Abstract
Diagnostic markers for non-alcoholic fatty liver disease (NAFLD) are still needed for screening individuals at risk. In recent years, the machine learning method was used to search for the diagnostic markers of multiple diseases. In this study, we developed and validated a machine learning model to diagnose NAFLD using laboratory indicators. NAFLD patients and non-NAFLD controls were recruited in the training and validation cohorts. The laboratory indicators of the participants in the training cohort were collected, and six indicators including alanine aminotransferase/aspartate aminotransferase (ALT/AST), white blood cells (WBC), alpha-L-fucosidase (AFU), hemoglobin (Hb), triglycerides (TG) and gamma-glutamyl transpeptidase (GGT) were screened out with higher weights by an integrate machine learning method. The areas under the receiver operating characteristic curves (AUROCs) for the selected indicators using logistic regression (LR), random forest (RF) and support vector machine (SVM) were 0.814, 0.837 and 0.810, respectively. Then the binary logistic regression was used to construct the predictive model. What’s more, the AUROC of the predicted model was 0.732 in the validation cohort of patients with NAFLD. And the combined AUROC of the six parameters was 0.716 in the mouse model fed with high-fat diet (HFD). In summary, we created a predictive model with six laboratory indicators for the diagnosis of NAFLD based on the machine learning method, which has the potential value for the diagnosis of the NAFLD.
Keywords: NAFLD, diagnosis, machine learning, laboratory indicator
Introduction
Non-alcoholic fatty liver disease (NAFLD) is one of the most common chronic liver diseases globally with an estimated prevalence of about 24% in North America and about 30% in Asia [1,2]. NAFLD is marked by excessive intrahepatic fat deposition [3]. The disease spectrum of NAFLD ranges from simple steatosis to non-alcoholic steatohepatitis (NASH), fibrosis, cirrhosis and even hepatocellular carcinoma (HCC) [4], which could bring heavy burdens on the health care system [5,6].
Although liver pathology was the gold standard for the diagnosis of NAFLD, it is invasive, which limites the wide application. Other diagnostic methods for NAFLD were based on imaging examinations such as B ultrasound, computed tomography (CT) or Fibroscan according to the Practice Guidance of the American Association for the Study of Liver Diseases (AASLD) Practice Guidelines [7]. However, CT and Fibroscan are not convenient for patients, and B ultrasound was limited in relatively low accuracy and specificity. Recently, blood indexes, which are convenient, low-cost and readily available, are ideal tools for the diagnosis of diseases. Several prediction models based on blood indexes have been constructed for the diagnosis of NAFLD. Elevated alanine aminotransferase (ALT) level is the predominant finding for the diagnosis of NAFLD, but elevated ALT level also occurred in other liver diseases such as hepatitis B virus (HBV) infection and intrahepatic cholangitis [8]. SteatoTest for the diagnosis of NAFLD is a logistic regression model containing 12 predicting indicators including a2-macroglobulin (A2M), apolipoprotein A1 (Apo A1), haptoglobin, gamma-glutamyl transpeptidase (GGT) levels, total bilirubin, cholesterol, triglycerides, glucose, age, gender, and body mass index [9], while it has not been proved as a practical model based on the combination of blood indexes with high sensitivity and specificity.
In recent years, machine learning methods including filter method, wrapper method and the embedded method were developed to select predicting parameters among all available indicators with maximum data and minimum bias to predict diseases. Machine learning could reveal the complex relationships among the indicators, which was useful for the diagnosis of diseases. Li et al. built multiparametric ultrasomics which could improve discrimination of significant fibrosis in chronic hepatitis B patients compared with mono or dual modalities [10]. What’s more, Liu et al. built an artificial neural network model that was useful for evaluating the probability of progression-free survival in patients with HCC [11].
In this study, we aimed to develop a predictive model for diagnosing NAFLD depending on the laboratory parameters using an integrated machine learning method and validated the diagnostic effects of the model for diagnosing NAFLD at the population and animal levels.
Methods
Research subjects
Inclusion criteria for NAFLD patients and non-NAFLD patients in this study: (1) 18-65 years of age; (2) Patients were negative for hepatitis B surface antigen, hepatitis C virus-RNA and hepatitis B virus DNA; (3) No clinical symptoms or signs of infection, no liver disorders or other critical diseases, and no fractures, osteoporosis, or tumors; (4) No pregnancy for women; (5) No drinks or no more than 70 g ethanol per week for women (about one standard drink daily), and 140 g for men (two standard drinks daily).
Exclusion criteria for NAFLD patients and non-NAFLD patients in this study: (1) Patients with an active implantable medical device (such as pacemaker or defibrillator); (2) Hematological diseases or diseases that may influence the parameters of blood cell counts; (3) Patients who had undergone liver transplantation, patients with cardiac failure and/or significant valvular disease.
Definition of NAFLD: Patients with and without NAFLD were first distinguished by pathology. If the pathology was not available, B ultrasound or CT was used. The diagnostic criteria of NAFLD were followed by the practice guidance from the American Association [12].
According to the criteria above, a total of 45 NAFLD patients and 53 non-NAFLD controls from Qingdao Municipal Hospital were recruited in the training group. Informed consent was signed by every participant. This study was approved by the Ethics Committee of the Qingdao Municipal Hospital (2019Y006). 81 NAFLD patients and 87 non-NAFLD controls from the Affiliated Hospital of Qingdao University were involved in the validation cohort. Informed consent was signed by every participant. This study was also approved by the Ethics Committee of the Affiliated Hospital of Qingdao University (QYFYWZLL26473).
Clinical data collection
The data for the clinical laboratory indicators of the first admission were collected from the Qingdao Municipal Hospital and the Affiliated Hospital of Qingdao University (Supplementary Tables 1 and 2). The data for the imaging examinations were also collected at the meantime.
Animal experiments
Eight-week-old male C57BL/6J mice were fed with high-fat diet (HFD) or chow diet (CD). Twelve weeks later, all mice were sacrificed. The hematoxylin-eosin (HE) staining of liver tissues was used to verify the successful establishment of the NAFLD mice model. WBC was calculated by XFA6030 Animal Blood Cell Analyzer (prolong, Beijing, China). ALT, aspartate aminotransferase (AST), triglycerides (TG), hemoglobin (Hb), and alpha-L-fucosidase (AFU) were tested by enzyme linked immunosorbent assay kit (Shanghai Enzyme-linked Biotechnology Co., Ltd., Shanghai, China). Animal experiments were approved by the Animal Experimental Ethical Committee of Qingdao University (AHQU-MAL20180504-1).
Statistical analysis
The weight values of candidate biomarkers were calculated by the method of integrated machine learning (Applied Protein Technology, Shanghai, China) [13-15]. To validate the effects of selected candidate biomarkers in classification models, three commercialized machine learning models including logistic regression (LR) [16], random forest (RF) [17] and support vector machine (SVM) [18] were used respectively. The diagnostic values of the indicators were evaluated by the receiver operating characteristic (ROC) curve and the areas under the receiver operating characteristic curves (AUROC). Pearson correlation coefficient (r) was calculated between the candidate biomarkers. When the value of r was more than 0.6, the correlation was defined as strong [19]. Data were analyzed by R version 3.6.1 (R Foundation for Statistical Computing, Vienna, Austria). Data were expressed as mean ± standard deviation (SD). Missing values were imputed using mean imputation. The difference between subgroups was analyzed by chi-square test for categorical parameters and Student’s t-test was used for continuous parameters.
Results
Patient characteristics
In the training cohort including 53 NAFLD patients and 45 non-NAFLD controls, the percentage of male was 60.38% in the NAFLD group, which was higher than that in the non-NAFLD controls (P<0.05). There were also significant differences in ALT/AST, ALT, white blood cells (WBC), AFU, Hb, TG, GGT, neutrophilic granulocyte count (NEUT), lymphocyte count (LYM), monocytes count (MON), basophils (BASO), uric acid (UA), high-density lipoprotein (HDL) and apolipoprotein A1/apolipoprotein B (APO A1/APO B) between the two groups (Table 1).
Table 1.
Partial differential characteristics of subjects with or without NAFLD in the training cohort
| Characters | NAFLD n=53 | No NAFLD n=45 | P value |
|---|---|---|---|
| Gender (male/female, n) | 32/21 | 25/20 | 0.02 |
| ALT (U/L) | 31.21±21.54 | 19.30±12.93 | <0.001 |
| ALT to AST | 1.33±0.64 | 0.90±0.29 | <0.001 |
| GGT (U/L) | 35.88±29.93 | 21.95±13.96 | 0.02 |
| AFU (U/L) | 26.45±7.91 | 23.32±7.12 | 0.046 |
| WBC (× 109/L) | 6.40±1.70 | 5.30±1.31 | <0.001 |
| TG (mmol/L) | 1.83±1.26 | 1.25±0.53 | <0.001 |
| Hb (g/L) | 144.98±11.69 | 138.22±15.76 | 0.03 |
| NEUT (109/L) | 3.40±1.11 | 2.90±0.94 | 0.02 |
| LYM (109/L) | 2.78±2.52 | 2.00±0.62 | 0.03 |
| MON (109/L) | 0.40±0.12 | 0.34±0.12 | 0.02 |
| BASO (109/L) | 0.04±0.02 | 0.03±0.02 | 0.04 |
| UA (μmol/L) | 360.34±86.08 | 312.26±70.99 | <0.001 |
| HDL (mmol/L) | 1.24±0.28 | 1.39±0.31 | 0.02 |
| APO A1/APO B | 1.35±0.39 | 1.62±0.70 | 0.04 |
Six parameters were selected using an integrated machine learning method
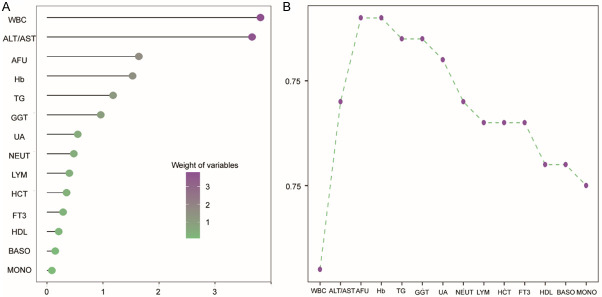
For the training cohort, the weight value of each biomarker was calculated by an integrated machine learning method. The higher value of the weight means the greater contribution of the biomarker in distinguishing the patients with NAFLD from the non-NAFLD controls. The top six candidate biomarkers were WBC, ALT to AST, AFU, Hb, TG and GGT (Figure 1A).
Figure 1.
Six parameters were selected using an integrated machine learning method in the training cohort of patients with NAFLD. A. Weight values of the variables; B. The cumulative AUC chart of the candidate biomarkers.
The cumulative AUROC chart was used to evaluate the impact of combined biomarkers and the sequence for the combination of indicators were determined according to the weight values. As shown in Figure 1B, the AUROC of the top six indicators was 0.772 and when the 7th indicator was added, the AUROC of the combined biomarkers was suddenly reduced. Consequently, we selected six parameters for further analysis.
Verification of the candidate biomarkers
ROC curves of three models including LR, RF and SVM demonstrated that the selected candidate biomarkers have excellent effects for classification, and the AUROC values of LR, RF, and SVM were 0.814, 0.837 and 0.810, respectively (Figure 2A). The importance coefficients of the candidate biomarkers were calculated by RF to compare the contribution of each biomarker in the model (Figure 2B). Accuracy, sensitivity, and specificity evaluated by LR, RF and SVM were revealed in Figure 2C. The results of verification demonstrated that the selected biomarkers have excellent effects of classification for NAFLD patients in the training cohort.
Figure 2.
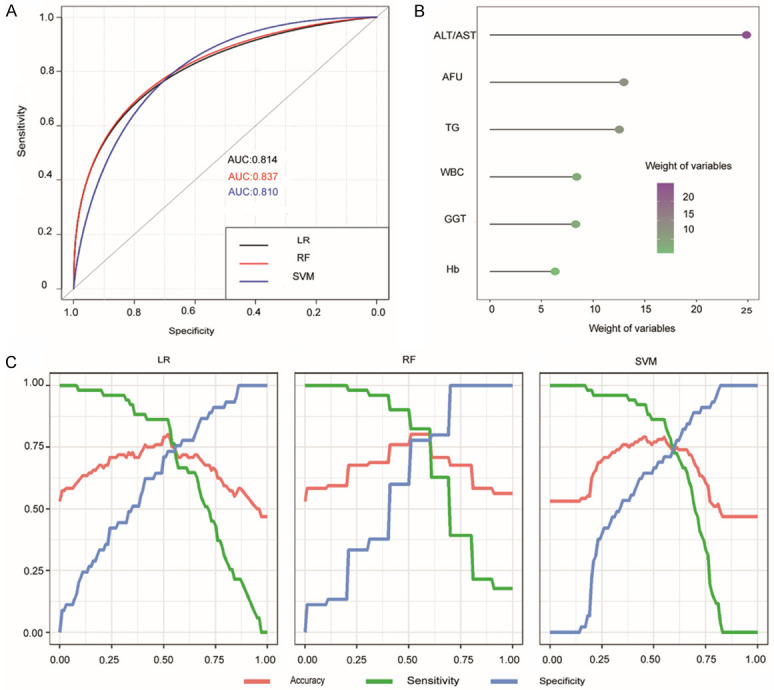
Verification of the candidate biomarkers in the training cohort of patients with NAFLD. A: The ROC curves of the candidate biomarkers for the diagnosis of NAFLD using LR, SVM and RF methods; B: The importance of the candidate biomarkers calculated by RF; C: Curves of accuracy, sensitivity, and specificity of the candidate biomarkers for the diagnosis of NAFLD evaluated by LR, RF and SVM.
Correlation of the candidate biomarkers
We conducted the correlation analysis among the selected biomarkers. If there was a strong correlation between the two biomarkers, one of them would be eliminated. As shown in Figure 3, no strong correlations between the selected biomarkers were observed, and none of the biomarkers in this study was eliminated.
Figure 3.
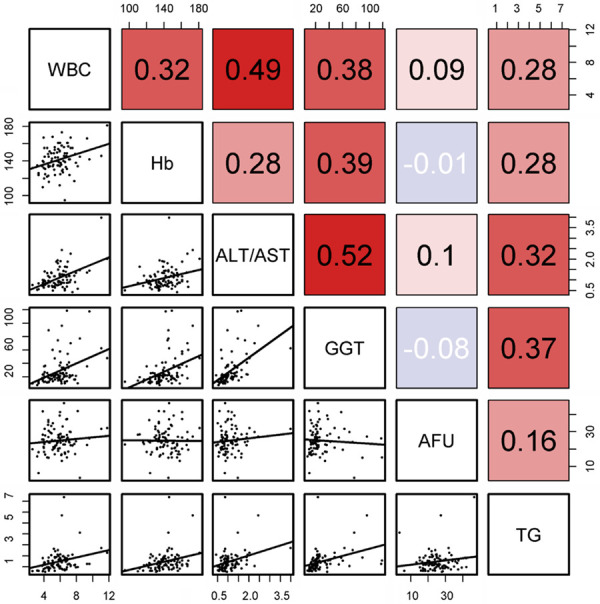
Correlations of the candidate biomarkers in the training cohort of patients with NAFLD. Pearson correlation coefficient (r) was used to evaluate the correlations.
Construction of the predicted model
The diagnostic panel of candidate biomarkers for NAFLD was built by logistic regression algorithm. The logical regression coefficients of WBC, ALT to AST, AFU, Hb, TG and GGT were 0.29, 2.23, 0.07, 0.01, 0.72 and 0.01, respectively (Supplementary Table 3). The equation was illustrated as following:
Risk score = exp (0.29 × WBC + 2.23 × ALT/AST + 0.07 × AFU + 0.01 × Hb + 0.72 × TG + 0.01 × GGT - 8.77)/[1 + exp (0.29 × WBC + 2.23 × ALT/AST + 0.07 × AFU + 0.01 × Hb + 0.72 × TG + 0.01 × GGT - 8.77)]
The best cutoff value of the risk score was 0.53 calculated by using Yoden Index (Supplementary Table 4). When the risk score was 0.53, the AUROC value, specificity and sensitivity for the predicted model were 0.821, 0.733 and 0.765, respectively. If the risk score of the equation was higher than the best cutoff value, the result would be positive.
The diagnostic evaluation of the predicted model in the validation cohort
81 NAFLD patients and 87 non-NAFLD controls were involved in the validation cohort from the Affiliated Hospital of Qingdao University. There were significant differences in the values of ALT, AST, ALT to AST, GGT, AFU, WBC, TG, LYM, MON, MON% BASO%, red blood cell volume distribution width (RDW), uric acid (UA), total cholesterol (TC), HDL, APO A1, APO B and APO A1/APO B between the two groups (Table 2). The predicted model kept its diagnostic efficacy in the validation group with the AUROC 0.732 (Figure 4).
Table 2.
Partial differential characteristics of subjects with or without NAFLD in the validation cohort
| Characters | NAFLD group n=81 | Control group n=87 | P value |
|---|---|---|---|
| ALT (U/L) | 31.45±19.24 | 23.27±15.86 | <0.001 |
| AST (U/L) | 21.82±8.52 | 19.22±6.69 | 0.03 |
| ALT to AST | 1.41±0.44 | 1.16±0.52 | <0.001 |
| GGT (U/L) | 33.78±14.53 | 18.20±6.47 | 0.02 |
| AFU (U/L) | 29.35±7.88 | 26.31±7.05 | 0.02 |
| WBC (× 109/L) | 6.46±1.69 | 5.80±1.25 | 0.01 |
| TG (mmol/L) | 2.03±1.89 | 1.39±1.14 | 0.02 |
| Hb (g/L) | 143.54±14.10 | 138.80±17.20 | 0.08 |
| LYM (109/L) | 2.19±0.75 | 1.91±0.59 | 0.01 |
| MON (109/L) | 0.46±0.15 | 0.37±0.13 | <0.001 |
| MON% | 7.32±2.02 | 6.42±2.02 | <0.001 |
| BASO% | 0.53±0.25 | 0.44±0.28 | 0.04 |
| RDW% | 12.37±0.55 | 22.65±13.79 | <0.001 |
| UA (μmol/L) | 366.70±78.65 | 306.36±80.34 | <0.001 |
| TC (mmol/L) | 4.54±0.54 | 4.15±1.60 | <0.001 |
| HDL (mmol/L) | 1.33±0.30 | 1.46±0.36 | 0.04 |
| APO A1 (g/L) | 1.38±0.24 | 1.52±0.22 | 0.01 |
| APO B (g/L) | 1.00±0.24 | 0.85±0.19 | 0.01 |
| APO A1/APO B | 1.48±0.51 | 1.88±0.53 | <0.001 |
Figure 4.
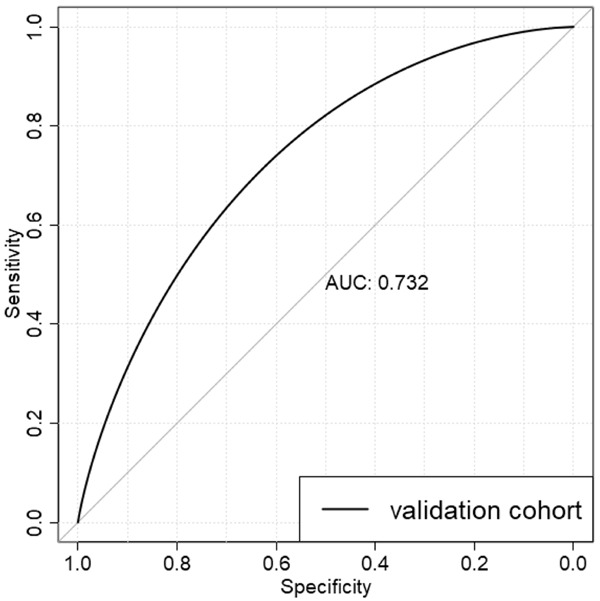
The ROC curve of the candidate biomarkers for the diagnosis of NAFLD in the validation cohort of NAFLD patients.
Validation of the selected parameters using the mouse model
Mice fed with HFD or CD were involved in animal experiments. The results of H&E and Oil Red O staining of liver tissue sections verified the construction of NAFLD mouse model (Figure 5A). Although there were no obvious differences in ALT, AST, ALT to AST, AFU, WBC, TG, Hb and GGT between the two groups (all P>0.05), the average values of all the selected indicators in HFD group were higher than those in CD group (Supplementary Table 5). Furthermore, the cumulative AUROC of the candidate biomarkers was up to 0.716 (Figure 5B).
Figure 5.
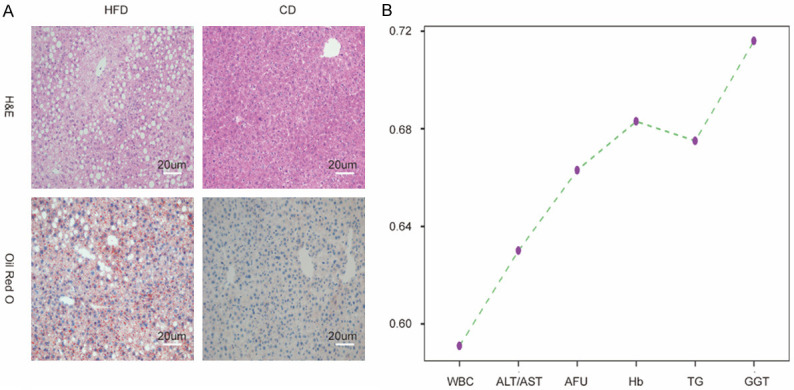
The Verification of the candidate parameters using the mouse model. A: H&E and Oil Red O stainings of representative liver sections in mice fed with HFD or CD. Scale bar: μm; B: The AUC Cumulative Chart of the selected parameters in the mouse model.
Discussion
In this study, our predictive model for NAFLD, which consisted of the common laboratory indicators in hospital, showed a brilliant performance. This study aimed to screen out the combination of laboratory indicators with higher sensitivity and specificity for clinical screening of NAFLD. Although there were no relevant mechanisms revealed in this study, the indicators screened out in this study were all classic indicators and there have been many reports on the relevant mechanisms for these indicators in NAFLD.
Both ALT and AST were mainly expressed in liver cells and their levels in plasma could indicate the damage and death of liver cells. When the liver injury occurred, ALT and AST are released from liver cells into the blood, leading to the increased serum ALT and AST levels [20]. Nanji et al. demonstrated that there was a significant correlation between the ALT/AST ratio and the degree of fatty accumulation of hepatic cells [21]. Long et al. also demonstrated that the ALT/AST ratio predicted hepatic steatosis better than either ALT or AST alone [22].
NAFLD was considered to be the liver manifestation of metabolic symptoms (MS) [23]. The relationship between WBC count and MS components had been demonstrated in some studies [24,25]. Moreover, WBC count was often used to evaluate inflammatory status [26] and inflammation plays a significant role in the development of NAFLD [3]. In view of the points above, WBC might reflect the occurrence and progression of NAFLD. Many studies have focused on the association between WBC count and the occurrence of NAFLD [27,28], and a previous study has clearly showed that the WBC count was a significant factor associated with NAFLD occurrence [29].
AFU is a lysosome enzyme expressed in all mammalian cells and hydrolyzes sugars containing L-fucose, and AFU levels are closely associated with the occurrence of HCC [30,31]. Blood AFU usually comes from lysosome leakage. Lipid peroxidation of liver cells modifies the functional characteristics of the cell membranes and membranes of intracellular organelles such as mitochondria and lysosomes, which might result in leakage of lysosome and the release of AFU [32,33]. As a result, AFU may have a close relationship with NAFLD. A previous study had attempted to explore the relationship between the AFU level and NAFLD occurrence. Lu et al. suggested that AFU levels were positively associated with NAFLD occurrence and might act as an independent risk factor for NAFLD. However, the AUROC of the only AFU levels for NAFLD diagnosis was only 0.606 [34].
Hb is an iron-containing metalloprotein. A previous study revealed that the iron depletion could increase the glucose uptake and insulin signaling in hepatic cells and improve liver function in NAFLD patients [35]. Bai et al. revealed that adults with high Hb levels (14.4 μg/dl for male and 13.2 μg/dl for female) were at the greatest risk for NAFLD [36]. Chung et al. demonstrated that serum Hb level was independently associated with the risk of developing incidental metabolic syndrome or NAFLD in men [37]. Recently, Giorgio et al. indicated that elevated Hb Level had obvious relationship with fibrosis in biopsy-diagnosed pediatric NAFLD patients [38]. Consistently, in a Mexican population study, there was an independent relationship between the serum Hb level and the steatosis severity [39].
GGT, which is a transmembrane protein generated in the microsome, could play an important role in maintaining the metabolism of glutathione and act as one of the most important antioxidants in human cells [40]. Meanwhile, oxidative stress could play an essential role in the development of NAFLD [40,41]. Jarčuška1 et al. found that about half of patients with NASH had elevated levels of GGT, and there was an obvious relationship between GGT and individual metabolic syndrome [42]. Furthermore, a previous study revealed that the GGT-to-platelet ratio (GPR) is better than aspartate transaminase-to-platelet ratio index and fibrosis index based on four factors (FIB-4) for diagnosing fibrosis and cirrhosis in NAFLD patients [43].
For patients with overnutrition and obesity, there is often a change of hepatic fatty acid metabolism, which could lead to the accumulation of triglycerides in hepatocytes and sometimes cause the occurrence of NAFLD [44].
Each indicator in this study has a certain tendency and limitation on the diagnosis of NAFLD, while the predictive model using the integrated machine learning method can overcome the limitations of a single indicator and improve the diagnostic ability. Some indicators which were not common in clinical laboratory were also reported to be the potential predictors for NAFLD. Kimura et al. demonstrated that serum thrombospondin 2 level was considered as a predictor of histological activity of NAFLD [45]. Mele et al. displayed that angiopoietin-like 8 has direct relationship with the presence and severity of NAFLD in patients with Prader-Willi Syndrome [46]. In further investigations, the uncommon indicators in clinical laboratory should be considered for the construction of predictive model.
In summary, we created a predictive model with laboratory indicators for the diagnosis of NAFLD using the machine learning method, which had the potential for the diagnosis of the NAFLD and provided the basis for the predictive model of the combination of indicators.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China [grant number: 31770837] and the Key Research and Development Program of Shandong Province [grant number: 2019GSF108148].
Disclosure of conflict of interest
None.
Abbreviations
- A/G
the ratio of ALB and GLOB
- APO A1
apolipoprotein A1
- APO B
apolipoprotein B
- AST
aspartate aminotransferase
- AUROC
the areas under the receiver-operating characteristic curve
- BASO
basophils
- BUN
blood urea nitrogen
- CK
creatine kinase
- CK-MB
creatine kinase isoenzyme-MB
- Cre
creatinine
- DB
direct bilirubin
- EOS
eosinophils count
- GLOB
globulin
- GLU
blood glucose
- HCT
hematocrit
- HDL
high-density lipoprotein
- IB
indirect bilirubin
- LDH
lactate dehydrogenase
- LDL
low-density lipoprotein
- LP
lipoproteins
- LR
logistic regression
- LYM
lymphocyte count
- MCH
mean corpsular hemoglobin
- MCHC
mean corpusular hemoglobin concerntration
- MCV
mean corpuscular volume
- MON
monocytes count
- MPV
mean platelet volume
- NAFLD
nonalcoholic fatty liver disease
- NEUT
the neutrophilic granulocyte count
- PCT
platelet hematocrit
- PDW
platelet distribution width
- P-LCR
platelet-larger cell ratio
- PLT
platelet count
- RBC
red blood cell
- RDW
red blood cell volume distribution width
- RF
random forest
- SVM
support vector machine
- TB
total bilirubin
- TBA
total bile acid
- TC
total cholesterol
- TP
total protein
- UA
uric acid
- WBC
white blood cell count
- HBDH
alpha-hydroxybutyrate dehydrogenase
- GGT
gamma-glutamyl transpeptidase
Supporting Information
References
- 1.Li J, Zou B, Yeo YH, Feng Y, Xie X, Lee DH, Fujii H, Wu Y, Kam LY, Ji F, Li X, Chien N, Wei M, Ogawa E, Zhao C, Wu X, Stave CD, Henry L, Barnett S, Takahashi H, Furusyo N, Eguchi Y, Hsu YC, Lee TY, Ren W, Qin C, Jun DW, Toyoda H, Wong VW, Cheung R, Zhu Q, Nguyen MH. Prevalence, incidence, and outcome of non-alcoholic fatty liver disease in Asia, 1999-2019: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2019;4:389–398. doi: 10.1016/S2468-1253(19)30039-1. [DOI] [PubMed] [Google Scholar]
- 2.Younossi Z, Tacke F, Arrese M, Chander Sharma B, Mostafa I, Bugianesi E, Wai-Sun Wong V, Yilmaz Y, George J, Fan J, Vos MB. Global perspectives on nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatology. 2019;69:2672–2682. doi: 10.1002/hep.30251. [DOI] [PubMed] [Google Scholar]
- 3.Lonardo A, Nascimbeni F, Maurantonio M, Marrazzo A, Rinaldi L, Adinolfi LE. Nonalcoholic fatty liver disease: evolving paradigms. World J Gastroenterol. 2017;23:6571–6592. doi: 10.3748/wjg.v23.i36.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fabbrini E, Sullivan S, Klein S. Obesity and nonalcoholic fatty liver disease: biochemical, metabolic, and clinical implications. Hepatology. 2010;51:679–689. doi: 10.1002/hep.23280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, George J, Bugianesi E. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15:11–20. doi: 10.1038/nrgastro.2017.109. [DOI] [PubMed] [Google Scholar]
- 6.Estes C, Anstee QM, Arias-Loste MT, Bantel H, Bellentani S, Caballeria J, Colombo M, Craxi A, Crespo J, Day CP, Eguchi Y, Geier A, Kondili LA, Kroy DC, Lazarus JV, Loomba R, Manns MP, Marchesini G, Nakajima A, Negro F, Petta S, Ratziu V, Romero-Gomez M, Sanyal A, Schattenberg JM, Tacke F, Tanaka J, Trautwein C, Wei L, Zeuzem S, Razavi H. Modeling NAFLD disease burden in China, France, Germany, Italy, Japan, Spain, United Kingdom, and United States for the period 2016-2030. J Hepatol. 2018;69:896–904. doi: 10.1016/j.jhep.2018.05.036. [DOI] [PubMed] [Google Scholar]
- 7.Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, Charlton M, Sanyal AJ. The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology. 2012;55:2005–2023. doi: 10.1002/hep.25762. [DOI] [PubMed] [Google Scholar]
- 8.Ma X, Liu S, Zhang J, Dong M, Wang Y, Wang M, Xin Y. Proportion of NAFLD patients with normal ALT value in overall NAFLD patients: a systematic review and meta-analysis. BMC Gastroenterol. 2020;20:10. doi: 10.1186/s12876-020-1165-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poynard T, Ratziu V, Naveau S, Thabut D, Charlotte F, Messous D, Capron D, Abella A, Massard J, Ngo Y, Munteanu M, Mercadier A, Manns M, Albrecht J. The diagnostic value of biomarkers (SteatoTest) for the prediction of liver steatosis. Comp Hepatol. 2005;4:10. doi: 10.1186/1476-5926-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W, Huang Y, Zhuang BW, Liu GJ, Hu HT, Li X, Liang JY, Wang Z, Huang XW, Zhang CQ, Ruan SM, Xie XY, Kuang M, Lu MD, Chen LD, Wang W. Multiparametric ultrasomics of significant liver fibrosis: a machine learning-based analysis. Eur Radiol. 2019;29:1496–1506. doi: 10.1007/s00330-018-5680-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X, Hou Y, Wang X, Yu L, Wang X, Jiang L, Yang Z. Machine learning-based development and validation of a scoring system for progression-free survival in liver cancer. Hepatol Int. 2020;14:567–576. doi: 10.1007/s12072-020-10046-w. [DOI] [PubMed] [Google Scholar]
- 12.Chalasani N, Younossi Z, Lavine JE, Charlton M, Cusi K, Rinella M, Harrison SA, Brunt EM, Sanyal AJ. The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American Association for the Study of Liver Diseases. Hepatology. 2018;67:328–357. doi: 10.1002/hep.29367. [DOI] [PubMed] [Google Scholar]
- 13.Abeel T, Helleputte T, Van de Peer Y, Dupont P, Saeys Y. Robust biomarker identification for cancer diagnosis with ensemble feature selection methods. Bioinformatics. 2010;26:392–398. doi: 10.1093/bioinformatics/btp630. [DOI] [PubMed] [Google Scholar]
- 14.Cai Z, Xu D, Zhang Q, Zhang J, Ngai SM, Shao J. Classification of lung cancer using ensemble-based feature selection and machine learning methods. Mol Biosyst. 2015;11:791–800. doi: 10.1039/c4mb00659c. [DOI] [PubMed] [Google Scholar]
- 15.He Z, Yu W. Stable feature selection for biomarker discovery. Comput Biol Chem. 2010;34:215–225. doi: 10.1016/j.compbiolchem.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 16.Hilbe JM. Logistic regression models. London: CRC press; 2009. [Google Scholar]
- 17.Breiman L. Random forests. In: Schapire , editor. Machine Learning. The Netherlands: Kluwer Academic Publishers; 2002. pp. 5–32. [Google Scholar]
- 18.Chang CC, Lin CJ. LIBSVM: a library for support vector machines. ACM Transactions on Intelligent Systems and Technology. 2011;2:1–39. [Google Scholar]
- 19.Perrier E, Rondeau P, Poupin M, Le Bellego L, Armstrong LE, Lang F, Stookey J, Tack I, Vergne S, Klein A. Relation between urinary hydration biomarkers and total fluid intake in healthy adults. Eur J Clin Nutr. 2013;67:939–943. doi: 10.1038/ejcn.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verslype C. Evaluation of abnormal liver-enzyme results in asymptomatic patients. Acta Clin Belg. 2004;59:285–289. doi: 10.1179/acb.2004.042. [DOI] [PubMed] [Google Scholar]
- 21.Nanji AA, French SW, Freeman JB. Serum alanine aminotransferase to aspartate aminotransferase ratio and degree of fatty liver in morbidly obese patients. Enzyme. 1986;36:266–269. doi: 10.1159/000469304. [DOI] [PubMed] [Google Scholar]
- 22.Long MT, Pedley A, Colantonio LD, Massaro JM, Hoffmann U, Muntner P, Fox CS. Development and validation of the framingham steatosis index to identify persons with hepatic steatosis. Clin Gastroenterol Hepatol. 2016;14:1172–1180. e1172. doi: 10.1016/j.cgh.2016.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tarantino G, Finelli C. What about non-alcoholic fatty liver disease as a new criterion to define metabolic syndrome? World J Gastroenterol. 2013;19:3375–3384. doi: 10.3748/wjg.v19.i22.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chao TT, Hsieh CH, Lin JD, Wu CZ, Hsu CH, Pei D, Chen YL, Liang YJ, Chang JB. Use of white blood cell counts to predict metabolic syndrome in the elderly: a 4 year longitudinal study. Aging Male. 2014;17:230–237. doi: 10.3109/13685538.2013.875989. [DOI] [PubMed] [Google Scholar]
- 25.Yang H, Fu YQ, Yang B, Zheng JS, Zeng XY, Zeng W, Fan ZF, Chen M, Wang L, Li D. Positive association between the metabolic syndrome and white blood cell counts in Chinese. Asia Pac J Clin Nutr. 2017;26:141–147. doi: 10.6133/apjcn.102015.13. [DOI] [PubMed] [Google Scholar]
- 26.Riley LK, Rupert J. Evaluation of patients with leukocytosis. Am Fam Physician. 2015;92:1004–1011. [PubMed] [Google Scholar]
- 27.Wang S, Zhang C, Zhang G, Yuan Z, Liu Y, Ding L, Sun X, Jia H, Xue F. Association between white blood cell count and non-alcoholic fatty liver disease in urban Han Chinese: a prospective cohort study. BMJ Open. 2016;6:e010342. doi: 10.1136/bmjopen-2015-010342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee YJ, Lee HR, Shim JY, Moon BS, Lee JH, Kim JK. Relationship between white blood cell count and nonalcoholic fatty liver disease. Dig Liver Dis. 2010;42:888–894. doi: 10.1016/j.dld.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 29.Chung GE, Yim JY, Kim D, Kwak MS, Yang JI, Chung SJ, Yang SY, Kim JS. Associations between white blood cell count and the development of incidental nonalcoholic fatty liver disease. Gastroenterol Res Pract. 2016;2016:7653689. doi: 10.1155/2016/7653689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stefaniuk P, Cianciara J, Wiercinska-Drapalo A. Present and future possibilities for early diagnosis of hepatocellular carcinoma. World J Gastroenterol. 2010;16:418–424. doi: 10.3748/wjg.v16.i4.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tangkijvanich P, Tosukhowong P, Bunyongyod P, Lertmaharit S, Hanvivatvong O, Kullavanijaya P, Poovorawan Y. Alpha-L-fucosidase as a serum marker of hepatocellular carcinoma in Thailand. Southeast Asian J Trop Med Public Health. 1999;30:110–114. [PubMed] [Google Scholar]
- 32.Ramm GA, Ruddell RG. Hepatotoxicity of iron overload: mechanisms of iron-induced hepatic fibrogenesis. Semin Liver Dis. 2005;25:433–449. doi: 10.1055/s-2005-923315. [DOI] [PubMed] [Google Scholar]
- 33.Yajima D, Motani H, Hayakawa M, Sato Y, Sato K, Iwase H. The relationship between cell membrane damage and lipid peroxidation under the condition of hypoxia-reoxygenation: analysis of the mechanism using antioxidants and electron transport inhibitors. Cell Biochem Funct. 2009;27:338–343. doi: 10.1002/cbf.1578. [DOI] [PubMed] [Google Scholar]
- 34.Lu ZY, Cen C, Shao Z, Chen XH, Xu CF, Li YM. Association between serum alpha-L-fucosidase and non-alcoholic fatty liver disease: cross-sectional study. World J Gastroenterol. 2016;22:1884–1890. doi: 10.3748/wjg.v22.i5.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dongiovanni P, Valenti L, Ludovica Fracanzani A, Gatti S, Cairo G, Fargion S. Iron depletion by deferoxamine up-regulates glucose uptake and insulin signaling in hepatoma cells and in rat liver. Am J Pathol. 2008;172:738–747. doi: 10.2353/ajpath.2008.070097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bai CH, Wu MS, Owaga E, Cheng SY, Pan WH, Chang JS. Relationship between hemoglobin levels and risk for suspected non-alcoholic fatty liver in Taiwanese adults. Chin J Physiol. 2014;57:286–294. doi: 10.4077/CJP.2014.BAD280. [DOI] [PubMed] [Google Scholar]
- 37.Chung GE, Yim JY, Kim D, Kwak MS, Yang JI, Chung SJ, Yang SY, Kim JS. Associations between hemoglobin concentrations and the development of incidental metabolic syndrome or nonalcoholic fatty liver disease. Dig Liver Dis. 2017;49:57–62. doi: 10.1016/j.dld.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 38.Giorgio V, Mosca A, Alterio A, Alisi A, Grieco A, Nobili V, Miele L. Elevated hemoglobin level is associated with advanced fibrosis in pediatric nonalcoholic fatty liver disease. J Pediatr Gastroenterol Nutr. 2017;65:150–155. doi: 10.1097/MPG.0000000000001614. [DOI] [PubMed] [Google Scholar]
- 39.Juárez-Hernández E, C Chávez-Tapia N, C Brizuela-Alcántara D, Uribe M, H Ramos-Ostos M, Nuño-Lámbarri N. Association between serum hemoglobin levels and non alcoholic fatty liver disease in a mexican population. Ann Hepatol. 2018;17:577–584. doi: 10.5604/01.3001.0012.0920. [DOI] [PubMed] [Google Scholar]
- 40.Whitfield JB. Gamma glutamyl transferase. Crit Rev Clin Lab Sci. 2001;38:263–355. doi: 10.1080/20014091084227. [DOI] [PubMed] [Google Scholar]
- 41.Satapati S, Kucejova B, Duarte JA, Fletcher JA, Reynolds L, Sunny NE, He T, Nair LA, Livingston KA, Fu X, Merritt ME, Sherry AD, Malloy CR, Shelton JM, Lambert J, Parks EJ, Corbin I, Magnuson MA, Browning JD, Burgess SC. Mitochondrial metabolism mediates oxidative stress and inflammation in fatty liver. J Clin Invest. 2015;125:4447–4462. doi: 10.1172/JCI82204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jarcuska P, Janicko M, Drazilova S, Senajova G, Veseliny E, Fedacko J, Siegfried L, Kristian P, Tkac M Jr, Pella D, Marekova M, Geckova AM, Jarcuska P HepaMeta Team. Gamma-glutamyl transpeptidase level associated with metabolic syndrome and proinflammatory parameters in the young Roma population in eastern Slovakia: a population-based study. Cent Eur J Public Health. 2014;22(Suppl):S43–50. doi: 10.21101/cejph.a3901. [DOI] [PubMed] [Google Scholar]
- 43.Li Q, Lu C, Li W, Huang Y, Chen L. The gamma-glutamyl transpeptidase to platelet ratio for non-invasive assessment of liver fibrosis in patients with chronic hepatitis B and non-alcoholic fatty liver disease. Oncotarget. 2017;8:28641–28649. doi: 10.18632/oncotarget.16162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svegliati-Baroni G, Pierantonelli I, Torquato P, Marinelli R, Ferreri C, Chatgilialoglu C, Bartolini D, Galli F. Lipidomic biomarkers and mechanisms of lipotoxicity in non-alcoholic fatty liver disease. Free Radic Biol Med. 2019;144:293–309. doi: 10.1016/j.freeradbiomed.2019.05.029. [DOI] [PubMed] [Google Scholar]
- 45.Kimura T, Tanaka N, Fujimori N, Yamazaki T, Katsuyama T, Iwashita Y, Pham J, Joshita S, Pydi SP, Umemura T. Serum thrombospondin 2 is a novel predictor for the severity in the patients with NAFLD. Liver Int. 2021;41:505–514. doi: 10.1111/liv.14776. [DOI] [PubMed] [Google Scholar]
- 46.Mele C, Crinò A, Fintini D, Mai S, Convertino A, Bocchini S, Di Paolo P, Grugni G, Aimaretti G, Scacchi M, Marzullo P. Angiopoietin-like 8 (ANGPTL8) as a potential predictor of NAFLD in paediatric patients with Prader-Willi Syndrome. J Endocrinol Invest. 2021;44:1447–1456. doi: 10.1007/s40618-020-01444-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.