Abstract
Cryptococcus neoformans is a fungal pathogen that causes life-threatening meningoencephalitis in lymphopenic patients. Pulmonary macrophages comprise the first line of host defense upon inhalation of fungal spores by aiding in clearance but can also potentially serve as a niche for their dissemination. Given that macrophages play a key role in the outcome of a cryptococcal infection, it is crucial to understand factors that mediate phagocytosis of C. neoformans. Since lipid rafts (high-order plasma membrane domains enriched in cholesterol and sphingomyelin [SM]) have been implicated in facilitating phagocytosis, we evaluated whether these ordered domains govern macrophages' ability to phagocytose C. neoformans. We found that cholesterol or SM depletion resulted in significantly deficient immunoglobulin G (IgG)-mediated phagocytosis of fungus. Moreover, repletion of macrophage cells with a raft-promoting sterol (7-dehydrocholesterol) rescued this phagocytic deficiency, whereas a raft-inhibiting sterol (coprostanol) significantly decreased IgG-mediated phagocytosis of C. neoformans. Using a photoswitchable SM (AzoSM), we observed that the raft-promoting conformation (trans-AzoSM) resulted in efficient phagocytosis, whereas the raft-inhibiting conformation (cis-AzoSM) significantly but reversibly blunted phagocytosis. We observed that the effect on phagocytosis may be facilitated by Fcγ receptor (FcγR) function, whereby IgG immune complexes crosslink to FcγRIII, resulting in tyrosine phosphorylation of FcR γ-subunit (FcRγ), an important accessory protein in the FcγR signaling cascade. Correspondingly, cholesterol or SM depletion resulted in decreased FcRγ phosphorylation. Repletion with 7-dehydrocholesterol restored phosphorylation, whereas repletion with coprostanol showed FcRγ phosphorylation comparable to unstimulated cells. Together, these data suggest that lipid rafts are critical for facilitating FcγRIII-mediated phagocytosis of C. neoformans.
Keywords: lipid raft, cholesterol, sphingomyelin, Fc-gamma receptor, phagocytosis, macrophage, fungi
Abbreviations: BSA, bovine serum albumin; bSMase, bacterial sphingomyelinase; DPBS, Dulbecco's PBS; DPH, diphenylhexatriene; EGFP, enhanced GFP; FBS, fetal bovine serum; FcRγ, FcR γ-subunit; FcγR, Fcγ receptor; GPMV, giant plasma membrane vesicle; GXM, glucuronoxylomannan; IgG, immunoglobulin G; IgGIC, IgG immune complex; ITAM, immunoreceptor tyrosine-based activation motif; MαCD, methyl-alpha-cyclodextrin; MβCD, methyl-beta-cyclodextrin; ODRB, octadecyl rhodamine B; PFA, paraformaldehyde; RT, room temperature; SM, sphingomyelin; TBST, Tris-buffered saline + Tween-20
Cryptococcus spp. are major opportunistic fungal pathogens that cause life-threatening meningoencephalitis in immunocompromised individuals (1, 2). Cryptococcus neoformans has been reported to cause approximately 223,100 cases of meningitis globally, resulting in 181,000 deaths every year (3). C. neoformans mediated host damage at the molecular and cellular level results in deficient immune effector cell function, disruption of endothelial barriers, and dissemination (4). Despite the substantial morbidity and mortality associated with cryptococcal meningoencephalitis, there is still much that is unknown about the delicate interactions between host immune cells and fungal pathogens like C. neoformans.
Pulmonary macrophages comprise the first line of host defense in response to C. neoformans, as infection begins with inhalation of fungal spores commonly found in urban environments. A healthy host can typically kill and clear the pathogen. However, the fungus can be a facultative intracellular pathogen that survives within macrophages in immunocompromised individuals (e.g., HIV and AIDS patients) (2, 5). In these cases, the macrophages can serve as a niche for the replication of the pathogen and may facilitate its dissemination to the central nervous system where the disease becomes fatal (6, 7, 8, 9). C. neoformans can impair mitochondrial function, alter protein synthesis, or undergo nonlytic exocytosis from macrophages (10, 11, 12, 13). Macrophages may even deliver the fungus directly into the meninges, helping the yeast to cross the blood brain barrier via the “Trojan horse” model (14, 15, 16, 17). Given that macrophages can determine the outcome of a cryptococcal infection, it is crucial to understand the factors that mediate phagocytosis of C. neoformans, a major function of macrophages.
Phagocytosis is a process by which extracellular entities are internalized by phagocytic cells. It is a key weapon in the immune system's arsenal to defend against pathogens, but the process may often be subverted by pathogens to allow for internalization and dissemination throughout the body (18). Phagocytosis is mediated by several signaling events that result in attachment and engulfment via rearrangements of the host cell's cytoskeleton. “Professional” phagocytes are able to recognize and bind to opsonins on the surface of the invading pathogen to signal for attachment and the formation of lamellipodia, which engulf the pathogen and form a phagosome (2). Macrophages are highly specialized cells that carry out protective functions that include seeking out and eliminating disease-causing agents, repairing damaged tissues, and mediating inflammation, most of these through the process of phagocytosis (18, 19).
Previous work in other pathogen systems point to lipid rafts formed by cholesterol and sphingomyelin (SM) as having an important role to play in phagocytosis (20, 21, 22, 23, 24, 25, 26). In fact, a recent study implicated lipid rafts in the phagocytic response to Aspergillus fumigatus, another opportunistic fungal pathogen (27). Cholesterol is the single most abundant lipid species in mammalian plasma membranes, comprising 25 to 50% of the cell membrane lipid content (28). It has been found to play a role in modulating the biophysical properties of membranes by changing their rigidity (29). Cholesterol and sphingolipids together form lipid microdomains within the membrane known as lipid rafts. Lipid rafts have been found to be involved in the formation of caveolae as well as activation of various signaling cascades, whereby domains favor specific protein–protein interactions (28, 29, 30). In fact, previous studies have implicated the role of rafts in immune receptor signaling (31). Given their small size, it is difficult to study lipid rafts in vivo. One useful way to study the role of lipid rafts is to alter their constituents. Cyclodextrins and sphingomyelinase have been found to deplete, replace, or alter lipids from host mammalian membranes and are commonly used to study the role of lipid rafts (30, 32, 33, 34, 35, 36, 37).
In this study, several approaches were employed to alter cholesterol or SM on the outer leaflet of the plasma membrane to examine how lipid rafts may play a role in phagocytosis of C. neoformans. We found that cholesterol and SM are critical for immunoglobulin G (IgG)-mediated phagocytosis of C. neoformans. Moreover, repletion with domain-promoting sterols like cholesterol or 7-dehydrocholesterol promoted efficient antibody-mediated phagocytosis of the fungal cells, whereas domain-inhibiting sterols like coprostanol significantly reduced it. Using a photoswitchable SM (AzoSM), we observed that the raft-promoting conformation (trans-AzoSM) resulted in efficient phagocytosis whereas raft-inhibiting conformation (cis-AzoSM) significantly blunted phagocytosis in a reversible manner. Mechanistically, we found that cholesterol and SM-enriched ordered domains may be important for the function of Fcγ receptors (FcγRs), the class of immune receptors activated by IgG immune complexes (IgGICs). These findings provide more directed insights into the role of cholesterol- and SM-rich lipid rafts in mediating FcγR activation and IgG-dependent phagocytosis of C. neoformans.
Results
Cholesterol or SM depletion affects IgG-mediated phagocytosis of C. neoformans
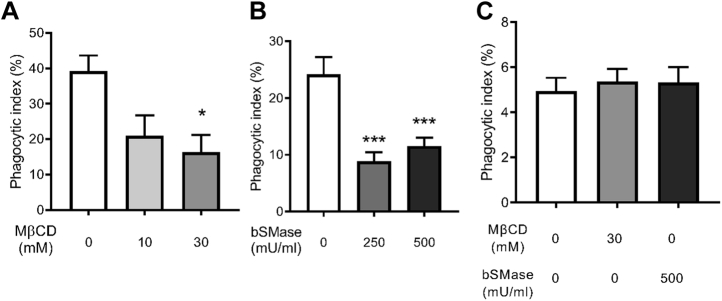
To investigate the role of cholesterol in phagocytosis of the fungal pathogen C. neoformans, we utilized methyl-beta-cyclodextrin (MβCD) to deplete cholesterol from the membrane of murine macrophages prior to infection with antibody-opsonized C. neoformans (38). We previously showed that treatment with 10 mM or 30 mM MβCD depletes approximately 50% or 75% of the total cholesterol in the cells, respectively (38). Furthermore, MβCD treatment did not significantly alter cell attachment or viability (38). Most importantly, cholesterol depletion resulted in a significant decrease in antibody-mediated phagocytosis of C. neoformans (38). This finding was recapitulated across two murine macrophage cell lines. Both alveolar (Fig. 1A) and peritoneally derived macrophages (J774.1; Fig. S1A) showed significant deficiency in phagocytosing C. neoformans cells opsonized with an anti-glucuronoxylomannan (GXM) IgG, an antibody specific to the cryptococcal capsule (39). When C. neoformans cells were instead opsonized with complement serum, phagocytosis was not affected (Fig. 1C).
Figure 1.
Cholesterol and sphingomyelin are important for antibody-mediated phagocytosis.A, macrophages (MH-S) treated with either 10 or 30 mM methyl-beta-cyclodextrin (MβCD) were coincubated with antibody-opsonized Cryptococcus neoformans H99 at a 1:1 ratio and allowed to interact for 2 h. Cells were then fixed and stained with Giemsa, and phagocytic index was calculated by microscopic observation (n = 4). B, macrophages (MH-S) treated with either 250 or 500 mU/ml bacterial sphingomyelinase (bSMase) were coincubated with antibody-opsonized C. neoformans H99 at a 1:1 ratio and allowed to interact for 2 h. Cells were then fixed and stained with Giemsa, and phagocytic index was calculated by microscopic observation (n = 3). C, macrophages (MH-S) treated with 30 mM MβCD or 500 mU/ml bSMase were coincubated with complement serum-opsonized C. neoformans H99 at a 1:1 ratio and allowed to interact for 3 h. Cells were then fixed and stained with Giemsa, and phagocytic index was calculated by microscopic observation (n = 4). Error bars represent the SEM, and statistical significance was determined using one-way ANOVA with Tukey's multiple comparisons test. ∗p < 0.05, ∗∗∗p < 0.001 compared with the untreated control. All p values were adjusted for multiplicity.
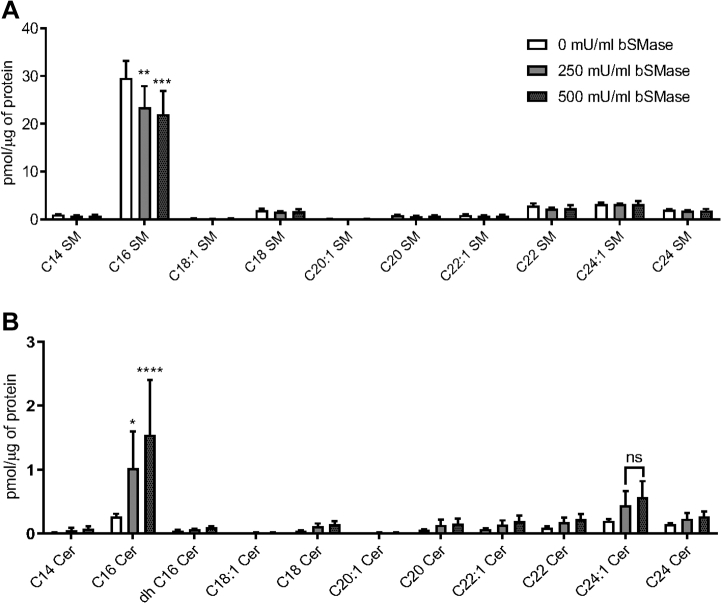
Given the collaborative role of cholesterol and SM in lipid rafts, we examined the effect of depleting SM on the plasma membrane of macrophages. One tool available for the study of SM is recombinant bacterial sphingomyelinase (bSMase) which directly probes for the role of SM on the plasma membrane, as the enzyme is too large to pass through the membrane (40). bSMase catalyzes the transformation of SM into ceramide (Fig. S2) and phosphorylcholine (40). To confirm SM depletion, cellular lipids were analyzed following treatment with 250 mU/ml or 500 mU/ml bSMase for 20 min. We found that treatment of macrophages with bSMase resulted in a significant decrease in C16 SM, the most abundant SM species detected in the cells, and a corresponding increase in C16 ceramide (Fig. 2). To assess the effect of bSMase treatment on phagocytosis, cells were coincubated with C. neoformans cells opsonized with anti-GXM IgG after bSMase treatment. We found a significant decrease in phagocytosis after bSMase treatment with both alveolar (MH-S; Fig. 1B) and peritoneally derived macrophages (J774.1; Fig. S1B), albeit in a non–dose-dependent fashion. This effect was not observed when complement serum was used as an opsonin (Fig. 1C). Together, these data suggest that cholesterol and SM are critical for IgG-mediated phagocytosis of C. neoformans. Moreover, our data suggest that this phenomenon is not cell line dependent.
Figure 2.
Measurement of sphingolipids after treatment with bacterial sphingomyelinase. Macrophages (J774.1) were treated with either 250 or 500 mU/ml bacterial sphingomyelinase (bSMase). Monolayers were then washed and subject to lipid extraction. Lipid extracts were then analyzed using LC-coupled MS and normalized to protein found in the cell extract using the Bradford assay. A, sphingomyelin (SM) species in the treated samples are shown (n = 3). B, ceramide (Cer) species in the treated samples are shown (n = 3). Error bars represent the SEM, and statistical significance was determined using ANOVA with Tukey's multiple comparisons test. Not significant (ns), ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, and ∗∗∗∗p < 0.0001 compared with the untreated control. All p values were adjusted for multiplicity.
Repletion with lipid raft–altering sterols affects IgG-mediated phagocytosis of C. neoformans
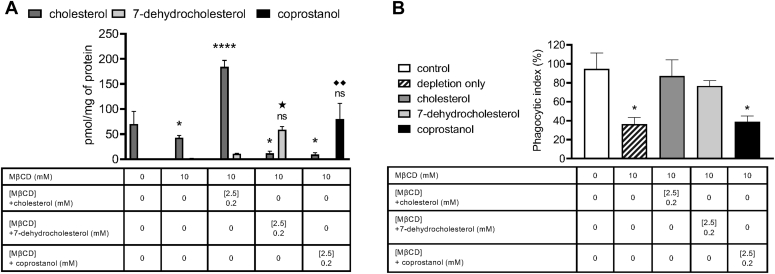
Given that multiple factors confound the use of MβCD to deplete cholesterol from cell membranes, various sterols were added back into the cholesterol-depleted macrophages to investigate the role of cholesterol in IgG-mediated phagocytosis of C. neoformans. MβCD may remove cholesterol from both raft and nonraft domains, alter the distribution of cholesterol between plasma and intracellular membranes, and nonspecifically extract phospholipids (41). To ascertain whether cholesterol sensitivity of IgG-dependent phagocytosis of C. neoformans could be attributed to lipid rafts, cholesterol-depleted macrophages were repleted with cholesterol, 7-dehydrocholesterol, or coprostanol (Fig. S2). 7-dehydrocholesterol has been shown to be significantly more domain promoting than cholesterol, whereas coprostanol strongly inhibits domain formation (36, 42, 43). We found that repletion with 0.2 mM cholesterol in 2.5 mM MβCD resulted in a significant increase in total cellular cholesterol compared with the untreated control. On the other hand, repletion with 0.2 mM 7-dehydrocholesterol or 0.2 mM coprostanol in 2.5 mM MβCD resulted in a significant decrease in cellular cholesterol and a marked increase in substituted sterol comparable to the cellular cholesterol for the untreated control (Fig. 3A). When treated macrophages were coincubated with C. neoformans cells opsonized with anti-GXM IgG, we found that we were able to restore phagocytosis by cholesterol or 7-dehydrocholesterol repletion, whereas coprostanol repletion did not restore phagocytosis (Fig. 3B).
Figure 3.
Repletion with raft-altering sterols affects phagocytosis.A, macrophages (J774.1) were pretreated with 10 mM methyl-beta-cyclodextrin (MβCD) to deplete cholesterol and then washed and incubated with 2.5 mM MβCD loaded with 0.2 mM of indicated sterol. Monolayers were then washed and subject to lipid extraction. Lipid extracts were then analyzed using GC-coupled MS and normalized to protein found in the cell extract by the Bradford assay (n = 3). B, macrophages were pretreated with 10 mM MβCD to deplete cholesterol and then washed and incubated with 2.5 mM MβCD loaded with 0.2 mM of indicated sterol. Monolayers were then washed and allowed to interact with antibody-opsonized Cryptococcus neoformans H99 at a 1:1 ratio for 2 h. Cells were then fixed and stained with Giemsa, and phagocytic index was calculated by microscopic observation (n = 3). Error bars represent SEM, and statistical significance was determined using one-way ANOVA with Tukey's multiple comparisons test. Not significant (ns), ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗∗p < 0.0001 compared with the cellular cholesterol for untreated control. ★p < 0.05 compared with the cellular 7-dehydrocholesterol for untreated control. ◆◆P < 0.01 compared with the cellular coprostanol for untreated control. All p values were adjusted for multiplicity.
Cholesterol, but not SM digestion, affects lipid nanodomain stability
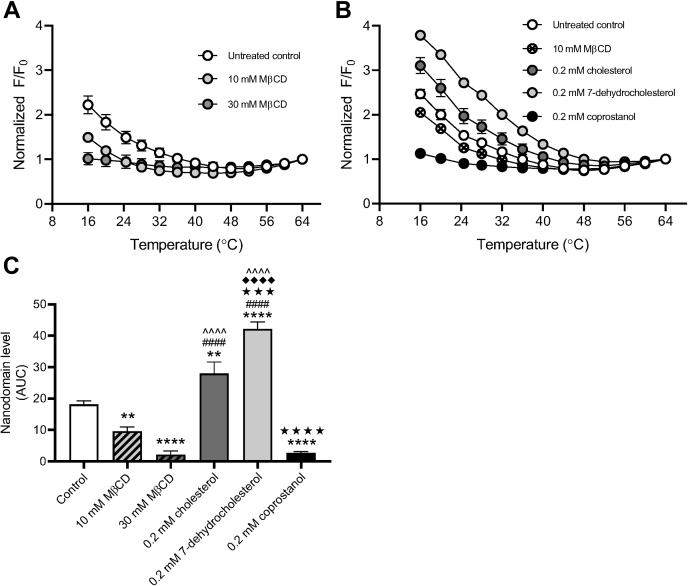
To assess how sterol depletion/repletion may affect lipid raft stability in macrophages, we evaluated ordered nanodomains (rafts) in giant plasma membrane vesicles (GPMVs) derived from macrophages through FRET. When rafts are present, FRET donor and FRET acceptor become partially segregated in different (raft and nonraft) domains, and FRET decreases so the FRET donor is more fluorescent (i.e., F/F0 increases) As previously, we used the temperature at which the value of F/F0 is a minimum as an approximate temperature for the upper limit of when rafts were present, that is, a measure of their thermal stability (36). We also used the total increase in F/F0 relative to the value at which F/F0 is a minimum in a sample as another rough measure of total raft formation over the entire temperature range.
GPMVs prepared from cholesterol-depleted macrophages showed a significant shift in nanodomain stability (Fig. 4A and Table S1, raw unnormalized F/F0 values for FRET data are shown in Fig. S3A). The presence of detectable ordered nanodomains significantly decreased, with up to 20 °C decrease in Tend, the temperature at which ordered domains are completely melted (36). Upon repletion with cholesterol, detectable ordered nanodomains significantly increased compared with the untreated and cholesterol-depleted macrophages with a corresponding recovery in Tend. As expected, repletion with 7-dehydrocholesterol resulted in significantly greater presence of detectable nanodomains as well as increased thermal stability (higher Tend compared with untreated control). On the other hand, repletion with coprostanol ablated both the presence of detectable ordered nanodomains and the GPMV thermal stability (Fig. 4B and Table S1; raw unnormalized F/F0 values for FRET data are shown in Fig. S3B).
Figure 4.
Repletion with raft-altering sterols affects nanodomain stability.A, macrophages (MH-S) were pretreated with either 10 or 30 mM methyl-beta-cyclodextrin (MβCD) to deplete cholesterol. Monolayers were then washed and subject to giant plasma membrane vesicle formation using 25 mM paraformaldehyde (PFA) and 2 mM DTT. Nanodomain stability was assessed via FRET with diphenylhexatriene (DPH) as the FRET donor and octadecyl rhodamine B (ODRB) as the FRET acceptor. The ratio of DPH fluorescence intensity in the presence versus absence of ODRB was calculated (F/F0). F/F0 values were normalized to the final F/F0 value at 64 °C (n = 3). B, macrophages were pretreated with 10 mM MβCD to deplete cholesterol and then washed and incubated with 2.5 mM MβCD loaded with 0.2 mM of indicated sterol. Monolayers were then washed and subject to giant plasma membrane vesicle formation using 25 mM PFA and 2 mM DTT. Nanodomain stability was assessed via FRET with DPH as the FRET donor and ODRB as the FRET acceptor. The ratio of DPH fluorescence intensity in the presence versus absence of ODRB was calculated (F/F0). F/F0 values were normalized to the final F/F0 value at 64 °C (n = 3). C, relative domain levels were estimated using the polynomial fits from (A) and (B) as described under the Experimental procedures section. Error bars represent SEM. Detectable nanodomains were compared using one-way ANOVA with Tukey's multiple comparisons test. ∗∗p < 0.01, ∗∗∗∗p < 0.0001 compared with the untreated control, ####p < 0.0001 compared with 10 mM MβCD, ˆˆˆˆp < 0.0001 compared with 30 mM MβCD, ★★★p < 0.001, ★★★★p < 0.0001 compared with 0.2 mM cholesterol, and ◆◆◆◆p < 0.0001 compared with 0.2 mM coprostanol. All p values were adjusted for multiplicity.
To ascertain whether SM depletion alters lipid raft stability in macrophages, we also evaluated nanodomain stability in GPMVs derived from bSMase-treated macrophages. We found that neither 250 nor 500 mU/ml bSMase treatment altered nanodomain stability, with Tend and GPMV levels comparable to the untreated control (Fig. S4 and Table S1; raw unnormalized F/F0 values for FRET data are shown in Fig. S4C). It should be noted that the lipid product of bSMase digestion, ceramide, is itself a raft-forming lipid but one that alters the lipid composition (e.g., resulting in displacement of cholesterol from rafts) and properties of lipid rafts (44, 45).
Together, these results not only suggest that the presence of ordered lipid domains mediate IgG-dependent phagocytosis of C. neoformans but that lipid domain stability is cholesterol dependent.
Cholesterol or SM depletion may affect structure of lipid rafts
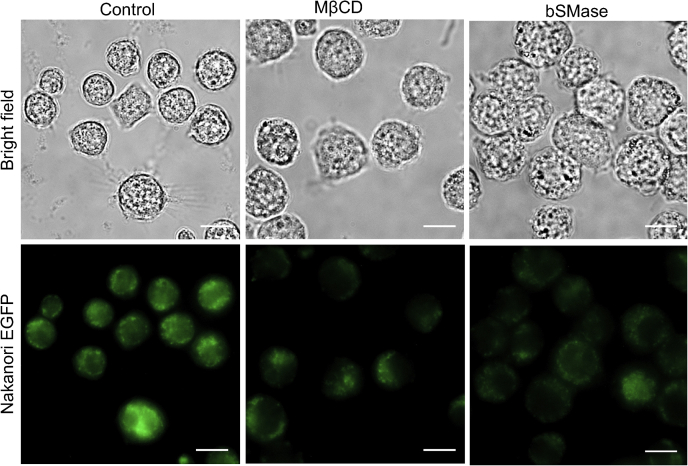
Enhanced GFP (EGFP)-nakanori is a protein that labels cell surface domains in an SM-dependent and cholesterol-dependent manner. More specifically, nakanori only binds pre-existing SM/cholesterol domains as it is unable to induce formation of SM–cholesterol complexes (46). To examine whether cholesterol depletion or SM depletion perturbs SM–cholesterol complexes, macrophages were either treated with MβCD or bSMase and labeled with EGFP-nakanori. Both cholesterol and SM depletion resulted in marked decrease in EGFP-nakanori binding, suggesting successful perturbation of SM–cholesterol complexes (Fig. 5). These results suggest that MβCD and bSMase treatment may potentially affect lipid raft structure independent of nanodomain stability.
Figure 5.
Nakanori labeling is affected by lipid-altering treatments. Macrophages (MH-S) were adhered onto glass bottomed microscopy dishes. After treatment with 10 mM methyl-beta-cyclodextrin (MβCD) or 250 mU/ml bacterial sphingomyelinase (bSMase) as described under the Experimental procedures section, live macrophages were labeled with recombinant nakanori-enhanced GFP and then fixed with 4% paraformaldehyde. The scale bar represents 10 μm.
AzoSM can reversibly modulate IgG-mediated phagocytosis of C. neoformans
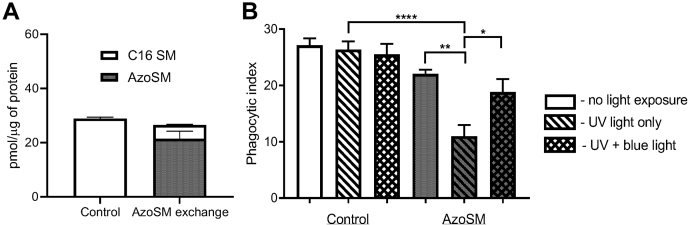
To better understand how SM facilitates IgG-dependent phagocytosis of C. neoformans, we utilized a photoswitchable SM. Photoswitchable lipids allow for the optical control of lipid function with high spatiotemporal resolution, enabling precise functional manipulations. They have been shown to modulate many aspects of membrane biophysics, including permeability, fluidity, lipid mobility, and domain formation (47, 48, 49). Recently, photoswitchable sphingolipids have emerged as potent tools to investigate cellular physiology using optical control, including receptor-mediated signaling and immune function (50, 51, 52, 53). We synthesized AzoSM, a C16:0 SM derivative containing an isosteric azobenzene photoswitch following a design principle called “azologization” (54). Optical control of photoswitchable sphingolipids is mediated through light exposure. UV light (λ = 365 nm) isomerizes the azobenzene group from a trans to cis conformation, whereas blue light (λ = 460 nm) reverts ∼80% of AzoSM from a cis to trans conformation (Fig. S2). UV light therefore introduces a kink into the fatty acyl tail to disrupt ordered domains, whereas blue light promotes domain formation (48, 49). Using methyl-alpha-cyclodextrin (MαCD), endogenous SM species in macrophages were exchanged for AzoSM. This method exchanges phospholipids but does not alter cell sterol (32). We found that MαCD effectively replaced ∼80% of endogenous C16:0 SM, the most abundant species of SM in MH-S murine macrophages, with AzoSM (Fig. 6A). When macrophages were coincubated with C. neoformans cells opsonized with anti-GXM IgG immediately following AzoSM exchange (no light exposure), we observed phagocytosis comparable to untreated control cells. Interestingly, we found that UV light treatment prior to coincubation with C. neoformans resulted in a significant decrease in phagocytosis. Moreover, UV light exposure immediately followed by blue light treatment prior to coincubation with fungal cells partially restored phagocytosis (Fig. 6B). Together, these data suggest that SM-dependent lipid domains may be critical for IgG-mediated phagocytosis of C. neoformans.
Figure 6.
Exclusion of sphingomyelin from rafts affects phagocytosis Cryptococcus neoformans.A, macrophages (MH-S) treated with the AzoSM exchange protocol using methyl-alpha-cyclodextrin (MαCD) as described in the Experimental procedures section. Cell extracts were then evaluated for endogenous C16 sphingomyelin (C16 SM) using LC–MS and AzoSM using HPLC. Lipid content from each sample was normalized by protein content as determined using the Bradford assay (n = 3). B, macrophages (MH-S) treated with the AzoSM exchange protocol were either unexposed, exposed to UV light, or exposed to UV light followed by blue light. Control samples were mock treated. Washed monolayers were then coincubated at a 1:1 ratio with C. neoformans H99 opsonized with 18B7 antibody and allowed to interact for 30 min. Cells were then fixed and stained with Giemsa, and phagocytic index was calculated by microscopic observation. Error bars represent the SEM, and statistical significance was determined using two-way ANOVA with Tukey's multiple comparisons test (n = 4). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗∗p < 0.0001. All p values were adjusted for multiplicity.
Alterations in rafts affect FcγR function
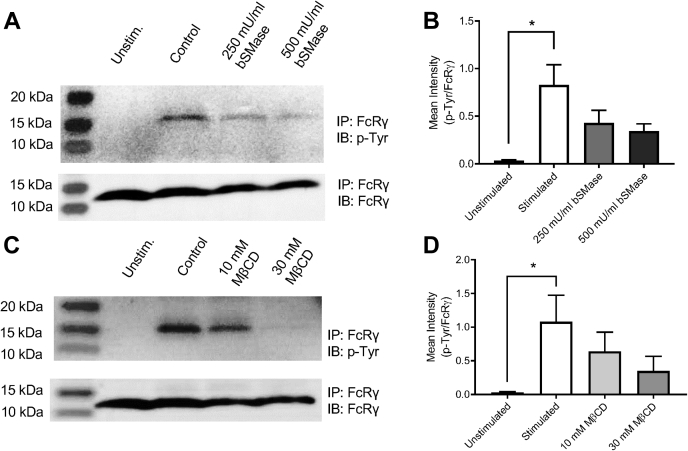
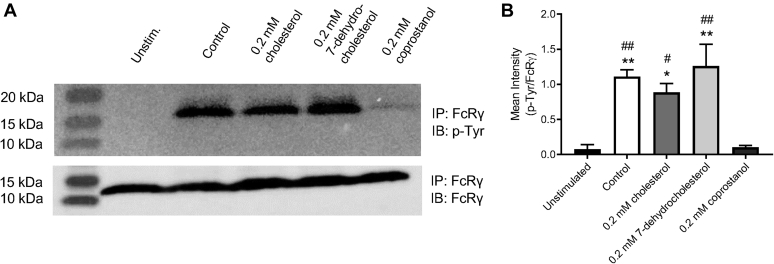
Since alterations in plasma membrane lipids only affected phagocytosis of C. neoformans opsonized with anti-GXM IgG, our data implicate a critical role for lipid rafts in FcγR-mediated phagocytosis. As the anti-GXM antibody falls in the IgG1κ subclass, antibody-opsonized C. neoformans binds to FcγRIIB and III, the low-affinity receptors for mouse IgG1 (39, 55). To assess whether lipid depletion treatments alter the amount of cell surface receptors, macrophages were probed for CD16/32, which recognizes FcγR II/III, and CD11b antibody, which recognizes complement receptor 3, known to mediate uptake of serum-opsonized C. neoformans (56). CD45 was used as a general leukocyte marker, which should not be affected by lipid-altering treatments. Flow cytometry showed that neither CD16/32 nor CD11b levels on the cell surface were affected by cholesterol depletion with MβCD or SM depletion with bSMase for either J774.1 or MH-S macrophages (Fig. S5). We next probed whether FcγR function rather than abundance on the plasma membrane was affected by lipid depletion. Murine FcγRIII requires the association of the FcR γ-subunit (FcRγ) following IgG immune complex and receptor aggregation at the plasma membrane. The FcRγ contains an immunoreceptor tyrosine-based activation motif (ITAM) that is required for cell activation (55). To investigate FcRγ activation following IgG immune complex and FcγR crosslinking, we utilized the approach by Rittirsch et al. (57). We observed tyrosine phosphorylation of FcRγ in macrophages stimulated with bovine serum albumin (BSA)–IgGICs within 5 min, with the phosphotyrosine levels subsiding 15 min after stimulation (Fig. S6). We then probed for tyrosine phosphorylation of FcRγ in macrophages following lipid-altering treatments. We observed that both SM depletion (Fig. 5, A and B) and cholesterol depletion (Fig. 5, C and D) resulted in reduced FcRγ activation upon IgGIC stimulation. Interestingly, repletion with cholesterol or 7-dehydrocholesterol showed comparable FcRγ activation to the untreated control, whereas repletion with coprostanol significantly ablated the FcRγ activation upon IgGIC stimulation (Fig. 6). Although nuanced, we also observed some decrease in FcRγ chain phosphorylation following UV light exposure post-AzoSM exchange, which was partially rescued upon blue light exposure (Fig. S7).
Together, these data suggest that cholesterol- and SM-dependent lipid rafts are critical for proper FcγR clustering upon IgG immune complex crosslinking in order to initiate the FcRγ-mediated signaling cascade essential for phagocytosis of IgG1-opsonized C. neoformans (58).
Discussion
In this study, we utilized various methods of lipid alteration to probe for the role of lipid rafts in phagocytosis of C. neoformans. Substituting either endogenous cholesterol with a raft-inhibiting sterol (coprostanol) or endogenous SM with domain-perturbing SM (cis-AzoSM) leads to a decrease in antibody-mediated phagocytosis of C. neoformans (Figure 7, Figure 8, Figure 9). Since depletion treatments perturbed SM–cholesterol complexes and significantly affected the FcγR-mediated cell signaling (as evidenced by decreased FcRγ ITAM activation) without altering the amount of FcγR IIB and III localized to the cell surface, it appears that SM and cholesterol-dependent lipid rafts are critical for facilitating FcγR-mediated cell signaling and function. Lipid rafts do not appear to be critical for phagocytosis mediated by complement serum opsonization, which was not affected by cholesterol depletion or bSMase treatment. The importance of cholesterol-dependent lipid rafts in FcγR function was further highlighted by the fact that repletion with cholesterol or 7-dehydrocholesterol (raft-promoting sterols) resulted in proper FcRγ ITAM activation, whereas repletion with coprostanol (raft-inhibiting sterol) significantly reduced FcRγ ITAM activation. Similarly, indeed, a recent review highlights the importance of plasma membrane domains on IgG Fc receptor function (59). However, studies of FcγRIII have been limited to the human FcγRIIIa expressed on natural killer cells. Furthermore, studies have been limited to cellular fractionation assays and confocal microscopy (60, 61). Although murine and human FcγR differ in terms of binding abilities and expression pattern, the function and signaling mechanism are conserved (55). As such, insights into how cholesterol and SM, two key components of lipid rafts, alter membrane properties and affect FcγR function can have broader implications.
Figure 7.
Lipid-depleting treatments affect Fcγ receptor–mediated signaling.A, macrophages (MH-S) were mock treated (control) or treated with bacterial sphingomyelinase (bSMase) and subsequently stimulated with bovine serum albumin (BSA)–IgG immune complex (IC) for 5 min. Fc receptor γ chain (FcRγ) was immunoprecipitated (IP) and evaluated for phosphorylation of the tyrosine residues. IP samples were probed for FcRγ or phosphotyrosine (p-Tyr). Representative immunoblots are shown. B, immunoblots were quantified using ImageJ software; p-Tyr was normalized by FcRγ (n = 3 experiments). Error bars represent the SEM, and statistical significance was determined using one-way ANOVA with Tukey's multiple comparisons test. ∗p < 0.05. All p values were adjusted for multiplicity using Bonferroni's correction. C, MH-S cells were mock treated (control) or treated with methyl-beta-cyclodextrin (MβCD) and subsequently stimulated with BSA–IgG IC for 5 min. FcRγ was IP and evaluated for phosphorylation of the tyrosine residue. IP samples were probed for FcRγ or p-Tyr. Representative immunoblots are shown. D, immunoblots were quantified using ImageJ software; p-Tyr was normalized by FcRγ (n = 3). Error bars represent the SEM, and statistical significance was determined using one-way ANOVA with Tukey's multiple comparisons test compared with the control. ∗p < 0.05. All p values were adjusted for multiplicity.
Figure 8.
Repletion with raft-altering sterols affects Fcγ receptor–mediated signaling.A, macrophages were mock treated (control) or pretreated with 10 mM methyl-beta-cyclodextrin (MβCD) to deplete cholesterol and then washed and incubated with 2.5 mM MβCD loaded with 0.2 mM of indicated sterol. Cells were then stimulated with bovine serum albumin (BSA)–IgG immune complex (IC) for 5 min. Fc receptor γ chain (FcRγ) was immunoprecipitated (IP) and evaluated for phosphorylation of the tyrosine residue. IP samples were probed for FcRγ or phosphotyrosine (p-Tyr). Representative immunoblots are shown. B, immunoblots were quantified using ImageJ software; p-Tyr was normalized by FcRγ (n = 3). Error bars represent the SEM, and statistical significance was determined using one-way ANOVA with Tukey's multiple comparisons test. ∗p < 0.05, ∗∗p < 0.01, compared with the untreated control. #p < 0.05, ##p < 0.01 compared with 0.2 mM coprostanol. All p values were adjusted for multiplicity.
Figure 9.
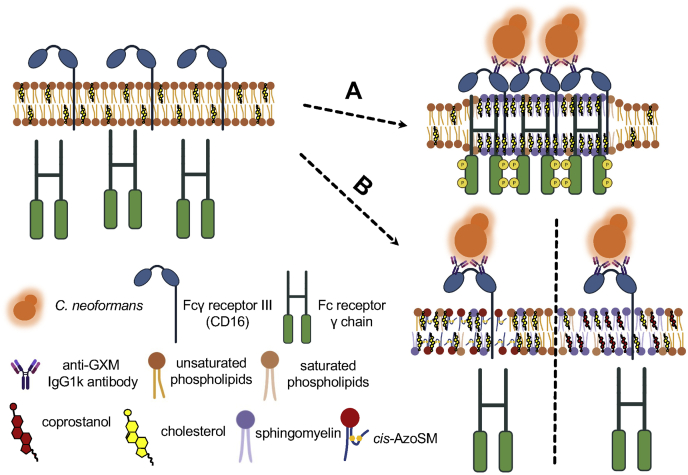
Cholesterol- and sphingomyelin (SM)-rich plasma lipid rafts are important for Fcγ receptor–mediated phagocytosis of Cryptococcus neoformans by macrophages. Upon binding IgG-based immune complexes comprised of antiglucuronoxylomannan (GXM) antibody-opsonized C. neoformans, Fcγ receptor III (CD16) localizes to the plasma lipid rafts. A, when lipid rafts are enriched with raft-forming sterols (i.e., cholesterol and 7-dehydrocholesterol; only cholesterol shown for simplicity) or SM (i.e., trans-AzoSM; only endogenous SM shown for simplicity), the Fc receptor γ chain is properly phosphorylated to initiate the signaling cascade associated with phagocytosis. B, when lipid rafts are enriched with raft-inhibiting sterols such as coprostanol (right) or raft-disrupting SM, cis-AzoSM (left), Fc receptor γ chain is not properly phosphorylated.
Lipid depletion is useful to investigate the role of key lipids in cellular processes. Given the physiological importance of cholesterol in the cell plasma membrane, MβCD has become the most common means of modifying the cholesterol content, mainly through cholesterol extraction. However, cyclodextrins may remove cholesterol from both raft and nonraft domains, alter the distribution of cholesterol between plasma and intracellular membranes, and simultaneously extract phospholipids (41). As such, cholesterol substitution with sterol structural analogs has been utilized in several studies to evaluate specific sterol–protein interactions or changes to the physical properties of the lipid bilayer (33, 42, 43, 62, 63). The key is substitution with an equivalent amount of another sterol without altering the total sterol level in cells (41). Although our cholesterol repletion increased total cellular sterol, it did not interfere with macrophages' ability to phagocytose or initiate FcγR signaling. When compared with 7-dehyrocholesterol or coprostanol repletion, nanodomain properties were consistent with raft formation.
bSMase is a useful tool to study the role of SM depletion as well as ceramide accumulation on the plasma membrane. The effects of SM digestion can be complex. Ceramides have been implicated in stabilizing SM-rich domains (43). They can also displace cholesterol from rafts and the plasma membrane (44, 64, 65). Modifying ordered domain properties, therefore, can have pleiotropic consequences. Previous work in other pathogen systems suggested that ceramide-rich membrane rafts resulting from bSMase activity are critical for host defense, including receptor clustering (66, 67). Interestingly, ceramide-rich membrane rafts have also been implicated in FcγR signaling. Indeed, the presence of C16 ceramide at the cell surface has been suggested to precede and control FcγRII clustering and phosphorylation in rafts, acting as a negative regulator of IgG-dependent phagocytosis (68, 69). FcγRII is an inhibitory immune receptor containing an immunoreceptor tyrosine-based inhibitory motif (70). Given our bSMase treatment resulted in C16 SM depletion and a corresponding accumulation of C16 ceramide (Fig. 2), aberrant FcγRII signaling may have contributed to the decrease in IgG-dependent phagocytosis of C. neoformans.
Given the complex and pleiotropic effects of bSMase treatment, AzoSM is a potent tool that permits direct control of lipid function and ordered domains with high spatiotemporal resolution. Together, sterol substitution and AzoSM exchange offer critical insights into the role of cholesterol and SM-dependent lipid domains in mediating membrane receptor signaling. Interestingly, protein tyrosine phosphatases have been reported to be excluded from lipid rafts during antigen-activated IgE signaling mediated by Fcε receptor I (71). In addition, a loss of insulin receptor accessibility to phosphatases when lipid raft levels are increased regulates its kinase activity (37). Consequently, exclusion of phosphatases from raft domains provides an amenable environment for efficient kinase activity and IgE signaling. Since both FcγRIII and Fcε receptor I require FcRγ phosphorylation to initiate the signaling cascade (72), raft disruption in our studies may have resulted in aberrant phosphatase activity on FcRγ to attenuate IgG signaling.
Together, these data demonstrate that lipid raft formation, high membrane order, and domain structure are critical for FcγRIII function, whereby disruption of lipid rafts interferes with the efficiency of FcγRIII signaling. Through this work, we provide direct evidence that cholesterol and SM play a critical role in coordinating a phagocytic response to C. neoformans.
Experimental procedures
Cells, media, and reagents for lipid alteration
C. neoformans var. grubii serotype A strain H99 was used in this study. Fungal cells were grown in yeast nitrogenous base medium containing 2% glucose, pH 7.2, for 16 to 18 h at 30 °C with shaking before experiments. Alveolar macrophage cell line MH-S (American Type Culture Collection CRL-2019) was cultured in complete RPMI1640 (supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin) at 37 °C + 5% CO2. Peritoneal macrophage cell line J774.1 (American Type Culture Collection TIB-67) was cultured in complete high-glucose Dulbecco's modified Eagle's medium (supplemented with 10% FBS and 1% penicillin/streptomycin). Macrophages were seeded at the following densities for experiments: 5 × 104 cells in 200 μl per well for 96-well plates (phagocytosis assay), 1 × 106 (lipid analysis) or 1.5 × 106 (flow cytometry) in 5 ml per well for 6-well plates, 1 × 106 (microscopy) in 5 ml per 35 mm glass bottom dish, and 8.6 × 106 (immunoprecipitation/Western blot), or 1.5 × 107 (FRET) in 15 ml for 100 mm dishes. Cells were allowed to adhere at 37 °C + 5% CO2 overnight prior to commencing experiments. AzoSM was synthesized as previously described (73). Sphingomyelinase from Staphylococcus aureus (Sigma), MαCD (Sigma), MβCD (Sigma), cholesterol (Sigma), 7-dehydrocholesterol (Sigma), and coprostanol (Cayman Chemicals) used for lipid-altering treatments were purchased.
Preparation of sterol-loaded MβCD
Sterol-loaded MβCD solutions were prepared as previously described (32, 33). Briefly, sterols dissolved in chloroform were dried under nitrogen gas (N-EVAP; Organomation) followed by high vacuum for at least 1 h (Savant SpeedVac; Thermo Fisher Scientific). They were then hydrated with 2.5 mM MβCD prepared in serum-free medium. Sterol-loaded MβCD solutions were then sonicated (Bransonic Ultrasonic Baths; Branson) for 5 min until no visible particles remained. Next, the sterol-loaded MβCD solutions were incubated at 55 °C for 30 min and then at 37 °C overnight with shaking. Solutions were filtered through a 0.22 μm polyethersulfone syringe filter (Millex-GP; Millipore Sigma) to remove undissolved sterol. Unless otherwise noted, the final solutions, assuming undissolved sterol, were negligible, contained 0.2 mM cholesterol, 7-dehydrocholesterol, or coprostanol mixed with 2.5 mM MβCD.
Cholesterol depletion/sterol repletion using MβCD
Cholesterol was depleted from macrophages using MβCD as previously described (38). Briefly, macrophages were seeded as described previously and allowed to adhere overnight at 37 °C + 5% CO2. Macrophage monolayers were washed twice with 1× Dulbecco's PBS (PBS). About 10 mM or 30 mM MβCD solutions were prepared in serum-free media (Dulbecco's modified Eagle's medium for J774.1 cells and RPMI1640 for MH-S cells) and added to macrophage monolayers as follows: 200 μl for 96-well plates, 2 ml for 6-well plates or 35 mm dishes, and 5 ml for 100 mm dishes. Cells were incubated in MβCD solution for 30 min at 37 °C + 5% CO2 with gentle shaking. After removing the MβCD solution, cells were washed three times with 1× DPBS before processing for analysis.
For sterol repletion, cholesterol was first depleted from macrophages using 10 mM MβCD in serum-free medium for 30 min at 37 °C + 5% CO2 with gentle shaking. The macrophage monolayers were then washed three times with 1× DPBS. Sterol-loaded MβCD solutions were added to the cells as follows: 200 μl for 96-well plates, 1 ml for 6-well plates or 35 mm dishes, and 2 ml for 100 mm dishes. Cells were incubated at 37 °C + 5% CO2 for 2 h with gentle shaking. Macrophage monolayers were then washed four times with 1× DPBS before processing for analysis.
SM depletion using bSMase
To reduce SM in the outer leaflet of the plasma membrane, macrophages were treated with bSMase to catalyze the transformation of SM into ceramide and phosphorylcholine as previously described (40). Briefly, macrophages were seeded as described previously and allowed to adhere overnight at 37 °C + 5% CO2. Macrophage monolayers were washed twice with 1× DPBS. About 50, 100, 250, or 500 mU/ml bSMase solutions were prepared in serum-free media and added to macrophage monolayers as follows: 200 μl for 96-well plates, 2 ml for 6-well plates or 35 mm dishes, and 5 ml for 100 mm dishes. Cells were incubated in bSMase solution for 20 min at 37 °C + 5% CO2 with gentle shaking. Macrophage monolayers were then washed three times with 1× DPBS before processing for analysis.
Preparation of AzoSM-loaded MαCD
AzoSM was loaded into MαCD for SM exchange as previously described (32, 36). Briefly, AzoSM dissolved in chloroform was dried under nitrogen gas (N-EVAP). The lipid was then resuspended in serum-free medium prewarmed to 70 °C, vortexed vigorously, and incubated at 70 °C for 5 min on a heat block. Next, the appropriate amount of MαCD was added to the solution to yield 40 mM MαCD. To load AzoSM into MαCD, the solution was then incubated for 30 to 45 min at 37 °C + 5% CO2 with constant shaking until no visible particles remained. The final solution, assuming undissolved AzoSM was negligible, contained 1.5 mM AzoSM mixed with 40 mM MαCD.
AzoSM exchange using MαCD and photoisomerization
Endogenous SM was exchanged for AzoSM using MαCD as previously described (36, 38). Briefly, macrophages were seeded as described previously and allowed to adhere overnight at 37 °C + 5% CO2. Macrophage monolayers were washed twice with 1× DPBS. AzoSM-loaded MαCD solution was then added to the cells as follows: 200 μl for 96-well plates, 1 ml for 6-well plates, and 2 ml for 100 mm dishes. Cells were incubated at 37 °C + 5% CO2 for 1 h with gentle shaking. Macrophage monolayers were then washed four times with 1× DPBS and incubated in serum-free media. To isomerize AzoSM from the dark-adapted trans conformation to the cis conformation, cells were treated with UV light (λ = 365 nm) for 20 s. To revert the isomerization (from cis conformation back to trans conformation), cells were subsequently treated with blue light (λ = 460 nm) for 40 s. Cells treated with UV light only subsequently remained in the dark to minimize isomerization back to trans.
In vitro phagocytosis assay
About 5 × 104 macrophages were seeded per well in 96-well plates. Cells were allowed to adhere overnight at 37 °C + 5% CO2. C. neoformans H99 cells were grown overnight in yeast nitrogenous base at 30 °C with shaking. Fungal cells were then washed twice with 1× PBS. For antibody-mediated phagocytosis, C. neoformans H99 cells were opsonized for 30 min at 37 °C + 5% CO2 with shaking using 1 μg of anti-GXM antibody (39) per 105 C. neoformans cells in 100 μl of complete media (10% FBS + 1% penicillin/streptomycin). Antibody-opsonized C. neoformans cells were added to washed macrophage monolayers at a 1:1 ratio in 200 μl and allowed to interact for 2 h at 37 °C + 5% CO2 unless noted otherwise. Macrophage monolayers were then washed three times with 1× DPBS to remove extracellular C. neoformans, fixed with ice-cold methanol for 15 min at room temperature (RT), and stained for 5 min at RT with Giemsa (Sigma) diluted 1:1 in deionized water. Stained cells were then washed three times with sterile deionized water and allowed to dry overnight at RT. For complement-mediated phagocytosis assays, C. neoformans H99 cells were opsonized for 1 h at 37 °C + 5% CO2 with shaking using 100 μl of complement-based media (20% mouse CD1 complement serum [Innovative Research] in serum-free medium). Complement-opsonized C. neoformans cells were added to washed macrophage monolayers at a 1:1 ratio with 10 units (1 unit = 0.1 ng/ml) of recombinant murine interferon gamma (Sigma) and 0.3 μg/ml of lipopolysaccharide (Sigma) in 200 μl and allowed to interact for 3 h at 37 °C + 5% CO2. Macrophage monolayers were then washed three times with 1× DPBS to remove extracellular C. neoformans, fixed with ice-cold methanol for 15 min at RT, and stained for 5 min at RT with Giemsa (Sigma) diluted 1:1 in deionized water. Stained cells were then washed three times with sterile deionized water and allowed to dry overnight at RT. Phagocytic index was calculated by microscopic observation. Micrographs were taken of at least four independent fields of view per well. At least 200 macrophage cells were counted per well. Ingested C. neoformans cells within macrophages were counted. Ingested C. neoformans cells were distinguished from attached cells by the stained macrophage cell membrane surrounding the cryptococcal capsule. Phagocytic index is defined as the percentage of yeast cells ingested per macrophage counted per well (9).
Lipid analysis
SM and ceramide species were analyzed as previously described (74). About 1 × 106 macrophages were seeded per well in 6-well plates. Cells were allowed to adhere overnight at 37 °C + 5% CO2. Macrophage monolayers were washed twice with 1× DPBS and treated for SM depletion as described previously. Following treatment, cells were washed three times with 1× DPBS and subsequently scraped in 1 ml ice-cold 1× DPBS. About 10 μl of cell suspension was set aside for protein quantification using the Bradford method. Remaining cells were pelleted at 1000g for 5 min at 4 °C. Lipids were extracted from each cell pellet in 2 ml isopropanol:water:ethyl acetate (30:10:60 by volume). Cell extracts were analyzed by reverse-phase high-pressure LC coupled to electrospray ionization and subsequent separation by MS. Analysis of ceramides and sphingomyelins was performed on a TSQ Quantum Ultra mass spectrometer (Thermo Fisher Scientific), operating in a multiple reaction monitoring positive ionization mode. Sphingolipid levels were normalized to the protein concentration estimated in the original cell extract. All experiments were performed in triplicates.
For analysis of sterols, 1 × 106 macrophages were seeded per well in 6-well plates. Cells were allowed to adhere overnight at 37 °C + 5% CO2. Macrophage monolayers were rinsed twice with 1× DPBS and treated for cholesterol depletion or sterol substitution as described previously. Following treatment, cells were washed three times with 1× DPBS and subsequently scraped in 1 ml ice-cold 1× DPBS. About 10 μl of cell suspension was set aside for protein quantification using the Bradford method. Remaining cells were pelleted at 1000g for 5 min at 4 °C. Sterol derivatization, detection, and analysis were performed by GC–MS as previously described (33, 75). Briefly, lipids were first extracted in 2 ml isopropanol:water:ethyl acetate (30:10:60 by volume), then subjected to mild alkaline base hydrolysis (0.6 M KOH in methanol) to remove the glycerol backbone containing lipid contaminants (74, 76). The extracted and base-hydrolyzed lipid samples were derivatized using 100 μl N,O-bis(trimethylsilyl) trifluoroacetamide/trimethylchlorosilane reagent at 85 °C for 90 min. Next, 50 μl n-hexane was added to the derivatized sample and vortexed. The samples were then analyzed using 30 m (0.25 μm) DB5-MS column on Agilent 7890 GC–MS (Agilent Technologies). The initial column temperature of 10 °C was held for 0.5 min, ramped up at 35 °C/min to 240 °C, then at 3 °C/min to 260 °C, and then at 1.5 °C/min to 305 °C with hold of 2 min. All EI-mass spectra were recorded at 70 eV with ion source temperature of 230 °C. Front inlet temperature was kept at 290 °C, and MSD transfer line temperature was kept at 280 °C. The retention time and mass spectral patterns of the appropriate sterol standards were used as a reference. Ergosterol (25 μg), a nonmammalian sterol, was added as an internal standard for these analyses prior to lipid extraction. Sterol species contents in each sample were quantified using standard calibration curves and normalized to protein concentration estimated in the original cell extract. All experiments were performed in triplicates.
For AzoSM analysis, 1 × 106 macrophages were seeded per well in 6-well plates. Cells were allowed to adhere overnight at 37 °C + 5% CO2. Macrophage monolayers were rinsed twice with 1× DPBS and treated for AzoSM exchange as described previously. Following treatment, cells were washed four times with 1× DPBS and subsequently scraped in 1 ml ice-cold 1× DPBS. About 10 μl of cell suspension was set aside for protein quantification using the Bradford method. Remaining cells were evenly split into two tubes and pelleted at 1000g for 5 min at 4 °C. One tube was set aside for SM analysis as described previously. For AzoSM analysis, cellular lipids were extracted from the second tube for each sample in 1.5 ml Mandala extraction buffer (77), followed by Bligh and Dyer extraction (78), and base hydrolysis (76). Cell extracts were each resuspended in 50 μl methanol and 10 μl applied to HPLC (Agilent Technologies). AzoSM was detected at 365 nm on a C-8 column with a flow rate of 0.5 ml/min in methanol/water (90:10 ratio) buffered with 1 mM ammonium formate and 0.2% formic acid (Fig. S8). Total AzoSM was calculated for each sample and then normalized to the protein concentration estimated in the original cell extract. All experiments were performed in triplicates.
FRET
Biological membrane nanodomain stability was assessed by measuring the temperature dependence of FRET using GPMVs as previously described (36). About 1.5 × 107 macrophages were seeded in 100 mm dishes. Cells were allowed to adhere overnight at 37 °C + 5% CO2. Macrophage monolayers were rinsed twice with 1× DPBS and treated for cholesterol depletion or sterol substitution as described previously. Following treatment, GPMVs were prepared using as previously described by Sezgin et al. (79) and Levental and Levental (79, 80). Briefly, paraformaldehyde (PFA) and DTT were freshly added to the GPMV buffer (10 mM Hepes, 150 mM NaCl, 2 mM CaCl2, and pH 7.4) at a final concentration of 24 mM PFA and 2 mM DTT when desired. Following cholesterol depletion or sterol substitution, macrophage monolayers were washed twice with 1× DPBS and then twice with GPMV buffer lacking PFA and DTT. Next, 2.5 ml of GPMV buffer with PFA and DTT was added and incubated at 37 °C + 5% CO2 with gentle shaking. The GPMV-containing buffer solution was gently harvested by pipetting. To remove intact cells, the solution was centrifuged at 100g for 5 min at RT. This GPMV-containing buffer solution was used for the FRET measurements. FRET measurements were made with diphenylhexatriene (DPH) (Sigma) as the FRET donor and octadecyl rhodamine B (ODRB) (Invitrogen) as the FRET acceptor. The “F sample” with FRET acceptor was prepared by adding 3.6 μl from 1.4 mM ODRB dissolved in ethanol to 1 ml of GPMVs, vortexed, and incubated at 37 °C for 1 h in the dark. The “Fo sample” lacking FRET acceptor was also incubated at 37 °C for 1 h with 3.6 μl ethanol. Before fluorescence measurements, DPH (1.2 μl from a 5 μM stock solution in ethanol) was added to both the “F sample” and “Fo sample” and incubated at RT for 5 min in the dark. Background fluorescence before adding DPH was negligible (<1% of samples containing DPH). To initiate measurements, samples were placed in a cuvette and transferred to a temperature-controlled sample spectrofluorometer (Horiba PTI Quantamaster) sample holder and cooled to 16 °C. DPH fluorescence (excitation λ = 358 nm and emission λ = 430 nm) was measured. Temperature was increased at 4 °C intervals every 5 min up to 64 °C. The ratio of DPH fluorescence intensity in the presence of ODRB to that in its absence (F/F0) was then calculated. All F/F0 values were normalized to the F/F0 value measured at 64 °C. Tend, the temperature above which segregation of lipids into ordered and disordered domains was fully lost (i.e., complete “melting” of ordered domains) was estimated by finding the minimum value of a polynomial fit applied to the F/F0 as previously described (36). The presence of detectable nanodomains between 16 °C and Tend was estimated by calculating the area between the polynomial fit and a horizontal line defined by Tend (i.e., y = y value at x = Tend for polynomial fit). All FRET experiments were performed three times.
Flow cytometry
Flow cytometry was used to assess the presence of FcγRs (CD16/CD32) and complement receptor (CD11b) on the cell surface following lipid-altering treatments. About 1 × 106 macrophages were seeded per well in 6-well plates. Cells were allowed to adhere overnight at 37 °C + 5% CO2. Macrophage monolayers were rinsed twice with 1× DPBS and treated for cholesterol or SM depletion as described previously. Following treatment, cells were washed three times with 1× DPBS and subsequently scraped in 1 ml ice-cold 1× DPBS. Cell concentrations were determined using a hemacytometer. About 1 × 106 cells were transferred into round-bottom test tubes (Falcon) per experimental sample and compensation control (i.e., unstained, single stained) and washed twice with 1× PBS +2% FBS. Cells were labeled with the following antibodies directly conjugated to the following fluorophores: CD45-PE/Cy7 (clone 30-F11; BioLegend), CD11b-BV510 (clone M1/70; BioLegend), and CD16/CD32-APC (clone 93; Invitrogen). Cells were also stained for viability using Alexa Fluor 700 carboxylic acid, succinimidyl ester (Thermo Fisher Scientific). Additional details for the antibodies/markers are available in Table S2. Cells were first incubated with CD16/CD32-APC at 4 °C for 30 min in the dark and then incubated with CD11b-BV510, CD45-PE/Cy7, and the viability stain at 4 °C for 30 min in the dark. All labelings were performed with shaking. Cells were then washed twice with 1× PBS + 2% FBS and fixed with Fluorofix Buffer (BioLegend) at RT for 30 min in the dark with shaking. Cells were washed once and resuspended in 400 μl 1× PBS + 2% FBS for analysis. Labeled cell suspensions were analyzed using the CytoFLEX Flow Cytometer (Beckman Coulter) and FlowJo Software, version 10.6.2 (Becton Dickinson), to measure the median fluorescence intensity of CD16/CD32-APC, CD11b-BV510, and CD45-PE/Cy7 for 50,000 live CD45+ cells. All experiments were performed in triplicates.
FcγR stimulation using IgGICs
To assess activation of the FcγR signaling cascade, macrophages were stimulated with BSA–IgGICs (57, 81). Briefly, IgGIC was prepared by incubating 500 μg of anti-BSA IgG (MP Biomedicals) per 100 μg of BSA (fatty acid free and protease free; Sigma) in 100 μl 1× PBS at RT for 30 min with shaking. IgGIC was diluted with complete media to a final concentration of 100 μg/ml. About 8.6 × 106 macrophages were seeded in 100 mm dishes. Cells were allowed to adhere overnight at 37 °C + 5% CO2. Macrophage monolayers were rinsed twice with 1× DPBS and treated for cholesterol depletion, sterol substitution, or SM depletion as described previously. Following treatment, cells were washed three times with 1× DPBS and incubated with 2.5 ml IgGIC at 37 °C + 5% CO2 with gentle shaking. Cells were then washed four times with 1× DPBS before processing for immunoprecipitation and immunoblotting.
Immunoprecipitation and immunoblotting
Following FcγR stimulation using IgGIC, FcRγ was immunoprecipitated and assessed for phosphorylation of tyrosine residues (57). Cells were lysed directly on the 100 mm dishes using ice-cold radioimmunoprecipitation assay buffer (Sigma) supplemented with 0.1% Triton X-100 (Sigma) and Halt Protease and Phosphatase Inhibitor Cocktail (sodium fluoride, sodium orthovanadate, β-glycerophosphate, sodium pyrophosphate, aprotinin, bestatin, E64, and leupeptin) (Thermo Fisher Scientific). Samples were incubated at 4 °C for 45 min with shaking. Samples were then centrifuged at 4 ˚C, 10,000 rpm for 5 min, and supernatants were transferred to prechilled microtubes. Protein concentration of each sample was determined by the Bradford assay. About 200 μg of each lysate was precleared with 20 μl of Protein A agarose beads (3 mg/ml; Roche) in 200 μl for 1 h at 4 ˚C on a Labquake Rotator (Barnstead Thermolyne). Precleared lysate was diluted to 500 μl with additional lysis buffer and incubated overnight with 40 μl Protein A agarose beads and 4 μg anti-FcRγ-subunit IgG (Millipore Sigma) at 4 °C on a Labquake Rotator. Beads were then centrifuged at 4 °C at 10,000 rpm for 5 min and rinsed five times with lysis buffer. Beads were then mixed with 40 μl of Laemmli Sample Buffer (Bio-Rad) containing 5% β-mercaptoethanol (Sigma) and boiled for 5 min at 95 ˚C. All samples were centrifuged briefly at 14,000 rpm before being loaded onto two SDS-polyacrylamide gel (4–15% gradient gel; Bio-Rad). Precision Plus Protein Kaleidoscope prestained protein ladder (Bio-Rad) was loaded along with the samples. After separation by electrophoresis at 100 V for 60 min in Tris–glycine/SDS buffer, proteins were transferred to Immobilon-PSQ polyvinylidene fluoride membrane (0.2 μm pore size; Millipore Sigma) by electrophoresis at 100 V for 30 min in Tris–glycine/methanol buffer. Nonspecific antibody binding was blocked using 10% nonfat dried milk or 10% BSA in 1× Tris-buffered saline + 0.05% Tween-20 (1× TBST).
After blocking with nonfat dried milk/1× TBST, FcRγ was probed with the anti-FcRγ IgG (Millipore Sigma) overnight at 4 °C with gentle shaking. After blocking with BSA/1× TBST, phosphor-tyrosine was probed with the anti-phosphotyrosine MultiMab mix (Cell Signaling) at 4 °C with gentle shaking. After incubation with primary antibody, membranes were washed five times with 1× TBST. Both membranes were subsequently incubated for 1 h at RT with the Clean-Blot IP Detection Reagent (HRP; Thermo Fisher Scientific). After washing the membrane three times with 1× TBST, the secondary antibody was detected using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific). Bands were visualized using ImageQuant LAS 500 (Cytiva). Bands were quantified using ImageJ. Phosphotyrosine bands were normalized by FcRγ bands. All experiments were performed three times. Additional details for the antibodies are available in Table S2.
Fluorescent lipid labeling
EGFP-conjugated nakanori (EGFP-nakanori), a protein that labels cell surface domains in a SM- and cholesterol-dependent manner (46), was used to label macrophages. Plasmid containing EGFP-nakanori was transformed into Escherichia coli strain BL21. Expression was induced in LB media containing isopropyl β-D-1-thiogalactopyranoside. The EGFP-nakanori protein was subsequently extracted and purified using HisTrap columns (Cytiva) and concentrated using an Amicon centrifugal unit.
Macrophages were subjected to either 10 mM MβCD or 250 mU/ml bSMase treatment as described previously. Live cells were subsequently coincubated with 50 μg EGFP-nakanori for 1 h, fixed with 4% formaldehyde, and imaged using the Axio Observer D1 Phase Contrast Fluorescence Microscope (Zeiss). Micrographs were captured using the AxioCam MR R3 camera (Zeiss).
Statistical analysis
For all statistical analyses, GraphPad Prism (GraphPad Software, Inc), version 8, software was used. Data are represented as mean ± SEM. α-level (type 1 error) was set at 0.05. Differences were considered significant when the probability of type 1 error was less than 5% (p < 0.05). To compare groups, one-way ANOVA with Tukey's multiple comparisons post hoc test was used. All p values from multiple comparisons post hoc tests were corrected for multiplicity using the Bonferroni adjustments.
Data availability
All data are contained within the article.
Supporting information
This article contains supporting information.
Conflict of interest
M. D. P. is a cofounder and Chief Scientific Officer of MicroRid Technologies, Inc. All the other authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We acknowledge Stony Brook Cancer Center (Biological Mass Spectrometry Shared Resource) for expert assistance with sphingolipid analysis and the technical support provided by the Research Flow Cytometry Laboratory at Stony Brook Medicine. We especially thank Izolda Mileva and Todd Rueb. We acknowledge Arturo Casadevall from Johns Hopkins University (Baltimore, MD, USA) for providing the anti-GXM antibody (clone 18B7) used for opsonization of C. neoformans and Yasushi Sako from RIKEN BioResource Research Center (Kyoto, Japan) for providing the EGFP-nakanori plasmid used for cell labeling. We would also like to acknowledge the contributions of Chiara Luberto for the insightful discussion and set up of the initial experiments.
Author contributions
A. M. B., J. K. Y., E. L., and M. D. P. conceptualization; A. M. B., J. K. Y., G. L., J. K., and A. M. F. methodology; G. L. validation; A. M. B., J. K. Y., G. L., J. K., A. M. F., E. L., and M. D. P. formal analysis; A. M. B., J. K. Y., G. L., A. S., J. M., D. T., N. P. d. S., T. G. N., and A. M. F. investigation; M. D. P. and E. L. resources; A. M. B., J. K. Y., A. S., A. M. F., E. L., and M. D. P. data curation; A. M. B., J. K. Y., G. L., and J. K. writing–original draft; A. M. B., J. K. Y., G. L., J. K., A. S., J. M., D. T., A. M. F., E. L., and M. D. P. writing–review and editing; E. L. and M. D. P. supervision; E. L. and M. D. P. project administration; M. D. P. funding acquisition.
Funding and additional information
This work was supported by the National Institutes of Health grants AI136934, AI116420, and AI125770 (to M. D. P.) and GM122493 (to E. L.), and the Merit Review Grant I01BX002924 from the Veterans Affairs Program (to M. D. P.). M. D. P. is a recipient of the Research Career Scientist Award (IK6 BX005386) and a Burroughs Welcome Investigator in Infectious Diseases at the Veterans Administration Medical Center in Northport, NY. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Dennis Voelker
Footnotes
Present address for Arielle M. Bryan: Ingenious Targeting Laboratory, Ronkonkoma, NY, USA.
Present address for Jeehyun Karen You: Case Western Reserve University, Cleveland, OH, USA.
Present address for JiHyun Kim: National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA.
Present address for Ashutosh Singh: Department of Biochemistry, University of Lucknow, Lucknow, Uttar Pradesh, India.
Present address for Johannes Morstein: Department of Cellular and Molecular Pharmacology and Howard Hughes Medical Institute, University of California, San Francisco, California, USA.
Present address for Amir M. Farnoud: Department of Chemical and Biomolecular Engineering, Ohio University, Athens, OH, USA.
Supporting information
References
- 1.Perfect J.R., Casadevall A. Cryptococcosis. Infect. Dis. Clin. North Am. 2002;16:837–874. doi: 10.1016/s0891-5520(02)00036-3. v-vi. [DOI] [PubMed] [Google Scholar]
- 2.Eisenman H.C., Casadevall A., McClelland E.E. New insights on the pathogenesis of invasive Cryptococcus neoformans infection. Curr. Infect. Dis. Rep. 2007;9:457–464. doi: 10.1007/s11908-007-0070-8. [DOI] [PubMed] [Google Scholar]
- 3.Rajasingham R., Smith R.M., Park B.J., Jarvis J.N., Govender N.P., Chiller T.M., Denning D.W., Loyse A., Boulware D.R. Global burden of disease of HIV-associated cryptococcal meningitis: An updated analysis. Lancet Infect. Dis. 2017;17:873–881. doi: 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casadevall A., Coelho C., Alanio A. Mechanisms of Cryptococcus neoformans-mediated host damage. Front. Immunol. 2018;9:855. doi: 10.3389/fimmu.2018.00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson B.N., Hawkins A.N., Wozniak K.L. Pulmonary macrophage and dendritic cell responses to Cryptococcus neoformans. Front. Cell Infect. Microbiol. 2020;10:37. doi: 10.3389/fcimb.2020.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kechichian T.B., Shea J., Del Poeta M. Depletion of alveolar macrophages decreases the dissemination of a glucosylceramide-deficient mutant of Cryptococcus neoformans in immunodeficient mice. Infect. Immun. 2007;75:4792–4798. doi: 10.1128/IAI.00587-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casadevall A. Cryptococci at the brain gate: Break and enter or use a Trojan horse? J. Clin. Invest. 2010;120:1389–1392. doi: 10.1172/JCI42949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chretien F., Lortholary O., Kansau I., Neuville S., Gray F., Dromer F. Pathogenesis of cerebral Cryptococcus neoformans infection after fungemia. J. Infect. Dis. 2002;186:522–530. doi: 10.1086/341564. [DOI] [PubMed] [Google Scholar]
- 9.Luberto C., Martinez-Marino B., Taraskiewicz D., Bolanos B., Chitano P., Toffaletti D.L., Cox G.M., Perfect J.R., Hannun Y.A., Balish E., Del Poeta M. Identification of App1 as a regulator of phagocytosis and virulence of Cryptococcus neoformans. J. Clin. Invest. 2003;112:1080–1094. doi: 10.1172/JCI18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ben-Abdallah M., Sturny-Leclere A., Ave P., Louise A., Moyrand F., Weih F., Janbon G., Memet S. Fungal-induced cell cycle impairment, chromosome instability and apoptosis via differential activation of NF-kappaB. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coelho C., Souza A.C., Derengowski Lda S., de Leon-Rodriguez C., Wang B., Leon-Rivera R., Bocca A.L., Goncalves T., Casadevall A. Macrophage mitochondrial and stress response to ingestion of Cryptococcus neoformans. J. Immunol. 2015;194:2345–2357. doi: 10.4049/jimmunol.1402350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma H., Croudace J.E., Lammas D.A., May R.C. Expulsion of live pathogenic yeast by macrophages. Curr. Biol. 2006;16:2156–2160. doi: 10.1016/j.cub.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 13.Alvarez M., Casadevall A. Cell-to-cell spread and massive vacuole formation after Cryptococcus neoformans infection of murine macrophages. BMC Immunol. 2007;8:16. doi: 10.1186/1471-2172-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu T.B., Perlin D.S., Xue C.Y. Molecular mechanisms of cryptococcal meningitis. Virulence. 2012;3:173–181. doi: 10.4161/viru.18685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McQuiston T.J., Williamson P.R. Paradoxical roles of alveolar macrophages in the host response to Cryptococcus neoformans. J. Infect. Chemother. 2012;18:1–9. doi: 10.1007/s10156-011-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Rodas R., Zaragoza O. Catch me if you can: Phagocytosis and killing avoidance by Cryptococcus neoformans. FEMS Immunol. Med. Microbiol. 2012;64:147–161. doi: 10.1111/j.1574-695X.2011.00871.x. [DOI] [PubMed] [Google Scholar]
- 17.Coelho C., Bocca A.L., Casadevall A. The intracellular life of Cryptococcus neoformans. Annu. Rev. Pathol. 2014;9:219–238. doi: 10.1146/annurev-pathol-012513-104653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarantis H., Grinstein S. Subversion of phagocytosis for pathogen survival. Cell Host Microbe. 2012;12:419–431. doi: 10.1016/j.chom.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 19.Rougerie P., Miskolci V., Cox D. Generation of membrane structures during phagocytosis and chemotaxis of macrophages: Role and regulation of the actin cytoskeleton. Immunol. Rev. 2013;256:222–239. doi: 10.1111/imr.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao M., Peachman K.K., Alving C.R., Rothwell S.W. Depletion of cellular cholesterol interferes with intracellular trafficking of liposome-encapsulated ovalbumin. Immunol. Cell Biol. 2003;81:415–423. doi: 10.1046/j.1440-1711.2003.01192.x. [DOI] [PubMed] [Google Scholar]
- 21.Sein K.K., Aikawa M. The prime role of plasma membrane cholesterol in the pathogenesis of immune evasion and clinical manifestations of falciparum malaria. Med. Hypotheses. 1998;51:105–110. doi: 10.1016/s0306-9877(98)90102-5. [DOI] [PubMed] [Google Scholar]
- 22.Pucadyil T.J., Tewary P., Madhubala R., Chattopadhyay A. Cholesterol is required for Leishmania donovani infection: Implications in leishmaniasis. Mol. Biochem. Parasitol. 2004;133:145–152. doi: 10.1016/j.molbiopara.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Kannan S., Audet A., Huang H., Chen L.J., Wu M. Cholesterol-rich membrane rafts and Lyn are involved in phagocytosis during Pseudomonas aeruginosa infection. J. Immunol. 2008;180:2396–2408. doi: 10.4049/jimmunol.180.4.2396. [DOI] [PubMed] [Google Scholar]
- 24.Wang M., Hajishengallis G. Lipid raft-dependent uptake, signalling and intracellular fate of Porphyromonas gingivalis in mouse macrophages. Cell Microbiol. 2008;10:2029–2042. doi: 10.1111/j.1462-5822.2008.01185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee J.-H., Kim D.Y., Lee S.H., Song D.-K., Bae J.-H., Im S.-S. LPS-induced mTORC1 signaling activates lipid raft-actin cytoskeletal interactions for phagocytosis through SREBP-1a. 2019;33:lb318. [Google Scholar]
- 26.Magenau A., Benzing C., Proschogo N., Don A.S., Hejazi L., Karunakaran D., Jessup W., Gaus K. Phagocytosis of IgG-coated polystyrene beads by macrophages induces and requires high membrane order. Traffic. 2011;12:1730–1743. doi: 10.1111/j.1600-0854.2011.01272.x. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt F., Thywissen A., Goldmann M., Cunha C., Cseresnyes Z., Schmidt H., Rafiq M., Galiani S., Graler M.H., Chamilos G., Lacerda J.F., Campos A., Jr., Eggeling C., Figge M.T., Heinekamp T., et al. Flotillin-dependent membrane microdomains are required for functional phagolysosomes against fungal infections. Cell Rep. 2020;32:108017. doi: 10.1016/j.celrep.2020.108017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crane J.M., Tamm L.K. Role of cholesterol in the formation and nature of lipid rafts in planar and spherical model membranes. Biophysical J. 2004;86:2965–2979. doi: 10.1016/S0006-3495(04)74347-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown D.A., London E. Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- 30.Simons K., Toomre D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 31.Rajendran L., Simons K. Lipid rafts and membrane dynamics. J. Cell Sci. 2005;118:1099–1102. doi: 10.1242/jcs.01681. [DOI] [PubMed] [Google Scholar]
- 32.Li G.T., Kim J., Huang Z., St Clair J.R., Brown D.A., London E. Efficient replacement of plasma membrane outer leaflet phospholipids and sphingolipids in cells with exogenous lipids. Proc. Natl. Acad. Sci. U. S. A. 2016;113:14025–14030. doi: 10.1073/pnas.1610705113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J., Singh A., Del Poeta M., Brown D.A., London E. The effect of sterol structure upon clathrin-mediated and clathrin-independent endocytosis. J. Cell Sci. 2017;130:2682–2695. doi: 10.1242/jcs.201731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diaz O., Mebarek-Azzam S., Benzaria A., Dubois M., Lagarde M., Nemoz G., Prigent A.F. Disruption of lipid rafts stimulates phospholipase d activity in human lymphocytes: Implication in the regulation of immune function. J. Immunol. 2005;175:8077–8086. doi: 10.4049/jimmunol.175.12.8077. [DOI] [PubMed] [Google Scholar]
- 35.Audi A., Soudani N., Dbaibo G., Zaraket H. Depletion of host and viral sphingomyelin impairs influenza virus infection. Front. Microbiol. 2020;11:612. doi: 10.3389/fmicb.2020.00612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li G., Wang Q., Kakuda S., London E. Nanodomains can persist at physiologic temperature in plasma membrane vesicles and be modulated by altering cell lipids. J. Lipid Res. 2020;61:758–766. doi: 10.1194/jlr.RA119000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suresh P., Miller W.T., London E. Phospholipid exchange shows insulin receptor activity is supported by both the propensity to form wide bilayers and ordered raft domains. J. Biol. Chem. 2021;297 doi: 10.1016/j.jbc.2021.101010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bryan A.M., Farnoud A.M., Mor V., Del Poeta M. Macrophage cholesterol depletion and its effect on the phagocytosis of Cryptococcus neoformans. J. Vis. Exp. 2014 doi: 10.3791/52432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Casadevall A., Cleare W., Feldmesser M., Glatman-Freedman A., Goldman D.L., Kozel T.R., Lendvai N., Mukherjee J., Pirofski L.A., Rivera J., Rosas A.L., Scharff M.D., Valadon P., Westin K., Zhong Z.J. Characterization of a murine monoclonal antibody to Cryptococcus neoformans polysaccharide that is a candidate for human therapeutic studies. Antimicrob. Agents Chemother. 1998;42:1437–1446. doi: 10.1128/aac.42.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Canals D., Jenkins R.W., Roddy P., Hernandez-Corbacho M.J., Obeid L.M., Hannun Y.A. Differential effects of ceramide and sphingosine 1-phosphate on ERM phosphorylation PROBING sphingolipid signaling at the outer plasma membrane. J. Biol. Chem. 2010;285:32476–32485. doi: 10.1074/jbc.M110.141028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zidovetzki R., Levitan I. Use of cyclodextrins to manipulate plasma membrane cholesterol content: Evidence, misconceptions and control strategies. Biochim. Biophys. Acta. 2007;1768:1311–1324. doi: 10.1016/j.bbamem.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu X., London E. The effect of sterol structure on membrane lipid domains reveals how cholesterol can induce lipid domain formation. Biochemistry. 2000;39:843–849. doi: 10.1021/bi992543v. [DOI] [PubMed] [Google Scholar]
- 43.Xu X., Bittman R., Duportail G., Heissler D., Vilcheze C., London E. Effect of the structure of natural sterols and sphingolipids on the formation of ordered sphingolipid/sterol domains (rafts). Comparison of cholesterol to plant, fungal, and disease-associated sterols and comparison of sphingomyelin, cerebrosides, and ceramide. J. Biol. Chem. 2001;276:33540–33546. doi: 10.1074/jbc.M104776200. [DOI] [PubMed] [Google Scholar]
- 44.Megha, London E. Ceramide selectively displaces cholesterol from ordered lipid domains (rafts): Implications for lipid raft structure and function. J. Biol. Chem. 2004;279:9997–10004. doi: 10.1074/jbc.M309992200. [DOI] [PubMed] [Google Scholar]
- 45.Megha, Bakht O., London E. Cholesterol precursors stabilize ordinary and ceramide-rich ordered lipid domains (lipid rafts) to different degrees. Implications for the bloch hypothesis and sterol biosynthesis disorders. J. Biol. Chem. 2006;281:21903–21913. doi: 10.1074/jbc.M600395200. [DOI] [PubMed] [Google Scholar]
- 46.Makino A., Abe M., Ishitsuka R., Murate M., Kishimoto T., Sakai S., Hullin-Matsuda F., Shimada Y., Inaba T., Miyatake H., Tanaka H., Kurahashi A., Pack C.G., Kasai R.S., Kubo S., et al. A novel sphingomyelin/cholesterol domain-specific probe reveals the dynamics of the membrane domains during virus release and in Niemann- Pick type C. FASEB J. 2017;31:1301–1322. doi: 10.1096/fj.201500075R. [DOI] [PubMed] [Google Scholar]
- 47.Morstein J., Impastato A.C., Trauner D. Photoswitchable lipids. Chembiochem. 2021;22:73–83. doi: 10.1002/cbic.202000449. [DOI] [PubMed] [Google Scholar]
- 48.Doroudgar M., Morstein J., Becker-Baldus J., Trauner D., Glaubitz C. How photoswitchable lipids affect the order and dynamics of lipid bilayers and embedded proteins. J. Am. Chem. Soc. 2021 doi: 10.1021/jacs.1c03524. [DOI] [PubMed] [Google Scholar]
- 49.Frank J.A., Franquelim H.G., Schwille P., Trauner D. Optical control of lipid rafts with photoswitchable ceramides. J. Am. Chem. Soc. 2016;138:12981–12986. doi: 10.1021/jacs.6b07278. [DOI] [PubMed] [Google Scholar]
- 50.Kol M., Williams B., Toombs-Ruane H., Franquelim H.G., Korneev S., Schroeer C., Schwille P., Trauner D., Holthuis J.C., Frank J.A. Optical manipulation of sphingolipid biosynthesis using photoswitchable ceramides. Elife. 2019;8 doi: 10.7554/eLife.43230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morstein J., Hill R.Z., Novak A.J.E., Feng S., Norman D.D., Donthamsetti P.C., Frank J.A., Harayama T., Williams B.M., Parrill A.L., Tigyi G.J., Riezman H., Isacoff E.Y., Bautista D.M., Trauner D. Optical control of sphingosine-1-phosphate formation and function. Nat. Chem. Biol. 2019;15:623–631. doi: 10.1038/s41589-019-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morstein J., Kol M., Novak A.J.E., Feng S., Khayyo S., Hinnah K., Li-Purcell N., Pan G., Williams B.M., Riezman H., Atilla-Gokcumen G.E., Holthuis J.C.M., Trauner D. Short photoswitchable ceramides enable optical control of apoptosis. ACS Chem. Biol. 2021;16:452–456. doi: 10.1021/acschembio.0c00823. [DOI] [PubMed] [Google Scholar]
- 53.Hartrampf N., Seki T., Baumann A., Watson P., Vepřek N.A., Hetzler B.E., Hoffmann-Röder A., Tsuji M., Trauner D. Optical control of cytokine production using photoswitchable galactosylceramides. Chemistry. 2020;26:4476–4479. doi: 10.1002/chem.201905279. [DOI] [PubMed] [Google Scholar]
- 54.Morstein J., Awale M., Reymond J.-L., Trauner D. Mapping the azolog space enables the optical control of new biological targets. ACS Cent. Sci. 2019;5:607–618. doi: 10.1021/acscentsci.8b00881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bruhns P. Properties of mouse and human IgG receptors and their contribution to disease models. Blood. 2012;119:5640–5649. doi: 10.1182/blood-2012-01-380121. [DOI] [PubMed] [Google Scholar]
- 56.Cross C.E., Collins H.L., Bancroft G.J. CR3-dependent phagocytosis by murine macrophages: Different cytokines regulate ingestion of a defined CR3 ligand and complement-opsonized Cryptococcus neoformans. Immunology. 1997;91:289–296. doi: 10.1046/j.1365-2567.1997.00238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rittirsch D., Flierl M.A., Day D.E., Nadeau B.A., Zetoune F.S., Sarma J.V., Werner C.M., Wanner G.A., Simmen H.P., Huber-Lang M.S., Ward P.A. Cross-talk between TLR4 and FcgammaReceptorIII (CD16) pathways. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yuan R., Clynes R., Oh J., Ravetch J.V., Scharff M.D. Antibody-mediated modulation of Cryptococcus neoformans infection is dependent on distinct Fc receptor functions and IgG subclasses. J. Exp. Med. 1998;187:641–648. doi: 10.1084/jem.187.4.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kara S., Amon L., Luhr J.J., Nimmerjahn F., Dudziak D., Lux A. Impact of plasma membrane domains on IgG Fc receptor function. Front. Immunol. 2020;11:1320. doi: 10.3389/fimmu.2020.01320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kondadasula S.V., Roda J.M., Parihar R., Yu J., Lehman A., Caligiuri M.A., Tridandapani S., Burry R.W., Carson W.E., 3rd. Colocalization of the IL-12 receptor and FcgammaRIIIa to natural killer cell lipid rafts leads to activation of ERK and enhanced production of interferon-gamma. Blood. 2008;111:4173–4183. doi: 10.1182/blood-2007-01-068908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Galandrini R., Tassi I., Mattia G., Lenti L., Piccoli M., Frati L., Santoni A. SH2-containing inositol phosphatase (SHIP-1) transiently translocates to raft domains and modulates CD16-mediated cytotoxicity in human NK cells. Blood. 2002;100:4581–4589. doi: 10.1182/blood-2002-04-1058. [DOI] [PubMed] [Google Scholar]
- 62.St Clair J.W., London E. Effect of sterol structure on ordered membrane domain (raft) stability in symmetric and asymmetric vesicles. Biochim. Biophys. Acta Biomembr. 2019;1861:1112–1122. doi: 10.1016/j.bbamem.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Delle Bovi R.J., Kim J., Suresh P., London E., Miller W.T. Sterol structure dependence of insulin receptor and insulin-like growth factor 1 receptor activation. Biochim. Biophys. Acta Biomembr. 2019;1861:819–826. doi: 10.1016/j.bbamem.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Slotte J.P., Hedstrom G., Rannstrom S., Ekman S. Effects of sphingomyelin degradation on cell cholesterol oxidizability and steady-state distribution between the cell surface and the cell interior. Biochim. Biophys. Acta. 1989;985:90–96. doi: 10.1016/0005-2736(89)90108-9. [DOI] [PubMed] [Google Scholar]
- 65.Ridgway N.D., Lagace T.A., Cook H.W., Byers D.M. Differential effects of sphingomyelin hydrolysis and cholesterol transport on oxysterol-binding protein phosphorylation and Golgi localization. J. Biol. Chem. 1998;273:31621–31628. doi: 10.1074/jbc.273.47.31621. [DOI] [PubMed] [Google Scholar]
- 66.Grassme H., Jendrossek V., Riehle A., von Kurthy G., Berger J., Schwarz H., Weller M., Kolesnick R., Gulbins E. Host defense against Pseudomonas aeruginosa requires ceramide-rich membrane rafts. Nat. Med. 2003;9:322–330. doi: 10.1038/nm823. [DOI] [PubMed] [Google Scholar]
- 67.Grassme H., Schwarz H., Gulbins E. Molecular mechanisms of ceramide-mediated CD95 clustering. Biochem. Biophys. Res. Commun. 2001;284:1016–1030. doi: 10.1006/bbrc.2001.5045. [DOI] [PubMed] [Google Scholar]
- 68.Suchard S.J., Hinkovska-Galcheva V., Mansfield P.J., Boxer L.A., Shayman J.A. Ceramide inhibits IgG-dependent phagocytosis in human polymorphonuclear leukocytes. Blood. 1997;89:2139–2147. [PubMed] [Google Scholar]
- 69.Abdel Shakor A.B., Kwiatkowska K., Sobota A. Cell surface ceramide generation precedes and controls FcgammaRII clustering and phosphorylation in rafts. J. Biol. Chem. 2004;279:36778–36787. doi: 10.1074/jbc.M402170200. [DOI] [PubMed] [Google Scholar]
- 70.Ravetch J.V., Lanier L.L. Immune inhibitory receptors. Science. 2000;290:84–89. doi: 10.1126/science.290.5489.84. [DOI] [PubMed] [Google Scholar]
- 71.Varshney P., Yadav V., Saini N. Lipid rafts in immune signalling: Current progress and future perspective. Immunology. 2016;149:13–24. doi: 10.1111/imm.12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Turner H., Kinet J.-P. Signalling through the high-affinity IgE receptor FcεRI. Nature. 1999;402:24–30. doi: 10.1038/35037021. [DOI] [PubMed] [Google Scholar]
- 73.Hartrampf N., Leitao S.M., Winter N., Toombs-Ruane H., Frank J.A., Schwille P., Trauner D., Franquelim H.G. Structural diversity of photoswitchable sphingolipids for optodynamic control of lipid raft microdomains. bioRxiv. 2021;2010 doi: 10.1101/2021.10.11.463883. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bielawski J., Pierce J.S., Snider J., Rembiesa B., Szulc Z.M., Bielawska A. Sphingolipid analysis by high performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) Adv. Exp. Med. Biol. 2010;688:46–59. doi: 10.1007/978-1-4419-6741-1_3. [DOI] [PubMed] [Google Scholar]
- 75.Singh A., MacKenzie A., Girnun G., Del Poeta M. Analysis of sphingolipids, sterols, and phospholipids in human pathogenic Cryptococcus strains. J. Lipid Res. 2017;58:2017–2036. doi: 10.1194/jlr.M078600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Clarke N.G., Dawson R.M. Alkaline O leads to N-transacylation. A new method for the quantitative deacylation of phospholipids. Biochem. J. 1981;195:301–306. doi: 10.1042/bj1950301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mandala S.M., Thornton R.A., Frommer B.R., Curotto J.E., Rozdilsky W., Kurtz M.B., Giacobbe R.A., Bills G.F., Cabello M.A., Martin I., Pelaez F., Harris G.H. The discovery of australifungin, a novel inhibitor of sphinganine N-acyltransferase from Sporormiella australis. Producing organism, fermentation, isolation, and biological activity. J. Antibiot. (Tokyo) 1995;48:349–356. doi: 10.7164/antibiotics.48.349. [DOI] [PubMed] [Google Scholar]
- 78.Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 79.Sezgin E., Kaiser H.J., Baumgart T., Schwille P., Simons K., Levental I. Elucidating membrane structure and protein behavior using giant plasma membrane vesicles. Nat. Protoc. 2012;7:1042–1051. doi: 10.1038/nprot.2012.059. [DOI] [PubMed] [Google Scholar]
- 80.Levental K.R., Levental I. Isolation of giant plasma membrane vesicles for evaluation of plasma membrane structure and protein partitioning. Methods Mol. Biol. 2015;1232:65–77. doi: 10.1007/978-1-4939-1752-5_6. [DOI] [PubMed] [Google Scholar]
- 81.Sun L., Guo R.F., Gao H., Sarma J.V., Zetoune F.S., Ward P.A. Attenuation of IgG immune complex-induced acute lung injury by silencing C5aR in lung epithelial cells. FASEB J. 2009;23:3808–3818. doi: 10.1096/fj.09-133694. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.