Abstract
Placental malaria infection is mediated by the binding of the malarial VAR2CSA protein to the placental glycosaminoglycan, chondroitin sulfate. Recombinant subfragments of VAR2CSA (rVAR2) have also been shown to bind specifically and with high affinity to cancer cells and tissues, suggesting the presence of a shared type of oncofetal chondroitin sulfate (ofCS) in the placenta and in tumors. However, the exact structure of ofCS and what determines the selective tropism of VAR2CSA remains poorly understood. In this study, ofCS was purified by affinity chromatography using rVAR2 and subjected to detailed structural analysis. We found high levels of N-acetylgalactosamine 4-O-sulfation (∼80–85%) in placenta- and tumor-derived ofCS. This level of 4-O-sulfation was also found in other tissues that do not support parasite sequestration, suggesting that VAR2CSA tropism is not exclusively determined by placenta- and tumor-specific sulfation. Here, we show that both placenta and tumors contain significantly more chondroitin sulfate moieties of higher molecular weight than other tissues. In line with this, CHPF and CHPF2, which encode proteins required for chondroitin polymerization, are significantly upregulated in most cancer types. CRISPR/Cas9 targeting of CHPF and CHPF2 in tumor cells reduced the average molecular weight of cell-surface chondroitin sulfate and resulted in a marked reduction of rVAR2 binding. Finally, utilizing a cell-based glycocalyx model, we showed that rVAR2 binding correlates with the length of the chondroitin sulfate chains in the cellular glycocalyx. These data demonstrate that the total amount and cellular accessibility of chondroitin sulfate chains impact rVAR2 binding and thus malaria infection.
Keywords: malaria, placental malaria, cancer, VAR2CSA, chondroitin sulfate, glycosaminoglycan, carbohydrate structure
Abbreviations: 2B6, anti-CS stub antibody 2B6; A-ChABC, Amsbio ChABC; AMAC, 2-aminoacridone; ChABC, chondroitinase ABC; ChAC, chondroitinase AC; ChB, Chondroitinase B; CHPF, chondroitin polymerizing factor; CHST14, carbohydrate sulfotransferase 14; CS, chondroitin sulfate; CZE, capillary zone electrophoresis; DS, dermatan sulfate; dp, degree of polymerization; CSPG, chondroitin sulfate proteoglycan; DSPE, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine; GAG, glycosaminoglycan; GalNAc, N-acetyl-D-galactosamine; GlcA, D-glucuronic acid; IdoA, iduronic acid; I-ofCS, Intact ofCS; L-ofCS, liberated ofCS; MW, molecular weight; ofCS, oncofetal chondroitin sulfate; P. falciparum, Plasmodium falciparum; PfEMP1, P. falciparum erythrocyte membrane protein 1; R-ofCS, retained ofCS; rVAR2, recombinant VAR2CSA; S-ChABC, Sigma ChABC; SpyC, SpyCatcher
Every year, more than 400,000 people die from Malaria infection (1). Ninety percent of these fatalities are caused by Plasmodium falciparum (P. falciparum), the most infectious of the four Plasmodium species (2). The P. falciparum parasites are especially virulent due to their unique ability to insert adhesins of parasite origin into the membrane of the infected erythrocytes, allowing them to adhere in the host microvasculature and thereby escape immune-mediated clearance in the spleen (3). Pregnant women remain susceptible to infection despite previously acquired immunity (4). Their susceptibility arises from a serologically distinct parasite that specifically sequesters in the placenta, leading to placental malaria (5).
The interaction of P. falciparum parasites with receptors in the placental vasculature is mediated by members of the P. falciparum erythrocyte membrane protein 1 (PfEMP1) family of proteins (6). Each PfEMP1 protein binds a different host receptor and therefore determines the parasitic tropism. In placental malaria, the parasites express the PfEMP1 called VAR2CSA, which allows the infected erythrocytes to adhere to chondroitin sulfate (CS) chains present on chondroitin sulfate proteoglycans (CSPGs) in the membrane of the syncytiotrophoblasts and in the intervillous space of the placenta (4, 7, 8, 9, 10). CS belongs to a group of polysaccharides termed glycosaminoglycan (GAGs), which are long linear polymers consisting of alternating N-acetyl-d-galactosamine (GalNAc) and d-glucuronic acid (GlcA) units. While the backbone structure is simple, an immense heterogeneity arises from variation in polymer length and differential sulfation of the sugar residues (11). CS can be O-sulfated at carbon-4 (4-O-Sulfation) and/or carbon-6 (6-O-Sulfation) of the N-acetylgalactosamine residue and at carbon-2 (2-O-sulfation) and more rarely at carbon-3 (3-O-sulfation) of the glucuronic acid residue. CS chains with high levels of 4-O-sulfation are referred to as chondroitin sulfate A (CSA), whereas 6-O-sulfated CS is termed chondroitin sulfate C (CSC) (11). Additionally, glucuronic acid residues may be epimerized into l-iduronic acid (IdoA) with or without subsequent 2-O-sulfation, giving rise to a dermatan/chondroitin sulfate hybrid (CS/DS), which contains both iduronic acid and glucuronic acid (12). The specific arrangement of sulfated residues and uronic acid epimers generates binding sites for CS/DS-binding proteins and is important for mediating a plethora of biological functions (13).
VAR2CSA expressing parasites adhere exclusively in the placenta (4, 9, 14), despite the fact that CS is omnipresent in the vasculature and organs of the human host (11). Surprisingly, most cancer cells express the distinct VAR2CSA-binding CS epitope normally restricted to trophoblastic cells in the placenta (15). This suggests that CS in placenta and cancer is distinct from CS in other tissues and that the VAR2CSA protein has been evolutionarily refined to bind selectively to this type of CS.
There are two components to a CSPG: the CS chain and the core protein to which it is attached. Only a small set of proteins contain CS/DS chains and can have different biological activities. We have previously shown that syndecan-1 is the primary placental receptor of P. falciparum malaria parasites (10). However, to investigate the repertoire of ofCS carrying CSPGs on various tumor types, we recently developed a glycoproteomics workflow coupled to VAR2CSA-based affinity-chromatography to analyze CSPGs (16). This study utilized a recombinant fragment of VAR2CSA (rVAR2), based on the ID1-DBL2-ID2a region, which we and others have shown to maintain the CS specificity of full-length native VAR2CSA (17, 18). Our results revealed the presence of at least 14 distinct core proteins carrying VAR2CSA reactive glycan chains in the placenta and tumor tissues, including both secreted and membrane-bound CSPGs. This large heterogeneity rules out the possibility of a single CSPG as the main receptor for VAR2CSA and points instead toward the existence of specific CS-motifs shared among many CSPGs that is crucial for VAR2CSA binding.
The absolute requirement of continuous CS stretches enriched in 4-O-sulfated N-acetylgalactosamine units in VAR2CSA binding has been reported in multiple studies using CS substrates derived from animal sources, as well as synthetic material (4, 15, 19, 20, 21). Initial findings suggested that the minimal length of the CS-binding motif is a dodecasaccharide (dp12) containing a minimum of 2–3 (19) or 4–5 (22) 4-O-sulfated N-acetylgalactosamine units. More recently we have confirmed the minimal requirement of a dp12, although longer oligosaccharides showed higher affinity binding (21). Contrary to early results suggesting a requirement for several nonsulfated sugars (19), we have shown that oligosaccharides with a high degree of 4-O-sulfation are more potent in inhibiting binding of rVAR2 to isolated CSPGs compared with those that were mostly nonsulfated (21). To expand this to cellular binding, we deconstructed the CS biosynthesis pathway by creating a library of enzyme knockouts using CRISPR/Cas9 in CHO cells (23). This revealed a strict requirement for 4-O-sulfated CS as inactivation of enzymes crucial for CS initiation, elongation, and 4-O sulfation completely abrogated rVAR2 binding.
Although it is clear that 4-O-sulfation is essential for VAR2CSA binding, the precise role of other CS modifications is less clear. Interestingly, DS-substrates display inhibitory activity although at slightly lower levels compared with purely 4-O-sulfated CS substrates (4, 20, 22, 24). Teasing out the direct contribution of the uronic acid epimers (glucuronic and iduronic acids) to VAR2CSA binding has been hampered by the fact that most CSPGs and DSPGs isolated from natural sources are in reality CS/DS copolymers with different uronic acid ratios (25). Importantly, most studies have used CS isolated from sources other than the most relevant placental ofCS, using indirect assays, such as inhibition of parasite adhesion.
In this study, we utilized an affinity chromatography approach using recombinant VAR2CSA and orthogonal analytical techniques to fully characterize the VAR2CSA of CS-binding motif in CS. We further developed cellular models to investigate the interaction within the glycocalyx. We show that the minimal CS-binding domain consists of a dp12 CS containing ∼85–90% 4-O-sulfated and ∼10–15% 6-O-sulfated N-acetylgalactosamine residues. We further show that the placenta and tumor tissues contain higher amounts of CS and CS of greater molecular weight (MW), compared with other tissues. Investigating rVAR2 binding to CS when part of a cellular glycocalyx containing a high density and quantity of cell surface glycans showed higher level of rVAR2 binding to high MW CS compared with low MW CS. This finding suggests that VAR2CSA tropism depends not only on the structure of the CS ligand, but also on its accessibility and presentation.
Results
Oncofetal CS is a highly 4-O-sulfated dp12
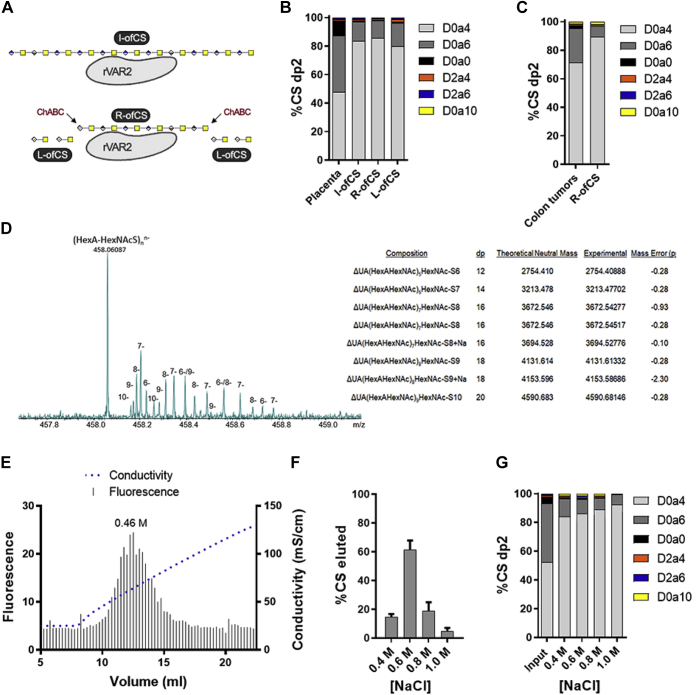
VAR2CSA selectively binds CS in the placenta and in malignant tumors, despite CS being ubiquitously expressed throughout the body (15). Several studies have attempted to explain the structure of the oncofetal CS (ofCS) that underlies this tissue tropism; however, the determinants that confer high specificity of VAR2CSA for ofCS remain elusive. To further characterize the structure and composition of ofCS, we developed an affinity chromatography method using immobilized VAR2CSA to isolate ofCS from purified placental and tumor CS. This assay utilized a recombinant fragment containing the minimal CS binding region in VAR2CSA, ID1-ID2a, which has been previously described to maintain the CS binding specificity of full-length VAR2CSA (17, 18). First, purified placental CS was loaded onto an rVAR2-conjugated column, washed with 0.25 M NaCl, and the intact chains containing the ofCS motif (I-ofCS) were then eluted with 2 M NaCl. In another setup we isolated the specific ofCS motif by an on-column digestion of rVAR2-bound I-ofCS with chondroitinase ABC (ChABC) to remove CS regions outside of the binding domain (liberated ofCS or L-ofCS). The ofCS-motifs that were retained on the rVAR2 column (retained ofCS or R-ofCS) were subsequently collected by elution with 2M NaCl (Fig. 1A). The I-, R-, and L-ofCS fractions were then analyzed by disaccharide analysis using high-performance liquid chromatography (LC) coupled with mass spectrometry (MS), as previously reported (26). From the 200 μg placental CS input, we obtained ∼12.5 μg I-ofCS, 3.9 μg R-ofCS, and 9.1 μg L-ofCS. The disaccharide composition of the total input placental CS was ∼50% 4-O-sulfated (D0a4), ∼40% 6-O-sulfated (D0a6), ∼10% non-sulfated (D0a0), and <1% disulfated CS units (Fig. 1B). All rVAR2 affinity purified fractions (I-, R- and L-ofCS) were highly enriched for 4-O-sulfated CS (∼80–85%) and depleted in both 6-O-sulfated (∼15–20%) and nonsulfated CS (∼1%) disaccharide units compared with the input material (Fig. 1B). Only minor differences were observed in these fractions, including slightly higher levels of 4-O-sulfation in R-ofCS (∼85%) compared with I- and L-ofCS (∼80%). To verify if the same structural motif was found in cancer tissues, CS isolated from human colon cancer tumors was subjected to the same analytical workflow. After rVAR2 affinity chromatography, a similar enrichment in 4-O-sulfation (from ∼70% to ∼90%) was observed, with the remaining disaccharides being primarily 6-O-sulfated (∼8%) (Fig. 1C). This suggests that rVAR2 prefers highly 4-O-sulfated CS in both placental and tumor tissues.
Figure 1.
Structural characterization of oncofetal CS.A, diagram representing intact oncofetal CS (I-ofCS), ChABC retained ofCS (R-ofCS), and ChABC liberated ofCS (L-ofCS) purified using rVAR2 affinity chromatography. B, disaccharide (dp2) structure analysis of placental CS and placental ofCS, analyzed by LC-MS (N = 3). C, dp2 analysis of colon tumor CS and R-ofCS (N = 1). D, MS analysis and composition of full-length R-ofCS (supplemental methods). E, AMAC-labeled R-ofCS was loaded on a rVAR2-column and eluted with a gradient of sodium chloride. R-ofCS elution was detected at 0.46 M NaCl. R-ofCS was fractionated with different concentrations of NaCl and subsequently (F) quantified and (G) structurally characterized by disaccharide analysis using LC-MS (N = 3). Error bars indicate mean ± SEM.
To estimate the average length of the rVAR2 minimal binding motif, R-ofCS was separated by capillary zone electrophoresis (CZE) and analyzed by mass spectrometry. The analysis identified species corresponding to dp12–dp20 oligosaccharides carrying one sulfate group per disaccharide unit (Fig. 1D), consistent with previous reports (19, 21, 22). The dominant charge state in the mass spectra of CS oligosaccharides with one sulfo group per disaccharide is equal to the number of disaccharide units (and also equal to the number of sulfo groups). The monoisotopic peak of (UA-GalNAcS – H)nn− is 458.0609, independent of the length of the oligosaccharide, yielding an abundant peak in the mass spectrum in Figure 1D that is a convolution of all chain lengths. However, the isotope peaks are separated from each other by 1/n, allowing the various charge states, dp's, and molecular weights to be assigned, as seen on the right side of Figure 1D. Analysis of intact GAG chains by mass spectrometry can be affected by in-source fragmentation, leading to a potential underestimation of the size and sulfation level of the GAG chains (26). Therefore, to further estimate the average MW of I- and R-ofCS, we labeled their reducing ends with fluorescent 2-aminoacridone (AMAC) and quantified the number of reducing ends by fluorescence detection and the total amount of CS by disaccharide analysis. The average MW of the I- and R-ofCS fractions was 15.2 kDa and 5.6 kDa, respectively, corresponding to an average length of dp33 and dp12. This supports the mass spectrometry analysis and validates the used fractionation scheme (Fig. 1A).
CS synthesis is a non-template-driven process and therefore leads to great structural heterogeneity in the resulting CS population. Given the high structural heterogeneity, it is possible that VAR2CSA can interact with multiple similar motifs with a graduation of overall affinity, binding strongest to the true high affinity ofCS site. To investigate this, we separated high and low affinity ofCS fractions by serial salt elution following immobilization on an rVAR2 conjugated affinity chromatography column. AMAC-labeled R-ofCS was loaded onto the rVAR2-column and eluted in a gradient of sodium chloride. The average concentration of salt required for complete elution was found to be 0.46 M NaCl, with a range of 0.35–0.85 M NaCl (Fig. 1E). To determine whether the low and high affinity fractions differed in structure, we fractionated unlabeled R-ofCS on the rVAR2 column, using placental CS as the input, and performed disaccharide analysis using LC-MS. MS showed that the majority of R-ofCS eluted in the 0.6 M NaCl fraction (Fig. 1F). The disaccharide analysis revealed a gradual increase in the proportion of 4-O-sulfation in the higher molar salt fractions, suggesting that high levels of 4-O-sulfation correlates with high affinity for rVAR2 (Fig. 1G).
Notably, the compositional analysis performed in this study confirmed that each disaccharide unit of ofCS is almost completely monosulfated, with only small amounts of nonsulfated units observed (less than 1%). This is in contrast to previous studies suggesting that nonsulfated units are critical for binding (19). In summary, these findings suggest that rVAR2 binds highly 4-O-sulfated CS in both placental and tumor tissues and that the minimal binding oligosaccharide is a dp12.
Placental ofCS are CS/DS copolymers
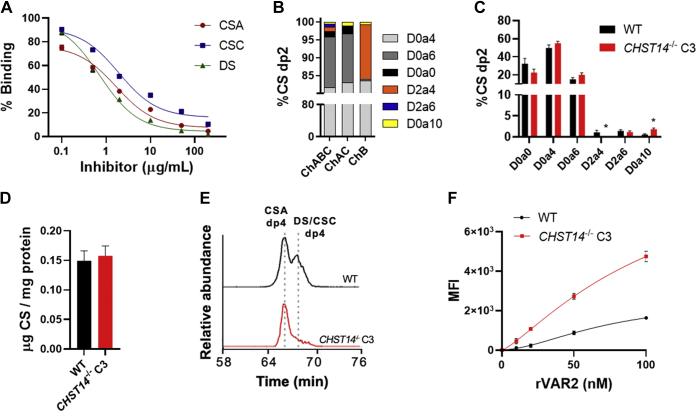
Previous studies have demonstrated that DS is capable of inhibiting parasite adhesion to placental tissue, albeit at lower levels than CSA (22, 27). A recent in-depth structural characterization of commercially available CSA, CSC, and DS substrates showed that both commercial preparations of CSA and DS were highly 4-O-sulfated (∼77% and ∼90%, respectively), whereas CSC was highly 6-O-sulfated (∼73%) and comparatively less 4-O-sulfated (∼16%) (27). In terms of uronic acid content, DS contained the highest proportion of iduronic acid (80%), whereas CSA and CSC contained mostly glucuronic acid (97% and 100%, respectively) (27). These CSA, CSC, and DS substrates were all able to inhibit rVAR2 binding to HeLa cells yielding IC50 values of 1.7, 1.9, and 0.7 μg/ml, respectively (Fig. 2A).
Figure 2.
Importance of iduronic acid for rVAR2 binding.A, inhibition of rVAR2 binding to HeLa cells using commercially available CSA, CSC, and DS substrates, performed by flow cytometry (N = 3), curve fitted with three parameter dose–response curve. B, disaccharide analysis of Sigma ChABC (S-ChABC), ChAC, and ChB digested R-ofCS, analyzed by LC-MS. C, structural analysis and (D) quantification of S-ChABC digested CS from HeLa WT and CHST14−/− cells, analyzed by LC-MS (N = 3). E, chromatograms of Amsbio ChABC (A-ChABC) resistant dp4 species generated from HeLa WT and CHST14−/− CS. F, titration of rVAR2 to HeLa WT and CHST14−/− cells, analyzed by flow cytometry (N = 3), and curve fitted with nonlinear 4PL regression. Error bars indicate mean ± SEM. ∗p < 0.05 by unpaired t test. CHST14, carbohydrate sulfotransferase.
To determine if iduronic acid enhances binding to rVAR2, we performed monosaccharide analysis using Dionex chromatography. Standard disaccharide compositional analysis after lyase treatment cannot differentiate between glucuronic acid and iduronic acid, since ChABC digestion results in the loss of uronic acid stereochemistry. Total placental CS contained ∼26% iduronic acid, whereas rVAR2-enriched I-ofCS contained ∼39% iduronic acid and R-ofCS contained ∼26% iduronic acid, suggesting that iduronic acid may be present in the rVAR2 binding motif. Digestion of R-ofCS with chondroitinase AC (ChAC), which only cleaves the hexosaminidic bond to glucuronic acid, showed that the liberated disaccharides were ∼85% 4-O-sulfated and ∼13% 6-O-sulfated (Fig. 2B). Chondroitinase B (ChB) digestion of R-ofCS, which cleaves the bond between N-acetylgalactosamine and iduronic acid, showed that the segments enriched in iduronic acid were ∼83% 4-O-sulfated and ∼15% 2,4-O-disulfated. However, the latter only amounted to <1% of the overall dp2 released by ChABC (Fig. 2B). In a dp12 oligosaccharide, this compositional analysis would correspond to 1–2 iduronic acid residues.
Digestion of CS with ChABC from Amsbio (A-ChABC) results in a mixture of dp2, dp4, and dp6 species, whereas ChABC from Sigma (S-ChABC) digests substrates to completion (27). These oligosaccharides can be separated and analyzed by LC-MS2 to distinguish between glucuronic acid- and iduronic acid-containing species. A-ChABC digestion of R-ofCS revealed two coeluting dp4 species and one or more dp6 species in the LC-MS chromatogram, in addition to the D0a4 and D0a6 dp2 (Fig. S1). MS2 analysis of the dp4 species in R-ofCS and input placental CS showed that the first eluting dp4 corresponds to an entirely 4-O-sulfated species (D0a4-G0a4), while the second eluting dp4 corresponds to an entirely 4-O-sulfated DS dp4 (D0a4-I0a4). Similarly, we identified two separated tri-sulfated dp6 species in A-ChABC digested R-ofCS, where MS2 analysis suggested one to be glucuronic acid-containing and the other iduronic acid-containing structures (Fig. S2). These data demonstrate that the rVAR2-binding oligosaccharides in placental ofCS contain both glucuronic acid and iduronic acid residues.
To obtain CS structures with similar levels of 4-O-sulfation and varying levels of iduronic acid, we generated CRISPR/Cas9-targeted clones bearing biallelic mutations in carbohydrate sulfotransferase 14 (CHST14) in HeLa cells (Fig. S3). CHST14 encodes for the dermatan 4-O-sulfotransferase, responsible for the 4-O-sulfation of GalNAc units to the reducing side of iduronic acid residues (28). The disaccharide analysis showed that the CHST14−/− clone C3 contained similar levels of overall CS and 4-O-sulfation as compared with wild type, with the expected reduction of D2a4 units and a small increase in disulfated disaccharide D0a10 (Fig. 2, C and D). Analysis of CS dp4 released by A-ChABC digestion showed a significant reduction in the level of DS/CSC dp4 species in the CHST14−/− cells, suggesting a reduction in the iduronic acid/glucuronic acid ratio (Fig. 2E). The CS/DS uronic acid content was determined by monosaccharide analysis to be 41% glucuronic acid and 59% iduronic acid in wild type and 100% glucuronic acid in the CHST14−/− clone C3. Thus, wild-type and CHST14−/− cells produce similar amounts of cell surface CS with a similar sulfation pattern and only differ in the composition of the uronic acids. Notably, rVAR2 binding was significantly higher to the CHST14−/− cells compared with the wild type (Fig. 2F) indicating that high levels of iduronic acid may actually interfere with rVAR2 binding. The binding was also significantly higher to a second CHST14−/− clone C4, which contained an intermediate amount of iduronic acid (21%) compared with the wild type and clone C3 (Fig. S4).
High-molecular-weight CS is upregulated in placental and tumor tissues
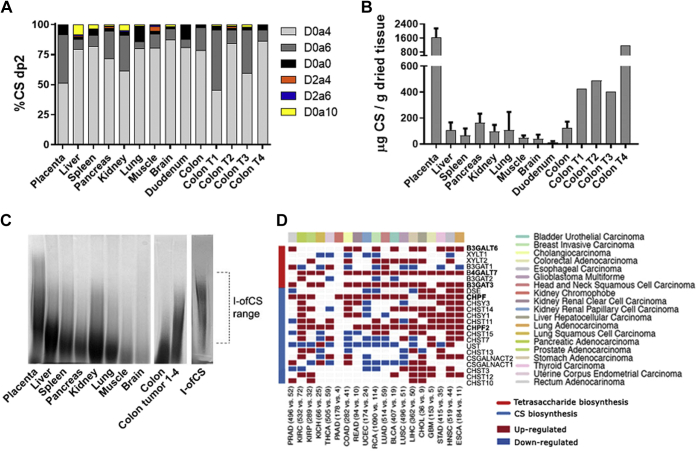
We next investigated the abundance and composition of CS in tissues and malignant tumors to determine if the level of CS might play a role in rVAR2 binding. CS was isolated from human liver, spleen, pancreas, kidney, lung, muscle, brain, and duodenum and compared with CS extracted from human placenta and several samples of patient colon tumors (Fig. 3). CS disaccharide analysis revealed high levels of 4-O-sulfation across all tissues (Fig. 3A). Interestingly, the placenta and two of the colon tumors had high level of 6-O-sulfation. To assess whether the rVAR2 ofCS binding motif can be found in all of these human tissues, we purified the I-ofCS fraction (Fig. S5). The highly 4-O-sulfated ofCS binding motif was found in all the organs, although at different levels (Fig. S5). Since rVAR2 does not bind these tissues (15), these results suggest that the amount and/or overall size of CS might vary in the different organs. Notably, placenta contained approximately 10-fold more CS per gram of dried tissue compared with other tissues (Fig. 3B). Similarly, the colon tumors expressed ≥3-fold more CS than healthy colon (Fig. 3B).
Figure 3.
CS structure and expression of CS biosynthesis enzymes in tissues.A, structural analysis (B) and quantification of CS disaccharides produced from S-ChABC digestion of human tissue CS (N = 3, except colon tumor (T) CS N = 1). Error bars indicate mean ± SEM. C, polyacrylamide gel electrophoresis (PAGE) analysis of human tissue CS, a mix of CS from colon tumor 1–4, and I-ofCS purified from placental CS, stained with Alcian blue. D, heat map illustrating RNA-Seq differential expression data of CS biosynthesis enzyme upregulated (red) and downregulated (blue) in tumor samples, compared with patient-matched healthy controls. Error bars indicate mean ± SEM.
To compare the size of CS across the tissues, samples from each organ and a mixture of CS from the four colon tumors were separated by polyacrylamide gel electrophoresis (PAGE) and stained with Alcian blue (Fig. 3C). Placental and colon tumor CS appeared to have a higher average MW than CS from any other tissue (Fig. 3C). Analysis of the length of rVAR2 purified I-ofCS chains suggested that the rVAR2-binding motif is primarily present in longer CS chains, which are found in low abundance in other tissues (Fig. 3C). To further investigate the difference in CS biology between tumors and healthy tissue, we compared the expression of CS biosynthesis enzymes across different tumor types with their healthy human control tissues (Fig. 3D). Enzymes involved in the biosynthesis of the CS tetrasaccharide linker region (beta-1,3-galactosyltransferase 6, beta-1,4-galactosyltransferase 7, and beta-1,3-glucuronyltransferase 3) and enzymes involved in the polymerization of the CS backbone (chondroitin polymerizing factor [CHPF] and CHPF2) were significantly upregulated in most types of tumors (Fig. 3D), suggesting that the enhanced expression of high MW CS could be a common tumor phenotype not specific to colon tumors.
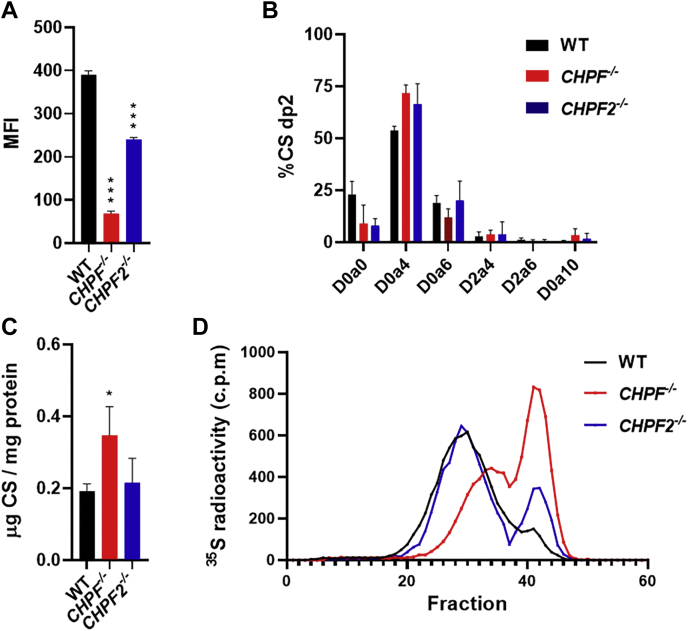
We next generated HeLa cells with inactivating mutations in CHPF and CHPF2 using CRISPR/Cas9 (Fig. S3). Interestingly, rVAR2 binding to both mutant cell lines was significantly reduced compared with WT, with an ∼80% reduction to CHPF−/− and a ∼40% reduction to CHPF2−/− (Fig. 4A). Disaccharide analysis showed no significant difference in the CS sulfation of the mutant cells (Fig. 4B) and a small increase in the overall quantity of CS (Fig. 4C) in the CHPF−/− cells. However, the average chain length was reduced in the mutants based on radiolabeling studies (Fig. 4D). Radiolabeled CS was purified from wild-type and mutant cell lines, grown in [35S]sulfate-containing media, and separated by size-exclusion chromatography. The average MW of CS from wild-type, CHPF−/−, and CHPF2−/− cells were determined to be 15.6 ± 0.2 kDa (∼dp34), 9.2 ± 0.1 kDa (∼dp19), and 13.7 ± 1.5 kDa (∼dp29), respectively. Thus, the CHPF−/− and CHPF2−/− cells expressed significantly shorter CS chains. Furthermore, the reduction in overall MW correlated to the difference seen in rVAR2 binding to these cells (Fig. 4A). Given that there was no difference in the overall amount of CS and its sulfation, the alteration in rVAR2 binding was most likely due to the length of CS produced by the cells.
Figure 4.
Impact of CHPF and CHPF2 inactivation on rVAR2 binding and CS chain length.A, rVAR2 binding to HeLa WT and mutant cell lines, analyzed by flow cytometry (N = 3). B, structural analysis and (C) quantification of cell surface CS from HeLa WT and mutant cell lines, analyzed by LC-MS (N = 3). D, size exclusion chromatogram of radiolabeled cell surface CS from WT and mutant cell lines, separated using a 1.5 × 50 cm column packed with Sepharose 6B (N = 2). Error bars indicate mean ± SEM. ∗∗∗p < 0.001 and ∗p < 0.05 by unpaired t test.
rVAR2 binds high MW CS chains in a cell-based model of a complex glycocalyx
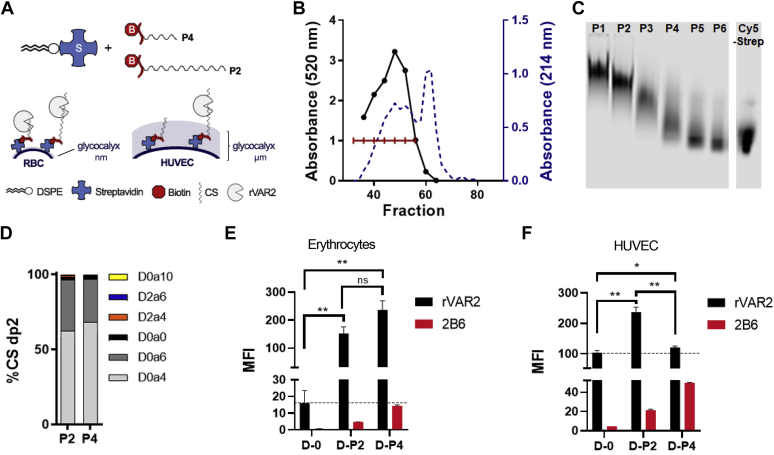
To investigate how CS chain length might dictate the VAR2CSA tropism, we developed a cell-based model to investigate rVAR2 binding in the context of a cellular glycocalyx of varying thickness (Fig. 5A). Erythrocytes are known to have a thin glycocalyx (∼10 nm), whereas human umbilical vein endothelial cells (HUVECs) have a much more complex and thicker glycocalyx reaching up to 2.5 μm (29, 30). Furthermore, both erythrocytes and HUVEC cells are ofCS negative and do not support rVAR2 binding (15). Thus, these cells represent two systems of varying glycocalyx complexity in which we can manipulate CS length and study its impact on the rVAR2 interaction.
Figure 5.
Cell-based glycocalyx model for the comparison of low and high MW CS.A, diagram showing the glycocalyx model setup. Erythrocytes and human umbilical vein endothelial cells (HUVECs) were used to compare the effect of a thin or dense glycocalyx, respectively, on rVAR2 binding to low MW placental CS (P4) and high MW placental CS (P2). The CS-peptide chains were biotinylated and integrated onto the cell membrane through a 1,2-Distearoyl-sn-glycero-3-phosphoethanolamine (DSPE) streptavidin conjugate. B, biotinylated placental CS was fractioned by size exclusion chromatography, and absorbance (214 nm) was measured on every other fraction, while the carbazole assay was performed on every four fractions for glucuronic acid detection. The individual fractions were combined into six fractions, as indicated, and designated as P1–P6. C, a portion of the P1–P6 fractions were conjugated to Cy5-Streptavidin, separated by polyacrylamide gel electrophoresis (PAGE), and detected at 700 nm. D, disaccharide analysis of the P2 and P4 CS fractions by LC-MS. Unconjugated DSPE (D-0), DSPE-P2 (D-P2), and DSPE-P4 (D-P4) were integrated into the cell membrane of (E) erythrocytes and (F) HUVECs. Half of the cells were stained with rVAR2, and the other half were ChABC digested and stained with 2B6 antibody, followed by analysis by flow cytometry (N = 3). Error bars indicate mean ± SEM. ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05 by unpaired t test.
ofCS presentation was achieved by inserting placental CS of varying MW directly into the cell membrane. Placental CS was biotinylated at the reducing end and separated by size-exclusion chromatography into large and short chains (P1-6, Fig. 5B). Samples of each fraction were conjugated to Cy5-Streptavidin and separated by PAGE, which showed the variation in MW of the CS chains (Fig. 5C). Structural analysis of the high MW CS (P2) and low MW CS (P4) revealed a similar sulfation pattern (Fig. 5D). Samples of P2 and P4 pools were then conjugated to a lipid anchor 1,2-distearoyl-sn-glycero-3-phosphoethanolamine (DSPE) streptavidin. Unconjugated DSPE (D-0), DSPE conjugated to the P2 fraction (D-P2), and DSPE conjugated to the P4 fraction (D-P4) readily incorporated into the membrane of erythrocytes, as measured by exhaustive ChABC treatment and staining with anti-CS stub antibody (2B6) that detects the tetrasaccharide linker (Fig. 4E, red bars). More D-P4 was incorporated compared with D-P2 since the cells were treated with an equal mass of CS-DSPE conjugate, and the P4 fraction is of lower MW. Erythrocytes treated with the DSPE conjugates were then incubated with rVAR2 and analyzed by flow cytometry. Incorporation of both the D-P2 and D-P4 fractions significantly increased rVAR2 binding compared with the control, with no apparent difference relating to MW (Fig. 5E, black bars). However, only the D-P2 increased rVAR2 binding to HUVEC cells, despite showing similar integration of both the D-P2 and D-P4 fractions (Fig. 5F). We interpret this finding to suggest that when a complex glycocalyx is present, longer CS chains may extend beyond this dense glycocalyx barrier and are more accessible to rVAR2 binding. Thus, CS chain length may contribute to the placenta and tumor selective tropism of VAR2CSA.
Discussion
This study aimed to characterize the structural and biochemical basis for the ofCS-specific tropism of the VAR2CSA malarial protein. Several studies have addressed the high selectivity of VAR2CSA expressing parasites for CS in the placenta (19, 20, 22, 31, 32). These studies mainly relied on parasite inhibition assays using purified CS fractions of different sulfation and length. We have further performed similar assays using rVAR2 (21). The conclusion from these studies was that VAR2CSA binding relies on a highly 4-O-sulfated dp12 oligosaccharide. However, this structure is not unique to the placenta or tumors, and thus the exact determinant of the VAR2CSA tropism has remained elusive. Here, we used immobilized rVAR2 to affinity purify ofCS and performed detailed structural characterization by mass spectrometry. In agreement with previous studies, we found a high enrichment in 4-O-sulfated CS, a depletion in 6-O-sulfated CS and no impact on iduronic acid content (4, 19, 22, 23, 33). In contrast with earlier findings suggesting that VAR2CSA expressing parasites interacted with an unusually low sulfated form of CS, we found the high-affinity rVAR2 purified fractions lacked nonsulfated CS. This discrepancy may be due to differences in CS preparations or methods used for purifying the materials (34). A previous study of placental CS may not have included membrane-bound CSPGs, whereas in the present study a large section of the placenta was homogenized and used for CS isolation. It is also possible that the differences result from isolate-specific variations among P. falciparum infected erythrocytes (20). We used ID1-ID2a for our studies rather than the full-length VAR2CSA protein, which might have slightly different specificities. However, we have previously shown that recombinant ID1-ID2a protein retains the high CS-binding affinity and specificity of the full length VAR2CSA protein and that both proteins specifically bind to CS present on placental syncytiotrophoblasts, in the villi stroma, and in the intervillous space (10, 18). Furthermore, in our previous studies we used this recombinant fragment to effectively target cancer cells in vivo and for the retrieval circulating tumor cells (15, 35). Our findings suggest that the minimal binding motif for rVAR2 is a dp12 containing ∼5 4-O-sulfated N-acetylgalactosamine residues and ∼1 6-O-sulfated N-acetylgalactosamine residues. Iduronic acid may be present as well, but studies of mutants with diminished iduronic acid did not affect binding. Additional studies are needed to investigate the exact arrangement of sulfated disaccharides within the dp12 oligosaccharides.
One hypothesis to account for the high specificity of VAR2CSA for placental and tumor-derived CS is that the ofCS motif contained a novel modification or a specific sequence of sulfated sugars (15). The evidence presented here suggests that this hypothesis may be oversimplified. Here, we provide compelling data suggesting that the specificity of VAR2CSA also relies on the accessibility of CS as well as overall CS amounts. The placental and tumor tissue, in general, contained more CS and CS of higher MW, compared with other tissues. In fact, the placenta contained ∼10-fold more CS than any other tissue, and this CS was of significantly higher MW. Interestingly, recent studies have reported high expression of the CS polymerizing factor CHPF in cancers, such as adenocarcinomas of the lung (36, 37), melanoma (38), and gliomas (39), and that CHPF knockdown inhibits proliferation and migration. Here, we found that several CS biosynthesis enzymes, including CHPF and CHPF2, were upregulated in a diverse set of cancer types. Furthermore, inactivation of CHPF, and to a lesser extent CHPF2, resulted in a reduction in CS chain length and in loss of rVAR2 binding. Collectively, these findings suggest that the placenta and malignant tumors express increased quantities of high MW CS. Whether a unique sequence of sulfation is also required remains to be fully determined.
The luminal surface of vascular endothelial cells is lined by a dense glycocalyx, consisting of various glycoproteins and proteoglycans, which act as a barrier to pathogen adhesion (40, 41). The thickness of this layer is approximately 0.5–5.0 μm depending on the cell type or tissue (42, 43, 44). Although we have shown that the minimal binding motif of rVAR2 is a dp12, parasite attachment may require longer CS chains that extend beyond the glycocalyx barrier. The cell-based glycocalyx model reported here allowed a comparison of rVAR2 binding to low and high MW placental CS in the context of different glycocalyces. We found that rVAR2 almost exclusively bound the high MW CS when imbedded in dense glycocalyx (HUVEC), suggesting that steric hindrance by glycocalyx components may prevent rVAR2 from adhering to other tissues. While CS is a component of the vascular endothelial glycocalyx, HS account for ∼80% of the sulfated GAGs found here (45). Furthermore, the permeability of the apical glycocalyx is largely determined by hyaluronan and allows for little to no access to infected erythrocytes (46). In contrast, staining experiments suggest that placental syncytiotrophoblasts and cytotrophoblasts produce very little hyaluronan, suggesting that infected erythrocytes could have greater access to interact with CS (47). The density of CSPGs is likely another major factor for the rVAR2 tropism, as high CSA density has been shown to correlate with increased VAR2CSA binding (48). Other, factors may play a role in attachment, such as shear stress and tumor/placental biology not considered in the single cell models described here. Several studies have used transmission electron microscopy for visualizing the endothelial glycocalyx; however, the main hurdle lies in preserving its native state (41). While newer methods have been developed to preserve the architecture of the glycocalyx, it is unclear whether these provide an accurate representation of the original tissues (49).
This study aimed at isolating and structurally characterizing ofCS in the human placenta and malignant tumors. The results suggest that the structural determinant for VAR2CSA binding is 4-O-sulfation and that the parasite tropism may be determined more by accessibility to the CS ligand at the cellular and tissue level than a specific structural motif. These results provide important insights into the biology of the VAR2CSA-ofCS interaction, with implications for both placental malaria prevention and cancer therapy.
Experimental procedures
Substrates and enzymes
Dp4 standards for LC-MS analysis were purchased from Iduron (Catalog #CSO04 and #DSO04) and dp2 standards were purchased from Sigma. Chondroitinase ABC was purchased from Sigma-Aldrich (S-ChABC) and from Amsbio (A-ChABC). IBEX chondroitinase AC (ChAC) and chondroitinase B (ChB) were kindly gifted by IBEX pharmaceuticals. All lyases were reconstituted as previously described (27), aliquoted, and snap-frozen immediately. Chondroitin sulfate A from bovine trachea and chondroitin sulfate C from shark cartilage were obtained from Sigma-Aldrich, and porcine intestinal mucosa dermatan sulfate was obtained from Celsus Laboratories Inc. As previously described, rVAR2 tagged with SpyTag was expressed in SHuffle T7 Express Competent E. coli (NEB) (15). SpyCatcher (SpyC) was produced in the E. coli BL21(DE3) strain and multibiotinylated using NHS biotin, as described (50). Both proteins were purified in two steps using (1) HisTrap columns from GE Healthcare and (2) cation exchange chromatography (rVAR2, HiTrap SP HP) or anion exchange chromatography (SpyC, HiTrap QHP) and formulated in PBS. Purity of the proteins was estimated by SDS-PAGE and tested for binding on Myla cells using flow cytometry. Free thiol groups were estimated using Ellman's Reagent (Thermo Scientific). rVAR2 fluorescent conjugate was generated by incubation of SpyC with AFDye 647 NHS Ester (Fluoroprobes) in a 1:1.2 M ratio in PBS at RT. After 1 h, the conjugation was deactivated with 2 mM ethanolamine for 10 min, and excess dye was washed away on a 10 kDa spin column (Sigma Aldrich). Prior to cell staining, the AFDye 647-SpyC was incubated with SpyTag-rVAR2 for 1 h at room temperature.
Patient samples
The collection of human tissue in this study abided by the Helsinki Principles and the General Data Protection Regulation (GDPR) of individual member states. Placental tissue was obtained from three pregnant women delivering at Rigshospitalet University Hospital, Copenhagen, Denmark. Human tissue stem was from individuals who had bequeathed their bodies to science and education at the Institute of Cellular and Molecular Medicine (ICMM), University of Copenhagen, according to Danish legislation (Health Law #546, §55 and §188). The study was approved by the head of the Body Donation Program at ICMM. Ethical committee permission is not necessary, as the individuals have donated their bodies for scientific purposes. Permission from the Data Protection Agency is also not necessary, as the donators are anonymized. The colon cancer samples were obtained from patients undergoing intended curative surgery at the Department of Surgery, Zealand University Hospital, Køge, Denmark. All participants provided informed consent for the tissue to be used for research purposes. The enrollment of patients was approved by the Regional Ethics Committee, Region of Zealand, Denmark (approval no. SJ-492), and the Danish Data Protection Agency (approval no. REG-141-2015). All samples were deidentified entirely before transfer to the researchers, and this did not need specific IRB approval.
GAG purification from tissues
Upon retrieval, tissues were washed in dPBS (Gibco) to remove residual blood and stored at −80 °C. The tissues were lyophilized until dry, powderized with a spatula, and resuspended in dPBS. The tissue was treated with 1 mg/ml Pronase (from Streptomyces griseus, Sigma Aldrich) in 10 mM CaCl2 and 0.1% Triton X-100 at 37 °C, overnight, shaking. The samples were centrifuged at 20,000g for 20 min, and supernatants were filtered through a 5 μm membrane filter (Sigma) and mixed 1:10 with equilibration buffer (50 mM sodium acetate, 0.2 M NaCl, 0.1% Triton X-100, pH 6.0). 0.5 ml bed volume DEAE Sephacel (GE healthcare) in disposable columns (2 ml bed volume Poly-Prep Chromatography Columns, Bio-Rad) were equilibrated in equilibration buffer, and samples were loaded onto the column. The columns were washed in 20 ml wash buffer (50 mM sodium acetate, 2.0 M NaCl, pH 6.0) and GAGs were eluded in 2.5 ml elution buffer (50 mM NaOAc, 0.2 M NaCl, pH 6.0). After elution, samples were added 99% ethanol saturated with sodium acetate (1:3, V:V) in a 1:3 ratio and stored at −20 °C overnight. GAGs were precipitated by centrifugation at 20,000g, 4 °C, for 20 min. Residual ethanol was evaporated in a centrifugal evaporator, and the remaining pellets were reconstituted in 100 uL DNase buffer (50 mM Tris, 50 mM NaCl, 2.5 mM MgCl2, 0.5 mM CaCl2, pH 8.0). The samples were digested with 20 kU/ml DNaseI (Deoxyribonuclease I from Bovine Pancreas, Sigma Aldrich) for 2 h at 37 °C, shaking, and supplemented with 10 μl 10× heparinase buffer (400 mM ammonium acetate and 33 mM calcium acetate, pH 7.0) and 200 mU/ml heparinase I–III + 20 U/ml hyaluronidase (from Streptomyces hyalurolyticus, Sigma Aldrich), following a 4 h incubation at 37 °C. The samples were β-eliminated overnight at 4 °C in 0.4 M NaOH and then neutralized with acetic acid. Finally, the CS was purified over a DEAE column and ethanol precipitated as described previously.
rVAR2 affinity chromatography
A HiTrap NHS HP 1 ml or 5 ml column (GE Healthcare) was activated according to vendor specifications and immobilized with 700 μg/ml SpyCatcher in dPBS. The column was inactivated with ethanolamine and washed according to vendor specifications. The column was equilibrated in five column volumes of dPBS and loaded with 1 mg/ml SpyTag-rVAR2 in dPBS. After 1 h incubation, the column was washed with five column volumes dPBS. 200 μg/ml purified tissue CS in dPBS was loaded onto the column and allowed to incubate for 15 min. The flow-through was reloaded an additional three passages and followed by a five column volume dPBS wash. For purification of on-column digested ofCS (R-ofCS), 200 mU/ml S-ChABC in digestion buffer (50 mM Tris, 50 mM ammonium acetate, pH 8.0) was loaded onto the column and incubated for 1.5 h. The column was washed with 3 ml digestion buffer, and this was collected for analysis of liberated ofCS (L-ofCS). For purification of intact ofCS (I-ofCS) the digestion step was omitted. Finally, the column was washed with five column volumes dPBS, three column volumes 0.25 M NaCl, and R- or I-ofCS were eluted with three column volumes 2 M NaCl. Eluents were precipitated by ethanol as described above.
For analytical chromatography analysis, AMAC-labeled R-ofCS was applied to the 1 ml HiTrap SpyCatcher-rVAR2 column. The column was washed with 5 ml dPBS and bound CS was eluted with a gradient of 0.15–2.0 M NaCl, containing 20 mM HEPES, pH 7.4, using a NGQ Quest 10 Plus chromatography system. Fluorescence intensity was detected on eluded fractions and plotted using GraphPad Prism v8. For fractionation followed by dp2 analysis, R-ofCS bound to a 1 ml HiTrap SpyCatcher-rVAR2 column was washed with 0.25 M NaCl and eluded in four fractions of 3 ml 0.4 M, 0.6 M, 0.8 M, and 1.0 M NaCl.
CS biotinylation and size-exclusion chromatography (SEC)
To generate biotinylated peptide-CS, CS was purified as previously described, but β-elimination was omitted. The peptide-CS was incubated with a 50 M excess of EZ-Link Sulfo-NHS-LC-Biotin (Thermo Scientific) in dPBS for 1 h. The biotin-CS was size-fractionated by SEC using a 1.5 × 50 cm column packed with Sepharose 6B (GE healthcare). The mobile phase consisted of the wash buffer described above. Five milligram sample was loaded onto the column and 80 1.5 ml fractions were collected under gravity. A214nm values were measured on every other fraction and the carbazole assay was performed on every four fractions.
Carbazole assay
CS fractions obtained from SEC were quantified by the carbazole assay (51). D-glucuronic acid standards (Sigma-Aldrich) were weighed out and dissolved in ddH2O at a concentration of 1 mg/ml. Duplicates of these, ranging from 0 to 10 μg, were diluted in wash buffer (100 μl), and 100 ul of every four GAG fractions were used for analysis. The samples were added 10 μl 4 M ammonium sulfamate (Sigma-Aldrich) and 0.5 ml 25 mM sodium tetraborate (Sigma Aldrich) in 98% sulfuric acid (Sigma-Aldrich). The samples were heated for 5 min in a 100 °C water bath, cooled, and mixed with 20 μl 0.1% carbazole (Eastman Organic Chemicals) in methanol. The samples were incubated at 100 °C for 15 min, cooled, and transferred to a 96-well plate. Absorbance at 520 nm was measured on an EnSpire Multimode Plate Reader (PerkinElmer), and a standard curve was derived by plotting absorbance of the uronic acid standards against their concentrations using GraphPad Prism v8 software.
AMAC labeling and length analysis
Labeling with 2-Aminoacridone (AMAC) and length analysis were done as previously described (27). Briefly, CS was labeled with AMAC (Sigma Aldrich) and reduced with sodium cyanoborohydride (Sigma Aldrich). The samples were lyophilized, dissolved, and precipitated with acetone over three rounds. The samples were reconstituted in 2% acetonitrile and fluorescence intensity was measured (excitation λ = 428 nm, emission λ = 525 nm). A standard curve was derived from the AMAC-labeled D0a4 standard (Sigma Aldrich). The ratio of the quantified CS, determined by MS analysis, to the concentration of chains, determined by AMAC labeling, was used to calculate the average molecular mass of the substrates.
GAG-preparation for LC-MS analysis
For CS/DS quantification and dp2 analysis, purified CS was digested with 50 mU/ml S-ChABC for 2 h at 37 °C in lyase buffer (50 mM Tris and 50 mM NaCl, pH 8.0). For tetrasaccharide analysis, the CS samples were digested with A-ChABC, and for CS and DS analysis, the samples were digested with ChAC and ChB, respectively. The samples were dried in a SpeedVac centrifuge and tagged by reductive amination with [12C6]aniline, as described (26).
LC-MS2 analysis with collision-induced dissociation
The [12C6]aniline tagged samples were mixed with 20 pmol of [13C6]aniline-labeled mixed dp2 standards and analyzed by LC-MS/MS with collision-induced dissociation (CID), as previously described (26, 27). The samples were separated using 5 mM of the ion-pairing agent dibutyl amine (Sigma-Aldrich) on a reverse-phase column (TARGA C18, 150 mm × 1.0 mm diameter, 5 μm beads, Higgins Analytical, Inc). The ions were monitored in negative mode using the same capillary temperature, gradient, and spray voltage as described (26). The analysis was done on a LTQ Orbitrap Discovery electrospray ionization mass spectrometer (Thermo Scientific) equipped with an Ultimate 3000 quaternary HPLC pump (Dionex). For CID analysis of CS tetrasaccharides (496.6, z = 2), ions were selected using a 2-atomic mass unit window and activated with 35% normalized collision energy.
Length analysis by Fourier transform mass spectrometry
Full-length R-ofCS GAG chains were analyzed on a 9.4 T Bruker Apex Qe Fourier transform mass spectrometer equipped with an Infinity cell. The instrument was externally calibrated to approximately 1 ppm prior to analysis. Samples were diluted to ∼0.1 mg/ml in 50:50 aqueous methanol prior to ionization by static nano-electrospray (nanoESI). Ions were externally accumulated for 0.5 s prior to detection over the range of 200–2000 m/z. Thirty-six, 1 M-word acquisitions were coadded to produce each mass spectrum. The assignment of R-ofCS chain compositions was based on accurate mass measurement and determined with in-house software.
Monosaccharide analysis
Monosaccharide analysis of CS/DS samples was performed as previously described (27). Briefly, samples were dissolved in Milli-Q water and 4 M trifluoroacetic acid, vortexed, and capped. The samples were hydrolyzed at 100 °C for 4 h and centrifuged, followed by evaporation of the supernates. When fully dried, the samples were dissolved in 50% isopropanol and solvents were evaporated. Finally, the samples were dissolved in Milli-Q water and analyzed by high-performance anion-exchange chromatography (HPAEC) with pulsed amperometric detection (PAD) using a Dionex ICS-3000 system equipped with a strong anion-exchange column (Dionex CarboPac PA1 column 4 mm × 250 mm, with a 4 mm × 50 mm Guard). The samples were monitored with a pulsed amperometer using quadruple potential waveform and using the same gradient as previously described (27).
Polyacrylamide gel electrophoresis (PAGE)
Ten microgram CS sample was dissolved in 40 μl 50% sucrose and 0.01% bromophenol blue (Sigma Aldrich) and loaded on a 10–20% (w/v) Tris-Glycine polyacrylamide gel (Invitrogen). Electrophoresis was performed at 150 V for 2 h in a 25 mM Tris-HCl/0.2 M glycine buffer. The gel was stained with 0.5% Alcian Blue 8GX (Sigma Aldrich) in 2% acetic acid for 2 h and subsequently destained with 2% acetic acid overnight before visualization. For the detection of biotinylated CS, the CS was preconjugated to Cy5-Streptavidin for 20 min, followed by PAGE, and the fluorescence signal was detected at 700 nm using an imaging system (Odyssey, Li-Cor Inc).
Tissue culture
HEK293 T cells (ATCC CRL-3216) and HeLa cells (ATCC CCL-2) were grown in DMEM medium containing 10% FBS and 100 IU/ml of penicillin and 100 μg/ml of streptomycin sulfate. HUVEC (ATCC CRL-1730) cells were grown in EGM-2 endothelial cell growth media (VWR International, LLC) containing 2% FBS and VEGF. The HUVEC cells were grown on gelatin-coated plates for ∼12 days before lifting for flow cytometry analysis. All cells were grown under an atmosphere of 5% CO2 and 95% air. Cells were passaged before 70–80% confluence was reached and seeded as explained for the individual assays. Turkey red blood cells (Lampire biological laboratories) were stored at 4 °C and used within a month of receipt.
Cell line generation
To generate a Cas9 lentiviral expression plasmid, 2.5 × 106 HEK293 T cells were seeded on a 10-cm diameter plate in DMEM/FBS. The following day, the cells were cotransfected with the psPAX2 packaging plasmid (Addgene plasmid #12260), pMD2.g envelope plasmid (Addgene plasmid #12259), and lenti-Cas9 plasmid (Addgene plasmid #52962) in DMEM supplemented with Fugene6 (Promega). Lentivirus-containing media was collected after 3 days and used to infect HeLa WT cells, which were subsequently cultured with 4 μg/ml blasticidin to select for stably transduced cells.
A single guide RNA (sgRNA) targeting CHST14 (5′-CTGTCCGCACACCGCCCGCA-3′), CHPF (5′-GGAACACC ACACGCTCCAGC-3′), or CHPF2 (5′-CAGTGAAGTAGAGT AACCGA-3′) was cloned into the lentiGuide-Puro plasmid (Addgene plasmid #52963) following published procedure (52). The lentiviral sgRNA construct was generated in HEK293T cells, using the same protocol as for the Cas9 expression plasmid, and used to infect HeLa-Cas9 cells and generate CRISPR/Cas9-mediated mutant cell lines. After infection, the cells were cultured with 2 μg/ml puromycin to select for cells with stably integrated lentivirus. After 7 days of selection, cells were serially diluted into 96-well plates for single colony expansion (Corning). DNA was extracted from individual clones using the DNeasy Blood and Tissue Kit (Qiagen). The DNA was sequenced (Genewiz Inc), and proper editing was analyzed using the online ICE tool from Synthego (Fig. S3).
GAG purification from cells
Cells were grown to 70–80% in a 10-cm diameter plate and washed with dPBS, lifted with 4 ml trypsin (Gibco), and centrifuged for 5 min at 500g. The trypsin supernatants were collected for GAG purification, and the cell pellets were washed in complete media, followed by a dPBS wash, and saved for BCA analysis (Pierce BCA Protein Assay Kit, Thermo Fisher), according to vendor specifications. The trypsin-released GAGs were Pronase digested and purified by DEAE-Sephacel chromatography, as described above. The GAGs were desalted using PD-10 columns (GE Healthcare), digested with DNase, β-eliminated, and purified over a second DEAE-Sephacel and PD-10 column.
Radiolabeling and CS length analysis
HeLa-Cas9 or CHPF/CHPF2 mutant cells (1 × 106) were radiolabeled with [35S]sulfate (250 μCi per plate) in F12 media containing 10% dialyzed FBS for 24 h at 37 °C. Cell surface CS was purified and analyzed by SEC using the methods described above. Radioactivity in each fraction was detected using the Beckman LS6500 instrument. CS average molecular mass was determined based on previous size determinations using a Sepharose 6B column (53).
Flow cytometry
HeLa cells were detached using 10 mM Ethylenediaminetetraacetic acid (EDTA, Sigma Aldrich) in dPBS and seeded in a 96-well U-bottom plate (100,000 cells/well). The cells were incubated with 20 nM AFDye 647-rVAR2 conjugate in 2% FBS in dPBS (PBS2) for 20 min at 4 °C. The cells were stained for 30 min at 4 °C, washed twice in PBS2, and analyzed by flow cytometry on a FACSCalibur (BD Biosciences) instrument. For inhibition assays, the rVAR2 conjugate was preincubated with the CS/DS substrate for 10 min at room temperature prior to cell staining. All experiments were performed in triplicates. The data were analyzed using Flowjo v7.6 software (BD Biosciences) and binding was quantified by the geometric mean of the fluorescence intensity (MFI).
DSPE-CS conjugation and binding analysis
0.5 μM 1,2-Distearoyl-sn-glycero-3-phosphoethanolamine (DSPE) streptavidin conjugate (Nanocs Inc) was incubated with or without biotinylated CS (1:2, molar ratio) in dPBS for 1 h. Erythrocytes, at a concentration of 5%/ml, and HUVECs, at a concentration of 107 cells/ml, were incubated with the DSPE conjugates in dPBS for 40 min at 37 °C, shaking. After two PBS2 washes, 105 HUVEC cells/well or 100 μl 0.5% erythrocytes/well were seeded into a 96-well plate. Half of the wells were incubated with 500 nM preconjugated AFDye 647-rVAR2, for 30 min at 4 °C. The other half were digested with 200 mU/ml S-ChABC in dPBS for 10 min at room temperature, washed, and stained with anti-CS Delta Di-4S supernatant (Clone 2B6, Amsbio) at 1:200 in PBS2, for 30 min at 4 °C. The 2B6-stained cells were washed twice and stained with FITC conjugated Goat Anti-Mouse IgG Polyclonal Antibody (Neta Scientific) at 1:1000 in PBS2, for 30 min at 4 °C. The cells were washed twice in PBS2 and analyzed by flow cytometry, as described above.
Differential gene expression analysis in cancer versus healthy RNA-seq data
We retrieved RNA-seq gene expression data for 19 cancer subtypes encompassing 17 cancer types by histology (bile duct cancer, bladder cancer, blood cancer, brain cancer, breast cancer, colorectal cancer, head and neck cancer, liver cancer, lung cancer, esophageal cancer, ovarian cancer, pancreatic cancer, prostate cancer, renal cell carcinoma, skin cancer, stomach cancer, thyroid cancer, and uterine cancer). Read count tables were retrieved for 16 of these subtypes from the Cancer Genome Atlas (TCGA) project (https://gdc-portal.nci.nih.gov/). RNA-seq data from primary tumor tissue samples and matched normal tissue samples were retrieved for each cancer subtype.
The gene expression data were preprocessed as previously described (54) using the limma R-package (55). Differential gene expression analysis to assess changes in transcriptional regulation between tumor and normal samples was performed separately for each cancer subtype using the voom R-package (56). For a given gene, changes in transcriptional regulation in either direction, e.g., upregulation or downregulation, from normal tissues were considered statistically significant if we observed a false discovery rate q < 0.001 and a minimum absolute fold-change in expression level >50%.
Statistics
Sample sizes are indicated in the figure legends (∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05; #p < 0). All statistical analysis were done using unpaired t test and performed in GraphPad Prism v8.
Data availability
All data generated in this study are contained within the manuscript (and its supporting information files).
Supporting information
This article contains supporting information (57, 58, 59, 60, 61, 62).
Conflict of interest
The University of California San Diego and J. D. E. have a financial interest in TEGA Therapeutics, Inc. The terms of this arrangement have been reviewed and approved by the University of California, San Diego, in accordance with its conflict-of-interest policies. A. S., T. M. C., and T. G. T. have financial interest in VAR2Pharmaceuticals that holds the rights to use VAR2CSA for cancer treatment and diagnostic purposes.
Acknowledgments
Author contributions
C. B. S., J. D. E., A. S., and T. M. C. conceptualization; C. B. S., J. D. E., A. S., and T. M. C. data curation; C. B. S., A. S., T. M. C., and J. D. E. funding acquisition; C. B. S., A. G. T., P. S., F. G., F. E. L., and T. M. C. investigation; C. B. S., A. G. T., P. S., F. G., F. E. L., and T. M. C. methodology; T. G., S. C., R. P. V., and I. G. resources; Y. M., T. G. T., and I. J. A. supervision; C. B. S., A. G. T., A. S., J. D. E., and T. M. C. writing–original draft; C. B. S., J. D. E. A. S., A. G. T., P. S., F. G., F. E. L., and T. M. C. writing–review and editing.
Funding and additional information
This work was supported in part by grants Gangstedfonden, Louis-Hansen Fonden, and Tømrermester Jørgen Holm og Hustru Elisa f. Hansens Mindelegat to C. B. S., the National Institutes of Health CA199292, HL131474 and GM33063 grants to J. D. E, the National Natural Science Foundation of China (91853131 and 81872786), and Guangdong Provincial Key Laboratory of Drug Nonclinical Evaluation and Research (2018B030323024) grants to Y. M., grants from the Alfred Benzon Foundation and Lundbeck Foundation to T. M. C.
Edited by Gerald Hart
Supporting information
References
- 1.WHO . World Health Organization; Geneva, Switzerland: 2020. World Malaria Report 2020. [Google Scholar]
- 2.Snow R.W., Craig M., Deichmann U., Marsh K. Estimating mortality, morbidity and disability due to malaria among Africa's non-pregnant population. Bull. World Health Organ. 1999;77:624–640. [PMC free article] [PubMed] [Google Scholar]
- 3.David P.H., Hommel M., Miller L.H., Udeinya I.J., Oligino L.D. Parasite sequestration in Plasmodium falciparum malaria: Spleen and antibody modulation of cytoadherence of infected erythrocytes. Proc. Natl. Acad. Sci. U. S. A. 1983;80:5075–5079. doi: 10.1073/pnas.80.16.5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fried M., Duffy P.E. Adherence of Plasmodium falciparum to chondroitin sulfate A in the human placenta. Science. 1996;272:1502–1504. doi: 10.1126/science.272.5267.1502. [DOI] [PubMed] [Google Scholar]
- 5.Fried M., Nosten F., Brockman A., Brabin B.J., Duffy P.E. Maternal antibodies block malaria. Nature. 1998;395:851–852. doi: 10.1038/27570. [DOI] [PubMed] [Google Scholar]
- 6.Gardner M.J., Hall N., Fung E., White O., Berriman M., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S., Paulsen I.T., James K., Eisen J.A., Rutherford K., Salzberg S.L., et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duffy M.F., Maier A.G., Byrne T.J., Marty A.J., Elliott S.R., O'Neill M.T., Payne P.D., Rogerson S.J., Cowman A.F., Crabb B.S., Brown G.V. VAR2CSA is the principal ligand for chondroitin sulfate A in two allogeneic isolates of Plasmodium falciparum. Mol. Biochem. Parasitol. 2006;148:117–124. doi: 10.1016/j.molbiopara.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Khunrae P., Dahlbäck M., Nielsen M.A., Andersen G., Ditlev S.B., Resende M., Pinto V.V., Theander T.G., Higgins M.K., Salanti A. Full-length recombinant Plasmodium falciparum VAR2CSA binds specifically to CSPG and induces potent parasite adhesion-blocking antibodies. J. Mol. Biol. 2010;397:826–834. doi: 10.1016/j.jmb.2010.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salanti A., Dahlbäck M., Turner L., Nielsen M.A., Barfod L., Magistrado P., Jensen A.T.R., Lavstsen T., Ofori M.F., Marsh K., Hviid L., Theander T.G. Evidence for the involvement of VAR2CSA in pregnancy-associated malaria. J. Exp. Med. 2004;200:1197–1203. doi: 10.1084/jem.20041579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pereira M.A., Clausen T.M., Pehrson C., Mao Y., Resende M., Daugaard M., Kristensen A.R., Spliid C., Mathiesen L., Knudsen L.E., Damm P., Theander T.G., Hansson S.R., Nielsen M.A., Salanti A. Placental sequestration of Plasmodium falciparum malaria parasites is mediated by the interaction between VAR2CSA and chondroitin sulfate A on syndecan-1. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Afratis N., Gialeli C., Nikitovic D., Tsegenidis T., Karousou E., Theocharis A.D., Pavão M.S., Tzanakakis G.N., Karamanos N.K. Glycosaminoglycans: Key players in cancer cell biology and treatment. FEBS J. 2012;279:1177–1197. doi: 10.1111/j.1742-4658.2012.08529.x. [DOI] [PubMed] [Google Scholar]
- 12.Silbert J.E., Sugumaran G. Biosynthesis of chondroitin/dermatan sulfate. IUBMB Life. 2002;54:177–186. doi: 10.1080/15216540214923. [DOI] [PubMed] [Google Scholar]
- 13.Esko J.D., H. Prestegard J., Linhardt R.J. Essentials of Glycobiology. 3rd Ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2015. Chapter 38: Proteins that bind sulfated glycosaminoglycans. [Google Scholar]
- 14.Salanti A., Staalsoe T., Lavstsen T., Jensen A.T.R., Sowa M.P.K., Arnot D.E., Hviid L., Theander T.G. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol. Microbiol. 2003;49:179–191. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- 15.Salanti A., Clausen T.M., Agerbæk M.Ø., Al Nakouzi N., Dahlbäck M., Oo H.Z., Lee S., Gustavsson T., Rich J.R., Hedberg B.J., Mao Y., Barington L., Pereira M.A., LoBello J., Endo M., et al. Targeting human cancer by a glycosaminoglycan binding malaria protein. Cancer Cell. 2015;28:500–514. doi: 10.1016/j.ccell.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toledo A.G., Pihl J., Spliid C.B., Persson A., Nilsson J., Pereira M.A., Gustavsson T., Choudhary S., Oo H.Z., Black P.C., Daugaard M., Esko J.D., Larson G., Salanti A., Clausen T.M. An affinity-chromatography and glycoproteomics workflow to profile the chondroitin sulfate proteoglycans that interact with malarial VAR2CSA in the placenta and in cancer. Glycobiology. 2020;30:989–1002. doi: 10.1093/glycob/cwaa039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srivastava A., Gangnard S., Dechavanne S., Amirat F., Lewit Bentley A., Bentley G.A., Gamain B. Var2CSA minimal CSA binding region is located within the N-terminal region. PLoS One. 2011;6 doi: 10.1371/journal.pone.0020270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clausen T.M., Christoffersen S., Dahlbäck M., Langkilde A.E., Jensen K.E., Resende M., Agerbæk M.Ø., Andersen D., Berisha B., Ditlev S.B., Pinto V.V., Nielsen M.A., Theander T.G., Larsen S., Salanti A. Structural and functional insight into how the Plasmodium falciparum VAR2CSA protein mediates binding to chondroitin sulfate A in placental malaria. J. Biol. Chem. 2012;287:23332–23345. doi: 10.1074/jbc.M112.348839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alkhalil A., Achur R.N., Valiyaveettil M., Ockenhouse C.F., Gowda D.C. Structural requirements for the adherence of Plasmodium falciparum-infected erythrocytes to chondroitin sulfate proteoglycans of human placenta. J. Biol. Chem. 2000;275:40357–40364. doi: 10.1074/jbc.M006399200. [DOI] [PubMed] [Google Scholar]
- 20.Beeson J.G., Andrews K.T., Boyle M., Duffy M.F., Choong E.K., Byrne T.J., Chesson J.M., Lawson A.M., Chai W. Structural basis for binding of Plasmodium falciparum erythrocyte membrane protein 1 to chondroitin sulfate and placental tissue and the influence of protein polymorphisms on binding specificity. J. Biol. Chem. 2007;282:22426–22436. doi: 10.1074/jbc.M700231200. [DOI] [PubMed] [Google Scholar]
- 21.Sugiura N., Clausen T.M., Shioiri T., Gustavsson T., Watanabe H., Salanti A. Molecular dissection of placental malaria protein VAR2CSA interaction with a chemo-enzymatically synthesized chondroitin sulfate library. Glycoconj. J. 2016;33:985–994. doi: 10.1007/s10719-016-9685-z. [DOI] [PubMed] [Google Scholar]
- 22.Chai W., Beeson J.G., Lawson A.M. The structural motif in chondroitin sulfate for adhesion of Plasmodium falciparum-infected erythrocytes comprises disaccharide units of 4-O-sulfated and non-sulfated N-acetylgalactosamine linked to glucuronic acid. J. Biol. Chem. 2002;277:22438–22446. doi: 10.1074/jbc.M111401200. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y.-H., Narimatsu Y., Clausen T.M., Gomes C., Karlsson R., Steentoft C., Spliid C.B., Gustavsson T., Salanti A., Persson A., Malmström A., Willén D., Ellervik U., Bennett E.P., Mao Y., et al. The GAGOme: A cell-based library of displayed glycosaminoglycans. Nat. Methods. 2018;15:881–888. doi: 10.1038/s41592-018-0086-z. [DOI] [PubMed] [Google Scholar]
- 24.Rogerson S.J., Chaiyaroj S.C., Ng K., Reeder J.C., Brown G.V. Chondroitin sulfate A is a cell surface receptor for Plasmodium falciparum-infected erythrocytes. J. Exp. Med. 1995;182:15–20. doi: 10.1084/jem.182.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakano T., Betti M., Pietrasik Z. Extraction, isolation and analysis of chondroitin sulfate glycosaminoglycans. Recent Patents Food Nutr. Agric. 2012;2:61–74. doi: 10.2174/2212798411002010061. [DOI] [PubMed] [Google Scholar]
- 26.Lawrence R., Olson S.K., Steele R.E., Wang L., Warrior R., Cummings R.D., Esko J.D. Evolutionary differences in glycosaminoglycan fine structure detected by quantitative glycan reductive isotope labeling. J. Biol. Chem. 2008;283:33674–33684. doi: 10.1074/jbc.M804288200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spliid C.B., Toledo A.G., Salanti A., Esko J.D., Clausen T.M. Beware, commercial chondroitinases vary in activity and substrate specificity. Glycobiology. 2021;31:103–115. doi: 10.1093/glycob/cwaa056. [DOI] [PubMed] [Google Scholar]
- 28.Mizumoto S., Kosho T., Hatamochi A., Honda T., Yamaguchi T., Okamoto N., Miyake N., Yamada S., Sugahara K. Defect in dermatan sulfate in urine of patients with Ehlers-Danlos syndrome caused by a CHST14/D4ST1 deficiency. Clin. Biochem. 2017;50:670–677. doi: 10.1016/j.clinbiochem.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 29.Honigfort D.J., Zhang M.H., Verespy S., Godula K. Engineering of spectator glycocalyx structures to evaluate molecular interactions at crowded cellular boundaries. Faraday Discuss. 2019;219:138–153. doi: 10.1039/c9fd00024k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barker A.L., Konopatskaya O., Neal C.R., Macpherson J.V., Whatmore J.L., Winlove C.P., Unwin P.R., Shore A.C. Observation and characterisation of the glycocalyx of viable human endothelial cells using confocal laser scanning microscopy. Phys. Chem. Chem. Phys. 2004;6:1006–1011. [Google Scholar]
- 31.Fried M., Lauder R.M., Duffy P.E. Plasmodium falciparum: Adhesion of placental isolates modulated by the sulfation characteristics of the glycosaminoglycan receptor. Exp. Parasitol. 2000;95:75–78. doi: 10.1006/expr.2000.4510. [DOI] [PubMed] [Google Scholar]
- 32.Achur R.N., Kakizaki I., Goel S., Kojima K., Madhunapantula S.V., Goyal A., Ohta M., Kumar S., Takagaki K., Gowda D.C. Structural interactions in chondroitin 4-sulfate mediated adherence of Plasmodium falciparum infected erythrocytes in human placenta during pregnancy-associated malaria. Biochemistry. 2008;47:12635–12643. doi: 10.1021/bi801643m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugiura N., Shioiri T., Chiba M., Sato T., Narimatsu H., Kimata K., Watanabe H. Construction of a chondroitin sulfate library with defined structures and analysis of molecular interactions. J. Biol. Chem. 2012;287:43390–43400. doi: 10.1074/jbc.M112.412676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Achur R.N., Valiyaveettil M., Alkhalil A., Ockenhouse C.F., Gowda D.C. Characterization of proteoglycans of human placenta and identification of unique chondroitin sulfate proteoglycans of the intervillous spaces that mediate the adherence of Plasmodium falciparum-infected erythrocytes to the placenta. J. Biol. Chem. 2000;275:40344–40356. doi: 10.1074/jbc.M006398200. [DOI] [PubMed] [Google Scholar]
- 35.Agerbæk M.Ø., Bang-Christensen S.R., Yang M.-H., Clausen T.M., Pereira M.A., Sharma S., Ditlev S.B., Nielsen M.A., Choudhary S., Gustavsson T., Sorensen P.H., Meyer T., Propper D., Shamash J., Theander T.G., et al. The VAR2CSA malaria protein efficiently retrieves circulating tumor cells in an EpCAM-independent manner. Nat. Commun. 2018;9:3279. doi: 10.1038/s41467-018-05793-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hou X., Zhang T., Da Z., Wu X. CHPF promotes lung adenocarcinoma proliferation and anti-apoptosis via the MAPK pathway. Pathol. Res. Pract. 2019;215:988–994. doi: 10.1016/j.prp.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Cao C., Liu Y., Wang Q., Zhao J., Shi M., Zheng J. Expression of CHPF modulates cell proliferation and invasion in lung cancer. Braz. J. Med. Biol. Res. 2020;53:19021. doi: 10.1590/1414-431X20209021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun W., Zhao F., Xu Y., Huang K., Guo X., Zheng B., Liu X., Luo Z., Kong Y., Xu M., Schadendorf D., Chen Y. Chondroitin polymerizing factor (CHPF) promotes development of malignant melanoma through regulation of CDK1. Cell Death Dis. 2020;11:496. doi: 10.1038/s41419-020-2526-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan Y.H., Xiao B., Lv S.G., Ye M.H., Zhu X.G., Wu M.J. Lentivirus-mediated knockdown of chondroitin polymerizing factor inhibits glioma cell growth in vitro. Oncol. Rep. 2017;38:1149–1155. doi: 10.3892/or.2017.5731. [DOI] [PubMed] [Google Scholar]
- 40.Frey A., Giannasca K.T., Weltzin R., Giannasca P.J., Reggio H., Lencer W.I., Neutra M.R. Role of the glycocalyx in regulating access of microparticles to apical plasma membranes of intestinal epithelial cells: Implications for microbial attachment and oral vaccine targeting. J. Exp. Med. 1996;184:1045–1059. doi: 10.1084/jem.184.3.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reitsma S., Slaaf D.W., Vink H., Van Zandvoort M.A.M.J., Oude Egbrink M.G.A. The endothelial glycocalyx: Composition, functions, and visualization. Pflugers Arch. Eur. J. Physiol. 2007;454:345–359. doi: 10.1007/s00424-007-0212-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vink H., Duling B.R. Identification of distinct luminal domains for macromolecules, erythrocytes, and leukocytes within mammalian capillaries. Circ. Res. 1996;79:581–589. doi: 10.1161/01.res.79.3.581. [DOI] [PubMed] [Google Scholar]
- 43.Nieuwdorp M., Van Haeften T.W., Gouverneur M.C.L.G., Mooij H.L., Van Lieshout M.H.P., Levi M., Meijers J.C.M., Holleman F., Hoekstra J.B.L., Vink H., Kastelein J.J.P., Stroes E.S.G. Loss of endothelial glycocalyx during acute hyperglycemia coincides with endothelial dysfunction and coagulation activation in vivo. Diabetes. 2006;55:480–486. doi: 10.2337/diabetes.55.02.06.db05-1103. [DOI] [PubMed] [Google Scholar]
- 44.Nieuwdorp M., Meuwese M.C., Mooij H.L., Ince C., Broekhuizen L.N., Kastelein J.J.P., Stroes E.S.G., Vink H. Measuring endothelial glycocalyx dimensions in humans: A potential novel tool to monitor vascular vulnerability. J. Appl. Physiol. 2008;104:845–852. doi: 10.1152/japplphysiol.00440.2007. [DOI] [PubMed] [Google Scholar]
- 45.Oohira A., Wight T.N., Bornstein P. Sulfated proteoglycans synthesized by vascular endothelial cells in culture. J. Biol. Chem. 1983;258:2014–2021. [PubMed] [Google Scholar]
- 46.Henry C.B.S., Duling B.R. Permeation of the luminal capillary glycocalyx is determined by hyaluronan. Am. J. Physiol. 1999;277:H508–H514. doi: 10.1152/ajpheart.1999.277.2.H508. [DOI] [PubMed] [Google Scholar]
- 47.Matejevic D., Neudeck H., Graf R., Müller T., Dietl J. Localization of hyaluronan with a hyaluronan-specific hyaluronic acid binding protein in the placenta in pre-eclampsia. Gynecol. Obstet. Invest. 2001;52:257–259. doi: 10.1159/000052986. [DOI] [PubMed] [Google Scholar]
- 48.Rieger H., Yoshikawa H.Y., Quadt K., Nielsen M.A., Sanchez C.P., Salanti A., Tanaka M., Lanzer M. Cytoadhesion of Plasmodium falciparum - infected erythrocytes to chondroitin-4-sulfate is cooperative and shear enhanced. Blood. 2015;125:383–391. doi: 10.1182/blood-2014-03-561019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ebong E.E., Macaluso F.P., Spray D.C., Tarbell J.M. Imaging the endothelial glycocalyx in vitro by rapid freezing/freeze substitution transmission electron microscopy. Arterioscler. Thromb. Vasc. Biol. 2011;31:1908–1915. doi: 10.1161/ATVBAHA.111.225268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bang-Christensen S.R., Pedersen R.S., Pereira M.A., Clausen T.M., Løppke C., Sand N.T., Ahrens T.D., Jørgensen A.M., Lim Y.C., Goksøyr L., Choudhary S., Gustavsson T., Dagil R., Daugaard M., Sander A.F., et al. Capture and detection of circulating glioma cells using the recombinant VAR2CSA malaria protein. Cells. 2019;8:998. doi: 10.3390/cells8090998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bitter T., Muir H.M. A modified uronic acid carbazole reaction. Anal. Biochem. 1962;4:330–334. doi: 10.1016/0003-2697(62)90095-7. [DOI] [PubMed] [Google Scholar]
- 52.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wasteson Å. A method for the determination of the molecular weight and molecular-weight distribution of chondroitin sulphate. J. Chromatogr. A. 1971;59:87–97. doi: 10.1016/s0021-9673(01)80009-1. [DOI] [PubMed] [Google Scholar]
- 54.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 56.Law C.W., Chen Y., Shi W., Smyth G.K. Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanderson P., Stickney M., Leach F.E., Xia Q., Yu Y., Zhang F., Linhardt R.J., Amster I.J. Heparin/heparan sulfate analysis by covalently modified reverse polarity capillary zone electrophoresis-mass spectrometry. J. Chromatogr. A. 2018;1545:75–83. doi: 10.1016/j.chroma.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin L., Liu X., Zhang F., Chi L., Amster I.J., Leach F.E., Xia Q., Linhardt R.J. Analysis of heparin oligosaccharides by capillary electrophoresis–negative-ion electrospray ionization mass spectrometry. Anal. Bioanal. Chem. 2017;409:411–420. doi: 10.1007/s00216-016-9662-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun L., Zhu G., Zhao Y., Yan X., Mou S., Dovichi N.J. Ultrasensitive and fast bottom-up analysis of femtogram amounts of complex proteome digests. Angew. Chem. Int. Ed. 2013;52:13661–13664. doi: 10.1002/anie.201308139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun L., Zhu G., Zhang Z., Mou S., Dovichi N.J. Third-generation electrokinetically pumped sheath-flow nanospray interface with improved stability and sensitivity for automated capillary zone electrophoresis-mass spectrometry analysis of complex proteome digests. J. Proteome Res. 2015;14:2312–2321. doi: 10.1021/acs.jproteome.5b00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ceroni A., Maass K., Geyer H., Geyer R., Dell A., Haslam S.M. GlycoWorkbench: A tool for the computer-assisted annotation of mass spectra of glycans. J. Proteome Res. 2008;7:1650–1659. doi: 10.1021/pr7008252. [DOI] [PubMed] [Google Scholar]
- 62.Domon B., Costello C.E. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj. J. 1988;5:397–409. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated in this study are contained within the manuscript (and its supporting information files).