Abstract
We present recently developed strategies to manipulate lipid levels in live cells by light. We focus on photo-removable protecting groups that lead to subcellular restricted localization and activation and discuss alternative techniques. We emphasize the development of organelle targeting of caged lipids and discuss recent advances in chromatic orthogonality of caging groups for future applications.
Introduction.
Compartmentalization of cellular events is a hallmark of eukaryotic cells.1 Classically, each compartment consists of a segregated membrane system forming a lumen, a luminal and a cytosolic membrane leaflet.2 In addition, the plasma membrane forms the boundary to the outer world, with strikingly different lipid compositions of the inner and outer leaflet.3 Recently, this picture was complicated by findings that the interaction of proteins with lipids on a kiss-and-run base differs strikingly from cases where the proteins are artificially transferred to a specific membrane, implying that any membrane surface might constitute its own “compartment”.4 In this respect, it becomes relevant that lipids are transported from one membrane side to the other by a variety of flipases.5 Further, there is constant membrane exchange between compartments via the transport of vesicles as well as lipid transport via transport proteins. For instance, COPI vesicles are released from the Golgi network and trafficked to the plasma membrane or the endoplasmic reticulum (ER) while COPII-decorated vesicles perform the retro-transport from the ER to the Golgi.6 These transport processes are both essential and quite rapid as inhibiting COPII transport by blocking of ER exit sites will exhaust the Golgi membrane system within 30 to 60 min.7 Apart from the exchange of entire membrane sections via vesicles, single lipid molecules such as cholesterol, phosphatidylinositols or ceramides are efficiently transferred via specific transport proteins, many of them shuttling lipids across membrane systems that are in close proximity, for instance at organelle-organelle contact sites.8,9,10 Despite this fast exchange, organelles seem to maintain a specific lipid composition most likely due to site-specific metabolism.11 The lipid composition is then not only responsible for organelle identity and function but also represents a signaling component that determines organelle transport and fusion events, for instance in the case of endosomes and their trafficking to either the plasma membrane (endosome recycling ) or the lysosome (for endolysosomal destruction).12,13
In order to study organelle function, organelle membrane identity, lipid transport, membrane composition and the corresponding cellular functions, there is a need for tools to alter lipid compositions specifically with high spatial resolution. An elegant option is to copy nature and alter the local lipid composition enzymatically by translocating a lipid-metabolizing enzyme specifically to the membrane of interest.14 One way to achieve this is by using chemically induced dimerization (CID) systems exploiting the formation of a tertiary protein complex (i.e. FRB:FKBP, eDHFR:FKBP, or SNAP:FKBP) and is induced by treatment with a small molecule (i.e. rapamycin, TMP-SLF, or rCD1/FK506, respectively). 15, 16, 17 Alternatively, several light-switchable genetically encoded translocation systems such as Cry2/CIB1, iLID and “Magnets” were compared as tools to switch lipid phosphatase activity.18 However, these switching systems require the introduction of the necessary enzyme, usually by transfection or viral transduction into the cell of interest, as well as the switch system. Another less invasive method is changing the lipid composition by small molecule addition and light. For this purpose, photo-activatable lipid derivatives, usually called “caged” lipids, or photo-switchable lipids based on light-induced cis-trans isomerization of azobenzenes are employed.19,20 Local release of a defined lipid species can be achieved by using highly focused light through the objective of a confocal microscope to remove a photo-sensitive protecting (“caging”) group and produce fairly well defined concentration changes.21 A photo-switchable lipid derivative will change from an inactive to an active form or vice versa, This technique has recently been reviewed in detail.22,23 The spatial resolution of these light-driven techniques is limited by the point-spread function of the laser light unless 2-photon excitation is used.24,25 It is important to note that 2-photon absorption (or non-linear absorption) is generally several orders of magnitude smaller than one-photon absorption. Consequently, not all photo-removable protecting groups will be sensitive to 2-photon uncaging.26 The second difficulty is to precisely hit a membrane or a small organelle in a 3-dimensional space. As a result, to date 2-photon uncaging was used to release glutamate in neuronal spines24 and in deep tissue27 but to our knowledge never for lipids in subcellular structures. A solution to these challenges might be to target caged lipids to one particular membrane system prior to the photo-release. This review covers the newest tools for achieving localized lipid release by light.
Caged lipid composition.
Photo-removable protecting groups are molecular leaving groups and their departure is triggered by light. They can be categorized on the bases of their chemical structures. The most common groups are derived from arylcarbonylmethyl, nitroaryl, coumarin-4-ylmethyl, or arylmethyl groups. 28 Coumarin-based photo-removable protecting groups have become popular in the field due to high uncaging yields and faster photo-releases than classical nitrobenzene derivatives, but also due to their ability of being fluorescent under a confocal microscope.29 In cells, the amount of coumarin-caged lipids can easily be quantified during the microscopy experiment by directly measuring the fluorescence emission of the sample. Typically, the release efficiency of caged lipids in cells is also determined by thin-layer chromatography (TLC) but the extraction efficiency needs to be accounted for. The ability of measuring the exact concentration of released species is critical to ensure good reproducibility between experiments. This becomes even more important when comparing the effect of a signaling lipid released from two different subcellular locations. Such experiments can only be reasonably performed if it is possible to demonstrate that the amounts of photo-released lipids are comparable in both places.
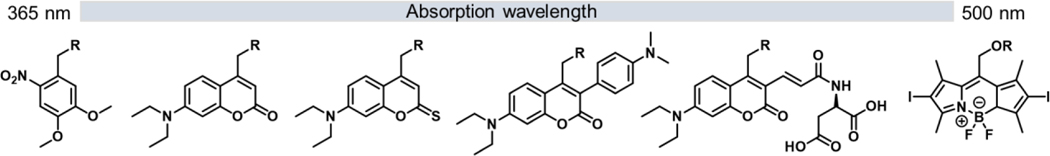
The primary goal of a photo-removable protecting group is to prevent the cellular machinery to recognize and respond to the signaling molecule of interest until removed by a flash of light. To disrupt recognition of a lipid, typically the head group with its functional and chemically reactive groups serves as carrier of the often bulky cage. Examples are the phosphates of phospholipids, 30 amino groups of sphingosines, 31 and hydroxy groups of diacylglycerols (DAGs).21 We recently developed a new chemical strategy for the caging of monoacylglycerols, which traditionally suffered from fatty acid migration and cross-activation with DAG by masking the two hydroxy groups with a photo-removable acetal.32,33 Coumarin and nitrobenzyl cages that are connected via carbamates or carbonates serve well for the protection and the rapid photo-release of most lipids in living cells (Figure 1).
Figure 1.
Example of photo-removable protecting groups expanding the spectral range of absorption for the uncaging of small molecules from UV to visible wavelengths.
How the cage influences location and activity.
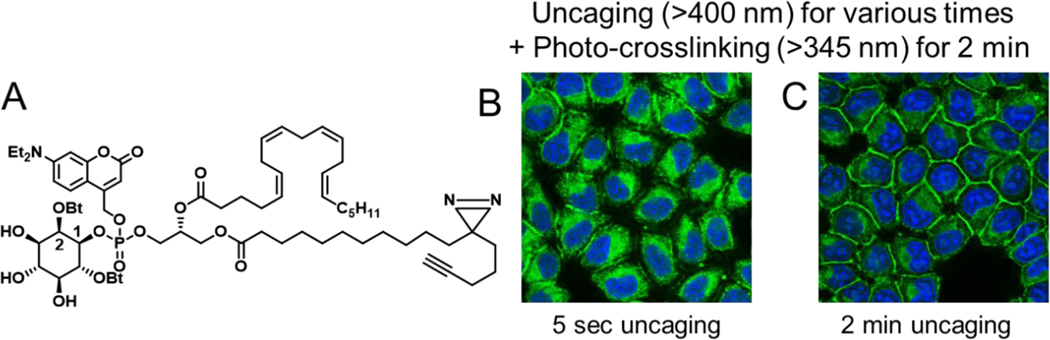
Due to the small size of lipids, the addition of large and aromatic fluorophores constitutes a major chemical alteration of their structure. This may dramatically affect their location and potentially the endogenous signaling properties. Fluorescently labelled lipids including those bearing photo-removable protecting groups, frequently mislocate in cells when compared to their endogenous counterparts.34 This feature may be turned into an advantage for using caged lipids. Especially neutral coumarin cages are highly lipophilic. Therefore, most coumarin-caged lipid species tend to accumulate in membranes, mostly endomembranes. 35 This is expected for all lipid derivatives with a single charge that is masked by the cage group. A good example is caged phosphatidylinositol (PI) (Figure 2A).36 PI is biosynthesized in ER membranes and coumarin-caged PI locates to exactly these membranes (Figure 2B). Upon uncaging, the PI derivative rapidly transferred to the plasma membrane (Figure 2C). The analysis of the PI location was made possible by two additional modifications: a diazirine group to photo-crosslink the lipid and prevent its wash-out during the cell fixation process and an alkyne group to tag the lipid after fixation via copper-catalyzed click chemistry. We refer to this combination of cage, diazirine and alkyne as trifunctional lipids. The concept is currently applied to many other lipids.37,38 It should be mentioned that even lipid derivatives with numerous charges on the headgroup, such as the higher phosphorylated phosphoinositides, will locate to endomembranes as long as an aromatic cage group is attached.39
Figure 2.
A. Chemical structure of tri-functional phosphatidylinositol (PI). Accumulation of PI in endo membranes after uncaging for 5 sec (B) or 2 min (C) followed by 2 min of photo-crosslinking. The location of the lipid was monitored by confocal microscopy after fixation and labelling the terminal alkyne of the lipid tail with an azide-bearing fluorophore via copper click chemistry.
For targeting lipids to certain membranes, the lipophilicity and interaction potential of the caging group can be appropriately modulated. Tuning the chemical properties of the cage will determine where the caged compounds will concentrate in the cellular environment. A simple introduction of charges to a coumarin cage in form of sulfonates led to an exclusive localization of caged arachidonic acid (AA) to the outer leaflet of the plasma membrane.40 Surprisingly, the photo-release of AA at the plasma membrane was shown to have profoundly different effects on intracellular calcium oscillations in the β-cell line MIN6 than a release from internal membranes. Likely, the extracellular AA predominantly triggered the fatty acid-specific GPR40 on the cell surface while AA inside cells seem to have acted on ion channels. This example demonstrated the feasibility and relevance of local lipid release. Furthermore, it illustrated that the same lipid can exert different functions at different location in the cell. Sulfonated cages were subsequently applied to caged diacylglycerols (DAGs).41 Schuhmacher et al successfully investigated the impact of the fatty acid composition on the interaction of DAGs with C1 domains as well as lipid trans-bilayer movement.
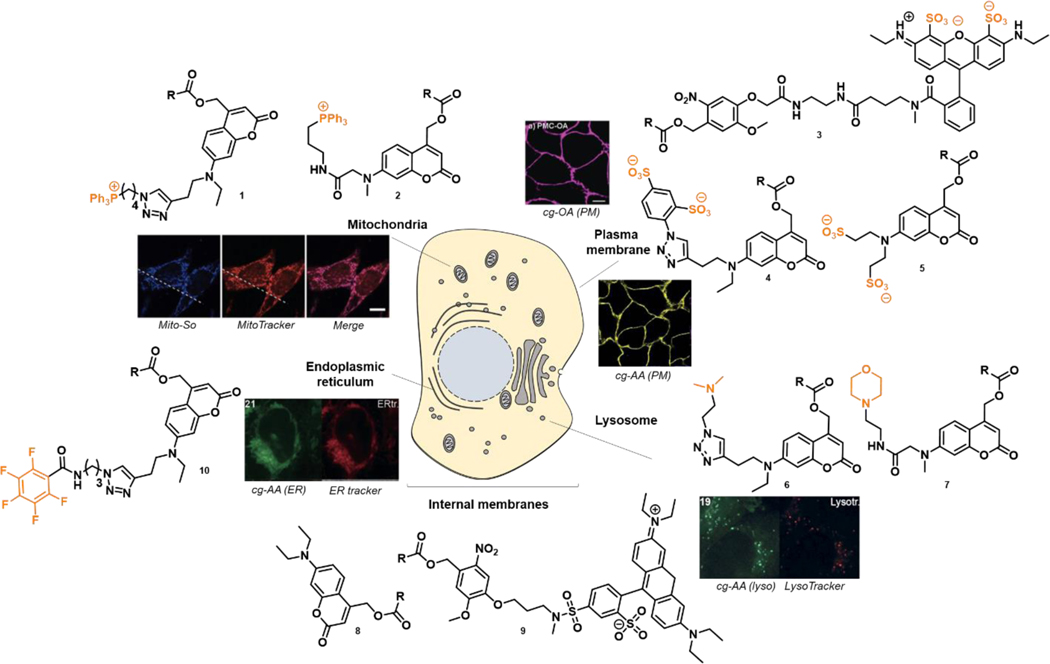
In the meantime, many more localized caged lipid tools have been introduced (Figure 3). An exciting application involved caged ceramides. 42, 43 The ceramide family of lipids have historically been quite difficult to add exogenously, due to their high hydrophobicity and toxicity. Only short-chain ceramides are readily applicable to cells. Kim et al were able to incorporate even C22 ceramide into HeLa cells by combining a coumarin cage with PEG groups for increased water solubility. 44 Interestingly, the fatty acid composition of the ceramide did affect the uncaging efficiency. Ceramide 1-phosphate (C1P) acts on several signaling pathways, both inside and at the cell surface, highlighting the importance of localization of signaling molecules. 7-Diethylaminocoumarin (DEAC)-caged C1P was compared with C1P added exogenously.45,46 While DEAC-C1P stimulated the proliferation of macrophages and caused the phosphorylation of ERK and Akt, only exogenous C1P was able to stimulate glucose uptake and cell migration.
Figure 3.
Chemical structures of photo-removable protecting groups targeting distinct cellular membranes. This allows local photo-release of signaling lipids in live cells. R refers to the lipid moiety.
As shown above, neutral cages penetrate non-selectively all endomembranes and negatively charged ones accumulate at the plasma membrane. A permanently positively charged triphenylphosphonium (TPP) group on a coumarin scaffold will concentrate the caged lipid inside mitochondria. Recently applied to subcellular manipulations of sphingosine, this strategy allowed investigating the distinct metabolic fates of the signaling lipid depending of its local accumulation in living cell.47 Stable isotope-labeled caged sphingosine precursors permitted to measure the conversion of sphingosine into ceramides and sphingomyelins after uncaging by mass spectrometry. Both caged sphingosines Mito-So and DEAC-So, once uncaged, were quickly phosphorylated to sphingosine-1-phosphate (S1P), but showed distinct patterns of incorporation into higher sphingolipids.47 Coumarin caged Sph (DEAC-So) was quickly incorporated into ceramide, but after 20 minutes this ceramide had already begun to disappear. To the contrary, sphingosine released from Mito-So continued to incorporate into ceramide even after 20 minutes and was incorporated into sphingomyelin for longer time periods than sphingosine from DEAC-So. Here, the mitochondria-specific photo-release of the lipid showed that sphingosine metabolism was highly dependent on its subcellular localization. A cytosolic increase of sphingosine was found to release calcium from lysosomes via TCP1 channels and activate the transcription factor TFEB.31 In contrast, a local increase of sphingosine levels in mitochondria was unable to induce calcium transients. Given the importance of lysosomal sphingosine metabolism in the neurodegenerative disease Niemann–Pick type C1 (NPC1), a lysosomal storage disorder, targeting the lysosome with a caged sphingosine derivative was a logical strategy. Feng et al attached a morpholino group to the coumarin molecule, leading to the accumulation of caged sphingosine (but also cholesterol) inside lysosomes of living cells. 48 Combined with the nonselective as well as the mitochondria-targeting uncaging, this toolset offered a sophisticated framework to study sphingolipid metabolism with a focus on the ceramide chain length. These subcellular manipulation approaches might be useful to apply to many other lipids in living cells.
Uniting these various directing strategies, Wagner et al conceptualized a coumarin-based cage which could be modified with various organelle directing groups.49 In this case, the amino group of DEAC was modified with a terminal alkyne group, to which a variety of azide-bearing directing groups were attached by copper-catalyzed click chemistry. The utility of this strategy was demonstrated again with both sphingosine and arachidonic acid, which were successfully targeted to the mitochondria (with the TPP group mentioned above), the lysosome (with a tertiary amino group), the plasma membrane (with a sulfonate group), or the ER (with a perfluorinated group).
The fluorescence of the coumarin molecule as a cage is often considered as an advantage, as it permits to monitor the cellular uptake and the proper subcellular localization of the caged lipid before uncaging. However, in some cases the sensitivity of the coumarin cage towards light is so high that imaging the caged compound in live cells – even at the lowest laser power – is sufficient to uncage a portion of the lipid and to perturb the experiment. To solve this limitation, Gaur et al dissociated the functions of uncaging and imaging into different units within the photo-removable group.50 The covalent binding of a photolabile ortho-nitrobenzyl (ONB) cage and a rhodamine dyes delivered fluorescent cages only sensitive to high-energy wavelength light below 400 nm. Using this approach, they designed both a globally neutral cage that enters cells and endomembranes non-specifically, and another bearing negatively charged sulfonate groups that selectively localized at the plasma membrane. These caged lipids were sensitive to 365 nm light uncaging and were proven to be resistant to other wavelengths.
Conclusion and outlook.
Caged lipids are powerful tools for investigating lipid signaling in live cell. They offer the opportunity to manipulate lipid levels without genetic or protein alterations and can therefore also be applied to primary cells. The many recent examples of targeting lipids to a distinct cellular membrane or organelle make them suitable tools for investigations at the subcellular level. The continuous development of new photo-removable protecting groups with targeting units therefore permits the precise manipulation of lipid species with exquisite spatio-temporal precision. In addition to the ability of selectively manipulating lipid concentration at subcellular levels, the diversification of the existing cages is showing premises for orthogonal, two-color uncaging (Figure 1).27,51,52 This will allow for sequential or simultaneous manipulation of a combination of different lipids or other signaling molecules in multiple organelles of the cell. By expanding the library of protecting groups for the caging of small molecules (Figure 1), recent studies provided photosensitive protecting groups that absorb photons in the visible part of the spectrum. Still based on a coumarin scaffold, DEAC-450 strongly absorbed blue light while being benign to ultra-violet light.53 This range of absorption is traditionally used for many ONB and DEAC caged compounds. Meso-substituted BODIPY are an alternative caging tool to coumarins.54,55 They were successfully applied to unmask carboxylic acids with green light excitation >500 nm in living cells. Using these new photo-removable protecting groups in combination with classical ONB or DEAC derivatives and with localization to different organelles in cells would pave a golden avenue for manipulating lipid levels with a unique degree of precision.
Acknowledgments.
S.F., A.L. and C.S. acknowledge financial support by OHSU. C.S. is grateful for funding by the NIH (R01GM127631 and R01 AI141549) and TRR186, funded by the German Research Council (DFG).
Footnotes
Competing interests.
The authors declare no competing interests for this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alberts Bruce, Johnson Alexander, Lewis Julian, Raff Martin, Roberts Keith, and Walter Peter. Molecular Biology of the Cell. 6th edition. New York: Garland Science; 2015. The Compartmentalization of Cells. Available from: https://www.ncbi.nlm.nih.gov/books/NBK26907/ [Google Scholar]
- 2.Harayama T, Riezman H. Understanding the diversity of membrane lipid composition. Nat Rev Mol Cell Biol. 2018. May;19(5):281–296. doi: 10.1038/nrm.2017.138. Epub 2018 Feb 7. Erratum in: Nat Rev Mol Cell Biol. 2019 Nov;20(11):715. [DOI] [PubMed] [Google Scholar]
- 3.Ingólfsson HI, Melo MN, van Eerden FJ, Arnarez C, Lopez CA, Wassenaar TA, Periole X, de Vries AH, Tieleman DP, Marrink SJ. Lipid organization of the plasma membrane. J Am Chem Soc. 2014. October 15;136(41):14554–9. doi: 10.1021/ja507832e. Epub 2014 Oct 1. [DOI] [PubMed] [Google Scholar]
- 4.van Meer G, de Kroon AI. Lipid map of the mammalian cell. J Cell Sci. 2011. January 1;124(Pt 1):5–8. doi: 10.1242/jcs.071233. [DOI] [PubMed] [Google Scholar]
- 5.Devaux PF, Herrmann A, Ohlwein N, Kozlov MM. How lipid flippases can modulate membrane structure. Biochim Biophys Acta. 2008. Jul-Aug;1778(7–8):1591–600. doi: 10.1016/j.bbamem.2008.03.007. Epub 2008 Apr 1. [DOI] [PubMed] [Google Scholar]
- 6.Gomez-Navarro N, Miller EA. COP-coated vesicles. Curr Biol. 2016. January 25;26(2):R54–R57. doi: 10.1016/j.cub.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 7.Dukhovny A, Papadopulos A, Hirschberg K. Quantitative live-cell analysis of microtubule-uncoupled cargo-protein sorting in the ER. J Cell Sci. 2008. March 15;121(Pt 6):865–76. doi: 10.1242/jcs.019463. Epub 2008 Feb 26. [DOI] [PubMed] [Google Scholar]
- 8.Wong LH, Gatta AT, Levine TP. Lipid transfer proteins: the lipid commute via shuttles, bridges and tubes. Nat Rev Mol Cell Biol. 2019. February;20(2):85–101. doi: 10.1038/s41580-018-0071-5. [DOI] [PubMed] [Google Scholar]
- 9.Saheki Y, De Camilli P. Endoplasmic Reticulum-Plasma Membrane Contact Sites. Annu Rev Biochem. 2017. June 20;86:659–684. doi: 10.1146/annurev-biochem-061516-044932. Epub 2017 Feb 23. [DOI] [PubMed] [Google Scholar]
- 10.Xu J, Huang X. Lipid Metabolism at Membrane Contacts: Dynamics and Functions Beyond Lipid Homeostasis. Front Cell Dev Biol. 2020. December 23;8:615856. doi: 10.3389/fcell.2020.615856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thiele C, Wunderling K, Leyendecker P. Multiplexed and single cell tracing of lipid metabolism. Nat Methods. 2019. November;16(11):1123–1130. doi: 10.1038/s41592-019-0593-6. Epub 2019 Oct 14. [DOI] [PubMed] [Google Scholar]
- 12.Posor Y, Eichhorn-Gruenig M, Puchkov D, Schöneberg J, Ullrich A, Lampe A, Müller R, Zarbakhsh S, Gulluni F, Hirsch E, Krauss M, Schultz C, Schmoranzer J, Noé F, Haucke V. Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature. 2013. July 11;499(7457):233–7. doi: 10.1038/nature12360. Epub 2013 Jul 3. [DOI] [PubMed] [Google Scholar]
- 13.Ketel K, Krauss M, Nicot AS, Puchkov D, Wieffer M, Müller R, Subramanian D, Schultz C, Laporte J, Haucke V. A phosphoinositide conversion mechanism for exit from endosomes. Nature. 2016. January 21;529(7586):408–12. doi: 10.1038/nature16516. Epub 2016 Jan 13. [DOI] [PubMed] [Google Scholar]
- 14.Varnai P, Thyagarajan B, Rohacs T, Balla T. Rapidly inducible changes in phosphatidylinositol 4,5-bisphosphate levels influence multiple regulatory functions of the lipid in intact living cells. J Cell Biol. 2006. November 6;175(3):377–82. doi: 10.1083/jcb.200607116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fegan A, White B, Carlson JC, Wagner CR. Chemically controlled protein assembly: techniques and applications. Chem Rev. 2010. June 9;110(6):3315–36. doi: 10.1021/cr8002888. [DOI] [PubMed] [Google Scholar]
- 16.Voß S, Klewer L, Wu YW. Chemically induced dimerization: reversible and spatiotemporal control of protein function in cells. Curr Opin Chem Biol. 2015. October;28:194–201. doi: 10.1016/j.cbpa.2015.09.003. Epub 2015 Sep 29. [DOI] [PubMed] [Google Scholar]
- 17.Ishida M, Watanabe H, Takigawa K, Kurishita Y, Oki C, Nakamura A, Hamachi I, Tsukiji S. Synthetic self-localizing ligands that control the spatial location of proteins in living cells. J Am Chem Soc. 2013. August 28;135(34):12684–9. doi: 10.1021/ja4046907. Epub 2013 Aug 14. [DOI] [PubMed] [Google Scholar]
- 18.Benedetti L, Barentine AES, Messa M, Wheeler H, Bewersdorf J, De Camilli P. Light-activated protein interaction with high spatial subcellular confinement. Proc Natl Acad Sci U S A. 2018. March 6;115(10):E2238–E2245. doi: 10.1073/pnas.1713845115. Epub 2018 Feb 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Höglinger D, Nadler A, Schultz C. Caged lipids as tools for investigating cellular signaling. Biochim Biophys Acta. 2014. August;1841(8):1085–96. doi: 10.1016/j.bbalip.2014.03.012. Epub 2014 Apr 5. [DOI] [PubMed] [Google Scholar]
- 20.Laguerre A, Schultz C. Novel lipid tools and probes for biological investigations. Curr Opin Cell Biol. 2018. August;53:97–104. doi: 10.1016/j.ceb.2018.06.013. Epub 2018 Jul 14. [DOI] [PubMed] [Google Scholar]
- 21.Nadler A, Reither G, Feng S, Stein F, Reither S, Müller R, Schultz C. The fatty acid composition of diacylglycerols determines local signaling patterns. Angew Chem Int Ed Engl. 2013. June 10;52(24):6330–4. doi: 10.1002/anie.201301716. Epub 2013 May 29. [DOI] [PubMed] [Google Scholar]
- 22.Morstein J, Impastato AC, Trauner D. Photoswitchable Lipids. Chembiochem. 2021. January 5;22(1):73–83. doi: 10.1002/cbic.202000449. Epub 2020 Nov 10. [DOI] [PubMed] [Google Scholar]
- 23.Morstein J, Trauner D. Photopharmacological control of lipid function. Methods Enzymol. 2020;638:219–232. doi: 10.1016/bs.mie.2020.04.025. Epub 2020 Apr 27. [DOI] [PubMed] [Google Scholar]
- 24.Matsuzaki M, Kasai H. Two-photon uncaging microscopy. Cold Spring Harb Protoc. 2011. May 1;2011(5):pdb.prot5620. doi: 10.1101/pdb.prot5620. [DOI] [PubMed] [Google Scholar]
- 25.Warther D, Gug S, Specht A, Bolze F, Nicoud JF, Mourot A, Goeldner M. Two-photon uncaging: New prospects in neuroscience and cellular biology. Bioorg Med Chem. 2010. November 15;18(22):7753–8. doi: 10.1016/j.bmc.2010.04.084. Epub 2010 Apr 29. [DOI] [PubMed] [Google Scholar]
- 26.Hagen V. (2006) Uncaging and Photoconversion/Activation. In: Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine. Springer, Berlin, Heidelberg. 10.1007/3-540-29623-9_5520 [DOI] [Google Scholar]
- 27.Olson JP, Kwon HB, Takasaki KT, Chiu CQ, Higley MJ, Sabatini BL, Ellis-Davies GC. Optically selective two-photon uncaging of glutamate at 900 nm. J Am Chem Soc. 2013. April 24;135(16):5954–7. doi: 10.1021/ja4019379. Epub 2013 Apr 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klán P, Šolomek T, Bochet CG, Blanc A, Givens R, Rubina M, Popik V, Kostikov A, Wirz J. Photoremovable protecting groups in chemistry and biology: reaction mechanisms and efficacy. Chem Rev. 2013. January 9;113(1):119–91. doi: 10.1021/cr300177k. Epub 2012 Dec 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bardhan A, Deiters A. Development of photolabile protecting groups and their application to the optochemical control of cell signaling. Curr Opin Struct Biol. 2019. August;57:164–175. doi: 10.1016/j.sbi.2019.03.028. Epub 2019 May 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walter AM, Müller R, Tawfik B, Wierda KD, Pinheiro PS, Nadler A, McCarthy AW, Ziomkiewicz I, Kruse M, Reither G, Rettig J, Lehmann M, Haucke V, Hille B, Schultz C, Sørensen JB. Phosphatidylinositol 4,5-bisphosphate optical uncaging potentiates exocytosis. Elife. 2017. October 25;6:e30203. doi: 10.7554/eLife.30203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Höglinger D, Haberkant P, Aguilera-Romero A, Riezman H, Porter FD, Platt FM, Galione A, Schultz C. Intracellular sphingosine releases calcium from lysosomes. Elife. 2015. November 27;4:e10616. doi: 10.7554/eLife.10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Laguerre A, Hauke S, Qiu J, Kelly MJ, Schultz C. Photorelease of 2-Arachidonoylglycerol in Live Cells. J Am Chem Soc. 2019. October 23;141(42):16544–16547. doi: 10.1021/jacs.9b05978. Epub 2019 Oct 10. * Laguerre et al. introduced a new chemical strategy for the caging of monoacylglycerols. They applied this technique to the caging of the 2-AG and measured the effect of the endocannabinoid’s photo-release in MIN6 beta-cells.
- 33.Laguerre A, Keutler K, Hauke S, Schultz C. Regulation of Calcium Oscillations in β-Cells by Co-activated Cannabinoid Receptors. Cell Chem Biol. 2021. January 21;28(1):88–96.e3. doi: 10.1016/j.chembiol.2020.10.006. Epub 2020 Nov 3. [DOI] [PubMed] [Google Scholar]
- 34.Neef AB, Schultz C. Selective fluorescence labeling of lipids in living cells. Angew Chem Int Ed Engl. 2009;48(8):1498–500. doi: 10.1002/anie.200805507. [DOI] [PubMed] [Google Scholar]
- 35.Laketa V, Zarbakhsh S, Traynor-Kaplan A, Macnamara A, Subramanian D, Putyrski M, Mueller R, Nadler A, Mentel M, Saez-Rodriguez J, Pepperkok R, Schultz C. PIP3 induces the recycling of receptor tyrosine kinases. Sci Signal. 2014. January 14;7(308):ra5. doi: 10.1126/scisignal.2004532. [DOI] [PubMed] [Google Scholar]
- 36. Müller R, Citir M, Hauke S, Schultz C. Synthesis and Cellular Labeling of Caged Phosphatidylinositol Derivatives. Chemistry. 2020. January 7;26(2):384–389. doi: 10.1002/chem.201903704. Epub 2019 Dec 6. * The use of a trifunctional lipid derivative permitted to visualize PI transport from the ER to the plasma membrane.
- 37.Höglinger D, Nadler A, Haberkant P, Kirkpatrick J, Schifferer M, Stein F, Hauke S, Porter FD, Schultz C. Trifunctional lipid probes for comprehensive studies of single lipid species in living cells. Proc Natl Acad Sci U S A. 2017. February 14;114(7):1566–1571. doi: 10.1073/pnas.1611096114. Epub 2017 Feb 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Höglinger D. Bi- and Trifunctional Lipids for Visualization of Sphingolipid Dynamics within the Cell. Methods Mol Biol. 2019;1949:95–103. doi: 10.1007/978-1-4939-9136-5_8. [DOI] [PubMed] [Google Scholar]
- 39.Mentel M, Laketa V, Subramanian D, Gillandt H, Schultz C. Photoactivatable and cell-membrane-permeable phosphatidylinositol 3,4,5-trisphosphate. Angew Chem Int Ed Engl. 2011. April 11;50(16):3811–4. doi: 10.1002/anie.201007796. Epub 2011 Mar 14. [DOI] [PubMed] [Google Scholar]
- 40.Nadler A, Yushchenko DA, Müller R, Stein F, Feng S, Mulle C, Carta M, Schultz C. Exclusive photorelease of signalling lipids at the plasma membrane. Nat Commun. 2015. December 21;6:10056. doi: 10.1038/ncomms10056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schuhmacher M, Grasskamp AT, Barahtjan P, Wagner N, Lombardot B, Schuhmacher JS, Sala P, Lohmann A, Henry I, Shevchenko A, Coskun Ü, Walter AM, Nadler A. Live-cell lipid biochemistry reveals a role of diacylglycerol side-chain composition for cellular lipid dynamics and protein affinities. Proc Natl Acad Sci U S A. 2020. April 7;117(14):7729–7738. doi: 10.1073/pnas.1912684117. Epub 2020 Mar 25. ** Schumacher et al. applied the negatively charged sulfonated coumarin cage to different diacylglycerols to investigate how the lipid composition would affect the trans-bilayer movements of DAGs and subsequent recruitment of DAG binding proteins.
- 42.Kim YA, Ramirez DM, Costain WJ, Johnston LJ, Bittman R. A new tool to assess ceramide bioactivity: 6-bromo-7-hydroxycoumarinyl-caged ceramide. Chem Commun (Camb). 2011. August 28;47(32):9236–8. doi: 10.1039/c1cc12345a. Epub 2011 Jul 15. [DOI] [PubMed] [Google Scholar]
- 43.Kim YA, Day J, Lirette CA, Costain WJ, Johnston LJ, Bittman R. Synthesis and photochemical properties of PEGylated coumarin-caged ceramides for cell studies. Chem Phys Lipids. 2016. January;194:117–24. doi: 10.1016/j.chemphyslip.2015.07.006. Epub 2015 Jul 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim YA, Day J, Lirette CA, Costain WJ, Johnston LJ, Bittman R. Synthesis and photochemical properties of PEGylated coumarin-caged ceramides for cell studies. Chem Phys Lipids. 2016. January;194:117–24. doi: 10.1016/j.chemphyslip.2015.07.006. Epub 2015 Jul 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lankalapalli RS, Ouro A, Arana L, Gómez-Muñoz A, Bittman R. Caged ceramide 1-phosphate analogues: synthesis and properties. J Org Chem. 2009. November 20;74(22):8844–7. doi: 10.1021/jo902076w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomez-Muñoz A, Gangoiti P, Rivera IG, Presa N, Gomez-Larrauri A, Ordoñez M. Caged ceramide 1-phosphate (C1P) analogs: Novel tools for studying C1P biology. Chem Phys Lipids. 2016. January;194:79–84. doi: 10.1016/j.chemphyslip.2015.07.019. Epub 2015 Jul 29. [DOI] [PubMed] [Google Scholar]
- 47.Feng S, Harayama T, Montessuit S, David FP, Winssinger N, Martinou JC, Riezman H. Mitochondria-specific photoactivation to monitor local sphingosine metabolism and function. Elife. 2018. January 29;7:e34555. doi: 10.7554/eLife.34555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng S, Harayama T, Chang D, Hannich JT, Winssinger N, Riezman H. Lysosome-targeted photoactivation reveals local sphingosine metabolism signatures. Chem Sci. 2018. December 26;10(8):2253–2258. doi: 10.1039/c8sc03614d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wagner N, Stephan M, Höglinger D, Nadler A. A Click Cage: Organelle-Specific Uncaging of Lipid Messengers. Angew Chem Int Ed Engl. 2018. October 1;57(40):13339–13343. doi: 10.1002/anie.201807497. Epub 2018 Sep 3. * Wagner et al. described clickable coumarin photoremovable protecting groups for the caging of signaling lipids. The alkyne on the coumarin permits the chemical introduction of organelle targeting groups that will serve the cage lipid to accumulate at a specific location inside cell before the uncaging.
- 50.Gaur P, Kucherak OA, Ermakova YG, Shvadchak VV, Yushchenko DA. Nitrobenzyl-based fluorescent photocages for spatial and temporal control of signalling lipids in cells. Chem Commun (Camb). 2019. October 10;55(82):12288–12291. doi: 10.1039/c9cc05602e. * [DOI] [PubMed] [Google Scholar]
- 51.Bardhan A, Deiters A. Development of photolabile protecting groups and their application to the optochemical control of cell signaling. Curr Opin Struct Biol. 2019. August;57:164–175. doi: 10.1016/j.sbi.2019.03.028. Epub 2019 May 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Weinstain R, Slanina T, Kand D, Klán P. Visible-to-NIR-Light Activated Release: From Small Molecules to Nanomaterials. Chem Rev. 2020. December 23;120(24):13135–13272. doi: 10.1021/acs.chemrev.0c00663. Epub 2020 Oct 30. ** Weinstain et al. provided an extensive review of photocaged compounds that enable noninvasive spatiotemporal photochemical control over the release of species of interest. This work describes the state-of-the-art.
- 53.Amatrudo JM, Olson JP, Lur G, Chiu CQ, Higley MJ, Ellis-Davies GC. Wavelength-selective one- and two-photon uncaging of GABA. ACS Chem Neurosci. 2014. January 15;5(1):64–70. doi: 10.1021/cn400185r. Epub 2013 Dec 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peterson JA, Wijesooriya C, Gehrmann EJ, Mahoney KM, Goswami PP, Albright TR, Syed A, Dutton AS, Smith EA, Winter AH. Family of BODIPY Photocages Cleaved by Single Photons of Visible/Near-Infrared Light. J Am Chem Soc. 2018. June 13;140(23):7343–7346. doi: 10.1021/jacs.8b04040. Epub 2018 May 29. [DOI] [PubMed] [Google Scholar]
- 55.Kand D, Liu P, Navarro MX, Fischer LJ, Rousso-Noori L, Friedmann-Morvinski D, Winter AH, Miller EW, Weinstain R. Water-Soluble BODIPY Photocages with Tunable Cellular Localization. J Am Chem Soc. 2020. March 18;142(11):4970–4974. doi: 10.1021/jacs.9b13219. Epub 2020 Mar 5. [DOI] [PMC free article] [PubMed] [Google Scholar]