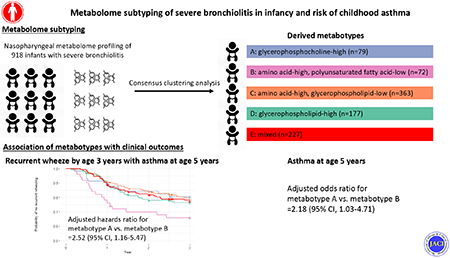
Abstract
Background:
Infants with bronchiolitis are at increased risk for developing asthma. Growing evidence suggests bronchiolitis is a heterogeneous condition.
Objective:
To identify biologically-distinct subgroups based on the metabolome signatures (metabotypes) in infants with severe bronchiolitis and to examine their longitudinal relationships with asthma development.
Methods:
In a multi-center prospective cohort study of infants (aged <12 months) hospitalized for bronchiolitis, the nasopharyngeal airway metabolome was profiled at hospitalization. Using a clustering approach, we identified mutually-exclusive metabotypes. We also examined their longitudinal association with the risk of developing asthma by age five years.
Results:
Of 918 infants hospitalized for bronchiolitis (median age, 3 months), we identified 5 distinct metabotypes—characterized by their nasopharyngeal metabolome profile: A) glycerophosphocholine-high; B) amino acid-high, polyunsaturated fatty acid (PUFA)-low; C) amino acid-high, glycerophospholipid-low; D) glycerophospholipid-high; and E) mixed. Compared with metabotype A infants (who clinically resembled “classic” bronchiolitis), metabotype B infants had a significantly higher risk for developing asthma (23% vs. 41%; ORadjusted, 2.22: 95%CI, 1.07-4.69). The pathway analysis showed that metabotype B had enriched amino acid (e.g., methionine, histidine, glutathione) and α-linolenic/linoleic acid metabolism pathways (all FDR<5<×10−14). Finally, the transcriptome analysis revealed that metabotype B infants had up-regulated interferon-α and interleukin-6/Janus kinase/signal transducer activator of transcription (IL-6/JAK/STAT3) pathways and down-regulated fatty acid metabolism pathways (both FDR<0.05).
Conclusion:
In this multicenter prospective cohort study of infant severe bronchiolitis, the clustering analysis of metabolome data identified biologically-distinct metabotypes, including one characterized by high inflammatory amino acids and low PUFAs that is at significantly increased risk for developing asthma.
Keywords: severe bronchiolitis, heterogeneity, subtype, asthma, metabolome, inflammatory amino acids, polyunsaturated fatty acids, transcriptome
Capsule Summary:
Among infant with severe bronchiolitis, clustering analysis of the nasopharyngeal airway metabolome identified biologically distinct metabotypes, including one characterized by high inflammatory amino acids and low PUFAs that is at increased risk for developing asthma.
Graphical Abstract
INTRODUCTION
Bronchiolitis is the most common lower respiratory tract infection among infants, accounting for ~110,000 hospitalizations (“severe bronchiolitis”) in the U.S. annually 1. In addition to the acute morbidity, its chronic morbidity burden is also substantial—e.g., of these infants with severe bronchiolitis, approximately 30% develop recurrent wheeze and asthma in childhood 2–7.
Although bronchiolitis was traditionally considered a single disease entity with similar pathobiology 7, there is growing evidence suggesting that bronchiolitis is a heterogenous condition 8–10. For example, recent epidemiological studies have identified 8 and validated 9 the presence of multiple phenotypes that have differential risks for developing recurrent wheeze. However, these phenotypes were identified only based on major clinical characteristics. The pathobiological mechanisms underlying the heterogeneity remains unclear. This insufficient understanding of pathobiology of bronchiolitis during infancy—a critical period of airway development—has hindered efforts to develop targeted bronchiolitis treatment and asthma prevention strategies. Metabolomics is well-suited to address this knowledge gap by broadly profiling small molecules (metabolites), which are a function of an individual’s genetic makeup and environmental factors 11. Regardless, no study has yet investigated the biologically-distinct subgroups of bronchiolitis based on metabolome signatures (“metabotypes” 12).
In this context, we aimed to identify distinct nasopharyngeal airway metabotypes by applying an unsupervised clustering approach to the metabolome data from a multicenter prospective study of infants with severe bronchiolitis. We also sought to investigate the longitudinal relationships of the metabotypes with recurrent wheeze and childhood asthma. This study provides further evidence that “bronchiolitis” represents several diseases with distinct biological mechanisms and differential risks of chronic airway morbidities in childhood.
METHODS
Study Design, Setting, and Participants
We analyzed data from a multicenter prospective cohort study of infants hospitalized for bronchiolitis—the 35th Multicenter Airway Research Collaboration (MARC-35) study 13, 14. MARC-35 is coordinated by the Emergency Medicine Network (EMNet, www.emnet-usa.org), an international research collaboration with 247 participating hospitals. Details of the study design, setting, participants, data collection, testing, and statistical analysis may be found in the Supplementary Methods. Briefly, MARC-35 investigators at 17 sites, across 14 U.S. states, enrolled infants (aged <1 year) who were hospitalized with an attending physician diagnosis of bronchiolitis during three bronchiolitis seasons (November 1 to April 30) from 2011 to 2014 (Table E1). The diagnosis of bronchiolitis was made according to the American Academy of Pediatrics bronchiolitis guidelines, defined as acute respiratory illness with a combination of rhinitis, cough, tachypnoea, wheezing, crackles, or retraction 15. We excluded infants with a preexisting heart and lung disease, immunodeficiency, immunosuppression, or gestational age of <32 weeks. All patients were treated at the discretion of the treating physicians. The institutional review board at each participating hospital approved the study with a written informed consent obtained from parent or guardian.
Data Collection
Clinical data (patients’ demographic characteristics, and family, environmental, and medical history, and details of the acute illness) were collected via structured interview and chart reviews using a standardized protocol 13, 14. After the index hospitalization for bronchiolitis, trained interviewers began interviewing parents/legal guardians by telephone at 6-month intervals in addition to medical record review by physicians. All data were reviewed at the Emergency Medicine Network Coordinating Center at Massachusetts General Hospital (Boston, MA, USA) 16. Nasopharyngeal specimens were collected within 24 hours of hospitalization using a standardized protocol 14. The details of the data collection and measurement methods are described in the Supplementary Methods.
Nasopharyngeal Metabolome Profiling
The details of metabolome profiling are described in Supplementary Methods. Briefly, the metabolome profiling was conducted by Metabolon (Durham, NC, USA) using liquid chromatography–tandem mass spectrometry. All specimens were blinded to the laboratory and processed in a random order. Instrument variability was 4%, as determined by calculating the median relative standard deviation for the internal standards. We used the metabolome data of 278 nasopharyngeal metabolites from 76 sub-pathways within seven super-pathways.
Nasopharyngeal Transcriptome Profiling
The details of RNA extraction, RNAseq, quality control, and transcriptome profiling are described in Supplementary Methods. Briefly, after total RNA extraction, DNase treatment, and rRNA reduction, we performed RNAseq with Illumina NovaSeq6000 (Illumina, San Diego, CA, USA). All RNAseq samples had high sequence coverage after quality control. The transcript abundances were estimated with Salmon 17 using the human genome (hg38) and the mapping-based mode.
Outcome Measures
The primary outcome was asthma at age five years. The secondary outcome was the development of recurrent wheeze by age three years stratified by asthma status, to account for the heterogeneity of recurrent wheeze according to prior research 16. The definition of asthma was based on a commonly-used epidemiologic definition of asthma 16, 18—physician-diagnosis of asthma by age five years, plus either asthma medication use (e.g., albuterol inhaler, inhaled corticosteroids, montelukast) or asthma-related symptoms in the preceding year. The definition of recurrent wheeze was based on the U.S. asthma guidelines—defined as having at least two corticosteroid-requiring exacerbations in six months or at least four wheezing episodes in one year that last at least one day and affect sleep 19.
Statistical Analysis
In the current study, our objectives were to identify biologically-distinct severe bronchiolitis subgroups based on the metabolome signatures (clustering/description) and to relate them to the outcomes (association). The analytic workflow is summarized in Figure 1.
Figure 1. Analytic workflow.
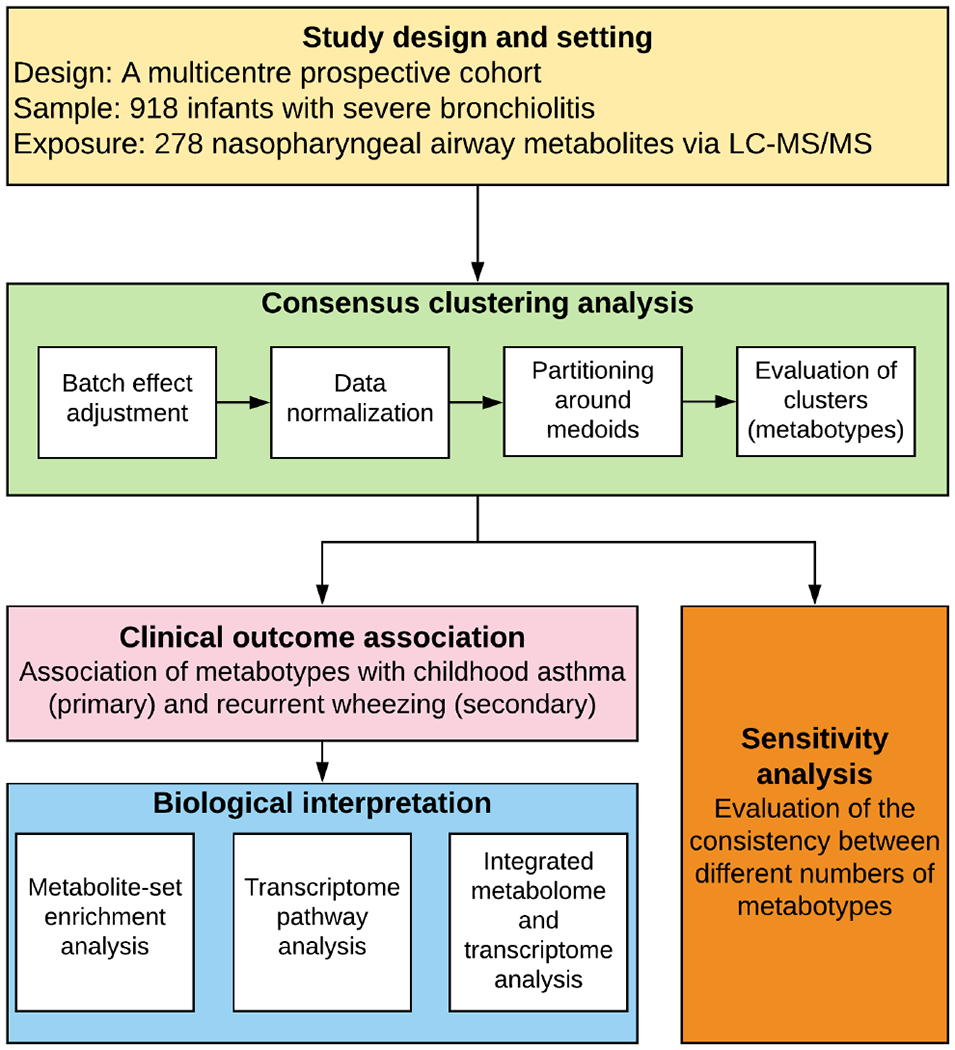
The analytical cohort consists of 918 infants hospitalized for bronchiolitis (severe bronchiolitis) in a multicenter prospective cohort study—the 35th Multicenter Airway Research Collaboration (MARC-35). The nasopharyngeal metabolome data were adjusted for the potential batch effect and normalized by total sum scaling before clustering analysis. To identify metabotypes, the partitioning around medoids algorithm was used in consensus clustering. To choose the optimal number of metabotypes, a combination of the average silhouette score, consensus matrix heatmap, and biological plausibility was used. The between-metabotype differences in the metabolome profile were visualized using a heatmap. The association between the metabotypes and the risk of developing childhood asthma (binary outcome) was estimated by fitting logistic regression models; the rate of developing recurrent wheeze (time-to-event outcome) was estimated by fitting Cox proportional hazards models. To examine the distinct function of each metabotype, 1) metabolite set enrichment analysis examining the metabolome data, 2) gene set enrichment analysis examining the nasopharyngeal transcriptome data, and 3) Wilcoxon pathway enrichment analysis integrating the metabolome and transcriptome data were conducted. In the sensitivity analysis, the concordance between the different numbers of metabotypes was also examined
Abbreviation: LC-MS/MS, liquid chromatography-tandem mass spectrometry
Data Normalization and Clustering to Identify Metabotypes
We first adjusted the metabolome data for potential batch effect by using empirical Bayes models (ComBat method) 20. Then, we normalized the data using total sum scaling method to account for potentially differential dilutions. To identify individuals with a similar metabolome signature (metabotype), we applied consensus clustering with partitioning around medoids algorithm. We determined the optimal number of clusters by a combination of the average silhouette score, consensus matrix heatmap (Figure E1), and biological plausibility 11, 21, 22.
Association of Metabotypes with Clinical Outcomes
To investigate the longitudinal association of metabotypes with asthma status at age five years (binary outcome), we fitted unadjusted and adjusted logistic regression models. To investigate the relation of metabotypes with the development of recurrent wheeze (time-to-event) outcomes, we constructed Cox proportional hazards models. Patients were censored at time of the recurrent wheeze development, loss to follow-up, or age 36-months, whichever occurred first. For both models, we adjusted for potential confounders (age, sex, parent history of asthma, number of previous breathing problems, respiratory syncytial virus infection and rhinovirus infection) based on a priori knowledge 13, 14, 16, 23, 24.
To examine the robustness of inferences, we conducted a series of sensitivity analyses. First, we repeated the outcome regression model by restricting the study sample to those without previous breathing problems. Second, we also examined the robustness of the metabotype-outcome associations by repeating the analysis with a different number of metabotypes. We also computed E-values to examine the robustness of the causal inference by using the R Evalue package 25. The E-value indicates the minimum strength of association that an unmeasured confounder(s) would need to have with both the exposure and outcome to fully explain away a specific association, conditional on the measured covariates 25. For example, an E-value of 2.5 means that unmeasured confounders that were associated with both the metabotypes and outcome by an odds ratio of 2.5 could explain away the estimate, but weaker confounding could not.
Pathway Analyses
To identify biologically-meaningful pathways, we conducted a metabolite-set enrichment analysis by comparing the reference metabotype with each of the other metabotypes using MetaboAnalyst 4.0 26 and the Small Molecule Pathway Database 27 library as the reference. To investigate whether gene expression for specific biological pathways are enriched, we conducted a functional class scoring analysis using R fgsea package 28. To detect differentially-expressed pathways by integrating the metabolome and transcriptome data, we performed a Wilcoxon pathway enrichment analysis using the Integrated Molecular Pathway Level Analysis (IMPaLA) method 29.
All statistical analyses were performed using R version 4.0.2. All P-values are two-tailed, with P<0.05 considered statistically significant. We further calculated the false discovery rate (FDR) that allows for the interpretation of statistical significance in the context of multiple hypothesis testing. The FDR value was controlled using Benjamini-Hochberg method 30.
RESULTS
Of 921 infants with severe bronchiolitis enrolled into this longitudinal cohort, the current study included 918 infants (>99%) who had information for the nasopharyngeal airway metabolome. Among the analytic cohort, the median age was 3 (IQR, 2-6) months, 40% were female, and 44% were non-Hispanic white. Subsequently, 32% developed recurrent wheeze by age 3 years and 27% developed asthma by age 5 years.
Identification of Metabotypes and Their Characteristics
By applying the consensus clustering approach on the nasopharyngeal airway metabolome data, we identified five distinct metabotypes (Figure E1): A) glycerophosphocholine (GPC)-high (8.6%); B) amino acid-high, polyunsaturated fatty acid (PUFA)-low (7.8%); C) amino acid-high, glycerophospholipid (GP)-low (39.5%); D) GP-high (19.3%); and E) mixed (24.7%) (Figure 2). Across these metabotypes, several clinical characteristics (e.g., age, number of previous breathing problems, and infecting virus) were significantly different (P<0.05; Table 1).
Figure 2. Abundance of selected nasopharyngeal metabolites between five metabotypes of severe bronchiolitis.
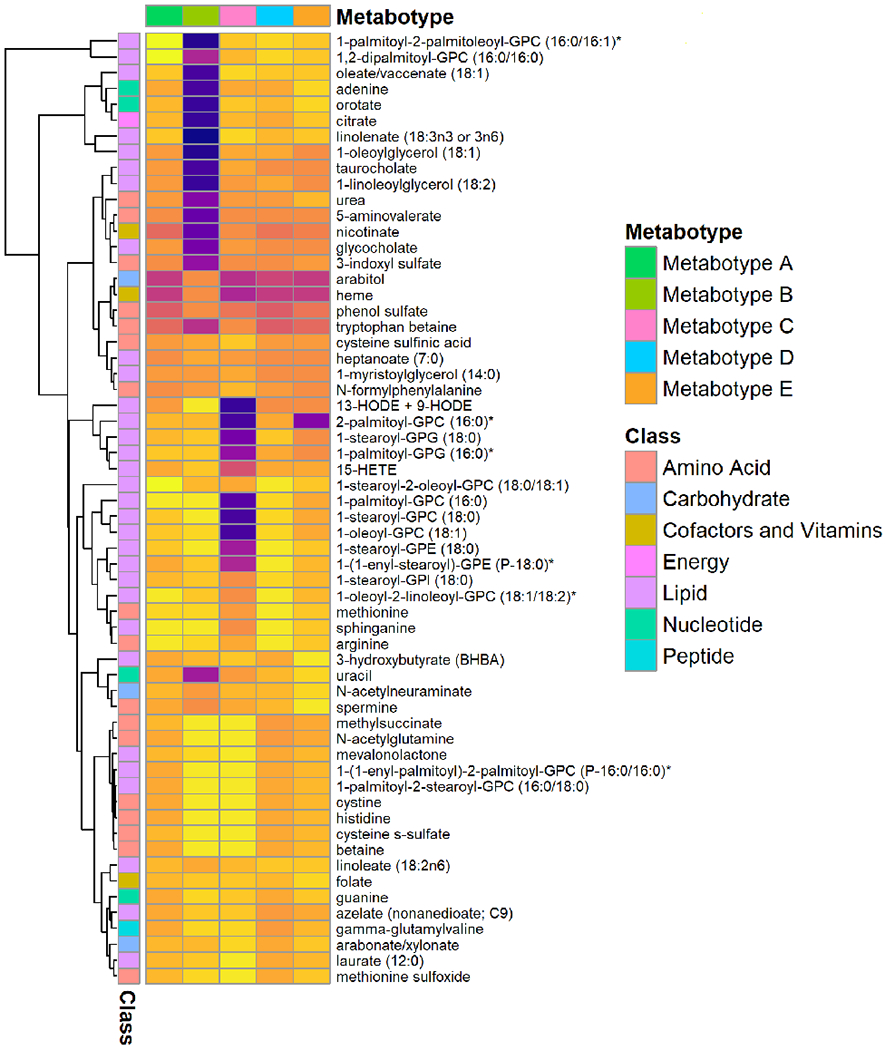
The normalized metabolomics data (50 metabolites with highest variance and top 10 metabolites in the metabolite-set enrichment analysis) are used to visualize the between-metabotype differences. The top row represents five different metabotypes (A-E). The first column on the left represents a major metabolite class (e.g., amino acids, lipids) for the corresponding metabolite. The tree on the left represents hierarchical clustering of metabolites. Each cell represents the median value of normalized metabolite data within each metabotype—the yellow color represents a high value, and dark purple color represents a low value. Asterisk on some metabolites indicates a compound that has not been confirmed based on a standard, but we are confident in its identity (by Metabolon, Inc).
Table 1.
Baseline characteristics and clinical course of infants, according to severe bronchiolitis metabotypes
| Characteristics | Metabotype A (n=79, 8.6%) | Metabotype B (n=72, 7.8%) | Metabotype C (n=363, 39.5%) | Metabotype D (n=177, 19.3%) | Metabotype E (n=227, 24.7%) | P-value |
|---|---|---|---|---|---|---|
| Demographics | ||||||
| Age (month), median (IQR) | 2 (1-3) | 3 (2-5) | 3 (2-6) | 3 (2-6) | 4 (2-7) | <0.001 |
| Female sex | 36 (45.6) | 27 (37.5) | 147 (40.5) | 74 (41.8) | 83 (36.6) | 0.63 |
| Race/ethnicity | 0.13 | |||||
| Non-Hispanic white | 28 (35.4) | 35 (48.6) | 167 (46.0) | 68 (38.4) | 101 (44.5) | |
| Non-Hispanic black | 15 (19.0) | 17 (23.6) | 89 (24.5) | 45 (25.4) | 44 (19.4) | |
| Hispanic | 34 (43.0) | 19 (26.4) | 90 (24.8) | 59 (33.3) | 73 (32.2) | |
| Other or unknown | 2 (2.5) | 1 (1.4) | 17 (4.7) | 5 (2.8) | 9 (4.0) | |
| Prematurity (32-37 weeks) | 14 (17.7) | 12 (16.7) | 67 (18.5) | 39 (22.0) | 39 (17.2) | 0.76 |
| Birth weight (kg), median (IQR) | 3.35 (2.95-3.60) | 3.33 (2.90-3.60) | 3.26 (2.90-3.58) | 3.20 (2.86-3.54) | 3.32 (2.98-3.63) | 0.51 |
| Mode of birth (caesarean delivery) | 24 (31.2) | 25 (35.2) | 126 (35.1) | 65 (36.9) | 71 (31.8) | 0.81 |
| Previous breathing problems (count) | 0.001 | |||||
| 0 | 69 (87.3) | 58 (80.6) | 293 (80.7) | 152 (85.9) | 160 (70.5) | |
| 1 | 6 (7.6) | 9 (12.5) | 61 (16.8) | 18 (10.2) | 51 (22.5) | |
| 2 | 4 (5.1) | 5 (6.9) | 9 (2.5) | 7 (4.0) | 16 (7.0) | |
| Previous ICU admission | 1 (1.3) | 2 (2.8) | 4 (1.1) | 2 (1.1) | 6 (2.6) | 0.56 |
| Child history of eczema | 5 (6.3) | 8 (11.1) | 57 (15.7) | 24 (13.6) | 43 (18.9) | 0.07 |
| Lifetime antibiotic use* | 24 (30.4) | 22 (30.6) | 111 (30.6) | 49 (27.7) | 87 (38.3) | 0.19 |
| Lifetime corticosteroid use* | 8 (10.1) | 13 (18.1) | 52 (14.3) | 27 (15.3) | 38 (16.7) | 0.61 |
| Ever attended daycare | 10 (12.7) | 18 (25.0) | 86 (23.7) | 31 (17.5) | 65 (28.6) | 0.02 |
| Cigarette smoke exposure at home | 7 (8.9) | 12 (16.7) | 55 (15.2) | 28 (15.8) | 35 (15.4) | 0.62 |
| Maternal smoking during pregnancy | 10 (13.0) | 11 (15.7) | 50 (13.9) | 26 (14.8) | 29 (12.9) | 0.97 |
| Parental history of asthma | 24 (30.4) | 25 (34.7) | 119 (32.8) | 54 (30.5) | 82 (36.1) | 0.77 |
| Parental history of eczema | 11 (13.9) | 14 (19.4) | 70 (19.3) | 39 (22.0) | 41 (18.1) | 0.64 |
| Clinical presentation | ||||||
| Weight (kg), median (IQR) | 4.99 (4.20-6.17) | 5.80 (4.70-7.15) | 6.10 (4.70-7.60) | 5.90 (4.80-7.60) | 6.90 (5.30-8.32) | <0.001 |
| Respiratory rate (per minute), median (IQR) | 48 (42-60) | 48 (39-60) | 48 (40-60) | 52 (42-62) | 50 (40-60) | 0.26 |
| Oxygen saturation | 0.50 | |||||
| <88% | 6 (7.7) | 7 (10.1) | 19 (5.3) | 10 (5.7) | 10 (4.5) | 0.50 |
| 88%-89.9% | 1 (1.3) | 5 (7.2) | 14 (3.9) | 7 (4.0) | 6 (2.7) | |
| 90%-93.9% | 9 (11.5) | 7 (10.1) | 51 (14.3) | 30 (17.2) | 38 (17.2) | |
| ≥94% | 62 (79.5) | 50 (72.5) | 272 (76.4) | 127 (73.0) | 167 (75.6) | |
| Blood eosinophilia (≥4%) | 9 (12.7) | 10 (17.9) | 38 (12.0) | 10 (6.2) | 15 (7.8) | 0.06 |
| Any IgE sensitization | 16 (20.3) | 15 (20.8) | 64 (17.6) | 47 (26.6) | 39 (17.2) | 0.13 |
| Aeroallergen sensitization | 0 (0.0) | 1 (1.4) | 6 (1.7) | 3 (1.7) | 4 (1.8) | 0.85 |
| Food allergen sensitization | 16 (20.3) | 14 (19.4) | 60 (16.5) | 44 (24.9) | 36 (15.9) | 0.14 |
| Clinical course | ||||||
| Positive pressure ventilation use† | 4 (5.1) | 2 (2.8) | 14 (3.9) | 14 (7.9) | 15 (6.6) | 0.23 |
| Intensive treatment use‡ | 11 (13.9) | 8 (11.1) | 49 (13.5) | 29 (16.4) | 44 (19.4) | 0.28 |
| Length-of-stay (day), median (IQR) | 2 (1-3) | 2 (1-3) | 2 (1-3) | 2 (1-4) | 2 (1-3) | 0.24 |
| Respiratory virus | ||||||
| Any RSV infection | 66 (83.5) | 61 (84.7) | 294 (81.0) | 155 (87.6) | 173 (76.2) | 0.05 |
| Any rhinovirus infection | 13 (16.5) | 9 (12.5) | 79 (21.8) | 27 (15.3) | 57 (25.1) | 0.04 |
| Rhinovirus-A | 6 (7.6) | 5 (6.9) | 31 (8.5) | 15 (8.5) | 31 (13.7) | 0.20 |
| Rhinovirus-B | 3 (3.8) | 0 (0.0) | 5 (1.4) | 2 (1.1) | 5 (2.2) | 0.36 |
| Rhinovirus-C | 6 (7.6) | 3 (4.2) | 41 (11.3) | 8 (4.5) | 23 (10.1) | 0.05 |
Abbreviations: ICU, intensive care unit; IgE, immunoglobulin E; IQR, interquartile range; RSV, respiratory syncytial virus
Data are no. (%) of infants unless otherwise indicated. Percentages may not equal 100, because of rounding and missingness.
Any systemic antibiotic or corticosteroid use from birth up to the index hospitalization for bronchiolitis.
Infants with severe bronchiolitis who underwent continuous positive airway ventilation and/or mechanical ventilation.
Infants with severe bronchiolitis who were admitted to ICU and/or who underwent positive pressure ventilation.
Descriptively, infants with a metabotype A were characterized by “classic” clinical presentation of bronchiolitis (e.g., young age and a low proportion of previous breathing problems, lifetime corticosteroid use, and parental asthma and eczema) and high abundance of GPC (Table 1, Figure 2). Metabotype B was characterized by a high proportion of lifetime corticosteroid use and parental asthma, a high abundance of amino acids and hydroxyoctadecadienoic acid (HODE) metabolites, and low abundance of PUFAs (Figures 2–3, Figure E2). Metabotype C was characterized by a high proportion of rhinovirus-C infection, high abundance of amino acids, and low abundance of GP (Figure 2, Figure E3). Metabotype D was characterized by a high proportion of parent history of eczema and IgE sensitization, and high abundance of GP (Figure 2, Figure E4). Lastly, the metabotype E was characterized by a high proportion of intensive treatment use, and a mixed metabolite profile (Figure 2, Figure E5).
Figure 3. Principal component analysis and functional pathway analyses in the metabotypes A vs. B comparison.
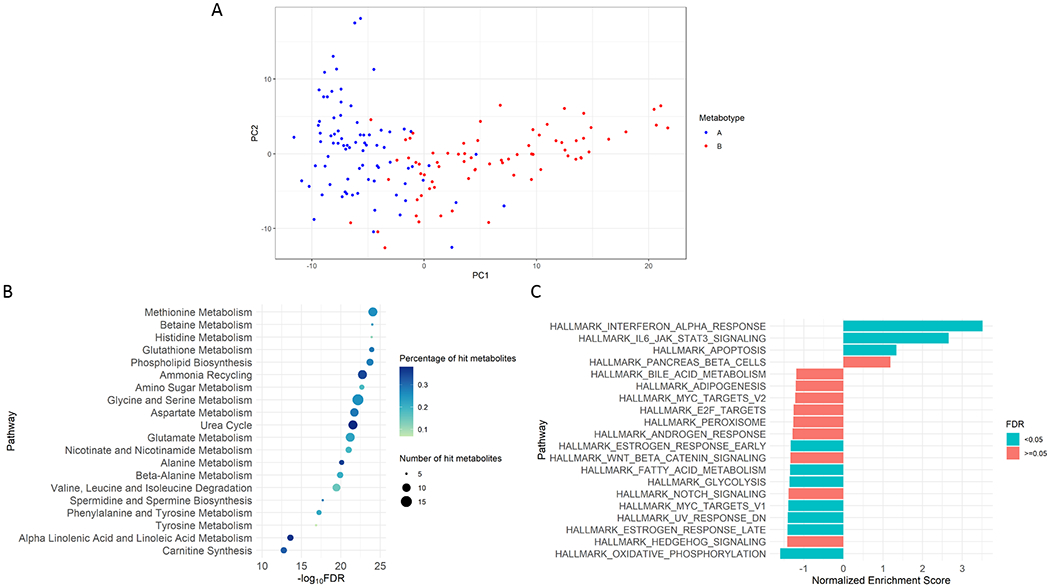
a. Principal component analysis
Principal component analysis of metabotypes A and B based on the first two principal components. The global metabolome profiles were distinct between the metabotypes A and B.
b. Metabolite-set enrichment analysis
Shown metabolite pathways (based on the Small Molecule Pathway Database [SMPDB]) are 20 pathways with a smallest false discovery rate (FDR) that have the number of hits per pathway of ≥5 nasopharyngeal metabolites. SMPDB is a 99-metabolite set based on normal human metabolism pathways. The colour of each dot represents the proportion of hit metabolites; the size of each dot represents the number of hit metabolites.
c. Functional class scoring analysis
Shown biological pathways (based on the Molecular Signatures Database [MSigDB] hallmark gene sets) are 20 pathways with a highest absolute value of normalized enriched score. MSigDB hallmark collection contains 50 gene sets, which summarize and represent specific well-defined biological states or processes and display coherent expression. The analysis is based on nasopharyngeal transcriptome (through RNAseq) data. The colour of bars represents FDR<0.05 (green) or ≥0.05 (orange).
Association of Metabotypes with Chronic Respiratory Outcomes
To investigate longitudinal association of the metabotypes with clinical outcomes, we compared the outcome risks between infants with a metabotype A (clinically “classic” bronchiolitis) and those with each of the other metabotypes (Table 2). Compared with metabotype A infants, metabotype B infants had a significantly higher risk of developing asthma (23.4% vs. 40.6% [binary outcome]; OR, 2.24; 95%CI, 1.10-4.63; P=0.03). This association remained significant after adjusting for the confounders (ORadj, 2.18; 95%CI, 1.03–4.71; P=0.04; E-value=2.32). By contrast, those with a metabotype C, D, or E did not have significantly differential risks.
Table 2.
Unadjusted and adjusted associations of severe bronchiolitis metabotypes in infants with development of childhood asthma
| Childhood asthma* |
||||
|---|---|---|---|---|
| Unadjusted model |
Adjusted model† |
|||
| Metabotypes | Odds ratio (95% CI) | P-value | Odds ratio (95% CI) | P-value |
| Metabotype A (GPC-high) | 1 [Reference] | – | 1 [Reference] | – |
| Metabotype B (amino acid-high, PUFA-low) | 2.24 (1.10-4.63) | 0.03 | 2.18 (1.03-4.71) | 0.04 |
| Metabotype C (amino acid-high, GP-low) | 1.14 (0.65-2.08) | 0.66 | 1.07 (0.58-2.02) | 0.84 |
| Metabotype D (GP-high) | 1.23 (0.66-2.34) | 0.52 | 1.25 (0.65-2.49) | 0.51 |
| Metabotype E (mixed) | 1.12 (0.62-2.11) | 0.72 | 0.87 (0.46-1.72) | 0.69 |
Abbreviations: CI, confidential interval; GP, glycerophospholipid; GPC, glycerophosphocholine; PUFA, polyunsaturated fatty acid.
Asthma was defined as physician-diagnosis of asthma by age 5 years, plus either asthma medication use (e.g., albuterol inhaler, inhaled corticosteroids, montelukast) or asthma-related symptoms in the preceding year. To examine the association between severe bronchiolitis metabotypes (metabotype A as the reference) and the risk of developing childhood asthma, logistic regression model was fit.
The multivariable logistic regression models adjusted for potential confounders, including patient’s age, sex, parent history of asthma, number of previous breathing problems, respiratory syncytial vims infection, and rhinovirus infection.
Likewise, compared with metabotype A infants, metabotype B infants had a significantly higher rate of recurrent wheeze that resulted in asthma (19.0% vs. 34.0%; HRadj, 2.52; 95%CI, 1.16–5.47; P=0.02; E-value=3.18; Table E2). By contrast, for the rate of recurrent wheeze that did not result in asthma, there were no significant differences between the metabotypes A and B (20.3% vs. 19.5%; HRadj, 0.88; 95%CI, 0.36–2.17; P=0.78).
Distinct Biological Characteristics of Metabotypes
Based on principal component analysis, the global metabolome profiles were distinct between the reference (metabotype A) and highest asthma risk (metabotype B) groups (Figure 3A). To better understand their functional difference, we conducted pathway analyses using the nasopharyngeal metabolome and transcriptome data. In the metabolite-set enrichment analysis, the differentially-enriched (FDR<0.05) pathways for metabotype B infants included amino acid (e.g., methionine, betaine, histidine, glutathione) and fatty acid (α-linolenic acid and linoleic acid) metabolism pathways (all FDR<5×10−14; Figure 3B). Additionally, in the gene-set enrichment analysis of the transcriptome data, infants with a metabotype B had differentially-enriched pathways, such as up-regulated interferon-α (IFN-α) and interleukin-6/Janus kinase/signal transducers and activators of transcription-3 (IL-6/JAK/STAT3) pathways as well as down-regulated fatty acid metabolism pathway. The integrated analysis of both metabolome and transcriptome data also identified differentially-enriched pathways in metabotype B, such as metabolism of lipids (FDR=5×10−27; Figure E2). For the comparisons of metabotype A vs. the other metabotypes (C, D, and E), the detailed functional differences are summarized in Figures E3–E5.
Sensitivity Analysis
In the subgroup analysis restricting the study sample to the infants without previous breathing problems, compared with metabotype A, metabotype B had a non-significantly higher risk of developing asthma (ORadj, 1.76; 95%CI, 0.75-4.16; P=0.19; Table E3). Second, we also examined different numbers of metabotypes. The alluvial plot (Figure E6) showed a consistency of the original 5 metabotypes (A-E) across the different numbers of metabotypes. With the use of 6 metabotypes, for example, the metabotype 1 had 98.6% concordance with the original metabotype A and the metabotype 2 had 96.5% concordance with the original metabotype B (Table E4). Similar to the primary analysis, the metabolite-set enrichment analysis demonstrated that, compared to metabotype 1 (which is concordant with metabotype A), metabotype 2 (which is concordant with metabotype B) infants also had differentially-enriched metabolism of amino acids and PUFAs (Figure E7). In addition, compared to metabotype 1, metabotype 2 infants had a non-significantly but consistently higher risk of asthma (23.9% vs. 41.8%; ORadj, 2.19; 95%CI, 0.97-5.01; P=0.06; Table E5) and significantly higher rate of recurrent wheeze that resulted in asthma (20.4% vs. 37.8%; HRadj, 2.55; 95%CI, 1.13-5.76; P=0.02; E-value=3.21; Table E6).
DISCUSSION
By applying a clustering approach to the nasopharyngeal airway metabolome data from a multicenter prospective cohort study of 918 infants with severe bronchiolitis, we identified five biologically-distinct metabotypes. In particular, compared to infants with metabotype A (who resemble “classic” bronchiolitis), those with a metabotype B (characterized by a high proportion of parental asthma, high abundance of amino acids, and low abundance of PUFAs) had a significantly higher risk for developing recurrent wheeze and asthma. To the best of our knowledge, this is the first study that has identified biologically-distinct airway metabotypes in infants with severe bronchiolitis and demonstrated their longitudinal relations with the risk of chronic respiratory outcomes.
Bronchiolitis has been traditionally viewed as a single disease condition with similar mechanisms. Indeed, the current U.S. guidelines for diagnosis and management of bronchiolitis rely on this assumption 15, 31. Yet, concordant with the results of the present study, a growing body of evidence supports the complexity of bronchiolitis, as reflected by heterogeneous clinical characteristics 8, 9, transcriptome 32–34, microRNA 33, cytokine 35, 36, and upper airway microbiome 37–39. Downstream of these transcriptional and translational processes, the metabolome—the global collection of small molecules (e.g., sugars, amino acids, lipids)—provides “proximal reports” of a system’s functional state 11. Recent studies have suggested the relationship of the metabolome with disease severity among infants with severe bronchiolitis 13, 14, 24, 40–42. For example, among 140 infants with severe bronchiolitis in the U.S., serum metabolome profiles (e.g., glutathione) was associated with a higher severity of illness 24. Furthermore, in a single-center study of 52 infants with bronchiolitis in Italy, urine amino acids (e.g., methionine) were associated with a higher risk of recurrent wheeze during the 2-year follow-up period 43. These earlier reports have supported the heterogeneity of bronchiolitis and discovered potential mechanisms that underlie the bronchiolitis-chronic respiratory outcomes link. The current study—with a sample size many times larger than any other prior study—corroborates earlier findings and extends them by identifying biologically-distinct severe bronchiolitis metabotypes that have differential risks of developing recurrent wheeze and asthma.
There are several potential mechanisms linking severe bronchiolitis—in particular metabotype B (amino acid-high, PUFA-low)—with subsequent respiratory outcomes. First, studies have shown inflammatory amino acids (such as betaine, methionine, and histidine) involved in DNA methylation in the asthma pathobiology. Specifically, betaine provides the methyl groups for synthesizing methionine, which further generates S-adenosylmethionine (SAM). SAM is a key methyl donor, which contributes to DNA methylation 44. Indeed, DNA methylation is vital in the activation of the immune response in patients with asthma. For example, a previous study showed an increased DNA methylation level at the IL-4 promoter in cells from patients with asthma after house dust mite stimulation 45. Second, the role of PUFAs (e.g., ω-3 PUFAs) as immune modulators has been demonstrated in relation to asthma 46. For example, α-linolenic acid (an ω-3 PUFA) is converted to eicosapentaenoic acid, which inhibits arachidonic acid thereby suppressing production of eicosanoid inflammatory mediators. Eicosapentaenoic acid also functions as a precursor for pro-resolving mediators (such as resolvins, protectins, and maresins) 47. These mediators have important anti-inflammatory properties with protective effects on asthma 46. Observational studies and randomized controlled trials have also shown a protective effect of PUFAs on development of asthma. For example, the supplement of ω-3 PUFAs during pregnancy are associated with a reduced risk of wheezing illness 48 and asthma 48, 49 in the offspring. Similarly, lower ω-3 PUFA intake is associated with a higher incidence of asthma in children 50. Third, in the current study, we also found a high level of 9- and 13-HODE in the metabotype B. Emerging evidence suggests that HODE plays a role in the pathobiology of asthma 51. For example, Panda et al. reported, in allergic asthma mouse and human bronchial epithelial cell models, that 13-HODE increases pNF-κB levels and reduces GR-α transcripts, thereby inducing corticosteroid resistance 51. Lastly, our transcriptome analysis also found that metabotype B had up-regulated IFN-α and IL-6/JAK/STAT3 pathways, which are involved in asthma pathobiology 52 Notwithstanding the complexity of these mechanisms, we believe that the identification of severe bronchiolitis subtypes based on metabolome data (or metabotypes) and their longitudinal relations with chronic respiratory outcomes are important. Our findings, in conjunction with the existent literature, should advance research into the development of subtype-specific strategies for bronchiolitis treatment and asthma prevention.
Our study has several potential limitations. First, bronchiolitis involves inflammation of the lower airways, in addition to the upper airways. While our data relied on nasopharyngeal specimens, research has shown that upper airway sampling provides reliable representation of inflammatory profiles in the lower airways 53. Furthermore, the use of upper airway specimens is preferable as bronchoscopy or other methods of lower airway sampling would be too invasive in young infants with bronchiolitis. Second, the nasopharyngeal specimens were obtained at a single timepoint. While longitudinal molecular data are also instrumental, the study objective was to identify metabotypes of severe bronchiolitis. Even with single-time point data, we successfully identified biologically-distinct metabotypes that are longitudinally associated with subsequent respiratory outcomes. Third, the current analysis was designed not to examine asthma phenotypes but to use the epidemiological definition of asthma at age five years 16, 18. The cohort is being followed up not only to minimize misclassification due to asthma development at a later age, but also to perform comprehensive phenotyping and endotyping of children who develop asthma. Fourth, our causal inference might have been confounded by unmeasured factors (e.g., host genetics, nutrition, inborn errors of metabolism). However, the relatively large E-values implied that confounders would have been strong to explain away our inference. Fifth, the current study used untargeted metabolomics procedure and it is important to conduct independent replication using targeted metabolomics technique to improve the profiling resolution and replicate our findings. Lastly, despite the study cohort consisting of racially/ethnically- and geographically-diverse infants, our inferences may not be generalizable to those with mild-to-moderate bronchiolitis, and hence warrant external validation. Nonetheless, our data remain directly relevant for the 110,000 infants hospitalized yearly in the U.S. 1, a population with substantial morbidity burden.
In conclusion, by applying an unsupervised clustering approach to the nasopharyngeal airway metabolome data from a multicenter prospective cohort study of 918 infants with severe bronchiolitis, we identified five biologically-distinct and clinically-meaningful metabotypes. Specifically, the metabotype characterized by a high abundance of inflammatory amino acids and low abundance of PUFAs had the highest risk for developing asthma. Our data lend significant support to the concept that “bronchiolitis” is a heterogenous syndrome with different biological mechanisms. These observations should facilitate further investigations into the development of metabotype-targeted strategies for bronchiolitis treatment and asthma prevention. Furthermore, future examinations of the relationship between bronchiolitis metabotypes and asthma endotypes (e.g., based on the genome, transcriptome, and metabolome) will also provide a new avenue for the development of endotypes-specific prevention strategies for asthma.
Supplementary Material
Figure 4. Kaplan-Meier curves for development of recurrent wheeze by age 3 years, according to severe bronchiolitis metabotypes.

a. Recurrent wheeze by age 3 years with asthma at age 5 years
Compared with metabotype A infants, the rate of developing recurrent wheeze was not significantly different in metabotype B, C, D, or E infants. However, the rate was significantly higher in metabotype B (amino acid-high/PUFA-low) infants (adjusted HR 2.52; 95% CI 1.16-5.47; P=0.02).
b. Recurrent wheeze by age 3 years without asthma at age 5 years
Compared with metabotype A infants, the rate of developing recurrent wheeze was not significantly different in metabotype B, C, D, or E. Corresponding hazards ratio estimates are presented in Table E2.
Key Messages:
Growing evidence suggests that bronchiolitis is a heterogeneous syndrome.
Using data of the nasopharyngeal airway metabolome from a multicenter cohort of infants with severe bronchiolitis, unsupervised clustering analysis identified five biologically distinct “metabotypes”.
These metabotypes were also clinically meaningful. For example, the metabotype characterized by high abundance of inflammatory amino acids and low abundance of PUFAs had the highest risk for developing asthma.
Acknowledgements:
We thank the MARC-35 study hospitals and research personnel for their ongoing dedication to bronchiolitis and asthma research (Table E1), and Ashley F. Sullivan, MS, MPH and Janice A. Espinola, MPH (Massachusetts General Hospital, Boston, MA) for their many contributions to the MARC-35 study. We also thank Alkis Togias, MD, at the National Institutes of Health (Bethesda, MD) for helpful comments about the study results.
Funding:
This study was supported by grants from the National Institutes of Health (Bethesda, MD): K01 AI-153558, U01 AI-087881, R01 AI-114552, R01 AI-127507, R01 AI-134940, R01 AI-137091, R01 AI-148338, and UG3/UH3 OD-023253. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funding organizations were not involved in the collection, management, or analysis of the data; preparation or approval of the manuscript; or decision to submit the manuscript for publication.
Abbreviations:
- DNA
Deoxyribonucleic acid
- FDR
False discovery rate
- GP
Glycerophospholipid
- GPC
Glycerophosphocholine
- GR-α
Glucocorticoid receptor-alpha
- HODE
Hydroxyoctadecadienoic acid
- IFN-α
Interferon-alpha
- IL-4
Interleukin-4
- IL-6
Interleukin-6
- IMPaLA
Integrated Molecular Pathway Level Analysis
- IQR
Interquartile range
- JAK
Janus kinase
- MARC-35
The 35th Multicenter Airway Research Collaboration
- NF-κB
Nuclear factor kappa-light-chain-enhancer of activated B cells
- OR
Odds ratio
- PUFA
Polyunsaturated fatty acid
- RSV
Respiratory syncytial virus
- SAM
S-adenosylmethionine
- STAT3
Signal transducers and activators of transcription-3
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interests statement: Dr. Zhu reports grants from National Institutes of Health during the conduct of the study. Dr. Camargo reports grants from National Institutes of Health during the conduct of the study. Dr. Woodruff reports personal fees from Sanofi, personal fees from Regeneron, personal fees from Astra Zeneca, personal fees from Glenmark Pharma, personal fees from Theravance, personal fees from NGM Pharma, personal fees from GSK, outside the submitted work. Dr. Hasegawa reports grants from National Institutes of Health during the conduct of the study; grants from Novartis, outside the submitted work. All other authors have indicated that they have no financial relationships relevant to this article to disclose.
REFERENCES
- 1.Fujiogi M, Goto T, Yasunaga H, Fujishiro J, Mansbach JM, Camargo CA Jr., et al. Trends in bronchiolitis hospitalizations in the united states: 2000-2016. Pediatrics 2019; 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kotaniemi-Syrjanen A, Vainionpaa R, Reijonen TM, Waris M, Korhonen K, Korppi M. Rhinovirus-induced wheezing in infancy--the first sign of childhood asthma? J Allergy Clin Immunol 2003; 111:66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll KN, Wu P, Gebretsadik T, Griffin MR, Dupont WD, Mitchel EF, et al. The severity-dependent relationship of infant bronchiolitis on the risk and morbidity of early childhood asthma. J Allergy Clin Immunol 2009; 123:1055–61, 61 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sigurs N, Aljassim F, Kjellman B, Robinson PD, Sigurbergsson F, Bjarnason R, et al. Asthma and allergy patterns over 18 years after severe rsv bronchiolitis in the first year of life. Thorax 2010; 65:1045–52. [DOI] [PubMed] [Google Scholar]
- 5.Bacharier LB, Cohen R, Schweiger T, Yin-Declue H, Christie C, Zheng J, et al. Determinants of asthma after severe respiratory syncytial virus bronchiolitis. J Allergy Clin Immunol 2012; 130:91–100 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Regnier SA, Huels J. Association between respiratory syncytial virus hospitalizations in infants and respiratory sequelae: Systematic review and meta-analysis. Pediatr Infect Dis J 2013; 32:820–6. [DOI] [PubMed] [Google Scholar]
- 7.Hasegawa K, Dumas O, Hartert TV, Camargo CA Jr. Advancing our understanding of infant bronchiolitis through phenotyping and endotyping: Clinical and molecular approaches. Expert Rev Respir Med 2016; 10:891–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dumas O, Mansbach JM, Jartti T, Hasegawa K, Sullivan AF, Piedra PA, et al. A clustering approach to identify severe bronchiolitis profiles in children. Thorax 2016; 71:712–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dumas O, Hasegawa K, Mansbach JM, Sullivan AF, Piedra PA, Camargo CA, Jr. Severe bronchiolitis profiles and risk of recurrent wheeze by age 3 years. J Allergy Clin Immunol 2019; 143:1371–9 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raita Y, Camargo CA Jr., Bochkov YA, Celedon JC, Gern JE, Mansbach JM, et al. Integrated-omics endotyping of infants with rhinovirus bronchiolitis and risk of childhood asthma. J Allergy Clin Immunol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu Z, Camargo CA Jr., Hasegawa K. Metabolomics in the prevention and management of asthma. Expert Rev Respir Med 2019; 13:1135–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reinke SN, Gallart-Ayala H, Gomez C, Checa A, Fauland A, Naz S, et al. Metabolomics analysis identifies different metabotypes of asthma severity. Eur Respir J 2017; 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart CJ, Mansbach JM, Ajami NJ, Petrosino JF, Zhu Z, Liang L, et al. Serum metabolome is associated with the nasopharyngeal microbiota and disease severity among infants with bronchiolitis. J Infect Dis 2019; 219:2005–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart CJ, Mansbach JM, Wong MC, Ajami NJ, Petrosino JF, Camargo CA Jr., et al. Associations of nasopharyngeal metabolome and microbiome with severity among infants with bronchiolitis. A multiomic analysis. Am J Respir Crit Care Med 2017; 196:882–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ralston SL, Lieberthal AS, Meissner HC, Alverson BK, Baley JE, Gadomski AM, et al. Clinical practice guideline: The diagnosis, management, and prevention of bronchiolitis. Pediatrics 2014; 134:e1474–502. [DOI] [PubMed] [Google Scholar]
- 16.Hasegawa K, Mansbach JM, Bochkov YA, Gern JE, Piedra PA, Bauer CS, et al. Association of rhinovirus c bronchiolitis and immunoglobulin e sensitization during infancy with development of recurrent wheeze. JAMA Pediatr 2019; 173:544–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 2017; 14:417–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camargo CA Jr., Ingham T, Wickens K, Thadhani R, Silvers KM, Epton MJ, et al. Cord-blood 25-hydroxyvitamin d levels and risk of respiratory infection, wheezing, and asthma. Pediatrics 2011; 127:e180–7. [DOI] [PubMed] [Google Scholar]
- 19.National Asthma Education and Prevention Program. Expert panel report 3 (epr-3): Guidelines for the diagnosis and management of asthma-summary report 2007. J Allergy Clin Immunol 2007; 120:S94–138. [DOI] [PubMed] [Google Scholar]
- 20.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012; 28:882–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Comhair SA, McDunn J, Bennett C, Fettig J, Erzurum SC, Kalhan SC. Metabolomic endotype of asthma. J Immunol 2015; 195:643–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly RS, Dahlin A, McGeachie MJ, Qiu W, Sordillo J, Wan ES, et al. Asthma metabolomics and the potential for integrative omics in research and the clinic. Chest 2017; 151:262–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abreo A, Gebretsadik T, Stone CA, Hartert TV. The impact of modifiable risk factor reduction on childhood asthma development. Clin Transl Med 2018; 7:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hasegawa K, Stewart CJ, Celedon JC, Mansbach JM, Tierney C, Camargo CA Jr., Serum 25-hydroxyvitamin d, metabolome, and bronchiolitis severity among infants-a multicenter cohort study. Pediatr Allergy Immunol 2018; 29:441–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.VanderWeele TJ, Ding P. Sensitivity analysis in observational research: Introducing the e-value. Ann Intern Med 2017; 167:268–74. [DOI] [PubMed] [Google Scholar]
- 26.Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, et al. Metaboanalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Research 2018; 46:W486–W94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frolkis A, Knox C, Lim E, Jewison T, Law V, Hau DD, et al. Smpdb: The small molecule pathway database. Nucleic Acids Res 2010; 38:D480–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korotkevich G, Sukhov V, Sergushichev A. Fast gene set enrichment analysis. bioRxiv 2019:060012. [Google Scholar]
- 29.Kamburov A, Cavill R, Ebbels TMD, Herwig R, Keun HC. Integrated pathway-level analysis of transcriptomics and metabolomics data with impala. Bioinformatics 2011; 27:2917–8. [DOI] [PubMed] [Google Scholar]
- 30.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995; 57:289–300. [Google Scholar]
- 31.National Institute for Health and Care Excellence. Bronchiolitis in children: Diagnosis and management. 2015. [PubMed]
- 32.de Steenhuijsen Piters WA, Heinonen S, Hasrat R, Bunsow E, Smith B, Suarez-Arrabal MC, et al. Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med 2016; 194:1104–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hasegawa K, Perez-Losada M, Hoptay CE, Epstein S, Mansbach JM, Teach SJ, et al. Rsv vs. Rhinovirus bronchiolitis: Difference in nasal airway microrna profiles and nfkappab signaling. Pediatr Res 2018; 83:606–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujiogi M, Camargo CA Jr., Bernot JP, Freishtat RJ, Harmon B, Mansbach JM, et al. In infants with severe bronchiolitis: Dual-transcriptomic profiling of nasopharyngeal microbiome and host response. Pediatr Res 2020; 88:144–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turi KN, Shankar J, Anderson LJ, Rajan D, Gaston K, Gebretsadik T, et al. Infant viral respiratory infection nasal immune-response patterns and their association with subsequent childhood recurrent wheeze. Am J Respir Crit Care Med 2018; 198:1064–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hasegawa K, Hoptay CE, Harmon B, Celedon JC, Mansbach JM, Piedra PA, et al. Association of type 2 cytokines in severe rhinovirus bronchiolitis during infancy with risk of developing asthma: A multicenter prospective study. Allergy 2019; 74:1374–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hasegawa K, Mansbach JM, Ajami NJ, Espinola JA, Henke DM, Petrosino JF, et al. Association of nasopharyngeal microbiota profiles with bronchiolitis severity in infants hospitalised for bronchiolitis. Eur Respir J 2016; 48:1329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mansbach JM, Hasegawa K, Henke DM, Ajami NJ, Petrosino JF, Shaw CA, et al. Respiratory syncytial virus and rhinovirus severe bronchiolitis are associated with distinct nasopharyngeal microbiota. J Allergy Clin Immunol 2016; 137:1909–13 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosas-Salazar C, Shilts MH, Tovchigrechko A, Schobel S, Chappell JD, Larkin EK, et al. Differences in the nasopharyngeal microbiome during acute respiratory tract infection with human rhinovirus and respiratory syncytial virus in infancy. J Infect Dis 2016; 214:1924–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hasegawa K, Stewart CJ, Celedon JC, Mansbach JM, Tierney C, Camargo CA, Jr. Circulating 25-hydroxyvitamin d, nasopharyngeal airway metabolome, and bronchiolitis severity. Allergy 2018; 73:1135–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fujiogi M, Camargo CA Jr., Raita Y, Bochkov YA, Gern JE, Mansbach JM, et al. Association of rhinovirus species with nasopharyngeal metabolome in bronchiolitis infants: A multicenter study. Allergy 2020; 75:2379–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujiogi M, Camargo CA Jr., Raita Y, Zhu Z, Celedon JC, Mansbach JM, et al. Integrated associations of nasopharyngeal and serum metabolome with bronchiolitis severity and asthma: A multicenter prospective cohort study. Pediatr Allergy Immunol 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barlotta A, Pirillo P, Stocchero M, Donato F, Giordano G, Bont L, et al. Metabolomic profiling of infants with recurrent wheezing after bronchiolitis. J Infect Dis 2019; 219:1216–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma S, Litonjua A. Asthma, allergy, and responses to methyl donor supplements and nutrients. J Allergy Clin Immunol 2014; 133:1246–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang CM, Chang CB, Chan MW, Wen ZH, Wu SF. Dust mite allergen-specific immunotherapy increases il4 DNA methylation and induces der p-specific t cell tolerance in children with allergic asthma. Cell Mol Immunol 2018; 15:963–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wendell SG, Baffi C, Holguin F. Fatty acids, inflammation, and asthma. J Allergy Clin Immunol 2014; 133:1255–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duvall MG, Levy BD. Dha- and epa-derived resolvins, protectins, and maresins in airway inflammation. Eur J Pharmacol 2016; 785:144–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bisgaard H, Stokholm J, Chawes BL, Vissing NH, Bjarnadottir E, Schoos AM, et al. Fish oil-derived fatty acids in pregnancy and wheeze and asthma in offspring. N Engl J Med 2016; 375:2530–9. [DOI] [PubMed] [Google Scholar]
- 49.Lumia M, Luukkainen P, Tapanainen H, Kaila M, Erkkola M, Uusitalo L, et al. Dietary fatty acid composition during pregnancy and the risk of asthma in the offspring. Pediatr Allergy Immunol 2011; 22:827–35. [DOI] [PubMed] [Google Scholar]
- 50.Lee-Sarwar K, Kelly RS, Lasky-Su J, Kachroo P, Zeiger RS, O’Connor GT, et al. Dietary and plasma polyunsaturated fatty acids are inversely associated with asthma and atopy in early childhood. J Allergy Clin Immunol Pract 2019; 7:529–38 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Panda L, Gheware A, Rehman R, Yadav MK, Jayaraj BS, Madhunapantula SV, et al. Linoleic acid metabolite leads to steroid resistant asthma features partially through nf-kappab. Sci Rep 2017; 7:9565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rincon M, Irvin CG. Role of il-6 in asthma and other inflammatory pulmonary diseases. Int J Biol Sci 2012; 8:1281–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poole A, Urbanek C, Eng C, Schageman J, Jacobson S, O’Connor BP, et al. Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. J Allergy Clin Immunol 2014; 133:670–8 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


