fundus examination performed by an ophthalmologist and was classified as non-proliferative diabetic retinopathy or proliferative diabetic retinopathy. Peripheral neuropathy was diagnosed by biothesiometer (non-perception of vibration sense at 15 or higher amperes at two or more sites on the feet) (Table 2).
Table 2.
Complications and current treatment for participants of the WellGen and ANDIS studies stratified by sex
| Complication/treatment | WellGen | ANDIS | ||||||
|---|---|---|---|---|---|---|---|---|
| Male participants | Female participants | Total (%) | p value | Male participants | Female participants | Total (%) | p value | |
| n (%) | 902 (56.0) | 710 (44.0) | 1612 (100) | 567 (58.9) | 395 (41.1) | 962 (100) | ||
| Duration of diabetes, years | 9.73 (8.57) | 9.70 (7.74) | 9.72 (8.21) | 4.27 (2.56) | 4.27 (2.34) | 4.27 (2.47) | ||
| Current treatment | ||||||||
| Diet only | 77 (8.5) | 58 (8.2) | 135 (8.4) | 0.791 | 34 (6.0) | 43 (10.9) | 77 (8.0) | 0.006a |
| OHA only | 558 (61.9) | 421 (59.3) | 979 (60.7) | 0.295 | 382 (67.4) | 247 (62.5) | 629 (65.4) | 0.121 |
| Only on insulin | 51 (5.7) | 34 (4.8) | 85 (5.3) | 0.440 | 28 (4.9) | 31 (7.8) | 59 (6.1) | 0.064 |
| Both OHA and insulin | 216 (23.9) | 197 (27.7) | 413 (25.6) | 0.083 | 122 (21.5) | 72 (18.2) | 194 (20.2) | 0.211 |
| Complications | ||||||||
| CVD (Coronary events and/or stroke)b | 89 (9.9) | 31 (4.4) | 120 (7.4) | 0.0001a | 27 (4.8) | 7 (1.8) | 34 (3.6) | 0.013 |
| Coronary event | 76 (8.4) | 24 (3.3) | 100 (6.2) | 0.0001a | 19 (3.4) | 4 (1.0) | 23 (2.4) | 0.019 |
| Stroke | 17 (1.9) | 9 (1.3) | 26 (1.7) | 0.329 | 8 (1.4) | 3 (0.8) | 11 (1.2) | 0.348 |
| Nephropathy (proteinuria and/or CKD)c | 361 (40.0) | 200 (28.2) | 561 (34.8) | 0.0001a | 181 (45.9) | 129 (45.7) | 310 (45.9) | 0.960 |
| Macroalbuminuriad | 167 (19.2) | 83 (12.3) | 250 (16.2) | 0.0001a | 61 (17.1) | 29 (11.4) | 90 (14.7) | 0.047 |
| CKD: eGFRe, f | 268 (32.3) | 145 (22.7) | 413 (28.1) | 0.0001a | 142 (26.0) | 109 (28.7) | 251 (27.1) | 0.359 |
| Early CKD (60–90 ml min−1 [1.73m]−2) | 208 (25.0) | 122 (19.1) | 330 (22.4) | 0.007 | 132 (24.6) | 105 (27.9) | 237 (26.0) | 0.257 |
| Moderate (30–60 ml min−1 [1.73m]−2) | 54 (6.5) | 20 (3.1) | 74 (5.0) | 0.003a | 7 (1.3) | 4 (1.1) | 11 (1.2) | 0.747 |
| Severe (<30 ml min−1 [1.73m]−2) | 6 (0.7) | 3 (0.5) | 9 (0.6) | 0.537 | 3 (0.5) | 0 (0.0) | 3 (0.3) | 0.148 |
| Retinopathy (n=657)g | 102 (29.4) | 78 (25.2) | 180 (27.4) | 0.225 | 34 (19.4) | 23 (20.0) | 57 (19.8) | 0.905 |
| NPDR | 93 (26.8) | 74 (23.9) | 167 (25.4) | 0.389 | 33 (19.0) | 22 (19.3) | 55 (19.1) | 0.944 |
| PDR | 9 (2.6) | 4 (1.3) | 13 (2.0) | 0.231 | 1 (0.6) | 1 (0.9) | 2 (0.7) | 0.764 |
| Neuropathyh | 359 (40.7) | 351 (50.6) | 710 (45.1) | 0.0001a | 14 (2.5) | 2 (0.5) | 16 (1.7) | 0.019 |
Values are n (%)
aBonferroni corrected significant p values
bCVD: data available for n=1612 (Male n=902, Female n=710) for WellGen, n=956 (Male n=563, Female n=393) for ANDIS
cNephropathy: data available for n=1612 (Male n=902, Female n=710) for WellGen, n=676 (Male n=394, Female n=282) for ANDIS
dMacroalbuminuria: data available for n=1544 (Male n=869, Female n=675) for WellGen, n=611(Male n=356, Female n=255) for ANDIS
eBased on MDRD formula
fCKD: eGFR: data available for n=1471(Male n=831, Female n=640) for WellGen and n=927 (Male n=547, Female n=380) for ANDIS
gDiabetic retinopathy: data available for n=657 (Male n=347, Female n=310) for WellGen and n=288 (Male n=174, Female n=114) for ANDIS
hNeuropathy: data available for n=1576 (Male n=883, Female n=693), diagnosed using Biothesiometry for WellGen, n=956 (Male n=563, Female n=393), ICD codes ICD-10 = E104 or E114 for ANDIS
p values were calculated by χ2 test
NPDR, non-proliferative diabetic retinopathy; OHA, oral glucose-lowering agent; PDR, proliferative diabetic retinopathy
Ahmedabad (Gujarat, western India)
Patients with a clinical diagnosis of type 2 diabetes visiting the DiaCare Clinic, Ahmedabad during 2018–2019 diagnosed below 45 years of age and duration of diabetes less than 2 years were invited to participate and 187 individuals consented. Measurements included anthropometry, HbA1c levels and a fasting measurement of plasma glucose and C-peptide (MAGLUMI C-peptide; CLIA, Shenzhen, China) (electronic supplementary material [ESM] Table 1).
PHENOEINDY-2 (Dibrugarh, Assam, North-east India)
Patients with clinical diagnosis of type 2 diabetes attending the medical outpatient department of Assam Medical College, Dibrugarh during 2017–2019 if diagnosed below 40 years of age were invited to participate and 205 individuals consented. Measurements included anthropometry, HbA1c levels, a fasting measurement of plasma glucose and C-peptide (ELISA, Diagnostic Biochem Canade, ON, Canada) (ESM Table 2).
ANDIS (Scania, Southern Sweden)
The ANDIS project comprises newly diagnosed diabetic individuals aged >18 years in Scania County, Sweden between 2008 and 2016 [2]. Biochemical and anthropometric measurements and presence of complications were recorded as described elsewhere [2]. For the current study, 962 individuals diagnosed with type 2 diabetes before 45 years of age were included. We excluded any person with known type 1 diabetes, monogenic diabetes and GAD antibody positivity (so-called SAID) to maintain concordance with the WellGen study. The prevalence of complications (diagnosed as described previously [2]; Table 2) was recorded ~4.2 years after diagnosis.
DIREVA (Vaasa, Western Finland)
DIREVA includes 5107 individuals with diabetes recruited from 2009 to 2014 in the Vaasa Hospital District. For the current study, 424 individuals with type 2 diabetes diagnosed below 45 years of age were included; exclusion criteria were similar to those for ANDIS (ESM Table 3). Biochemical and anthropometric measurements have been described elsewhere [2]. No treatment or complication data from DIREVA have been included in the current study.
Ethics statement
All studies were approved by the local/regional Institutional Ethics Committees, and all participants gave written informed consent.
Statistical methods
Participants with measurements above or below 5 SDs from the mean for the clustering parameters were excluded from the analysis and values outside the limits for HOMA2 calculation (fasting glucose <3 mmol/l or >25 mmol/l; C-peptide <0.2 ng/ml or >3.5 ng/ml) were capped to the proximal upper or lower limits. To perform supervised clustering in relation to the European-derived cluster coordinates, phenotypes (age at diagnosis, HbA1c, HOMA2-B, HOMA2-IR and BMI) were scaled using the same scaling parameters (mean and SD) as described previously [2]. Due to the unavailability of GAD autoantibody data in the Indian study (WellGen), we only included clusters 2–5 (SIDD, SIRD, MOD and MARD). Participants were assigned to the predetermined clusters on the basis of which ANDIS cluster they were most similar to, calculated as their Euclidean distance from the nearest cluster centre derived from ANDIS coordinates. The nearest centroid method was used to find the nearest centroid (as measured with Euclidean distances) for each individual. This resulted in each participant being assigned to any of the four clusters: 2/SIDD; 3/SIRD; 4/MOD; or 5/MARD. Given the wide range of duration of diabetes in WellGen, we performed a sensitivity analysis by separately assessing the type 2 diabetes subgroups among those within 5 years of diagnosis and those above.
To perform unsupervised clustering, all previously mentioned variables were used in a separate analysis. Given that the results from the supervised clustering analysis showed a strong bias towards the 2/SIDD and 4/MOD cluster, and the silhouette analysis indicated that two was the most stable number of clusters, we performed k-means clustering into two clusters. All phenotypes were scaled to have a mean of 0 and an SD of 1, this time with scaling parameters derived from the data itself. k-means clustering was then performed separately for female and male participants using the k-means runs algorithm from the fpc package version 2.1–11.1 (https://CRAN.R-project.org/package=fpc) in R version 3.4 [22].
We investigated differences between groups (male and female participants or between four subgroups) by t test or ANOVA for clinical characteristics and by χ2 test for complications. To compare differences in the rate of complications between subgroups, we also used logistic regression with adjustment for age at diagnosis, sex and duration of diabetes. We used SPSS version 22.0 for these analyses.
Type 1 diabetes genetic risk scores
In the absence of GAD autoantibody data, we applied a previously validated ‘type 1’ genetic risk score (GRS) (ESM Table 4) to 560 WellGen participants with available data to estimate the proportion of those carrying autoimmune risk alleles. The characteristics of these individuals did not differ from the characteristics of those for whom genotyping was not available (ESM Table 5). A positive control group comprised 261 individuals with type 1 diabetes, as described previously [23]. A negative control group comprised 461 participants with normal glucose tolerance (75 g OGTT; WHO 1999 criteria) from the Pune Maternal Nutrition Study (PMNS) [24].
Genotyping
Genome-wide genotyping data was generated on WellGen and PMNS participants using Affymetrix SNP 6.0 Chips (Affymetrix, CA, USA) and the Infinium Global Screening Array V1 B37 (Illumina, San Diego, CA, USA) for the type 1 diabetes cohort. Quality control and imputation were performed as described in ESM Methods.
GRS
A previously described set of nine SNPs was used for type 1 diabetes GRS calculations (ESM Table 4) [23, 25, 26]. In the absence of genotyping data for rs7454108, a proxy SNP rs3957146 (linkage disequilibrium [LD]: r2 = 1, D′ = 1) was used. LD is the non-random association of alleles at multiple DNA markers resulting from their close proximity to one another within a chromosome and are therefore inherited together. Classical LD measures include D′ and r2, where D′ and r2 values >0.8 indicate a higher degree of co-inheritance. The haplotype was constructed using rs2187668 + rs3957146 as described previously [23] and GRS scores were computed on PLINK 1.09 (http://pngu.mgh.harvard.edu/~purcell/plink/) [27] using weighted scores. Logistic regression was performed to assess the discriminatory power of GRS between type 1 diabetes and other subgroups.
Results
We first sought to investigate the subgroups of individuals with young-onset type 2 diabetes in the Indian WellGen study and compare them with Swedish subgroups from the ANDIS study.
The WellGen study comprised individuals diagnosed with type 2 diabetes before 45 years of age, and all relevant data required for clustering were available for 1624 participants. After applying exclusion criteria, 1612 individuals (56% men) with mean age at diagnosis of 37 years, duration of diabetes ~10 years and BMI 26.18 kg/m2 were included (Table 1 and ESM Fig. 1).
For comparison, we selected 962 participants with type 2 diabetes (58.9% men, mean age at diagnosis 38.83 years) from the ANDIS study, after excluding 577 individuals belonging to cluster 1 (SAID) [2]. The Indian participants were younger at diagnosis, had lower BMI, higher fasting plasma glucose and lower fasting C-peptide, HOMA2-B and HOMA2-IR compared with the ANDIS sub-cohort (Table 1). The proportion of participants receiving lifestyle management alone, glucose-lowering oral agents and insulin treatment was broadly similar in both cohorts (Table 2). In the WellGen study, men had a higher prevalence of cardiovascular events and nephropathy compared with women, whereas the prevalence of neuropathy was higher in women. There was no difference with respect to these complications between men and women in the ANDIS study (Table 2). We did not compare the complication rates between the two cohorts because of the difference in duration of diabetes.
SIDD predominates in India, MOD in Sweden
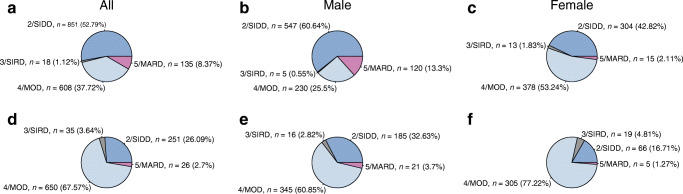
In the absence of GAD autoantibody data, we obtained the four expected clusters, albeit with different proportions. In the WellGen study, the SIDD cluster was the largest subgroup (52.8%), followed by the mild obesity-related MOD (37.7%), while severe insulin-resistant SIRD (1.1%) and mild age-related MARD (8.4%) were less common (Fig. 1, Table 3). In a sensitivity analysis, with increasing duration of diabetes (from <5 years to >5 years), the proportion of participants in the SIDD subgroup increased (from 45.5% to 56.9%) while that in MOD group decreased (from 44.7% to 33.8%) (ESM Table 6).
Fig. 1.
Distribution of participants from the WellGen and ANDIS study in the predefined clusters. (a–c) Distribution of WellGen participants, showing all participants (n=1612) (a), men with diabetes (n=902) (b) and women with diabetes (n=710) (c). (d–f) Distribution of ANDIS participants, showing all participants (n=962) (d), men with diabetes (n=567) (e) and women with diabetes (n=395) (f)
Table 3.
Characteristics of participants enrolled in WellGen and ANDIS study by clusters for all participants, and for male and female participants
| Characteristic | WellGen | ANDIS | WellGen vs ANDIS | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SIDD | SIRD | MOD | MARD | p value | p1 value | SIDD | SIRD | MOD | MARD | p value | p1 value | p value for SIDD | p value for MOD | |
| All participants (N = 1612 for WellGen, N = 962 for ANDIS) | ||||||||||||||
| Participants in subroup, n (%) | 851 (52.79) | 18 (1.12) | 608 (37.72) | 135 (8.37) | 251 (26.09) | 35 (3.64) | 650 (67.57) | 26 (2.7) | ||||||
| Age at diagnosis, years | 36.86 (5.81) | 38.83 (4.93) | 36.25 (5.77) | 40.84 (3.98) | 0.005a | <0.0001a | 38.98 (6.24) | 39.94 (5.87) | 38.56 (5.9) | 42.5 (2.79) | <0.0001a | <0.0001a | <0.0001a | <0.0001a |
| Duration of diabetes, years | 10.49 (8.25) | 6.41 (7.16) | 8.72 (8.03) | 9.83 (8.44) | 0.001a | -- | -- | -- | -- | -- | -- | -- | -- | -- |
| BMI, kg/m2 | 25.02 (3.45) | 28.37 (5.27) | 28.4 (4.03) | 23.22 (2.44) | <0.0001a | <0.0001a | 28.29 (5.43) | 36.62 (6.28) | 34.92 (6.41) | 24.11 (2.33) | <0.0001a | <0.0001a | <0.0001a | <0.0001a |
| Fasting glucose, mmol/l | 10.77 (3.39) | 6.86 (1.89) | 7.85 (2.37) | 6.50 (1.47) | <0.0001a | <0.0001a | 11.75 (3.67) | 7.05 (2.22) | 7.97 (2.25) | 6.75 (1.26) | <0.0001a | <0.0001a | <0.0001a | 0.06 |
| HbA1c, mmol/mol | 87.01 (19.34) | 56.7 (13.1) | 58.33 (12.92) | 47.25 (10.37) | <0.0001a | <0.0001a | 100.6 (19.62) | 55.19 (13.79) | 58.87 (17.64) | 45.85 (7.41) | <0.0001a | <0.0001a | <0.0001a | 0.53 |
| HbA1c, % | 10.1 (3.9) | 7.3 (3.3) | 7.5 (3.3) | 6.5 (3.1) | <0.0001a | <0.0001a | 11.4 (4.0) | 7.2 (3.4) | 7.5 (3.8) | 6.3 (2.8) | <0.0001a | <0.0001a | <0.0001a | 0.53 |
| Fasting C-peptide, nmol/l | 0.68 (0.39) | 1.01 (0.62) | 0.89 (0.48) | 0.58 (0.29) | <0.0001a | <0.0001a | 0.85 (0.4) | 2.53 (0.61) | 1.29 (0.48) | 0.73 (0.24) | <0.0001a | <0.0001a | <0.0001a | <0.0001a |
| HOMA2-B | 37.66 (22.24) | 214.99 (41.01) | 77.49 (42.02) | 73.74 (31.16) | <0.0001a | <0.0001a | 41.03 (28.34) | 968.71 (56.43) | 94.79 (38.53) | 80.15 (27.62) | <0.0001a | <0.0001a | 0.048 | <0.0001a |
| HOMA2-IR | 2.08 (1.41) | 4.73 (1.56) | 2.28 (1.29) | 1.42 (0.76) | 0.314 | <0.0001a | 2.67 (1.42) | 6.17 (1.86) | 3.31 (1.42) | 1.78 (0.63) | <0.0001a | <0.0001a | 0.043 | <0.0001a |
| Male participants (n = 902 for WellGen, n = 567 for ANDIS) | ||||||||||||||
| Participants in subgroup, n (%) | 547 (60.6) | 5 (0.55) | 230 (25.50) | 120 (13.30) | 185 (32.6) | 16 (2.82) | 345 (60.85) | 21 (3.7) | ||||||
| Age at diagnosis, years | 36.96 (5.67) | 42.07 (10.27) | 36.59 (5.43) | 40.7 (4.12) | <0.0001a | <0.0001a | 38.91 (6.08) | 40.56 (6.19) | 39.0 (5.73) | 42.05 (2.87) | 0.08 | 0.089 | <0.0001a | <0.0001a |
| Duration of diabetes, years | 10.55 (8.70) | 8.03 (6.59) | 7.71 (7.98) | 9.98 (8.56) | 0.005a | -- | ||||||||
| BMI, kg/m2 | 24.44 (3.08) | 28.75 (3.54) | 28.77 (3.67) | 23.41 (2.34) | <0.0001a | <0.0001a | 27.45 (3.9) | 36.74 (6.01) | 35.44 (5.9) | 24.72 (2.11) | <0.0001a | <0.0001a | <0.0001a | <0.0001a |
| Fasting glucose, mmol/l | 10.29 (3.22) | 5.75 (1.0) | 7.63 (2.43) | 6.52 (1.53) | <0.0001a | <0.0001a | 11.80 (3.5) | 7.58 (3.04) | 8.16 (2.53) | 6.77 (2.53) | <0.0001a | <0.0001a | <0.0001a | 0.013 |
| HbA1c, mmol/mol | 84.67 (20.57) | 49.07 (10.27) | 59.94 (13.35) | 47.55 (10.02) | <0.0001a | <0.0001a | 99.58 (20.31) | 57.33 (13.82) | 64.59 (18.46) | 45.30 (7.11) | <0.0001a | <0.0001a | <0.0001a | 0.001a |
| HbA1c, % | 9.9 (4.0) | 6.6 (3.1) | 7.6 (3.3) | 6.5 (3.1) | <0.0001a | <0.0001a | 11.3 (4.0) | 7.4 (3.4) | 8.1 (3.8) | 6.3 (2.8) | <0.0001a | <0.0001a | <0.0001a | 0.001a |
| Fasting C-peptide, nmol/l | 0.66 (0.38) | 2.17 (0.44) | 1.08 (0.51) | 0.60 (0.30) | <0.0001a | <0.0001a | 0.82 (0.35) | 2.65 (0.61) | 1.36 (0.49) | 0.72 (0.49) | <0.0001a | <0.0001a | <0.0001a | <0.0001a |
| HOMA2-B | 39.33 (22.5) | 229.16 (40.98) | 92.27 (42.49) | 75.58 (31.92) | <0.0001a | <0.0001a | 39.15 (25.31) | 200.56 (74.37) | 98.19 (40.98) | 78.57 (27.41) | <0.0001a | <0.0001a | 0.92 | 0.094 |
| HOMA2-IR | 1.96 (1.34) | 4.99 (1.23) | 2.76 (1.41) | 1.47 (0.78) | 0.092 | <0.0001a | 2.59 (1.27) | 6.67 (2.23) | 3.56 (1.48) | 1.77 (0.65) | <0.0001a | <0.0001a | <0.0001a | <0.0001a |
| Female participants (n = 710 for WellGen, n = 395 for ANDIS) | ||||||||||||||
| Participants in subgroup, n (%) | 304 (42.82) | 13 (1.83) | 378 (53.24) | 15 (2.11) | 66 (16.7) | 19 (4.8) | 305 (77.2) | 5 (1.26) | ||||||
| Age at diagnosis, years | 36.68 (6.06) | 37.51 (4.92) | 36.05 (5.96) | 41.98 (2.51) | 0.722 | 0.001a | 39.2 (6.72) | 39.42 (5.71) | 38.07 (6.06) | 44.4 (1.34) | 0.03 | 0.045 | 0.0028a | <0.0001a |
| Duration of diabetes, years | 10.38 (7.37) | 5.79 (7.53) | 9.34 (8.00) | 8.57 (7.53) | 0.070 | -- | ||||||||
| BMI, kg/m2 | 26.07 (3.82) | 28.23 (5.92) | 28.18 (4.22) | 21.69 (2.75) | <0.0001a | <0.0001a | 30.65 (7.92) | 36.51 (6.67) | 34.33 (6.90) | 21.54 (1.19) | <0.0001a | <0.0001a | <0.0001a | <0.0001a |
| Fasting glucose, mmol/l | 11.64 (3.53) | 5.89 (1.44) | 7.98 (2.33) | 6.33 (0.97) | <0.0001a | <0.0001a | 11.61 (4.11) | 6.63 (1.08) | 7.76 (1.88) | 6.66 (1.62) | <0.0001a | <0.0001a | 0.92 | 0.19 |
| HbA1c, mmol/mol | 91.22 (16.11) | 59.63 (13.21) | 57.34 (12.57) | 44.85 (12.98) | <0.0001a | <0.0001a | 103.48 (17.35) | 53.39 (13.88) | 52.41 (14.12) | 48.15 (9.04) | <0.0001a | <0.0001a | <0.0001a | <0.0001a |
| HbA1c, % | 10.5 (3.6) | 7.6 (3.4) | 7.4 (3.3) | 6.3 (3.3) | <0.0001a | <0.0001a | 11.7 (3.7) | 7.0 (3.4) | 6.9 (3.4) | 6.6 (3.0) | <0.0001a | <0.0001a | <0.0001a | <0.0001a |
| Fasting C-peptide, nmol/l | 0.71 (0.41) | 1.98 (0.63) | 0.77 (0.42) | 0.42 (0.20) | 0.479 | <0.0001a | 0.90 (0.5) | 2.43 (0.61) | 1.19 (0.46) | 0.76 (0.23) | <0.0001a | <0.0001a | 0.0013a | <0.0001a |
| HOMA2-B | 34.67 (21.47) | 209.54 (41.32) | 68.5 (39.13) | 59.05 (19.27) | <0.0001a | <0.0001a | 46.29 (35.19) | 193.47 (37.16) | 90.94 (35.22) | 86.78 (30.71) | <0.0001a | <0.0001a | 0.00052a | <0.0001a |
| HOMA2-IR | 2.29 (1.49) | 4.63 (1.71) | 1.98 (1.12) | 1.03 (0.47) | <0.0001a | <0.0001a | 2.9 (1.78) | 5.74 (1.39) | 3.03 (1.30) | 1.84 (0.61) | <0.0001a | <0.0001a | 0.0043a | <0.0001a |
Note: Values are mean (SD) unless otherwise indicated
aBonferroni corrected significant p values
p value by ANOVA, p1 adjusted for duration of diabetes
In the sex-stratified analysis, SIDD (60.6%) remained the predominant cluster in men whereas the main subgroup was MOD (53.2%) in women; MARD was more common in men (13.3% vs 2.1%) (Fig. 1, Table 3).
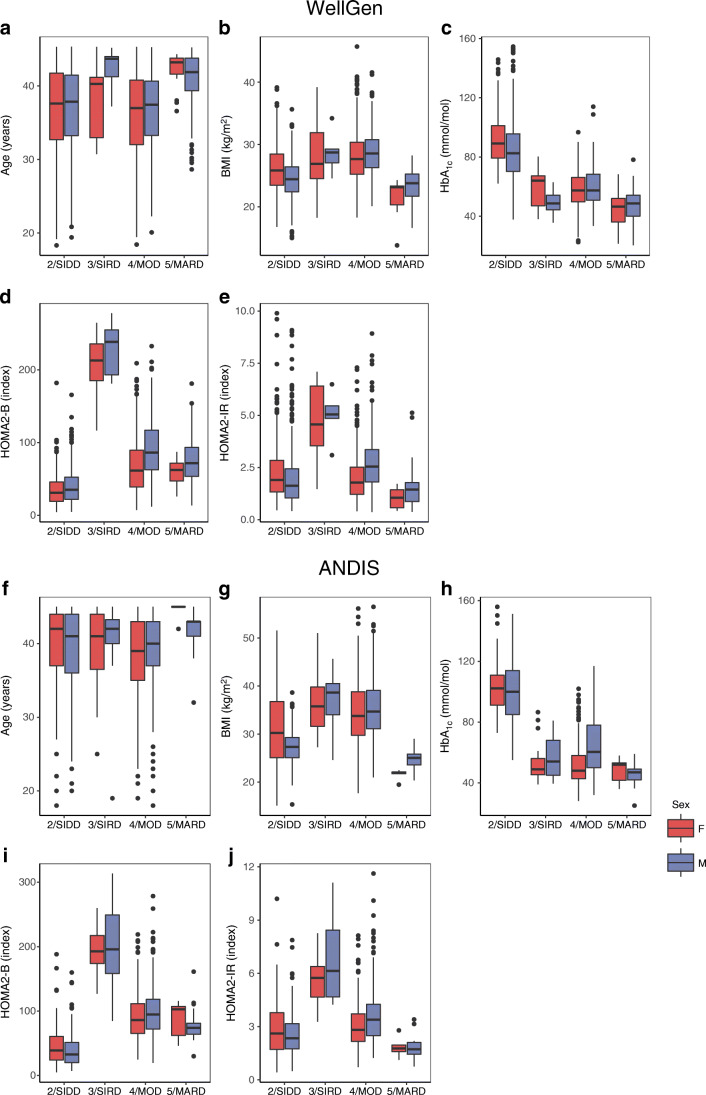
Concordant with the diabetes subgroups in ANDIS, among the two predominant subgroups, Indian individuals in the SIDD subgroup had the lowest insulin secretion (HOMA2-B) and the highest blood glucose levels while those in the MOD subgroup had the highest BMI. Individuals in the SIRD subgroup were the most insulin resistant, whereas those in the MARD subgroup were the oldest at diagnosis, with the lowest blood glucose levels and lowest degree of insulin resistance (Fig. 2, Table 3). These results support the pathophysiological basis for subclassification in a population which has a different genetic and socioeconomic background compared with the Swedish population.
Fig. 2.
Box plot of cluster characteristics in the WellGen and ANDIS studies. Distribution of age at diagnosis (a, f), BMI (b, g), HbA1c (c, h), HOMA2-B (d, i) and HOMA2-IR (e, j) in the WellGen (a–e) and ANDIS (f–j) studies for each cluster. The central line within each box represents the median and the upper and lower limits of the box represent the IQR. The whiskers are the most extreme values within 1.5× the IQR from the first and second quartiles. k-means clustering was done separately for men and women; data are shown for each sex separately. F, female sex; M, male sex
The distribution of subgroups in ANDIS differed from that of WellGen; MOD was the most predominant cluster (67.57%), followed by SIDD (26.09%), SIRD (3.64%) and MARD (2.70%) (Fig. 1, Table 3). These distributions were similar in men and women. The pathophysiological characteristics of these Swedish individuals with young-onset type 2 diabetes were similar to those in the parent ANDIS cohort (Fig. 2, Table 3).
Treatment
In both WellGen and ANDIS, insulin treatment (alone or in combination with OHAs) was most commonly prescribed to individuals in the SIDD subgroup (38.3% in WellGen, 51.0% in ANDIS) (Table 4).
Table 4.
Treatment and complications by cluster in the WellGen and ANDIS study
| Treatment/complication | WellGen (N=1612) | ANDIS (N=962) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SIDD | SIRD | MOD | MARD | p value | p1 value | p2 value | SIDD | SIRD | MOD | MARD | p value | p1 value | p2 value | |
| No. of participants in subgroup | 851 | 18 | 608 | 135 | 251 | 35 | 650 | 26 | ||||||
| Current treatment | ||||||||||||||
| Diet only | 68 (8.0) | 4 (22.2) | 54 (8.9) | 9 (6.7) | 0.146 | 0.544 | 0.922 | 4 (1.6) | 5 (14.3) | 60 (9.2) | 8 (30.8) | 0.0001a | 0.0001a | 0.002a |
| OHA only (SU + metformin ± thiazolidinedione) | 457 (53.7) | 13 (72.2) | 409 (67.3) | 100 (74.1) | 0.0001a | 0.0001a | 0.0001a | 117 (46.6) | 33 (74.3) | 470 (72.3) | 16 (61.5) | 0.0001a | 0.0001a | 0.0001a |
| Insulin only | 58 (6.8) | 0 (0) | 19 (3.1) | 8 (5.9) | 0.013 | 0.002a | 0.004a | 36 (14.3) | 0 (0) | 23 (3.5) | 0 (0) | 0.0001a | 0.0001a | 0.0001a |
| Both OHA+ insulinb | 268 (31.5) | 1 (5.6) | 126 (20.7) | 18 (13.3) | 0.0001a | 0.0001a | 0.0001a | 92 (36.7) | 4 (11.4) | 96 (18.8) | 2 (7.7) | 0.0001a | 0.0001a | 0.0001a |
| Complications | ||||||||||||||
| Cardiovascular disease (coronary events and/or stroke)c | 61 (7.2) | 0 (0) | 39 (6.4) | 20 (14.8) | 0.004a | 0.574 | 0.242 | 10 (4.0) | 3 (8.6) | 20 (3.1) | 1 (3.8) | 0.375 | 0.502 | 0.633 |
| Coronary event | 51 (6.0) | 0 (0) | 32 (5.3) | 17 (12.6) | 0.009 | 0.553 | 0.257 | 7 (2.8) | 3 (8.6) | 13 (2.0) | 0 (0) | 0.075 | 0.476 | 0.493 |
| Stroke | 13 (1.6) | 0 (0) | 9 (1.5) | 4 (3.1) | 0.582 | 0.942 | 0.746 | 3 (1.2) | 0 (0) | 7 (1.1) | 1 (3.8) | 0.522 | 0.884 | 0.980 |
| Nephropathy (macroalbuminuria and/or CKD)d | 340 (40.0) | 6 (33.3) | 147 (24.2) | 68 (50.4) | 0.0001a | 0.0001a | 0.0001a | 76 (40.0) | 19 (70.4) | 206 (46.6) | 9 (52.9) | 0.022 | 0.126 | 0.087 |
| Macroalbuminuriae | 172 (21.1) | 0 (0) | 66 (11.3) | 12 (9.4) | 0.0001a | 0.0001a | 0.001a | 25 (13.8) | 5 (5.0) | 59 (14.9) | 1 (7.1) | 0.484 | 0.731 | 0.635 |
| CKD: eGFRf,g | 247 (31.7) | 6 (35.3) | 98 (17.6) | 62 (52.1) | 0.0001a | 0.0001a | 0.0001a | 62 (25.4) | 16 (47.1) | 165 (26.4) | 8 (33.3) | 0.049 | 0.765 | 0.475 |
| Early CKD (60–90 ml min−1 [1.73m]−2) | 190 (24.4) | 6 (35.3) | 85 (15.3) | 49 (41.2) | 0.0001a | 0.0001a | 0.006a | 56 (23.5) | 16 (47.1) | 157 (25.4) | 8 (33.3) | 0.025 | 0.561 | 0.831 |
| Moderate CKD (30–60 ml min−1 [1.73m]−2) | 52 (6.7) | 0 (0) | 10 (1.8) | 12 (10.1) | 0.0001a | 0.0001a | 0.002a | 5 (2.1) | 0 (0) | 6 (1.0) | 0 (0) | 0.471 | 0.196 | 0.162 |
| Severe CKD (<30 ml min−1 [1.73m]−2) | 5 (0.6) | 0 (0) | 3 (0.5) | 1 (0.8) | 0.966 | 0.811 | 0.898 | 1 (0.4) | 0 (0) | 2 (0.3) | 0 (0) | 0.970 | 0.893 | 0.935 |
| Diabetic retinopathy (n=657)h | 112 (36.8) | 4 (57.1) | 51 (18.0) | 13 (21.0) | 0.0001a | 0.0001a | 0.0001a | 23 (24.7) | 4 (44.4) | 30 (16.6) | 0 (0) | 0.050 | 0.11 | 0.13 |
| NPDR | 105 (34.5) | 4 (57.1) | 46 (16.2) | 12 (19.4) | 0.0001a | 0.0001a | 0.0001a | 22 (23.9) | 5 (44.4) | 29 (16.1) | 0 (0) | 0.050 | 0.050 | 0.225 |
| PDR | 7 (2.3) | 0 (0) | 5 (1.8) | 1 (1.6) | 0.936 | 0.642 | 0.847 | 1 (1.1) | 0 (0) | 1 (0.6) | 0 (0) | 0.948 | 0.948 | 0.821 |
| Neuropathyi | 322 (38.8) | 9 (50.0) | 323 (53.8) | 56 (43.4) | 0.0001a | 0.0001a | 0.003a | 12 (4.8) | 1 (2.9) | 3 (0.5) | 0 (0) | 0.0001a | 0.0001a | 0.002a |
Values are n (%)
aBonferroni corrected significant p values
bData for metformin + insulin only for ANDIS clusters
cCVD: data available for all (n=1612) for WellGen, n=956 (SIDD n=250, SIRD n=35, MOD n=645, MARD n=26) for ANDIS
dNephropathy: data available for all (n=1612) for WellGen, n=676 (SIDD n=190, SIRD n=27, MOD n=442, MARD n=17) for ANDIS
eMacroalbuminuria: data available for n=1544 (SIDD n=816, SIRD n=18, MOD n=582, MARD n=128) for WellGen, n=611 (SIDD n=181, SIRD n=20, MOD n=396, MARD n=14) for ANDIS
fBased on MDRD formula
gCKD: eGFR: data available for n=1471 (SIDD n=779, SIRD n=17, MOD n=556, MARD n=119) for WellGen, n=927 (SIDD n=244, SIRD n=34, MOD n=625, MARD n=24) for ANDIS
hDiabetic retinopathy: data available for n=657 (SIDD n=304, SIRD n=7, MOD n=284, MARD n=62) for WellGen and n=288 (SIDD n=92, SIRD n=9, MOD n=180, MARD n=7) for ANDIS
iNeuropathy: data available for n=1576 (SIDD n=829, SIRD n=18, MOD n=600, MARD n=129), diagnosed using Biothesiometry for WellGen, n=956 (SIDD n=250, SIRD n=35, MOD n=645, MARD n=26), ICD codes ICD-10 = E104 or E114 for ANDIS
p value was calculated by χ2 test and p1 value by χ2 test for SIDD and MOD clusters only; p2 value was calculated by logistic regression, adjusted for duration of diabetes and sex for SIDD and MOD clusters
NPDR, non-proliferative diabetic retinopathy; OHA, oral glucose-lowering agent; PDR, proliferative diabetic retinopathy; SU, sulfonylurea
Complications
In WellGen, we compared the prevalence of complications in the two major subtypes, SIDD and MOD. Small numbers in the SIRD and MARD groups precluded comparison of complications. Retinopathy and nephropathy were most common in the SIDD subgroup whereas neuropathy was more prevalent in MOD (Table 4). The prevalence of macrovascular complications was similar in these two subtypes. Of the less common subgroups, SIRD had a high prevalence of retinopathy while MARD had a high prevalence of nephropathy and macrovascular disease.
In the ANDIS cohort, nephropathy (70.4%) and retinopathy (44.4%) prevalence were highest in SIRD whereas neuropathy was most common in SIDD (Table 4). Consistent with previous findings, SIRD also showed the highest prevalence of CKD (47.1%) while macroalbuminuria (14.9%) was most common in MOD.
De novo clusters show a high degree of concordance with SIDD and MOD
We also applied the de novo k-means clustering to assess the subgroups obtained in the Indian study population and compared them with those obtained using the previously published algorithm. Two was the optimum number of clusters (ESM Fig. 2a); cluster 1 had a prevalence of 66.6% while cluster 2 had a prevalence of 33.4% (ESM Table 7). Cluster 1 showed 88.8% concordance with SIDD (86.8% in male participants, 92.4% in female participants) while cluster 2 had an overlap of 62.5% with MOD (83.9% in male participants, ~49.5% in female participants) (ESM Fig. 2b).
Both clusters had the same cluster characteristics as seen using the centroid method, thereby providing a technical replication (ESM Table 7). The similarity also extended to complication rates, with nephropathy and retinopathy being prevalent in cluster 1 compared with cluster 2 whereas neuropathy was more prevalent in cluster 2 (ESM Table 8).
Low prevalence of genetic type 1 diabetes in WellGen type 2 diabetes subgroups
In the absence of GAD autoantibody data, a previously established type 1 diabetes GRS [25] comprising nine SNPs (ESM Table 4), which was validated in the Indian population [23], was applied to estimate the proportion of participants with autoimmune diabetes in a subset of the WellGen cohort (ESM Table 5). The GRS was associated with the positive control participants with type 1 diabetes compared with SIDD and MOD. The same GRS did not associate with either SIDD or MOD compared with non-diabetic control individuals (ESM Fig. 3, ESM Table 9). The proportion of participants with GRS ≥90% and 80% was ~5% and 28.7%, respectively, in type 1 diabetes (positive controls) whereas it was 0% and 1.4% in SIDD, 0% and 4.7% in MOD, and 0% and 1% in control participants. The same GRS was associated with SAID in young ANDIS participants (β = 7.3 ± 0.72, p < 2 × 10−16).
Indian diabetes subgroups are similar to subgroups in European cohort DIREVA with longer diabetes duration
Given the difference in duration of diabetes in ANDIS and WellGen, we compared the WellGen subgroups with those in the Finnish cohort DIREVA (n = 420) with duration of diabetes 14.4 years (ESM Table 10). The differences in proportion of subgroups and cluster characteristics between WellGen and DIREVA were similar to those between WellGen and ANDIS. Similar to ANDIS, SIDD was less common compared with MOD in DIREVA (23.6% vs 70.8%) (ESM Table 10, ESM Fig. 4).
Subgroups of diabetes in other regions of India
We applied the Swedish algorithm [2] to two studies from different geographical regions in India, Ahmedabad, Gujarat, western India (N = 187) and Dibrugarh, Assam, north-eastern India (N = 205; PHENOEINDY-2 cohort) (ESM Tables 11, 12). Concordant with findings in WellGen, the Ahmedabad cohort had the highest proportion of participants in the SIDD subgroup (56.68%) followed by MOD (33.15%) and MARD (10.16%) (ESM Table 11). The similarity extended to the subgroup distributions in the two sexes: the SIDD subgroup was most prevalent in male participants (61.59%) while MOD was most prevalent in female participants (53.06%) (ESM Table 11, ESM Fig. 5). The PHENOEINDY-2 cohort was the youngest and the thinnest cohort of all; the proportion of SIDD (66.66%) was the highest, followed by MOD (23.20%), MARD (7.72%) and SIRD (1.40%) (ESM Table 12, ESM Fig. 6).
Discussion
We showed that the clusters described in the newly diagnosed unselected European type 2 diabetes patients [2, 3] are also seen in the younger and thinner Indians. SIDD and MOD were the two predominant subgroups, while MARD was less common and SIRD the least common in both populations. SIDD was the predominant cluster in Indians whereas MOD was the most prevalent in Europeans. The predominance of SIDD was replicated in two independent, geographically distinct Indian cohorts of individuals with young-onset type 2 diabetes.
The distribution of the clusters suggests that deficient insulin secretion, rather than the often-purported insulin resistance, is the driver of young-onset type 2 diabetes in India. In contrast, in the young Swedish and Finnish type 2 diabetes populations, obesity and insulin resistance seemed to be the primary pathophysiological drivers. The proposed prominent role of insulin resistance was based on previous demonstrations of higher insulin resistance in Indians compared with Europeans at a given BMI possibly due to relatively more adipose body composition [7, 28, 29]. Despite the differences in age, BMI and duration of diabetes, the characteristics of the clusters themselves in Indians broadly reflected those in the European studies [2] Our new findings suggest a paradigm shift for the understanding of the pathophysiology of type 2 diabetes in young Indians, albeit they do not preclude the role of insulin resistance.
De novo k-means subclassification validated the two major diabetes subgroups obtained from the European-derived centroids. The concordance was greater in men for both subgroups, while it was lower in women for the newly obtained cluster 2 with MOD. While this increases our confidence in the classification, reclassification of a proportion of women with MOD to a SIDD equivalent cluster 1 highlights the role of insulin deficiency in the pathogenesis of type 2 diabetes in young Indians.
We applied the European-derived centroids to two smaller cohorts of individuals with young-onset type 2 diabetes from western (Ahmedabad, Gujarat) and north-eastern (Dibrugarh, Assam) India. The proportion of subclasses in the Ahmedabad participants was nearly identical to those in Pune, while the proportion of individuals in the SIDD subgroup was highest in Dibrugarh. Gujarat is a more affluent state while Assam has a lower development index and high prevalence of undernutrition.
Physicians in India have long realised the phenotypic differences of Indian diabetes patients compared with those described in patients of European origin [30, 31]. Interestingly, lean type 2 diabetes has been prominently reported in the impoverished states of Orissa and north-east India, where malnutrition-related diabetes (MRDM) has been described [32]. A proportion of SIDD patients from Assam could well be characterised similarly. There is an increasing recognition that early-life undernutrition could lead to smaller beta cell mass and insulin secretion defects demonstrable from early childhood in serially studied birth cohorts and could manifest as prediabetes or type 2 diabetes in young adulthood [33, 34]. Animal studies have clearly demonstrated poor beta cell development and islet dysfunction in offspring born to malnourished pregnant mothers [35–37]. It is intriguing that the highest rise in the prevalence of type 2 diabetes in India over the last 25 years has been demonstrated in states that have suffered chronic environmental, socioeconomic and nutritional deficits [38]. On such a background of intergenerational deprivation, a relatively small socioeconomic development appears enough to precipitate diabetes at a young age. It is of note that the prevalence of diabetes in those above 20 years of age has increased from 5.5% to 7.5% between 1990 and 2016 in the state of Assam.
Individuals in the diabetes subgroups displayed different sensitivities to micro- and macrovascular complications. Microvascular disorders of the retina and kidney were more prevalent in SIDD compared with MOD, while peripheral nerve damage was more prevalent in MOD. Possible reasons for these differences may lie in the pathophysiological mechanisms driving the subgroups and this needs to be studied further. Prevalence of macrovascular disease was similar in two subgroups. In the original Swedish classification, the SIRD subgroup generated a lot of interest given the high propensity of affected individuals for developing nephropathy. SIRD was the smallest subgroup in Indians with high insulin resistance as well as insulin secretion, although it was heterogeneous between the Indian cohorts. Intriguingly, the MARD subgroup had a strikingly high rate of macrovascular disease. The unique profiles of these subgroups could well represent population-specific differences and highlight the need for customisation of the clustering algorithm.
Other studies have investigated the heterogeneity of type 2 diabetes in Indians. The INSPIRED study from a chain of private diabetes clinics in India reported four subclasses, two of which were similar to the Swedish study (SIDD and MARD) [5] and two of which were new (insulin-resistant obese diabetes [IROD] and combined insulin resistance and deficient diabetes [CIRDD]) [14]. However, the clustering parameters were different and therefore not directly comparable with our study. The MASALA-MESA study reported subclasses in a mixed population in the USA, including migrant South Asian Indians (n = 217) [15]. They found an excess of younger, thinner and severe hyperglycaemic individuals among South Asian Indians, supporting our findings.
This study has strengths and limitations. This is the first attempt in India to subclassify patients with a diagnosis of type 2 diabetes at a young age. The presence of subgroups in our study comparable with those in a genetically and historically distinct European population validates the subgroups. While the power of the study is limited, validation by de novo clustering increases confidence in classification. The Indian patients are clinic-based and enrolled many years after diabetes diagnosis while on glucose-lowering treatment, which may affect the proportions of subclasses. We cannot rule out the possibility that some individuals might shift to different subgroups over time; however, only a small proportion did so in another study [3]. A sensitivity analysis in WellGen showed that the proportions of the subgroups varied with increasing diabetes duration, although SIDD remained the predominant subgroup (45.5%) even in those with less than 5 years of diagnosis. Observations in other two Indian cohorts further validated these findings. Another limitation is that this study is an opportunistic comparison of existing data and therefore laboratory measurements are not fully harmonised between cohorts. However, C-peptide measurements in different cohorts were calibrated against the same WHO standard, facilitating comparisons. Given the lack of GAD autoantibody data, it might be suspected that the SIDD group in WellGen includes individuals with autoimmune diabetes (latent autoimmune diabetes in adults [LADA]). However, the low prevalence of individuals with high type 1 diabetes GRS scores in WellGen deems a large contribution of autoimmune diabetes extremely unlikely.
In summary, we demonstrate the applicability of a European algorithm for subclassifying type 2 diabetes in young Indian patients. Our results demonstrate a prominent role for insulin secretion defects in the pathophysiology of diabetes in this group. These results could potentially influence treatment strategies for achieving optimal metabolic control, with possible benefits for long-term health [4, 39]. Translation to personalised medicine will come from carefully designed prospective studies including genetic and epigenetic investigations to elucidate pathophysiological mechanisms underlying the subgroups.
Supplementary information
(PDF 837 kb)
Acknowledgements
We thank all patients and healthcare providers for their support and willingness to participate. We thank A. Hattersley (University of Exeter Medical School, UK), G. Chandak and D. Tallapragada (Genomic Research on Complex Diseases [GRC], CCMB, India), and S. Kale, S. Sardeshpande, K. Meenakumari, P. Yajnik, R. Ladkat, D. Raut, M. Deshmukh and C. Hole (Diabetes Unit, Kamalnayan Bajaj Diabetology Research Centre, King Edward Memorial Hospital and Research Centre, Pune, India) for their invaluable contribution to the WellGen study. We gratefully acknowledge J. Postma, J. Hultman, J. Kravic, G. Gremsperger, Y. Wessman and U. Blom-Nilsson (Department of Clinical Sciences, Diabetes and Endocrinology, CRC, Lund University, Malmö, Sweden) for excellent technical and administrative support and thank the ANDIS steering committee for their support.
Authors’ relationships and activities
The authors declare that there are no relationships or activities that might bias, or be perceived to bias, their work.
Abbreviations
- CKD
Chronic kidney disease
- GRS
Genetic risk score
- MARD
Mild age-related diabetes
- MDRD
Modification of Diet in Renal Disease
- MOD
Mild obesity-related diabetes
- SAID
Severe autoimmune diabetes
- SIDD
Severe insulin-deficient diabetes
- SIRD
Severe insulin-resistant diabetes
Contribution statement
RBP, CSY and LG contributed to the conception of the work. CSY, SRS, DB and SP contributed to data collection in Pune, India, MP, MS and BS in Ahmedabad, India, AD and SK in Dibrugarh, India, RBP, EA, OA and LG in Sweden, and AK and TT in Finland. RPB, OA, SRS, EA, RW and PK contributed to the data analysis. RBP, CSY and LG drafted the article. All authors contributed to the interpretation of data and critical revision of the article. All authors gave final approval of the version to be published. RBP and CSY are the guarantors of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Funding
Open access funding provided by Lund University. This study was supported by grants from the Indo-Swedish joint network grant from the Swedish Research Council and the Department of Science and Technology, India (Dnr/Reg. nr: 2015-06722 to RBP and C/3019/IFD/2018-2019 to CSY), Wellcome Trust (to CSY), Swedish Research Council (2017-02688), Diabetes Wellness Sweden (25-420 PG) and the Novo Nordisk Foundation (NNF18OC0034408 to EA), Swedish Research Foundation (2015-2558 to LG), the Swedish Heart and Lung Foundation, the Crafoord Foundation, the Swedish Diabetes Foundation and the Albert Påhlsson Research Foundation. The DIREVA study is supported by the Vasa Hospital district, The Academy of Finland (grants no. 263401, 267882, 312063 to LG, 312072 to TT), University of Helsinki, Ollqvist Foundation, Finnish Diabetes Research Foundation, Helsinki University Central Hospital Research Foundation and Jakobstadsnejdens heart foundation (to AK). CSY was a visiting Professor to Danish Diabetes Academy (supported by Novo Nordisk Foundation) and Southern University of Denmark during 2016-2019.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Footnotes
The original online version of this article was revised: The name Malay Parekh was spelt incorrectly in the author list.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
11/22/2021
A Correction to this paper has been published: 10.1007/s00125-021-05620-2
Contributor Information
Rashmi B. Prasad, Email: rashmi.prasad@med.lu.se
Chittaranjan S. Yajnik, Email: csyajnik@gmail.com
References
- 1.WHO Study Group on Diabetes Mellitus & World Health Organization (1985) Diabetes mellitus. Report of a WHO Study Group. World Health Organ Tech Rep Ser 727. WHO, Geneva
- 2.Ahlqvist E, Storm P, Karajamaki A, et al. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol. 2018;6:361–369. doi: 10.1016/S2213-8587(18)30051-2. [DOI] [PubMed] [Google Scholar]
- 3.Zaharia OP, Strassburger K, Strom A, et al. Risk of diabetes-associated diseases in subgroups of patients with recent-onset diabetes: a 5-year follow-up study. Lancet Diabetes Endocrinol. 2019;7:684–694. doi: 10.1016/S2213-8587(19)30187-1. [DOI] [PubMed] [Google Scholar]
- 4.Prasad RB, Groop L. Precision medicine in type 2 diabetes. J Intern Med. 2019;285:40–48. doi: 10.1111/joim.12859. [DOI] [PubMed] [Google Scholar]
- 5.Ahlqvist E, Prasad RB, Groop L. Subtypes of Type 2 Diabetes Determined From Clinical Parameters. Diabetes. 2020;69:2086–2093. doi: 10.2337/dbi20-0001. [DOI] [PubMed] [Google Scholar]
- 6.Shelgikar KM, Hockaday TD, Yajnik CS. Central rather than generalized obesity is related to hyperglycaemia in Asian Indian subjects. Diabet Med. 1991;8:712–717. doi: 10.1111/j.1464-5491.1991.tb01689.x. [DOI] [PubMed] [Google Scholar]
- 7.Yajnik CS. The insulin resistance epidemic in India: fetal origins, later lifestyle, or both? Nutr Rev. 2001;59:1–9. doi: 10.1111/j.1753-4887.2001.tb01898.x. [DOI] [PubMed] [Google Scholar]
- 8.Yajnik CS, Yudkin JS. The Y-Y paradox. Lancet. 2004;363:163. doi: 10.1016/S0140-6736(03)15269-5. [DOI] [PubMed] [Google Scholar]
- 9.Narayan KMV, Kondal D, Daya N, et al. Incidence and pathophysiology of diabetes in South Asian adults living in India and Pakistan compared with US blacks and whites. BMJ Open Diabetes Res Care. 2021;9:e001927. doi: 10.1136/bmjdrc-2020-001927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narayan KMV, Kanaya AM. Why are South Asians prone to type 2 diabetes? A hypothesis based on underexplored pathways. Diabetologia. 2020;63:1103–1109. doi: 10.1007/s00125-020-05132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watts G. Lalit Dandona: surveying the burden of disease in India. Lancet. 2017;390:2428. doi: 10.1016/S0140-6736(17)32861-1. [DOI] [PubMed] [Google Scholar]
- 12.Anjana RM, Deepa M, Pradeepa R, et al. Prevalence of diabetes and prediabetes in 15 states of India: results from the ICMR-INDIAB population-based cross-sectional study. Lancet Diabetes Endocrinol. 2017;5:585–596. doi: 10.1016/S2213-8587(17)30174-2. [DOI] [PubMed] [Google Scholar]
- 13.Samal KC, Tripathy BB. Malnutrition related diabetes. J Assoc Physicians India. 1987;35:170. [PubMed] [Google Scholar]
- 14.Anjana RM, Baskar V, Nair ATN, et al. Novel subgroups of type 2 diabetes and their association with microvascular outcomes in an Asian Indian population: a data-driven cluster analysis: the INSPIRED study. BMJ Open Diabetes Res Care. 2020;8:e001506. doi: 10.1136/bmjdrc-2020-001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bancks MP, Bertoni AG, Carnethon M, et al. Association of Diabetes Subgroups With Race/Ethnicity, Risk Factor Burden and Complications: The MASALA and MESA Studies. J Clin Endocrinol Metab. 2021;106:e2106–e2115. doi: 10.1210/clinem/dgaa962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ministry of Health and Family Welfare (MoHFW) GoI, UNICEF, (CNNS) aPCCNNS, Delhi. NRN (2016-2018) Comprehensive National Nutrition Survey. https://www.popcouncil.org/uploads/pdfs/2019RH_CNNSreport.pdf Accessed on 12 Feb 2020
- 17.Golden SH, Yajnik C, Phatak S, Hanson RL, Knowler WC. Racial/ethnic differences in the burden of type 2 diabetes over the life course: a focus on the USA and India. Diabetologia. 2019;62:1751–1760. doi: 10.1007/s00125-019-4968-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandak GR, Janipalli CS, Bhaskar S, et al. Common variants in the TCF7L2 gene are strongly associated with type 2 diabetes mellitus in the Indian population. Diabetologia. 2007;50:63–67. doi: 10.1007/s00125-006-0502-2. [DOI] [PubMed] [Google Scholar]
- 19.Yajnik CS, Joglekar CV, Lubree HG, et al. Adiposity, inflammation and hyperglycaemia in rural and urban Indian men: Coronary Risk of Insulin Sensitivity in Indian Subjects (CRISIS) Study. Diabetologia. 2008;51:39–46. doi: 10.1007/s00125-007-0847-1. [DOI] [PubMed] [Google Scholar]
- 20.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 21.Levy JC, Matthews DR, Hermans MP. Correct homeostasis model assessment (HOMA) evaluation uses the computer program. Diabetes Care. 1998;21:2191–2192. doi: 10.2337/diacare.21.12.2191. [DOI] [PubMed] [Google Scholar]
- 22.R Development Core Team R: A language and environment for statistical computing. In. R Foundation for Statistical Computing, Vienna, Austria
- 23.Harrison JW, Tallapragada DSP, Baptist A, et al. Type 1 diabetes genetic risk score is discriminative of diabetes in non-Europeans: evidence from a study in India. Sci Rep. 2020;10:9450. doi: 10.1038/s41598-020-65317-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao S, Yajnik CS, Kanade A, et al. Intake of micronutrient-rich foods in rural Indian mothers is associated with the size of their babies at birth: Pune Maternal Nutrition Study. J Nutr. 2001;131:1217–1224. doi: 10.1093/jn/131.4.1217. [DOI] [PubMed] [Google Scholar]
- 25.Oram RA, Patel K, Hill A, et al. A Type 1 Diabetes Genetic Risk Score Can Aid Discrimination Between Type 1 and Type 2 Diabetes in Young Adults. Diabetes Care. 2016;39:337–344. doi: 10.2337/dc15-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winkler C, Krumsiek J, Buettner F, et al. Feature ranking of type 1 diabetes susceptibility genes improves prediction of type 1 diabetes. Diabetologia. 2014;57:2521–2529. doi: 10.1007/s00125-014-3362-1. [DOI] [PubMed] [Google Scholar]
- 27.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharp PS, Mohan V, Levy JC, Mather HM, Kohner EM. Insulin resistance in patients of Asian Indian and European origin with non-insulin dependent diabetes. Horm Metab Res. 1987;19:84–85. doi: 10.1055/s-2007-1011745. [DOI] [PubMed] [Google Scholar]
- 29.Ramachandran A, Snehalatha C, Viswanathan V, Viswanathan M, Haffner SM. Risk of noninsulin dependent diabetes mellitus conferred by obesity and central adiposity in different ethnic groups: a comparative analysis between Asian Indians, Mexican Americans and Whites. Diabetes Res Clin Pract. 1997;36:121–125. doi: 10.1016/S0168-8227(97)00040-5. [DOI] [PubMed] [Google Scholar]
- 30.Ramachandran A, Snehalatha C. Epidemiology of Diabetes in Developing Countries: The Scenario in Asia. In: Chandalia HB, editor. RSSDI - Text Book of Diabetes Mellitus. New Delhi: Jaypee Brothers Medical Publishers; 2014. pp. 171–180. [Google Scholar]
- 31.Das S, Panda JK. Islets Β Cell Response and Glucose Homeostasis in Low Body Weight Type 2 Diabetes. J Diabetes Metab. 2014;5:1–6. [Google Scholar]
- 32.Rao RH, Yajnik CS. Commentary: time to rethink malnutrition and diabetes in the tropics. Diabetes Care. 1996;19:1014–1017. doi: 10.2337/diacare.19.9.1014. [DOI] [PubMed] [Google Scholar]
- 33.Memane NS, Bhat D, Raut DA, et al. Pregnancy Glycemia Reflects Life Course Glycemia of the Mother. Diabetes. 2018;67:118-OR. doi: 10.2337/db18-118-OR. [DOI] [Google Scholar]
- 34.India State-Level Disease Burden Initiative Malnutrition Collaborators (2019) The burden of child and maternal malnutrition and trends in its indicators in the states of India: the Global Burden of Disease Study 1990-2017. Lancet Child Adolesc Health [DOI] [PMC free article] [PubMed]
- 35.Snoeck A, Remacle C, Reusens B, Hoet JJ. Effect of a low protein diet during pregnancy on the fetal rat endocrine pancreas. Biol Neonate. 1990;57:107–118. doi: 10.1159/000243170. [DOI] [PubMed] [Google Scholar]
- 36.Fernandez-Twinn DS, Wayman A, Ekizoglou S, Martin MS, Hales CN, Ozanne SE. Maternal protein restriction leads to hyperinsulinemia and reduced insulin-signaling protein expression in 21-mo-old female rat offspring. Am J Physiol Regul Integr Comp Physiol. 2005;288:R368–R373. doi: 10.1152/ajpregu.00206.2004. [DOI] [PubMed] [Google Scholar]
- 37.Kumar KA, Lalitha A, Reddy U, Chandak GR, Sengupta S, Raghunath M. Chronic maternal vitamin B12 restriction induced changes in body composition & glucose metabolism in the Wistar rat offspring are partly correctable by rehabilitation. PLoS One. 2014;9:e112991. doi: 10.1371/journal.pone.0112991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.India State-Level Disease Burden Initiative Collaborators Nations within a nation: variations in epidemiological transition across the states of India, 1990-2016 in the Global Burden of Disease Study. Lancet. 2017;390:2437–2460. doi: 10.1016/S0140-6736(17)32804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prasad RB, Ahlqvist E, Groop L. Heterogeneity of diabetes - An Indian perspective. Diabetes Metab Syndr. 2019;13:3065–3067. doi: 10.1016/j.dsx.2018.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 837 kb)
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.