Abstract
Background
Stereotactic radiosurgery (SRS) may cause radiation necrosis (RN) that is difficult to distinguish from tumor progression (TP) by conventional MRI. We hypothesize that MRI-based multiparametric radiomics (mpRad) and machine learning (ML) can differentiate TP from RN in a multi-institutional cohort.
Methods
Patients with growing brain metastases after SRS at 2 institutions underwent surgery, and RN or TP were confirmed by histopathology. A radiomic tissue signature (RTS) was selected from mpRad, as well as single T1 post-contrast (T1c) and T2 fluid-attenuated inversion recovery (T2-FLAIR) radiomic features. Feature selection and supervised ML were performed in a randomly selected training cohort (N = 95) and validated in the remaining cases (N = 40) using surgical pathology as the gold standard.
Results
One hundred and thirty-five discrete lesions (37 RN, 98 TP) from 109 patients were included. Radiographic diagnoses by an experienced neuroradiologist were concordant with histopathology in 67% of cases (sensitivity 69%, specificity 59% for TP). Radiomic analysis indicated institutional origin as a significant confounding factor for diagnosis. A random forest model incorporating 1 mpRad, 4 T1c, and 4 T2-FLAIR features had an AUC of 0.77 (95% confidence interval [CI]: 0.66–0.88), sensitivity of 67% and specificity of 86% in the training cohort, and AUC of 0.71 (95% CI: 0.51–0.91), sensitivity of 52% and specificity of 90% in the validation cohort.
Conclusions
MRI-based mpRad and ML can distinguish TP from RN with high specificity, which may facilitate the triage of patients with growing brain metastases after SRS for repeat radiation versus surgical intervention.
Keywords: brain metastasis, machine learning, magnetic resonance imaging, multiparametric radiomics, radiation necrosis
Key Points.
A growing lesion after stereotactic radiosurgery is difficult to diagnose on conventional MRI.
Multiparametric radiomics and machine learning can provide accurate, noninvasive diagnosis.
Importance of the Study.
Stereotactic radiosurgery (SRS) is a key treatment option for patients with brain metastases. It may cause radiation necrosis, which can manifest clinically and radiographically indistinguishable from tumor progression on conventional MRI. Using the largest, pathologically confirmed, multi-institutional collection of post-SRS radiation necrosis and progression, we identified and validated a novel multiparametric radiomic tissue signature for radiation necrosis. Our study demonstrates the utility of combining multiparametric radiomics and machine learning in ascertaining individual diagnosis. Furthermore, our results highlight important potential confounding factors in the research of imaging biomarkers, namely different clinical practices and imaging parameters employed by different institutions. The multiparametric radiomic signature identified in our study has the important clinical utility of helping to triage patients for repeat radiation versus surgical sampling in the presence of lesion growth after SRS.
Stereotactic radiosurgery (SRS) is a standard of care in patients with brain metastasis.1,2 SRS provides excellent local control, but one of the most common side effects is radiation necrosis (RN). The incidence of RN after SRS can range between 5% and 25%, depending on the definition, diagnostic approach, and whether pathologic confirmation is obtained.3 However, tumor progression (TP) can often manifest with similar imaging characteristics on conventional MRI (cMRI) with T1 post-contrast (T1c) and T2 fluid-attenuated inversion recovery (FLAIR) sequences.4,5 Clinically, accurate diagnosis of a growing lesion is imperative, because early salvage intervention, such as repeat SRS, may offer superior oncologic outcomes for patients with progressive tumor, whereas RN may often stabilize or resolve spontaneously, obviating the need for invasive procedures. Accumulating evidence indicates that repeat SRS may be safely delivered in cases of confirmed TP.6–8 Moreover, in patients with RN who are asymptomatic or minimally symptomatic, surgical interventions may interfere with the quality of life.9,10 Therefore, noninvasive diagnostic approaches may allow selection of the subset of patients who may benefit from repeat radiation and reduce the need for surgical intervention.
Currently, lesion growth after SRS often requires confirmation by advanced MRI or surgical pathology, as cMRI has shown limited clinical utility in differentiating TP from RN.11 Reports have indicated wide variation in sensitivity (43%–91%) and specificity (33%–88%) using qualitative cMRI features.12–14 In contrast, radiomics can extract quantitative texture features, which can serve as noninvasive biomarkers for various disease processes.15–17 CT and MRI radiomic features have been shown to correlate with benign versus malignant processes.18–20 Furthermore, newer methods, such as multiparametric radiomics (mpRad), offer more complete tissue characterization.20,21 MpRad has outperformed single radiomic parameters in predicting breast cancer recurrence when correlated with Oncotype gene array methods.22 Moreover, radiomics combined with machine learning (ML) have been applied in patients with brain metastases to distinguish RN from TP with moderate success.23–26 However, previous studies only included small, single-institution cohorts, with limited pathologic information and little to no external validation.
We previously published an MRI radiomic tissue signature (RTS-1) consisting of 6 T1c and 4 T2-FLAIR features, which showed an area under the curve (AUC) of 0.81 for the receiver operating characteristics (ROC) using an Isomap SVM (IsoSVM) algorithm in a single-institution series.27 The aim of this study is to test whether the addition of mpRad features can improve the diagnostic accuracy of the ML classifier in a large, multi-institutional patient cohort of post-SRS RN and TP, using surgical pathology as the gold standard.
Methods and Materials
Patients
This study was approved by the Institutional Review Board. Patients with brain metastases treated with SRS at 2 independent institutions (Johns Hopkins University [JH] and Wake Forest University [WF]) from 2003 through 2017 were included. Patients whose index lesions were previously treated with whole brain radiotherapy (WBRT) or surgery were eligible. The JH cohort was included in a previous single-institution study.27 Brain metastases that increased in size on cMRI subsequent to SRS, generally through several scans and with associated symptoms, were surgically sampled for therapeutic benefit, pathologic confirmation, and/or symptom relief. A small subset of cases (5 lesions) where the growing lesions subsequently stabilized or regressed were also included as RN as per prior study.27 Lesions that had mixed RN and TP on pathology were classified as TP.27
Neuroradiologist Interpretation
The pre- and post-SRS MRIs were reviewed by a board-certified neuroradiologist (D.L.) with 20 years of post-fellowship experience blinded to the histopathologic diagnosis. Radiographic diagnoses of either RN or TP were made based on cMRI lesion characteristics. Mixed lesions, for which at least part of the lesion on cMRI was interpreted as tumor, were classified as progression to be consistent with pathologic interpretation. The JH cohort was previously reviewed,27 and was re-reviewed in the same manner as the WF cases for the current study to ensure consistency between the 2 cohorts.
MRI Acquisition and Radiomic Feature Extraction
MRI acquisition parameters are listed in Supplementary Table S1. There were significant variations in the MRI scanners and imaging parameters used. This study followed the previously published radiomics workflow.27 In brief, T2-FLAIR images were rigidly registered to the T1c sequence based on bony anatomical references using the Velocity AI software (Varian Medical Systems). The lesions were segmented manually based on the single largest diameter seen on the T1c sequence. Fifty-six single radiomic features (for each MRI sequence) were extracted using an in-house software developed with MATLAB (The Math Works). In addition, 39 mpRad features from the combined T1c and T2-FLAIR images were also extracted as previously described.20 The full list of single and mpRad features are listed in Supplementary Table S2.
Exploratory Data Analysis
To visualize the heterogeneity and clustering tendency of the combined dataset, we performed t-distributed stochastic neighbor embedding (tSNE) and hierarchical clustering using “Rtsne” and “ComplexHeatmap” packages in R software (version 3.6.3).28,29 The tSNE is a nonlinear dimensionality reduction technique to visualize complex data in a 2-dimensional space.28 Radiomic features were first normalized by centering the means at 0 and scaling by standard deviation. The parameters for tSNE analysis were determined empirically as follows: dimension 2, perplexity 30, max iteration 10,000, and learning rate 5. Hierarchical clustering was performed in a bottom up manner using complete linkage method, with 4 clusters of cases split by institutional origins and pathologic diagnoses (Supplementary Figure S1).
Radiomic Tissue Signature (RTS-2) Selection
Seventy percent of the combined dataset was randomly assigned to training (N = 95) and the remaining 30% to validation (N = 40). All feature selection and model optimization procedures were performed in the training dataset to avoid information leakage using the “Caret” package in R software. Pathologically confirmed RN and TP lesions were compared using 2-sided t-tests, and those with P values <.25 were further selected by recursive elimination (Supplementary Table S3, RN vs TP). The threshold used here was iteratively selected to improve inclusiveness of radiomic features and model performance. Given the potential confounding factor of institutional origin, 16 features that were strongly correlated with institutional origin (JH vs WF P < 3.60 × 10-4 on t-tests with Bonferroni correction) were excluded from further feature selection (Supplementary Table S3, features labeled with *). Finally, all features from the previous RTS-1 was also included in recursive elimination.27 This resulted in 19 T1c, 18 T2-FLAIR, and 5 mpRad features for recursive elimination, which was performed by iterative elimination of one feature from the training dataset and maximizing the AUC of a random forest model based on the remaining features in each iteration.
Statistical Analyses
All statistical analyses were performed in the R software. Clinical characteristics were compared using Fisher’s exact tests for categorical variables or Wilcoxon rank sum tests for continuous variables. Radiographic diagnoses by the neuroradiologist were compared to pathologic diagnoses, and the sensitivity and specificity were calculated using a confusion matrix. Intra-observer reliability was assessed in the JH cohort by comparing the initial radiographic diagnoses for the previous study to the diagnoses from re-review in the current study using kappa test. Radiomic features were compared between institutional origins (JH vs WF) and between diagnoses (RN vs TP) using 2-sided t-tests. Detailed description of machine learning methods can be found in the Supplementary Material. Performance of ML models was evaluated using AUCs in the training and testing datasets. The optimal sensitivity and specificity were calculated by maximizing the Youden index. Wilcoxon rank sum tests were used to compare the median AUCs of the 2 RTSs and the sampling techniques (none vs random oversampling vs SMOTE). DeLong’s tests were used to compare ROCs of the 4 ML algorithms. Statistical significance was defined as P < .05.
Results
Patients and Treatment Characteristics
A total of 135 lesions (37 pure RN, 78 pure TP, 20 mixed) from 109 patients were evaluated. Table 1 lists the baseline patient and treatment characteristics. The WF dataset contained more cases of pure TP (69.8 vs 50.0%, P = .01), fewer lesions in the frontal lobe (9.4 vs 35.4%) and more in the cerebellum (28.3 vs 17.1%, P = .009). The JH dataset contained more postoperative cavities (41.5 vs 5.7%, P < .001) and fewer patients who received prior WBRT (18.3 vs 41.5%, P = .005). The majority of the SRS treatments at JH were delivered using a robotic system (70.7%) or linear accelerator (LINAC, 19.5%), while all SRS treatments at WF were delivered using a Cobalt-60 system (P < .001). Thirty-seven patients in the JH cohort (45.1%) and none in the WF cohort received multi-fraction SRS in 3–5 fractions. The remaining clinical characteristics were not significantly different between JH and WF cohorts, including primary histology (most commonly non-small cell lung cancer and breast, P = .06), lesion volume (median 3.7 vs 2.5cm3, P = .72), WBRT dose (median 35 vs 35Gy, P = .08), SRS marginal dose (median 20 vs 18Gy, P = .05), and days from SRS to surgery (median 278 vs 321 days, P = .32).
Table 1.
Patient and Treatment Characteristics
| Total N = 135 |
JH N = 82 |
WF N = 53 |
P values JH vs WF |
|
|---|---|---|---|---|
| Male (%) | 50 (37.0) | 30 (36.6) | 20 (37.7) | 1 |
| Pathology/N (%) | .01 | |||
| RN | 37 (27.4) | 30 (36.6) | 7 (13.2) | |
| TP | 78 (57.8) | 41 (50.0) | 37 (69.8) | |
| Mixed | 20 (14.8) | 11 (13.4) | 9 (17.0) | |
| Primary histology | .06 | |||
| NSCLC | 41 (30.4) | 28 (34.1) | 13 (24.5) | |
| Breast | 36 (26.7) | 16 (19.5) | 20 (37.7) | |
| Melanoma | 27 (20.0) | 21 (25.6) | 6 (11.3) | |
| SCLC | 12 (8.9) | 6 (7.3) | 6 (11.3) | |
| Other | 19 (14.1) | 11 (13.4) | 8 (15.1) | |
| Tumor Characteristics | ||||
| Lesion location (%) | .009 | |||
| Frontal | 34 (25.2) | 29 (35.4) | 5 (9.4) | |
| Parietal | 33 (24.4) | 16 (19.5) | 17 (32.1) | |
| Temporal | 24 (17.8) | 14 (17.1) | 10 (18.9) | |
| Occipital | 15 (11.1) | 9 (11.0) | 6 (11.3) | |
| Cerebellar | 29 (21.5) | 14 (17.1) | 15 (28.3) | |
| Post-op cavity | 37 (27.4) | 34 (41.5) | 3 (5.7) | <.001 |
| Lesion volume/cm3 | 3.3 [0.04–52.8] |
3.7 [0.04–36.4] |
2.5 [0.18–52.8] |
.72 |
| Radiation Parameters | ||||
| Prior WBRT | 37 (27.4) | 15 (18.3) | 22 (41.5) | .005 |
| WBRT dose/Gy | 35 [25–40] | 35 [25–37] | 35 [30–40] | .08 |
| SRS technique | <.001 | |||
| Robotic | 58 (43.0) | 58 (70.7) | 0 (0.0) | |
| Cobalt-60 | 61 (45.2) | 8 (9.8) | 53 (100.0) | |
| LINAC | 16 (11.9) | 16 (19.5) | 0 (0.0) | |
| SRS marginal dose/Gy | 20 [10–25] | 20 [14–25] | 18 [10–22] | .05 |
| SRS fractions | 1 [1–5] | 1 [1–5] | 1 [1–1] | <.001 |
| SRS prescription isodose line (%) | 63 [42–95] | 68.5 [50–95] | 50 [42–80] | <.001 |
| Days from SRS to surgery | 307 [21–1351] | 278 [21–1351] | 321 [65–1226] | .32 |
Summary statistics are presented as number (percentage) for categorical variables, and median [range] for continuous variables.
JH, Johns Hopkins cases; LINAC, linear accelerator; NSCLC, non-small cell lung cancer; RN, radiation necrosis; SCLC, small cell lung cancer; SRS, stereotactic radiosurgery; TP, tumor progression; WBRT, whole brain radiotherapy; WF, Wake Forest cases.
Neuroradiologist Interpretation
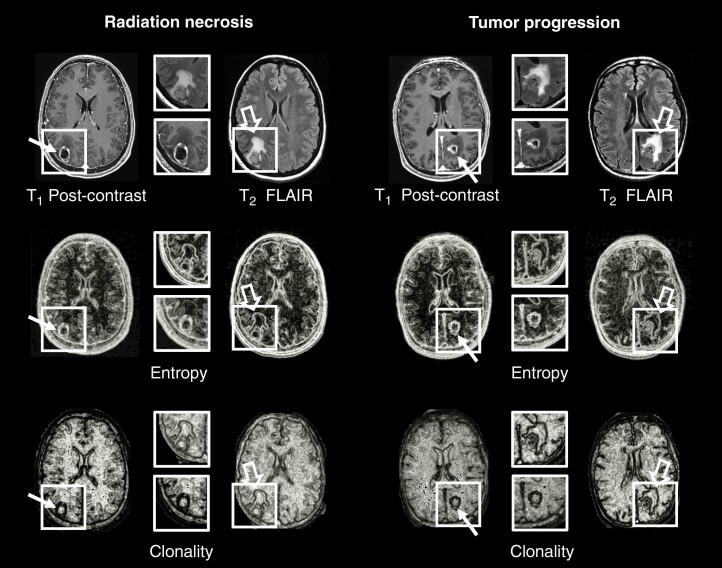
Representative MRI images and radiomic features are shown in Figure 1. Both RN (Figure 1, top left panels) and TP (Figure 1, top right panels) demonstrated ring-enhancement around the periphery on T1c images and significant vasogenic edema on T2-FLAIR images. The radiomic images (single radiomic Entropy, middle panels; mpRad Clonality, bottom panels) clearly demarcate the heterogeneity of the lesion and surrounding tissue in both RN and TP. All cases were reviewed by an experienced neuroradiologist to render an interpretation. Radiographic diagnoses were concordant with pathology in 90 of the 135 cases (67% accuracy), with a sensitivity of 69% and specificity of 59%. Within the JH cohort, which have been re-reviewed for the current study, there was poor intra-observer agreement (72% concordant between previous and current interpretation), with a kappa statistic of 0.0727 (P = .48), indicating tremendous uncertainty in radiographic diagnosis based on cMRI alone.
Figure 1.
Representative examples of radiation necrosis (Left) and tumor progression (Right) in metastatic brain lesions (boxes) treated with stereotactic radiosurgery. The original T1 post-contrast and T2-FLAIR images are shown in the top row. Radiomic images of entropy (middle row) and mpRad clonality (bottom row) demonstrate heterogeneity of the lesions (line arrows) and surrounding edema (open arrows).
Multiparametric Radiomics and Feature Selection
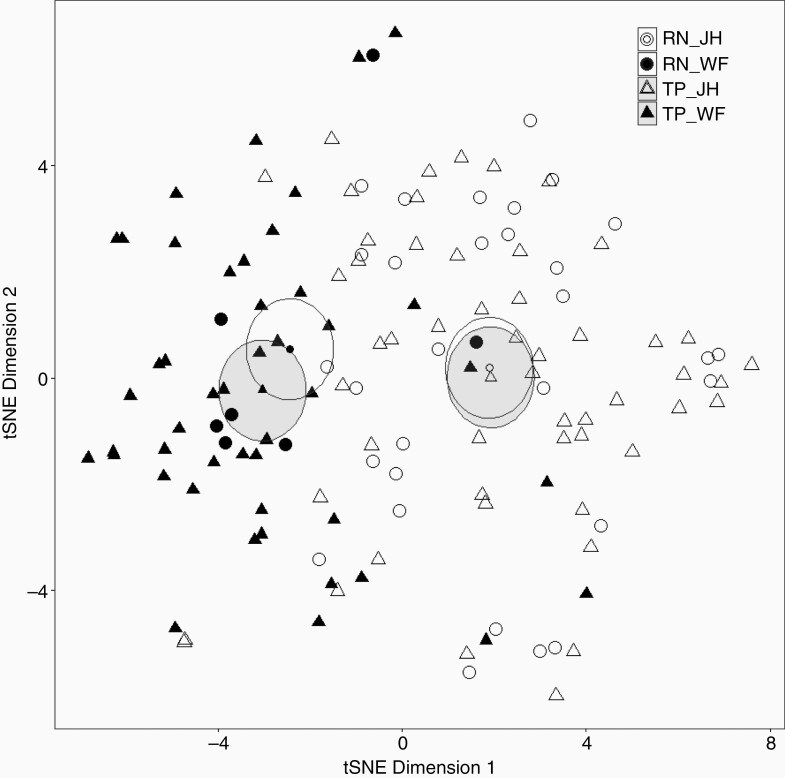
The initial radiomic feature space consisted of 139 variables. We performed tSNE analysis to explore the heterogeneity within the combined dataset. As shown in Figure 2, JH (open dots) and WF (closed dots) cases clearly separated into 2 groups with different group means (represented by the smaller dots in the center of the ellipses). RN (individual lesions represented by circles and 95% confidence intervals [CI] of the mean represented by white ellipses) and TP (triangles, with gray ellipses representing 95% CIs) did not clearly separate. Supplementary Figure S1 shows the heatmap and hierarchical clustering of all normalized radiomic features (x-axis) and cases (y-axis) by diagnosis and institutional origins. Some radiomic features (marked by the yellow boxes) were strongly associated with institutional origin rather than diagnosis, suggesting that case origin may be a significant confounding factor for diagnoses. Those features that strongly correlated with institutional origins were excluded from further feature selection (Supplementary Table S3, features labeled with *).
Figure 2.
T-distributed stochastic neighbor embedding (tSNE) analysis showing separation of cases from the 2 institutions. The high-dimensional radiomic feature space containing 139 single and multiparametric features and 135 cases is represented in 2-dimensional space with arbitrary axes using the tSNE. Individual cases are represented by circles (radiation necrosis, RN, N = 37) and triangles (tumor progression, TP, N = 98). Institutional origins are represented by open (Johns Hopkins cases, JH) or closed (Wake Forest cases, WF) dots. Ellipses (RN, white; TP, gray) represent 95% confidence intervals of group means (smaller dots in the center of the ellipses).
After recursive elimination, RTS-2 included one mpRad feature (mpRad Minimum) and eight single radiomic features (4 T1c and 4 T2-FLAIR). Table 2 compares the current radiomic signature (RTS-2) to the previously published signature (RTS-1), stratified by institutional origin and diagnosis. In addition to mpRad Minimum, new features in RTS-2 included T1c Cluster Tendency, T2-FLAIR Informational Measure of Correlation 2, and T2-FLAIR Short and Long Run High Gray-Level Emphasis, all of which showed significant or near significant association with diagnosis (Table 2). Among the 6 features from RTS-1 that were not selected for RTS-2, 5 (T1c NGTDM Texture Strength, T1c Grey Level Nonuniformity, T1c Run Percentage, T2-FLAIR NGTDM Texture Strength, T2-FLAIR NGTDM Coarseness) showed significant or near significant association with institutional origin, suggesting that case origin may be a confounding factor for RTS-1. Since no mpRad features were included in the previous study, RTS-1 with or without mpRad Minimum was tested separately for its predictive performance and compared with RTS-2.
Table 2.
Summary of Radiomic Features (mean ± standard deviation) in the 2 Radiomic Tissue Signatures (RTS)
| RTS-1 | RTS-2 | JH vs WF | RN vs TP | |||||
|---|---|---|---|---|---|---|---|---|
| JH N = 82 |
WF N = 53 |
P value | RN N = 37 |
TP N = 98 |
P value | |||
| MpRad Minimum | N | Y | 0.09 ± 0.06 | 0.08 ± 0.06 | .392 | 0.11 ± 0.06 | 0.08 ± 0.07 | .036 |
| T1c Minimum | Y | Y | 158.16 ± 118.44 | 900.51 ± 961.05 | <.001 | 315.95 ± 374.05 | 500.06 ± 792.74 | .178 |
| T1c Cluster Tendency | N | Y | 162.75 ± 106.17 | 176.37 ± 143.92 | .529 | 141.47 ± 96.32 | 178.15 ± 129.47 | .12 |
| T1c Fractal Dimension | Y | Y | 1.41 ± 0.14 | 1.43 ± 0.11 | .29 | 1.38 ± 0.15 | 1.43 ± 0.12 | .047 |
| T1c NGTDM Texture Strength | Y | N | 94.76 ± 72.32 | 70.81 ± 64.98 | .053 | 114.64 ± 81.33 | 74.30 ± 62.58 | .003 |
| T1c NGTDM Coarseness | Y | Y | 0.03 ± 0.02 | 0.02 ± 0.01 | .004 | 0.03 ± 0.02 | 0.02 ± 0.02 | .001 |
| T1c Grey Level Nonuniformity | Y | N | 751.57 ± 501.93 | 1441.08 ± 991.96 | <.001 | 688.53 ± 474.12 | 1148.26 ± 868.17 | .003 |
| T1c Run Percentage | Y | N | 0.46 ± 0.15 | 0.64 ± 0.21 | <.001 | 0.44 ± 0.16 | 0.56 ± 0.20 | .002 |
| T2-FLAIR Minimum | Y | Y | 3487.84 ± 2303.25 | 3250.70 ± 1932.44 | .603 | 4091.22 ± 1875.55 | 3160.93 ± 2267.18 | .041 |
| T2-FLAIR Kurtosis | Y | N | 3.37 ± 1.22 | 3.67 ± 1.58 | .285 | 3.11 ± 1.36 | 3.59 ± 1.31 | .08 |
| T2-FLAIR NGTDM Texture Strength | Y | N | 253.08 ± 360.44 | 123.26 ± 80.98 | .043 | 328.66 ± 496.18 | 172.32 ± 189.14 | .016 |
| T2-FLAIR NGTDM Coarseness | Y | N | 0.04 ± 0.03 | 0.02 ± 0.01 | .001 | 0.05 ± 0.04 | 0.03 ± 0.02 | <.001 |
| T2-FLAIR Informational Measure of Correlation 2 | N | Y | 0.87 ± 0.07 | 0.91 ± 0.04 | .002 | 0.86 ± 0.10 | 0.89 ± 0.05 | .045 |
| T2-FLAIR SRHGE | N | Y | 0.02 ± 0.02 | 0.02 ± 0.03 | .383 | 0.01 ± 0.01 | 0.02 ± 0.03 | .114 |
| T2-FLAIR LRHGE | N | Y | 1475.00 ± 1011.33 | 1870.48 ± 852.17 | .05 | 1418.46 ± 637.62 | 1654.04 ± 1081.33 | .25 |
Y and N indicate features included or not included in each RTS, respectively. P values were from 2-sided T-tests.
JH, Johns Hopkins cohort; LRHGE, Long Run High Gray-Level Emphasis; MpRad, multiparametric radiomic feature; NGTDM, Neighborhood Greytone Difference Matrix; RN, radiation necrosis; SRHGE, Short Run High Gray-Level Emphasis; TP, tumor progression; WF, Wake Forest cohort.
Distinguishing Tumor Progression from Radiation Necrosis Using Radiomics-Based ML
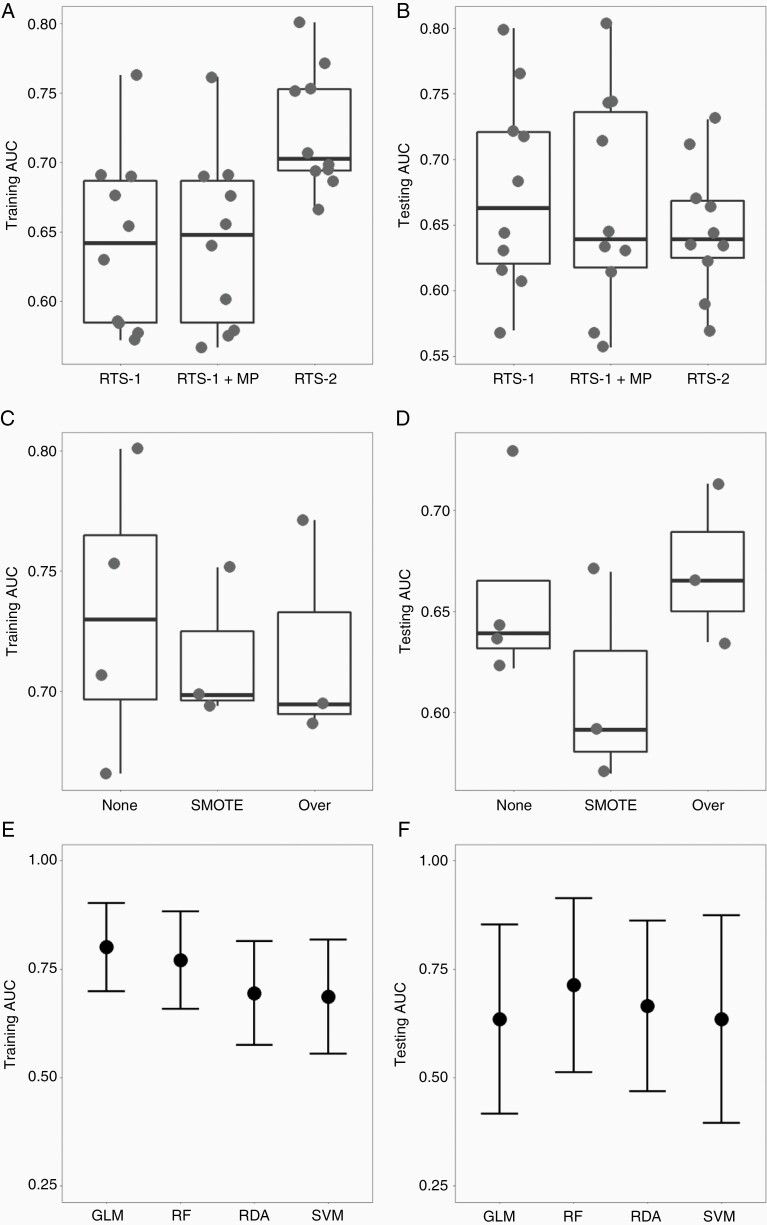
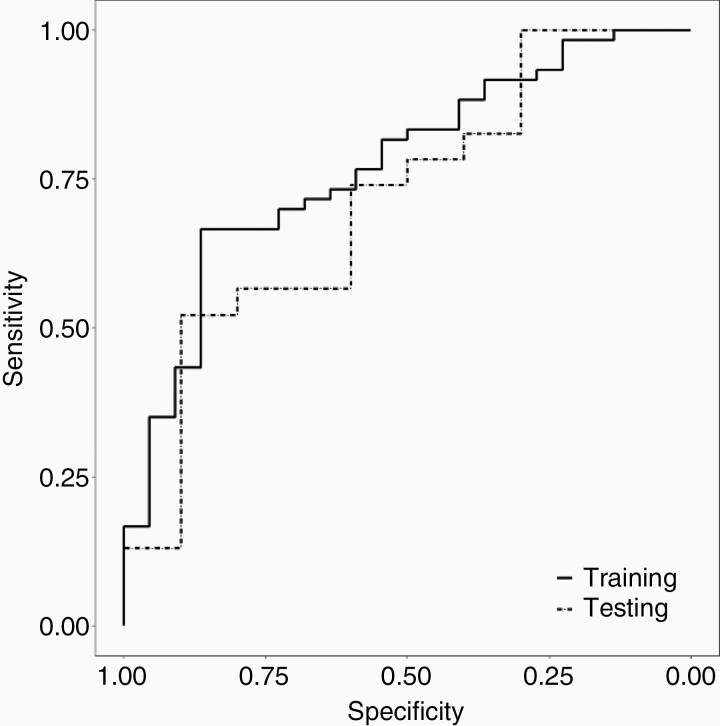
Figure 3 and Supplementary Table S4 summarize the predictive performance of RTS-2, in comparison with RTS-1 and RTS-1 plus mpRad Minimum. The AUCs of all models were plotted in Figure 3A (training) and 3B (validation). RTS-2 had higher median AUC in the training cohort than RTS-1 (P = .004) and RTS-1 plus mpRad Minimum (P = .004, Supplementary Table S5). In the validation cohort, RTS-2 had more consistent performance (AUC range 0.59–0.73) than RTS-1 (0.57–0.80, F test for variance P = .12). Figure 3C (training) and 3D (validation) demonstrate the performance of RTS-2-based models stratified by sampling techniques to mitigate the effect of class imbalance. There was a trend of better performance for models using random oversampling compared to SMOTE in the validation cohort (P = .20, Figure 3D, Supplementary Table S5). Finally, using RTS-2 and random oversampling, the AUCs from 4 ML algorithms (GLM, RF, RDA, and SVM) were compared in Figure 3E (training) and 3F (validation). The RF model showed a trend for better performance in both the training (AUC = 0.77, 95% CI 0.66–0.88) and validation cohort (AUC = 0.71, 95% CI 0.51–0.91). Figure 4 shows the ROC curves of the final RF model. The sensitivity and specificity (for TP) were 67% and 86% in the training cohort, and 52% and 90% in the validation cohort, resulting in a positive likelihood ratio of 4.8 and 5.2, respectively.
Figure 3.
Comparisons of radiomic tissue signatures (RTS), sampling techniques and machine learning algorithms. RTS-1 with or without mpRad Minimum (MP), and RTS-2 were entered into supervised machine learning models using general linear model (GLM), random forest (RF), regularized discriminant analysis (RDA), and support vector machine (SVM) algorithms, with no oversampling, Synthetic Minority Oversampling Technique (SMOTE), or random oversampling (Over) of RN cases. Each dot represents the area under the curve (AUC) of the receiver operating characteristics in the training (A, C and E) or testing (B, D and F) cohorts. Panels A and B compare RTS-1, RTS-1 with MP and RTS-2. Panels C and D compare RTS-2-based models with no oversampling (None), SMOTE or random oversampling (Over). The box plots indicate the median and interquartile range, while the whiskers mark min and max. Panels E and F compare AUCs (central dot, whiskers indicate 95% confidence intervals) of the 4 algorithms using RTS-2 and random oversampling.
Figure 4.
Receiver operating characteristics (ROC) of the final random forest model (RTS-2 with random oversampling of minority cases for class imbalance) for distinguishing tumor progression from radiation necrosis in the training (solid line) and validation (dashed line) cohorts.
Discussion
Distinguishing RN from TP represents a major clinical challenge in optimizing the management of patients with brain metastases treated with SRS. To address this challenge, we have employed novel mpRad for improved brain tissue characterization. This study represents the largest, multi-institutional collection of cases of TP and RN, in which individual diagnoses were confirmed by surgical pathology. We demonstrated the feasibility of distinguishing TP from RN using advanced mpRad and ML techniques, and identified a new radiomic signature and predictive model with improved performance in a heterogeneous cohort of patients. Furthermore, we systematically evaluated ML algorithms and adjusted for class imbalance to improve the rigor of feature selection and modeling. The final optimized classifier was able to identify TP with high specificity. Our methodology and results will further advance the utilization of artificial intelligence and radiomics in radiation oncology and medicine in general.
Stereotactic radiosurgery provides excellent oncologic outcomes, with 12-month local control up to 75%–90% and preservation of neurocognitive functions in several large prospective randomized trials.30–32 However, radiation necrosis can occur at a similar rate to local recurrence, in 5%–25% of lesions.33,34 Recognized risk factors of RN include lesion size and SRS dose.35,36 A large retrospective study showed a crude rate of RN of 25.8% at a median follow-up of 17 months, 67% of which were symptomatic.35 Another multi-institutional study of 1533 brain metastases treated with single-fraction SRS showed a 2-year cumulative incidence of only 8.9% for RN.36 The large variation in the reported rates of RN may be partly due to significant diagnostic uncertainty, especially without pathologic confirmation. In a report of 15 cases of imaging-diagnosed RN, only 7 cases were confirmed to be pure RN on histopathology, while the remaining 8 cases represented tumor progression based on pathology.37 Our own data underlined such uncertainty, in that the intra-observer agreement of an experienced neuroradiologist reviewing the same images was only 72%, and the concordance between radiographic and pathologic diagnoses was only 67%. Thus, better quantitative biomarkers are needed to improve the interpretation of cMRI changes after SRS.
Clinically, an integrated imaging approach using advanced MRI methods coupled with quantitative radiomic metrics would be a major advancement in providing optimal, individualized care for patients with brain metastases. Imaging confirmation of TP may guide the use of repeat radiation without invasive diagnostic procedures and minimize the need for surgical intervention, which would improve patients’ quality of life.6–8 In this study, our best radiomic-based model demonstrates similar sensitivity to a neuroradiologist for identifying tumor progression (67% vs 69%), but with substantially higher specificity (86% vs 59%). The high positive likelihood ratio (4.8–5.2) of this model suggests that it may be able to identify a subset of patients for whom progression is more likely and repeat radiation can be safely delivered without the need for surgical confirmation. For those patients whose diagnoses remain ambiguous by imaging or who have significant symptom burden, surgical sampling and pathologic confirmation should still be the gold standard for diagnosis, as there is concern about inadvertently irradiating existing necrosis. Therefore, the radiomic tissue signature and machine learning model identified in this study may be useful in complementing radiologists’ interpretation and triaging patients for noninvasive versus invasive management pathways.
Various advanced imaging modalities, such as positron emission tomography (PET), MR perfusion and MR spectroscopy (MRS), have been investigated for distinguishing RN from TP.38–41 Recent studies combining PET and radiomics analysis have demonstrate encouraging results in single-institution studies with small patient cohorts, although these will require independent validation.42,43 However, there is currently no consensus on how to incorporate these imaging modalities into routine clinical practice.3 Furthermore, these techniques may not be widely available, and their interpretation may be subject to local technical expertise, variations in scanning techniques, and radiologists’ experience. Several cMRI features have been proposed to distinguish tumor from treatment effect. A retrospective study of 49 patients with 52 brain metastases treated by SRS showed that perilesional edema may differentiate TP from RN with positive predictive value of 92%, although this has not been independently validated.44 In another cohort of 32 patients with growing lesions after SRS, lesion quotient, or the ratio between the lesion seen on T2 images to the total enhancing area on T1 post-contrast images, was most effective in distinguishing TP from RN, with a sensitivity of 100% but specificity of only 32%.12 However, this feature failed to provide accurate diagnoses in an external validation cohort.14
In contrast to qualitative radiographic biomarkers, unbiased quantitative metrics, such as radiomic features, may improve the diagnostic accuracy with cMRI.17,45–47 Radiomics can extract the different contrast and inherent texture of MRI images, which may be related to the underlying biology of the tissue.20,47 Several machine learning algorithms, combined with radiomics, have been used to distinguish RN versus TP. Tiwari et al. described a procedure of minimum redundancy and maximum relevance feature-selection and feed-forward selection in combination with SVM to distinguish RN and TP in a small cohort of patients (22 in training and 4 in validation cohort).23 T2-FLAIR features showed an AUCs of 0.79 in the training dataset, but an accuracy of only 50% in the external validation set.23 Similarly, another single-institution study employed recursive feature elimination and SVM, resulting in an AUC of >0.9. However, only 10% of the lesions in this cohort had pathologic confirmation, and there was no external validation.24 Finally, Zhang et al. used concordance correlation coefficients for feature selection and RUSBoost, a decision tree–based ensemble algorithm, to classify RN versus TP in a single-institution cohort of 87 patients with pathologic confirmation.25 The model produced an AUC of 0.73 by leave-one-out cross validation, but this study also did not contain an external validation set.25 None of the previous reports presented full radiomic images that could be compared to the mpMRI data in the current study.
Recently, a meta-analysis reviewed all published studies of conventional MRI radiomics in diagnosing progression of brain metastasis after treatment, and found that all studies to date were single-institution and most were without pathologic confirmation. Therefore, our study represents a major step forward in this area.48 Our study represents one of the largest reported collections (109 patients with 135 individual lesions from 2 independent institutions) of pathologically confirmed radiation necrosis. Given the heterogeneity of this combined dataset, we systematically evaluated several steps of the radiomics and machine learning process. First, we conducted careful feature selection using t-tests and recursive feature elimination, while excluding the features that may be confounded by case origin. Second, we incorporated the newly discovered mpRad features, and demonstrated improved predictive performance compared to the previously published radiomic signature. Third, since RN represents a substantial minority, we deliberately evaluated random oversampling and SMOTE during model optimization to circumvent the potential model instability caused by class imbalance. Finally, we compared 4 supervised ML algorithms and demonstrated that a random forest model trained with RTS-2 and random oversampling achieved the highest AUC in the validation cohort. In summary, since no single algorithm can universally fit all situations and data types, our work adds tremendous insight into the complexity of the machine learning workflow and may help to improve radiomics-based classification studies in the future.
One of the limitations of radiomics-based machine learning is the potential for model overfitting.16,21 This may be mitigated by careful feature selection and the use of external validation data. The heterogeneity within the combined radiomic dataset may be related to variations in treatment techniques, imaging parameters, and clinical practice, such as the threshold to recommend surgery and the types of surgery performed. This inherent heterogeneity, evidenced by the clear segregation of cases with respect to institutional origins, may improve the external applicability of our results.
There are several potential caveats to our study. Despite the extensive effort to collect cases, our combined dataset included only 109 patients, partly due to the fact that the vast majority of patients with imaging evidence of tumor growth did not require surgical intervention. We considered it essential to include patients with pathologic confirmation, as standard MRI diagnosis of necrosis versus progressive tumor is unreliable. However, there may be subtle differences in the imaging characteristics or tumor/host biology between patients with RN that require surgical intervention and those that do not. Lastly, differences in institutional practices may add unknown biases due to subtle differences in how patients are recommended for surgery between the 2 centers. Future validation of our results in an independent dataset will be necessary, and requires multilateral collaboration.
Conclusions
In a large, diverse patient cohort from 2 institutions, our data highlight the diagnostic uncertainty of radiation necrosis in patients with brain metastases treated with stereotactic radiosurgery. A new radiomic tissue signature incorporating mpRad features demonstrated improved diagnostic accuracy over a previous signature using single radiomic features. Systematic evaluation of multiple steps in the radiomics and machine learning workflow resulted in improved model performance in the validation dataset. This model may be of important clinical utility by selecting for the patients who may benefit from salvage SRS without the need for surgical sampling. Our methodology and results should be validated prospectively in future clinical trials.
Supplementary Material
Funding
This study was supported by the National Institutes of Health (5P30CA006973, U01CA140204) and Nicholl Family Foundation.
Conflict of interest statement. M.A.J. and V.P. have patents US patents, nos. 20170112459 and 20160171695, issued relevant to the machine learning algorithms applied in this work. M.A.J. reports nonfinancial support from NVIDIA Corporation during the conduct of the study. L.R.K. received research grants from Novocure, Arbor and Accuray; has performed consulting for Novocure and Accuray; and is on the advisory board for Novocure, outside the submitted work. All other authors declare no conflict of interest.
Authorship Statement. Conceptualization: X.C., L.P., M.A.J., L.R.K. Methodology: X.C., V.S.P., M.A.J., L.R.K. Resources: M.D.C., K.J.R., M.S., E.M., L.R.K. Formal Analysis: X.C., V.S.P., D.L. Writing (original draft): X.C., L.R.K. Writing (review & editing): X.C., V.S.P., L.P., M.D.C., K.J.R., M.S., E.M., D.L., M.A.J., L.R.K. Supervision: M.A.J., L.R.K.
References
- 1. Sahgal A, Aoyama H, Kocher M, et al. Phase 3 trials of stereotactic radiosurgery with or without whole-brain radiation therapy for 1 to 4 brain metastases: individual patient data meta-analysis. Int J Radiat Oncol Biol Phys. 2015;91(4):710–717. [DOI] [PubMed] [Google Scholar]
- 2. Kotecha R, Gondi V, Ahluwalia MS, Brastianos PK, Mehta MP. Recent advances in managing brain metastasis. F1000Res. 2018;7:1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vellayappan B, Tan CL, Yong C, et al. Diagnosis and management of radiation necrosis in patients with brain metastases. Front Oncol. 2018;8:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kang TW, Kim ST, Byun HS, et al. Morphological and functional MRI, MRS, perfusion and diffusion changes after radiosurgery of brain metastasis. Eur J Radiol. 2009;72(3):370–380. [DOI] [PubMed] [Google Scholar]
- 5. Suh CH, Kim HS, Jung SC, Choi CG, Kim SJ. Comparison of MRI and PET as potential surrogate endpoints for treatment response after stereotactic Radiosurgery in patients with brain metastasis. AJR Am J Roentgenol. 2018;211(6):1332–1341. [DOI] [PubMed] [Google Scholar]
- 6. Iorio-Morin C, Mercure-Cyr R, Figueiredo G, Touchette CJ, Masson-Côté L, Mathieu D. Repeat stereotactic radiosurgery for the management of locally recurrent brain metastases. J Neurooncol. 2019;145(3):551–559. [DOI] [PubMed] [Google Scholar]
- 7. Kim IY, Jung S, Jung TY, et al. Repeat stereotactic radiosurgery for recurred metastatic brain tumors. J Korean Neurosurg Soc. 2018;61(5):633–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McKay WH, McTyre ER, Okoukoni C, et al. Repeat stereotactic radiosurgery as salvage therapy for locally recurrent brain metastases previously treated with radiosurgery. J Neurosurg. 2017;127(1): 148–156. [DOI] [PubMed] [Google Scholar]
- 9. Stockham AL, Suh JH, Chao ST, Barnett GH. Management of recurrent brain metastasis after radiosurgery. Prog Neurol Surg. 2012;25:273–286. [DOI] [PubMed] [Google Scholar]
- 10. Hong CS, Deng D, Vera A, Chiang VL. Laser-interstitial thermal therapy compared to craniotomy for treatment of radiation necrosis or recurrent tumor in brain metastases failing radiosurgery. J Neurooncol. 2019;142(2):309–317. [DOI] [PubMed] [Google Scholar]
- 11. Moravan MJ, Fecci PE, Anders CK, et al. Current multidisciplinary management of brain metastases. Cancer. 2020;126(7):1390–1406. [DOI] [PubMed] [Google Scholar]
- 12. Dequesada IM, Quisling RG, Yachnis A, Friedman WA. Can standard magnetic resonance imaging reliably distinguish recurrent tumor from radiation necrosis after radiosurgery for brain metastases? A radiographic-pathological study. Neurosurgery. 2008;63(5):898–903; discussion 904. [DOI] [PubMed] [Google Scholar]
- 13. Kano H, Kondziolka D, Lobato-Polo J, Zorro O, Flickinger JC, Lunsford LD. T1/T2 matching to differentiate tumor growth from radiation effects after stereotactic radiosurgery. Neurosurgery. 2010; 66(3):486–491; discussion 491–482. [DOI] [PubMed] [Google Scholar]
- 14. Stockham AL, Tievsky AL, Koyfman SA, et al. Conventional MRI does not reliably distinguish radiation necrosis from tumor recurrence after stereotactic radiosurgery. J Neurooncol. 2012;109(1):149–158. [DOI] [PubMed] [Google Scholar]
- 15. Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJWL. Machine learning methods for quantitative radiomic biomarkers. Sci Rep. 2015;5:13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–762. [DOI] [PubMed] [Google Scholar]
- 17. Parekh V, Jacobs MA. Radiomics: a new application from established techniques. Expert Rev Precis Med Drug Dev. 2016;1(2):207–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alilou M, Orooji M, Beig N, et al. Quantitative vessel tortuosity: a potential CT imaging biomarker for distinguishing lung granulomas from adenocarcinomas. Sci Rep. 2018;8(1):15290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beig N, Khorrami M, Alilou M, et al. Perinodular and intranodular radiomic features on lung CT images distinguish adenocarcinomas from granulomas. Radiology. 2019;290(3):783–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Parekh VS, Jacobs MA. Multiparametric radiomics methods for breast cancer tissue characterization using radiological imaging. Breast Cancer Res Treat. 2020;180(2):407–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parekh VS, Jacobs MA. Deep learning and radiomics in precision medicine. Expert Rev Precis Med Drug Dev. 2019;4(2):59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jacobs MA, Umbricht CB, Parekh VS, et al. Integrated multiparametric radiomics and informatics system for characterizing breast tumor characteristics with the OncotypeDX Gene Assay. Cancers (Basel). 2020;12(10):2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tiwari P, Prasanna P, Wolansky L, et al. Computer-extracted texture features to distinguish cerebral radionecrosis from recurrent brain tumors on multiparametric MRI: a feasibility study. AJNR Am J Neuroradiol. 2016;37(12):2231–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Larroza A, Moratal D, Paredes-Sánchez A, et al. Support vector machine classification of brain metastasis and radiation necrosis based on texture analysis in MRI. J Magn Reson Imaging. 2015;42(5):1362–1368. [DOI] [PubMed] [Google Scholar]
- 25. Zhang Z, Yang J, Ho A, et al. A predictive model for distinguishing radiation necrosis from tumour progression after gamma knife radiosurgery based on radiomic features from MR images. Eur Radiol. 2018;28(6):2255–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hettal L, Stefani A, Salleron J, et al. Radiomics method for the differential diagnosis of radionecrosis versus progression after fractionated stereotactic body radiotherapy for brain oligometastasis. Radiat Res. 2020;193(5):471–480. [DOI] [PubMed] [Google Scholar]
- 27. Peng L, Parekh V, Huang P, et al. Distinguishing true progression from Radionecrosis after stereotactic radiation therapy for brain metastases with machine learning and Radiomics. Int J Radiat Oncol Biol Phys. 2018;102(4):1236–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 29. Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–2849. [DOI] [PubMed] [Google Scholar]
- 30. Aoyama H, Shirato H, Tago M, et al. Stereotactic radiosurgery plus whole-brain radiation therapy vs stereotactic radiosurgery alone for treatment of brain metastases: a randomized controlled trial. JAMA. 2006;295(21):2483–2491. [DOI] [PubMed] [Google Scholar]
- 31. Brown PD, Jaeckle K, Ballman KV, et al. Effect of Radiosurgery Alone vs Radiosurgery with whole brain radiation therapy on cognitive function in patients with 1 to 3 brain metastases: a randomized clinical trial. JAMA. 2016;316(4):401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Churilla TM, Chowdhury IH, Handorf E, et al. Comparison of local control of brain metastases with stereotactic Radiosurgery vs Surgical Resection: a secondary analysis of a randomized clinical trial. JAMA Oncol. 2019;5(2):243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chin LS, Ma L, DiBiase S. Radiation necrosis following gamma knife surgery: a case-controlled comparison of treatment parameters and long-term clinical follow up. J Neurosurg. 2001;94(6):899–904. [DOI] [PubMed] [Google Scholar]
- 34. Minniti G, Clarke E, Lanzetta G, et al. Stereotactic radiosurgery for brain metastases: analysis of outcome and risk of brain radionecrosis. Radiat Oncol. 2011;6:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kohutek ZA, Yamada Y, Chan TA, et al. Long-term risk of radionecrosis and imaging changes after stereotactic radiosurgery for brain metastases. J Neurooncol. 2015;125(1):149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moraes FY, Winter J, Atenafu EG, et al. Outcomes following stereotactic radiosurgery for small to medium-sized brain metastases are exceptionally dependent upon tumor size and prescribed dose. Neuro Oncol. 2019;21(2):242–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Telera S, Fabi A, Pace A, et al. Radionecrosis induced by stereotactic radiosurgery of brain metastases: results of surgery and outcome of disease. J Neurooncol. 2013;113(2):313–325. [DOI] [PubMed] [Google Scholar]
- 38. Verma N, Cowperthwaite MC, Burnett MG, Markey MK. Differentiating tumor recurrence from treatment necrosis: a review of neuro-oncologic imaging strategies. Neuro Oncol. 2013;15(5):515–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Parvez K, Parvez A, Zadeh G. The diagnosis and treatment of pseudoprogression, radiation necrosis and brain tumor recurrence. Int J Mol Sci. 2014;15(7):11832–11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Imani F, Boada FE, Lieberman FS, Davis DK, Mountz JM. Molecular and metabolic pattern classification for detection of brain glioma progression. Eur J Radiol. 2014;83(2):e100–e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu Y, Ma Y, Sun M, Jiang W, Yuan T, Tong D. Meta-analysis of the diagnostic performance of diffusion magnetic resonance imaging with apparent diffusion coefficient measurements for differentiating glioma recurrence from pseudoprogression. Medicine (Baltimore). 2020;99(23):e20270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hotta M, Minamimoto R, Miwa K. 11C-methionine-PET for differentiating recurrent brain tumor from radiation necrosis: radiomics approach with random forest classifier. Sci Rep. 2019;9(1):15666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lohmann P, Kocher M, Ceccon G, et al. Combined FET PET/MRI radiomics differentiates radiation injury from recurrent brain metastasis. Neuroimage Clin. 2018;20:537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Leeman JE, Clump DA, Flickinger JC, Mintz AH, Burton SA, Heron DE. Extent of perilesional edema differentiates radionecrosis from tumor recurrence following stereotactic radiosurgery for brain metastases. Neuro Oncol. 2013;15(12):1732–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cai J, Zheng J, Shen J, et al. A radiomics model for predicting the response to bevacizumab in brain necrosis after radiotherapy. Clin Cancer Res. 2020;26:5438–5447. [DOI] [PubMed] [Google Scholar]
- 46. Korfiatis P, Kline TL, Coufalova L, et al. MRI texture features as biomarkers to predict MGMT methylation status in glioblastomas. Med Phys. 2016;43(6):2835–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Parekh VS, Jacobs MA. Integrated radiomic framework for breast cancer and tumor biology using advanced machine learning and multiparametric MRI. NPJ Breast Cancer. 2017;3:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim HY, Cho SJ, Sunwoo L, et al. Classification of true progression after radiotherapy of brain metastasis on MRI using artificial intelligence: a systematic review and meta-analysis. Neurooncol Adv. 2021;3(1):vdab080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.