Abstract
In type 1 diabetes, autoimmune β-cell destruction may be favored by neoantigens harboring posttranslational modifications (PTMs) such as citrullination. We studied the recognition of native and citrullinated glucose-regulated protein (GRP)78 peptides by CD8+ T cells. Citrullination modulated T-cell recognition and, to a lesser extent, HLA-A2 binding. GRP78-reactive CD8+ T cells circulated at similar frequencies in healthy donors and donors with type 1 diabetes and preferentially recognized either native or citrullinated versions, without cross-reactivity. Rather, the preference for native GRP78 epitopes was associated with CD8+ T cells cross-reactive with bacterial mimotopes. In the pancreas, a dominant GRP78 peptide was instead preferentially recognized when citrullinated. To further clarify these recognition patterns, we considered the possibility of citrullination in the thymus. Citrullinating peptidylarginine deiminase (Padi) enzymes were expressed in murine and human medullary epithelial cells (mTECs), with citrullinated proteins detected in murine mTECs. However, Padi2 and Padi4 expression was diminished in mature mTECs from NOD mice versus C57BL/6 mice. We conclude that, on one hand, the CD8+ T cell preference for native GRP78 peptides may be shaped by cross-reactivity with bacterial mimotopes. On the other hand, PTMs may not invariably favor loss of tolerance because thymic citrullination, although impaired in NOD mice, may drive deletion of citrulline-reactive T cells.
Introduction
Islet-reactive CD8+ T cells are held as the final mediators of β-cell destruction (1). Surprisingly, they circulate at similar frequencies in most individuals, irrespective of type 1 diabetes status (2–4). This is true for known islet antigens and for novel ones, e.g., urocortin 3 (UCN3), secretogranin-5, and proconvertase-2 (3,4). The frequency of most of these islet epitope-reactive T cells falls in a relatively narrow range of ∼1–50/106 CD8+ T cells (0.0001–0.005%), which overlaps with that of naïve CD8+ T cells recognizing viral peptides in virus seronegative donors (5,6). In agreement with this frequency overlap, circulating islet-reactive CD8+ T cells are largely naïve in both healthy donors and donors with type 1 diabetes (2–4), suggesting a limited recruitment in the autoimmune process. The fraction engaged in this process seems instead sequestered in the pancreas, where the same T cells are enriched in patients with type 1 diabetes and display a CD45RO+ phenotype, compatible with a prior antigen encounter (2,3). Thus, a universal state of “benign” islet autoimmunity exists for circulating CD8+ T cells, to a much larger extent than previously appreciated (1,7). In line with recent reports (6), we demonstrated that this benign autoimmunity is imprinted in the thymus due to a marginal deletion of autoreactive CD8+ T cells, since the presence or absence of ectopic islet antigen presentation in thymic medullary epithelial cells (mTECs) does not significantly modulate this process (2,3).
It is unknown whether this novel paradigm of benign autoimmunity applies to β-cell antigens undergoing posttranslational modifications (PTMs). PTMs occurring in peripheral tissues, especially under inflammatory conditions, may favor priming of T cell precursors that have escaped thymic deletion, due to their selection against native peptides with different physicochemical properties (8). One posttranslationally modified antigen is glucose-regulated protein (GRP)78, which is citrullinated on arginine (R) residues in inflamed mouse and human islets (9,10) by peptidyl arginine deiminase enzymes (PADI; Padi in the mouse). GRP78 (also known as BiP and HSPA5) is an endoplasmic reticulum (ER) chaperone of the heat shock protein 70 family. It is involved in anabolic pathways, i.e., protein (re)folding and translocation of secretory proteins, and catabolic pathways, i.e., the import of polypeptides—especially hydrophobic ones—inside the ER, the ER-associated degradation pathway, peptide export outside the ER for proteasome degradation, and the regulation of the unfolded protein response (11). Citrullinated GRP78 is translocated to the plasma membrane and secreted, triggering β-cell apoptosis (9,12).
We first reported that the murine ortholog of GRP78 (Grp78) is targeted by T cells and autoantibodies (aAb) in NOD mice (9). While interferon-γ (IFN-γ) T cell responses were only observed against citrullinated Grp78, aAb were detected against both native and citrullinated Grp78, but the latter was preferentially recognized, with higher aAb titers in NOD mice than in nonautoimmune strains (9). Translating to humans, a CD4+ T-cell line grown from type 1 diabetic islets responded to a GRP78292–305 peptide only when carrying an arginine-to-citrulline (R→X) modification (13). aAb against both native GRP78 (R-GRP78) and citrullinated (X-GRP78) GRP78 were detected at increased titers in individuals with type 1 diabetes versus age-matched healthy individuals, but again X-GRP78 was preferentially recognized (10). CD4+ T cells reactive to an HLA-DR4 binding X-GRP78498–512 peptide displayed higher frequencies in donors with type 1 diabetes versus healthy donors, but these frequencies were similar to those of CD4+ T cells recognizing R-GRP78498–512. Importantly, this GRP78498–512 epitope features an R510 residue that is citrullinated in human islets exposed in vitro to inflammatory cytokines (10). Collectively, these results suggest that CD4+ T cells recognize preferentially, but not exclusively, X-GRP78 over its native version.
We therefore studied the recognition of R- and X-GRP78 peptides restricted for HLA-A*02:01 (HLA-A2) by the CD8+ T cells of donors with type 1 diabetes and healthy donors. Our results show that the preferential recognition of X-GRP78 versions is not the rule and that this recognition may be shaped by T cell cross-reactivity with bacterial mimotopes and by PADI-mediated citrullination in the thymus and islets.
Research Design and Methods
HLA-A2 Binding Measurements
Experimental binding of peptides (>90% purity; SynPeptide) to HLA-A2 was measured by flow cytometry using biotin-tagged HLA-A2 monomers (immunAware) assembled as previously described (14), captured on streptavidin-coated beads (Spherotech), and revealed by an anti-β2m monoclonal antibody (Research Resource Identifier [RRID] AB_626748) and an Alexa Fluor (AF)488-conjugated goat anti-mouse IgG (RRID AB_2728715).
Donors and Specimens
Human studies were authorized under ethics approval no. S52697 (UZ Leuven), and written informed consent was obtained from all participants. Heparinized blood was drawn from HLA-A*02:01+ donors: 16 aAb+, insulin-treated adults with type 1 diabetes (median age 27 years [range 19–47]; 9 female and 7 male; median disease duration 4.3 years [1.7–6.0]); 11 healthy donors (median age 37 years [26–60]; 7 female and 4 male); and 8 adolescents with type 1 diabetes (median age 16 years [12–17]; 3 female and 5 male; median disease duration 4.2 years [1.0–12.0]). Ficoll-purified peripheral blood mononuclear cells (PBMCs) were stored in liquid nitrogen until use. Pancreatic tissue sections were provided by the Network for Pancreatic Organ donors with Diabetes (nPOD).
HLA-A2 Multimer Staining
Our combinatorial HLA-A2 multimer (MMr) assays, previously described (3,4), allow us to multiplex the detection of 15 different T-cell epitope specificities in the same PBMC sample. It is based on the use of unique fluorochrome pairs to label each peptide-loaded MMr, with subsequent detection of double-labeled MMr+CD8+ T cells with increased specificity. This format was modified so that each R-GRP78 MMr shared one fluorochrome with its corresponding X-GRP78 MMr, thus allowing selective gating of CD8+ T cells recognizing only the R or X version (i.e., double MMr+) or both (i.e., triple MMr+). For example, the GRP78-261R and GRP78-261X peptides were loaded on phycoerythrin (PE)/BV786- and PE/PE-CF594–labeled MMr pairs, respectively, thus sharing the PE fluorochrome. GRP78-261R, -261X, and -261R/X MMr+CD8+ T cells were, hence, defined as PE+/BV786+, PE+/PECF594+, and PE+/BV786+/PECF594+, respectively. MMr staining and acquisition were performed as previously described (3). HLA-A2 binding epitope candidates without appreciable MMr staining provided negative controls. Data were analyzed with FlowJo software as detailed in Supplementary Figs. 1 and 2. In situ HLA-A2 MMr immunohistochemistry staining was performed as previously described (3).
mTEC Isolation
For quantitative RT-PCR (RT-qPCR) and proteomics, thymi from 6- to 8-week-old C57BL/6 and NOD mice were minced; gently agitated for release of excess thymocytes; digested with Liberase; filtered; panned on anti-CD90.2–coated plates; stained with Fixable Viability Dye eFluor 780, CD45-PE/Cy7, MHC class II (MHC-II)–AF488, EPCAM-BV421, BP-1–APC, Ulex europaeus agglutinin-1 (UEA-1–biotin), and streptavidin-PE; and sorted on a BD Influx instrument.
For RNA sequencing (RNAseq), thymi were digested with collagenase D and collagenase/dispase; depleted with CD45 MicroBeads; stained with CD45-PerCPCy5.5, Ly51-PE, and MHC-II–APC; and sorted on a BD FACSAria III instrument. These procedures are described in Supplementary Material.
RT-qPCR, Proteomics, and RNAseq
Mouse studies were performed under authorization no. P076/2020 (UZ Leuven). Islets were isolated from 6- and 10-week-old C57BL/6 and NOD mice as previously described (15). Total RNA was extracted from sorted mTECs or islets, with the Single Cell RNA Purification Kit (Norgen Biotek) and cDNA synthesized with SuperScript VILO (Invitrogen). RT-qPCR was performed on a StepOnePlus Real-Time PCR System (Applied Biosystems). The relative fold gene expression was calculated using the ΔΔCt method.
Liquid chromatography–tandem mass spectrometry and manual verification of citrullinated residues were performed essentially as previously described (16). RNAseq procedures have previously been published (17). These procedures are detailed in Supplementary Material.
Statistics
Significance was assessed with two-tailed tests with a cutoff value of α = 0.05, as detailed for each figure and in Supplementary Material.
Data and Resource Availability
The mTEC RNAseq data sets have been deposited in the Gene Expression Omnibus (GEO) under accession nos. GSE140815, GSE140683, and GSE178456. All raw data are available from the corresponding author upon reasonable request. The study does not involve any noncommercial reagent or tool.
Results
Citrullination of GRP78 Peptides Does Not Decrease HLA-A2 Binding Affinity
We selected six candidate epitopes by screening the whole GRP78 sequence for predicted HLA-A2 binders carrying R residues (Fig. 1A): GRP789–18 (GRP78-9 from hereon), GRP78261–269 (GRP78-261), GRP78298–307 (GRP78-298), GRP78360–368 (GRP78-360), GRP78435–443 (GRP78-435), and GRP78502–511 (GRP78-502).
Figure 1.
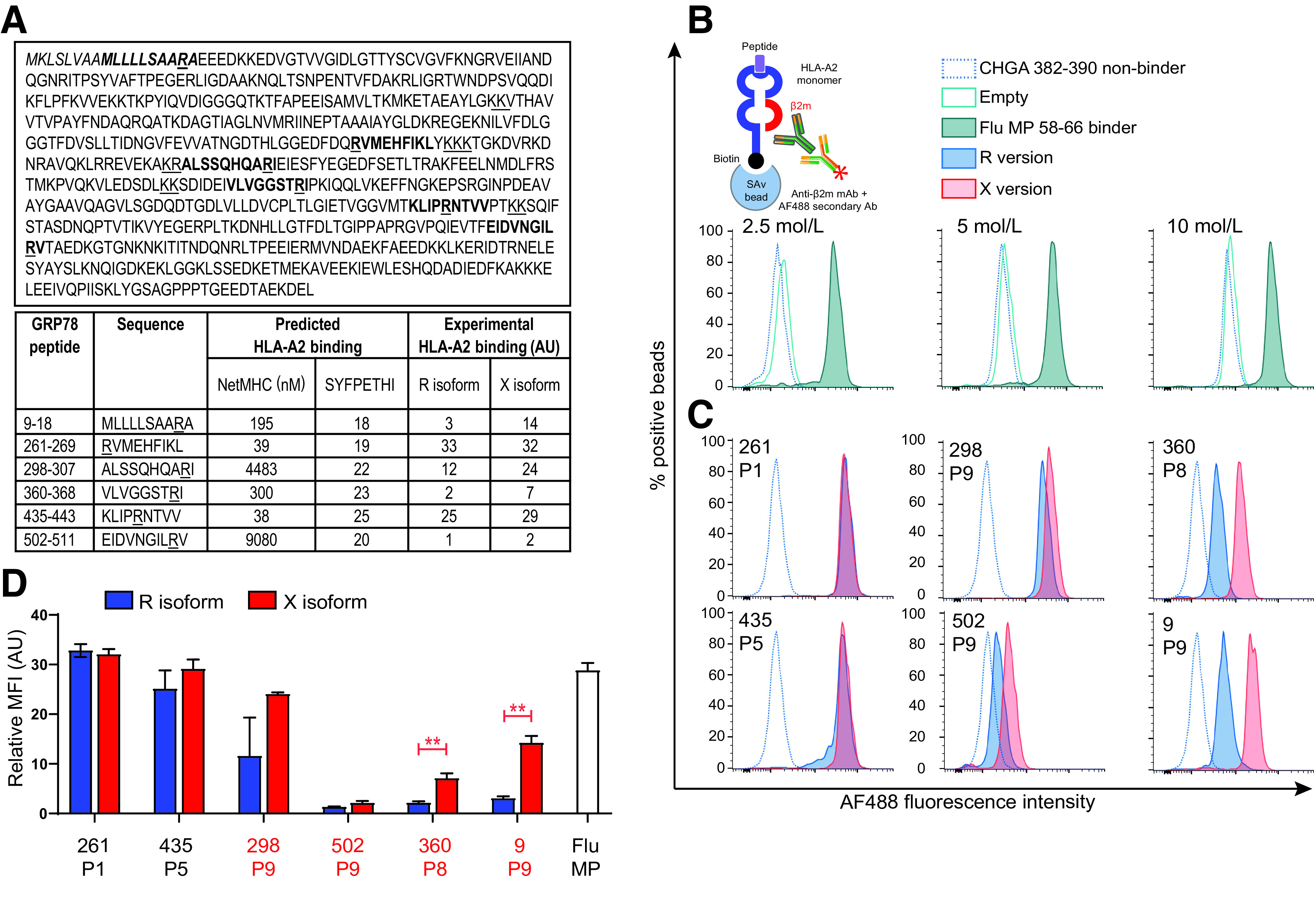
HLA-A2 binding of native (R) and citrullinated (X) GRP78 epitopes. A: Epitope candidates (shown in bold within the GRP78 sequence, UniProt identifier P11021) were selected in silico based on a NetMHC 4.0–predicted KD ≤300 nmol/L or a SYFPETHI score ≥20 (www.cbs.dtu.dk/services/NetMHC and www.syfpeithi.de, respectively). For length variants of the same peptide, the one with the best binding affinity was retained. R residues comprised within the selected candidates and dibasic cleavage motifs are underlined. The GRP78 signal peptide is indicated in italics. The experimental HLA-A2 binding affinity measured for the R and X versions in the experiments shown in subsequent panels is reported in arbitrary units (AU). B: Schematic view and setup of the assay. Biotinylated monomeric HLA-A2 molecules are incubated with peptide and β2m. After capturing on streptavidin-coated beads, an anti-β2m antibody is added, followed by an AF488-labeled secondary antibody. The bead-associated fluorescence is therefore detected only if the test peptide supports the folding of the HLA-A2/β2m complex. Histograms (gated on single AF488+ beads) depict representative flow cytometry staining obtained with increasing concentrations of HLA-A2 molecules folded with the nonbinding CHGA382–390 (HPVGEADYF) and the binding Flu MP58–66 (GILGFVFTL) peptides (negative and positive control, respectively). The 2.5 nmol/L concentration was retained for subsequent experiments based on the best separation of the positive and negative peaks. C: Representative staining of the tested GRP78 peptides in their native (blue) or citrullinated versions (red). The R/X position within the peptide sequence, either outside (P1, P5) or inside (P8, P9) the COOH-terminal region, is indicated in each histogram. D: Relative median fluorescence intensity (MFI) AU values for peptide–HLA-A2 complexes at 2.5 nmol/L, normalized to the ChgA382–390 negative control peptide. Data are expressed as median ± SD of triplicate measurements, and the R/X position is indicated for each peptide. Ab, antibody; mAb, monoclonal antibody. **P ≤ 0.003 by Student t test.
Since binding affinities were predicted for the native peptide sequence without citrullination, we experimentally verified binding using monomeric HLA-A2 molecules and a flow cytometry assay detecting peptide/HLA-A2/β2m complexes (4) (Fig. 1B). First, this assay verified the proper folding of the peptide/HLA-A2 monomers subsequently used for MMr synthesis. Second, it revealed strong (GRP78-261, -435, -298), moderate (GRP78-360, -9), and weak (GRP78-502) binders (Fig. 1C and D). The HLA-A2 binding of the citrullinated (X) versus native (R) versions was similar (GRP78-261, -435), marginally increased (GRP78-298, -502), or significantly increased (GRP78-360, -9) but never decreased. The increase in HLA-A2 binding was observed for those peptides in which the R→X substitution was introduced at the COOH-terminal P8-P9 HLA anchor positions.
Collectively, these data show that citrullination does not negatively affect HLA-A2 binding for the candidate epitopes identified.
Blood CD8+ T Cells Preferentially Recognize Either Native or Citrullinated Peptide Versions and Display Similar Frequencies in Donors With Type 1 Diabetes and Healthy Donors
We subsequently set up combinatorial HLA-A2 MMr assays (3,4), with two purposes: first, to define the ex vivo frequency and naïve/antigen-experienced phenotype of GRP78-reactive CD8+ T cells in donors with type 1 diabetes and healthy donors, and second, to investigate whether the native and citrullinated versions of any given peptide are recognized by the same T cell clonotypes. For this second purpose, we modified our previous assays so that the MMr pairs loaded with each R version shared one fluorochrome with the MMr pairs loaded with its corresponding X version (Supplementary Table 1), thus allowing selectively gating of CD8+ T cells recognizing only the R or X version or both (Fig. 2A and B). A representative staining is shown in Supplementary Fig. 1, and the analysis strategy is detailed in Supplementary Fig. 2.
Figure 2.
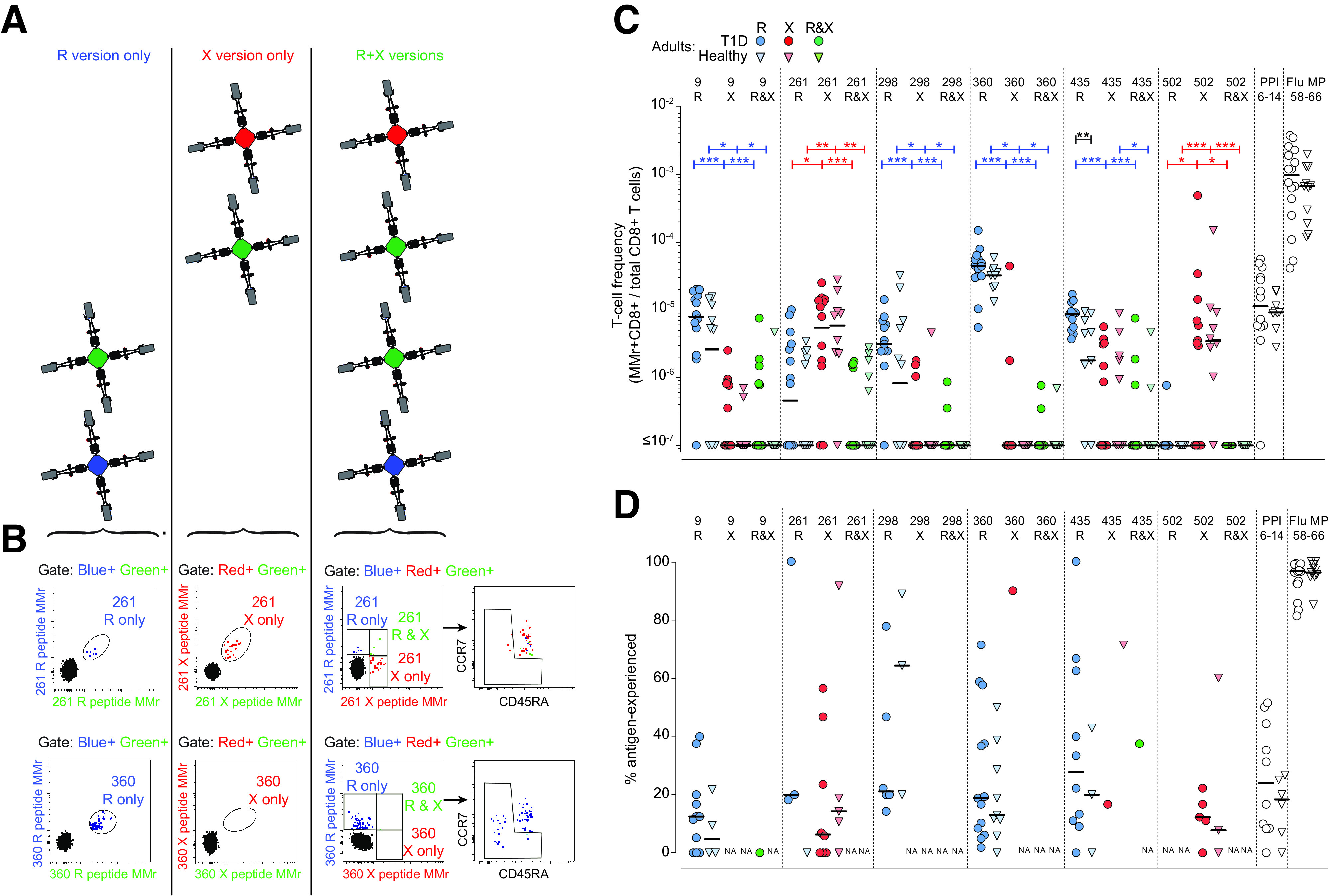
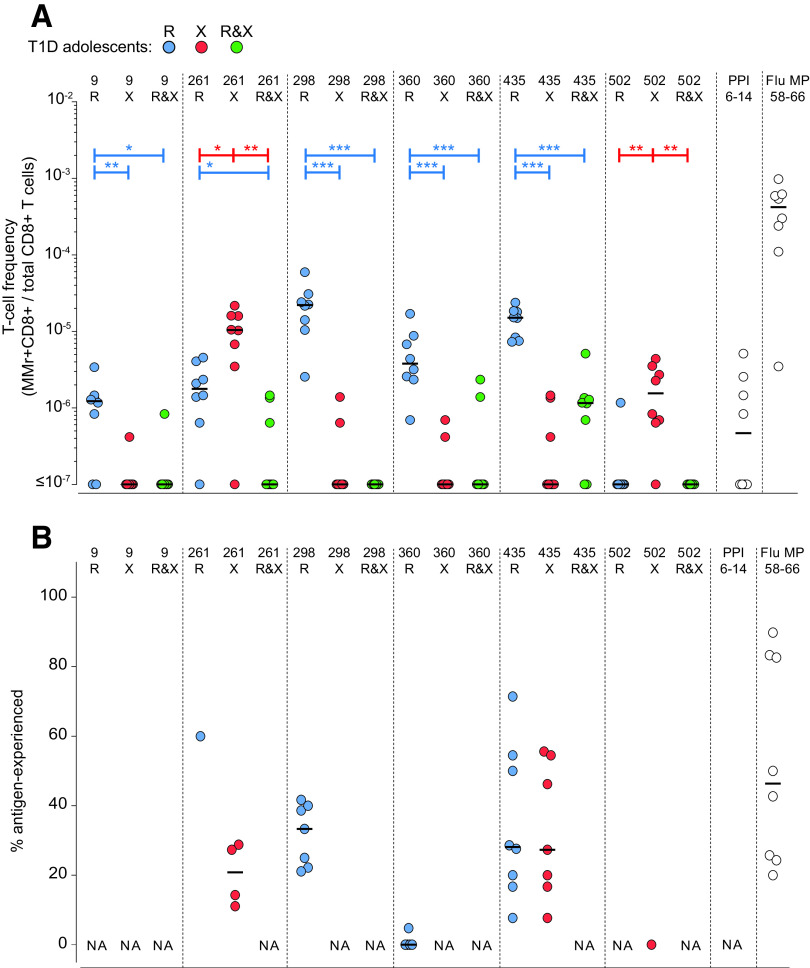
Blood CD8+ T cells recognize either R- or X-GRP78 peptide versions and display similar frequencies in adults with type 1 diabetes and healthy adults. A: Schematic view of the combinatorial analysis applied to GRP78 MMr+CD8+ T cells. Each peptide is loaded onto HLA-A2 MMrs coupled with two different fluorochromes. Six fluorochromes are used to obtain 15 unique pairs. Consequently, each peptide-reactive cell population is stained with a unique pair of fluorochromes. Native (R) and citrullinated (X) versions of the same GRP78 peptide are stained with fluorochrome pairs that share one fluorochrome (green). With gating on double-MMr+ events and exclusion of all other fluorochromes, cells recognizing only the R version or X version are visualized. With gating on triple-MMr+ events, cells recognizing the R only, X only, or both versions are visualized. Subsequent staining with CD45RA and CCR7 allows characterization of the naïve/memory phenotype of each population. B: Representative examples for the GRP78-261 (top) and -360 (bottom) peptide. This strategy is detailed in Supplementary Figs. 1 and 2. C: Frequencies of MMr+CD8+ cells reactive exclusively to the GRP78-R (blue symbols) or GRP78-X (red symbols) peptides, or to both (green symbols), in HLA-A2+ adults with type 1 diabetes (T1D) (circles) (n = 16) and healthy adults (triangles) (n = 11). PPI6–14 and Flu MP58–66 peptides were included as controls. At least 0.13 × 106 CD8+ T cells were counted for each donor (median 5.4 × 106 [range 0.13–280 × 106]). D: Percentage of antigen-experienced cells out of total MMr+ cells for the GRP78 peptides depicted in C. Data points with <5 MMr+ cells were excluded (median 13 MMr+ cells [range 5–524] for GRP78 peptides). Bars show median values. NA, not available (i.e., <5 MMr+ cells counted). *P < 0.05, **P < 0.01, ***P < 0.001 by Mann-Whitney U test.
The results from adult donors are summarized in Fig. 2C. Recognition of the R and X versions was rather exclusive, with very few donors recognizing both. The native R version was preferentially recognized for peptides GRP78-9, -298, -360, and -435, while GRP78-261 and -502 were preferred in their X version. The R version was also preferred for those peptides whose citrullination at the COOH terminus enhanced HLA-A2 binding (GRP78-9, -360, and, to a lesser extent, -298), thus excluding that this preference was simply driven by HLA-A2 binding affinity. Despite some differences in frequency, the preferred and exclusive recognition of the R or X version according to individual peptides was similar in adolescents with type 1 diabetes (Fig. 3).
Figure 3.
Blood CD8+ T cells recognize either R- or X-GRP78 peptide versions in adolescents with type 1 diabetes. A: Frequencies of MMr+CD8+ cells reactive exclusively to the GRP78-R (blue symbols) or GRP78-X (red symbols) peptides, or to both (green symbols), in HLA-A2+ adolescents with type 1 diabetes (T1D) (n = 8). PPI6–14 and Flu MP58–66 peptides were included as controls. At least 0.39 × 106 CD8+ T cells were counted for each donor (median 1.1 × 106 [range 0.39–2.4 × 106]). B: Percentage of antigen-experienced cells out of total MMr+ cells for the GRP78 peptides depicted in A. Data points with <5 MMr+ cells were excluded (median 15 MMr+ cells [range 5–115] for GRP78 peptides). Bars show median values. NA, not available (i.e., <5 MMr+ cells counted). *P < 0.05, **P < 0.01, ***P < 0.001 by Mann-Whitney U test.
As for other islet epitopes (2–4,18–20), the frequency of GRP78-reactive CD8+ T cells was not higher in adults with type 1 diabetes than in healthy adults (Fig. 2C), with one exception noted for GRP78-435R. All positive GRP78-reactive CD8+ T cell fractions displayed frequencies similar to those observed for the control preproinsulin (PPI)6–14 islet epitope, confirming the reported range of ∼1–50/106 CD8+ T cells (2–4).
Collectively, these results show that, depending on the individual peptide, CD8+ T cells exclusively recognize either the native or citrullinated version, with no significant cross-reactivity. Moreover, they circulate at similar frequencies in adults with type 1 diabetes and healthy adults.
GRP78 Peptides Preferentially Recognized in Their Native Version Are Cross-reactive With Gut Bacterial Mimotopes
Another recurrent feature of circulating CD8+ T cells recognizing other islet epitopes is their largely naïve phenotype (2–4). GRP78-reactive CD8+ T cells were also largely (>80%) naïve in most donors, but substantial antigen-experienced fractions were observed in some. This was noteworthy for epitopes GRP78-298R, -360R, and -435R preferentially recognized in their native version, which displayed >30% antigen-experienced fractions in >30% of adult donors (Fig. 2D). We therefore hypothesized that these R-GRP78–reactive CD8+ T cells may cross-react with foreign antigens. A sequence homology search (Table 1) retrieved homologous bacterial sequences (mimotopes) for these three epitopes and for GRP78-502, which was instead predominantly recognized as citrullinated. Bacterial sequences were identical for GRP78-435R and, notably, GRP78-360R, which displayed particularly high cognate CD8+ T-cell frequencies (Fig. 2C). These mimotopes mapped to proteins expressed by gut commensal bacteria (Enterobacteriaceae, Proteobacteria, Clostridia, and Escherichia coli), including the well-known DnaK chaperone (21). While the GRP78-360R– and GRP78-435R–identical bacterial sequences (“homotopes”) are by definition cross-reactive, we investigated whether this was also the case for the GRP78-298R and -502X mimotopes. To this end, we used a combinatorial MMr strategy similar to the one applied to define R- and X-GRP78 cross-reactivity (Fig. 4A). Each of three PBMC aliquots was stained with two MMr pairs, and each MMr pair was loaded with the GRP78, mimotope, or control GRP78-360R or Flu MP58–66 peptide. The two MMr pairs in each tube shared one fluorochrome with each other for visualization of cross-reactive T cells as triple-labeled MMr+ events. A sizable fraction (24 ± 4% in healthy donors, 25 ± 14% in donors with type 1 diabetes) of CD8+ T cells recognizing the GRP78-298R epitope was cross-reactive with its bacterial mimotope (Fig. 4B, left), while this was not the case in analyses of cross-reactivity with the control GRP78-360R or Flu epitope (Fig. 4B, middle). As a positive control, GRP78-360R/GRP78-360R or Flu/Flu triple-MMr+ events were correctly visualized as largely cross-reactive (Fig. 4B, right). This cross-reactivity was instead lower for CD8+ T cells recognizing the GRP78-502X epitope (4 ± 3% of total in healthy donors, 13 ± 4% in donors with type 1 diabetes) (Fig. 4C).
Table 1.
GRP78 mimotopes from human intestinal commensal bacteria
| GRP78-298R (R version preferred) | ALSSQHQARI |
| Enterobacteriaceae bacterium acetyl-CoA C-acyltransferase | ALSSQH–ARI |
| UniProt A0A3C2BKS9 | ALSSQHKARI |
| GRP78-360R (R version preferred) | VLVGGSTRI |
| Proteobacteria DnaK chaperone | VLVGGSTRI |
| UniProt A0A2G6J6V3 | VLVGGSTRI |
| GRP78-435R (R version preferred) | KLIPRNTVV |
| Clostridium species restriction endonuclease subunit S | KLIPRNTVV |
| UniProt A0A3R6J648, R7CB01, A0A373M041 | KLIPRNTVV |
| GRP78-502X (X version preferred) | EIDVNGILXV |
| E. coli DnaK chaperone | +ID+–GIL–V |
| UniProt P0A6Y8 | DIDADGILHV |
Homologous bacterial sequences were searched with Basic Local Alignment Search Tool (BLAST). The alignment with GRP78 epitope sequences (in boldface type) is shown in italics, where – and + indicate nonconservative and conservative substitutions, respectively. The human GRP78 sequence is from UniProt P11021.
Figure 4.
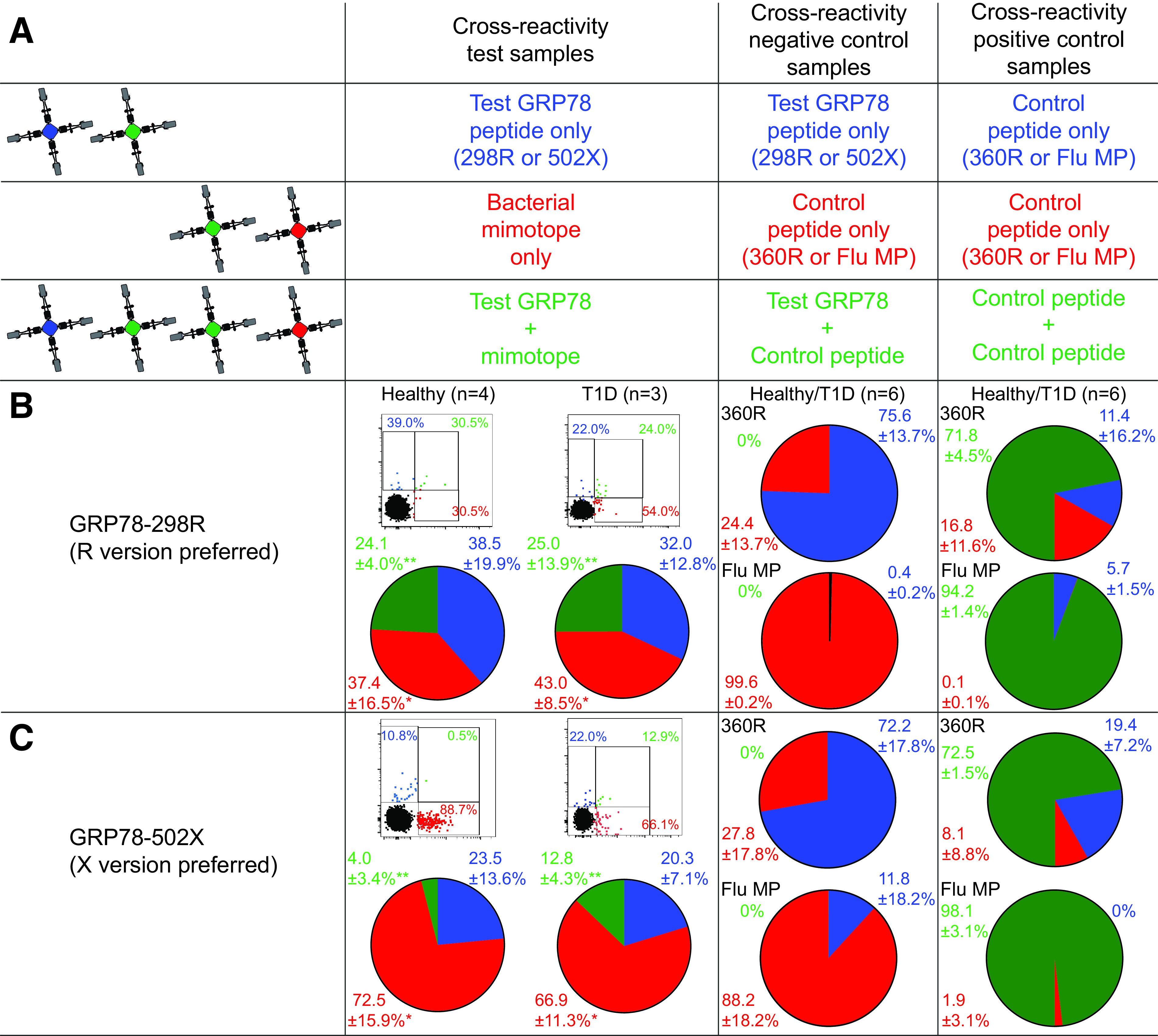
GRP78 peptides preferentially recognized in their native R version are cross-reactive with gut bacterial mimotopes. A: Schematic view of the combinatorial analysis applied. With a strategy similar to the one described in Fig. 2A, three PBMC aliquots were stained with two MMr pairs sharing one fluorochrome (green). With gating on double or triple MMr+ events, cells recognizing only one peptide or both are visualized, respectively. B and C: Representative dot plots and cumulative results from healthy donors and donors with type 1 diabetes (pie charts) obtained for GRP78-298R (B), GRP78-502X (C), and their respective peptide mimotopes. The first column shows the results obtained on the first T cell aliquot (test sample) by crossing MMrs loaded with GRP78 and bacterial mimotope peptides, with the T cell fraction recognizing both highlighted in green. The second column displays the cross-reactivity between GRP78 peptide and GRP78-360R or Flu MP58–66 epitope (negative control; second PBMC aliquot). The third column displays the cross-reactivity between GRP78-360R or Flu MP58–66 peptides charged on both MMr pairs (positive control; third PBMC aliquot). Dot plots and pie charts display the percentage of MMr+ cells binding either MMr (blue or red) or both (green). In dot plots, the MMr‒ population is displayed in black for visualization of the position of MMr+ events relative to it. **P = 0.016 and *P = 0.031 by Wilcoxon signed rank test for the comparison of cross-reactive (green) fractions and of mimotope-reactive (red) fractions, respectively, between GRP78-298R and -502X (pooled healthy donors and donors with type 1 diabetes). T1D, type 1 diabetes.
Collectively, these results suggest that the preferential CD8+ T cell recognition of some R-GRP78 versions is influenced by cross-reactivity with bacterial mimotopes.
Pancreas-Infiltrating CD8+ T Cells Preferentially Recognize the Citrullinated GRP78-9X Epitope
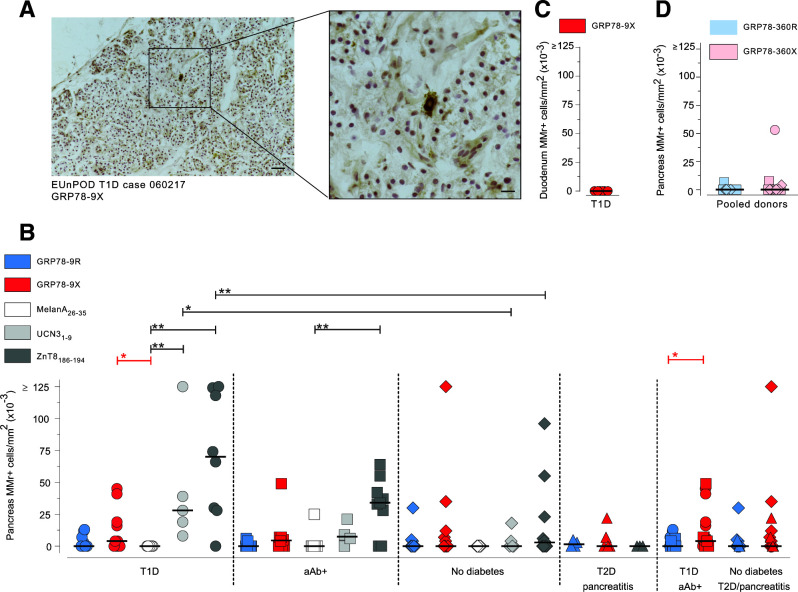
While the preferential recognition of GRP78 native versions in peripheral blood may be favored by bacterial cross-reactivity, the situation may be different in the pancreas, where citrullination and its catalyzing enzymes PADIs are upregulated by islet inflammation (9,10). We therefore performed in situ MMr staining on pancreas sections from nPOD donors (Supplementary Table 2) using the prototype epitope GRP78-9R/X, whose R version was preferentially recognized in peripheral blood. A representative GRP78-9X MMr staining on pancreas sections is shown in Fig. 5A. As reported for other islet antigen specificities (2,3,22), scattered GRP78-9X MMr+ cells were detected in the islet vicinity and in the exocrine tissue. In donors with type 1 diabetes, they displayed significantly higher densities than control MMr+ cells recognizing the melanocyte epitope MelanA26–35 (Fig. 5B). However, GRP78-9X MMr+ cells were not enriched in patients with type 1 diabetes compared with donors without diabetes, suggesting no disease specificity. The density of these GRP78-9X MMr+ cells was lower than that of ZnT8186–194 MMr+ cells and similar to that of the UCN31–9 MMr+ cells. GRP78-9X MMr+ cells were significantly more abundant than their GRP78-9R MMr+ counterparts in pooling of donors with type 1 diabetes and aAb+ donors but not in pooling of control donors without diabetes and donors with type 2 diabetes. They were instead absent in duodenal mucosa sections from those with type 1 diabetes (Fig. 5C), despite the presence of CD8+ T cells (not shown), thus confirming pancreas specificity. Analysis of a more limited set of samples for the epitope GRP78-360R/X, which, like GRP78-9R/X, was preferentially recognized in its R version in peripheral blood, also detected recognition of the X version in the pancreas of one donor with type 1 diabetes (Fig. 5D).
Figure 5.
Pancreas-infiltrating CD8+ T cells preferentially recognize the citrullinated GRP78-9X epitope. Pancreas sections from nPOD cases (Supplementary Table 2) were immunohistochemically stained in situ with MMrs loaded with the indicated GRP78 peptides, with a negative control melanocyte MelanA26–35 peptide, and with positive control UCN31–9 and ZnT8186–194 islet epitopes (low and high reactivity, respectively). A: Representative staining with GRP78-9X MMrs (scale bar 100 µm) and higher magnification of the dotted area (scale bar 33 µm). B–D: Number of MMr+ cells/mm2 section area of pancreas (B and D) and duodenal mucosa (C). Each point represents an individual case; bars indicate median values. T1D, type 1 diabetes; T2D, type 2 diabetes. *P < 0.05, **P ≤ 0.01 by Mann-Whitney U test.
Collectively, these data indicate that citrullinated GRP78-9X rather than its native GRP78-9R version is recognized by CD8+ T cells in the pancreas and that this pancreas-specific reactivity is not enriched in those with type 1 diabetes.
The Citrullinating Padi2 Enzyme Is Expressed in Murine mTECs Under the Control of Aire and Is Diminished in NOD Mice
To summarize, we observed that blood CD8+ T cells preferentially recognize the native R-GRP78 versions for several epitopes, including GRP78-9R, while the citrullinated GRP78-9X version is preferentially recognized in the pancreas. Besides the potential effect of bacterial cross-reactivity, we asked how to reconcile these findings with the expected impact of central and peripheral tolerance mechanisms in shaping these responses. It is currently held that PTM recognition should be favored because PTMs may take place under inflammatory conditions in the periphery and not in the thymus (8). Hence, T cells recognizing citrullinated epitopes may be more likely to escape thymic deletion and may then be preferentially primed in the inflammatory milieu of insulitis that promotes citrullination.
To address this possibility, we compared the gene expression of Padi2, which is the predominant islet isoform (23), in the islets and thymus between NOD and nonautoimmune C57BL/6 mice. As previously reported (9), islet Padi2 expression was negligible in C57BL/6 mice and higher in NOD mice (Fig. 6A). This expression increased with age and correlated with a difference in the Padi enzymatic activity (Fig. 6B). In mTECs (Fig. 6C and D; gating strategy in Supplementary Fig. 3A and B), Padi2 and Padi4 were substantially upregulated with maturation in C57BL/6 but not NOD mice and lower in NOD versus C57BL/6 mice for the mature MHC-IIhi mTECs driving T cell deletion. Mature MHC-IIhi mTECs were also less represented in NOD mice (Supplementary Fig. 3C). The Padi enzymatic activity from whole thymic tissue was not significantly different between the two strains (Fig. 6B), possibly reflecting the mixture of mTECs (both MHC-IIlo and MHC-IIhi) and other cells in these tissue preparations. Proteomics analysis of mTECs revealed citrullinated R residues in three proteins (Supplementary Fig. 4) (3 of 920 unique proteins detected [0.33%]). PADI gene expression was also observed in human mTECs (Supplementary Fig. 5) and was dominated by the PADI2 isoform.
Figure 6.
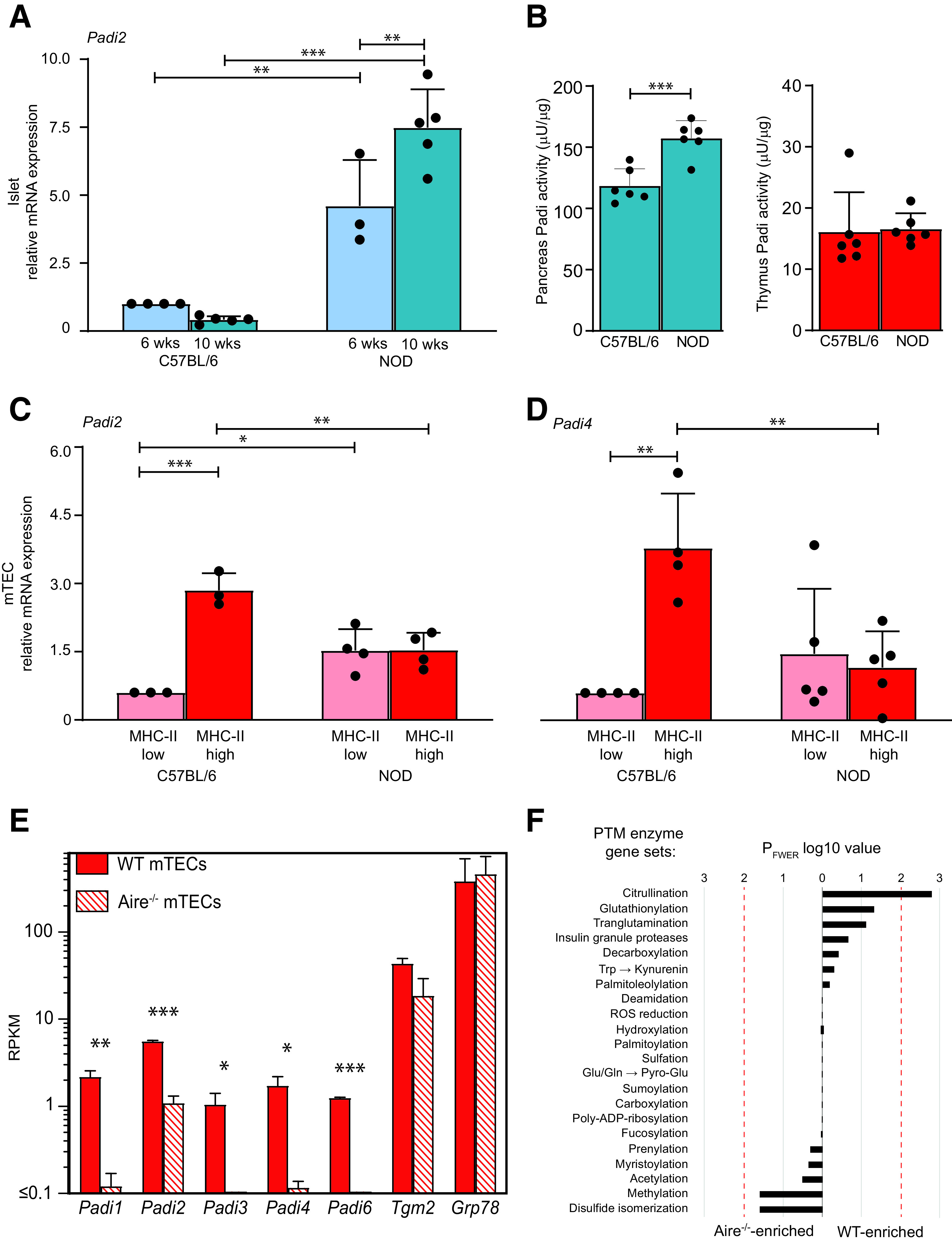
Gene expression of Padi citrullinating enzymes in murine mTECs and islets. A, C, and D: RT-qPCR expression (mean ± SD) of Padi2 in islets from 6-week-old and 10-week-old mice (A) and of Padi2 (C) and Padi4 (D) in mTECs from 6- to 8-week-old C57BL/6 and NOD mice; *P ≤ 0.03, **P ≤ 0.005, ***P < 0.001 by one-way ANOVA. B: Padi enzymatic activity (mean ± SD) in whole pancreata and thymi of 6-week-old C57BL/6 and NOD mice; ***P < 0.001 by Student t test. E: Expression (median ± range) of Padi isoforms in WT and Aire−/− C57BL/6 mice. Murine mature MHC-IIhigh mTECs were prepared from 4- to 6-week-old mice (n = 3/each) and analyzed by RNAseq; *P ≤ 0.03, **P ≤ 0.003, ***P < 0.001 by Welch t test. F: Enrichment of PTM enzyme gene sets in MHC-IIhigh mTECs from WT vs. Aire−/− mice. Red lines indicate the 0.01 family-wise error rate P (PFWER) cutoff value for statistical significance (for citrullination, PFWER = 0.0016, corresponding to a WT-normalized enrichment score of 2.4). ROS, reactive oxygen species; RPKM, reads per kilobase per million mapped reads; wks, weeks.
We next asked whether mTECs express Padi isoforms under the control of the autoimmune regulator (Aire) transcription factor. Expression of all Padi isoforms was detected by RNAseq in MHC-IIhi mTECs isolated from wild-type (WT) C57BL/6 mice (Fig. 6E) but significantly diminished or absent in Aire−/− mice, suggesting an Aire-dependent/enhanced expression. As in islets, the predominant mTEC isoform was Padi2, which was less impacted by Aire knockout in C57BL/6 mice. The enrichment of Padi2 and Padi4 in MHC-IIhi mTECs from WT versus Aire−/− NOD mice (Supplementary Fig. 6A) further supported an Aire-dependent/enhanced expression. No modulation by Aire was detected for Grp78 or the tissue transglutaminase gene Tgm2 responsible for the PTM transglutamination. The Assay for Transposable-Accessible Chromatin with high-throughput sequencing (ATAC-seq) and chromatin immunoprecipitation sequencing (ChIP-seq) profiles of Padi genes (Supplementary Fig. 7) were also in line with an Aire-dependent expression mechanism at an epigenetic level (24), i.e., closed transcription start sites, near absence of activating chromatin modifications (H3K4Ac, H3K9Ac), and presence of repressive modifications (H3K27me3).
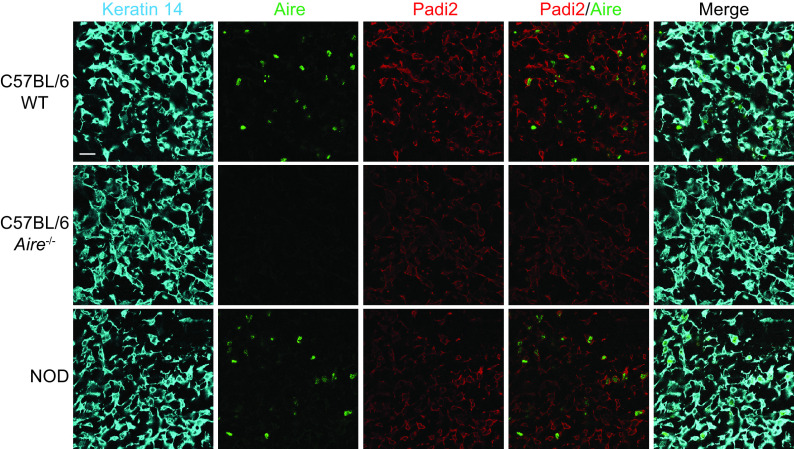
Further analysis of gene sets specific for modifying enzymes (Supplementary Table 3) revealed that, among the 22 enzymatic PTMs analyzed, citrullination was the only one under the control of Aire (Fig. 6F; Supplementary Fig. 6B for NOD mice) and enriched in mature MHC-IIhigh versus immature MHC-IIlow mTECs of WT mice (Supplementary Fig. 8). Finally, reduced Padi2 expression in the mTECs of NOD and Aire−/− C57BL/6 mice compared with WT C57BL/6 mice was confirmed at the protein level with confocal microscopy (Fig. 7).
Figure 7.
Padi2 protein expression in murine mTECs. Confocal microscopy of frozen thymic tissue section from 6- to 8-week-old C57BL/6 WT and Aire−/− mice and NOD mice (scale bar 20 µm). mTECs are visualized by keratin-14 staining.
Collectively, these observations suggest that Padi gene expression and enzymatic activity may catalyze citrullination not only in the islets but also in the thymus. In the NOD mice, the increased, age-dependent expression of Padi2 in the islets may augment the availability of citrullinated peptides for T cell recognition. On the other hand, the reduced Padi2 and Padi4 expression in the mature MHC-IIhigh mTECs of NOD mice may favor the escape of T cells reactive to citrullinated peptides.
Discussion
It is currently posited that posttranslationally modified epitopes may be more prone to loss of tolerance (8). This is based on the assumption that these epitopes may not be presented in the thymus, as PTMs are usually induced in a tissue-specific fashion and upregulated by inflammation. This assumption has been experimentally verified in elegant mouse models for glycosylated collagen type II (25), which is expressed in cartilages (including those from healthy donors) but not in the thymus. By analyzing GRP78 citrullination, we here document that this tenet may not always apply. Indeed, either native or citrullinated epitopes were preferentially recognized by circulating CD8+ T cells. Moreover, this recognition was not promiscuous, since most T cells recognized exclusively one epitope version or the other.
These observations prompt two questions. First, how can GRP78 be recognized in its native form, despite its ubiquitous expression? We suggest that one driver for such recognition may be cross-reactivity with gut bacterial sequences, as mimotopes—and even homotopes—were detected for some GRP78 peptides preferred in their R version but not for those preferentially recognized as citrullinated. Of note, these R-GRP78 versions were also those displaying significant fractions of antigen-experienced CD8+ T cells in some individuals and, for those with an identical bacterial match (GRP78-360R and -435R), particularly high frequencies. The final outcome of this bacterial cross recognition is likely to be tolerance, which may, however, be lost with gut dysbiosis, as recently described for IGRP microbial mimotopes (26,27).
Second, why are citrullinated GRP78 peptides not universally preferred by CD8+ T cells? This may reflect the presentation of citrullinated peptides in the thymus, as both murine and human mTECs robustly expressed citrullinating Padi enzymes, most notably Padi2. Padi expression in murine mTECs has previously been reported along with the presence of citrullination (28). The finding of Padi2 expression and Padi enzymatic activity in the murine thymus and of citrullinated proteins in purified mTECs provides direct evidence that this gene expression can translate into PTMs. We further show that Padi expression is under the control of Aire in murine mTECs. The effect of Aire on promiscuous antigen expression may thus extend to citrullinated proteins. Hence, CD8+ T cells recognizing native and citrullinated peptides may undergo a similar, albeit incomplete (29), thymic deletion. Interestingly, NOD mice displayed lower Padi2 and Padi4 expression in mature mTECs and higher expression in islets (likely in both β-cells and immune infiltrates [30]), which may exert a synergistic effect on the loss of central and peripheral tolerance against citrullinated epitopes, respectively. In line with this possibility, we previously showed that spleen T cells from NOD mice, but not from C57BL/6 mice, recognize the citrullinated version of the Grp78 protein but not its native version (9). Moreover, the lack of Padi2/Padi4 upregulation between MHC-IIlo (and Aire‒) and MHC-IIhi (Aire+) mTECs in NOD mice suggests that the Aire-driven control is impaired in this strain. The NOD thymus also harbored fewer MHC-IIhi mTECs, which may further contribute to defective central tolerance. Although interspecies differences are possible, it will be relevant to define whether an equivalent of this Padi2 expression pattern in the mTECs and islets of NOD mice exists in patients with type 1 diabetes.
The finding of a similar frequency of GRP78-reactive CD8+ T cells in the blood of individuals with type 1 diabetes and healthy individuals mirrors previous observations with other islet epitopes (2–4,18,20). These observations are likely to reflect the sequestration of the fraction engaged in the disease in the pancreas, where the same T cells were enriched in donors with type 1 diabetes versus nondiabetic donors (2,3). Given the notion that citrullination is enhanced in the inflammatory milieu of insulitis (30), we hypothesized that X-GRP78–reactive T cells may preferentially accumulate in the type 1 diabetic pancreas. However, despite the fact that the native GRP78-9R peptide recognized by circulating CD8+ T cells was preferred in its citrullinated version in the pancreas, these T cells were not specific for type 1 diabetes.
This notion of tissue but not disease specificity echoes our recent report documenting that citrullination is also detectable in noninfiltrated islets of nonautoimmune C57BL/6 mice (30). It is possible that citrullination, although upregulated by inflammation (9,10), may be, to a certain extent, physiological in the pancreas, as is the case in other tissues (31) and for other PTMs (25), e.g., iodinated thyroglobulin and glycosylated collagen. At the steady state, the outcome of this physiological citrullination may be peripheral tolerance, which may be broken under conditions of inflammation and ER stress that also upregulate GRP78 expression (11). Whether citrullination results in the presentation of citrullinated peptides by HLA class I molecules on β-cells or antigen-presenting cells remains an open question. Despite the presence of GRP78 peptides, immunopeptidomics studies on human and mouse islets (3,32) did not retrieve citrullinated peptides. It is noteworthy that, despite the notion of protein citrullination in inflamed cartilages (10,30,33), direct evidence of HLA-eluted citrullinated peptides is missing even for rheumatoid arthritis. While this may reflect a technical limitation of immunopeptidomics strategies, it invites the question of whether the diabetogenic role of citrullination (30) relies on T cells recognizing X-GRP78 peptides. We documented that Padi inhibition by BB-Cl-amidine treatment protects NOD mice from diabetes and is associated with decreased citrullination of GRP78 and other proteins in the pancreas, reduced anti–X-GRP78 aAb titers, and diminished formation of neutrophil extracellular traps (30). Interestingly, diabetes protection was observed upon treatment from 8 weeks of age, when insulitis is already present, suggesting an effect on disease amplification rather than priming. This is consistent with the higher titers of aAb against X-GRP78 detected in long-standing patients with type 1 diabetes (10). As citrullination is involved in the control of several biological processes (31), both physiological and pathological (e.g., neutrophil extracellular trap formation [34]), pathogenic effects independent of antigen presentation and T-cell recognition are possible. For instance, circulating aAbs against native or citrullinated GRP78 are described in patients with cancers (35), rheumatoid arthritis (36), or atherosclerosis (37). aAb binding to surface GRP78 activates endothelial cells (37) and monocytes/macrophages (36), upregulating inflammatory cytokines and adhesion molecules. Whether a similar process takes place in β-cells in the course of type 1 diabetes deserves further investigation.
This study carries some limitations. First, we did not provide evidence of natural processing and presentation of the GRP78 epitopes studied. Most of them are in close proximity to dibasic cleavage sites, which may favor their processing by the proteasome and other proteases. For the GRP78-502 epitope, which is exclusively recognized as citrullinated, we reported direct proteomic evidence for citrullination at position 510R in cytokine-treated human islets (10) and rat INS1E β-cells (9). Interestingly, this peptide overlaps with the citrullinated GRP78498–512 epitope targeted by circulating CD4+ T cells that are enriched in donors with type 1 diabetes (10). Second, the limited availability of human tissues allowed us to verify the absence of GRP78-9X–reactive CD8+ T cells outside the pancreas only in the duodenum.
In conclusion, our study shows that GRP78 is targeted by circulating CD8+ T cells that do not always prefer citrullinated peptides. Their recognition pattern may be shaped by cross-reactivity with gut microbial mimotopes and by thymic PADI expression. These findings mitigate the current claims that citrullinated epitopes are invariably more immunogenic than their native counterparts.
Article Information
Acknowledgments. The authors thank C. Maillard, Cochin Institute, Paris, France, for technical assistance, the Cochin Institute CYBIO and KU Leuven Flow Cytometry Core Facilities for assistance with cell analysis and sorting, and J. Perez-Hernandez, Cochin Institute, Paris, France, for critical reviewing of the manuscript.
Funding. This work was supported by JDRF grants 2-SRA-2016-164-Q-R (to R.M.), 2-SRA-2015-52-Q-R (to C.M. and L.O.), and 2-SRA-2018-480-S-B (to M.N.) and postdoctoral fellowship 3-PDF-2020-942-A-N (to Z.Z.); The Leona M. and Harry B. Helmsley Charitable Trust (1901-03689); Fondation Francophone pour la Recherche sur le Diabète; the European Foundation for the Study of Diabetes (EFSD/JDRF/Lilly European Programme in Type 1 Diabetes Research 2015); Agence Nationale de la Recherche (ANR-19-CE15-0014-01) and Fondation pour la Recherche Medicale (EQU20193007831) (to R.M.); and the National Institute of Diabetes and Digestive and Kidney Diseases (R01DK099317, R01DK032083 to M.N.). M.B. and A.Cal. were supported by a fellowship from the Flemish Research Foundation (12R0719N and 1189518N, respectively). F.D., D.L.E., C.M., L.O., and R.M. received funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreements 115797 and 945268 (INNODIA and INNODIA HARVEST), which receive support from the EU Horizon 2020 program, JDRF, and The Leona M. & Harry B. Helmsley Charitable Trust. This research was performed with the support of the nPOD (RRID SCR_014641), a collaborative type 1 diabetes research project supported by JDRF (nPOD: 5-SRA-2018-557-Q-R) and The Leona M. & Harry B. Helmsley Charitable Trust (grant 2018PG-T1D053). Organ procurement organizations (OPO) partnering with nPOD to provide research resources are listed at www.jdrfnpod.org/for-partners/npod-partners.
The content and views expressed are the responsibility of the authors and do not necessarily reflect an official view of nPOD.
Duality of Interest. INNODIA and INNODIA HARVEST also receive support from the European Federation of Pharmaceutical Industries and Associations. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. F.S., M.G., C.M., L.O., and R.M. contributed to study conceptualization. M.E.A., F.S., M.B., L.N., N.B., A.Cal., M.G., M.I., A.I.L., A.Car., Z.Z., B.B., M.L.C., G.S., F.D., M.N., D.L.E., S.Y., S.P., M.J.M., Y.V., J.V., S.B., C.M., L.O., and R.M. contributed to development of study methodology. M.E.A., F.S., M.B., L.N., N.B., A.Cal., M.G., M.I., A.I.L., A.Car., G.A., Z.Z., B.B., M.L.C., G.S., M.N., L.O., and R.M. contributed to data generation. M.E.A., F.S., M.B., L.N., N.B., A.Cal., M.G., A.Car., M.L.C., G.S., M.J.M., L.O., and R.M. contributed to data curation. M.E.A., F.S., M.B., M.G., L.O., and R.M. contributed to writing the manuscript. M.E.A., F.S., M.B., L.N., N.B., A.Cal., M.G., M.I., L.O., and R.M. contributed to data visualization. L.O. and R.M. contributed to study supervision. S.Y., C.M., L.O., and R.M. contributed to funding acquisition. R.M. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the nPOD 12th Annual Scientific Meeting, Tampa, FL, 23–26 February 2020.
Footnotes
M.E.A. and F.S. contributed equally to this work.
This article contains supplementary material online at https://doi.org/10.2337/figshare.16654669.
References
- 1. Carré A, Richardson SJ, Larger E, Mallone R. Presumption of guilt for T cells in type 1 diabetes: lead culprits or partners in crime depending on age of onset? Diabetologia 2021;64:15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Culina S, Lalanne AI, Afonso G, et al.; ImMaDiab Study Group . Islet-reactive CD8+ T cell frequencies in the pancreas, but not in blood, distinguish type 1 diabetic patients from healthy donors. Sci Immunol 2018;3:eaao4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gonzalez-Duque S, Azoury ME, Colli ML, et al. Conventional and neo-antigenic peptides presented by β cells are targeted by circulating naïve CD8+ T cells in type 1 diabetic and healthy donors. Cell Metab 2018;28:946–960.e6 [DOI] [PubMed] [Google Scholar]
- 4. Azoury ME, Tarayrah M, Afonso G, et al. Peptides derived from insulin granule proteins are targeted by CD8+ T cells across MHC class I restrictions in humans and NOD mice. Diabetes 2020;69:2678–2690 [DOI] [PubMed] [Google Scholar]
- 5. Alanio C, Lemaitre F, Law HK, Hasan M, Albert ML. Enumeration of human antigen-specific naive CD8+ T cells reveals conserved precursor frequencies. Blood 2010;115:3718–3725 [DOI] [PubMed] [Google Scholar]
- 6. Yu W, Jiang N, Ebert PJ, et al. Clonal deletion prunes but does not eliminate self-specific αβ CD8(+) T lymphocytes. Immunity 2015;42:929–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mallone R, Eizirik DL. Presumption of innocence for beta cells: why are they vulnerable autoimmune targets in type 1 diabetes? Diabetologia 2020;63:1999–2006 [DOI] [PubMed] [Google Scholar]
- 8. James EA, Pietropaolo M, Mamula MJ. Immune recognition of β-cells: neoepitopes as key players in the loss of tolerance. Diabetes 2018;67:1035–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rondas D, Crèvecoeur I, D’Hertog W, et al. Citrullinated glucose-regulated protein 78 is an autoantigen in type 1 diabetes. Diabetes 2015;64:573–586 [DOI] [PubMed] [Google Scholar]
- 10. Buitinga M, Callebaut A, Marques Câmara Sodré F, et al. Inflammation-induced citrullinated glucose-regulated protein 78 elicits immune responses in human type 1 diabetes. Diabetes 2018;67:2337–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang J, Lee J, Liem D, Ping P. HSPA5 gene encoding Hsp70 chaperone BiP in the endoplasmic reticulum. Gene 2017;618:14–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vig S, Buitinga M, Rondas D, et al. Cytokine-induced translocation of GRP78 to the plasma membrane triggers a pro-apoptotic feedback loop in pancreatic beta cells. Cell Death Dis 2019;10:309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Babon JA, DeNicola ME, Blodgett DM, et al. Analysis of self-antigen specificity of islet-infiltrating T cells from human donors with type 1 diabetes. Nat Med 2016;22:1482–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Leisner C, Loeth N, Lamberth K, et al. One-pot, mix-and-read peptide-MHC tetramers. PLoS One 2008;3:e1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Casteels K, Waer M, Laureys J, et al. Prevention of autoimmune destruction of syngeneic islet grafts in spontaneously diabetic nonobese diabetic mice by a combination of a vitamin D3 analog and cyclosporine. Transplantation 1998;65:1225–1232 [DOI] [PubMed] [Google Scholar]
- 16. Callebaut A, Derua R, Vig S, Delong T, Mathieu C, Overbergh L. Identification of deamidated peptides in cytokine-exposed MIN6 cells through LC-MS/MS using a shortened digestion time and inspection of MS2 spectra. J Proteome Res 2021;20:1405–1414 [DOI] [PubMed] [Google Scholar]
- 17. Guyon C, Jmari N, Padonou F, et al. Aire-dependent genes undergo Clp1-mediated 3'UTR shortening associated with higher transcript stability in the thymus. eLife 2020;9:e52985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wiedeman AE, Muir VS, Rosasco MG, et al. Autoreactive CD8+ T cell exhaustion distinguishes subjects with slow type 1 diabetes progression. J Clin Invest 2020;130:480–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yeo L, Pujol-Autonell I, Baptista R, et al. Circulating β cell-specific CD8+ T cells restricted by high-risk HLA class I molecules show antigen experience in children with and at risk of type 1 diabetes. Clin Exp Immunol 2020;199:263–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Skowera A, Ladell K, McLaren JE, et al. β-Cell-specific CD8 T cell phenotype in type 1 diabetes reflects chronic autoantigen exposure. Diabetes 2015;64:916–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Genest O, Wickner S, Doyle SM. Hsp90 and Hsp70 chaperones: collaborators in protein remodeling. J Biol Chem 2019;294:2109–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bender C, Rodriguez-Calvo T, Amirian N, Coppieters KT, von Herrath MG. The healthy exocrine pancreas contains preproinsulin-specific CD8 T cells that attack islets in type 1 diabetes. Sci Adv 2020;6:eabc5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Crèvecoeur I, Gudmundsdottir V, Vig S, et al. Early differences in islets from prediabetic NOD mice: combined microarray and proteomic analysis. Diabetologia 2017;60:475–489 [DOI] [PubMed] [Google Scholar]
- 24. Handel AE, Shikama-Dorn N, Zhanybekova S, et al. Comprehensively profiling the chromatin architecture of Tissue restricted antigen expression in thymic epithelial cells over development. Front Immunol 2018;9:2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Raposo B, Merky P, Lundqvist C, et al. T cells specific for post-translational modifications escape intrathymic tolerance induction. Nat Commun 2018;9:353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hebbandi Nanjundappa R, Ronchi F, Wang J, et al. A gut microbial mimic that hijacks diabetogenic autoreactivity to suppress colitis. Cell 2017;171:655–667.e17 [DOI] [PubMed] [Google Scholar]
- 27. Tai N, Peng J, Liu F, et al. Microbial antigen mimics activate diabetogenic CD8 T cells in NOD mice. J Exp Med 2016;213:2129–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Engelmann R, Biemelt A, Cordshagen A, Johl A, Kuthning D, Müller-Hilke B. The prerequisites for central tolerance induction against citrullinated proteins in the mouse. PLoS One 2016;11:e0158773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Davis MM. Not-So-negative selection. Immunity 2015;43:833–835 [DOI] [PubMed] [Google Scholar]
- 30. Sodré FMC, Bissenova S, Bruggeman Y, et al. Peptidylarginine deiminase inhibition prevents diabetes development in NOD mice. Diabetes 2021;70:516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alghamdi M, Alasmari D, Assiri A, et al. An overview of the intrinsic role of citrullination in autoimmune disorders. J Immunol Res 2019;2019:7592851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wan X, Vomund AN, Peterson OJ, Chervonsky AV, Lichti CF, Unanue ER. The MHC-II peptidome of pancreatic islets identifies key features of autoimmune peptides. Nat Immunol 2020;21:455–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tilvawala R, Nguyen SH, Maurais AJ, et al. The rheumatoid arthritis-associated citrullinome. Cell Chem Biol 2018;25:691–704.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Papayannopoulos V. Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol 2018;18:134–147 [DOI] [PubMed] [Google Scholar]
- 35. Mintz PJ, Kim J, Do KA, et al. Fingerprinting the circulating repertoire of antibodies from cancer patients. Nat Biotechnol 2003;21:57–63 [DOI] [PubMed] [Google Scholar]
- 36. Lu MC, Lai NS, Yu HC, Huang HB, Hsieh SC, Yu CL. Anti-citrullinated protein antibodies bind surface-expressed citrullinated Grp78 on monocyte/macrophages and stimulate tumor necrosis factor alpha production. Arthritis Rheum 2010;62:1213–1223 [DOI] [PubMed] [Google Scholar]
- 37. Crane ED, Al-Hashimi AA, Chen J, et al. Anti-GRP78 autoantibodies induce endothelial cell activation and accelerate the development of atherosclerotic lesions. JCI Insight 2018;3:e99363. [DOI] [PMC free article] [PubMed] [Google Scholar]