Abstract
The pathophysiological mechanisms of postoperative delirium (POD) are still not clear, and no reliable biomarker is available to differentiate those with and without POD. Pre- and post-surgery blood from epilepsy subjects undergoing neurosurgery were collected. DNA methylation (DNAm) levels of the TNF gene, IL1B gene, and IL6 gene by the Illumina EPIC array method, and DNAm levels of the TNF gene by pyrosequencing, were analyzed. Blood from 37 subjects were analyzed by the EPIC array method, and blood from 27 subjects were analyzed by pyrosequencing. Several CpGs in the TNF gene in preoperative blood showed a negative correlation between their DNAm and age both in the POD group and in the non-POD group. However, these negative correlations were observed only in the POD group after neurosurgery. Neurosurgery significantly altered DNAm levels at 17 out of 24 CpG sites on the TNF gene, 8 out of 14 CpG sites on the IL1B gene, and 4 out of 14 CpG sites on the IL6 gene. Furthermore, it was found that the Inflammatory Methylation Index (IMI), which was based on the post-surgery DNAm levels at the selected five CpG sites, can be a potential detection tool for delirium with moderate accuracy; area under the curve (AUC) value was 0.84. The moderate accuracy of this IMI was replicated using another cohort from our previous study, in which the AUC was 0.79. Our findings provide further evidence of the potential role of epigenetics and inflammation in the pathophysiology of delirium.
Subject terms: Epigenetics and behaviour, Molecular neuroscience
Introduction
Delirium is an acute and devastating illness that can be caused by various medical and surgical conditions or drugs. Postoperative delirium (POD) occurs in up to 50–70% of high-risk patient groups [1] and is associated with prolonged hospital stay [1–3], higher rates of mortality [1–4], and institutionalization after discharge [5, 6]. Although prevention, early detection, and effective treatment for delirium are important, their effectiveness is limited. Part of the reason is due to a lack of a clear understanding of the pathophysiological mechanisms of delirium, and no reliable biomarker is available to differentiate those with and without delirium. Thus, there is an urgent need to identify biomarkers of delirium risk and to better elucidate the pathophysiology of delirium.
Accumulating evidence suggests that systemic and neuroinflammation play a key role in the development of delirium [7–10]. Many human studies have investigated inflammatory cytokines in delirium subjects [11, 12]. Although a meta-analysis showed increased levels of IL-6 in blood from POD patients, the evidence for other inflammatory markers is still unclear [11, 12].
We have previously reported several epigenetic studies that investigated DNA methylation (DNAm) levels of delirium subjects [13–16]. For example, it was shown that DNAm levels in several CpG sites on the TNF gene were negatively correlated with age only in delirium subjects [16]. We also identified a genome-wide significant CpG at the LDLRAD4 gene [14]. Furthermore, an enrichment analysis demonstrated significant differences related to the immune/inflammatory response pathways between patients with delirium and non-delirium controls [14]. These evidences suggest that DNAm levels, especially at inflammatory-related genes, can be a potential biomarker for delirium. However, no pre- and post-delirium sample to compare was available in the previous studies. Thus, no information was obtainable if those DNAm changes were present before the onset of delirium (as an indicator of their baseline vulnerable state), or if those DNAm alterations occurred after delirium or etiological disease conditions such as infection or surgery (as a marker of disease state). Also, the data mentioned above was based on delirium subjects from diverse medical/surgical conditions and thus prone to have many uncontrollable confounders.
Here, we hypothesized that surgical invasion alters DNAm levels in inflammatory-related genes, which causes systemic inflammation followed by POD, and that altered inflammation-related DNAm levels can be used as a biomarker to detect POD. In order to test this hypothesis and overcome the challenges from our previous studies, in the present study, we collected blood from pre- and post-neurosurgery patients and examined levels of DNAm in inflammatory-related genes, the TNF, IL1B, and IL6, at each time point pre- and post-surgery. We investigated correlations between age and DNAm levels. We also compared DNAm levels of POD patients and ones of non-POD subjects to find a biomarker for POD. Furthermore, we investigated how neurosurgery altered DNAm levels to explore epigenetic mechanisms of POD. Finally, by using significant DNAm differences at several CpG sites, we construct a score to differentiate POD versus non-POD, and tested against an independent sample from our previous cohort to test the performance of such a scoring method.
Patients and methods
Participants
Thirty-nine subjects with medically intractable epilepsy undergoing neurosurgery were recruited between April 2015 and September 2020 at the University of Iowa Hospitals and Clinics. Eighteen of them were subjects who were recruited in our previous studies [17, 18]. Participants formed a consecutive series. This study was approved by the University of Iowa Human Subjects Office Institutional Review Board. We obtained written informed consent from all participants.
Clinical assessment and case definition
Briefly, we obtained demographic information from electronic medical records (EMRs) and patient interviews. We reviewed data on each subject using EMRs to capture if there was evidence of delirium after the surgery. POD-positive cases were identified based on if they tested positive on the Confusion Assessment Method for the Intensive Care Unit (CAM-ICU) [19] or had clinical documentation of altered mental status or confusion consistent with delirium from the EMR. The CAM-ICU was administered by nursing staff. A board-certified consultation-liaison psychiatrist (G.S.), who was blinded to methylation status, reviewed each patient record for final determination of case categorization in question.
Sample collection and processing
Preoperative blood samples were collected in the operating room at the beginning of surgery preparation when intravenous lines were placed following our standard protocol. Thus, this was certainly before the start of the surgical procedure. Postoperative blood samples were again collected in the operating room after the procedure but before awakening from anesthesia. We collected whole blood samples in EDTA tubes. Blood cells were stored at −80 °C until DNA extraction.
DNA extraction and bisulfite conversion
As previously described [14–16, 18], DNA was isolated from blood with the MasterPureTM DNA extraction kit (Epicenter, MCD85201) following the respective protocols. DNA quality was assessed with NanoDrop spectrometry and quantity was assessed with the Qubit™ dsDNA Broad Range Assay Kit (ThermoFisher Scientific, Q32850). DNA was stored at −80 °C until bisulfite conversion. DNA was bisulfite-converted using the EZ DNA Methylation™ Kit (Zymo Research, D5002). Bisulfite-converted DNA was stored at −80 °C until epigenetics analysis. DNA was used for EPIC analysis first, and the remainder of the DNA was used for pyrosequencing. Pyrosequencing was not conducted when enough DNA was not available after EPIC analysis.
A genome-wide analysis using Illumina EPIC array
DNAm of bisulfite-converted blood DNA samples were analyzed with the Infinium HumanMethylationEPIC BeadChip™ Kit (WG-317-1002, Illumina) as previously described [14–16, 18]. The samples were placed on EPIC array chips randomly to avoid potential confounding batch effects. Thus, there were no distortion of sample placement across those chips. DNAm data of 24 CpG sites on the TNF gene, 14 CpG sites on the IL1B gene, and 14 CpG sites on the IL6 gene were extracted and analyzed.
Nested polymerase chain reaction (PCR) and pyrosequencing
Thirty-two CpG sites in the TNF gene were targeted in our pyrosequencing analyses as previously described [16]. Nested PCRs were conducted using bisulfite-converted DNA samples as previously described [16]. After nested PCRs, pyrosequencing was performed using PyroMark Q96 (Qiagen, #972804) and PyroMark Q24 Pyrosequencer (Qiagen, #9001514) to detect levels of DNA methylation.
Statistical analysis
Statistical analyses were performed using R [20]. The student’s t-test was performed to compare DNAm levels between the POD group and the non-POD group. Multiple regression analysis was also performed to compare POD and non-POD; results of the EPIC analysis were corrected for age, sex, and cell distribution, and results of pyrosequencing were corrected for age and sex. Paired t-test was performed to compare DNAm levels pre-surgery and post-surgery. Pearson’s correlation analysis was performed to calculate correlations between age and DNAm levels of each CpG sites. After correction for multiple testing, P values <0.05 (EPIC; P < 0.05/52 = 0.00096, pyrosequencing; P < 0.05/32 = 0.00156) were considered statistically significant.
The receiver operating characteristic (ROC) curve with area under the curve (AUC) were used to analyze the relationship between DNAm level from post-surgery blood and POD. Five CpG sites from each gene were chosen based on the post-neurosurgery results and scores were given based on their best cut-off scores. A total score named “Inflammatory Methylation Index (IMI)” (ranging from 0 to 5) was obtained for each subject, and AUC based on the ROC curve was calculated to evaluate the relationship between IMI and POD. IMI using the same five CpG sites was also obtained from our previous delirium study cohort [14, 16], and a ROC curve was made to test the performance of this scoring method for the detection of delirium. Furthermore, IMIs using other five CpG sites were obtained from DNAm level from pre-surgery blood and amount of change in DNAm level after surgery, and ROC curves were made to test the performance of this scoring method for prediction of POD. Details of IMI calculation are described in the supplementary method.
Results
Study subject demographics
Totally, 39 subjects were enrolled in this study; 37 subjects (10 POD subjects and 27 non-POD subjects) were included in the EPIC array analysis, and 27 subjects (7 POD subjects and 20 non-POD subjects) were included in pyrosequencing analysis. Demographic information is shown in Supplementary Table 1. Mean age was higher in the POD group than in the non-POD group. Sex, race, alcohol usage, tobacco usage, American Society of Anesthesiologists (ASA) physical status, body mass index (BMI), anesthesia time, procedure time, or amount of bleeding was not different between the POD group and non-POD group (Supplementary Table 1). There were no significant differences in CD8 T cells, CD4 T cells, natural killer cells, B cells, and monocytes between the POD group and the non-POD group (Supplementary Table 2).
DNA methylation before surgery
TNF gene
Data from EPIC analysis demonstrated that there was no difference of DNAm level in the TNF gene before surgery between the POD group and the non-POD group (Supplementary Table 3). Negative correlations between age and DNAm level were observed in several CpG sites on the TNF gene with statistical significance (Table 1). These negative correlations were observed both in the POD group and in the non-POD group (Table 1). Pyrosequencing analysis demonstrated the same tendency. There was no difference in DNA methylation level before surgery between the POD group and the non-POD group (Supplementary Table 3). Negative correlations between age and DNAm were observed in several CpG sites on the TNF gene with nominal significances (Table 1). These negative correlations were observed both in the POD group and in a non-POD group (Table 1).
Table 1.
Correlations between age and pre-surgery DNAm in the TNF gene.
| EPIC | EPIC | EPIC | pyrosequencing | pyrosequencing | pyrosequencing | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| all | POD | non-POD | all | POD | non-POD | ||||||||
| N = 37 | N = 10 | N = 27 | N = 27 | N = 7 | N = 20 | ||||||||
| cg | location | r | p | r | p | r | p | r | p | r | p | r | p |
| chr6: 31542460 | −0.43 | 0.03 | −0.57 | 0.18 | −0.29 | 0.22 | |||||||
| cg08639424 | chr6: 31542556 | −0.18 | 0.28 | −0.21 | 0.56 | −0.17 | 0.39 | ||||||
| cg19978379 | chr6: 31542671 | 0.28 | 0.10 | 0.01 | 0.98 | 0.40 | 0.04 | ||||||
| cg24452282 | chr6: 31542741 | 0.22 | 0.18 | 0.14 | 0.70 | 0.27 | 0.17 | ||||||
| chr6: 31543170 | −0.47 | 0.01 | −0.62 | 0.14 | −0.42 | 0.06 | |||||||
| chr6: 31543176 | −0.46 | 0.02 | −0.70 | 0.08 | −0.41 | 0.07 | |||||||
| chr6: 31543178 | −0.54 | 0.004 | −0.62 | 0.14 | −0.39 | 0.09 | |||||||
| chr6: 31543193 | −0.51 | 0.006 | −0.56 | 0.19 | −0.49 | 0.028 | |||||||
| cg21370522 | chr6: 31543219 | −0.17 | 0.32 | −0.03 | 0.93 | −0.29 | 0.15 | −0.49 | 0.009 | −0.57 | 0.18 | −0.47 | 0.035 |
| cg19648923 | chr6: 31543266 | −0.40 | 0.01 | −0.32 | 0.37 | −0.44 | 0.02 | −0.38 | 0.051 | −0.54 | 0.22 | −0.31 | 0.18 |
| cg01569083 | chr6: 31543290 | −0.39 | 0.02 | −0.29 | 0.41 | −0.47 | 0.01 | −0.53 | 0.005 | −0.64 | 0.12 | −0.46 | 0.040 |
| cg03037030 | chr6: 31543300 | −0.47 | 0.004 | −0.43 | 0.21 | −0.47 | 0.01 | −0.40 | 0.04 | −0.53 | 0.23 | −0.37 | 0.11 |
| chr6: 31543430 | −0.48 | 0.01 | −0.41 | 0.36 | −0.41 | 0.08 | |||||||
| chr6: 31543487 | −0.45 | 0.02 | −0.33 | 0.46 | −0.42 | 0.07 | |||||||
| cg12681001 | chr6: 31543541 | −0.52 | *0.0009 | −0.42 | 0.22 | −0.58 | 0.0014 | −0.38 | 0.049 | −0.13 | 0.78 | −0.37 | 0.10 |
| cg21222743 | chr6: 31543546 | −0.60 | *<0.0001 | −0.66 | 0.04 | −0.57 | 0.0018 | −0.42 | 0.03 | −0.20 | 0.67 | −0.42 | 0.07 |
| cg10717214 | chr6: 31543558 | −0.58 | *0.0002 | −0.70 | 0.02 | −0.54 | 0.0038 | −0.02 | 0.94 | 0.11 | 0.83 | −0.07 | 0.79 |
| cg04425624 | chr6: 31543566 | −0.50 | 0.0016 | −0.62 | 0.06 | −0.50 | 0.0081 | −0.24 | 0.26 | −0.22 | 0.68 | −0.30 | 0.24 |
| cg21467614 | chr6: 31543638 | −0.64 | *<0.0001 | −0.70 | 0.03 | −0.66 | *0.0002 | ||||||
| cg08553327 | chr6: 31543647 | −0.57 | *0.0003 | −0.63 | 0.05 | −0.56 | 0.0023 | ||||||
| cg26729380 | chr6: 31543655 | −0.62 | *<0.0001 | −0.71 | 0.02 | −0.59 | 0.0013 | ||||||
| cg10650821 | chr6: 31543686 | −0.46 | 0.0039 | −0.37 | 0.29 | −0.52 | 0.0056 | ||||||
| chr6: 31544695 | −0.03 | 0.89 | 0.11 | 0.82 | 0.01 | 0.98 | |||||||
| chr6: 31544752 | −0.12 | 0.56 | −0.04 | 0.92 | −0.24 | 0.31 | |||||||
| chr6: 31544800 | −0.05 | 0.80 | −0.20 | 0.67 | −0.18 | 0.46 | |||||||
| chr6: 31544822 | −0.05 | 0.82 | 0.13 | 0.78 | −0.24 | 0.31 | |||||||
| chr6: 31544848 | −0.24 | 0.22 | −0.76 | 0.047 | −0.34 | 0.14 | |||||||
| cg01360627 | chr6: 31544931 | −0.09 | 0.60 | −0.11 | 0.75 | −0.08 | 0.70 | −0.11 | 0.60 | −0.19 | 0.68 | −0.29 | 0.22 |
| cg23384708 | chr6: 31544934 | −0.01 | 0.96 | 0.06 | 0.87 | −0.27 | 0.17 | −0.01 | 0.96 | −0.03 | 0.95 | −0.24 | 0.31 |
| cg20477259 | chr6: 31544960 | 0.01 | 0.97 | 0.15 | 0.69 | −0.13 | 0.53 | 0.03 | 0.89 | 0.01 | 0.98 | −0.03 | 0.89 |
| chr6: 31545253 | −0.02 | 0.92 | 0.52 | 0.23 | −0.20 | 0.39 | |||||||
| chr6: 31545258 | −0.11 | 0.58 | 0.27 | 0.56 | −0.36 | 0.11 | |||||||
| cg15989608 | chr6: 31545322 | −0.21 | 0.20 | −0.31 | 0.38 | −0.25 | 0.21 | −0.11 | 0.57 | 0.25 | 0.59 | −0.40 | 0.08 |
| chr6: 31545267 | −0.09 | 0.66 | 0.13 | 0.80 | −0.38 | 0.11 | |||||||
| cg26736341 | chr6: 31545343 | −0.18 | 0.28 | −0.24 | 0.51 | −0.26 | 0.20 | 0.28 | 0.17 | 0.51 | 0.30 | 0.28 | 0.24 |
| chr6: 31545432 | 0.10 | 0.63 | 0.23 | 0.62 | −0.12 | 0.62 | |||||||
| cg04472685 | chr6: 31545474 | −0.11 | 0.53 | −0.15 | 0.68 | −0.14 | 0.49 | −0.32 | 0.15 | 0.18 | 0.77 | −0.46 | 0.06 |
| cg19124225 | chr6: 31545836 | −0.08 | 0.63 | 0.02 | 0.96 | 0.01 | 0.97 | ||||||
| cg02137984 | chr6: 31545899 | −0.34 | 0.04 | −0.40 | 0.25 | −0.21 | 0.29 | ||||||
| cg06825478 | chr6: 31546068 | 0.10 | 0.54 | −0.17 | 0.63 | 0.14 | 0.48 | 0.21 | 0.29 | 0.44 | 0.32 | 0.15 | 0.52 |
| chr6: 31546086 | 0.00 | 0.99 | 0.22 | 0.64 | −0.10 | 0.67 |
underline nominal significance (P < 0.05).
*Significant after correction for multiple testing level (EPIC; P < 0.05/52 = 0.00096, pyrosequencing; P < 0.05/32 = 0.00156).
IL1B and IL6
There was no difference in DNA methylation level before surgery between the POD group and the non-POD group (Supplementary Table 4). No CpG site showed a correlation between age and DNAm level with statistical significance (Supplementary Table 5).
DNA methylation after surgery
TNF gene
The EPIC analysis demonstrated that there was no significant difference in DNAm level after surgery between the POD group and the non-POD group (Supplementary Table 6). Moderate negative correlations between age and DNAm level, observed in pre-surgery samples, were observed in several CpG sites (from chr6: 31543546 to chr6: 31543655) on the TNF gene in the POD group, but not in the non-POD group (Table 2).
Table 2.
Correlations between age and post-surgery DNAm in the TNF gene.
| EPIC | EPIC | EPIC | pyrosequencing | pyrosequencing | pyrosequencing | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| all | POD | non-POD | all | POD | non-POD | ||||||||
| N = 37 | N = 10 | N = 27 | N = 27 | N = 7 | N = 20 | ||||||||
| cg | location | r | p | r | p | r | p | r | p | r | p | r | p |
| chr6: 31542460 | 0.09 | 0.67 | 0.01 | 0.98 | 0.13 | 0.60 | |||||||
| cg08639424 | chr6: 31542556 | −0.03 | 0.84 | −0.50 | 0.14 | −0.08 | 0.69 | ||||||
| cg19978379 | chr6: 31542671 | 0.17 | 0.33 | −0.14 | 0.70 | 0.19 | 0.34 | ||||||
| cg24452282 | chr6: 31542741 | 0.19 | 0.25 | 0.00 | 0.99 | 0.17 | 0.40 | ||||||
| chr6: 31543170 | −0.33 | 0.10 | −0.44 | 0.32 | −0.54 | 0.01 | |||||||
| chr6: 31543176 | –0.43 | 0.02 | -0.33 | 0.47 | −0.53 | 0.02 | |||||||
| chr6: 31543178 | −0.21 | 0.29 | −0.08 | 0.87 | −0.22 | 0.34 | |||||||
| chr6: 31543193 | −0.49 | 0.01 | −0.61 | 0.14 | −0.45 | 0.046 | |||||||
| cg21370522 | chr6: 31543219 | 0.26 | 0.13 | 0.23 | 0.51 | 0.28 | 0.16 | −0.34 | 0.08 | −0.44 | 0.32 | −0.46 | 0.04 |
| cg19648923 | chr6: 31543266 | 0.00 | 0.98 | 0.00 | 0.99 | 0.02 | 0.91 | −0.26 | 0.18 | −0.39 | 0.39 | −0.33 | 0.16 |
| cg01569083 | chr6: 31543290 | 0.06 | 0.71 | −0.09 | 0.80 | 0.18 | 0.36 | −0.47 | 0.01 | −0.30 | 0.51 | −0.55 | 0.01 |
| cg03037030 | chr6: 31543300 | −0.03 | 0.84 | −0.18 | 0.61 | 0.07 | 0.73 | −0.29 | 0.14 | 0.03 | 0.96 | −0.40 | 0.08 |
| chr6: 31543430 | −0.44 | 0.02 | −0.60 | 0.16 | −0.41 | 0.07 | |||||||
| chr6: 31543487 | −0.38 | 0.05 | −0.48 | 0.28 | −0.33 | 0.16 | |||||||
| cg12681001 | chr6: 31543541 | −0.02 | 0.89 | 0.09 | 0.81 | 0.02 | 0.93 | −0.12 | 0.54 | −0.18 | 0.69 | −0.17 | 0.47 |
| cg21222743 | chr6: 31543546 | −0.36 | 0.03 | −0.84 | 0.003 | −0.22 | 0.28 | −0.33 | 0.09 | −0.18 | 0.69 | −0.40 | 0.08 |
| cg10717214 | chr6: 31543558 | −0.29 | 0.08 | −0.57 | 0.09 | −0.22 | 0.27 | 0.37 | 0.09 | 0.24 | 0.70 | 0.34 | 0.18 |
| cg04425624 | chr6: 31543566 | −0.16 | 0.35 | −0.34 | 0.33 | −0.04 | 0.85 | 0.10 | 0.65 | −0.85 | 0.07 | 0.30 | 0.25 |
| cg21467614 | chr6: 31543638 | −0.33 | 0.04 | −0.56 | 0.09 | −0.22 | 0.27 | ||||||
| cg08553327 | chr6: 31543647 | −0.24 | 0.15 | −0.37 | 0.30 | −0.13 | 0.53 | ||||||
| cg26729380 | chr6: 31543655 | −0.37 | 0.03 | −0.51 | 0.13 | −0.26 | 0.20 | ||||||
| cg10650821 | chr6: 31543686 | −0.16 | 0.35 | 0.03 | 0.93 | −0.24 | 0.22 | ||||||
| chr6: 31544695 | 0.02 | 0.93 | −0.25 | 0.59 | 0.21 | 0.37 | |||||||
| chr6: 31544752 | 0.06 | 0.77 | 0.65 | 0.11 | −0.06 | 0.79 | |||||||
| chr6: 31544800 | 0.27 | 0.18 | 0.28 | 0.55 | 0.05 | 0.82 | |||||||
| chr6: 31544822 | 0.18 | 0.36 | 0.36 | 0.43 | 0.15 | 0.53 | |||||||
| chr6: 31544848 | −0.07 | 0.72 | −0.11 | 0.82 | −0.33 | 0.16 | |||||||
| cg01360627 | chr6: 31544931 | −0.05 | 0.79 | −0.21 | 0.57 | −0.09 | 0.67 | 0.16 | 0.44 | 0.04 | 0.92 | 0.05 | 0.85 |
| cg23384708 | chr6: 31544934 | 0.16 | 0.35 | 0.31 | 0.38 | 0.06 | 0.77 | 0.29 | 0.14 | 0.64 | 0.12 | 0.13 | 0.58 |
| cg20477259 | chr6: 31544960 | 0.07 | 0.69 | 0.08 | 0.82 | 0.01 | 0.96 | 0.25 | 0.21 | 0.36 | 0.43 | 0.13 | 0.57 |
| chr6: 31545253 | 0.09 | 0.67 | −0.29 | 0.53 | 0.22 | 0.34 | |||||||
| chr6: 31545258 | 0.01 | 0.95 | 0.33 | 0.48 | −0.12 | 0.62 | |||||||
| chr6: 31545267 | 0.16 | 0.43 | 0.17 | 0.72 | 0.20 | 0.39 | |||||||
| cg15989608 | chr6: 31545322 | 0.04 | 0.80 | −0.26 | 0.47 | 0.05 | 0.81 | 0.10 | 0.60 | 0.52 | 0.23 | 0.16 | 0.51 |
| cg26736341 | chr6: 31545343 | 0.05 | 0.78 | −0.36 | 0.31 | 0.10 | 0.61 | 0.06 | 0.77 | 0.53 | 0.22 | −0.18 | 0.44 |
| chr6: 31545432 | 0.07 | 0.73 | −0.28 | 0.54 | 0.05 | 0.83 | |||||||
| cg04472685 | chr6: 31545474 | 0.00 | 0.99 | 0.20 | 0.57 | −0.08 | 0.68 | −0.05 | 0.84 | −0.44 | 0.39 | 0.07 | 0.80 |
| cg19124225 | chr6: 31545836 | 0.07 | 0.70 | 0.51 | 0.13 | −0.20 | 0.32 | ||||||
| cg02137984 | chr6: 31545899 | −0.03 | 0.87 | −0.24 | 0.50 | −0.02 | 0.91 | ||||||
| cg06825478 | chr6: 31546068 | −0.15 | 0.37 | −0.12 | 0.75 | −0.22 | 0.27 | 0.31 | 0.12 | 0.38 | 0.40 | 0.30 | 0.20 |
| chr6: 31546086 | 0.18 | 0.38 | 0.24 | 0.61 | 0.17 | 0.48 |
underline nominal significance (P < 0.05).
Pyrosequencing analysis did not demonstrate any DNA methylation differences between the POD group and the non-POD group (Supplementary Table 6). In the pyrosequencing analysis, negative correlations between age and DNAm were observed in several CpG sites on the TNF gene both in the POD group and in the non-POD group (Table 2).
IL1B and IL6
There was no difference in DNA methylation level after surgery between the POD group and the non-POD group (Supplementary Table 7). No CpG site showed a correlation between age and DNA methylation with statistical significance (Supplementary Table 8).
Change of DNA methylation by neurosurgery
Among 24 CpG sites on the TNF gene measured by EPIC analysis, 11 CpG sites showed a significant increase of DNA methylation and six CpG sites showed a significant decrease of DNA methylation after neurosurgery in all subjects (Table 3). The same patterns of DNAm change were shown in pyrosequencing. Eight CpG sites showed a significant increase of DNAm level and one CpG site showed a significant decrease of DNA methylation after neurosurgery (Table 3). DNAm in most of CpG sites upstream from chr6: 31542741 and downstream from chr6: 31544695 increased after neurosurgery. Otherwise, DNAm in CpG sites from chr6: 31543219 to chr6: 31543686 decreased after neurosurgery. However, the degree of DNAm changes by the neurosurgery between the POD group and non-POD group were not different with statistical significance either in EPIC analysis or in pyrosequencing (Supplementary Table 9).
Table 3.
Changes of DNAm in TNF gene after neurosurgery.
| EPIC | EPIC | EPIC | pyrosequencing | pyrosequencing | pyrosequencing | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N = 37 | N = 10 | N = 27 | N = 27 | N = 7 | N = 20 | ||||||||||||||||||||
| all | POD | non-POD | all | POD | non-POD | ||||||||||||||||||||
| pre | post | paired-t | pre | post | paired-t | pre | post | paired-t | pre | post | paired-t | pre | post | paired-t | pre | post | paired-t | ||||||||
| cg | location | mean | mean | diff | p value | mean | mean | diff | p value | mean | mean | diff | p value | mean | mean | diff | p value | mean | mean | diff | p value | mean | mean | diff | p value |
| chr6: 31542460 | 74.0 | 78.1 | 4.1 | *0.0004 | 72.7 | 78.1 | 5.4 | 0.10 | 74.5 | 78.1 | 3.6 | 0.00158 | |||||||||||||
| cg08639424 | chr6: 31542556 | 77.9 | 86.2 | 8.3 | *<0.0001 | 77.7 | 88.0 | 10.3 | 0.001 | 78.0 | 85.5 | 7.5 | *<0.0001 | ||||||||||||
| cg19978379 | chr6: 31542671 | 71.8 | 81.0 | 9.2 | *<0.0001 | 73.0 | 81.9 | 8.9 | 0.001 | 71.3 | 80.6 | 9.3 | *<0.0001 | ||||||||||||
| cg24452282 | chr6: 31542741 | 73.5 | 84.5 | 11.1 | *<0.0001 | 73.8 | 86.9 | 13.1 | *<0.0001 | 73.3 | 83.6 | 10.3 | *<0.0001 | ||||||||||||
| chr6: 31543170 | 7.0 | 5.7 | −1.4 | 0.02 | 6.5 | 6.4 | −0.1 | 0.94 | 7.2 | 5.4 | −1.8 | 0.007 | |||||||||||||
| chr6: 31543176 | 8.9 | 7.5 | −1.3 | 0.03 | 8.0 | 7.5 | −0.5 | 0.57 | 9.2 | 7.6 | −1.6 | 0.04 | |||||||||||||
| chr6: 31543178 | 3.3 | 2.6 | −0.7 | 0.15 | 2.0 | 2.3 | 0.3 | 0.84 | 3.8 | 2.7 | −1.1 | 0.03 | |||||||||||||
| chr6: 31543193 | 5.2 | 4.0 | −1.3 | 0.01 | 4.6 | 3.5 | −1.1 | 0.32 | 5.5 | 4.1 | −1.3 | 0.02 | |||||||||||||
| cg21370522 | chr6: 31543219 | 13.2 | 14.1 | 0.9 | 0.34 | 13.6 | 14.4 | 0.8 | 0.64 | 13.0 | 13.9 | 1.0 | 0.41 | 4.2 | 3.5 | −0.8 | 0.02 | 3.8 | 3.6 | −0.2 | 0.75 | 4.4 | 3.4 | −0.9 | 0.008 |
| cg19648923 | chr6: 31543266 | 10.6 | 8.1 | −2.4 | *0.0002 | 10.3 | 7.8 | −2.5 | 0.005 | 10.6 | 8.2 | −2.4 | 0.004 | 2.9 | 2.3 | −0.6 | 0.02 | 2.7 | 2.4 | −0.3 | 0.63 | 3.0 | 2.3 | −0.8 | 0.02 |
| cg01569083 | chr6: 31543290 | 18.9 | 17.4 | −1.5 | 0.14 | 19.0 | 16.8 | −2.2 | 0.12 | 18.8 | 17.7 | −1.2 | 0.36 | 3.3 | 2.6 | −0.7 | 0.02 | 2.9 | 2.5 | −0.4 | 0.55 | 3.5 | 2.7 | −0.8 | 0.02 |
| cg03037030 | chr6: 31543300 | 6.3 | 4.9 | −1.4 | 0.008 | 5.6 | 4.5 | −1.1 | 0.06 | 6.6 | 5.0 | −1.6 | 0.03 | 9.0 | 7.8 | −1.2 | 0.02 | 8.5 | 7.6 | −0.9 | 0.44 | 9.1 | 7.8 | −1.3 | 0.02 |
| chr6: 31543430 | 1.9 | 2.0 | 0.0 | 0.97 | 1.5 | 1.9 | 0.4 | 0.42 | 2.1 | 2.0 | −0.1 | 0.55 | |||||||||||||
| chr6: 31543487 | 6.3 | 5.3 | −1.0 | 0.046 | 5.3 | 4.9 | −0.4 | 0.60 | 6.6 | 5.4 | −1.2 | 0.06 | |||||||||||||
| cg12681001 | chr6: 31543541 | 9.0 | 7.3 | −1.8 | 0.002 | 8.8 | 6.7 | −2.0 | 0.04 | 9.1 | 7.5 | −1.7 | 0.02 | 10.4 | 9.1 | −1.3 | 0.052 | 9.1 | 9.5 | 0.4 | 0.67 | 10.9 | 9.0 | −1.9 | 0.02 |
| cg21222743 | chr6: 31543546 | 6.6 | 4.0 | −2.6 | *<0.0001 | 5.8 | 3.4 | −2.4 | 0.003 | 6.9 | 4.2 | −2.7 | *0.0002 | 8.8 | 7.3 | −1.5 | 0.01 | 7.7 | 7.1 | −0.6 | 0.57 | 9.2 | 7.4 | −1.8 | 0.01 |
| cg10717214 | chr6: 31543558 | 9.3 | 6.4 | −2.9 | *<0.0001 | 8.3 | 6.0 | −2.2 | 0.002 | 9.7 | 6.5 | −3.1 | *0.0002 | 33.5 | 32.7 | −0.8 | 0.47 | 33.6 | 35.0 | 1.4 | 0.48 | 33.5 | 32.0 | −1.5 | 0.34 |
| cg04425624 | chr6: 31543566 | 17.4 | 13.1 | −4.3 | *0.0003 | 17.2 | 12.0 | −5.2 | 0.01 | 17.5 | 13.5 | −3.9 | 0.006 | 21.2 | 20.8 | −0.4 | 0.86 | 20.8 | 21.6 | 0.8 | 0.57 | 21.3 | 20.5 | −0.8 | 0.62 |
| cg21467614 | chr6: 31543638 | 17.7 | 14.7 | −3.0 | *<0.0001 | 17.1 | 14.1 | −3.0 | *0.0005 | 17.9 | 15.0 | −2.9 | 0.003 | ||||||||||||
| cg08553327 | chr6: 31543647 | 16.5 | 13.1 | −3.4 | 0.001 | 15.4 | 11.6 | −3.8 | 0.003 | 16.9 | 13.7 | −3.2 | 0.02 | ||||||||||||
| cg26729380 | chr6: 31543655 | 11.5 | 7.9 | −3.7 | *<0.0001 | 10.6 | 6.4 | −4.2 | 0.005 | 11.9 | 8.4 | −3.5 | 0.001 | ||||||||||||
| cg10650821 | chr6: 31543686 | 11.6 | 10.1 | −1.5 | 0.007 | 11.4 | 10.0 | −1.4 | 0.10 | 11.7 | 10.2 | −1.5 | 0.03 | ||||||||||||
| chr6: 31544695 | 65.4 | 72.4 | 7.1 | 0.007 | 63.2 | 69.4 | 6.2 | 0.36 | 66.1 | 73.5 | 7.4 | 0.008 | |||||||||||||
| chr6: 31544752 | 77.2 | 85.9 | 8.8 | *<0.0001 | 78.2 | 86.2 | 8.0 | 0.10 | 76.8 | 85.9 | 9.0 | *0.0004 | |||||||||||||
| chr6: 31544800 | 22.7 | 24.4 | 1.7 | 0.01 | 24.0 | 26.3 | 2.3 | 0.25 | 22.3 | 23.8 | 1.5 | 0.02 | |||||||||||||
| chr6: 31544822 | 56.6 | 62.2 | 5.6 | *0.0005 | 58.1 | 62.5 | 4.4 | 0.13 | 56.0 | 62.1 | 6.1 | 0.002 | |||||||||||||
| chr6: 31544848 | 13.3 | 13.3 | 0.0 | 0.95 | 14.6 | 14.0 | −0.6 | 0.51 | 12.8 | 13.0 | 0.2 | 0.56 | |||||||||||||
| cg01360627 | chr6: 31544931 | 70.0 | 78.6 | 8.7 | *<0.0001 | 69.8 | 80.2 | 10.3 | *<0.0001 | 70.0 | 78.1 | 8.0 | *<0.0001 | 25.9 | 27.3 | 1.4 | 0.03 | 27.4 | 28.9 | 1.5 | 0.26 | 25.3 | 26.7 | 1.4 | 0.09 |
| cg23384708 | chr6: 31544934 | 64.1 | 72.2 | 8.1 | *<0.0001 | 67.4 | 74.0 | 6.7 | 0.004 | 63.0 | 71.6 | 8.6 | *<0.0001 | 74.0 | 82.5 | 8.5 | *0.0001 | 77.3 | 83.9 | 6.6 | 0.18 | 72.8 | 82.0 | 9.2 | *0.0004 |
| cg20477259 | chr6: 31544960 | 61.6 | 71.8 | 10.2 | *<0.0001 | 62.5 | 73.4 | 10.9 | *<0.0001 | 61.3 | 71.3 | 10.0 | *<0.0001 | 67.9 | 74.7 | 6.8 | *0.0004 | 68.9 | 76.4 | 7.5 | 0.09 | 67.5 | 74.1 | 6.6 | 0.003 |
| chr6: 31545253 | 88.1 | 91.9 | 3.8 | 0.003 | 87.9 | 92.3 | 4.4 | 0.17 | 88.2 | 91.8 | 3.6 | 0.010 | |||||||||||||
| chr6: 31545258 | 87.7 | 91.6 | 3.8 | 0.009 | 88.7 | 92.2 | 3.5 | 0.25 | 87.4 | 91.3 | 3.9 | 0.02 | |||||||||||||
| chr6: 31545267 | 82.0 | 87.8 | 5.8 | *0.0005 | 84.1 | 87.8 | 3.7 | 0.33 | 81.3 | 87.8 | 6.5 | *0.0005 | |||||||||||||
| cg15989608 | chr6: 31545322 | 71.3 | 79.6 | 8.4 | *<0.0001 | 72.2 | 80.8 | 8.7 | 0.001 | 70.9 | 79.1 | 8.2 | *<0.0001 | 90.4 | 93.6 | 3.3 | 0.01 | 92.1 | 91.9 | −0.2 | 0.83 | 89.8 | 94.2 | 4.4 | *0.0006 |
| cg26736341 | chr6: 31545343 | 70.0 | 79.4 | 9.4 | *<0.0001 | 71.4 | 80.6 | 9.3 | *0.0008 | 69.5 | 79.0 | 9.4 | *<0.0001 | 78.9 | 81.0 | 2.0 | 0.04 | 78.0 | 82.4 | 4.4 | 0.28 | 79.2 | 80.4 | 1.3 | 0.10 |
| chr6: 31545432 | 87.0 | 89.7 | 2.7 | 0.17 | 89.9 | 91.4 | 1.5 | 0.44 | 85.9 | 89.1 | 3.2 | 0.23 | |||||||||||||
| cg04472685 | chr6: 31545474 | 81.4 | 87.5 | 6.1 | *<0.0001 | 82.0 | 87.9 | 6.0 | 0.002 | 81.2 | 87.4 | 6.2 | *<0.0001 | 84.0 | 86.8 | 2.8 | 0.008 | 83.4 | 87.1 | 3.6 | 0.31 | 84.2 | 86.7 | 2.5 | 0.02 |
| cg19124225 | chr6: 31545836 | 95.6 | 96.2 | 0.6 | 0.15 | 94.8 | 96.5 | 1.7 | 0.06 | 95.9 | 96.1 | 0.2 | 0.67 | ||||||||||||
| cg02137984 | chr6: 31545899 | 94.7 | 96.6 | 1.8 | *0.0002 | 93.7 | 96.8 | 3.1 | 0.006 | 95.1 | 96.5 | 1.4 | 0.01 | ||||||||||||
| cg06825478 | chr6: 31546068 | 90.2 | 94.3 | 4.1 | *<0.0001 | 91.0 | 94.7 | 3.6 | 0.01 | 89.9 | 94.1 | 4.2 | 0.0009 | 91.8 | 94.3 | 2.5 | *<0.0001 | 91.8 | 94.5 | 2.6 | 0.02 | 91.8 | 94.2 | 2.4 | *0.0014 |
| chr6: 31546086 | 85.1 | 89.3 | 4.2 | *<0.0001 | 85.2 | 89.4 | 4.3 | 0.09 | 85.1 | 89.3 | 4.2 | *0.0004 | |||||||||||||
diff = post - pre.
underline nominal significance (P < 0.05).
*Significant after correction for multiple testing level (EPIC; P < 0.05/52 = 0.00096, pyrosequencing; P < 0.05/32 = 0.00156).
DNAm at eight out of 14 CpG sites on the IL1B gene and four out of 14 CpG sites on the IL6 gene changed, most of them decreased, significantly after neurosurgery (Table 4). However, the degree of DNAm changes by the neurosurgery between the POD group and the non-POD group were not different with statistical significance (Supplementary Table 10).
Table 4.
Changes of DNAm in the IL1B and IL6 gene after neurosurgery.
| all | POD | non-POD | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N = 37 | N = 10 | N = 27 | ||||||||||
| pre | post | post-pre | paired-t | pre | post | post-pre | paired-t | pre | post | post-pre | paired-t | |
| cg | mean | mean | diff | p value | mean | mean | diff | p value | mean | mean | diff | p value |
| IL1B | ||||||||||||
| cg01290568 | 26.4 | 16.1 | −10.3 | *<0.0001 | 26.2 | 12.7 | −13.4 | *<0.0001 | 26.5 | 17.3 | −9.1 | *0.0001 |
| cg02596281 | 84.3 | 82.6 | −1.7 | 0.01 | 84.4 | 81.8 | −2.6 | 0.15 | 84.2 | 82.9 | −1.3 | 0.05 |
| cg07250315 | 37.7 | 33.2 | −4.5 | *<0.0001 | 39.9 | 33.0 | −6.9 | *0.0004 | 36.9 | 33.3 | −3.6 | 0.004 |
| cg07935264 | 9.0 | 6.5 | −2.6 | *0.0001 | 8.8 | 5.5 | −3.2 | 0.02 | 9.2 | 6.8 | −2.4 | 0.002 |
| cg10486274 | 93.7 | 93.1 | −0.6 | 0.09 | 93.9 | 93.1 | −0.7 | 0.40 | 93.6 | 93.1 | −0.5 | 0.15 |
| cg14117934 | 86.3 | 85.5 | −0.8 | 0.37 | 86.7 | 85.8 | −0.9 | 0.41 | 86.1 | 85.4 | −0.7 | 0.53 |
| cg15218327 | 92.1 | 88.6 | −3.5 | *<0.0001 | 92.4 | 89.6 | −2.8 | 0.01 | 92.0 | 88.2 | −3.7 | *<0.0001 |
| cg15836722 | 26.3 | 20.1 | −6.2 | *<0.0001 | 27.3 | 17.1 | −10.1 | *0.0001 | 26.0 | 21.2 | −4.7 | 0.003 |
| cg18635064 | 94.6 | 94.3 | −0.3 | 0.36 | 93.2 | 92.4 | −0.8 | 0.26 | 95.1 | 95.0 | −0.1 | 0.76 |
| cg18773937 | 7.2 | 4.8 | −2.3 | *0.0001 | 6.8 | 4.2 | −2.5 | 0.03 | 7.3 | 5.1 | −2.3 | 0.002 |
| cg19890119 | 15.6 | 8.8 | −6.8 | *<0.0001 | 15.9 | 6.7 | −9.2 | *<0.0001 | 15.5 | 9.6 | −5.9 | *0.0004 |
| cg20157753 | 5.4 | 4.8 | −0.6 | 0.06 | 5.9 | 4.2 | −1.7 | 0.02 | 5.2 | 5.0 | −0.2 | 0.56 |
| cg20983042 | 87.1 | 87.0 | 0.0 | 0.95 | 87.5 | 86.8 | −0.6 | 0.69 | 86.9 | 87.1 | 0.2 | 0.82 |
| cg23149881 | 14.8 | 8.4 | −6.3 | *<0.0001 | 14.1 | 6.8 | −7.4 | 0.003 | 15.0 | 9.1 | −6.0 | *0.0001 |
| IL6 | ||||||||||||
| cg00087425 | 6.4 | 6.6 | 0.2 | 0.62 | 6.3 | 6.2 | −0.1 | 0.84 | 6.5 | 6.8 | 0.3 | 0.54 |
| cg00175482 | 89.0 | 86.8 | −2.2 | 0.002 | 88.6 | 85.7 | −2.9 | 0.047 | 89.1 | 87.3 | −1.9 | 0.02 |
| cg01770232 | 7.1 | 7.6 | 0.5 | 0.09 | 7.7 | 7.7 | 0.1 | 0.90 | 6.9 | 7.5 | 0.6 | 0.04 |
| cg02335517 | 93.4 | 92.7 | −0.7 | 0.02 | 93.3 | 93.0 | −0.3 | 0.42 | 93.4 | 92.5 | −0.9 | 0.03 |
| cg03601896 | 3.8 | 3.0 | −0.8 | 0.02 | 4.0 | 2.7 | −1.3 | 0.03 | 3.7 | 3.1 | −0.6 | 0.14 |
| cg05265849 | 20.2 | 17.6 | −2.5 | *0.0001 | 21.4 | 16.9 | −4.5 | *0.0005 | 19.7 | 17.9 | −1.8 | 0.02 |
| cg07998387 | 69.5 | 70.2 | 0.6 | 0.28 | 69.8 | 68.6 | −1.3 | 0.31 | 69.4 | 70.7 | 1.4 | 0.04 |
| cg10140158 | 87.7 | 87.9 | 0.2 | 0.70 | 86.8 | 87.0 | 0.2 | 0.79 | 88.0 | 88.2 | 0.2 | 0.77 |
| cg13104385 | 39.9 | 39.1 | −0.8 | 0.15 | 41.1 | 38.4 | −2.6 | 0.02 | 39.5 | 39.4 | −0.1 | 0.84 |
| cg15703690 | 9.6 | 5.8 | −3.8 | *<0.0001 | 10.0 | 4.6 | −5.4 | 0.001 | 9.5 | 6.2 | −3.2 | *0.0003 |
| cg17067544 | 78.7 | 83.3 | 4.6 | *<0.0001 | 79.3 | 85.0 | 5.7 | 0.01 | 78.5 | 82.7 | 4.2 | *<0.0001 |
| cg20509117 | 62.3 | 61.9 | −0.4 | 0.68 | 62.2 | 62.5 | 0.2 | 0.90 | 62.3 | 61.6 | −0.7 | 0.59 |
| cg21785978 | 5.4 | 5.1 | −0.3 | 0.44 | 5.2 | 4.4 | −0.8 | 0.27 | 5.5 | 5.3 | −0.1 | 0.78 |
| cg23731304 | 37.6 | 34.5 | −3.1 | *0.0002 | 38.7 | 33.5 | −5.2 | 0.005 | 37.2 | 34.9 | −2.3 | 0.01 |
underline nominal significance (P < 0.05).
*Significant after correction for multiple testing level (EPIC; P < 0.05/52 = 0.00096).
Inflammatory methylation index (IMI) and ROC curve
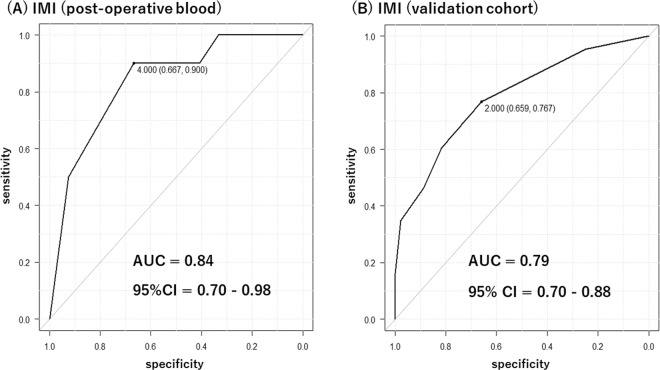
We found that levels of DNAm at multiple CpG sites converged to a certain narrow range after surgery (Supplementary Fig. 1). Then, we calculated IMI (ranging from 0 to 5) based on the postoperative DNAm levels at the selected five CpG sites: one CpG site from the TNF gene (cg08639424), two CpG sites from the IL1B gene (cg15836722 and cg23149881), and two CpG sites from the IL6 gene (cg15703690 and cg17067544). The AUC based on IMI was 0.84 (95% CI: 0.70–0.98) (Fig. 1A). We also calculated the IMI from the same five CpG sites using our previous delirium study cohort, including 87 subjects (43 delirium subjects and 44 control subjects) [14, 16], and tested the performance of the scoring using the ROC curve. The AUC based on the IMI was 0.79 (95% CI: 0.70–0.88) (Fig. 1B). Furthermore, the same approach was used to calculate the IMI from the preoperative DNAm (pre-IMI) and the amount of change in DNAm level after surgery (diff-IMI). Five different CpG sites were selected to optimize the ROC (Supplementary method and Supplementary Fig. 1). The AUC based on the pre-IMI was 0.92 (95% CI: 0.82–1.00) (Supplementary Fig. 2A), and the AUC based on the diff-IMI was 0.87 (95% CI: 0.73–1.00) (Supplementary Fig. 2B). IMI distributions are shown in Supplementary Fig. 3.
Fig. 1. ROC curves based on IMI.
(A) ROC curve based on IMI in the current dataset. (B) ROC curve based on IMI in validation dataset from our previous study.
Discussion
In this study, we investigated DNAm levels in inflammation-related genes using blood samples from patients who had neurosurgery. We examined correlations between age and DNAm level, compared peri-operative DNAm levels and postoperative ones, and compared DNAm levels in POD subjects with ones in non-POD subjects. We also constructed IMI aiming to differentiate POD from non-POD, validated with an independent cohort from our previous delirium study.
We have reported the relationship between delirium and DNAm [13–16]. The data from the Grady Trauma Project cohort demonstrated negative correlations between age and DNAm in several CpG sites on the TNF gene, but not in CpG sites on other inflammatory-related genes [13]. We also recently reported that delirium patients showed negative correlations between age and DNAm level in several CpG sites on the TNF gene [16]. In this current study, we demonstrated that DNAm levels in several CpG sites only on the TNF gene correlated with age using blood samples from all preoperative patients, consistent with our previous study [13]. Furthermore, negative correlations between age and DNAm levels on the TNF gene were observed after neurosurgery only in the POD group in EPIC analysis. This result is consistent with our previous study demonstrating negative correlations between age and DNAm level in several CpG sites on the TNF gene only in delirium subjects [16]. On the other hand, pyrosequencing showed negative correlations between age and DNAm levels after neurosurgery in both the POD group and the non-POD group. The discrepancy between these two results by different measurement methods requires further study.
A recent study from Sadahiro et al. showed that various major surgeries cause changes in DNAm at sites annotated to immune system genes [21]. In this current study, we also demonstrated that neurosurgery alters DNAm levels at many CpG sites on inflammation-related genes. When focusing on the TNF gene, levels of DNAm at CpG sites located on the promoter region decreased after neurosurgery [16, 22]. By contrast, levels of DNAm at CpG sites in other regions increased after neurosurgery. In the IL1B and the IL6 gene, levels of DNAm decreased in several CpG sites after neurosurgery. Several studies reported that inflammatory cytokines, including TNF-alpha, IL-1beta, and IL-6, increase after surgery [23, 24]. It is possible that changes of DNAm level after surgery observed in our data are the initial molecular mechanism that causes such elevation of inflammatory cytokines, leading to neuroinflammation, and thus increased risk of delirium [10, 25].
When focusing on each CpG site, there was no significant difference between the POD group and the non-POD group in terms of preoperative DNAm level, postoperative DNAm level, or degree of change by neurosurgery. This might be partly due to the limited sample size. However, in the POD group, we observed at multiple CpG sites that the DNAm level, which varied before surgery, accumulated to a certain narrow range after surgery (Supplementary Fig. 1). Furthermore, it was suggested that scoring of these multiple CpG sites by DNAm level of postoperative blood (IMI) may be useful for the diagnosis of POD (Fig. 1A). The detection ability by IMI was well validated by our independent data set (Fig. 1B). On the other hand, pre-IMI score and diff-IMI score, which were based on DNAm level of preoperative blood and amount of change in DNAm level after surgery, also showed high AUCs, although these methods were not validated as we did not have samples from independent cohorts to validate. Although there are numberless studies investigating delirium biomarkers, those results do not support the use of any one biomarker [26]. This might be because protein level and RNA expression can be easily altered depending on the timing and method of sample collection, storage, and downstream multiple-step laboratory techniques. On the other hand, it is known that DNAm is relatively stable even among various epigenetics mechanisms. Thus, it might be suggested that DNAm can be a potential detection tool for delirium.
Although we analyzed the level of DNAm in peripheral blood samples, it is not clear whether similar changes occur in the brain. In our previous study investigating the relationship between DNAm levels measured at peripheral tissues and brain tissue resected from epilepsy patients, relatively high levels of overall genome-wide DNAm correlation between brain and blood were observed [18]. However, when focusing specifically on the TNF, IL1B, and IL6 genes, correlations between brain tissue and blood were not strong. This discrepancy might be due to the fact that the whole brain tissue instead of sorted cells were used to develop the correlation database, “IMAGE-CpG”, in the study [18]. In our other study, all CpGs in the TNF gene showed negative correlations between DNAm level and age in glia, but not in neurons [13]. These evidences suggest that it is possible that if we assess the DNAm of each cell type in the brain, especially glial cell, separately, we can better understand the epigenetic status involved in delirium and neuroinflammation, and its relationships to that observed in the blood.
A strength of this work is that it is the first prospective-designed study investigating DNAm from subjects who developed POD from one type of surgery. This is an advantage because potential confounders from different baseline illness types and diverse surgical incisions were avoided. Furthermore, the detection performance of the IMI was validated with two independent cohorts.
We acknowledge several limitations of this study. First, our definition of POD solely depended on retrospective chart reviews of EMRs. Thus, there are certain possibilities of false-positive and false-negative cases in our dataset. However, even with the potential false classification of cases, we observed intriguing signals as outlined above. Second, we have not examined protein levels or mRNA levels of these cytokines, thus the DNAm level discussed above and the subsequent change in cytokine levels and inflammation is speculation, although based on well-accepted molecular processes, and clinical phenotype of POD was associated with this epigenetics data. Third, participants in this study were limited to epilepsy subjects undergoing neurosurgery and the sample size was very small. This was because the number of patients undergoing surgery who could participate in the study was limited. Further study is needed to confirm the present findings using another cohort. Fourth, this study was conducted at a single institution, and greater than 95% of study subjects were non-Hispanic white. Thus, generalizability requires confirmation with a more diverse ethnic population. However, even with these limitations, the present data support evidence of a potential epigenetic contribution to the pathophysiology of POD in the pro-inflammatory cytokine gene.
In summary, this is the first study demonstrating the role of DNAm change among POD patients using both pre- and post-surgery samples. Our findings provide further evidence of the potential role of epigenetics and inflammation in the pathophysiology of delirium.
Supplementary information
Acknowledgements
The authors thank the patients who participated in this study. Also, we would like to thank Paul J. Casella for English language editing. This work was supported by research grants from the National Institute of Mental Health, United States (R01 MH119165). Gen Shinozaki receives research grant support from National institute of health (NIH), National Science Foundation (NSF), and Fujitsu Laboratories LTD.
Role of the sponsors
The supporters had no role in the design, analysis, interpretation, or publication of this study.
Author contributions
T.Y. analyzed data and wrote the initial draft of the manuscript. T.N. collected samples and clinical data. N.E.W. collected clinical data. P.S.M., K.J.C., and M.M.H. analyzed data. A.A.M., A.J.A., and S.S.J. processed samples. H.K. participated in its design, collected samples, and helped coordination. C.G.H., P.P.P., M.A.H., M.I., and M.M.H. critically reviewed the manuscript. G.S. conceived ideas of the study, planned its design and coordination, and edited the manuscript for the final form.
Data availability
The data that support the findings of this study are available from the corresponding author, G.S., upon reasonable request.
Conflict of interest
G.S. has pending patents “Epigenetic Biomarker of Delirium Risk” in the PCT Application No. PCT/US19/51276, and in US Provisional Patent No. 62/731,599. The remaining authors declared no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Takehiko Yamanashi, Email: yamatake@stanford.edu.
Gen Shinozaki, Email: gens@stanford.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41398-021-01752-6.
References
- 1.Jin Z, Hu J, Ma D. Postoperative delirium: perioperative assessment, risk reduction, and management. Br J Anaesth. 2020;125:492–504. doi: 10.1016/j.bja.2020.06.063. [DOI] [PubMed] [Google Scholar]
- 2.Scholz AF, Oldroyd C, McCarthy K, Quinn TJ, Hewitt J. Systematic review and meta-analysis of risk factors for postoperative delirium among older patients undergoing gastrointestinal surgery. Br J Surg. 2016;103:e21–8. doi: 10.1002/bjs.10062. [DOI] [PubMed] [Google Scholar]
- 3.Maniar HS, Lindman BR, Escallier K, Avidan M, Novak E, Melby SJ, et al. Delirium after surgical and transcatheter aortic valve replacement is associated with increased mortality. J Thorac Cardiovasc Surg. 2016;151:815–23.e2. doi: 10.1016/j.jtcvs.2015.10.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamilton GM, Wheeler K, Di Michele J, Lalu MM, McIsaac DI. A systematic review and meta-analysis examining the impact of incident postoperative delirium on mortality. Anesthesiology. 2017;127:78–88. doi: 10.1097/ALN.0000000000001660. [DOI] [PubMed] [Google Scholar]
- 5.Gleason LJ, Schmitt EM, Kosar CM, Tabloski P, Saczynski JS, Robinson T, et al. Effect of delirium and other major complications on outcomes after elective surgery in older adults. JAMA Surg. 2015;150:1134–40. doi: 10.1001/jamasurg.2015.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huded CP, Huded JM, Sweis RN, Ricciardi MJ, Malaisrie SC, Davidson CJ, et al. The impact of delirium on healthcare utilization and survival after transcatheter aortic valve replacement. Catheter Cardiovasc Interv. 2017;89:1286–91. doi: 10.1002/ccd.26776. [DOI] [PubMed] [Google Scholar]
- 7.Simone MJ, Tan ZS. The role of inflammation in the pathogenesis of delirium and dementia in older adults: a review. CNS Neurosci Ther. 2011;17:506–13. doi: 10.1111/j.1755-5949.2010.00173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerejeira J, Firmino H, Vaz-Serra A, Mukaetova-Ladinska EB. The neuroinflammatory hypothesis of delirium. Acta Neuropathol. 2010;119:737–54. doi: 10.1007/s00401-010-0674-1. [DOI] [PubMed] [Google Scholar]
- 9.Alam A, Hana Z, Jin Z, Suen KC, Ma D. Surgery, neuroinflammation and cognitive impairment. EBioMedicine. 2018;37:547–56. doi: 10.1016/j.ebiom.2018.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunningham C, Maclullich AM. At the extreme end of the psychoneuroimmunological spectrum: delirium as a maladaptive sickness behaviour response. Brain Behav Immun. 2013;28:1–13. doi: 10.1016/j.bbi.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X, Yu Y, Zhu S. Inflammatory markers in postoperative delirium (POD) and cognitive dysfunction (POCD): a meta-analysis of observational studies. PLoS ONE. 2018;13:e0195659. doi: 10.1371/journal.pone.0195659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noah AM, Almghairbi D, Evley R, Moppett IK. Preoperative inflammatory mediators and postoperative delirium: systematic review and meta-analysis. Br J Anaesth. 2021;127:424–34. doi: 10.1016/j.bja.2021.04.033. [DOI] [PubMed] [Google Scholar]
- 13.Shinozaki G, Braun PR, Hing BWQ, Ratanatharathorn A, Klisares MJ, Duncan GN, et al. Epigenetics of delirium and aging: potential role of DNA methylation change on cytokine genes in glia and blood along with aging. Front Aging Neurosci. 2018;10:311. doi: 10.3389/fnagi.2018.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito T, Toda H, Duncan GN, Jellison SS, Yu T, Klisares MJ, et al. Epigenetics of neuroinflammation: immune response, inflammatory response and cholinergic synaptic involvement evidenced by genome-wide DNA methylation analysis of delirious inpatients. J Psychiatr Res. 2020;129:61–5. doi: 10.1016/j.jpsychires.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saito T, Braun PR, Daniel S, Jellison SS, Hellman M, Shinozaki E, et al. The relationship between DNA methylation in neurotrophic genes and age as evidenced from three independent cohorts: differences by delirium status. Neurobiol Aging. 2020;94:227–35. doi: 10.1016/j.neurobiolaging.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamanashi T, Saito T, Yu T, Alario A, Comp K, Crutchley KJ, et al. DNA methylation in the TNF-alpha gene decreases along with aging among delirium inpatients. Neurobiol Aging. 2021;105:310–17. doi: 10.1016/j.neurobiolaging.2021.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Braun PR, Tanaka-Sahker M, Chan AC, Jellison SS, Klisares MJ, Hing BW, et al. Genome-wide DNA methylation investigation of glucocorticoid exposure within buccal samples. Psychiatry Clin Neurosci. 2019;73:323–30. doi: 10.1111/pcn.12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braun PR, Han S, Hing B, Nagahama Y, Gaul LN, Heinzman JT, et al. Genome-wide DNA methylation comparison between live human brain and peripheral tissues within individuals. Transl Psychiatry. 2019;9:47. doi: 10.1038/s41398-019-0376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ely EW, Inouye SK, Bernard GR, Gordon S, Francis J, May L, et al. Delirium in mechanically ventilated patients: validity and reliability of the confusion assessment method for the intensive care unit (CAM-ICU) JAMA. 2001;286:2703–10. doi: 10.1001/jama.286.21.2703. [DOI] [PubMed] [Google Scholar]
- 20.Team RC. R: a language and environment for statistical computing. 2017. https://www.Rproject.org.
- 21.Sadahiro R, Knight B, James F, Hannon E, Charity J, Daniels IR, et al. Major surgery induces acute changes in measured DNA methylation associated with immune response pathways. Sci Rep. 2020;10:5743. doi: 10.1038/s41598-020-62262-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu Y, Ishii KK, Munakata Y, Saitoh T, Kaku M, Sasaki T. Regulation of tumor necrosis factor alpha promoter by human parvovirus B19 NS1 through activation of AP-1 and AP-2. J Virol. 2002;76:5395–403. doi: 10.1128/JVI.76.11.5395-5403.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, Wang T, Wang G, Yang M, Zhou Y, Yuan Y. Effects of dexmedetomidine on postoperative delirium and expression of IL-1β, IL-6, and TNF-α in elderly patients after hip fracture operation. Front Pharmacol. 2020;11:678. doi: 10.3389/fphar.2020.00678. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Tang C, Hu Y, Zhang Z, Wei Z, Wang H, Geng Q, et al. Dexmedetomidine with sufentanil in intravenous patient-controlled analgesia for relief from postoperative pain, inflammation and delirium after esophageal cancer surgery. Biosci Rep. 2020;40:5. doi: 10.1042/BSR20193410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inouye SK, Westendorp RG, Saczynski JS. Delirium in elderly people. Lancet. 2014;383:911–22. doi: 10.1016/S0140-6736(13)60688-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunne SS, Coffey JC, Konje S, Gasior S, Clancy CC, Gulati G, et al. Biomarkers in delirium: a systematic review. J Psychosom Res. 2021;147:110530. doi: 10.1016/j.jpsychores.2021.110530. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, G.S., upon reasonable request.