Abstract
A system based on PCR and restriction endonuclease analysis was developed to distinguish the seven currently recognized Malassezia species. Seventy-eight strains, including authentic culture collection strains and routine clinical isolates, were investigated for variation in the ribosomal DNA repeat units. Two genomic regions, namely, the large subunit of the ribosomal gene and the internal transcribed spacer (ITS) region, were amplified by PCR, and products were digested with restriction endonucleases. The patterns generated were useful in identification of five out of seven Malassezia species. M. sympodialis was readily distinguishable in that its ITS region yielded a 700-bp amplified fragment, whereas the other six species yielded an 800-bp fragment. M. globosa and M. restricta were very similar in the regions studied and could be distinguished only by performing a hot start-touchdown PCR on primers for the β-tubulin gene. Primers based on the conserved areas of the Candida cylindracea lipase gene, which were used in an attempt to amplify Malassezia lipases, yielded an amplification product after annealing at 55°C only with M. pachydermatis. This specific amplification may facilitate the rapid identification of this organism.
Within the past decade, reviews on emerging yeast infections have repeatedly mentioned members of the Malassezia furfur complex as opportunistic yeasts of increasing importance (1, 18, 27, 31, 33). Malassezia (Pityrosporum) species are lipophilic yeasts commonly recognized as commensals of the skin of warm-blooded vertebrates that can become pathogenic under certain conditions, usually by causing the skin condition tinea (pityriasis) versicolor. Several exogenous and endogenous factors such as high temperature, high relative humidity, greasy skin, corticosteroid treatment, and immunodeficiency can influence these yeasts to become pathogenic (15).
Prior to 1990, only three Malassezia species were recognized. These were M. furfur (Robin) Baillon, M. pachydermatis (Weidman) C. W. Dodge, and M. sympodialis (Simmons and Guého). With the development of molecular techniques, new species have been segregated within Malassezia (16). The group of lineages formerly regarded as M. furfur (sensu lato [i.e., in the broad sense]) has now been divided into six species on the basis of genomic and ribosomal sequence comparisons of a large number of human and animal isolates (14). Four new taxa that have been added to Malassezia are M. globosa, M. obtusa, M. restricta, and M. slooffiae.
Molecular biological studies of Malassezia yeasts initially consisted of determining the G+C content of chromosomal DNA (13) and direct rRNA sequencing (14, 16). Pulsed-field gel electrophoresis studies have confirmed the robustness of the new taxonomic structure of Malassezia, with all Malassezia species characterized by their individual karyotypes (5, 6, 20). Beside karyotyping, molecular differentiation of Malassezia species has also been attempted by PCR fingerprinting (4), restriction analysis (2), and randomly amplified polymorphic DNA analysis (6). Boekhout et al. (6) reported that although Malassezia species could be distinguished by randomly amplified polymorphic DNA typing, the varying amounts of heterogeneity observed within the species renders this method unreliable for species identification. Thus, pulsed-field gel electrophoresis is the only technique that can reliably differentiate between all seven currently known Malassezia species. While karyotyping is very robust, its time-consuming and labor-intensive nature necessitates the development of alternative molecular methods. A rapid and reliable molecular system for identification of Malassezia species is needed to facilitate epidemiological and related research studies and may also be of potential utility in reference laboratories.
Comparative studies of nucleotide sequences of rRNA genes have been used extensively in molecular studies of fungi, as they provide a means for analyzing phylogenetic relationships over a wide range of taxonomic levels (7, 21, 35). Polymorphisms in the internal transcribed spacer (ITS) region and intergenic spacer of fungal ribosomal DNA repeat units, at both the inter- and intraspecific levels, have provided practical epidemiological markers for typing a range of clinically important species (8, 19, 22, 25, 26, 29). Guillot and Guého (16) had also used direct rRNA sequencing to delineate different Malassezia species. This method, however, cannot be used for routine analysis and diagnosis because it requires relatively large amounts of RNA and is very time-consuming. However, given that there is variability in this region, a PCR-based analysis and specific amplification of the target region would be advantageous.
In this paper we have used PCR-restriction endonuclease analysis (PCR-REA) to differentiate between the seven currently recognized Malassezia species. Universal fungal primers from the ITS region and specific primers designed from the published partial sequences of the large subunits (LSUs) of ribosomal genes of Malassezia species were used to develop a rapid and reliable PCR-restriction fragment length polymorphism (RFLP)-based system for identification of Malassezia species.
MATERIALS AND METHODS
Yeast strains.
The sources and origins of the 78 strains investigated in this study are listed in Table 1. Of the 78 strains investigated, 64 strains were isolated from routine specimens sent to the Mycology Laboratory, Laboratories Branch, Ontario Ministry of Health, Toronto, Ontario, Canada, for fungal analysis. Among the remaining 14 strains, 6 strains were obtained from the authentic culture collections (Centraalbureau voor Schimmelcultures, Baarn, The Netherlands, and American Type Culture Collection, Manassas, Va.), 5 strains were received as a gift from Gillian Midgley (London, United Kingdom) and Jan Faergemann (Göteberg, Sweden), and 3 strains were isolated from skin scrapings of pityriasis versicolor patients residing in Hawaii and South Africa. Before molecular analysis was conducted, identification of different Malassezia species among all authentic and clinical strains was performed on the basis of macro- and microscopic features and physiological characteristics as described by Guého et al. (14) and Guillot et al. (17).
TABLE 1.
Sources, origins, and multilocus genotypes of 78 strains from seven Malassezia species
| Malassezia species | Straina | Origin | Complex PCR-REA typed |
|---|---|---|---|
| M. furfur(n = 13) | CBS 1878, NTb | Dandruff | A′CFN |
| JF 04 | Sweden | ACFP | |
| GM 551 | London, United Kingdom | ACFN | |
| 97 FR-1272 | Blood culture, PHLOg | ACFP | |
| 97 FR-3007f | Bronchial wash, PHLO | ACFN | |
| 97 F-661f | Toenail swab, PHLO | ACFN | |
| 98 F-3617 | Neck, PHLO | ACF– | |
| 97 F-8817 | Left arm, PHLO | AC–P | |
| 98 F-5399f | Trunk, PHLO | ACFP | |
| 98 F-10017 | Right index nail, PHLO | ACFN | |
| 99 F-542 | PHLO | A′CFN | |
| 99 FR-178 | PHLO | ACFN | |
| M. furfure | 99 F-1436 | PHLO | ACFN |
| M. globosa(n = 10) | 98 F-3552 | Anticubital area, PHLO | BDG– |
| 98 F-4888 | Back, PHLO | BDGN | |
| 98 F-6443 | Body, PHLO | BDG– | |
| 98 F-7317f | Chest, PHLO | BDGN | |
| 98 F-8304f | Scalp hair, Canada | BDG– | |
| 99 F-160 | PHLO | BD–N | |
| YKM 48 | Forehead, PHLO | B–GN | |
| YKM 58 | Forehead, PHLO | BD–N | |
| YKM 45 | Forehead, PHLO | BD–N | |
| SF7 | Arm, South Africa | BDGN | |
| M. obtusa(n = 3) | 98 F-3529 | Neck, Canada | BCGP |
| M. obtusae | 98 F-8316 | Toenail, PHLO | ACGP |
| M. obtusae | WF 7 | Trunk, Hawaii | B–GN |
| M. pachydermatis(n = 3) | ATCC 14521 | BCF– | |
| CBS 1879, NT | Dog with otitis externa, Sweden | BCFN | |
| M. pachydermatise | GM 420 | London, United Kingdom | ACFP |
| M. restricta (n = 5) | CBS 7877, Tc | Skin, United Kingdom | BDGP |
| YKM 8 | Forehead, PHLO | BDGP | |
| YKM 31 | Scalp, PHLO | BDGP | |
| YKM 32 | Scalp, PHLO | BDGP | |
| YKM 53 | Scalp, PHLO | BDGP | |
| M. slooffiae(n = 7) | CBS 7956, T | Healthy ear of pig | AC′GP |
| JF 06 | Sweden | AC′GN | |
| TV1f | Skin, PHLO | A′–GN | |
| 98 F-4721f | Scalp, PHLO | AC′GP | |
| 98 F-5360 | Neck, PHLO | AC′GP | |
| 98 F-6419 | Groin, PHLO | A–GN | |
| 99 F-411 | PHLO | A′C′–N | |
| M. sympodialis(n = 37) | CBS 7222, T | Human ear, United States | AEHN |
| GM 323 | London, United Kingdom | AD′–N | |
| 97FR-2125 | Skin from chest, PHLO | AEHN | |
| 97 F-8615f | Upper chest, PHLO | AEHN | |
| 98 F-4202 | Trunk, PHLO | AEHN | |
| 98 F-4769 | Back, PHLO | AEHP | |
| 98 F-4784 | Trunk, PHLO | AEHP | |
| 98 F-4941 | Abdomen, PHLO | AEG′P | |
| 98 F-5394 | Back, PHLO | AEHN | |
| 98 F-5413 | Back, PHLO | AEHN | |
| 98 F-5763 | Neck/back, PHLO | AEHP | |
| 98 F-5850f | Left breast, PHLO | AEHP | |
| 98 F-6388 | Body, PHLO | AEHN | |
| 98 F-7582 | Chest, PHLO | AEHN | |
| 98 F-7807 | PHLO | AEHN | |
| 98 F-8046 | Trunk, PHLO | AEHN | |
| 98 F-8139 | Upper back, PHLO | AEHN | |
| 98 F-9662 | Skin of scalp, PHLO | AEHN | |
| 98 F-9846 | Back, PHLO | AE-N | |
| 98 F-9925 | Trunk, PHLO | AEHN | |
| 98 F-10007 | Back, PHLO | AEHN | |
| 98 F-10770 | PHLO | AEHN | |
| 98 F-10143 | PHLO | AEHN | |
| 98 SF-4645 | CVP line swab, PHLO | AEHP | |
| 99 F-440 | PHLO | AEG′N | |
| 99 F-869 | PHLO | AD′–P | |
| 99 F-902 | PHLO | AEHN | |
| 99 F-944 | PHLO | AEHN | |
| 99 F-946 | PHLO | AE-N | |
| 99 F-1072 | PHLO | AEHN | |
| 99 F-1181 | PHLO | AEHN | |
| 99 F-1192 | PHLO | AEHN | |
| 99 F-1407 | PHLO | AEHN | |
| YKM 56 | London, Ontario, Canada | AEHN | |
| YKM 73 | London, Ontario, Canada | AD′HP | |
| SF4 | Back, South Africa | AEHN |
CBS, strains bought from the Centraalbureau voor Schimmelcultures; ATCC, strains bought from the American Type Culture Collection; GM, strains obtained as a gift from Gillian Midgley, Department of Medical Mycology, St. John's Institute of Dermatology, London, United Kingdom; JM, strains obtained as a gift from Jan Faergemann, Department of Dermatology and Venereology, Göteberg University, Göteberg, Sweden; 97/98/99 F, 97/98/99 FR, and 97/98/99 SF, strains that were sent for mycological analysis to the Medical Mycology Laboratory, Public Health Laboratories of Ontario, Etobicoke, Ontario, Canada; YKM, strains isolated by Yatika Kohli from contact plates; SF, strains isolated from specimens obtained from South Africa; WF, strains isolated from specimens obtained from Hawaii.
NT, neotype for Pityrosporum ovale (CBS 1878) or Pityrosporum pachydermatis (CBS 1879).
T, ex-type isolate.
The multilocus genotype represents the alleles observed for each of the strains analyzed by PCR-RFLP. The first letter designates one of the three alleles observed at the LSU locus with the restriction enzyme AvaI. The second letter designates one of the five alleles observed at the ITS locus with the restriction enzyme EcoRI. The third letter designates one of the four alleles observed at the ITS locus with restriction enzyme NcoI. The fourth letter designates the presence (P) or absence (N) of a fragment amplified after hot start-TD PCR with primers for the β-tubulin gene. –, missing data.
Some ambiguity remains regarding the correct identification of this strain (see Results).
Strain accessioned in the Centraalbureau voor Schimmelcultures yeast culture collection.
PHLO, Public Health Laboratories of Ontario, Etobicoke, Ontario, Canada.
DNA extraction.
The fastidious growth and requirement of lipid supplements in the culture medium makes it very difficult to obtain protoplasts for DNA isolation from some strains. For successful extraction, the DNA isolation protocol was modified from the method of Sansinforiano and coworkers (34) for Cryptococcus neoformans and was optimized for Malassezia. The yeasts were grown on Leeming-Notman agar (23) for 2 to 5 days at 32°C (14). Cultures were harvested and diluted in sterile saline (0.85%) to ∼109 CFU/ml. The cells were pelleted by centrifugation at 8,000 × g for 10 min and then suspended in TE–β-mercaptoethanol buffer (280 μl of TE buffer [100 mM Tris, 100 mM EDTA, pH 8.0], 300 μl of deionized H2O, 3 μl of β-mercaptoethanol), incubated at 30°C for 45 min, and pelleted by centrifugation at 8,000 × g for 10 min. They were then suspended in 1 ml of urea lysis buffer (8 M urea, 0.5 M NaCl, 20 mM Tris, 20 M EDTA, and 2% sodium dodecyl sulfate, pH 8.0), and the suspension was incubated at 37°C for at least 3 h with occasional mixing by vortexing prior to DNA extraction. The protoplasts were pelleted by centrifugation at 8,000 × g for 10 min. They were resuspended in 500 to 600 μl of lysis buffer (the same as described above but without the urea), and DNA was extracted with phenol-chloroform-isoamyl alcohol (25:24:1) as described by Sambrook et al. (32), followed by isopropanol precipitation and treatment with RNase at a final concentration of 10 μg/ml. RNase treatment was followed by a chloroform-isoamyl alcohol (24:1) extraction, and the DNA was precipitated in 2 volumes of cold ethanol with high-speed centrifugation at 4°C for 15 min. The nucleic acid pellet was rinsed with cold 70% ethanol, and the air-dried pellet was suspended in 50 μl of TE (10 mM Tris, 1 mM EDTA). Two microliters of the TE-suspended DNA was used as a template for the PCR.
This method of DNA extraction and preparation of protoplasts was also compared with extraction methods using enzymatic digestion and homogenization with glass beads for cell disruption.
Primers for PCR.
Four genomic regions of Malassezia were amplified by PCR, namely, the LSU of the ribosomal gene, the ITS, the β-tubulin gene, and the lipase gene. The details of the primers used and the genomic regions amplified in this study are provided in Table 2.
TABLE 2.
Primers used to screen for interspecific variation in Malassezia species
| Locus or region | Primer set | Primer sequence (5′-3′) | Product size (bp) | Reference |
|---|---|---|---|---|
| Large-subunit rRNA gene | 26S-Sa | GCTGAACTTAAG | 16 | |
| CATATCAT | ||||
| 26S-Ab | TAGACGTTAGAC | 642 | 11 | |
| TCCTTGGT | ||||
| ITS | ITS 1 | TCCGTAGGTGAA | ∼800/700 | 35 |
| CCTGCGG | ||||
| ITS 4 | TCCTCCGCTTATT | |||
| GATATGC | ||||
| β-Tubulin gene | Bt-2a | GGTAACCAAATC | ∼550 | 12 |
| GGTGCTGCTTTC | ||||
| Bt-2b | ACCCTCAGTGTA | |||
| GTGACCCTTGGC | ||||
| Lipase genec | LIP-F | GTGTTGGCGTAC | ∼530 | 24 |
| CCGTCGTT | ||||
| LIP-R | CGAGGTCGTTGG | |||
| CAAACGCA |
The primer designed correspond to nucleotides 1 to 20 of the partial sequence of M. furfur (CBS 1878; accession no. AF063214).
The antisense primers for the LSU rRNA gene were designed from the sequence of Saccharomyces cerevisiae (accession no. J01355).
Primers for the lipase region were designed from the sequence of the LIP2 gene of C. cylindracea (accession no. X64704). A 700-bp fragment that was conserved among the five accessioned sequences of lipase genes from C. cylindracea was used for designing primers.
Conventional PCR.
Conventional PCR was generally performed in a 50-μl reaction volume containing 25 μl of Taq PCR master mix (the mix contains 2.5 U of Taq DNA polymerae, 200 μM each deoxynucleoside triphosphate, and 1× PCR buffer with 1.5 mM MgCl2) (QIAGEN Inc., Mississauga, Ontario, Canada), 0.5 μM each primer, and 2 μl of DNA template. DNA amplifications were carried out in a 9600 thermocycler (Perkin-Elmer, Norwalk, Conn.) programmed for initial denaturation at 95°C for 3 min followed by 40 cycles of 95, 50, and 72°C for 1 min each, with a final extension of 10 minutes at 72°C. Amplification at 55°C was also performed, but sometimes this did not yield an amplification product (see Results). Amplified products were electrophoresed through a 0.8% agarose gel in 1× Tris-acetate-EDTA buffer, and ethidium bromide-stained gels were visualized with a UV transilluminator (λ = 320 nm).
PCR-REA.
DNAs that were PCR amplified using primer sets 26S-A–26S-S and ITS 1-ITS 4 (Table 2) were subjected to further restriction endonuclease analysis. The partial sequence of LSU rRNA of M. furfur (GenBank accession no. AF063214) (16) reveals restriction sites for two endonucleases, AvaI and NcoI. Additionally the four-base cutters HaeIII and MspI were also used for the LSU region of Malassezia. To screen for interspecies variation in the ITS region of Malassezia, the following restriction endonucleases were used: AvaI, BamHI, EcoRI, MspI, NcoI, and PstI (obtained from New England Biolabs, Mississauga, Ontario, Canada).
Hot start-TD PCR.
To reduce mispriming and to increase the efficiency and specificity of amplicons obtained by conventional PCR, touchdown (TD) PCR was performed for amplification with primers for the β-tubulin gene (3). The amplification mixture (50 μl) contained 200 μM each deoxynucleoside triphosphate, 1× PCR buffer with 1.5 mM MgCl2, 2.5 U of Taq DNA polymerase (added as Taq PCR master mix) (QIAGEN Inc.), and 0.2 μM each primer. Following a hot start, the thermocycler (9600; Perkin-Elmer) was programmed for the first cycle at 94, 67, and 72°C for 3, 1, and 1 min each. The second cycle was set at 94, 65, and 72°C for 30 s, 1 min, and 1 min each. The annealing temperature was lowered by 2°C in each of the following steps, with a final annealing temperature of 55°C. Thirty cycles were subsequently run (94°C for 30 s, 55°C for 1 min, 72°C for 1 min), ending with a final 5-min extension at 72°C.
RESULTS
DNA extraction.
We compared three different methods for DNA extraction and preparation of protoplasts, using enzymatic digestion, homogenization with glass beads, and lysis in urea buffer. The last method was found to be most successful (34). The basic requirement for a good yield of DNA, however, is freshly grown (2- to 5-day-old) yeast cells. In most cases the DNA was good enough for a successful amplification, but for some strains the impurities in DNA interfered with the primers for the ITS region in PCRs. Repurification of DNA by an additional ethanol precipitation step, however, generally resulted in a positive amplification.
PCR-REA of LSU and ITS regions.
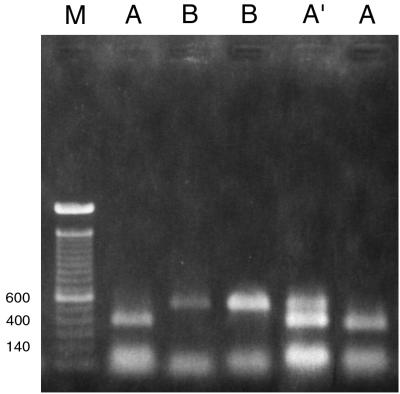
PCR amplification for the LSU region with the designed primers (26S-S and 26S-A [Table 2]) produced an amplicon of the expected size (∼640 bp) for all Malassezia species. Restriction analysis of the amplified product was useful only with AvaI and not with NcoI. This resulted in three PCR-REA types, A, A′, and B (Fig. 1; Table 3). PCR-REA of the LSU region divided the seven Malassezia species into two major groups. All strains of M. sympodialis and most strains of M. furfur and M. slooffiae showed PCR-REA type A. Only 2 out of 11 strains of M. furfur and 2 out of 7 strains of M. slooffiae showed PCR-REA type A′ (the restriction pattern is a combination of those for PCR-REA types A and B [Fig. 1]). PCR-REA type B was characteristic of M. pachydermatis, M. globosa, M. restricta, and M. obtusa (Fig. 1). All of the strains were consistently amplified with these primers at an annealing temperature of 50°C. The use of a higher annealing temperature of 55°C sometimes resulted in the loss of product for some strains of M. sympodialis. All strains of each of the seven species gave the same restriction pattern repeatedly. A double digest of amplified product with restriction endonucleases HaeIII and MspI resulted in four different patterns (data not shown). With these enzymes, more than one restriction pattern was observed among strains of M. furfur and M. globosa. Also, the same pattern was shared by more than two Malassezia species (data not shown). These enzymes are being evaluated further for elucidating intraspecific variation.
FIG. 1.
Restriction patterns obtained by AvaI digestion of amplified product from the LSU region (primers 26S-S and 26S-A). A, B, and A′, PCR-REA type; M, 100-bp ladder used as a size marker. Strains loaded in lanes from left to right, JF4 (M. furfur), 99F 160 (M. globosa), ATCC 14521 (M. pachydermatis), CBS 1878 (M. furfur), and 97F 8615 (M. sympodialis). A polaroid picture of the gel was scanned in the computer, and the relevant lanes were aligned together to create this image. Numbers on the left are base pairs.
TABLE 3.
Molecular differentiation of Malassezia species by PCR-REA
| Genomic region | Restriction endonuclease | PCR-REA types (Malassezia speciesa) |
|---|---|---|
| 26S rRNA gene (LSU) | AvaI | A (Mf, Msy, Msl); A′ (Mf, Msl); B (Mg, Mr, Mo, Mp) |
| TS (ITS 1-ITS 4) | EcoRI | C (Mf, Mp, Mo); C′ (Msl); D (Mg, Mr); D′b, E (Msy) |
| NcoI | F (Mf, Mp); G (Mg, Mr, Mo, Msl); G′c, H (Msy) | |
| β-Tubulin gene | P (Mr, Mf, Msy, Msl, Mp, Mo)d; N (Mg, Mf, Msy, Msl, Mp)e |
Mf, M. furfur; Msl, M. slooffiae; Msy, M. sympodialis; Mp, M. pachydermatis; Mo, M. obtusa; Mg, M. globosa; Mr, M. restricta.
Only 3 of 37 M. sympodialis strains had this PCR type (see Fig. 2B).
Only 2 of 37 M. sympodialis strains had this PCR type. Restriction with NcoI produced two bands of 500 and 200 bp (PCR-REA type G′) instead of one 350-bp band of double intensity (PCR-REA type H).
Positive amplification with primers for β-tubulin gene.
Negative amplification with primers for β-tubulin gene.
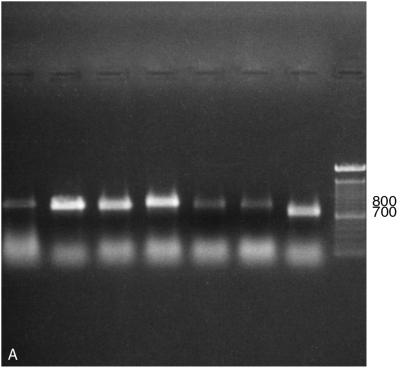
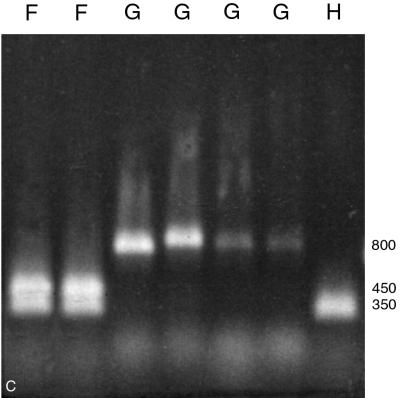
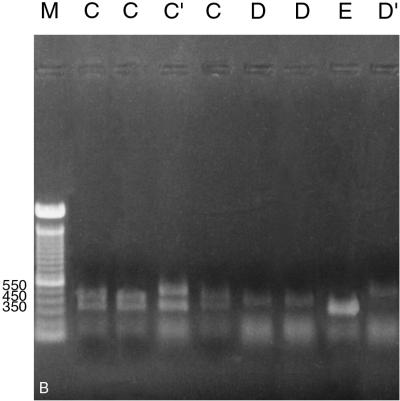
PCR amplification of the ITS region with primers ITS 1 and ITS 4 (Table 2) readily distinguished M. sympodialis from other Malassezia species by its smaller amplified fragment, a 700-bp product rather than the 800-bp product produced by the others (Fig. 2A). For further species distinction with this amplicon, two restriction endonucleases proved useful. EcoRI divided the seven Malassezia species into five PCR-REA types, designated types C, C′, D, D′, and E (Fig. 2B; Table 3). One Malassezia group, showing restriction pattern type C, comprised three species, i.e., M. furfur, M. pachydermatis, and M. obtusa (Fig. 2B). A unique PCR-REA type, C′, characterized M. slooffiae. PCR-REA type D comprised two species, M. globosa and M. restricta (Fig. 2B). Most strains of M. sympodialis showed PCR-REA type E (Fig. 2B), whereas 3 of 37 strains showed PCR-REA type D′ (with a single fragment ∼200 bp larger than that of type E) (Fig. 2B). NcoI divided the seven Malassezia species into four PCR-REA types, represented as F, G, G′, and H (Fig. 2C; Table 3). As with EcoRI, M. furfur and M. pachydermatis showed the same restriction pattern, in this case type F. M. obtusa and M. slooffiae, however, showed a different restriction pattern, type G, that was the same as for M. globosa and M. restricta (Fig. 2C). Again, M. sympodialis was unique, with either type G′ or type H (Fig. 2C).
FIG. 2.
(A) Amplification of seven Malassezia species using primers for the ITS region. Lanes (left to right): M. furfur (CBS 1878), M. pachydermatis (ATCC 14521), M. slooffiae (CBS 7956), M. obtusa (98F 3529), M. globosa (98F 6443), M. restricta (YKM 31), M. sympodialis (CBS 7222), and 100-bp ladder. Numbers on the right are base pairs. (B) Restriction patterns obtained by EcoRI digestion of amplified products from the ITS region (primers ITS 1 and ITS 4). Letters represent PCR-REA types. Lanes (left to right): M. furfur (CBS 1878), M. pachydermatis (ATCC 14521), M. slooffiae (CBS 7956), M. obtusa (98F 3529), M. globosa (98F 6443), M. restricta (YKM 31), M. sympodialis (CBS 7222), and M. sympodialis (GM 323). This image was generated by scanning a polaroid picture of the gel in the computer. Numbers on the left are base pairs. (C) Restriction patterns obtained by NcoI digestion of amplified products from the ITS region (primers ITS 1 and ITS 4). Letters represent PCR-REA types. Lanes (left to right): M. furfur (CBS 1878), M. pachydermatis (ATCC 14521), M. slooffiae (CBS 7956), M. obtusa (98F 3529), M. globosa (98F 6443), M. restricta (YKM 31), and M. sympodialis (CBS 7222). This image was generated by scanning a polaroid picture of the gel in the computer. Numbers on the right are base pairs.
Five of the seven Malassezia species could thus be distinguished using the combination of the LSU and ITS regions. Multilocus genotypes of seven Malassezia species were constructed using these two regions and the commonly represented PCR-REA types (Table 4). A similarity index was calculated for all Malassezia species in a pairwise comparison on the basis of number of matches (Table 4).
TABLE 4.
Similarity matrix of seven Malassezia species on the basis of PCR-REA types of the LSU and ITS regions
| Malassezia species | Multilocus genotypea | Similarity Indexb with:
|
|||||
|---|---|---|---|---|---|---|---|
| M. pachydermatis | M. sympodialis | M. slooffiae | M. obtusa | M. globosa | M. restricta | ||
| M. furfur | 101000100 | 0.77 | 0.55 | 0.55 | 0.55 | 0.33 | 0.33 |
| M. pachydermatis | 011000100 | 0.33 | 0.33 | 0.77 | 0.55 | 0.55 | |
| M. sympodialis | 100001001 | 0.55 | 0.33 | 0.33 | 0.33 | ||
| M. slooffiae | 100100010 | 0.55 | 0.55 | 0.55 | |||
| M. obtusa | 011000010 | 0.77 | 0.77 | ||||
| M. globosa | 010010010 | 1.00c | |||||
| M. restricta | 010010010 | ||||||
PCR-REA types that resulted from the analysis of the LSU of the rRNA gene and the ITS have been used to construct these multilocus genotypes. Only the common representative PCR-REA types for each Malassezia species are included. The sequence of PCR-REA types is ABCC′DEFGH; 1 represents the presence and 0 represents the absence of a particular PCR-REA type.
Similarity indices are calculated on the basis of the number of matches between all pairs of Malassezia species. Either the presence or absence of a particular PCR-REA type is considered a match.
M. globosa and M. restricta could be distinguished only by TD-PCR, using primers for the β-tubulin gene.
Although in most cases the results of molecular and nonmolecular tests for the strains under investigation were in agreement, there were four ambiguous cases (Table 1). First, strain 99F-1436 was identified as M. globosa on the basis of microscopic examination and utilization of tween compounds (Tween 20, 40, 60, and 80) (17), but the molecular features of this strain were characteristic of M. furfur. Second, while strain 98F-8316 was identified as M. obtusa on the basis of microscopic observation and physiological tests, this strain had a PCR-REA type A rather than the usual type B for the LSU region. Third, strain WF7, which had all the microscopic, physiological, and molecular features characteristic of M. obtusa, showed similar growth at both 32 and 40°C (M. obtusa is not known to grow well at 40°C) (14). Lastly, strain GM 420, although identified as M. pachydermatis on the basis of physiological properties, had molecular features consistent with M. furfur. For this strain, however, the PCR amplification with primers for the lipase gene resulted in a product similar to that of M. pachydermatis strains (see below).
TD-PCR for β-tubulin gene.
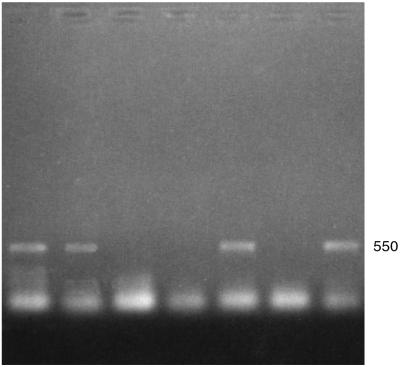
Initial screening by conventional PCR with primers for the β-tubulin gene (Table 2) (12), suggested that this region might be useful in differentiating Malassezia species (data not shown). However, consistent bands were not obtained in repeated PCRs for the same strains done under the same conditions. A hot start-TD PCR was performed for this region to determine if it would yield more consistent results. TD-PCR, by starting the annealing process at 12 to 15°C above the calculated Tm and gradually decreasing the annealing temperature, has been shown to increase the efficiency and specificity of amplicons obtained by conventional PCR (10). Primers for the β-tubulin gene proved useful in differentiating M. globosa and M. restricta by the presence or absence of an amplified fragment following a TD-PCR procedure. All strains of M. restricta were consistently amplified, yielding an ∼550-bp fragment, whereas those of M. globosa were never amplified (Fig. 3). Other species gave inconsistent results (Tables 1 and 3).
FIG. 3.
Hot start-TD PCR amplification product obtained using primers for the β-tubulin gene (12). Lanes (left to right): YKM 31 (M. restricta), CBS 7877 (M. restricta), 98F 9925 (M. sympodialis), 98F-7317 (M. globosa), JF4 (M. furfur); 98F-8316 (M. obtusa), and CBS 7956 (M. slooffiae). Positive amplification of a band of ∼550 bp is seen in lanes 1, 2, 5, and 7.
Lipase gene.
The lipase gene region was of primary interest to us because of basic differences in the lipid requirements among Malassezia species. While six of the seven Malassezia species require exogenous lipids for growth, M. pachydermatis can grow in media without additional lipids. Since the lipase gene of Malassezia has not been characterized molecularly, we designed primers from the published sequence of the LIP2 gene of Candida cylindracea (24). Lipase sequences of C. cylindracea were used because the lipase activities and pH profiles of M. furfur and C. cylindracea lipases have been shown to be similar (30). On annealing at 50°C, these primers resulted in several nonspecific bands for different Malassezia species that were not reproducible, whereas fragments of ∼850 and <1,200 bp were consistently amplified for the two M. pachydermatis strains included in this study. Annealing at 55°C resulted in a product of ∼560 bp (close to the expected size [Table 2]) from two M. pachydermatis strains, CBS 1879 and ATCC 14521, as well as from strain GM420. At 55°C annealing, the one M. globosa strain tested (98F-8304) revealed a product of <1,200 bp, whereas for the remaining five Malassezia species, no amplification product was observed (data not shown). These preliminary results suggest that this region might be useful in identification of M. pachydermatis and M. globosa strains.
DISCUSSION
The data presented in this study conform to the present nomenclature of Malassezia yeasts. Our results show that PCR-REA may provide a rapid and reliable technique for molecular differentiation of Malassezia species. However, the status of a small number of isolates remained ambiguous after PCR-REA typing, and these techniques should be employed as a useful adjunct to conventional testing until additional study resolves the correct placement of the anomalous isolates.
M. furfur, M. sympodialis, and M. slooffiae are physiologically very similar, and ambiguity remained regarding correct identification of these species on the basis of tests for utilization of tween compounds in the simple media (17). Recently, Mayser et al. (28) have reported the use of additional tests, such as addition of cremophor EL in diffusion plates and characterization of β-glucosidase activity, to resolve this ambiguity. In the present study, PCR-RFLP analysis of only one genomic region, the ITS region, proved sufficient to resolve the ambiguity between the three physiologically similar species. Similarly, M. obtusa, which is similar in growth requirements to M. globosa and M. restricta, could be differentiated from these species by variation in the ITS region. M. globosa and M. restricta, however, which are readily distinguishable by differences in cell morphology and catalase activity (14), were difficult to distinguish on the basis of variation in ribosomal DNA.
Although we believe that the PCR-REA system is generally very reliable, we recommend that a microscopic examination of yeast cells and a catalase test should also be done for an accurate identification. The four discrepancies observed among species identifications on the basis of physiological and molecular tests suggest that for some ambiguous cases it may become necessary to conduct additional tests, such as a test for growth on Sabouraud agar to distinguish M. pachydermatis from M. furfur. The presence of more than one PCR-REA type, in most cases, can be explained by either gain or loss of a restriction site, with the only exception being type A′ of the LSU region, which is indicative of a recombinant strain. The presence of more than one PCR-REA type for M. furfur, M. sympodialis, and M. slooffiae is suggestive of intraspecific variation, but such intraspecific variation, if present, has to be confirmed further with additional screening.
The results of this study are comparable with those of Guillot and Guého (16), who, in an rRNA sequence study of Malassezia species, observed unique sequences for M. sympodialis. In the present study as well, M. sympodialis was readily distinguishable from other species on the basis of a smaller amplicon (∼700 bp) for the ITS region. Guillot and Guého (16) found that when the most variable D2 region of the ribosomal gene was examined, maximal divergence was observed between sequences for M. furfur and M. restricta. We also found that M. furfur was most divergent from M. restricta and M. globosa, with only 33% similarity among these species on the basis of the molecular markers screened.
The neotype strain of Pityrosporum ovale (CBS 1878), currently identified as M. furfur, is atypical culturally and serologically (9) as well as karyotypically (6). Its anomalous nature is confirmed in this study. PCR amplification of CBS 1878 with primers for LSU region followed by restriction analysis revealed in a PCR-REA type that was different from that of the other representative strains of M. furfur.
Karyotyping, although very reliable for molecular differentiation of Malassezia species, takes at least 60 to 72 h for analysis of each sample after culture growth and isolation of protoplasts. In contrast, the PCR-RFLP analysis reported here can be completed in less than 15 h after DNA extraction. Once the extraction protocol is optimized for direct DNA extractions from skin scrapings, the 2 to 5 days required for appreciable yeast growth in culture can be eliminated. It may thus be possible, after further development, to provide reliable species identifications back to the physician in less than 3 days. Thus, the PCR-RFLP procedure may ultimately prove to be preferable to both karyotyping and culture analysis for identification of Malassezia species.
PCR-RFLP analysis for other regions or protein-encoding genes may provide further insight into intraspecific variation and thus be very useful in epidemiological studies.
REFERENCES
- 1.Anaissie E, Bodey G P. Nosocomial fungal infections: old problems and new challenges. Infect Dis Clin N Am. 1989;3:867–882. [PubMed] [Google Scholar]
- 2.Anthony R M, Howell S A, Pinters L. Application of DNA typing methods to the study of the epidemiology of Malassezia pachydermatis. Microb Ecol Health Dis. 1994;7:161–168. [Google Scholar]
- 3.Ault G S, Ryschkewitsch C F, Stoner G L. Type-specific amplification of viral DNA using touchdown and hotstart PCR. J Virol Methods. 1994;46:145–156. doi: 10.1016/0166-0934(94)90099-x. [DOI] [PubMed] [Google Scholar]
- 4.Belkum A, Boekhout T, Bosboom R. Monitoring spread of Malassezia infections in a neonatal intensive care unit by PCR-mediated genetic typing. J Clin Microbiol. 1994;32:2528–2532. doi: 10.1128/jcm.32.10.2528-2532.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boekhout T, Bosboom R W. Karyotyping of Malassezia yeasts: taxonomic and epidemiological implications. Syst Appl Microbiol. 1994;17:147–153. [Google Scholar]
- 6.Boekhout T, Kamp M, Guého E. Molecular typing of Malassezia species with PFGE and RAPD. Med Mycol. 1998;36:365–372. doi: 10.1080/02681219880000581. [DOI] [PubMed] [Google Scholar]
- 7.Carbone I, Kohn L M. Ribosomal DNA sequence divergence within internal transcribed spacer 1 of the Sclerotiniaceae. Mycologia. 1993;85:415–427. [Google Scholar]
- 8.Carlotti A, Chaib P, Couble A, Bourgeois N, Blanchard V, Villard J. Rapid identification and fingerprinting of Candida krusei by PCR-based amplification of the species-specific repeated polymorphic sequence CKRS-1. J Clin Microbiol. 1997;35:1337–1343. doi: 10.1128/jcm.35.6.1337-1343.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunningham A C, Leeming J P, Ingham E, Gowland G. Differentiation of three serovars of Malassezia furfur. J Appl Bacteriol. 1990;68:439–446. doi: 10.1111/j.1365-2672.1990.tb02894.x. [DOI] [PubMed] [Google Scholar]
- 10.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. Touchdown PCR to circumvent spurious priming gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geogiev O I, Nikolaev N, Hadjiolov A A, Skryabin K G, Zakharyev N M, Bayev A A. The structure of the yeast ribosomal genes. 4. Complete sequence of the 25S rRNA gene from Saccharomyces cerevisiae. Nucleic Acids Res. 1981;9:6953–6958. doi: 10.1093/nar/9.24.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glass N L, Donaldson G C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guého E, Meyer S A. A reevaluation of the genus Malassezia by means of genome comparison. Antonie Leeuwenhock. 1989;55:245–251. doi: 10.1007/BF00393853. [DOI] [PubMed] [Google Scholar]
- 14.Guého E, Midgley G, Guillot J. The genus Malassezia with description of four new species. Antonie Leeuwenhoek. 1996;69:337–355. doi: 10.1007/BF00399623. [DOI] [PubMed] [Google Scholar]
- 15.Guého E, Boekhout T, Ashbee H R, Guillot J, Van Belkum A, Faegermann J. The role of Malassezia species in the ecology of human skin and as pathogens. Med Mycol. 1998;36:220–229. [PubMed] [Google Scholar]
- 16.Guillot J, Guého E. The diversity of Malassezia yeasts confirmed by rRNA sequence and nuclear DNA comparisons. Antonie Leeuwenhoek. 1995;67:297–314. doi: 10.1007/BF00873693. [DOI] [PubMed] [Google Scholar]
- 17.Guillot J, Guého E, Lesourd M, Midgley G, Chévrier G, Dupont B. Identification of Malassezia species: a practical approach. J Mycol Med. 1996;6:103–110. [Google Scholar]
- 18.Hazen K C. New and emerging yeast pathogens. Clin Mirobiol Rev. 1995;8:462–478. doi: 10.1128/cmr.8.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson C, Barton R C, Evans G V. Species identification and strain differentiation of dermatophyte fungi by analysis of ribosomal-DNA intergenic spacer regions. J Clin Microbiol. 1999;37:931–936. doi: 10.1128/jcm.37.4.931-936.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiuchi A, Taharaguchi S, Hanazawa R, Hara M, Ikeda T, Tabuchi K. Chromosome-sized DNA of Malassezia pachydermatis by pulsed-field gel electrophoresis. J Vet Med Sci. 1992;54:1219–1220. doi: 10.1292/jvms.54.1219. [DOI] [PubMed] [Google Scholar]
- 21.Leclerc M C, Philippe H, Guého E. Phylogeny of dermatophytes and dimorphic fungi based on large subunit ribosomal RNA sequence comparisons. J Med Vet Mycol. 1994;32:331–341. doi: 10.1080/02681219480000451. [DOI] [PubMed] [Google Scholar]
- 22.Lee C-H, Helweg-Larsen J, Tang X, Jin S, Li B, Barlett M S, Liu J-J. Update on Pneumocystis carinii f. sp. hominis typing based on nucleotide sequence variations in internal transcribed spacer regions of rRNA genes. J Clin Microbiol. 1998;36:734–741. doi: 10.1128/jcm.36.3.734-741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leeming J P, Notman F H. Improved methods for isolation and enumeration of Malassezia furfur from human skin. J Clin Microbiol. 1987;25:2017–2019. doi: 10.1128/jcm.25.10.2017-2019.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Longhi S, Fuseth F, Gordon R, Lotti M, Vanoni M, Alberghina L. Cloning and nucleotide sequences of two Lip genes from Candida cylindracea. Biochim Biophys Acta. 1992;1131:227–232. doi: 10.1016/0167-4781(92)90085-e. [DOI] [PubMed] [Google Scholar]
- 25.Makimura K, Murayama S Y, Yamaguchi H. Detection of a wide range of medically important fungi by polymerase chain reaction. J Med Microbiol. 1994;40:358–364. doi: 10.1099/00222615-40-5-358. [DOI] [PubMed] [Google Scholar]
- 26.Makimura K, Tamura Y, Mochizuki T, Hasegawa A, Tajiri Y, Hanazawa R, Uchida K, Saito H, Yamaguchi H. Phylogenetic classification and species identification of dermatophyte strains based on DNA sequences of nuclear ribosomal internal transcribed spacer 1 regions. J Clin Microbiol. 1999;37:920–924. doi: 10.1128/jcm.37.4.920-924.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcon M J, Powell D A. Human infections due to Malassezia spp. Clin Microbiol Rev. 1992;5:101–119. doi: 10.1128/cmr.5.2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayser P, Haze P, Papavassilis C, Pickel M, Gruender K, Guého E. Differentiation of Malassezia species: selectivity of cremophor EL, castor oil and ricinoleic acid for M. furfur. Br J Dermatol. 1997;137:208–213. doi: 10.1046/j.1365-2133.1997.18071890.x. [DOI] [PubMed] [Google Scholar]
- 29.Radford S A, Johnson E M, Leeming J P, Millar M R, Cornish J M, Foot A B M, Warnock D W. Molecular epidemiological study of Aspergillus fumigatus in a bone marrow transplantation unit by PCR amplification of ribosomal intergenic spacer sequences. J Clin Microbiol. 1998;36:1294–1299. doi: 10.1128/jcm.36.5.1294-1299.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ran Y, Yoshike T, Ogawa H. Lipase of Malassezia furfur: some properties and their relationship to cell growth. J Med Vet Mycol. 1993;31:77–85. doi: 10.1080/02681219380000081. [DOI] [PubMed] [Google Scholar]
- 31.Rinaldi M G. Emerging opportunists. Infect Dis Clin N Am. 1989;3:65–76. [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Samonis G, Bafaloukos D. Fungal infections in cancer patients: an escalating problem. In Vivo. 1992;6:183–194. [PubMed] [Google Scholar]
- 34.Sansinforiano M E, Padilla J A, de Mendoza H, de Mendoza M H, Fernandez-Garcia J L, Martinez-Trancon M, Rabasco A, Parejo J C. Rapid and easy method to extract and preserve DNA from Cryptococcus neoformans and other pathogenic yeasts. Mycoses. 1998;41:195–198. doi: 10.1111/j.1439-0507.1998.tb00323.x. [DOI] [PubMed] [Google Scholar]
- 35.White T J, Burns T, Lee S, Taylor J W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 315–322. [Google Scholar]