Abstract
p53 is a well-established critical cell cycle regulator. By inducing transcription of the gene encoding p21, p53 inhibits cyclin-dependent kinase (CDK)-mediated phosphorylation of cell cycle inhibitor retinoblastoma (RB) proteins. Phosphorylation of RB releases E2F transcription factor proteins that transactivate cell cycle–promoting genes. Here, we sought to uncover the contribution of p53, p21, CDK, RB, and E2F to the regulation of ferroptosis, an oxidative form of cell death. Our studies have uncovered unexpected complexity in this regulation. First, we showed that elevated levels of p53 enhance ferroptosis in multiple inducible and isogenic systems. On the other hand, we found that p21 suppresses ferroptosis. Elevation of CDK activity also suppressed ferroptosis under conditions where p21 suppressed ferroptosis, suggesting that the impact of p21 must extend beyond CDK inhibition. Furthermore, we showed that overexpression of E2F suppresses ferroptosis in part via a p21-dependent mechanism, consistent with reports that this transcription factor can induce transcription of p21. Finally, deletion of RB genes enhanced ferroptosis. Taken together, these results show that signals affecting ferroptotic sensitivity emanate from multiple points within the p53 tumor suppressor pathway.
Keywords: ferroptosis, retinoblastoma protein, cyclin E, cyclin-dependent kinase
Abbreviations: CDK, cyclin-dependent kinase; GPX, GSH peroxidase; MEFs, mouse embryo fibroblasts; MOI, multiplicity of infection; RB/p105RB, retinoblastoma protein; ROS, reactive oxygen species; RSL, ras-selective lethal; SH, safe harbor; TKO, triple knock out
Through an interconnected series of pathways, external growth factors, DNA damage, and other stresses regulate effector proteins that control progression through the cell cycle (1, 2). Mutations in the components of these pathways occur in cancer to drive uncontrolled proliferation (2). The p53 tumor suppressor controls a major pathway that coordinates the cellular response to DNA damage (3). p53 induced in response to DNA damage upregulates the transcription of p21, an inhibitor of cyclin-dependent kinases (CDKs) (4). Because CDKs phosphorylate retinoblastoma (RB) to modulate its activity, this p53/p21/CDK/RB pathway links DNA damage to cell cycle arrest by modulating RB phosphorylation. Phosphorylation of RB by CDKs reduces RB binding to E2F resulting in increased transcription of E2F targets encoding cell cycle proteins (2). Recent studies have added additional complexity to the understanding of phosphoregulation of RB. For example, CDK4/6 bound to D-cyclins phosphorylate RB at single sites to create a heterogeneous mixture of monophosphorylated RB during early G1. Monophosphorylated RB proteins still interact with a host of cellular proteins and regulate transcription of multiple targets (5). As cells pass through G1, CDK2 phosphorylates RB on up to 14 sites to release E2F for cell cycle entry (5, 6). Thus, the effect of phosphorylation on RB depends on the exact phosphorylation state and the specific RB-regulated activity in question.
Cell cycle entry results in elevated rates of oxidative ATP synthesis, which generates reactive oxygen species (ROS) as a byproduct (7). Mechanisms to avoid cellular damage caused by ROS include superoxide dismutases, catalase, and a family of GSH peroxidases (GPX) (8). When ROS generation outpaces antioxidant capacity, damage to membrane lipids and cell lysis occurs. In a number of biological systems, the process of ROS-mediated lipid peroxidation and cell lysis requires iron and has been called ferroptosis (9, 10, 11). Ferroptosis can be induced by blocking the uptake of cystine needed for the synthesis of GSH, a reducing agent used by GPX4 to limit peroxidation of membrane lipids (10, 11). Ferroptosis can also be induced by small-molecule GPX4 inhibitors (12). Therefore, cells maintain a homeostatic balance between ROS generation via metabolic activity and antioxidant capacity. Inhibiting antioxidants allows ROS to accumulate to toxic levels.
Despite intense interest in the regulation of ferroptosis by p53 and its downstream pathways, this area of biology is incompletely understood. In fact, whether p53 enhances (13, 14) or suppresses (15, 16) ferroptosis differs under seemingly similar conditions. In one recent study, p53 delayed ferroptosis in HT1080 fibrosarcoma cells (15). In contrast, our studies indicate p53 enhances ferroptosis in multiple cell types with inducible p53 alleles. Furthermore, we find that Nutlin-3a, which is commonly used to study p53 responses, independently inhibits ferroptosis by an unknown mechanism. Less yet is known about how RB modulates ferroptosis with one article showing a potential role (17). Here, we show that p21, CDKs, RB, and E2F modulate sensitivity to ferroptosis but not in a manner that can be explained by operation of a linear pathway culminating in E2F activation that is well documented in cell cycle research. Thus, modulation of ferroptosis appears to emanate from multiple points within the p53/p21/CDK/RB/E2F pathway.
Results
Regulation of ferroptosis by p53
In a number of reports documenting the effects of p53 on ferroptosis, several apparent inconsistencies exist. For example, p53 was found to induce ferroptosis through enhancing lipid peroxidation enzymes and suppressing SLC7A11 (13, 14). In another set of studies, p53 was proposed to inhibit ferroptosis in certain cell types including HT1080 fibrosarcoma cells (15, 16). To better understand the role of p53 in ferroptosis, we tested multiple different systems with inducible p53 or consisting of isogenic cells differing in p53 status. For most of our studies, we used a novel small molecule from a series of analogs we have named CETZOLES (analogs sharing a common 4-cyclopentenyl-2-ethynylthiazole skeleton) (Fig. 1A) (18, 19). CETZOLE 1 blocks cystine uptake leading to loss of GSH, elevation of total cellular ROS and lipid peroxidation (19). Cell death is blocked by iron chelators and antioxidants and is elevated in mesenchymal cancer cells (18, 19). Thus, the CETZOLEs show many of the hallmarks of inducing ferroptosis. Suppression of cystine uptake and glutamate secretion suggest that these compounds inhibit the function of the system xc− transporter. In some experiments, we compared CETZOLE 1 with Erastin, a well-documented inhibitor of cystine uptake. Erastin also targets mitochondria by binding to VDAC2, which may contribute to its ability to induce ferroptosis (20). We also repeated some of our studies with the GPX4 inhibitor ras-selective lethal 3 (RSL3).
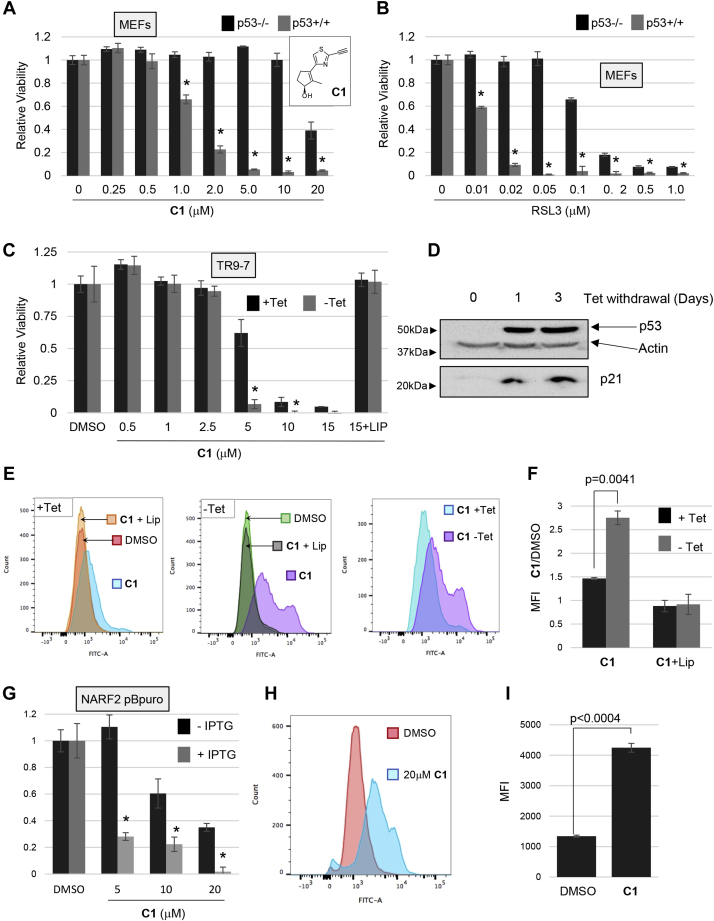
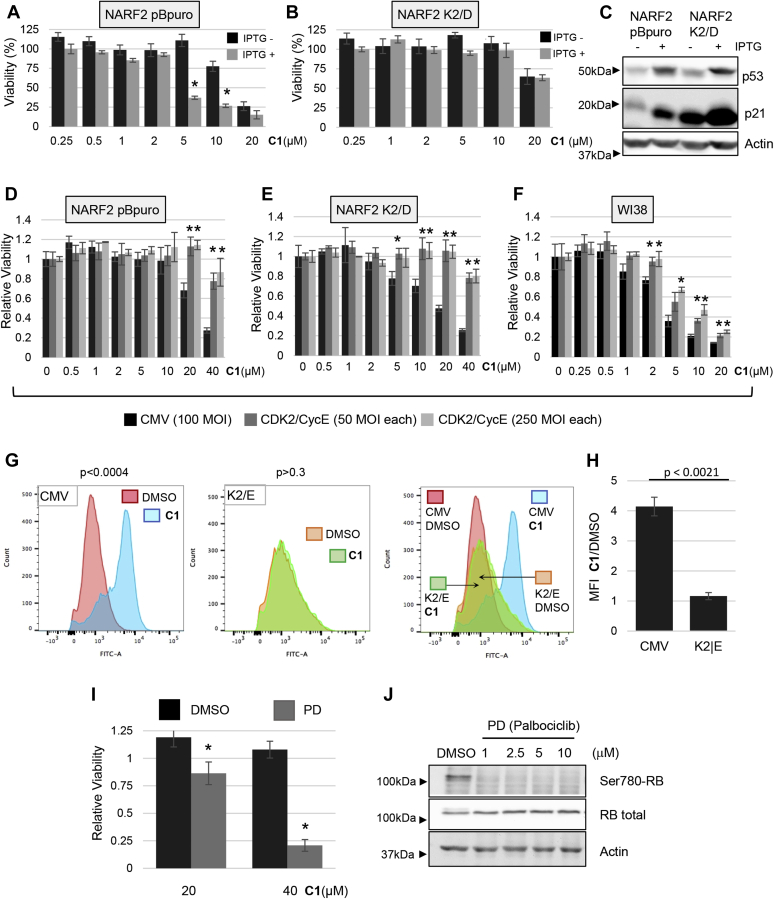
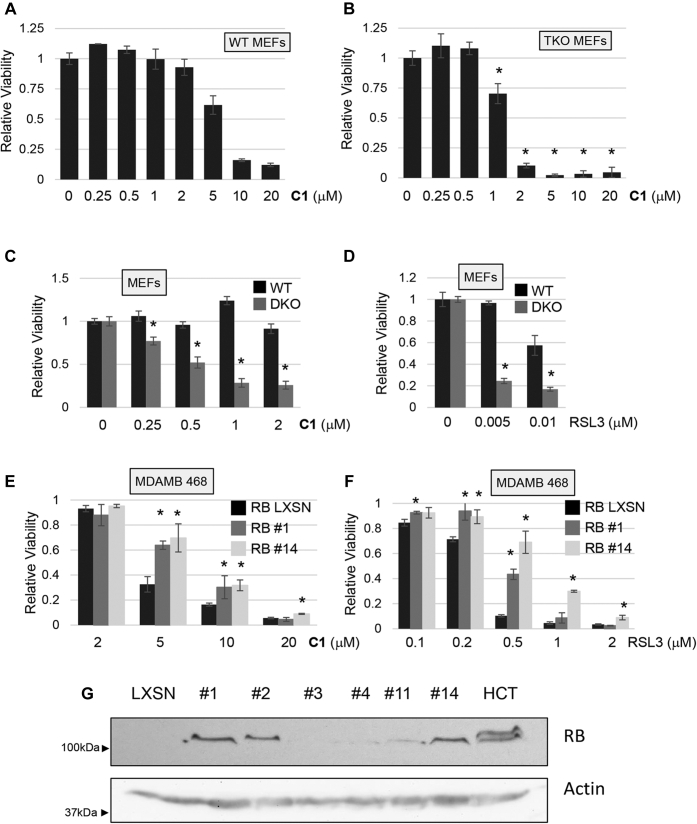
Figure 1.
p53 enhances ferroptosis. Cells differing in p53 status or with inducible p53 were tested for response to ferroptosis inducers. A and B, p53−/− and p53+/+ MEFs were plated and exposed to cystine uptake inhibitor CETZOLE 1 (C1) or a GPX-4 inhibitor RSL3 the next day. Cell viability was determined 72 h after treatment. Structure of CETZOLE 1 is shown in the inset to panel A. C, TR9-7 cells were grown with or without tetracycline for 3 days and then exposed to CETZOLE 1 for 48 h followed by viability analysis. Lipid ROS scavenger Liproxstatin-1 (Lip: 0.25 μM) was added along with 15 μM C1 to assess whether death of TR9-7 cells was consistent with ferroptosis. D, expression of p53 in TR9-7 cells upon tetracycline withdrawal was detected by Western blotting. E, analysis of lipid peroxidation using BODIPY-C11. TR9-7 cells were incubated with or without tetracycline for 3 days and then exposed to 6 μM CETZOLE 1 (C1) for 24 h. BODIPY-C11 was also added to cultures together with CETZOLE 1 followed by analysis by flow cytometry. In some samples, CETZOLE 1 was combined with 0.25 μM liproxstatin-1. F, the mean fluorescence intensity (MFI) of BODIPY-C11 plots obtained as in “E” are shown as a ratio of CETZOLE 1/DMSO (n = 3). G, viability of NARF2 cells with IPTG-inducible p14ARF. Cells were exposed to IPTG for 24 h and then to CETZOLE 1 (20 μM) for 24 h followed by analysis of viability. H and I, lipid peroxidation in NARF2 cells determined by staining with BODIPY-C11. Cells were exposed to IPTG for 24 h to induce p14ARF and p53. CETZOLE 1 or DMSO was added for 24 h along with BODIPY-C11 dye. Representative flow cytometry profiles are shown in (H) and MFI in (I) with and without CETZOLE 1. GPX, GSH peroxidase; MEFs, mouse embryo fibroblasts; ROS, reactive oxygen species; RSL3, ras-selective lethal 3. ∗ indicates p<0.05 using a student's t-test comparing two adjacent bars.
p53−/− mouse embryo fibroblasts (MEFs) (21) were more resistant to CETZOLE 1 and RSL3 than WT MEFs (Fig. 1, A and B). Next, we tested a human cell line expressing a tetracycline-regulated p53 allele. TR9-7 cells were derived from MDAH041 cells, a p53-null immortalized adult human fibroblast cell line from a patient with Li-Fraumeni syndrome (22, 23). WT p53 was reconstituted using a tetracycline-regulated system (24). TR9-7 cells incubated without tetracycline for 3 days showed elevated p53 and p21 and exhibited enhanced ferroptosis when exposed to CETZOLE 1 (Fig. 1, C and D). Moreover, liproxstatin-1, a lipid ROS scavenger, eliminated killing of TR9-7 cells by CETZOLE 1 consistent with ferroptosis as a mode of death in this cell line (Fig. 1C). Analysis of membrane lipid peroxidation using the redox-sensitive dye BODIPY-C11 showed that induction of p53 in TR9-7 cells significantly elevated membrane damage by CETZOLE 1 (Fig. 1, E and F). Lipid peroxidation is a hallmark of ferroptosis. To expand these observations, we analyzed NARF2 cells derived from osteosarcoma cell line U2OS with WT p53. NARF2 express an IPTG-inducible p14ARF gene. p14ARF protein disrupts the p53-MDM2 interaction leading to elevated p53 upon IPTG addition (25). Addition of IPTG to NARF2 cells for 24 h enhanced ferroptosis in response to CETZOLE 1 and RSL3 (Fig. 1G and our unpublished results). BODIPY-C11 oxidation was induced by CETZOLE 1 in NARF2 cells pretreated with IPTG, confirming that this cell type undergoes ferroptosis under these conditions (Fig. 1, H and I). Together, these systems show that ferroptosis induced by CETZOLE 1 and RSL3 is enhanced by p53.
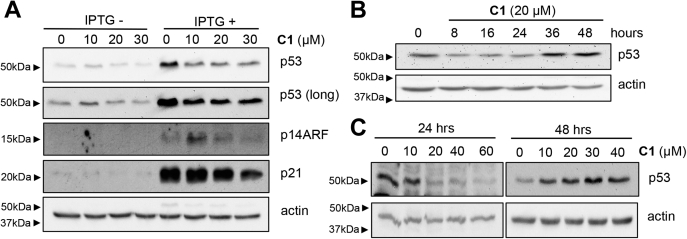
As expected, p14ARF, p53, and p21 were elevated in NARF2 cells exposed to IPTG (Fig. 2A). Exposure of NARF2 cells to CETZOLE 1 for 24 h caused a dose-dependent reduction in the level of p53 (Fig. 2, A and C). To assess the time course of this response, NARF2 without IPTG were exposed to CETZOLE 1 because most cells were killed at later time points when CETZOLE 1 was combined with IPTG. The effect of CETZOLE 1 was biphasic with p53 levels rebounding above untreated levels after 36 and 48 h of treatment (Fig. 2, B and C). p53 participates in a negative feedback loop that causes oscillations in p53 levels (26, 27). Specifically, Mdm2 is both a transcriptional target of p53 and an E3 ligase targeting p53 for degradation. Whether Mdm2 contributes to the biphasic p53 response in CETZOLE 1-treated cells is unknown.
Figure 2.
Western blot analysis of p53, p21, and p14ARF in NARF2 cells.A, NARF2 cells were pretreated with or without IPTG for 24 h and exposed to CETZOLE 1 for another 24 h at indicated concentrations. Western blotting was used to analyze p53, p21, p14ARF, and actin as a loading control. B, time course of p53 expression in NARF2 cells after CETZOLE 1 treatment. Cells were incubated without IPTG and then exposed to CETZOLE 1 (20 μM) for the times indicated. C, p53 expression in cells exposed to different doses of CETZOLE 1 at two time points (24 h and 48 h). NARF2 cells without IPTG were exposed to CETZOLE 1 as indicated and analyzed by Western blotting.
Nutlin-3a inhibits ferroptosis
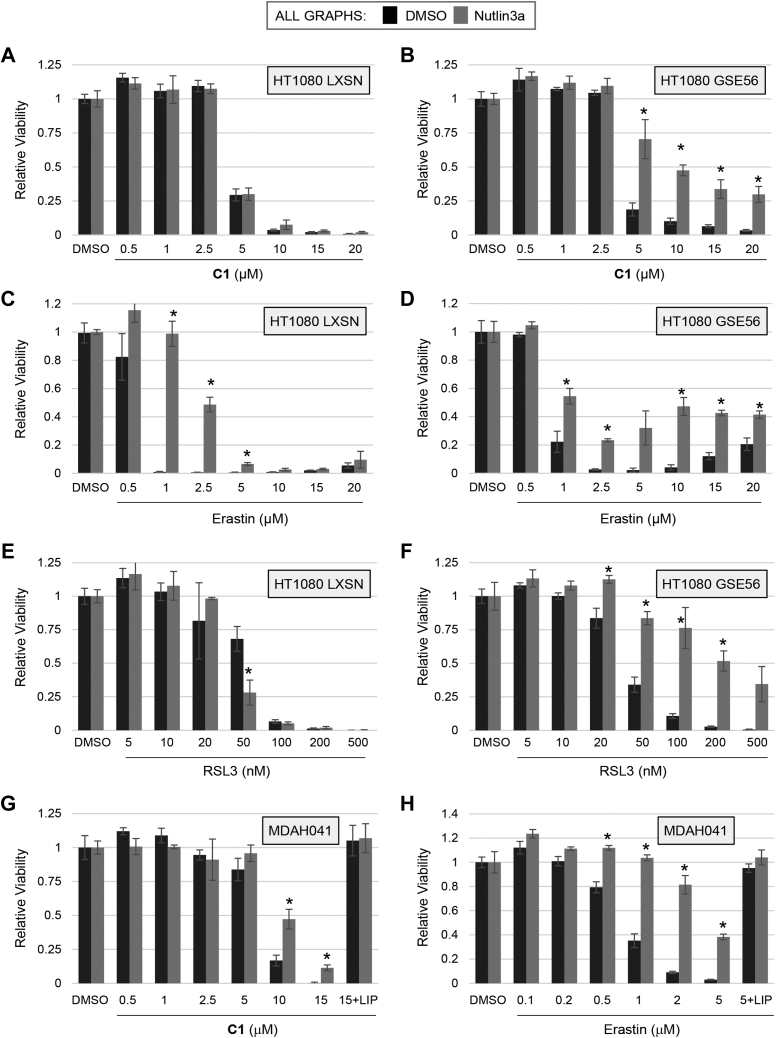
In a recent report, p53 was found to suppress ferroptosis in HT1080 fibrosarcoma cells (15). In some of those experiments, p53 was induced by the small molecule Nutlin-3a that binds to Mdm2, blocking its interaction with p53 (28, 29). Our observations using inducible systems did not appear to be consistent with this finding. Therefore, we tested the effect of Nutlin-3a on CETZOLE 1 sensitivity in HT1080. To test the role of p53, we analyzed HT1080 fibrosarcoma cells expressing a dominant-negative p53 fragment (GSE56) (30) and compared with parental cells with WT p53 expressing the empty retroviral vector (LXSN).
Surprisingly, Nutlin-3a did not affect the sensitivity of HT1080-LXSN cells to CETZOLE 1 but instead protected HT1080-GSE56 cells from this compound (Fig. 3, A and B). Nutlin-3a reduced ferroptosis induced by Erastin in both cell types (Fig. 3, C and D). Nutlin-3a enhanced RSL3 toxicity in HT1080-LXSN but only at the 50 nM dose (Fig. 3E). In contrast, Nutlin-3a protected HT1080-GSE cells at multiple doses of RSL3 (Fig. 3F). These observations suggested that Nutlin-3a inhibited ferroptosis in a p53-independent manner because the effect was most evident in cells expressing the dominant-negative GSE56 fragment. To confirm this idea, we analyzed the effect of Nutlin-3a in p53-deficient MDAH041 cells. MDAH041 are an immortalized human fibroblast cell line derived from a patient with Li-Fraumeni syndrome (22, 23). One p53 allele contains a frame-shift mutation at position 148 and the other allele was deleted during establishment of the cell line (31). p53 protein is undetectable in this cell line (24, 32, 33, 34). Nutlin-3a inhibited ferroptosis in MDAH041 cells treated with either CETZOLE 1 or with Erastin (Fig. 3, G and H). As expected, our Western blot analysis showed no detectable p53 or p21 in MDAH041 cells (Fig. 4A). Together, these results indicate that Nutlin-3a inhibits ferroptosis in a p53-independent manner.
Figure 3.
Effects of Nutlin-3a and p53 on ferroptosis in HT1080 and MDAH041 cells.A–F, HT1080 LXSN (HT-LXSN) and HT1080 GSE56 (HT-GSE56) cells were pretreated with Nutlin-3a (10 μM) for 24 h and then exposed to CETZOLE 1 (A and B), Erastin (C and D), or RSL3 (E and F). Viability was determined using methylene blue staining 48 h later. G and H, p53-deficient MDAH041 fibroblasts were pretreated with Nutlin-3a (10 μM) for 24 h and then exposed to CETZOLE 1 for 48 h followed by analysis of viability. Liproxstatin (LIP: 0.25 μM) was added in samples indicated. In all cases, cell viability was determined using methylene blue staining (absorbance at 668 nm). Bars represent the mean, and error bars represent the SD. ∗p < 0.05 in all cases generated using Student's t test to compare adjacent bars. RSL3, ras-selective lethal 3.
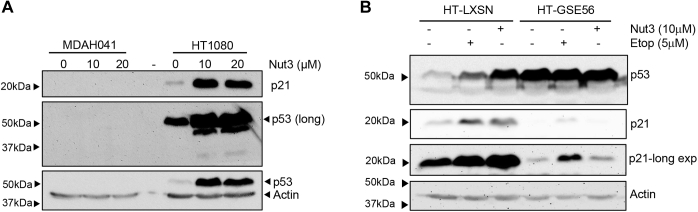
Figure 4.
Expression of p53 and p21 in MDAH01 and HT1080 cells. Western blot analysis of p53 and p21 expression. A, Western blot of MDAH041 cells exposed to Nutlin-3a (Nut) for 24 h. HT1080 were included as a positive control for p53 and p21 expression. Actin was assessed to determine loading. B, Western blot analysis of HT1080-LXSN and HT1080-GSE56 cells exposed to either etoposide or Nutlin-3a (Nut3) for 24 h.
Western blotting confirmed that both Nutlin-3a and etoposide upregulate p53 and p21 in HT1080-LXSN cells (Fig. 4B). As we have previously observed, p53 levels were already elevated in untreated HT1080-GSE56 cells, but p21 levels were diminished (Fig. 4B) (30). Although expressed at a lower level, p21 was still induced by etoposide in HT1080-GSE56 cells, whereas Nutlin-3a had no effect on p21 in these cells (Fig. 4B).
Elevated CDK activity suppresses ferroptosis
Our studies using inducible systems indicate that p53 enhances ferroptosis. To investigate downstream effects of p53, we analyzed NARF2 cells expressing a constitutive CDK2–cyclin D1 fusion protein (NARF2 K2/D). As a control, we analyzed NARF2 containing the empty retroviral vector (NARF2-pBpuro). Without IPTG, NARF2 K2/D cells were slightly more resistant to ferroptosis than NARF2 pBpuro cells (Fig. 5, A and B). Furthermore, IPTG had nearly no effect on ferroptotic sensitivity in NARF2 K2/D cells (Fig. 5, A and B). Western blotting confirmed induction of p53 and p21 after IPTG addition (Fig. 5C). We also noted that basal levels of p21 (minus IPTG) were elevated in NARF2 K2/D compared with the control NARF2 pBpuro (Fig. 5C).
Figure 5.
Elevated CDK activity suppresses ferroptosis.A, NARF2 pBpuro and (B) NARF2 K2/D expressing a constitutive CDK2–cyclin D1 fusion protein were exposed to IPTG, and then, 1 day later, exposed to CETZOLE 1 (C1) to trigger ferroptosis. Viability was determined 48 h after CETZOLE 1 treatment. Average values with SDs are shown. ∗p < 0.05 to compare IPTG+ and IPTG- samples. C, Western blot of p53 and p21 in the cell lines indicated 24 h after adding IPTG. D, NARF2 pBpuro, (E) NARF2 K2/D, and (F) WI-38 cells were plated and transduced with empty (CMV) or CDK2 and cyclin E adenoviruses at indicated MOIs the next day. Forty-eight hours later, cells were treated with CETZOLE 1, and viability was measured. ∗p < 0.05 when compared with CMV infected cells. G and H, lipid peroxidation analyzed using BODIPY-C11. NARF2 cells were infected with adenoviruses to express CDK2 and cyclin E, or with empty CMV virus (MOI-100 each). CETZOLE 1 and BODIPY-C11 dye were added 48 h later. After another 48 h, cells were analyzed by flow cytometry. Representative profiles are shown in (G) and the mean fluorescence intensity (MFI) ratios in (H), with p value from a Student's t test indicated. I, NARF2 pBpuro cells were pretreated with 1 μM PD 0332991 (Palbociclib) for 48 h, and then, DMSO or CETZOLE 1 was added as indicated. Forty-eight later, cells were harvested. ∗p < 0.05 when compared with DMSO. J, NARF2 pBpuro cells were exposed to different concentrations of PD 0332991, and Western blotting was used to measure p-RB (ser780), total RB, and actin as a loading control. Cell viability was measured using methylene blue staining. p values were generated using the Student's t test. CDK, cyclin-dependent kinase; MOI, multiplicity of infection.
To confirm the effect of CDKs on ferroptosis, we overexpressed CDK2 and cyclin E using recombinant adenoviruses. An empty adenovirus (adeno-CMV) was used as a negative control. CDK2/cyclin E diminished ferroptosis induced by CETZOLE 1 when expressed in NARF2 pBpuro and NARF2 K2/D cells (Fig. 5, D and E). CDK2/cyclin E also inhibited CETZOLE 1-induced ferroptosis when expressed in human embryonic lung fibroblasts (WI-38) (Fig. 5F). Similarly, CDK2/cyclin E overexpression suppressed RSL3- or Erastin-induced cell death in NARF2 pBpuro cells (Fig. S1, A and B). BODIPY-C11 staining indicated that CETZOLE 1 had no effect on lipid peroxidation in NARF2 pBpuro cells expressing adenoviral CDK2 and cyclin E. In fact, the staining profile of CETZOLE 1-treated CDK2/cyclin E-expressing cells overlapped with untreated cells and CMV-infected cells (Fig. 5, G and H).
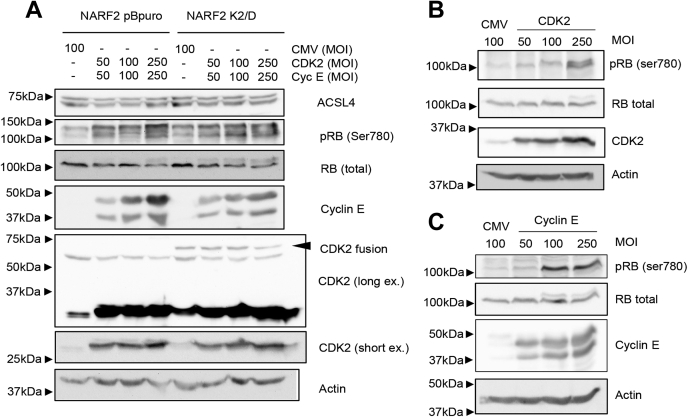
Our results suggest that elevated activity of CDK–cyclin complexes protects from ferroptosis. To test this idea, we used a small-molecule CDK4/6 inhibitor PD 0332991 (Palbociclib). Pretreatment of NARF2 cells with PD 0332991 for 48 h sensitized them to ferroptosis when exposed to CETZOLE 1 (Fig. 5I). Phosphorylation of RB at S780 was reduced upon PD 0332991 treatment, confirming the effect of this compound on CDK activity (Fig. 5J). Western blotting confirmed the expression of the ectopic CDK2 and cyclin E in NARF2 derived from recombinant adenoviruses (Fig. 6). We also observed elevated phosphorylation of RB after expression of CDK2/cyclin E together (Fig. 6A) or on their own in NARF2 cells (Fig. 6, B and C). These experiments show that elevated CDK activity suppresses ferroptosis.
Figure 6.
Western blot analysis of CDK, cyclin, and RB. Western blotting was used to measure levels of CDK2, cyclin E, p-RB (ser780), total RB, ACSL4, and actin. NARF2 pBpuro or K2/D cells were infected with adenoviruses that express CDK2, cyclin E, or an empty vector (CMV) at indicated MOIs and incubated for 48 h before Western blot analysis. A, CDK2 and cyclin E were expressed together. B and C, these proteins were expressed separately in NARF2 pBpuro cells for 48 h before Western blot analysis. CDK, cyclin-dependent kinase; RB, retinoblastoma; MOI, multiplicity of infection.
p21 suppresses ferroptosis
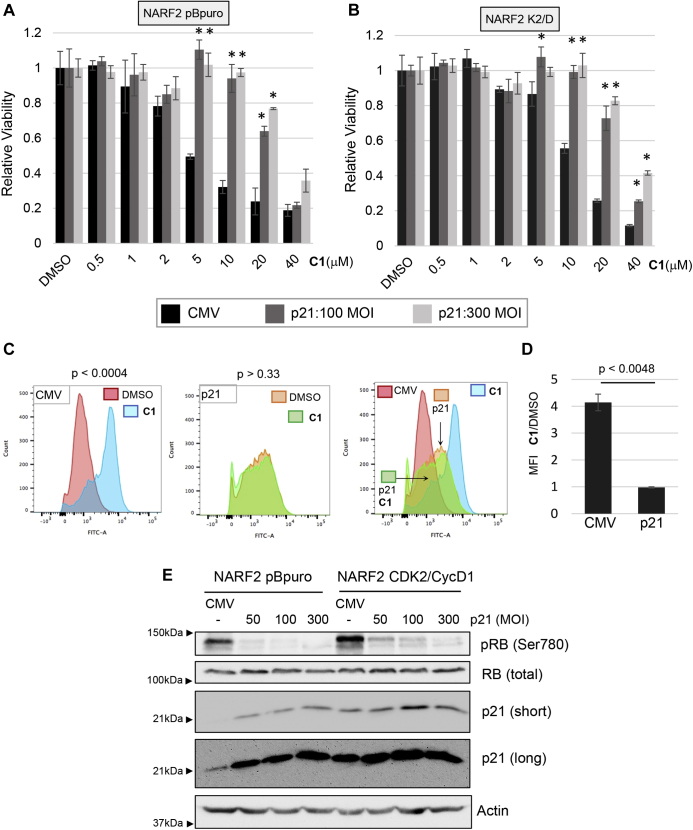
Cells that overexpress the CDK2–cyclin D1 fusion show increased expression of p21 coincident with resistance to ferroptosis (Fig. 5C). In a previous study, p21 was found to inhibit ferroptosis (15). On the other hand, we observed that p53 enhances ferroptosis under conditions where p21 is induced. Therefore, we reassessed the effect of p21 on ferroptosis. Overexpression of p21 using recombinant adenoviruses protected NARF2 cells from ferroptosis induced by CETZOLE 1 (Fig. 7, A and B). This effect was observed in NARF2 pBpuro cells containing an empty vector and in NARF2 cells expressing CDK2/CycD fusion protein (Fig. 7, A and B). CETZOLE 1 had no effect on BODIPY-C11 staining in cells expressing p21, suggesting that this protein has blocked ferroptosis (Fig. 7, C and D). Interestingly, p21 expression consistently elevated BODIPY-C11 staining compared with cells infected with empty CMV virus (Fig. 7C). This elevated staining was intermediate between untreated and CETZOLE 1-treated cells. This observation is consistent with a previous report showing that p21 elevates the cellular ROS level and can induce senescence independently of p53 (35). ROS elevation by p21 was correlated with its ability to induce expression of PIG3, an oxidoreductase (35). We hypothesize that p21 must have additional targets that allow cells to survive elevated ROS. For example, p21 is known to bind to Nrf2 to protect cells from ROS (36).
Figure 7.
Overexpressed p21 inhibits ferroptosis.A and B, NARF2 pBpuro and NARF2 K2/D expressing constitutive CDK2-cyclin D were transduced with p21or CMV (empty) recombinant adenoviruses at different MOIs as indicated. Forty-eight hours later, the cells were treated with CETZOLE 1 (C1) and cell viability was determined after 48 h by methylene blue staining. ∗p < 0.05 when compared with CMV infected cells. C and D, lipid peroxidation as assessed using BODIPY-C11. NARF2-pBpuro cells in the absence of IPTG were infected with adenoviral vectors to overexpress p21 (MOI-100). CETZOLE 1 was added 48 h later along with BODIPY-C11 dye. Flow cytometry was used to assess BODIPY oxidation after 48 h. These p21 overexpression experiments were done simultaneously with CDK2/cyclin E experiments shown earlier. Therefore, in (C), the two left-most panels (CMV, DMSO, and C1) are identical to those in Figure 5G. The mean fluorescence intensity ratios from triplicate samples are shown in (D). p value from a Student's t test is shown. E, Western blot analysis depicts the protein levels of p21, p-RB (ser780), total RB, and actin as a loading control after infection with adenovirus to overexpress p21. CDK, cyclin-dependent kinase; RB, retinoblastoma.
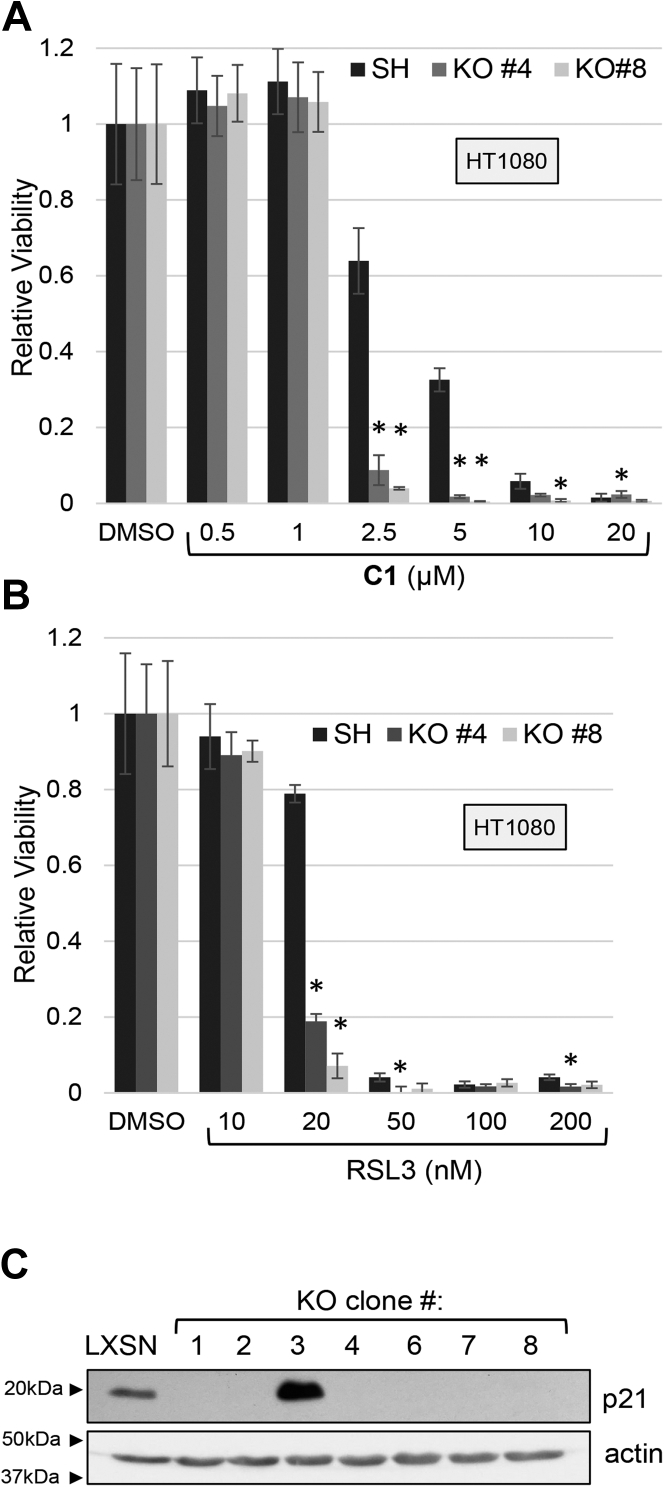
Overexpression of p21 reduced the phosphorylation of RB at S780 (Fig. 7E). Importantly, the small-molecule CDK inhibitor Palbociclib also reduced RB phosphorylation but enhanced ferroptosis (Fig. 5, I and J), unlike overexpressing p21. This suggests that p21 has a role in ferroptosis independent of its ability to inhibit CDK activity. To further probe the role of p21 in ferroptosis, we used CRISPR-CAS9 to disrupt the locus in HT1080 cells with WT p53 and p21. Two independent clones of p21 −/− cells showed enhanced sensitivity to CETZOLE 1 and RSL3 compared with cells expressing the safe-harbor (SH) sgRNA (Fig. 8, A–C). The effects of p21 illustrate part of the complexity of ferroptosis modulation by core cell cycle regulators. For example, p53 enhances ferroptosis at the same time that it induces p21, an inhibitor of ferroptosis. The simplest explanation is that additional p53 targets are strongly proferroptotic such that they overwhelm any inhibitory effect of p21.
Figure 8.
p21-null cells are sensitized to ferroptosis. HT1080 cells lacking p21 were generated with CRISPR-CAS9 using a safe-harbor sgRNA as a negative control. Control SH and KO cells were exposed to either (A) CETZOLE 1 (C1) or (B) RSL3 for 48 h followed by analysis of viability using methylene blue staining. ∗p < 0.05 when compared with parental cell line expressing the safe-harbor sgRNA and CAS9. C, Western blot analysis to confirm p21 KO. Membrane was probed with p21 and actin antibodies. LXSN are p21+/+ HT1080 cells transduced with an empty LXSN retrovirus. p values were generated using the Student's t test. SH, safe harbor; RSL3, ras-selective lethal 3.
Role of RB and E2F in ferroptosis
One of major targets of CDK activity is the RB family consisting of p130, p107, and p105RB. RB family regulates redox homeostasis suggesting that it may affect ferroptosis (37). Strikingly, we found that MEFs lacking all three RB family proteins (triple knock out [TKO]) (38) were more sensitive to ferroptosis than WT MEFs (Fig. 9, A and B). This result is similar to a previous finding in which RB-deficient cells were more sensitive to ferroptosis induced by sorafenib (17). We observed that MEFs lacking p107 and p130 (39, 40) were also more sensitive to ferroptosis than WT MEFs from littermate controls when exposed to either CETZOLE 1 or the RSL3 inhibitor (Fig. 9, C and D). MDA MB 468 (breast cancer) cells do not express endogenous p105RB; thus, we transfected WT RB and isolated individual clones. Two independent clones re-expressing p105RB were more resistant to ferroptosis induced by either CETZOLE 1 or RSL3 than control MDA MB 468 LXSN cells (Fig. 9, E–G). These findings show that the RB family inhibits ferroptosis.
Figure 9.
RB inhibits ferroptosis. The cells indicated were exposed to either CETZOLE 1 (C1) or RSL3 to induce ferroptosis. Viability was determined 48 h later using methylene blue staining. WT and RB family triple KO (TKO) cells are shown in (A) and (B), whereas cells lacking p107 and p130 (DKO) are shown in (C) and (D). ∗p < 0.05 when comparing KO with the corresponding WT cell line. E–G, MDA MB 468 cells which normally lack p105RB were reconstituted with WT p105RB by stable transfection. A negative control cell line was prepared by stable transfection with empty LXSN plasmid. Clones #1 and #14 expressing RB were exposed to (E) CETZOLE 1 or (F) RSL3 for 72 h and analyzed for viability. ∗p < 0.05 when comparing RB expressing clones with control LXSN cells. G, Western blotting was used to identify clones expressing p105RB. Actin was used as a loading control. p values were generated using the Student's t test. DKO, double KO; RB, retinoblastoma; RSL3, ras-selective lethal 3.
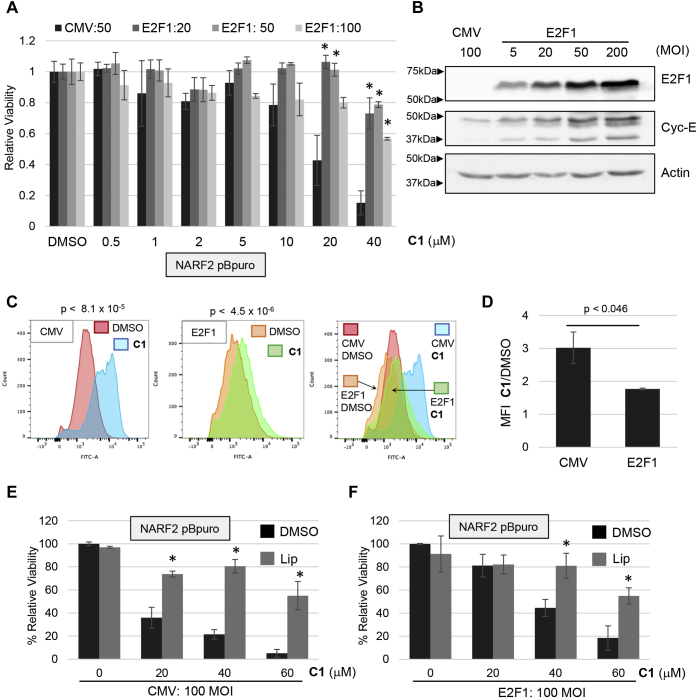
One important function of RB is to regulate the E2F family, itself consisting of nine members. Therefore, we expressed human E2F1 using a recombinant empty adenovirus (adeno-CMV) as a negative control and tested ferroptosis. Overexpression of E2F1 inhibited killing of NARF2 cells by CETZOLE 1 (Fig. 10, A and B). CETZOLE 1 was no longer able to elevate lipid peroxides after overexpression of E2F1 as determined using BODIPY-C11 staining (Fig. 10, C and D). CETZOLE 1-induced cell death upon CMV or E2F1 adenoviral transduction was rescued by liproxstatin-1 (Fig. 10, E and F). Thus, E2F1 inhibits ferroptosis. E2F2 also inhibited ferroptosis but less effectively than E2F1 (our unpublished data). Once again, comparison of the effects of the RB family and E2F1 overexpression reveal complexities in the regulation of ferroptosis. For example, deletion of RB elevates E2F-dependent transcription of many cell cycle genes. However, RB deletion and overexpression of E2F1 have opposite effects on ferroptosis.
Figure 10.
E2F1 inhibits ferroptosis.A, NARF2 pBpuro cells were infected E2F1 expressing adenovirus at the indicated MOI. CETZOLE 1 was added 2 days later and viability determine 48 h later using methylene blue staining. B, Western blotting was used to measure E2F1 and cyclin E in NARF2 pBpuro cells 48 h after infection with adenoviruses at the indicated MOI. Actin was used as a loading control. ∗p < 0.05 when compared with CMV infected using the Student's t test. C and D, BODIPY-C11 staining to assess lipid peroxidation. NARF2 pBpuro cells were infected with recombinant adenoviruses for 48 h (MOI-100). CETZOLE 1 (20 μM) was cotreated with BODIPY stain for another 48 h. Flow cytometry profiles are shown in (C) and mean fluorescence intensity (MFI) ratios in (D). E and F, NARF2 cells were cotreated with CETZOLE 1 and Liproxsatin-1 (Lip) upon E2F1 or CMV (MOI-100) transduction. Cell viability was determined after 48-h treatment. p value is from a Student's t test. MOI, multiplicity of infection.
Inhibition of ferroptosis by E2F1 depends on p21
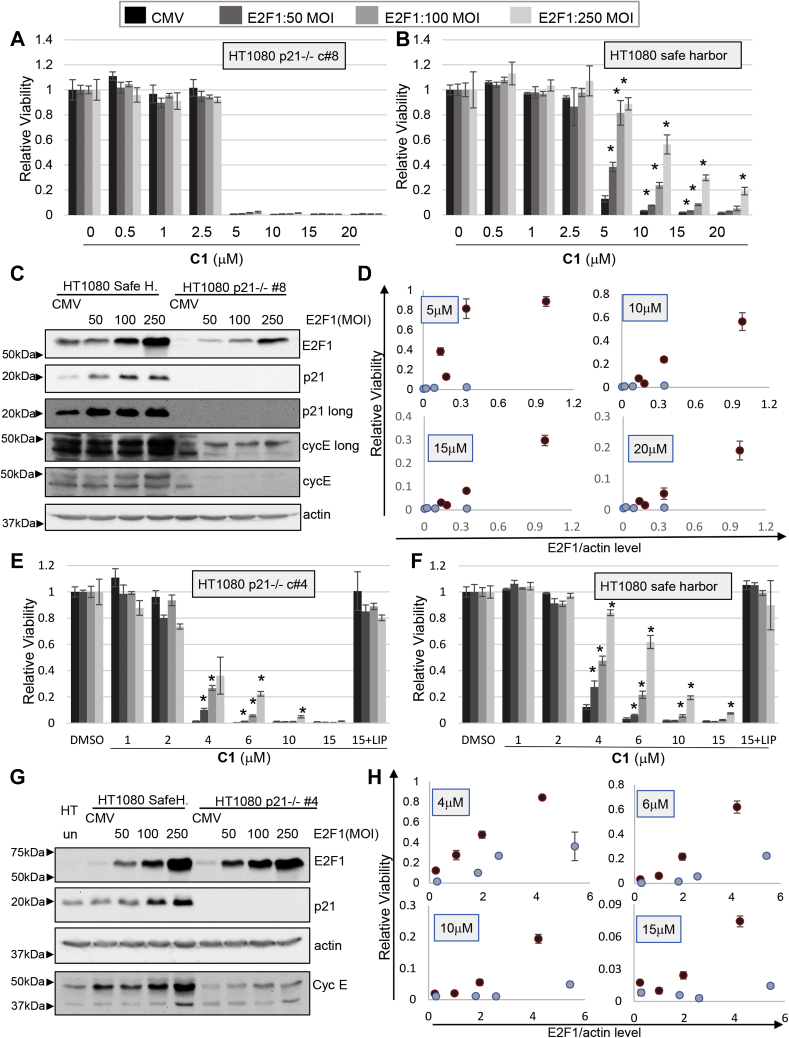
NARF2 cells expressing the constitutive CDK2/cyclin D1 fusion had elevated levels of p21 (Figs. 5C and 7E). One explanation is based on the previous identification of functionally important E2F-binding sites in the p21 promoter (41). Thus, constitutive CDK2/cyclin D1 may elevate E2F leading to increased p21 transcription. Furthermore, the ability of E2F1 to suppress ferroptosis might in fact be mediated by p21. To test this idea, we overexpressed E2F1 using recombinant adenoviruses in p21-KO HT1080 cells from two independent clones (clone #4 and #8). E2F1 had no effect on ferroptosis induced by CETZOLE 1 in p21 KO clone #8 (Fig. 11, A and B). As predicted, p21 was induced after overexpression of E2F1 (Fig. 11, C and G). However, we observed that the basal level of E2F1 in clone #8 cells was lower than the SH control cell line (Fig. 11C). As a consequence, the total amount of E2F1 after adenovirus transduction is higher in SH cells than clone #8 cells. Therefore, an alternative explanation is that adenoviral E2F1 does not protect p21-null cells because expression is too low. To test this idea, we quantified E2F1 expression levels using Western blots visualized with a digital imager. By plotting E2F1 levels versus relative viability, we observed that when E2F1 levels were comparable, p21-null cells were still less protected from ferroptosis (Fig. 11D). For example, the expression level of E2F1 after infection of clone #8 cells with 250 multiplicity of infection (MOI) of adenovirus is similar to the level of E2F1 using 100 MOI of virus in SH HT1080 cells. Under these conditions, survival of p21−/− cells is substantially lower than SH cells (Fig. 11D).
Figure 11.
E2F1 inhibition of ferroptosis requires p21. HT1080 cells lacking p21 or controls cells expressing the safe-harbor gRNA were infected with recombinant adenoviruses to express E2F1. Two KO clones ([clone #8: A–D] and [clone #4: E and F]) were analyzed. Ferroptosis was induced by exposure to CETZOLE 1 (C1), which was added 48 h after virus infection. Cell viability was determined 48 h after adding CETZOLE 1 using methylene blue staining. Viability is presented as average relative viability + SD. ∗p < 0.05 when compared with CMV-infected cells at the same dose of CETZOLE 1. Multiplicity of infection (MOI) is indicated. A and B, viability in response to CETZOLE 1 and E2F1 expression in safe-harbor and clone #8 cells. C, Western blot analysis shows the level of p21, E2F1, cyclin E, and actin proteins upon E2F1 transduction in p21 −/− clone #8 cells. D, relative viability is plotted as a function of E2F1 expression level determined by quantifying Western blot signals. Each symbol represents average viability and E2F1 expression values at a particular MOI. Clone #8 cells (blue symbols) express less E2F1 than safe harbor cells (red symbols) so viability should be compared at similar E2F1 values and not equal MOI. Error bars represent the SD of cell viability of each dose. E and F, viability analysis in safe-harbor and clone #4 cells. Liproxstatin-1 (0.25 μM) was added along 15 μM CETZOLE 1 to assess the contribution of lipid oxidation to cell death. G, Western blot analysis after E2F1 transduction in clone #4 cells. H, plot of viability versus E2F1 expression levels measured from Western blots. Clone #4 cells (blue symbols) express similar levels of E2F1 as safe-harbor cells (red symbols) when infected at the same viral MOI.
Next, we tested a second independent p21−/− clone (#4). In clone #4 cells, E2F1 expression after adenoviral delivery was similar in KO and control cells (Fig. 11G). In clone #4 cells, E2F1 retained a partial ability to block CETZOLE 1 killing; however, the effect is still more robust in the SH cells (Fig. 11, E, F, and H). These observations suggest that E2F1 protects from ferroptosis via both p21-dependent and p21-independent mechanisms.
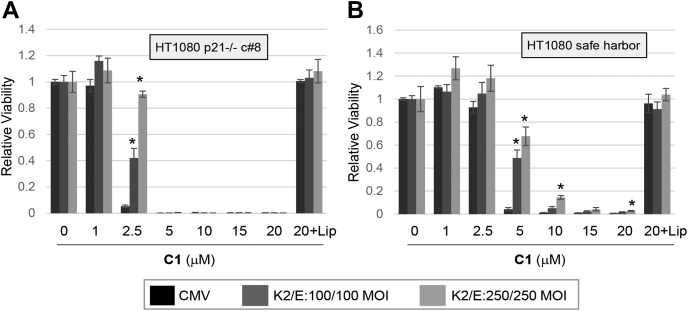
In contrast to E2F1, overexpression of CDK2 and cyclin E inhibited ferroptosis in p21−/− cells to a similar extent as SH cells (Fig. 12, A and B). Thus, although constitutive CDKs elevate p21 expression, this effect does not appear to be a major factor in ferroptosis suppression by CDKs. We also observed that both p21−/− clones expressed reduced levels of cyclin E (Fig. 11, C and G), possibly linked to the activity of p21 as a CDK/cyclin assembly factor (42).
Figure 12.
CDK2/cyclin E inhibition of ferroptosis does not require p21.A, p21 KO HT080 cells (clone #8) or (B) controls expressing the safe-harbor gRNA were infected with recombinant adenoviruses at the indicated MOI (multiplicity of infection) to express CDK2 and cyclin E. Ferroptosis was induced by adding CETZOLE 1 (C1) 48 h after virus infection. Cell viability was determined 48 h after adding CETZOLE 1 using methylene blue staining. Average values with SDs are shown. ∗p < 0.05 when compared with CMV-infected cells at the same dose of CETZOLE 1.
Discussion
Tumor suppressor p53 has been implicated as both a positive and negative modulator of ferroptosis (14, 16, 43, 44, 45, 46). In a series of studies, p53 enhanced ferroptosis by transcriptionally repressing SLC7A11, leading to the activation of arachidonate 12 lipoxygenase (13, 14). In other reports, however, p53 prevented ferroptosis potentially by inhibiting dipeptidyl-peptidase-4 (15, 16). As a consequence, the role of p53 in ferroptosis is in need of clarification. To gain more insight into the role of p53 in ferroptosis, we used CETZOLE 1 discovered in our laboratory (18). Cell death induced by CETZOLE 1 has many of the hallmarks of ferroptosis including elevated lipid ROS, loss of reduced GSH, and reversal by ferroptotic inhibitors liproxstatin and ferrostatin (19, 47). Some of the studies reported herein were repeated using the GPX4 inhibitor RSL3 and the well-documented ferroptosis compound Erastin. Using a variety of inducible systems, we observed that p53 enhanced ferroptosis induced by CETZOLE 1. This effect was observed in mouse as well as human cells. All of the cell systems we used were of mesenchymal origin and included fibroblasts (MEFs, TR97, MDAH041, WI38), fibrosarcoma (HT1080), and osteosarcoma cells (NARF2). Mesenchymal cell lines are more susceptible to ferroptosis than epithelial cells (11).
Experiments in HT1080 cells uncovered a p53-independent effect of Nutlin-3a, a small molecule commonly used to induce p53. Nutlin-3a reduced cell killing by CETZOLE 1, RSL3, and Erastin in HT1080 cells expressing GSE56, a dominant-negative targeting p53. Furthermore, suppression of ferroptosis by Nutlin-3a was more effective in HT1080-GSE56 cells that express lower levels of p21 than the parental HT1080-LXSN cells. This suggests that the inhibitory effect of Nutlin-3a does not operate through p21. Suppression of ferroptosis was also observed in p53-deficient Li-Fraumeni fibroblast MDAH041 providing additional evidence for a p53/p21-independent effect (Fig. 13). Nutlin-3a did not elevate CETZOLE-induced killing in p53-proficient HT1080-LXSN cells. While Nutlin-3a can induce p53 in this cell line, we hypothesize that the proferroptotic p53 signal is antagonized by the additional antiferroptotic effect of Nutlin-3a. When GSE56 is expressed, the proferroptotic p53 signal is presumably diminished, and thus, Nutlin-3a shows enhanced protection. These and other observations suggest that multiple proferroptotic and antiferroptotic signals interact to determine the ultimate outcome.
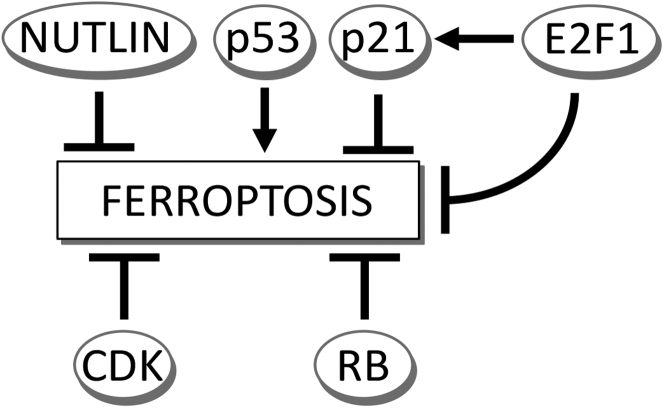
Figure 13.
Multiple proferroptotic and antiferroptotic signals emanate from core cell cycle regulators. We provide evidence that p53 regulates ferroptosis independently of p21. E2F1 functions via p21-dependent and p21-independent mechanisms to suppress ferroptosis. Nutlin-3a inhibits ferroptosis in a p53-independent manner. Both CDKs and RB family modulate ferroptosis. It is possible that CDKs generate phosphorylated RB species to modulate ferroptosis. The classical cell cycle pathway linking these proteins is not shown but includes induction of p21 by p53, inhibition of CDKs by p21, phosphorylation of RB by CDKs, and release of E2F1. All of these regulatory linkages are expected to influence the ultimate ferroptotic outcome. CDKs, cyclin-dependent kinases; RB, retinoblastoma.
The mechanism responsible for p53-independent inhibition of ferroptosis by Nutlin-3a is unknown. In preliminary tests, we found no effect of Nutlin-3a on the absorbance of 2,2-diphenyl-1-(2,4,6-trinitrophenyl)-hydrazyl, suggesting that it is not a general radical scavenger (our unpublished data). Furthermore, fluorescence of Calcein AM in NARF2 cells was unaffected by Nutlin-3a, suggesting that it is not an iron chelator (our unpublished data). Nutlin-3a binds to Bcl-2 or Bcl-XL (48) in addition to Mdm2. Furthermore, Mdm2 enhances ferroptosis independently of p53 (49). Therefore, Nutlin-3a may influence ferroptosis via one of these or other intracellular targets.
p21 is an essential target of p53 in the context of cell cycle control (50). Also, p21 can inhibit ferroptosis (15). While we also observed inhibition of ferroptosis by p21, p53 enhanced ferroptosis under very similar conditions. Thus, p53 must also engage p21-independent mechanisms to regulate ferroptosis. Furthermore, because p53 induces p21 under conditions where ferroptosis is elevated, we hypothesize that proferroptotic signals emanating from p53 must be stronger than antiferroptotic signals from p21. In some contexts, the mode of p53 induction can influence cellular outcomes. For example, dynamic analysis of p53 including single cell measurements showed that induction of this protein in response to stress is oscillatory (26, 27, 51, 52). The mechanism involves induction of the E3 ligase Mdm2 by p53. Because p53 induces an E3 that targets its own degradation, p53 levels, and subsequently Mdm2 levels, will diminish after some time, setting the stage for oscillatory behavior (26, 27). Furthermore, cells that show p53 oscillation recovered from DNA damage, whereas cells that show steady upregulation of p53 underwent senescence (52). In our experiments, induction of p21 using adenovirus would not be expected to be oscillatory because Mdm2 is not involved. However, elevation of p21 in response to p53 might involve Mdm2-dependent oscillations. Therefore, it is possible that the mode of p21 induction can affect ferroptotic outcomes. Currently, we have no direct evidence to address this idea.
Our analysis of the role of p53 in ferroptosis included the cell line NARF2 with IPTG-inducible p14ARF gene. IPTG elevated p53 and increased ferroptosis. IPTG no longer elevated ferroptosis in NARF2 cells expressing a constitutive CDK2/cyclin D fusion protein. This fusion protein was originally created to assess the functional significance of cyclin D1–Cdk2 complexes in cancer cells with cyclin D1 overexpression (53). The antiferroptotic effect of elevated CDK activity was confirmed by overexpressing CDK2 and cyclin E in NARF2 cells and WI38 human fibroblasts using recombinant adenoviruses. Furthermore, NARF2 pBpuro cells were more sensitive to CETZOLE 1 when exposed to the CDK4/6 inhibitor PD 0332991. These observations indicate that both CDK2 and CDK4/6 have a role in regulating sensitivity to ferroptosis.
CDKs phosphorylate many proteins involved in cell cycle progression with RB proteins coordinating many of the transcriptional responses (54). In addition to regulating cell cycle genes, RB proteins modulate pathways linked to ROS metabolism and ferroptotic sensitivity (46, 55). We observed that TKO cells lacking p105RB, p107, and p130 and double KO cells lacking p107 and p130 were more sensitive to ferroptosis. Furthermore, MDA MB 468 reconstituted with p105RB were more resistant to CETZOLE 1 or RSL3 than control MDA MB 468 cells. Taken together, these effects on ferroptosis cannot be explained by CDKs simply inactivating RB via phosphorylation. If this were the case, RB deletion should protect from ferroptosis similarly to CDK overexpression. In addition, because unphosphorylated RB family proteins sequester E2F, a simply linear model would predict that RB deletion would have similar effects as E2F1 overexpression with respect to ferroptosis. Clearly, this is simplification because RB family proteins show some separation function as do the nine members of the E2F family (2, 56). In addition, RB has numerous binding partners in addition to E2Fs. Furthermore, depending on the phase of the cell cycle, multiple monophosphorylated forms of RB exist (5, 6). Monophosphorylated RB still binds to many of its direct binding partners and regulates transcription of downstream genes depending on the actual phosphorylation state (5). The detailed operation of the CDK/RB/E2F pathway likely holds the clues to understanding how these proteins regulate ferroptosis.
Interestingly, we observed that the NARF2 K2/D cell line expresses higher levels of p21 than control cells without constitutive CDK activity. Previous studies identified E2F binding sites in the p21 promoter and documented induction of p21 by E2F (41). Therefore, NARF2 K2/D cells may express elevated p21 as a result of high E2F activity (due to elevated RB phosphorylation). In fact, we did observe elevation of p21 levels after overexpressing E2F1. Furthermore, E2F1 was less able to inhibit ferroptosis in cells lacking p21 providing evidence linking these two proteins in this cellular response. Returning to CDKs, we also tested the idea that these proteins suppress ferroptosis by elevating p21, possibly by releasing E2F. However, CDKs still suppressed ferroptosis in p21-null cells pointing to additional CDK targets, perhaps RB itself.
In summary, the linear p53/p21/CDK/RB/E2F pathway that explains many cell cycle–dependent phenomena is insufficient to explain the effects of these proteins in the regulation of ferroptosis. Instead, multiple proferroptotic and antiferroptotic signals emanate from the individual components of this pathway (Fig. 13). We show that p53 enhances ferroptosis in a number of contexts. It is likely that multiple direct and indirect targets of p53 modulate ferroptotic sensitivity. The suppressive effects of p21 on ferroptosis appears to occur independently of its ability to inhibit CDKs. Otherwise, small-molecule CDK inhibitor PD 0332991 would also inhibit ferroptosis when in fact this chemical enhances ferroptosis. Elevation of CDK activity inhibited ferroptosis, whereas RB deletion enhanced ferroptosis. Whether these two effects are related is unknown. Elevation of E2F suppressed ferroptosis, and we show that this effect depends in part on p21. In contrast, p21 is dispensable for inhibition of ferroptosis by CDKs.
Experimental procedures
Cell lines and culture conditions
Cell lines were cultured in a humidified atmosphere containing 10% CO2 in Dulbecco's modified Eagle's medium (Mediatech, Inc) supplemented with 10% fetal bovine serum (Atlanta Biologicals). Cell types used are p53 +/+ or p53 −/− MEFs, TR9-7 (human adult fibroblast; parental MDAH041), HT1080-LXSN (human fibrosarcoma cells expressing empty vector LXSN), HT1080-GSE56 (human fibrosarcoma expressing dominant negative C-terminus fragment of p53), NARF2 pBpuro (osteosarcoma; expressing pBpuro), NARF2 K2/D cell line (osteosarcoma expressing CDK2–cyclin D fusion protein), WI-38 (human embryonic lung fibroblast), NeoWT#5 (WT mouse embryonic fibroblast), TKO (mouse embryonic fibroblast lacking of three RB family proteins (p105RB, p107, and p130)), HO75-E9 (WT MEFs), double KO (MEFs lack of p107 and p130), and MDA MB 468 (breast cancer).
To determine viability throughout the study, 20,000 cells were plated per well of a 24-well plate and drugs added 1 day later. For adenoviral transduction, 4000 NARF2 or MEF or HT1080 cells or 6000 WI38 cells were plated per well into 24-well plates. Next day, cells were infected with viruses and 1 day later, fresh media were replaced. Drugs were added 48 h after adenoviral infection. Cells were stained 1 to 3 days later with a saturated solution of methylene blue in 50% ethanol. Plates were rinsed and retained dye quantified by spectrophotometry. Absorbance was normalized to DMSO and given as 1 or 100% for cell viability. We have recently shown that, in the context of ferroptosis, methylene blue staining results are very similar to 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (47). Results are representative of at least two independent experiments. Statistical significance was assessed using the Student's t test. All commercially available chemicals were obtained from Cayman Chemicals unless otherwise noted. Novel compound CETZOLE 1 was synthesized as we have described in (18).
Stable transfection
Seventy-five thousand MDA MB 468 cells were seeded per well into a 24-well plate. Next day, 1-μg plasmid expressing WT-RB gene (Addgene #58905) (6) was cotransfected in 3:1 ratio with a retroviral empty vector plasmid pLXSN (0.33 μg) or pLXSN was transfected as a control using the Lipofectamine 3000 reagent (Thermo Fisher) according to manufacturer's instructions. After 18 h, transfected cells were expanded into separate 10-cm plates. Selection drug G418 was added at 400 μg/ml 48 h after transfection. Ten to 14 days later, Individual clones were isolated, expanded, grown in a selection media, and confirmed by Western blot analysis.
p21-KO HT1080 cells
To KO p21 (CDKN1A), we used a single lentivirus expressing a specific sgRNA and CAS9. CDKN1A sgRNA was a gift from Wen Xue (Addgene plasmid # 138189; http://n2t.net/addgene:138189; RRID: Addgene_138189) (57). For control, we used a lentivirus expressing a gRNA targeting an SH region of the genome (Chr 4: 58,110,237–58,110,808; GRCH38.p2) (58) that was coinfected with a second lentivirus to express CAS9. First, lentiviral particles were packaged: HEK 293 cells were plated to be at 90% confluency on the day of transduction into a 6-well plate. The cells were transfected with CDKN1A sgRNA/CAS9 plasmid (2 μg) and packaging and envelope proteins expressing plasmids pSPAX (2 μg) and pMD.2 G (1 μg) using Lipofectamine 3000 according to manufacturer's instructions. After 8 h, media were replaced. Generated lentiviruses were collected after 48 h after transfection. HT1080 cells from 24-well plates were infected with 100 μl of 48-hr viruses separately along with polybrene (4 μg/ml). Next day, cells were expanded into 10-cm plates separately. p21 (CDKN1A) KO cells were grown in culture media containing hygromycin (200 μg/ml), whereas control cells were grown in culture media containing blasticidin (5 μg/ml) and puromycin (1 μg/ml). After 10 to 14 days, individual colonies were selected, expanded, grown, and confirmed by Western blot analysis.
Western blotting
Cells were harvested by scraping and lysed in a buffer solution containing: 50 mM Tris (pH 7.4), 150 mM NaCl, 0.5% NP-40, 1 μg/ml aprotinin, 2 μg/ml leupeptin, 1 μg/ml pepstatin A, 1 mM DTT, 1 mM PMSF, 5 mM sodium fluoride, and 2 mM sodium vanadate for 20 min on ice. Insoluble debris was removed by centrifugation at 16,000g for 20 min at 4 °C. Equal amounts of protein for each sample (determined using BCA protein assay kit–Pierce) were separated by SDS-PAGE. Gels were transferred to polyvinylidene difluoride membranes (Millipore), blocked in a solution containing 5% (w/v) nonfat dry milk dissolved in PBS containing 0.05% (v/v) Tween 20, and probed with antibodies as indicated. For phospho-specific antibodies, membranes were blocked in 5% (w/v) bovine serum albumin in Tween 20 containing Tris buffered saline.
Antibodies were generally diluted in the blocking solution at 1:1000. Primary antibodies recognizing p-ser780-RB (Cell signaling technology #9307S), RB (Thermo Fisher #MS-107-P0), cyclin-E (BD Pharmingen #BD-51-1459GR), p21 (Santa Cruz #C-19/HRP), p53 (Santa Cruz #D0-1/HRP), p14ARF (Cell Signaling Technology #2407, 1:250 dilution), E2F1 (Santa Cruz #56661), ACSL4 (Invitrogen #PA5-27137), CDK2 (Santa Cruz #M2), and Actin (Abcam #3280) were generally incubated at 4 ° C overnight (or 1.5 h at room temperature). Signals were detected using horseradish peroxidase–conjugated secondary antibodies (Bio-Rad) and enhanced chemiluminescence (Bio-Rad). Western blot images were mostly taken by a ChemiDoc, and the digital images were analyzed using ImageJ software.
Lipid ROS measurement
NARF2 pBpuro cells were seeded at 1.5 × 105 cells per 9-cm plate and the next day transduced with CMV, CDK2/cyclin E, p21, or E2F1 adenoviruses at 100 MOI. Fresh complete media were replaced overnight. After 48-h transduction, infected cells were treated with 20 μM CETZOLE 1 along with BODIPY 581/591-C11 dye (0.5 μM) (Thermo Fisher). Forty-eight hours after treatment, cells were collected by trypsinization, washed with 1× PBS, and resuspended in 2% FBS containing 1× PBS. Cells were analyzed in FITC channel using a BD LSR Fortessa FACScanner. Twenty thousand events per condition were analyzed from three independent samples. The experiments were performed twice, each time with triplicate samples (n = 6). Collected data were processed with FlowJo v10 software. Lipid ROS measurement in TR9-7 or NARF2 cells was performed similarly at the indicated conditions as mentioned in the respective figure legends. Where appropriate, dead cells were excluded by gating. When gates were applied, they were identical across samples within an experiment.
Determining MOI of adenovirus
HEK293 cells were plated in a 10-cm plate and infected with second-generation recombinant adenoviruses the next day. Generated viruses were collected 2 to 3 days later from both the supernatant and cells by freeze-thawing process three times (frozen at dry ice and thawed at 37 °C). To determine viral titers, HEK293 cells were seeded in 96-well plates, and the next day, the virus suspension was diluted at 1:100. Later, 10-μl virus supernatant was added to cells in 90-μl culture media. One week later, the cytopathic effect was assessed by phase-contrast microscopy. Alternatively, infected cells were fixed in 100% methanol at −20 °C for 20 min. Cells were blocked by 1% PBSP. One hour later, cells were probed with FITC-conjugated hexon antibody at 1:5000 in the blocking buffer for 1 h. Cells were washed 3× with PBSP, and infected cells were visualized by fluorescence microscopy using an EVOS microscope. Titers determined using the cytopathic effect or hexon immunofluorescence were similar.
Data availability
Data that support the findings of this study are available within the article and in supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
L. M. V. T. and W. R. T. appear as inventors on patents covering the compound CETZOLE 1 described in this article. N. K. and S. D. declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors thank Dylan Launder and Radhika Koranne for help with flow cytometry.
Author contributions
N. K., S. D., L. M. V. T., and W. R. T. methodology; N. K. and S. D. validation; N. K. formal analysis; N. K. and S. D. investigation; N. K. writing–original draft; N. K., L. M. V. T., and W. R. T. visualization; L. M. V. T. and W. R. T. conceptualization; L. M. V. T. and W. R. T. resources; L. M. V. T. and W. R. T. writing–review and editing; L. M. V. T. and W. R. T. supervision; L. M. V. T. and W. R. T. project administration; L. M. V. T. and W. R. T. funding acquisition.
Funding and additional information
This project was supported by National Institute of Health grants: NIH-R15CA213185 to L. M. V. T. and NIH-R15GM120712 and NIH-R15GM141712 to W. R. T.
Edited by Eric Fearon
Contributor Information
L. M. Viranga Tillekeratne, Email: ltillek@utnet.utoledo.edu.
William R. Taylor, Email: william.taylor3@utoledo.edu.
Supporting information
References
- 1.Friend S.H., Bernards R., Rogelj S., Weinberg R.A., Rapaport J.M., Albert D.M., Dryja T.P. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323:643–646. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- 2.Giacinti C., Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220–5227. doi: 10.1038/sj.onc.1209615. [DOI] [PubMed] [Google Scholar]
- 3.Agarwal M.L., Taylor W.R., Chernov M.V., Chernova O.B., Stark G.R. The p53 network. J. Biol. Chem. 1998;273:1–4. doi: 10.1074/jbc.273.1.1. [DOI] [PubMed] [Google Scholar]
- 4.el-Deiry W.S., Tokino T., Velculescu V.E., Levy D.B., Parsons R., Trent J.M., Lin D., Mercer W.E., Kinzler K.W., Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 5.Sanidas I., Morris R., Fella K.A., Rumde P.H., Boukhali M., Tai E.C., Ting D.T., Lawrence M.S., Haas W., Dyson N.J. A code of mono-phosphorylation modulates the function of RB. Mol. Cell. 2019;73:985–1000.e1006. doi: 10.1016/j.molcel.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Narasimha A.M., Kaulich M., Shapiro G.S., Choi Y.J., Sicinski P., Dowdy S.F. Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. Elife. 2014;3 doi: 10.7554/eLife.02872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sosa V., Moline T., Somoza R., Paciucci R., Kondoh H., ME L.L. Oxidative stress and cancer: An overview. Ageing Res. Rev. 2013;12:376–390. doi: 10.1016/j.arr.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Schieber M., Chandel N.S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014;24:R453–R462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang W.S., Kim K.J., Gaschler M.M., Patel M., Shchepinov M.S., Stockwell B.R. Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E4966–E4975. doi: 10.1073/pnas.1603244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., Patel D.N., Bauer A.J., Cantley A.M., Yang W.S., Morrison B., 3rd, Stockwell B.R. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Viswanathan V.S., Ryan M.J., Dhruv H.D., Gill S., Eichhoff O.M., Seashore-Ludlow B., Kaffenberger S.D., Eaton J.K., Shimada K., Aguirre A.J., Viswanathan S.R., Chattopadhyay S., Tamayo P., Yang W.S., Rees M.G., et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547:453–457. doi: 10.1038/nature23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang W.S., SriRamaratnam R., Welsch M.E., Shimada K., Skouta R., Viswanathan V.S., Cheah J.H., Clemons P.A., Shamji A.F., Clish C.B., Brown L.M., Girotti A.W., Cornish V.W., Schreiber S.L., Stockwell B.R. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang L., Kon N., Li T., Wang S.J., Su T., Hibshoosh H., Baer R., Gu W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu B., Kon N., Chen D., Li T., Liu T., Jiang L., Song S., Tavana O., Gu W. ALOX12 is required for p53-mediated tumour suppression through a distinct ferroptosis pathway. Nat. Cell Biol. 2019;21:579–591. doi: 10.1038/s41556-019-0305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarangelo A., Magtanong L., Bieging-Rolett K.T., Li Y., Ye J., Attardi L.D., Dixon S.J. p53 suppresses metabolic stress-induced ferroptosis in cancer cells. Cell Rep. 2018;22:569–575. doi: 10.1016/j.celrep.2017.12.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie Y., Zhu S., Song X., Sun X., Fan Y., Liu J., Zhong M., Yuan H., Zhang L., Billiar T.R., Lotze M.T., Zeh H.J., 3rd, Kang R., Kroemer G., Tang D. The tumor suppressor p53 limits ferroptosis by blocking DPP4 activity. Cell Rep. 2017;20:1692–1704. doi: 10.1016/j.celrep.2017.07.055. [DOI] [PubMed] [Google Scholar]
- 17.Louandre C., Marcq I., Bouhlal H., Lachaier E., Godin C., Saidak Z., Francois C., Chatelain D., Debuysscher V., Barbare J.C., Chauffert B., Galmiche A. The retinoblastoma (Rb) protein regulates ferroptosis induced by sorafenib in human hepatocellular carcinoma cells. Cancer Lett. 2015;356:971–977. doi: 10.1016/j.canlet.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 18.Fedorka S.R., So K., Al-Hamashi A.A., Gad I., Shah R., Kholodovych V., Alqahtani H.D., Taylor W.R., Tillekeratne L.M.V. Small-molecule anticancer agents kill cancer cells by harnessing reactive oxygen species in an iron-dependent manner. Org. Biomol. Chem. 2018;16:1465–1479. doi: 10.1039/c7ob03086j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor W.R., Fedorka S.R., Gad I., Shah R., Alqahtani H.D., Koranne R., Kuganesan N., Dlamini S., Rogers T., Al-Hamashi A., Kholodovych V., Barudi Y., Junk D., Rashid M.S., Jackson M.W., et al. Small-molecule ferroptotic agents with potential to selectively target cancer stem cells. Sci. Rep. 2019;9:5926. doi: 10.1038/s41598-019-42251-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yagoda N., von Rechenberg M., Zaganjor E., Bauer A.J., Yang W.S., Fridman D.J., Wolpaw A.J., Smukste I., Peltier J.M., Boniface J.J., Smith R., Lessnick S.L., Sahasrabudhe S., Stockwell B.R. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature. 2007;447:864–868. doi: 10.1038/nature05859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donehower L.A., Harvey M., Slagle B.L., McArthur M.J., Montgomery C.A., Jr., Butel J.S., Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 22.Little J.B., Nove J., Dahlberg W.K., Troilo P., Nichols W.W., Strong L.C. Normal cytotoxic response of skin fibroblasts from patients with Li-Fraumeni familial cancer syndrome to DNA-damaging agents in vitro. Cancer Res. 1987;47:4229–4234. [PubMed] [Google Scholar]
- 23.Bischoff F.Z., Yim S.O., Pathak S., Grant G., Siciliano M.J., Giovanella B.C., Strong L.C., Tainsky M.A. Spontaneous abnormalities in normal fibroblasts from patients with Li-Fraumeni cancer syndrome: Aneuploidy and immortalization. Cancer Res. 1990;50:7979–7984. [PubMed] [Google Scholar]
- 24.Agarwal M.L., Agarwal A., Taylor W.R., Stark G.R. p53 controls both the G2/M and the G1 cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc. Natl. Acad. Sci. U. S. A. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson M.W., Agarwal M.K., Yang J., Bruss P., Uchiumi T., Agarwal M.L., Stark G.R., Taylor W.R. p130/p107/p105Rb-dependent transcriptional repression during DNA-damage-induced cell-cycle exit at G2. J. Cell Sci. 2005;118:1821–1832. doi: 10.1242/jcs.02307. [DOI] [PubMed] [Google Scholar]
- 26.Bose I., Ghosh B. The p53-MDM2 network: From oscillations to apoptosis. J. Biosci. 2007;32:991–997. doi: 10.1007/s12038-007-0103-3. [DOI] [PubMed] [Google Scholar]
- 27.Lahav G. Oscillations by the p53-Mdm2 feedback loop. Adv. Exp. Med. Biol. 2008;641:28–38. doi: 10.1007/978-0-387-09794-7_2. [DOI] [PubMed] [Google Scholar]
- 28.Vassilev L.T., Vu B.T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., Fotouhi N., Liu E.A. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 29.Tovar C., Rosinski J., Filipovic Z., Higgins B., Kolinsky K., Hilton H., Zhao X., Vu B.T., Qing W., Packman K., Myklebost O., Heimbrook D.C., Vassilev L.T. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: Implications for therapy. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clifford B., Beljin M., Stark G.R., Taylor W.R. G2 arrest in response to topoisomerase II inhibitors: The role of p53. Cancer Res. 2003;63:4074–4081. [PubMed] [Google Scholar]
- 31.Yin Y., Tainsky M.A., Bischoff F.Z., Strong L.C., Wahl G.M. Wild-type p53 restores cell cycle control and inhibits gene amplification in cells with mutant p53 alleles. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]
- 32.Xiao G., Chicas A., Olivier M., Taya Y., Tyagi S., Kramer F.R., Bargonetti J. A DNA damage signal is required for p53 to activate gadd45. Cancer Res. 2000;60:1711–1719. [PubMed] [Google Scholar]
- 33.Choudhuri T., Pal S., Agwarwal M.L., Das T., Sa G. Curcumin induces apoptosis in human breast cancer cells through p53-dependent Bax induction. FEBS Lett. 2002;512:334–340. doi: 10.1016/s0014-5793(02)02292-5. [DOI] [PubMed] [Google Scholar]
- 34.Agarwal M.L., Agarwal A., Taylor W.R., Chernova O., Sharma Y., Stark G.R. A p53-dependent S-phase checkpoint helps to protect cells from DNA damage in response to starvation for pyrimidine nucleotides. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14775–14780. doi: 10.1073/pnas.95.25.14775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macip S., Igarashi M., Fang L., Chen A., Pan Z.Q., Lee S.W., Aaronson S.A. Inhibition of p21-mediated ROS accumulation can rescue p21-induced senescence. EMBO J. 2002;21:2180–2188. doi: 10.1093/emboj/21.9.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen W., Sun Z., Wang X.J., Jiang T., Huang Z., Fang D., Zhang D.D. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol. Cell. 2009;34:663–673. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macleod K.F. The role of the RB tumour suppressor pathway in oxidative stress responses in the haematopoietic system. Nat. Rev. Cancer. 2008;8:769–781. doi: 10.1038/nrc2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sage J., Mulligan G.J., Attardi L.D., Miller A., Chen S., Williams B., Theodorou E., Jacks T. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 2000;14:3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wise D.R., DeBerardinis R.J., Mancuso A., Sayed N., Zhang X.Y., Pfeiffer H.K., Nissim I., Daikhin E., Yudkoff M., McMahon S.B., Thompson C.B. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hurford R.K., Jr., Cobrinik D., Lee M.H., Dyson N. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- 41.Hiyama H., Iavarone A., Reeves S.A. Regulation of the cdk inhibitor p21 gene during cell cycle progression is under the control of the transcription factor E2F. Oncogene. 1998;16:1513–1523. doi: 10.1038/sj.onc.1201667. [DOI] [PubMed] [Google Scholar]
- 42.LaBaer J., Garrett M.D., Stevenson L.F., Slingerland J.M., Sandhu C., Chou H.S., Fattaey A., Harlow E. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 43.Jennis M., Kung C.P., Basu S., Budina-Kolomets A., Leu J.I., Khaku S., Scott J.P., Cai K.Q., Campbell M.R., Porter D.K., Wang X., Bell D.A., Li X., Garlick D.S., Liu Q., et al. An African-specific polymorphism in the TP53 gene impairs p53 tumor suppressor function in a mouse model. Genes Dev. 2016;30:918–930. doi: 10.1101/gad.275891.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang S.J., Li D., Ou Y., Jiang L., Chen Y., Zhao Y., Gu W. Acetylation is crucial for p53-mediated ferroptosis and tumor suppression. Cell Rep. 2016;17:366–373. doi: 10.1016/j.celrep.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ou Y., Wang S.J., Li D., Chu B., Gu W. Activation of SAT1 engages polyamine metabolism with p53-mediated ferroptotic responses. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E6806–E6812. doi: 10.1073/pnas.1607152113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao M., Monian P., Quadri N., Ramasamy R., Jiang X. Glutaminolysis and transferrin regulate ferroptosis. Mol. Cell. 2015;59:298–308. doi: 10.1016/j.molcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuganesan N., Dlamini S., McDaniel J., Tillekeratne V.L.M., Taylor W.R. Identification and initial characterization of a potent inhibitor of ferroptosis. J. Cell Biochem. 2021;122:413–424. doi: 10.1002/jcb.29870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shin J.S., Ha J.H., He F., Muto Y., Ryu K.S., Yoon H.S., Kang S., Park S.G., Park B.C., Choi S.U., Chi S.W. Structural insights into the dual-targeting mechanism of Nutlin-3. Biochem. Biophys. Res. Commun. 2012;420:48–53. doi: 10.1016/j.bbrc.2012.02.113. [DOI] [PubMed] [Google Scholar]
- 49.Venkatesh D., O'Brien N.A., Zandkarimi F., Tong D.R., Stokes M.E., Dunn D.E., Kengmana E.S., Aron A.T., Klein A.M., Csuka J.M., Moon S.H., Conrad M., Chang C.J., Lo D.C., D'Alessandro A., et al. MDM2 and MDMX promote ferroptosis by PPARalpha-mediated lipid remodeling. Genes Dev. 2020;34:526–543. doi: 10.1101/gad.334219.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wierod L., Rosseland C.M., Lindeman B., Oksvold M.P., Grosvik H., Skarpen E., Huitfeldt H.S. Activation of the p53-p21(Cip1) pathway is required for CDK2 activation and S-phase entry in primary rat hepatocytes. Oncogene. 2008;27:2763–2771. doi: 10.1038/sj.onc.1210937. [DOI] [PubMed] [Google Scholar]
- 51.Batchelor E., Loewer A., Mock C., Lahav G. Stimulus-dependent dynamics of p53 in single cells. Mol. Syst. Biol. 2011;7:488. doi: 10.1038/msb.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Purvis J.E., Karhohs K.W., Mock C., Batchelor E., Loewer A., Lahav G. p53 dynamics control cell fate. Science. 2012;336:1440–1444. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chytil A., Waltner-Law M., West R., Friedman D., Aakre M., Barker D., Law B. Construction of a cyclin D1-Cdk2 fusion protein to model the biological functions of cyclin D1-Cdk2 complexes. J. Biol. Chem. 2004;279:47688–47698. doi: 10.1074/jbc.M405938200. [DOI] [PubMed] [Google Scholar]
- 54.Lim S., Kaldis P. Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Development. 2013;140:3079–3093. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- 55.Reynolds M.R., Lane A.N., Robertson B., Kemp S., Liu Y., Hill B.G., Dean D.C., Clem B.F. Control of glutamine metabolism by the tumor suppressor Rb. Oncogene. 2014;33:556–566. doi: 10.1038/onc.2012.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Du W., Pogoriler J. Retinoblastoma family genes. Oncogene. 2006;25:5190–5200. doi: 10.1038/sj.onc.1209651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kwan S.Y., Sheel A., Song C.Q., Zhang X.O., Jiang T., Dang H., Cao Y., Ozata D.M., Mou H., Yin H., Weng Z., Wang X.W., Xue W. Depletion of TRRAP induces p53-independent senescence in liver cancer by down-regulating mitotic genes. Hepatology. 2020;71:275–290. doi: 10.1002/hep.30807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Phelps M.P., Bailey J.N., Vleeshouwer-Neumann T., Chen E.Y. CRISPR screen identifies the NCOR/HDAC3 complex as a major suppressor of differentiation in rhabdomyosarcoma. Proc. Natl. Acad. Sci. U. S. A. 2016;113:15090–15095. doi: 10.1073/pnas.1610270114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data that support the findings of this study are available within the article and in supporting information.