Abstract
Osteosarcoma is a primary malignant bone tumor that occurs frequently in children and adolescents and has a propensity for drug resistance, recurrence, and metastasis. The purpose of this study was to identify potential target genes to predict metastasis and survival in patients with osteosarcoma. We analyzed gene expression profiles and corresponding clinical data of patients with osteosarcoma in the Gene Expression Omnibus database and identified 202 genes that were differentially expressed between osteosarcoma cells and normal osteoblasts. Univariate and multivariable Cox regression analyses identified four risk genes that affected osteosarcoma prognosis: MCAM, ENPEP, LRRC1, and CPE. Independent prognostic analyses and clinical correlation studies showed that the four risk genes constituted an independent prognostic signature that correlated with survival and clinical parameters including age and distant metastasis. In a single-sample Gene Set Enrichment Analysis, risk scores based on the prognostic signature correlated with tumor infiltration by immune cells and immune functions in osteosarcoma. A subsequent analysis showed that the expression levels of the four genes in the prognostic signature were predictive of overall survival and metastasis-free survival of patients with osteosarcoma. Furthermore, Human Cancer Metastasis Database and qRT-PCR analyses demonstrated that the four risk genes are overexpressed in osteosarcoma tissues and cell lines. In summary, we developed and validated a four-gene prognostic signature that may be useful in osteosarcoma diagnosis and metastasis prediction.
Keywords: Osteosarcoma, prognosis, survival analysis, metastasis, bioinformatic analysis
Introduction
Osteosarcoma is a common primary and malignant bone tumor that originates from mesenchymal tissue and typically affects the fast-growing metaphysis of long bones, including the distal femur, proximal tibia, or proximal humerus [1]. Osteosarcoma has a high incidence among children and adolescents [2,3], with a predilection for drug resistance, recurrence, and distant metastasis, mainly in the lungs [4]. A combination of neoadjuvant chemotherapy, surgery, chemotherapy, and biological therapy has led to significant progress in the treatment efficacy of osteosarcoma, resulting in a gradual increase in the overall survival (OS) of patients with non-metastatic osteosarcoma from less than 20% to as high as 70% [5-8]. Despite the progress, patients with metastatic or recurrent osteosarcoma still have a poor outlook, with a 5-year OS rate of only 20% [7]. In order to improve the early diagnosis and treatment of osteosarcoma, novel biomarkers and therapeutic targets are needed.
In recent years, analysis of public bioinformatics databases such as the Gene Expression Omnibus (GEO) to explore target genes as predictors of metastasis and survival has provided new insights into the treatment of osteosarcoma [9-11]. For example, Niu et al. [12] integrated three datasets (GSE14827, GSE21257, and GSE32981) with robust rank aggregation to examine tumor infiltration by immune cells in osteosarcoma and found that CXCL10 and MYC expression levels were significant predictors of patients’ OS. Similarly, Xu et al. [13] investigated the association between expression of lncRNA SNHG4 and clinicopathological features and prognosis in patients with osteosarcoma and found that high SNHG4 expression was linked to large tumor size and high patient mortality. In another study, a bioinformatics analysis indicated that N6-Methyladenosine (m6A) regulators may be involved in osteosarcoma metastasis and prognosis through the humoral immune response and cell cycle pathways [14]. An analysis of the relationship between osteosarcoma development and the tumor microenvironment using an algorithm known as ESTIMATE showed that the polarization of M0 to M1 or M2 macrophages might be associated with improved prognosis, and that patients with increased immune cell infiltration in the tumor microenvironment had significantly better prognosis than patients with less immune cell infiltration [15]. A seven-gene signature related to energy metabolism in osteosarcoma showed promise for osteosarcoma diagnosis, treatment, and prediction [16], although the molecular mechanisms of these genes in osteosarcoma need to be further explored. The previous success of projects to identify prognostic target genes in osteosarcoma suggests that it may be possible to uncover more molecular mechanisms in osteosarcoma and develop novel therapeutic targets through a combination of bioinformatics analysis and experimental validation.
We used microarray data from the GEO database for differential expression analysis and identified 202 differentially expressed genes (DEGs) in osteosarcoma and normal osteoblasts. The potential biological activities of co-expressed DEGs were investigated using Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analyses. We then constructed a four-gene signature to predict osteosarcoma outcomes. We validated the signature by evaluating the association between the expression of the four genes and clinical characteristics and immune cell infiltration in osteosarcoma tumors. Finally, we used the Human Cancer Metastasis Database (HCMDB) and quantitative real-time PCR (qRT-PCR) to determine the expression levels of the four risk genes in osteosarcoma, so as to evaluate these four risk genes as possible markers for osteosarcoma diagnostics and prognostics.
Materials and methods
Microarray data
We pooled three microarray expression datasets (GSE12885, GSE14359, GSE21257) from the GEO database (https://www.ncbi.nlm.nih.gov/gds). Table 1 lists the information from three selected GEO datasets that we used in the study. This information is freely available online.
Table 1.
Characteristics of the GEO datasets selected in this study
| GEO ID | Author, year | Platform | Country | Samples (T:N) |
|---|---|---|---|---|
| GSE14359 | Guenther et al., 2010 | GPL96 | Germany | 18:2 |
| GSE12885 | Sadikovic et al., 2009 | GPL6244 | Canada | 52:6 |
| GSE21257 | Kuijjer et al., 2011 | GPL10295 | Norway | 53:0 |
GEO: Gene Expression Omnibus; T: patients with osteosarcoma; N: normal controls.
Identification of DEGs
We used the GEO2R online analysis tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/) to identify DEGs in osteosarcoma samples compared with normal human osteoblasts in the GSE14359 and GSE12885 datasets [15]. Statistically significant DEGs had an absolute value of the log2 fold change (|log2FC|) ≥2 and an adjusted P-value (adj. P) <0.05. In addition, we visualized the DEGs in a volcano plot using “ggplot2”. We used Venn Diagram webtool (bioinformatics.psb.ugent.be/webtools/Venn/) to identify DEGs that overlapped both datasets.
GO and KEGG pathway enrichment analysis of DEGs
To get a deeper understanding of the significant functional terms and biological pathways of these overlapping DEGs, we performed GO and KEGG pathway enrichment analyses. GO terms indicate biological processes, molecular functions, and cellular components associated with genes and are widely used for gene annotation and functional genomics research. The KEGG database (http://www.genome.jp/) connects genetic information with knowledge about biological processes, chemicals, diseases, and medicines. We used the software packages “clusterProfiler”, “org.Hs.eg.db”, “ggplot2”, and “enrichment plot” for annotation and visualization. Results with a false discovery rate less than 0.05 were considered statistically significant.
Protein-protein interaction (PPI) network construction and module analysis
We created a PPI network of DEGs using the Search Tool for the Retrieval of Interacting Genes (STRING, version 11.0, http://string-db.org/) with minimum required interaction score >0.4. STRING is an online database that provides information on known and anticipated protein interactions, as well as insights into disease onset and development processes. We visualized the network using Cytoscape (http://www.cytoscape.org/). We used Molecular Complex Detection (MCODE), a plugin for Cytoscape, to cluster the genes and find a densely connected area according to the following criteria: degree cut-off = 2, score >10, node >10, haircut on, node score cut-off = 0.2, max depth = 100, and k-score = 2.
Construction and validation of the Cox regression signature
We downloaded all clinical data from the GSE21257 GEO dataset and analyzed survival and outcomes among a total of 53 patients with osteosarcoma. We conducted univariate Cox regression analysis with the “survival” package based on the patient characteristics in the GSE21257 data (P-value <0.05 was used as a screening criterion). We classified genes with a hazard ratio (HR) greater than 1 as high-risk genes and those with an HR less than 1 as low-risk genes. Multivariate Cox regression analysis was used to further filter prognosis-related DEGs in order to develop a prognostic signature, and the risk score per patient was calculated as: risk score = (coefficientgene1 × expressiongene1) + (coefficientgene2 × expressiongene2) + … + (coefficientgenex × expressiongenex), where x corresponds to the total number of genes present in this signature. We then divided the patients into a low-risk group and a high-risk group based on the median risk score among the patients. We created a Kaplan-Meier survival curve using the R “survival” and “survminer” packages to determine the OS rates of the two groups, and determined the specificity and sensitivity of this prognostic signature by constructing receiver operating characteristic (ROC) curves with the “SurvivalROC” package. We considered predictions excellent if the area under the curve (AUC) was greater than 0.75. The expression heat map of the genes in the risk signature, the survival status, the survival time, and the risk score distribution were used to evaluate prognostic models. In addition, we identified copy-number alterations and performed mutation analysis of the risk genes using the cBioportal database (http://www.cbioportal.org/).
Correlation analysis between prognostic model and immune features
We applied the “gsva” software package to perform ssGSEA analysis of the high-risk and low-risk patient groups in order to compare tumor infiltration by immune cells and immune functions. Using analysis of the TIMER database (https://cistrome.shinyapps.io/timer), we examined the effects of the risk genes on the levels of infiltrating immune cells in sarcomas and compared the correlations between copy-number alteration and immune cell infiltration.
Independent prognostic analysis and clinical correlation analysis
We implemented the “survival” package to conduct both univariate and multivariate Cox regression analysis to assess the independence of the prognosis model from other clinical variables. The R “RMS” package was then used to generate nomograms to predict survival in patients with osteosarcoma over the course of 1, 3, and 5 years. In addition, patients with osteosarcoma were separated into two cohorts based on their age (≤20 or >20 years old), Huvos grade (Huvos grade I or II versus Huvos grade III or IV), gender (male or female), and metastasis status (metastatic, non-metastatic) [17,18]. The R “Beeswarm” package was used to assess the relationship between the signature-based risk genes and clinical variables mentioned above.
Survival analysis and expression level verification of risk genes
A survival analysis of the four risk genes in the prognostic signature was performed using the osteosarcoma microarray dataset from the R2 (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi?option=kaplan_main) database. In addition, the expression of the four risk genes in osteosarcoma tissues was determined using the Human Cancer Metastasis Database (HCMDB; http://hcmdb.i-sanger.com/index) [19].
Cell culture and qRT-PCR assays
We purchased the human osteosarcoma cell lines U-20S, 143B, and MNNG/HOS and the osteoblast cell line hFOB 1.19 from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). We cultured the cells in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, USA) containing 10% fetal bovine serum (FBS, Gibco, USA) and 1% antibiotics (penicillin/streptomycin) at 37°C with 5% CO2 and replacement of the culture medium approximately every 2 to 3 days.
For qRT-PCR, we inoculated the osteosarcoma cell lines on six-well plates and subjected them to the indicated treatments for 24 h. We then isolated total RNA with Trizol (Cwbio, Jiangsu, China) and reverse-transcribed it into cDNA. We performed gene amplification and detection using a Step One Plus Real-Time PCR system (Life Technologies, Foster, CA, USA). We analyzed the resulting data by the comparative 2-ΔΔCt method to quantify the mRNA levels of risk genes relative to that of GAPDH as the internal reference. Table 2 lists the primer sequences for each gene.
Table 2.
Primers for qRT-PCR
| Primer | Direction | Sequence (5’-3’) |
|---|---|---|
| MCAM | Forward | GATGGCATTCAAGGAGAGGA |
| Reverse | GAGTCTGGTGTGAGGGTGGT | |
| ENPEP | Forward | AGAGGGCTCTAAGAGATACTGC |
| Reverse | CCACGGCAAGTCCCACTATT | |
| LRRC1 | Forward | AGTGGCCTGACTTCATTAACG |
| Reverse | GATCCACCTTCAAGATTGACAGT | |
| CPE | Forward | GTGAACTCAAGGACTGGTTTGT |
| Reverse | TCACGTACACTATCCTATCCAGG |
Statistical analysis
Measurement data were expressed as the mean ± standard deviation (SD). GraphPad Prism 5.0 (GraphPad Software, Inc., USA) was used to for data analysis. Comparison between two groups was performed using Student’s t-tests. Statistical significance was determined as *P<0.05 or **P<0.01.
Results
DEGs in human osteosarcoma and osteoblasts
By using two threshold criteria (adj. P and log2FC), 368 up-regulated genes and 417 down-regulated genes were identified in the human osteosarcoma samples compared with the normal osteoblast samples in the GSE12885 dataset. In the GSE14359 dataset, 457 up-regulated genes and 256 down-regulated genes were identified in the osteosarcoma samples (Figure 1A and 1B). Subsequent Venn analysis identified 120 and 82 overlapping DEGs that were up-regulated and down-regulated, respectively, in the osteosarcoma samples in both datasets (Figure 1C and 1D).
Figure 1.
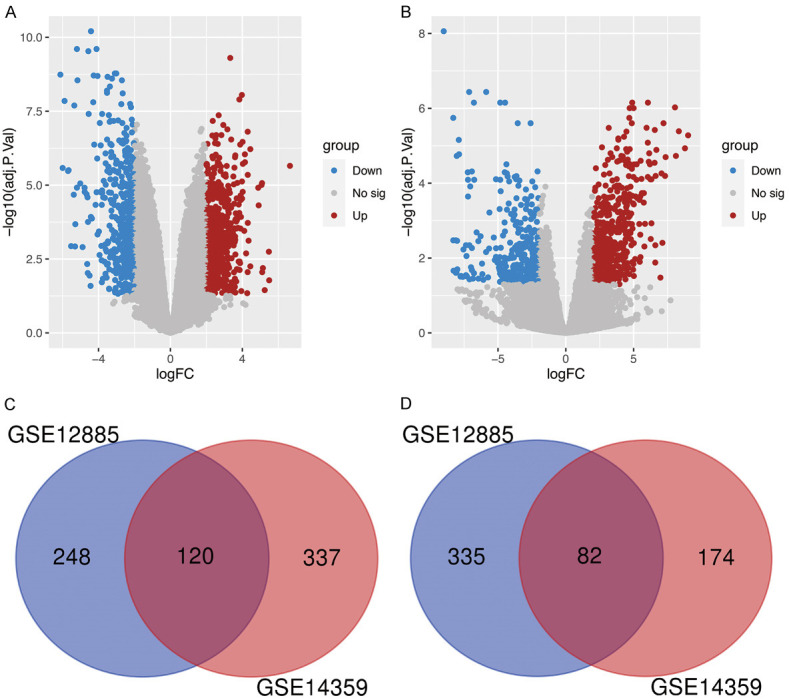
Identification of DEGs between osteosarcoma and osteoblasts. (A, B) Volcano plots of DEGs in dataset GSE12885 (A) and GES14359 (B). DEGs with red, blue and gray dots indicate up-regulated, down-regulated and no significant differences. (C) Venn diagram of upregulated DEGs common to GSE12885 and GES14359. (D) Venn diagram of downregulated DEGs common to GSE12885 and GES14359. DEGs, differentially expressed genes.
KEGG and GO enrichment analyses of the overlapping DEGs
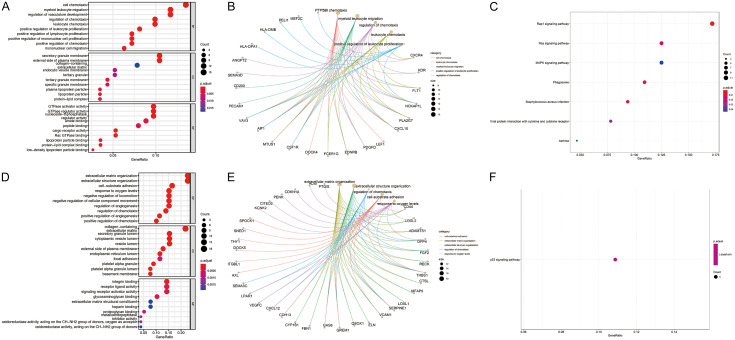
GO analysis revealed that the most common biological processes associated with the up-regulated overlapping DEGs were cell chemotaxis, positive regulation of leukocyte proliferation, and regulation of chemotaxis. The cellular compartments most commonly associated with the up-regulated overlapping DEGs were the secretory granule membrane, tertiary granule membrane, and lipoprotein particle. The molecular functions most commonly associated with the up-regulated overlapping DEGs were GTPase activator activity, GTPase regulator activity, and cargo receptor activity (Figure 2A). The up-regulated DEGs included in the top five significantly enriched biological process terms are presented in Figure 2B. According to the KEGG pathway analysis, the up-regulated overlapping DEGs were frequently involved in pathways related to Rap1 (Ras-associated protein 1) signaling pathway, staphylococcus aureus infection, Ras signaling pathway, and phagosomes (Figure 2C).
Figure 2.
GO function and KEGG pathway enrichment analyses of DEGs. A. GO enrichment analysis of upregulated DEGs, including BP, CC, and MF. B. The cnetplot of upregulated DEGs in the top five significantly enriched BP terms. C. KEGG pathway enrichment analysis of upregulated DEGs. D. GO enrichment analysis of downregulated DEGs, including BP, CC, and MF. E. The cnetplot of downregulated DEGs in the top five significantly enriched BP terms. F. KEGG pathway enrichment analysis of downregulated DEGs. DEGs, differentially expressed genes. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, biological processes; CC, cell component; MF, molecular function.
The most common biological processes associated with the down-regulated overlapping DEGs were cell-substrate adhesion, response to oxygen levels, and regulation of chemotaxis. The cellular compartments commonly associated with the down-regulated overlapping DEGs were the collagen-containing extracellular matrix, cytoplasmic vesicle lumen and basement membrane. The most common molecular functions associated with the down-regulated overlapping DEGs were integrin binding, proteoglycan binding, and glycosaminoglycan binding (Figure 2D). The down-regulated overlapping DEGs significantly enriched in the top five biological process categories are shown in Figure 2E. Moreover, the co-expressed DEGs that were downregulated primarily had associations with the p53 signaling pathway, according to KEGG pathway enrichment analysis (Figure 2F).
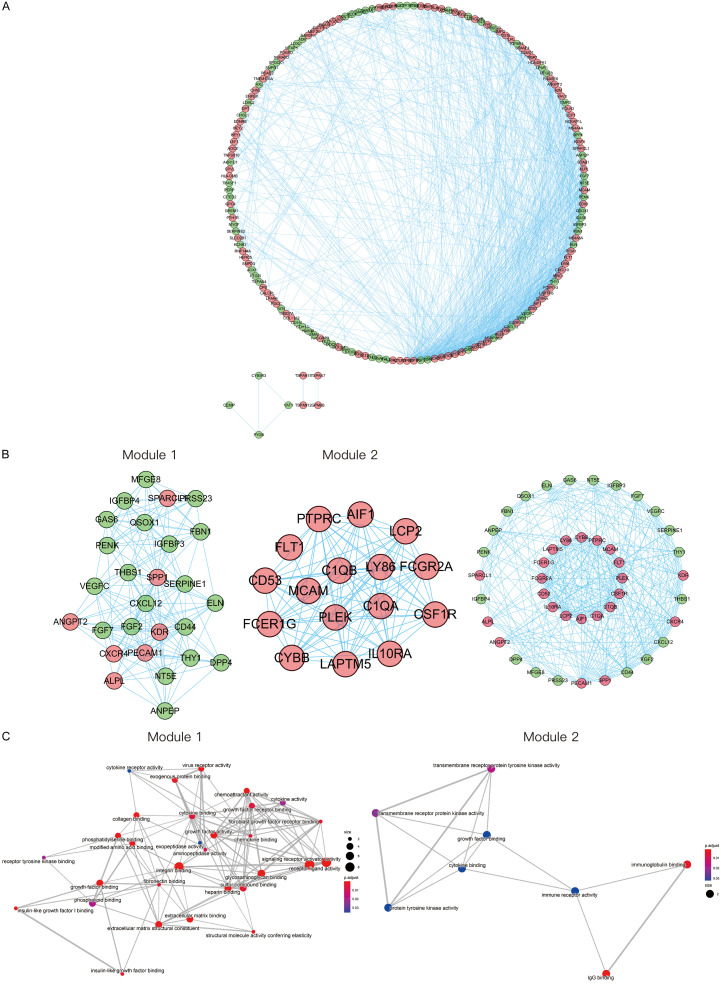
PPI network and modules analysis
As shown in Figure 3A, the whole PPI network consisted of 178 nodes and 934 edges. We screened out the two most important modules of the network using MCODE. Module 1 had the highest clustering score and consisted of 27 core proteins and 171 edges. Module 2 had the second highest clustering score and consisted of a total of 16 nodes and 93 edges that formed a network (Figure 3B). Genes in the above two modules were analyzed by GO enrichment analyses respectively. The results are shown in Figure 3C.
Figure 3.
Construction of PPI network and modules analysis. A. PPI network of DEGs. The red nodes indicate upregulated DEGs and the green nodes indicate downregulated DEGs. B. The two most important modules of the PPI network. C. GO-MF enrichment analysis of the two most important modules. DEGs, differentially expressed genes. GO, Gene Ontology; MF, molecular function.
Construction and validation of the Cox regression signature
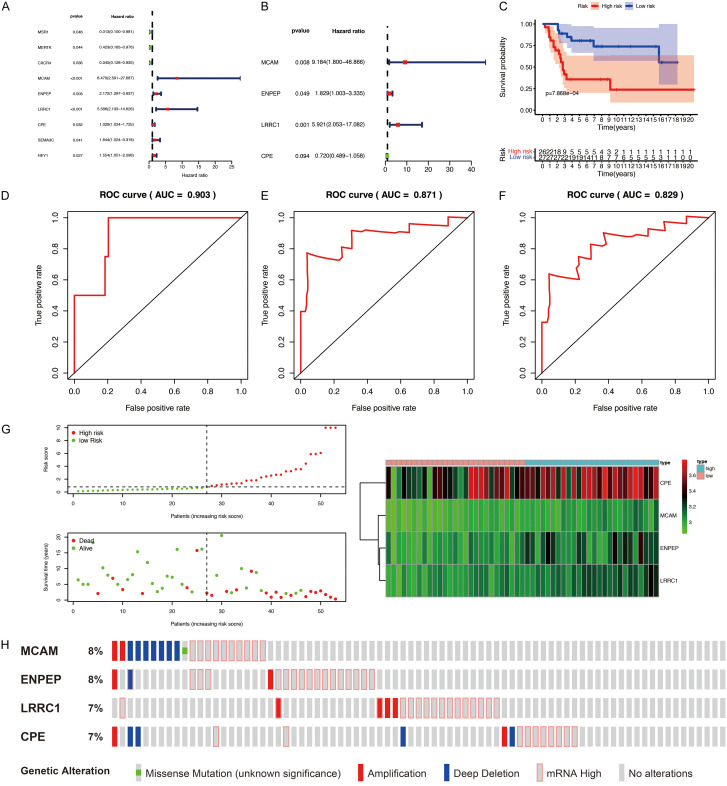
To further screen candidate prognostic DEGs from the PPI network, univariate Cox regression analysis of the 178 DEGs in the PPI network was completed using clinical data from the GSE21257 dataset. The results revealed 9 candidate prognostic DEGs (Figure 4A). Subsequent multivariate stepwise Cox regression analysis indicated that four of the candidate DEGs were independent predictors of patient outcomes (Figure 4B). Of the four prognostic DEGs, MCAM, ENPEP, and LRRC1 were regarded as high-risk genes based on their HRs, whereas CPE was regarded as a low-risk gene.
Figure 4.
Construction and validation of the four-gene prognostic signature for osteosarcoma. A, B. Cox regression analysis of DEGs from PPI network. A. Univariate Cox regression analysis. B. Multivariate Cox regression analysis. C. Survival analyses for low-risk and high-risk groups using Kaplan-Meier curves. D-F. Time-dependent ROC analysis for evaluating the prognostic performance of the signature. D. 1-year (AUC = 0.903). E. 3-year (AUC = 0.871). F. 5-year (AUC = 0.829). G. The distributions of risk scores, survival status, survival time, and expression heatmap of the four risk genes. H. Genomic alterations of four risk genes. DEGs, differentially expressed genes. ROC, receiver operating characteristic; AUC, area under the ROC curve.
We constructed a composite risk score based on the four prognostic DEGs (risk Score = (2.2175×ExpMCAM) + (0.6038×ExpENPEP) + (1.7786×ExpLRRC1) + (-0.3291×Exp CPE)) and divided the patients in the GSE21257 dataset into high-risk and low-risk groups on the basis of the median risk score. Kaplan-Meier curves revealed that the patients in the high-risk group had worse OS than the patients in the low-risk group (P<0.05; Figure 4C). A time-dependent ROC curve was then constructed to determine the reliability of this four-gene prognostic signature. The AUC was 0.903 for 1-year OS, 0.871 for 3-year OS, and 0.829 for 5-year OS, providing evidence that the four-gene prognostic model performed well as a predictor of OS (Figure 4D-F). The risk scores, survival status, survival time, and expression heatmap of the four risk genes in the two groups of patients are shown in Figure 4G. As a result, we found that patients with higher risk scores had shorter OS and worse prognoses than patients with lower risk scores, and that there were significant differences in gene expression levels between the two groups of patients. Analyses of copy-number alterations and mutations of the four risk genes revealed several missense mutations, amplifications, deep deletions, and high levels of mRNA expression (Figure 4H).
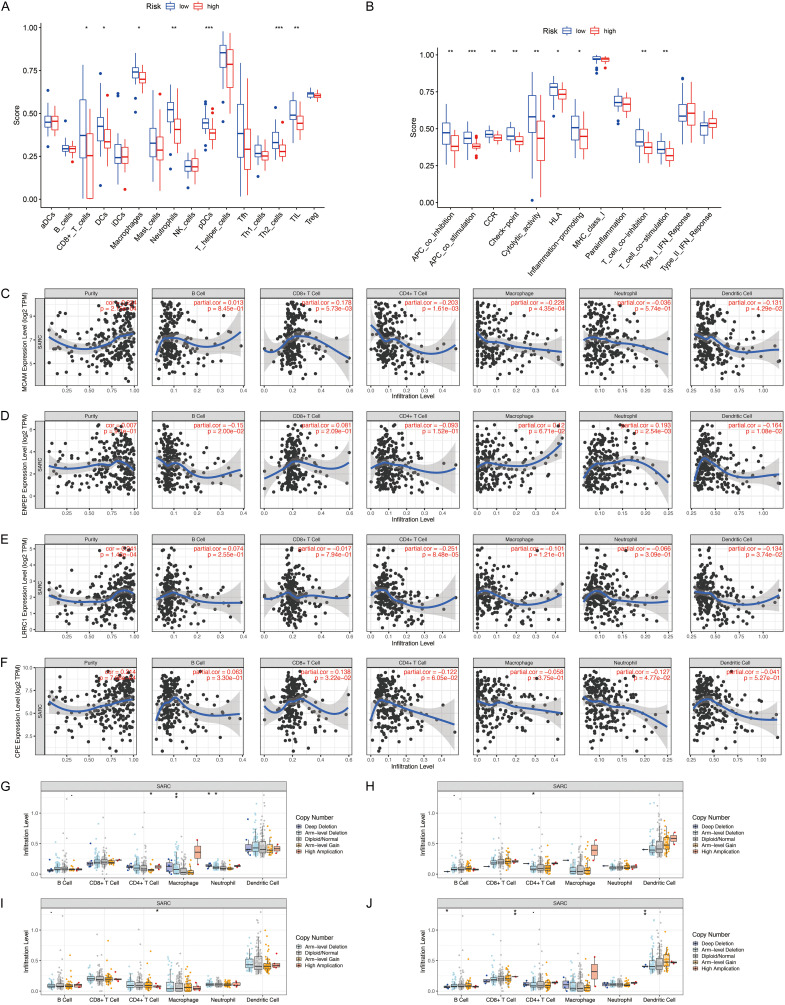
The risk score is related to immune characteristics in osteosarcoma
The results of the analysis using the ssGSEA algorithm indicated that compared with the patients in the low-risk group, the patients in the high-risk group had lower levels of tumor infiltration by immune cells, including CD8+ T cells, dendritic cells, plasmacytoid dendritic cells, macrophages, tumor-infiltrating lymphocytes, neutrophils, and T helper 2 cells (Figure 5A). Moreover, in contrast to the Type I interferon (IFN) response, major histocompatibility complex class I response, para-inflammation, and Type II IFN response, the other nine immune mechanisms were all down-regulated in the patients in the high-risk group in comparison with the patients in the low-risk group (Figure 5B). These results indicated that the signature-based risk score was directly related to immune characteristics, and that immune activity in the patients in the low-risk group may exert some antitumor effect.
Figure 5.
Immune characteristics analysis of the prognostic signature. (A) Comparisons of immune cell infiltration levels between different risk groups. (B) Comparisons of immune functions between different risk groups. (C-F) Correlation analysis of MCAM (C), ENPEP (D), LRRC1 (E), and CPE (F) expression with immune cell infiltration levels. (G-J) Correlation analysis of copy-number alteration of MCAM (G), ENPEP (H), LRRC1 (I), and CPE (J) with immune cell infiltration levels. (*P<0.05, **P<0.01, ***P<0.001).
To investigate the implications of the expression levels of this four risk genes on immune infiltration levels, we performed a comprehensive analysis using the TIMER database. As shown in Figure 5C, MCAM expression was positively correlated with CD8+ T cell levels but negatively correlated with dendritic cell, CD4+ T cell, and macrophage levels. The expression of ENPEP was positively correlated with neutrophil levels but negatively correlated with B cell and dendritic cell levels (Figure 5D). Figure 5E shows that LRRC1 expression was negatively correlated with CD4+ T cell and dendritic cell levels. The expression of CPE was positively correlated with CD8+ T cell levels but negatively correlated with neutrophil levels (Figure 5F). On the other hand, except for ENPEP, the expression of the other three genes was significantly correlated with tumor purity (Figure 5C-F). The impact of copy-number alterations of the signature genes on immune cell infiltration is shown in Figure 5G-J.
Independent prognostic value of the signature
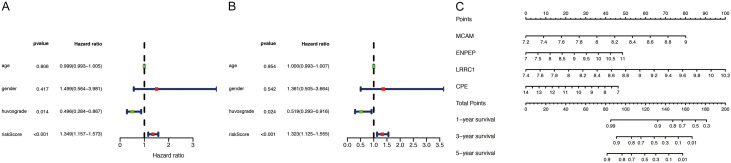
The risk score was independently associated with patient outcomes when it was assessed along with other clinical data, including gender, Huvos classification, and age (Table 3). The Huvos grade (P<0.05) and risk score (P<0.001) had statistical significance in a single-factor independent prognosis analysis (Figure 6A). Furthermore, in a multivariate independent prognostic analysis, Huvos grade (P<0.05, HR = 0.519, 95% confidence interval (CI) = 0.293-0.916) and risk score (P<0.001, HR = 1.323, 95% CI = 1.125-1.555) remained independent prognostic factors in patients with osteosarcoma (Figure 6B). We also predicted survival rates over 1 year, 3 years, and 5 years to help guide clinical decision-making (Figure 6C).
Table 3.
Independent prognostic analysis of the signature
| Clinical factors | Univariate Cox | Multivariate Cox | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| HR | 95% CI | P value | HR | 95% CI | P value | |
| age | 0.999 | 0.993-1.005 | 0.806 | 1.000 | 0.993-1.007 | 0.954 |
| gender | 1.499 | 0.564-3.981 | 0.417 | 1.361 | 0.505-3.664 | 0.542 |
| Huvos grade | 0.496 | 0.284-0.867 | 0.014 | 0.519 | 0.293-0.916 | 0.024 |
| Risk Score | 1.349 | 1.157-1.573 | <0.001 | 1.323 | 1.125-1.555 | 0.001 |
Figure 6.
Independent prognosis analysis and the nomogram for predicting probabilities of osteosarcoma patient’s overall survival. A. Single-factor prognosis analysis. B. Multi-factor prognosis analysis. C. The nomograms for predicting 1-year, 3-year, and 5-year overall survival of patients with osteosarcoma.
Clinical utility of the signature
To better evaluate the predictive value of the model for patients with osteosarcoma, we investigated the relationship between the four-gene signature and clinical variables (Table 4). The results indicated that males had a higher prevalence of LRRC1 expression than females, and as MCAM, ENPEP, LRRC1, and CPE expression increased, the M stage in patients with osteosarcoma also increased (Figure 7A-E). Additionally, patients with higher risk scores were younger (Figure 7F, P<0.05) and had an increased risk of metastasis (Figure 7G, P<0.01) than patients with lower risk scores. These findings indicated that both the composite risk score and the expression levels of the four risk genes were significantly associated with metastasis of osteosarcoma, supporting the accuracy of the model.
Table 4.
Correlation analysis between prognostic signature and clinical parameters for osteosarcoma
| id | age (≤20, >20) t(p) | gender (Female, Male) t(p) | Huvos grade (I/II, III/IV) t(p) | M Stage (M0, M1) t(p) |
|---|---|---|---|---|
| MCAM | 1.515 (0.147) | -0.585 (0.562) | 0.197 (0.845) | -3.468 (0.001) |
| ENPEP | 0.804 (0.432) | -0.356 (0.724) | -0.283 (0.779) | -3.302 (0.002) |
| LRRC1 | 0.356 (0.726) | -2.184 (0.038) | 1.16 (0.252) | -2.367 (0.023) |
| CPE | 0.594 (0.562) | -1.456 (0.154) | -0.223 (0.825) | -2.979 (0.005) |
| Risk Score | 2.241 (0.030) | -1.56 (0.126) | 1.317 (0.194) | -3.617 (0.001) |
t: t value from Student’s t test; p: P-value from Student’s t test.
Figure 7.
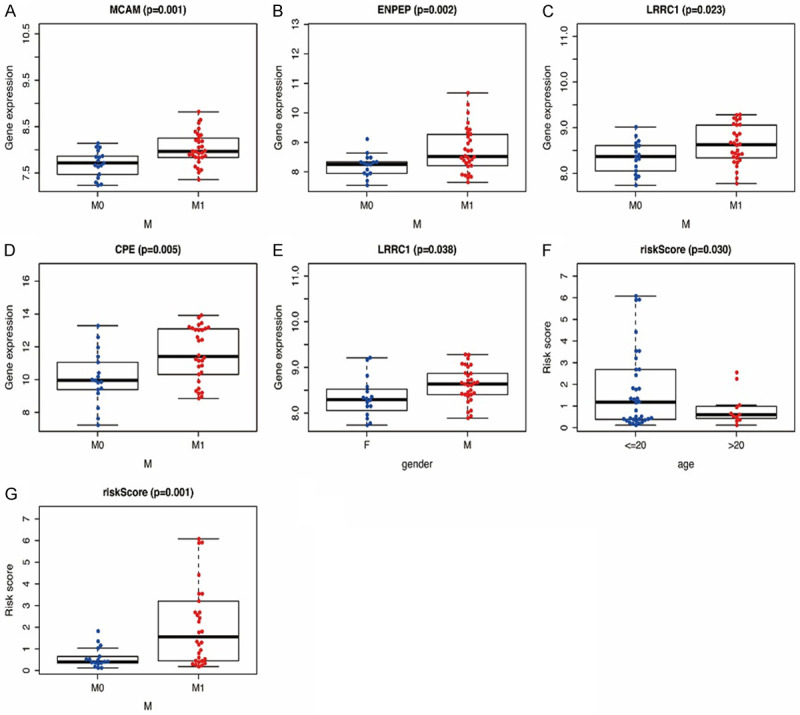
Correlation between the prognostic signature and clinical factors in osteosarcoma. (A-D) Relationship between MCAM (A), ENPEP (B), LRRC1 (C), CPE (D), and metastasis status. (E) Relationship between LRRC1 and gender category. (F) Relationship between risk score and age category. (G) Relationship between risk score and metastasis status.
The prognostic value and expression level verification of risk genes
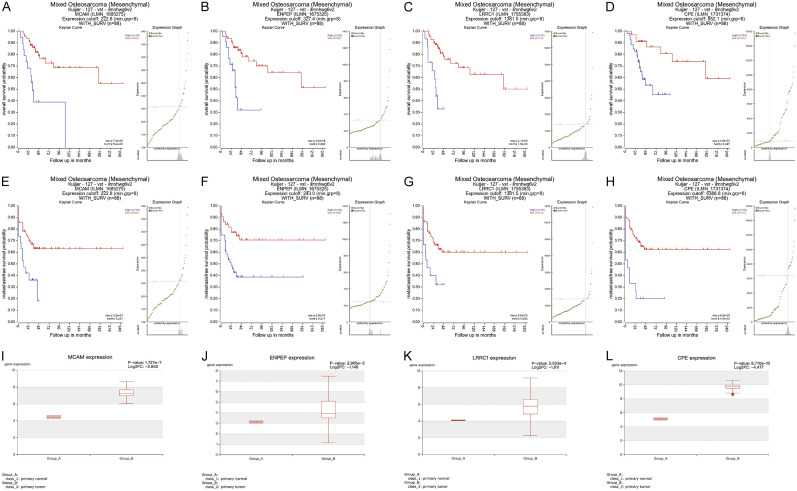
We analyzed the osteosarcoma dataset from an independent database, the R2 dataset, to further evaluate the prognostic value of the four risk genes. The results indicated that higher expression levels of MCAM (P<0.001), ENPEP (P<0.001), LRRC1 (P<0.001), and CPE (P<0.05) were correlated with shorter OS among patients with osteosarcoma (Figure 8A-D). Higher levels of MCAM, ENPEP, LRRC1, and CPE expression were also associated with shorter metastasis-free survival (MFS; Figure 8E-H). In addition, we analyzed the expression levels of the four risk genes in osteosarcoma tissues in the HCMDB database. Consistent with our other results, the expression levels of all four risk genes were higher in osteosarcoma tissues than in the control samples (Figure 8I-L).
Figure 8.
The survival analysis and expression level verification of risk genes. (A-D) Analysis of the association between MCAM (A), ENPEP (B), LRRC1 (C), and CPE (D) expression and OS in patients with osteosarcoma. (E-H) Analysis of the association between MCAM (E), ENPEP (F), LRRC1 (G), and CPE (H) expression and MFS in patients with osteosarcoma. (I-L) The expression level verification of MCAM (I), ENPEP (J), LRRC1 (K), and CPE (L) in osteosarcoma tissues. OS, overall survival; MFS, metastasis-free survival.
Verification of expression levels in osteosarcoma cell lines
We performed quantitative RT-PCR to determine the expression levels of the four risk genes in 143B, MNNG/HOS, and U2OS osteosarcoma cells and hFOB 1.19 osteoblasts. The results showed that MCAM, ENPEP, LRRC1, and CPE were all up-regulated in each osteosarcoma cell line relative to their expression levels in hFOB 1.19 cells (Figure 9A-D, P<0.05).
Figure 9.
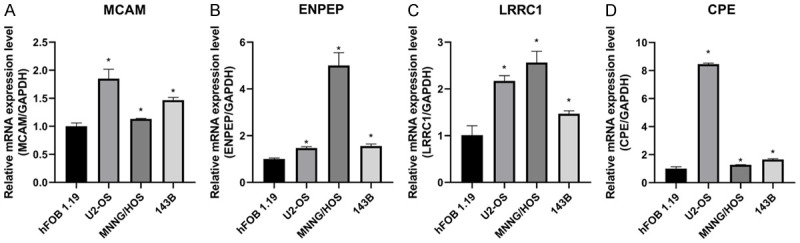
Expression of four risk genes in osteosarcoma cell lines (143B, U2OS, and MNNG/HOS) and osteoblast cell line (hFOB 1.19). (A-D) Differential expression of MCAM (A), ENPEP (B), LRRC1 (C), and CPE (D) in the human osteosarcoma cell lines and osteoblastic cell line (*P<0.05).
Discussion
Osteosarcomas are among the most aggressive malignancies of the skeleton, with 15-20% of patients having pulmonary metastases when first diagnosed [20,21]. Despite recent developments that have improved the prognosis of osteosarcoma, overall survival in patients with metastatic or recurrent osteosarcoma remains poor [8]. Therefore, novel diagnostic and prognostic biomarkers for osteosarcoma should be identified to provide new molecular targets to improve patient’s outcomes. Increasing evidence indicates that abnormal copy-number alterations and mutations of genes, such as TP53, epidermal growth factor receptor (EGFR), c-myc, and RB, play a crucial role in the occurrence and progression of osteosarcoma [22-24]. However, the exact mechanisms are still not fully understood.
In recent years, high-throughput sequencing technologies and bioinformatics analysis have enabled exploration of genetic alterations in osteosarcoma and provided an effective method to identify potentially beneficial markers for other cancer types. In this study, we established and validated a four-gene prognostic model which could be regarded as an independent predictor variable and provide more accurate prediction of OS in patients with osteosarcoma than age, gender, and Huvos grade. In addition, risk scores based on the signature, and also the expression levels of the four genes in the signature, were significantly associated with metastasis of osteosarcoma. We investigated the expression of the four signature genes in osteosarcoma cell lines to provide a reference for further basic research on osteosarcoma.
We analyzed two mRNA microarray datasets to detect co-expressed DEGs in osteosarcoma and human primary osteoblasts. Overall, there were 202 overlapping DEGs, in which 120 were upregulated and 82 were downregulated. GO and KEGG enrichment analyses of the identified DEGs revealed that Rap1 and the Ras signaling pathway were consistently up-regulated in osteosarcoma. Rap1 and Ras signaling pathways appear to be crucial in the course and maintenance of many tumor types. Studies have shown that Ras genes are mutated and abnormally activated in a large proportion of leukemias and solid tumors and are involved in cancer cell growth, invasion, migration, programmed cell death, and neovascularization [25,26]. Previous studies using osteosarcoma cell lines confirmed that 143B cells were more tumorigenic and metastatic than MNNG/HOS cells because of K-Ras transformation [27], and that MNNG/HOS cells transgenic for the K-Ras gene had increased metastatic potential compared with wild-type MNNG/HOS cells [28]. Wang et al. also reported that osteosarcomas proliferate and migrate primarily as a result of the Ras/MAPK kinase cascade initiated by macrophage migration inhibitory factor [29]. In another study, Lakshmikanthan et al. [30] showed that Rap1 was involved in various integrin-mediated biological processes, including angiogenesis and activation of vascular endothelial growth factor receptor 2. Rap1 was also shown to play a role in cell matrix adhesion and cell migration in head and neck cancer via the Rap1/Rac1 signaling axis rather than individual functions of α5 or β1 integrin [31]. In addition, Rap1 and Ras share similar binding partners and play a synergistic role in initiating and maintaining ERK signals [32]. Another pathway that significantly enriched is the p53 signaling pathway, which has always been a hotspot in cancer research. The p53 signaling pathway is essential for many cellular processes including apoptosis, senescence, DNA repair, and metabolism. Mutation of p53 is associated with lung metastasis in osteosarcoma mediated through the ONZIN-CXCL5-MAPK signaling pathway [33]. In addition, p53 plays a pivotal role in regulating bone remodeling associated with osteosarcoma, and loss of p53 function is associated with abnormal osteogenic differentiation of mesenchymal stem cells (MSCs) [34]. These established mechanistic links between osteosarcoma and Rap1/Ras signaling and p53 may explain how the DEGs identified in the expression analysis could affect the development of osteosarcoma.
Through univariate and multivariate Cox regression analyses, we identified four key DEGs: MCAM, ENPEP, LRRC1, and CPE, that are associated with prognosis in osteosarcoma. We then used those DEGs to construct a prognostic signature. Past studies revealed the involvement of the signature genes in cancer development. MCAM was shown to be the target gene of KDM3A in Ewing Sarcoma and an important effector of KDM3A in promoting metastasis [35]. Consistent with the findings of our study, Wang et al. observed that the MCAM protein was up-regulated in osteosarcoma compared with normal osteoblasts, and that the expression of MCAM on the surface of osteosarcoma cells was generally at cell-to-cell contact sites [36]. ENPEP is a coronavirus receptor when it is overexpressed and is also a risk factor for colorectal cancer progression [37]. In another study, Feliciano et al. also reported that ENPEP protein was upregulated in 56% of breast cancer patients, and miR-125b directly targeted ENPEP to induce cell cycle arrest and inhibit tumor growth [38]. Furthermore, LRRC1 was shown to exert its effects via the Wnt/β-Catenin pathway in genetically modified breast carcinoma stem cells, and the expression of LRRC1 was also correlated with stem cell characteristics in normal and neoplastic breast epithelium [39]. In addition, MSC-derived exosomes with up-regulated miR-193a and down-regulated LRRC1 reduced cisplatin resistance in non-small cell lung cancer cells [40]. Silencing of CPE led to G0/G1 cell cycle stagnation in an osteosarcoma cell line and inhibited osteosarcoma cell growth, migration, and invasion in vivo by reducing expression of cyclin D1 [41].
To evaluate the independent predictive ability and clinical utility of the model, we analyzed the signature-based risk score along with several clinical variables. We found that the risk score and Huvos grade were independent prognostic factors. Previous studies found that Huvos grade is a reliable predictor of patient survival in osteosarcoma, and that the percentage of necrotic tissue after chemotherapy classified using the Huvos system reflects the effectiveness of the treatment [42,43]. Further comparisons indicated that the prognostic signature was superior to the Huvos grade in predicting osteosarcoma outcomes, which suggests that the four-gene signature may be reliable. In addition, patients with metastatic osteosarcoma had higher expression levels of the four signature genes and higher risk scores than patients with non-metastatic osteosarcoma. An independent analysis of the R2 osteosarcoma dataset likewise showed that higher MCAM, ENPEP, LRRC1, and CPE expression was associated with shorter OS and MFS. These results suggest that the four risk genes in the prognostic model are reliable as a tool to predict metastasis and patient survival.
Infiltrating immune cells are a critical factor in cancer initiation and progression and have been studied extensively in recent years [44]. We found immune-related clues based on the four genes in the prognostic signature. Patients with low risk scores based on the four-gene signature had higher levels of tumor-infiltrating immune cells than patients with high risk scores, and immunity was impaired overall in the latter group. We speculate that reduced levels of antitumor immunity in patients with high risk scores may be associated with poor survival outcomes. Moreover, the expression levels of the four risk genes showed a significant correlation with tumor purity and immune cell infiltration.
Conclusion
We designed and validated a four-gene prognostic signature as an independent predictive marker for osteosarcoma, which may contribute to early diagnosis and treatment decision-making. The roles of the four risk genes; MCAM, ENPEP, LRRC1, and CPE; in osteosarcoma are still not fully elucidated. Further studies in clinical cohorts and other databases are needed to validate our findings.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81601931; 81672229), the Natural Science Foundation of Jiangsu Province (BK20150475), the Youth Medical Key Talent Project of Jiangsu (QNRC2016844), “Six One Projects” for high-level health professionals in Jiangsu Province Top Talent Project (LGY2019089), Jiangsu Provincial key research and development program (BE2020679), and the Shenzhen Science and Technology Program (No. KQTD20170810154011370).
Disclosure of conflict of interest
None.
References
- 1.Brown HK, Tellez-Gabriel M, Heymann D. Cancer stem cells in osteosarcoma. Cancer Lett. 2017;386:189–195. doi: 10.1016/j.canlet.2016.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Gianferante DM, Mirabello L, Savage SA. Germline and somatic genetics of osteosarcoma-connecting aetiology, biology and therapy. Nat Rev Endocrinol. 2017;13:480–491. doi: 10.1038/nrendo.2017.16. [DOI] [PubMed] [Google Scholar]
- 3.Wu Y, Jin Y, Yamamoto N, Takeuchi A, Miwa S, Tsuchiya H, Yang Z. MSX2 inhibits the growth and migration of osteosarcoma cells by repressing SOX2. Am J Transl Res. 2021;13:5851–5865. [PMC free article] [PubMed] [Google Scholar]
- 4.Yang C, Tian Y, Zhao F, Chen Z, Su P, Li Y, Qian A. Bone microenvironment and osteosarcoma metastasis. Int J Mol Sci. 2020;21:6985. doi: 10.3390/ijms21196985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whelan JS, Davis LE. Osteosarcoma, chondrosarcoma, and chordoma. J. Clin. Oncol. 2018;36:188–193. doi: 10.1200/JCO.2017.75.1743. [DOI] [PubMed] [Google Scholar]
- 6.Chen C, Xie L, Ren T, Huang Y, Xu J, Guo W. Immunotherapy for osteosarcoma: fundamental mechanism, rationale, and recent breakthroughs. Cancer Lett. 2021;500:1–10. doi: 10.1016/j.canlet.2020.12.024. [DOI] [PubMed] [Google Scholar]
- 7.Harrison DJ, Geller DS, Gill JD, Lewis VO, Gorlick R. Current and future therapeutic approaches for osteosarcoma. Expert Rev Anticancer Ther. 2018;18:39–50. doi: 10.1080/14737140.2018.1413939. [DOI] [PubMed] [Google Scholar]
- 8.Isakoff MS, Bielack SS, Meltzer P, Gorlick R. Osteosarcoma: current treatment and a collaborative pathway to success. J. Clin. Oncol. 2015;33:3029–3035. doi: 10.1200/JCO.2014.59.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin CD, Hou YL, Liu XR, He YS, Wang XP, Li CJ, Tan XH, Liu J. Development of an immune-related prognostic index associated with osteosarcoma. Bioengineered. 2021;12:172–182. doi: 10.1080/21655979.2020.1864096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu K, Zhang P, Zhang J, Quan H, Wang J, Liang Y. Identification of potential micro-messenger RNAs (miRNA-mRNA) interaction network of osteosarcoma. Bioengineered. 2021;12:3275–3293. doi: 10.1080/21655979.2021.1947065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang WT, Liu AG, Cai KT, He RQ, Li Z, Wei QJ, Chen MY, Huang JY, Yan WY, Zhou H, Chen G, Ma J. Exploration and validation of downregulated microRNA-199a-3p, downstream messenger RNA targets and transcriptional regulation in osteosarcoma. Am J Transl Res. 2019;11:7538–7554. [PMC free article] [PubMed] [Google Scholar]
- 12.Niu J, Yan T, Guo W, Wang W, Zhao Z, Ren T, Huang Y, Zhang H, Yu Y, Liang X. Identification of potential therapeutic targets and immune cell infiltration characteristics in osteosarcoma using bioinformatics strategy. Front Oncol. 2020;10:1628. doi: 10.3389/fonc.2020.01628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu R, Feng F, Yu X, Liu Z, Lao L. LncRNA SNHG4 promotes tumour growth by sponging miR-224-3p and predicts poor survival and recurrence in human osteosarcoma. Cell Prolif. 2018;51:e12515. doi: 10.1111/cpr.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Rao B, Yang J, Liu L, Huang M, Liu X, Cui G, Li C, Han Q, Yang H, Cui X, Sun R. Dysregulated m6A-related regulators are associated with tumor metastasis and poor prognosis in osteosarcoma. Front Oncol. 2020;10:769. doi: 10.3389/fonc.2020.00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang C, Zheng JH, Lin ZH, Lv HY, Ye ZM, Chen YP, Zhang XY. Profiles of immune cell infiltration and immune-related genes in the tumor microenvironment of osteosarcoma. Aging (Albany NY) 2020;12:3486–3501. doi: 10.18632/aging.102824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu N, Hou J, Ma G, Guo S, Zhao C, Chen B. Co-expression network analysis identifies a gene signature as a predictive biomarker for energy metabolism in osteosarcoma. Cancer Cell Int. 2020;20:259. doi: 10.1186/s12935-020-01352-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang JS, Duan MY, Zhong YS, Li XD, Du SX, Xie P, Zheng GZ, Han JM. Investigating age-induced differentially expressed genes and potential molecular mechanisms in osteosarcoma based on integrated bioinformatics analysis. Mol Med Rep. 2019;19:2729–2739. doi: 10.3892/mmr.2019.9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu Y, Zhang H, Ren T, Huang Y, Liang X, Wang W, Niu J, Han Y, Guo W. Development of a prognostic gene signature based on an immunogenomic infiltration analysis of osteosarcoma. J Cell Mol Med. 2020;24:11230–11242. doi: 10.1111/jcmm.15687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng G, Ma Y, Zou Y, Yin A, Li W, Dong D. HCMDB: the human cancer metastasis database. Nucleic Acids Res. 2018;46:D950–D955. doi: 10.1093/nar/gkx1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritter J, Bielack SS. Osteosarcoma. Ann Oncol. 2010;21(Suppl 7):vii320–325. doi: 10.1093/annonc/mdq276. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Yu X, Long X, Pu Q. MIR205 host gene (MIR205HG) drives osteosarcoma metastasis via regulating the microRNA 2114-3p (miR-2114-3p)/twist family bHLH transcription factor 2 (TWIST2) axis. Bioengineered. 2021;12:1576–1586. doi: 10.1080/21655979.2021.1920326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Czarnecka AM, Synoradzki K, Firlej W, Bartnik E, Sobczuk P, Fiedorowicz M, Grieb P, Rutkowski P. Molecular biology of osteosarcoma. Cancers (Basel) 2020;12:2130. doi: 10.3390/cancers12082130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji XL, He M. Sodium cantharidate targets STAT3 and abrogates EGFR inhibitor resistance in osteosarcoma. Aging (Albany NY) 2019;11:5848–5863. doi: 10.18632/aging.102193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen S, Yao T, Xu Y, Zhang D, Fan S, Ma J. CircECE1 activates energy metabolism in osteosarcoma by stabilizing c-Myc. Mol Cancer. 2020;19:151. doi: 10.1186/s12943-020-01269-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 26.Bullinger L, Döhner K, Döhner H. Genomics of acute myeloid leukemia diagnosis and pathways. J. Clin. Oncol. 2017;35:934–946. doi: 10.1200/JCO.2016.71.2208. [DOI] [PubMed] [Google Scholar]
- 27.Luu HH, Kang Q, Park JK, Si W, Luo Q, Jiang W, Yin H, Montag AG, Simon MA, Peabody TD, Haydon RC, Rinker-Schaeffer CW, He TC. An orthotopic model of human osteosarcoma growth and spontaneous pulmonary metastasis. Clin Exp Metastasis. 2005;22:319–329. doi: 10.1007/s10585-005-0365-9. [DOI] [PubMed] [Google Scholar]
- 28.Tome Y, Tsuchiya H, Hayashi K, Yamauchi K, Sugimoto N, Kanaya F, Tomita K, Hoffman RM. In vivo gene transfer between interacting human osteosarcoma cell lines is associated with acquisition of enhanced metastatic potential. J Cell Biochem. 2009;108:362–367. doi: 10.1002/jcb.22259. [DOI] [PubMed] [Google Scholar]
- 29.Wang C, Zhou X, Li W, Li M, Tu T, Ba X, Wu Y, Huang Z, Fan G, Zhou G, Wu S, Zhao J, Zhang J, Chen J. Macrophage migration inhibitory factor promotes osteosarcoma growth and lung metastasis through activating the RAS/MAPK pathway. Cancer Lett. 2017;403:271–279. doi: 10.1016/j.canlet.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 30.Lakshmikanthan S, Sobczak M, Chun C, Henschel A, Dargatz J, Ramchandran R, Chrzanowska-Wodnicka M. Rap1 promotes VEGFR2 activation and angiogenesis by a mechanism involving integrin αvβ3. Blood. 2011;118:2015–2026. doi: 10.1182/blood-2011-04-349282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu M, Banerjee R, Rossa C Jr, D’Silva NJ. RAP1-RAC1 signaling has an important role in adhesion and migration in HNSCC. J Dent Res. 2020;99:959–968. doi: 10.1177/0022034520917058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shah S, Brock EJ, Ji K, Mattingly RR. Ras and Rap1: a tale of two GTPases. Semin Cancer Biol. 2019;54:29–39. doi: 10.1016/j.semcancer.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Hu Q, Li G, Li L, Liang S, Zhang Y, Liu J, Fan Z, Li L, Zhou B, Ruan Y, Yang X, Chen S, Mu T, Wang G, Xiong S. ONZIN upregulation by mutant p53 contributes to osteosarcoma metastasis through the CXCL5-MAPK signaling pathway. Cell Physiol Biochem. 2018;48:1099–1111. doi: 10.1159/000491976. [DOI] [PubMed] [Google Scholar]
- 34.Velletri T, Huang Y, Wang Y, Li Q, Hu M, Xie N, Yang Q, Chen X, Chen Q, Shou P, Gan Y, Candi E, Margherita AP, Agostini M, Yang H, Melino G, Shi Y, Wang Y. Loss of p53 in mesenchymal stem cells promotes alteration of bone remodeling through negative regulation of osteoprotegerin. Cell Death Differ. 2021;28:156–169. doi: 10.1038/s41418-020-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sechler M, Parrish JK, Birks DK, Jedlicka P. The histone demethylase KDM3A, and its downstream target MCAM, promote Ewing sarcoma cell migration and metastasis. Oncogene. 2017;36:4150–4160. doi: 10.1038/onc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schiano C, Grimaldi V, Casamassimi A, Infante T, Esposito A, Giovane A, Napoli C. Different expression of CD146 in human normal and osteosarcoma cell lines. Med Oncol. 2012;29:2998–3002. doi: 10.1007/s12032-012-0158-3. [DOI] [PubMed] [Google Scholar]
- 37.Yuan Y, Chen J, Wang J, Xu M, Zhang Y, Sun P, Liang L. Development and clinical validation of a novel 4-gene prognostic signature predicting survival in colorectal cancer. Front Oncol. 2020;10:595. doi: 10.3389/fonc.2020.00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feliciano A, Castellvi J, Artero-Castro A, Leal JA, Romagosa C, Hernández-Losa J, Peg V, Fabra A, Vidal F, Kondoh H, Ramón Y Cajal S, Lleonart ME. miR-125b acts as a tumor suppressor in breast tumorigenesis via its novel direct targets ENPEP, CK2-α, CCNJ, and MEGF9. PLoS One. 2013;8:e76247. doi: 10.1371/journal.pone.0076247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopez Almeida L, Sebbagh M, Bertucci F, Finetti P, Wicinski J, Marchetto S, Castellano R, Josselin E, Charafe-Jauffret E, Ginestier C, Borg JP, Santoni MJ. The SCRIB paralog LANO/LRRC1 regulates breast cancer stem cell fate through WNT/β-catenin signaling. Stem Cell Reports. 2018;11:1040–1050. doi: 10.1016/j.stemcr.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu H, Mu X, Liu L, Wu H, Hu X, Chen L, Liu J, Mu Y, Yuan F, Liu W, Zhao Y. Bone marrow mesenchymal stem cells-derived exosomal microRNA-193a reduces cisplatin resistance of non-small cell lung cancer cells via targeting LRRC1. Cell Death Dis. 2020;11:801. doi: 10.1038/s41419-020-02962-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan S, Li X, Li L, Wang L, Du Z, Yang Y, Zhao J, Li Y. Silencing of carboxypeptidase E inhibits cell proliferation, tumorigenicity, and metastasis of osteosarcoma cells. Onco Targets Ther. 2016;9:2795–2803. doi: 10.2147/OTT.S98991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mintz MB, Sowers R, Brown KM, Hilmer SC, Mazza B, Huvos AG, Meyers PA, Lafleur B, McDonough WS, Henry MM, Ramsey KE, Antonescu CR, Chen W, Healey JH, Daluski A, Berens ME, Macdonald TJ, Gorlick R, Stephan DA. An expression signature classifies chemotherapy-resistant pediatric osteosarcoma. Cancer Res. 2005;65:1748–1754. doi: 10.1158/0008-5472.CAN-04-2463. [DOI] [PubMed] [Google Scholar]
- 43.Meyers PA, Gorlick R, Heller G, Casper E, Lane J, Huvos AG, Healey JH. Intensification of preoperative chemotherapy for osteogenic sarcoma: results of the Memorial Sloan-Kettering (T12) protocol. J. Clin. Oncol. 1998;16:2452–2458. doi: 10.1200/JCO.1998.16.7.2452. [DOI] [PubMed] [Google Scholar]
- 44.Meng Y, Yang Y, Zhang Y, Yang X, Li X, Hu C. The role of an immune signature for prognosis and immunotherapy response in endometrial cancer. Am J Transl Res. 2021;13:532–548. [PMC free article] [PubMed] [Google Scholar]