Abstract
In this study, the mechanism of mammalian gene replacement was investigated. The system is based on detecting homologous recombination between transferred vector DNA and the haploid, chromosomal immunoglobulin μ-δ region in a murine hybridoma cell line. The backbone of the gene replacement vector (pCμCδpal) consists of pSV2neo sequences bounded on one side by homology to the μ gene constant (Cμ) region and on the other side by homology to the δ gene constant (Cδ) region. The Cμ and Cδ flanking arms of homology were marked by insertions of an identical 30-bp palindrome which frequently escapes mismatch repair when in heteroduplex DNA (hDNA). As a result, intermediates bearing unrepaired hDNA generate mixed (sectored) recombinants following DNA replication and cell division. To monitor the presence and position of sectored sites and, hence, hDNA formation during the recombination process, the palindrome contained a unique NotI site that replaced an endogenous restriction enzyme site at each marker position in the vector-borne Cμ and Cδ regions. Gene replacement was studied under conditions which permitted the efficient recovery of the product(s) of individual recombination events. Analysis of marker segregation patterns in independent recombinants revealed that extensive hDNA was formed within the Cμ and Cδ regions. In several recombinants, palindrome markers in the Cμ and Cδ regions resided on opposite DNA strands (trans configuration). These results are consistent with the mammalian gene replacement reaction involving two crossing-over events in homologous flanking DNA.
The introduction of predetermined alterations in chromosomal sequences by homologous recombination with transferred DNA (gene targeting) is a powerful technology for modifying gene structure and function. It has applications that include the study of gene expression in its normal chromosomal environment and the creation of animal models of human genetic diseases and, ultimately, it has the potential to be an effective form of gene therapy (3, 33, 35). In addition, the ability to manipulate the transforming DNA makes gene targeting a valuable model system in the study of homologous recombination mechanisms.
Gene targeting can be performed with either insertion (“ends-in” or O-type) vectors or replacement (“ends-out” or Ω-type) vectors. In an insertion vector, a double-strand break is introduced within the homology region creating DNA ends that invade cognate chromosomal sequences. In Saccharomyces cerevisiae, gene targeting using an insertion vector is consistent with the double-strand-break repair (DSBR) model (24, 32). Like yeast, targeted vector insertion in mammalian cells also has features consistent with DSBR (16, 17, 23, 34). In a gene replacement vector, the region of homology is interrupted by a selectable genetic marker. Since the ends of the vector are discontinuous with the chromosome, recombination replaces a region of the chromosome with vector sequences. The mechanism of homologous recombination with a gene replacement vector is unknown. In principle, it might occur by assimilation of a single strand of the vector into the chromosome, as was proposed to explain the replacement of a chromosomal allele with linear duplex DNA in S. cerevisiae (15). However, single-strand assimilation might be impeded by the heterology encoded by the selectable marker. Nevertheless, Negritto et al. (21) found at least a 10-fold increase in marker incorporation in an msh2 mutant even when all flanking markers were identical. Thus, marker assimilation may occur and be corrected by a process that involves mismatch repair (MMR) genes (15, 21). Gene replacement by single strand assimilation predicts that markers in flanking heteroduplex DNA (hDNA) will reside in a cis configuration. Alternatively, gene replacement might involve two crossing-over events in the homologous DNA flanking the selectable marker. This could explain how chromosomal deletions are engineered in the yeast genome using replacement vectors in which the selectable marker is flanked by two very distant homology regions (10, 26, 31). In this instance, markers in flanking hDNA would reside in a trans configuration. Thus, the models make testable predictions about the configuration of hDNA in the recombinants.
An important contribution to the study of hDNA formation during homologous recombination came from studies in S. cerevisiae, where a small palindrome was shown to avoid MMR when encompassed within hDNA, likely as a consequence of it forming a small hairpin structure (20). Semiconservative DNA replication of unrepaired hDNA, followed by cell division, generates a mixed (sectored) recombinant. Thus, the positions of sectored sites mark the location of hDNA formed during recombination. Earlier, we showed that a small palindrome in hDNA is also poorly repairable by the mammalian MMR system (16), a feature that provided some important insights into the formation of hDNA during targeted vector insertion in mammalian cells (16, 17). To obtain information about the presence and position of hDNA formed during mammalian gene replacement, we constructed a gene replacement vector in which the flanking arms of homology were marked by insertions of the small palindrome. Among the several independent recombinants in which hDNA was present in both flanking arms of homology, the palindrome markers were predominantly in a trans configuration. This result is consistent with the mammalian gene replacement reaction involving two crossing-over events.
MATERIALS AND METHODS
Recipient hybridoma cells.
The haploid, chromosomal immunoglobulin μ-δ heavy chain locus in the wild-type murine hybridoma cell line, Sp6/HL, serves as the target for gene replacement (see Fig. 2). The origin of Sp6/HL and the methods used for hybridoma cell culture have been described previously (13, 14).
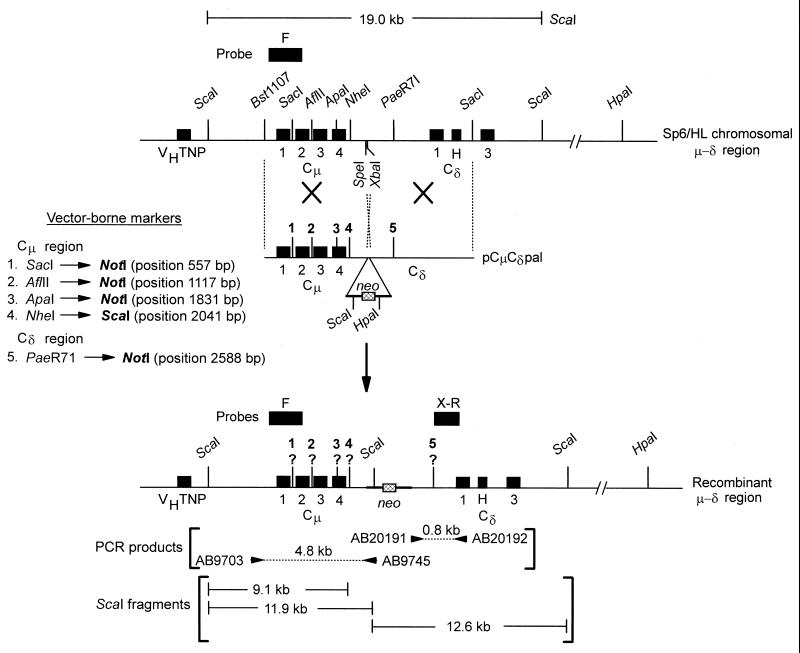
FIG. 2.
Restriction enzyme maps of the Cμ and Cδ region in the recombinants. (A) Diagram of the 4.8-kb PCR product generated from the recombinant Cμ region using primer pair AB9703-AB9745 and the fragment sizes expected if the indicated positions bear either the vector-borne or chromosomal restriction enzyme site markers. (B) Illustration of the Southern blot analysis used to resolve whether the vector-borne palindrome NotI site or chromosomal PaeR71 site resides in the Cδ region. Vector-borne markers are denoted in bold typeface. The diagrams are not drawn to scale.
Gene replacement vector.
The 13.1-kb omega (Ω)-form, enhancer-trap gene replacement vector, pCμCδpal (Fig. 1) was used in these studies. The backbone of pCμCδpal consists of a 5.4-kb segment of pSV2neo (28) from which the 372-bp NsiI/NdeI fragment encompassing the simian virus 40 early-region enhancer responsible for neo gene expression was removed. To effect gene replacement, the flanking arms of homology in pCμCδpal consisted of a 4.2-kb Bst1107/XbaI Cμ region fragment and a 3.5-kb SpeI/SacI Cδ region fragment positioned to the left and right of pSV2neo, respectively. Both fragments were derived from cloned, wild-type Sp6/HL genomic DNA, and their alignment with the Sp6/HL chromosomal μ-δ region is indicated by the dashed lines in Fig. 1. The Cμ and Cδ homology regions share a slight (63-bp) overlap between the SpeI and XbaI sites located 3′ of Cμ.
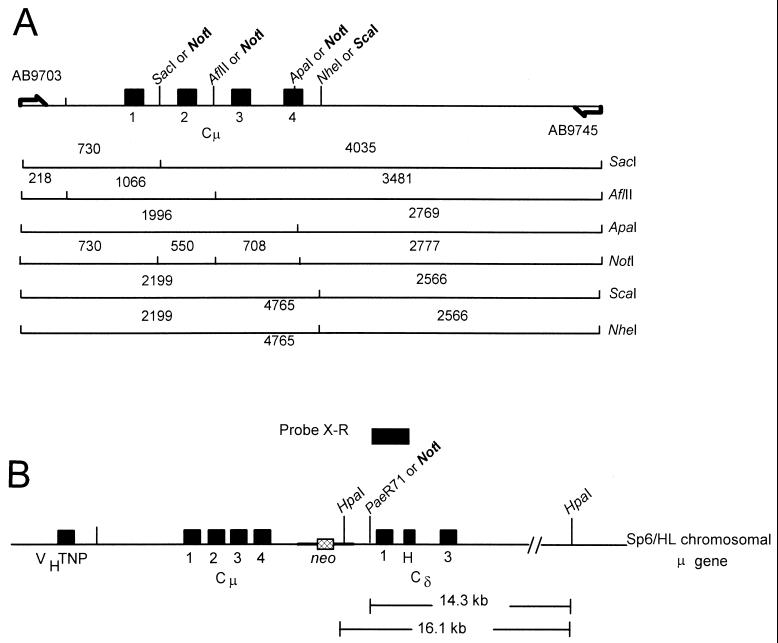
FIG. 1.
Gene replacement at the chromosomal immunoglobulin μ-δ locus. The structure of the haploid, chromosomal immunoglobulin heavy chain μ-δ region in the recipient wild-type mouse hybridoma cell line, Sp6/HL, is presented, along with this locus in a recombinant hybridoma cell line generated as a result of gene replacement with a single copy of the vector, pCμCδpal. In pCμCδpal, endogenous restriction enzyme sites in the Cμ and Cδ regions (denoted in normal typeface) have been replaced with the indicated genetic markers (denoted in bold typeface). The vector-borne NotI site is contained within a perfect, 30-bp palindrome insertion. In the Cμ region, marker positions are designated relative to the Bst1107 site that defines the 5′ end of this segment, while the single marker position in the Cδ region is numbered relative to the SacI site that marks the 3′ end of this segment. For details relating to vector construction, refer to Materials and Methods. Gene replacement has the potential to generate different marker combinations in the various recombinant hybridoma cell lines. Therefore, the five marker positions in the recombinant Cμ and Cδ regions are designated by a question mark. The primer pair AB9703-AB9745 binds outside the Cμ region of homology in the replacement vector and generates a specific 4.8-kb PCR product from the recombinant Cμ region as shown. The sequence of the primers and where they bind have been presented previously (17, 23). The primer pair AB20191-AB20192 binds within the Cδ region of homology and generates a 0.8-kb product. The sequence of the Cδ primers and their binding sites are presented in Materials and Methods. Probe fragments: Cμ-specific probe fragment F is an 870-bp XbaI/BamHI fragment; probe X-R is a 913-bp XhoI/EcoRI fragment from the Cδ region. Abbreviations: Cμ, μ gene constant region; Cδ, δ gene constant region; VHTNP, TNP-specific heavy-chain variable region; neo, neomycin phosphotransferase gene. The thickened line represents the vector, pSV2neo (28). The figure is not drawn to scale.
The Cμ and Cδ homology regions bear specific genetic markers. The Cμ region contains a 30-bp palindrome (5′-GTACTGTATGTGCGGCCGCACATACAGTAC-3′) inserted into the endogenous SacI, AflII, and ApaI sites at bp 557, 1117, and 1831. Palindrome insertion replaces each endogenous site with the novel NotI site in the palindrome (indicated in boldface type in the sequence above). In Fig. 1, the positions of the vector-borne and chromosomal markers are numbered relative to the Bst1107 site that marks the beginning of the vector-borne Cμ region according to the published sequence (4, 7). Originally, the Cμ region genetic markers were constructed for use in other gene targeting vectors, and the relevant details are presented in Li and Baker (16) and Ng and Baker (23). For this study, standard molecular cloning procedures (27) were used to move the markers into the Cμ region of pCμCδpal. As a consequence of the choice of restriction enzyme sites used for subcloning, the vector-borne Cμ region also contained a ScaI site replacing the endogenous NheI site at bp 2041. The Cδ region in pCμCδpal contains a single NotI-palindrome genetic marker that replaces the endogenous PaeR71 site located 2,588 bp from the Cδ terminus. For marker insertion, the Cδ region was digested with PaeR71, and the cohesive ends were ligated to the following oligonucleotide (5′-tcgacGTACTGTATGTGCGGCCGCACATACAGTACg-3′). The oligonucleotide contains the same 30-bp palindrome sequence as indicated above (denoted in uppercase letters) with the embedded diagnostic NotI site (denoted in boldface type). To permit ligation, the oligonucleotide was synthesized with the terminal nucleotides, 5′-tcga-3′. Nucleotides c and g (lowercase underlined) were included at the indicated positions to prevent recreation of the endogenous PaeR71 site. With the exception of the inserted genetic markers, the vector-borne Cμ and Cδ regions were isogenic with the corresponding chromosomal regions in the recipient Sp6/HL hybridoma cell line.
Vector transfer and isolation of independent G418R transformants.
The pCμCδpal vector (8.7 pmol) was transferred to 2 × 107 recipient Sp6/HL hybridoma cells by electroporation as described elsewhere (1). Since the enhancer-trap vector significantly reduces the frequency of random transformants but not targeted recombinants (2, 22), independent G418R transformants could be isolated according to the plating procedure described previously (16, 23).
Recombinant identification and genetic marker analysis.
Genomic DNA was prepared from individual G418R transformants by the method of Gross-Bellard et al. (8). To identify recombinants, individual DNA samples were screened by a PCR assay utilizing the primer pair AB9703-AB9745 that amplifies a specific 4.8-kb Cμ product from recombinant cells as illustrated in Fig. 1. The primer sequences and their binding sites have been reported elsewhere (17, 23). Hybridoma cell lines identified as being recombinants were further characterized by Southern analysis to verify the gene replacement event as detailed in Results. For Southern analysis, restriction enzymes were purchased from New England Biolabs, Inc. (Beverley, Mass.), Amersham Pharmacia Biotech, Inc. (Baie d'Urfé, Québec, Canada), and Canadian Life Technologies, Inc. (Burlington, Ontario, Canada) and used in accordance with the manufacturer's specifications. Gel electrophoresis, transfer of DNA onto nitrocellulose membrane, 32P-labeled probe preparation, and hybridization were all performed according to standard procedures (27).
For determination of the genetic markers residing in the Cμ region of the recombinants, the specific 4.8-kb PCR product was digested separately with enzymes diagnostic of the various vector-borne and chromosomal markers. As described in Results, each enzyme produces diagnostic fragment sizes which can be resolved by standard gel electrophoresis, thus permitting genetic marker assignment to the correct positions. To determine whether the endogenous PaeR71 site or the vector-borne palindrome NotI site resided in the Cδ region, a combination of Southern and PCR analysis was used as explained in Results. The Cδ region PCR made use of primer AB20191 (5′-GAATAGAGCCTAGGAACTGG-3′) that binds to the coding strand at Cδ genomic position bp 15566 and primer AB20192 (5′-CAGGTCCTCCTCTCAATGTA-3′) that binds to the noncoding strand at Cδ genomic position bp 16413. As shown in Fig. 1, this primer pair generates an 848-bp product that spans the PaeR71/NotI site and, as explained in the text below, can be tested for resistance or sensitivity to cleavage with either PaeR71 or NotI.
RESULTS
Recombinant identification.
The purpose of this study was to determine whether genetic markers form hDNA in a cis or a trans configuration during mammalian gene replacement. The gene targeting system is based on the wild-type hybridoma cell line, Sp6/HL, which bears a single copy of the chromosomal immunoglobulin μ-δ region that serves as the target for homologous recombination with the omega (Ω)-form of the enhancer-trap replacement vector, pCμCδpal (Fig. 1). As reported previously (22, 23), enhancer-trap vectors permit efficient isolation of targeted recombinants at the chromosomal μ-δ locus. The 4.2-kb Cμ and 3.5-kb Cδ flanking arms of homology in pCμCδpal were distinguishable from the corresponding regions of the chromosome at several positions as a consequence of inclusion of a 30-bp palindrome containing a unique NotI restriction enzyme site. The palindrome genetic marker is poorly repaired by the mammalian MMR system (16). Thus, following DNA replication and cell division in a recombinant bearing unrepaired hDNA, a sectored (mixed) recombinant is generated. A plating (sectoring) assay was used to recover independent recombinants. As described previously (16, 23), the procedure ensures that each recombinant represents the progeny of a single G418R cell and that the G418R product(s) of recombination is retained for molecular analysis.
Two separate vector transfers were performed. In each, 8.7 pmol of pCμCδpal was introduced into 2 × 107 recipient Sp6/HL hybridoma cells by electroporation as described earlier (1). Within 1 h following electroporation, 0.1-ml aliquots of the culture, each containing ∼6,000 hybridoma cells, were distributed to 3,333 individual wells of 96-well tissue culture plates. Trypan blue staining revealed that, on average, ∼50% of the hybridoma cells survived electroporation. Therefore, each culture well received ∼3,000 viable hybridoma cells. After G418 selection, the number of G418R transformants was recorded. A total of 281 independent G418R transformants were generated from the ∼2 × 107 cells surviving electroporation in the two experiments, giving a transformation frequency of ∼1.4 × 10−5 G418R transformants/cell. Genomic DNA was prepared from 217 independent G418R transformants and screened by PCR using primer pair AB9703-AB9745 for the specific 4.8-kb Cμ region product predicted by the gene replacement reaction (Fig. 1). This screening identified 12 and 16 hybridoma cell lines from the first and second electroporations, respectively, that bore the 4.8-kb Cμ region PCR product. No PCR product was evident in the remaining 189 G418R transformants, suggesting that the endogenous μ-δ region was not targeted (data not shown).
The 28 independent hybridoma cell lines were further characterized by Southern analysis to confirm the gene replacement event. As shown in Fig. 1, linkage of vector-borne and chromosomal sequences is expected to replace the endogenous Sp6/HL 19.0-kb ScaI fragment with ScaI fragments of 11.9 and 12.6 kb detected with probe fragments F and X-R, respectively. In the event, the chromosomal NheI and vector-borne ScaI Cμ region sites are encompassed within hDNA and the mismatch repaired to the ScaI site, the 11.9-kb ScaI fragment detected with probe F will be replaced with a 9.1-kb ScaI fragment as shown. These digests revealed that 26 of the 28 hybridoma cell lines contained the expected ScaI fragment sizes (data not shown). In one of the 26 cell lines (recombinant 34/2), in addition to the expected fragment sizes, additional ScaI fragments consisting of the endogenous 19.0-kb μ-δ region and a novel 15.5-kb fragment were also visible. This suggested that hybridoma cell line 34/2 might actually be derived from two cells; one cell in which the endogenous μ-δ region is targeted and a second cell in which the replacement vector has integrated randomly in the genome and where the endogenous μ-δ region is unaltered. As described below, this interpretation was verified following analysis of hybridoma 34/2 subclones. The fragment sizes observed in 2 of the 28 hybridoma cell lines (37/1 and 8/2) did not fit the expected pattern entirely and are still under investigation.
In summary, of the 28 hybridoma cell lines identified by PCR screening, 26 (or ∼12% of the total G418R transformants analyzed) were verified by Southern analysis as being correct gene replacement events. When expressed as a frequency of the number of hybridoma cells surviving electroporation, the absolute mean frequency of gene replacement was 1.8 × 10−6 G418R recombinants/cell. Since the recovery of recombinants among the 6,666 wells plated in the two electroporations is expected to follow the Poisson distribution, the probability that any of the G418R recombinants actually derived from more than one independent recombinant is ∼0.002.
Genetic marker determination.
For determination of the Cμ region genetic markers, the 4.8-kb PCR product was tested for its resistance or sensitivity to cleavage with restriction enzymes specific for the vector-borne markers, namely, NotI and ScaI, or the corresponding enzymes specific for chromosomal markers, namely, SacI, AflII, ApaI, and NheI (Fig. 1). As shown in Fig. 2A, the various restriction enzymes generate diagnostic Cμ fragment sizes that can be conveniently analyzed by agarose gel electrophoresis. To determine which genetic marker resided in the Cδ region of each recombinant, Southern analysis was utilized. As illustrated in Fig. 2B, digestion of genomic DNA with the HpaI-NotI combination and, in a separate digest, the combination of HpaI and PaeR71 followed by hybridization with probe X-R distinguishes whether the Cδ marker is the vector-borne NotI palindrome or the corresponding endogenous PaeR71 site.
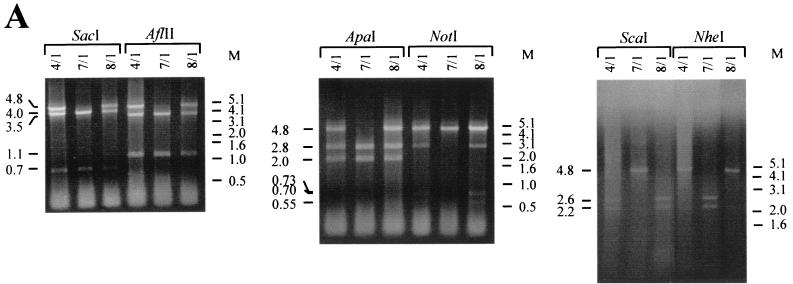
As an example of this analysis, Fig. 3A and B present the determination of the Cμ and Cδ region markers, respectively, for recombinants 4/1, 7/1, and 8/1. With respect to the Cμ region (Fig. 3A), in recombinant 7/1, a single 4.8-kb fragment was visible following NotI digestion. This is the expected size for the undigested PCR product, indicating the absence of NotI sites in the Cμ region of this recombinant. However, the Cμ region PCR product was sensitive to cleavage with SacI, AflII, and ApaI producing the expected fragment sizes in each case. The Cμ region from recombinant 7/1 was also completely sensitive to digestion with NheI, producing the expected 2.2- and 2.6-kb fragments (Fig. 2A) but was resistant to cleavage with ScaI, as evidenced by the presence of the uncut 4.8-kb PCR product. Thus, recombinant 7/1 contains the endogenous SacI, AflII, ApaI, and NheI sites in the Cμ region. In contrast, the Cμ region in recombinants 4/1 and 8/1 was partially sensitive to digestion with the SacI, AflII, and ApaI enzymes diagnostic of the chromosomal markers, as well as NotI specific for the vector-borne palindrome marker, as evidenced by the presence of the expected cleavage products for each enzyme tested as well as residual, uncut 4.8-kb fragment. When the Cμ region PCR product of both recombinants was digested separately with NheI and ScaI, the results revealed complete cleavage with ScaI. Thus, recombinants 4/1 and 8/1 both bear the ScaI marker at Cμ position bp 2041 but are heterogeneous for the remaining Cμ marker positions.
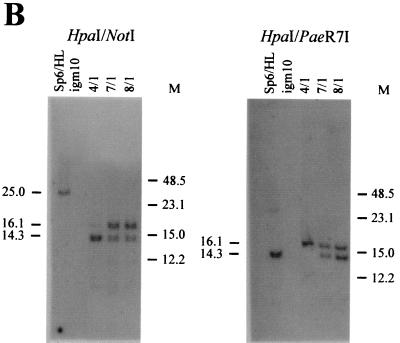
FIG. 3.
Analysis of Cμ and Cδ region marker patterns in recombinants 4/1, 7/1, and 8/1. The results in panels A and B present the analysis of the Cμ and Cδ region genetic markers, respectively, in recombinants 4/1, 7/1, and 8/1. The sizes of fragments of interest are presented on the left of each figure, while the sizes of relevant DNA marker bands (denoted M) are presented on the right.
With respect to the Cδ region (Fig. 3B), Southern analysis of genomic DNA from recombinant 4/1 digested with the combination HpaI-NotI and hybridized with probe X-R revealed the 14.3-kb fragment. When this DNA was digested with HpaI-PaeR71, only the uncut 16.1-kb fragment was visible. Therefore, recombinant 4/1 bears the NotI-palindrome at this site in the Cδ region. In contrast, HpaI/NotI-digested genomic DNA from recombinants 7/1 and 8/1 revealed both 14.3- and 16.1-kb fragments. The 14.3-kb fragment indicates that a portion of the DNA contains the NotI palindrome at the expected position, while the continued presence of the 16.1-kb fragment reveals that, in the remaining DNA, the NotI site is absent. This suggests that these recombinants are heterogeneous at this Cδ marker position, and this was confirmed following digestion with the combination HpaI-PaeR71. Here, a portion of the DNA is sensitive to cleavage with PaeR71, generating the 14.3-kb fragment, while the remaining DNA is insensitive to cleavage with this enzyme, yielding the 16.1-kb fragment (Fig. 3A).
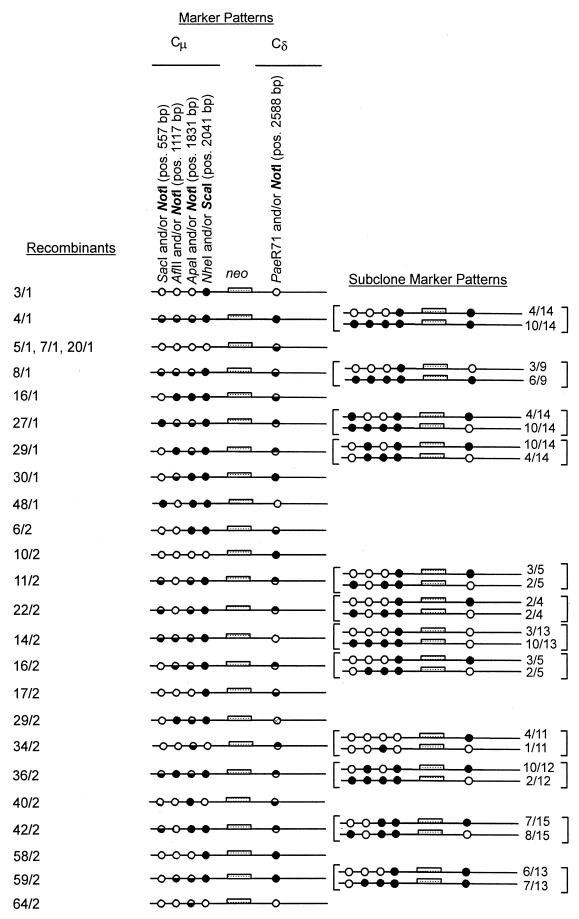
The same analysis was performed on each of the remaining recombinants (data not shown), and the results are summarized in Fig. 4. Recombinants identified from the two separate electroporations are indicated by the coding n/1 or n/2, respectively. In a single recombinant (29/2), neither the vector-borne NotI-palindrome nor the chromosomal PaeR71 marker was present in the Cδ region, suggesting mutation of this site (as indicated by the cross-hatched circle). In the event this was a deletion, it must have been small because the Cδ region ScaI fragment in this recombinant was of the expected size of 12.6 kb (data not shown).
FIG. 4.
Marker segregation patterns in independent recombinants. The results of the Cμ and Cδ region marker analysis for the recombinants are presented. The symbols used to denote the various genetic markers are explained in the text. The results of subcloning analysis of recombinants displaying >1 sectored site, along with the observed frequencies of the individual cell types, are presented in brackets. In the diagrams, the pSV2neo sequences separating the Cμ and Cδ regions are denoted as neo. Positions bearing the vector-borne NotI-palindrome are indicated by a filled circle (●); those with a chromosomal marker are indicated by an open circle (○), while those sites that are sensitive to clevage with restriction enzymes diagnostic of both the chromosome and vector-borne markers (i.e., heterogeneous sites) are indicated by half-filled circles (◒). The cross-hatched circle with recombinant 29/2 indicates that neither the vector-borne NotI-palindrome nor the chromosomal PaeR71 marker was present in the Cδ region.
Clonal analysis of sectored recombinants.
As described above, the recombinant isolation procedure ensures that the individual product(s) of each gene replacement event is confined to a single culture well. Consequently, the marker configurations in the cell populations comprising each sectored recombinant reflect those present in each strand of the hDNA intermediate. As is evident from Fig. 4, recombinants 4/1, 8/1, 27/1, 29/1, 11/2, 22/2, 14/2, 16/2, 34/2, 36/2, 42/2, and 59/2 were sectored for more than one position in the Cμ and/or Cδ region. Therefore, for these recombinants, it was necessary to establish marker linkage. This was accomplished by cloning each recombinant at 0.1 cell/well in 96-well tissue culture plates and repeating the Cμ and Cδ marker determinations on several independent subclones (data not shown). As for the parental recombinants, PCR and gel analysis methods were used to determine the Cμ region markers in the subclones. For the Cδ region, Southern blotting was utilized as described above. In addition, a more convenient PCR-gel analysis assay was developed. As illustrated in Fig. 1, the primer pair AB20192-AB20191 lies within the vector-borne (and chromosomal) Cδ region of homology. These primers amplify a specific 848-bp PCR product that spans the potential mismatch created by the endogenous PaeR71 and vector-borne NotI-palindrome markers. If either PaeR71 or NotI sites are present in the recombinants, the PCR product will be cleaved to yield diagnostic fragments of 644 and 203 bp. Using these procedures, the Cδ region marker patterns in the subclones of the various parental recombinants was determined. Equivalent results were obtained with either Southern or PCR assays. The parental recombinants, the subclone marker patterns, and the frequency of the various subclone types are presented in brackets in Fig. 4.
Subclones of recombinants 4/1, 14/2, and 59/2 contained either the vector-borne NotI-palindrome or the corresponding chromosomal marker at those Cμ region positions that were originally sectored as expected for replication and cell division of a hDNA intermediate. In recombinants 8/1, 27/1, 29/1, 11/2, 22/2, 16/2, 34/2, 36/2, and 42/2, the sectored sites resided in both the Cμ and the Cδ regions and the marker linkage was of particular interest. For all recombinants except 8/1, the markers associated with the sectored sites resided in a trans configuration. That is, in one population of cells, the vector-borne NotI marker in a Cμ region position(s) was linked to the chromosomal PaeR71 site in the Cδ region, while in the other cell population, the vector-borne NotI marker in the Cδ region was linked to a chromosomal marker(s) in the Cμ region. In the single exception (recombinant 8/1) the markers in the sectored sites were linked in a cis configuration. The significance of the marker linkage pattern in these recombinants will be explained in the Discussion.
Southern analysis suggested that recombinant 34/2 may have been derived from two cells, one cell in which a targeted gene replacement has occurred and a second cell in which the replacement vector has integrated randomly. The subcloning results support this interpretation with the evidence as follows. As for the other recombinants, in the single cell undergoing gene replacement, hDNA was formed but not repaired. Thus, following DNA replication and cell division, two cell populations were generated in which the Cμ and Cδ markers were in a trans configuration in the proportions indicated in Fig. 4. The presence of a second cell with an unmodified endogenous μ-δ region and a random vector integration was revealed during subcloning analysis as a third population of cells comprising 6 of the 11 subclones studied. In these cells, the Cμ region could not be amplified with primer pair AB9703-AB9745, a result expected if gene replacement had not occurred (Fig. 1). Random vector integration was revealed by production of the 848-bp Cδ region PCR product with primers AB20192 and AB20191 and by its sensitivity to cleavage with both NotI (expected for the randomly integrated vector) and PaeR71 (expected for the unmodified endogenous locus).
In a few of the parental recombinants, the frequency of subclone types appeared to deviate somewhat from the equivalence expected for replication of the hDNA intermediate (Fig. 4). Likely reasons for this include the small subclone sample size and, perhaps, the loss of some progeny during expansion of the single recombinant cell rather than to any difference between these recombinants and the others with respect to the mechanism of gene replacement.
Evidence for extensive hDNA formation and of MMR during gene replacement.
The positions of sectored sites revealed that hDNA formation during mammalian gene replacement was extensive (Fig. 4). In recombinants 4/1, 8/1, and 14/2, all positions up to and including the ApaI/NotI-palindrome mismatch located 1,831 bp from the beginning of the Cμ region were sectored, suggesting that hDNA had formed over this distance. In recombinants 5/1, 7/1, 20/1, and several others, the PaeR71/NotI-palindrome mismatch located 2,588 bp from the Cδ terminus was sectored, suggesting that a long hDNA tract had spanned this region. Also, as indicated in the previous section, sectoring was observed for internal markers in both the Cμ and Cδ region of several recombinants. These results indicate that in mammalian cells long regions of hDNA are formed in both flanking arms of homology in the replacement vector.
Several examples of repair of mismatches involving the palindrome were evident. In recombinants 11/2, 22/2, and 36/2, sectoring was observed at Cμ marker positions bp 557 and bp 1831 with the internal marker at position bp 1117 being converted to the chromosomal AflII site (recombinants 11/2 and 22/2) or restored to the vector-borne NotI site in recombinant 36/2). In recombinants 27/1, 29/1, and 29/2, a sectored site(s) in the Cμ region was preceded by the vector-borne NotI palindrome marker. This suggested that an hDNA tract spanning at least these marker positions was subject to MMR generating the observed restoration events. In recombinants 34/2, 42/2, and 64/2 a sectored site was followed by a chromosomal marker, suggesting that the original hDNA tract spanning at least these sites was partially converted. In recombinant 48/1, the hDNA tract must have spanned at a minimum the first two Cμ markers with MMR generating the gene conversion at position bp 1117. Finally, in recombinant 40/2, the hDNA tract spanned at least the last two Cμ markers at positions bp 1831 and 2041, with the mismatch at the latter site undergoing gene conversion. During the various subcloning steps that were involved in constructing pCμCδpal, the Cμ region of homology in the vector also contained an additional polymorphism, a simple 4-bp insertion loop that created a ScaI site in place of the endogenous NheI site at position (2041 bp). In contrast to mismatches involving the palindrome, sectoring at the ScaI/NheI polymorphism was never observed. If this site was encompassed within hDNA, as suggested by the data above, then the lack of sectoring is not surprising since our studies have shown that these simple mismatches are usually well repaired prior to DNA replication (23).
As is evident from examination of Fig. 4, of the total of 130 Cμ and Cδ marker positions, 50 of 130 bore a chromosomal marker, 36 of 130 bore the vector-borne NotI palindrome marker and, with the exception of the single Cδ site in recombinant 29/2 where there was no marker, the remaining 43 of the 130 sites were sectored. Of the total of 86 sites that bore either a chromosomal or vector-borne marker, the slightly higher frequency of sites bearing a chromosomal marker was not significantly different according to a χ2 test (P = 0.13).
DISCUSSION
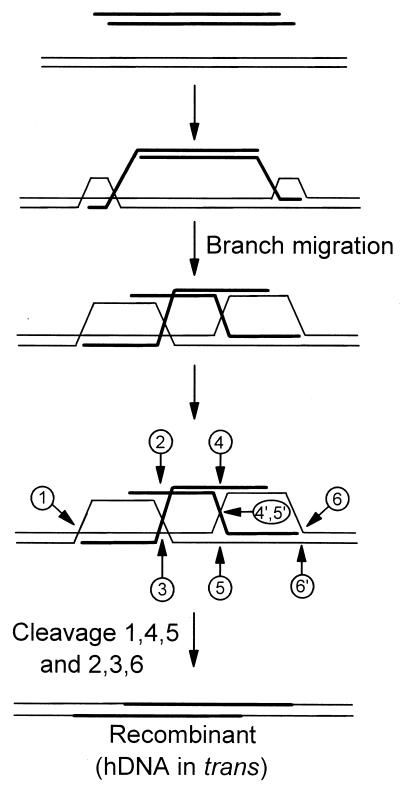
Eight recombinants (8/1, 27/1, 29/1, 11/2, 22/2, 16/2, 34/2, 36/2, and 42/2) were sectored in both flanking arms of homology. Genetic marker analysis in subclones derived from each recombinant revealed that in seven (27/1, 29/1, 11/2, 22/2, 16/2, 34/2, 36/2, and 42/2), vector-borne and chromosomal markers in the two homology regions were linked in a trans configuration. The trans configuration of markers in hDNA is inconsistent with assimilation of a single strand of the vector into the chromosome but, rather, strongly supports a mechanism of gene replacement in mammalian cells that involves two crossing-over events in homologous flanking DNA. A proposed model is presented in Fig. 5. Essentially, this model is the alternative to the single-strand assimilation model as presented in Fig. 3C of a study by Leung et al. (15). Recombination initiates when the two ends of the vector invade the chromosome and pair with their complementary sequences. For simplicity, invasion by single-stranded (perhaps, 3′) ends of opposite DNA strands is shown (30). An alternative, unconventional model might involve invasion of both ends of the same vector strand. Our data do not permit this distinction to be made. In either case, Holliday junction branch migration results in the formation of extensive hDNA in both flanking regions of homology. The entire vector is incorporated into the chromosome by crossover resolution of the two Holliday junctions as a consequence of cutting the DNA strands at positions numbered 1, 4, and 5 and positions number 2, 3, and 6 as indicated. Following strand ligation, genetic markers form hDNA in the trans configuration. The cis configuration of hDNA in the single recombinant (8/1) can also be explained if cuts in the DNA strands are made at the alternate positions of 1 and 6′ and of 2, 3, 4′, and 5′ as shown in the figure. Thus, this simple model readily accounts for the generation of the recombinants. Notable features include pairing between complementary strands of the vector and the chromosomal target, the formation of long regions of hDNA, and the requirement for resolution of two Holliday junctions for vector incorporation into the chromosome. Thus, in these respects the model bears resemblance to the DSBR mechanism of recombination (24, 32), a model that appears consistent with targeted vector insertion in mammalian cells (16, 17, 23, 34).
FIG. 5.
Proposed model for mammalian gene replacement. Recombination is depicted as initiating when different DNA strands from the two homologous arms of the vector invade the chromosome. Gene replacement results from crossing-over in each flanking arm and is accompanied by extensive formation of hDNA. For further mechanistic details, refer to the text.
The finding that mammalian gene replacement occurs by two crossing-over events is of importance in view of current models of gene replacement in yeast in which strand assimilation leading to gene conversion is proposed to occur predominantly (15). In fact, based on the yeast studies, it was suggested that the preferential repair of hDNA in favor of the chromosome might frequently eliminate vector-borne heterologies, including the selectable genetic marker, and thus constitute an efficient barrier to recombination with the Ω-form gene replacement vectors in mammalian cells (15). Thus, the yeast data predict that gene replacement with a linear fragment of DNA contiguous with the chromosome should be much more efficient than that with a replacement vector. Thus, it was surprising that, in our previous study (18), we observed that gene targeting with a linear genomic fragment was only slightly more efficient (∼2-fold) than with an Ω-form replacement vector bearing a similar amount of overall homology. The above data suggest that the mammalian gene replacement reaction is not frequently overcome by the cellular MMR machinery. In support of this, a biased elimination of vector-borne markers as a consequence of MMR was not clearly evident in this or our previous gene targeting studies (18, 23). Also, there was no evidence that inclusion of genetic markers in the vector-borne homology regions reduced the efficiency of gene targeting compared to vectors bearing a completely wild-type, isogenic sequence (18, 23). The differences between yeast and mammalian cells can be reconciled if, as suggested by this study, mammalian cells mediate the gene replacement reaction primarily by two crossing-over events. Thus, in contrast to yeast, a factor(s) other than biased action of the cell's MMR machinery may be responsible for the low efficiency of gene targeting relative to random integration in the mammalian genome.
The gene replacement mechanism in Fig. 5 depends on the two ends of the replacement vector locating the correct target sequence and undergoing strand invasion. Since the vector ends point away from each other, there would appear to be a requirement for them to behave independently for this to be accomplished. Therefore, strand invasion may be an efficient process in mammalian gene targeting. In contrast, strand invasion may be inherently inefficient in yeast, as suggested previously (15). Thus, it would be unlikely that both ends of a linear fragment would independently engage the target sequence and undergo strand invasion before strand assimilation from one end spans the entire fragment.
In the gene replacement vector used in this study, the vector ends were completely homologous to the chromosomal target, a feature which presumably made the strand invasion step relatively straightforward. This is not always the case as replacement vectors designed for use in positive-negative selection (PNS) schemes (19) terminate in heterologous sequences that encode a counterselectable marker(s) (for example, the herpes simplex virus thymidine kinase gene). Therefore, in order for these vector designs to undergo correct gene replacement, the terminal nonhomology must be removed. In S. cerevisiae, removal of nonhomologous DNA ends during recombination depends on the activity of the nucleotide excision repair endonuclease Rad1 or Rad10 and also on the mismatch repair proteins Msh2 and Msh3 (25, 29). In mammalian cells, equivalent proteins acting in the same manner may fulfill this role. Terminal nonhomology may be removed prior to any homologous pairing, thus creating homologous ends that can undergo strand invasion. However, it is also possible that pairing between homologous vector-borne and chromosomal sequences occurs first between more internal sequences in each flanking arm with subsequent removal of the terminal “flaps” and resynthesis, as suggested by the ectopic recombination data of Inbar and Kupiec (12).
The positions of sectored sites in several recombinants provided strong evidence for extensive formation of hDNA in both flanking arms of homology during mammalian gene replacement. This observation is novel and pertinent to the model presented in Fig. 5. Long hDNA tracts as revealed by the positions of sectored sites generated by failure to repair mismatches involving the palindrome have also been reported for targeted vector insertion in mammalian cells (16). The formation of long hDNA tracts during mammalian gene targeting provides a conceptual basis for the gene conversion tracts reported previously by us (18, 23) and others (5, 6, 11) which were suggested to have resulted from MMR of hDNA. Similarly, in the present study, the results suggested that some mismatches in hDNA were also subject to repair undergoing either restoration or gene conversion. However, in some cases, there are difficulties in distinguishing MMR of hDNA from other possible explanations. For example, although MMR of hDNA provides an explanation for the presence of vector-borne markers in proximity to the pSV2neo sequences such as in recombinants 3/1, 4/1, 8/1, 16/1, and others, they are also consistent with the possibility of a crossover event in the Cμ region just prior to the markers. Another example is the presence of chromosomal markers in the Cμ region of several recombinants (3/1, 5/1, 7/1, 20/1, and others), a pattern that, while consistent with gene conversion resulting from MMR of hDNA, might also result from deletion of vector sequences from the ends of the transferred DNA followed by their replacement with chromosomal sequences. While several studies in mammalian cells have suggested that, in general, degradation from the ends of transfected DNA is probably not extensive (6, 10, 12, 16, 23, 26, 27, 30), studies in yeast show that there can be extensive 5′-3′ exonucleolytic resection of 5′-ending strands to expose long single-stranded regions (9, 30). If such extensive resection is a component of the mammalian gene replacement reaction, it would support the two-stranded invasion model of gene replacement depicted in Fig. 5 and may contribute substantially to the conversion and/or restoration events observed in the recombinants.
ACKNOWLEDGMENTS
This work was supported by an operating grant from the Canadian Institutes of Health Research (CIHR) (MOP-14416) to M.D.B. and a CIHR Post-Doctoral Fellowship award to J.L.
We thank Steven Raynard, Patricia Bell, and Richard McCulloch, members of our laboratory, for their helpful comments during the course of this work. We thank Erin Wever for excellent technical assistance.
REFERENCES
- 1.Baker M D, Pennell N, Bosnoyan L, Shulman M J. Homologous recombination can restore normal immunoglobulin production in a mutant hybridoma cell line. Proc Natl Acad Sci USA. 1988;85:6432–6436. doi: 10.1073/pnas.85.17.6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bautista D, Shulman M J. A hit-and-run system for introducing mutations into the immunoglobulin heavy chain locus of hybridoma cells by homologous recombination. J Immunol. 1993;151:1950–1958. [PubMed] [Google Scholar]
- 3.Bertling W M. Gene targeting. In: Vega M A, editor. Gene targeting. Boca Raton, Fla: CRC Press, Inc.; 1995. pp. 1–44. [Google Scholar]
- 4.Bilofsky H S, Burks C, Fickett J W, Goad W B, Lewitter F I, Rindone W P, Swindell C D, Tung C-S. The Gen-Bank® genetic sequence databank. Nucleic Acids Res. 1986;14:1–4. doi: 10.1093/nar/14.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donoho C, Jasin M, Berg P. Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cells. Mol Cell Biol. 1998;18:4070–4078. doi: 10.1128/mcb.18.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliot B, Richardson C, Winderbaum J, Nickoloff J A, Jasin M. Gene conversion tracts from double-strand break repair in mammalian cells. Mol Cell Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldberg G I, Vanin E F, Zrolka A M, Blattner F R. Sequence of the gene for the constant region of the μ chain of Balb/c mouse. Gene. 1981;15:33–42. doi: 10.1016/0378-1119(81)90102-5. [DOI] [PubMed] [Google Scholar]
- 8.Gross-Bellard M, Qudet P, Chambon P. Isolation of high-molecular weight DNA from mammalian cells. Eur J Biochem. 1973;36:32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- 9.Haber J. In vivo biochemistry: physical monitoring of recombination induced by site-specific endonucleases. Bioessays. 1995;17:609–620. doi: 10.1002/bies.950170707. [DOI] [PubMed] [Google Scholar]
- 10.Hastings P J, McGill C, Shafer B, Strathern J N. Ends-in vs. ends-out recombination in yeast. Genetics. 1993;135:973–980. doi: 10.1093/genetics/135.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hasty P, Rivera-Perez J, Bradley A. Gene conversion during vector insertion in embryonic stem cells. Mol Cell Biol. 1995;12:2464–2474. [Google Scholar]
- 12.Inbar O, Kupiec M. Homology search and choice of homologous partner during mitotic recombination. Mol Cell Biol. 1999;19:4134–4142. doi: 10.1128/mcb.19.6.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Köhler G, Potash M J, Lehrach H, Shulman M J. Deletions in immunoglobulin mu chains. EMBO J. 1982;1:555–563. doi: 10.1002/j.1460-2075.1982.tb01208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Köhler G, Shulman M J. Immunoglobulin M mutants. Eur J Immunol. 1980;10:467–476. [Google Scholar]
- 15.Leung W-Y, Malkova A, Haber J E. Gene targeting by linear duplex DNA frequently occurs by assimilation of a single strand that is subject to preferential mismatch correction. Proc Natl Acad Sci USA. 1997;94:6851–6856. doi: 10.1073/pnas.94.13.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Baker M D. Use of a small palindrome genetic marker to investigate mechanisms of double-strand-break repair in mammalian cells. Genetics. 2000;154:1281–1289. doi: 10.1093/genetics/154.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Baker M D. Formation and repair of heteroduplex DNA on both sides of the double-strand break during mammalian gene targeting. J Mol Biol. 2000;295:505–516. doi: 10.1006/jmbi.1999.3400. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Baker M D. Mechanisms involved in targeted gene replacement in mammalian cells. Genetics. 2000;156:809–821. doi: 10.1093/genetics/156.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mansour S L, Thomas K R, Capecchi M R. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 20.Nag D K, White M A, Petes T D. Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature. 1989;340:318–320. doi: 10.1038/340318a0. [DOI] [PubMed] [Google Scholar]
- 21.Negritto M T, Wu X, Kuo T, Chu S, Bailis A M. Influence of DNA sequence identity on efficiency of targeted gene replacement. Mol Cell Biol. 1997;17:278–286. doi: 10.1128/mcb.17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng P, Baker M D. High efficiency, site-specific modification of the chromosomal immunoglobulin locus by gene targeting. J Immunol Methods. 1998;214:81–96. doi: 10.1016/s0022-1759(98)00033-7. [DOI] [PubMed] [Google Scholar]
- 23.Ng P, Baker M D. Mechanisms of double-strand-break repair during gene targeting in mammalian cells. Genetics. 1999;151:1127–1141. doi: 10.1093/genetics/151.3.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orr-Weaver T L, Szostak J W, Rothstein R J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci USA. 1981;78:6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pâques F, Haber J E. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rothstein R. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, F. E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 28.Southern P J, Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Appl Mol Genet. 1981;1:327–341. [PubMed] [Google Scholar]
- 29.Sugawara N, Pâques F, Colaiacovo M, Haber J E. Role of Msh2 and Msh3 repair proteins in double-strand break induced recombination. Proc Natl Acad Sci USA. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun H, Treco D, Szostak J W. Extensive 3′-overhanging, single-stranded DNA associated with meiosis-specific double strand breaks at the ARG4 recombination initiation site. Cell. 1991;64:1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- 31.Surosky R T, Tye B K. Site-directed chromosomal rearrangements in yeast. Methods Enzymol. 1987;153:243–253. doi: 10.1016/0076-6879(87)53057-9. [DOI] [PubMed] [Google Scholar]
- 32.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 33.Waldman A S. Targeted homologous recombination in mammalian cells. Crit Rev Oncol Hematol. 1992;12:49–64. doi: 10.1016/1040-8428(92)90064-w. [DOI] [PubMed] [Google Scholar]
- 34.Valancius V, Smithies O. Double-strand gap repair in a mammalian gene targeting reaction. Mol Cell Biol. 1991;11:4389–4397. doi: 10.1128/mcb.11.9.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vega M A. Gene targeting in human gene therapy. In: Vega M A, editor. Gene targeting. Boca Raton, Fla: CRC Press, Inc.; 1995. pp. 211–229. [Google Scholar]