Abstract
In Saccharomyces cerevisiae the subcellular distribution of Bcy1 is carbon source dependent. In glucose-grown cells, Bcy1 is almost exclusively nuclear, while it appears more evenly distributed between nucleus and cytoplasm in carbon source-derepressed cells. Here we show that phosphorylation of its N-terminal domain directs Bcy1 to the cytoplasm. Biochemical fractionation revealed that the cytoplasmic fraction contains mostly phosphorylated Bcy1, whereas unmodified Bcy1 is predominantly present in the nuclear fraction. Site-directed mutagenesis of two clusters (I and II) of serines near the N terminus to alanine resulted in an enhanced nuclear accumulation of Bcy1 in ethanol-grown cells. In contrast, substitutions to Asp led to a dramatic increase of cytoplasmic localization in glucose-grown cells. Bcy1 modification was found to be dependent on Yak1 kinase and, consequently, in ethanol-grown yak1 cells the Bcy1 remained nuclear. A two-hybrid screen aimed to isolate genes encoding proteins that interact with the Bcy1 N-terminal domain identified Zds1. In ethanol-grown zds1 cells, cytoplasmic localization of Bcy1 was largely absent, while overexpression of ZDS1 led to increased cytoplasmic Bcy1 localization. Zds1 does not regulate Bcy1 modification since this was found to be unaffected in zds1 cells. However, in zds1 cells cluster II-mediated, but not cluster I-mediated, cytoplasmic localization of Bcy1 was found to be absent. Altogether, these results suggest that Zds1-mediated cytoplasmic localization of Bcy1 is regulated by carbon source-dependent phosphorylation of cluster II serines, while cluster I acts in a Zds1-independent manner.
Throughout the eukaryotic kingdom cyclic AMP (cAMP)-dependent protein kinases (PKAs) play important and diverse roles in signal transduction (for reviews, see references 2, 7, and 28 and references therein). Structurally, PKAs are conserved, consisting of two catalytic subunits that bind, in their inactive configuration, to a regulatory subunit homodimer. Binding of cAMP to the regulatory subunit results in dissociation, and thereby activation, of the catalytic subunits (7, 28). The multitude of intracellular PKA substrates and their different subcellular distribution raises important questions about the specificity, timing, and substrate targeting of PKA-mediated signaling. One regulatory level to ensure proper signal transduction is specific targeting of signaling components to subcellular compartments. In multicellular eukaryotes A-kinase anchor proteins (AKAPs) have been identified that target type I or type II (RI or RII) PKA-regulatory subunits to their effector substrates localized in various subcellular compartments (for recent reviews, see references 5 and 6 and references therein). AKAPs possess a site for constitutive avid binding of RI or RII and a targeting domain that complexes with subcellular structures. Directing PKA to specific microenvironments facilitates phosphorylation of colocalized effector molecules.
In contrast to cells from multicellular organisms, yeast cells possess one isoform of the PKA regulatory subunit, Bcy1, which controls three catalytic subunits which are functionally partly redundant. Bcy1 responds to glucose, the only extracellular signal identified so far, that triggers an increase in cAMP levels (8, 31). Yeast Tpk proteins control a wide variety of important intracellular processes at both the transcriptional and the posttranscriptional levels (for a recent review, see reference (29) and references therein).
Localization studies on the budding yeast PKA catalytic subunit Tpk1 and the regulatory subunit Bcy1 revealed that the subcellular localization of both types of subunits is regulated (12). cAMP, whose production is controlled by glucose, affects the subcellular distribution of Tpk1. The localization of Bcy1 was also found to be dependent on the available carbon source. In glucose-grown cells the regulatory subunit is strongly concentrated in the nucleus. In carbon source-derepressed cells, however, it is present in both the nuclear and the cytoplasmic compartments. The increased cytoplasmic localization of Bcy1 in such cells might be important for the proper regulation of cytoplasmic substrates, e.g., metabolic enzymes that are regulated by PKA. Characterization of mutants with mislocalized Bcy1 revealed that proper localization of Bcy1 is important for efficient sporulation, viability in the stationary phase, and the rapid reproliferation of stationary-phase cells (12).
Although Bcy1 has an architecture characteristic for type II PKA-regulatory subunits (31), structural yeast AKAP homologs do not seem to exist (5; unpublished results) suggesting that different mechanisms control the subcellular distribution of Bcy1. Considering the wide variety of PKA-controlled substrates, their different subcellular distribution and the carbon source-dependent localization of PKA in budding yeast, we surmise that the subcellular targeting of Bcy1, as for RI and RII, provides an additional level of regulation of intracellular signaling. The present study concentrates on the molecular mechanisms of Bcy1 localization. We provide evidence that the subcellular distribution of Bcy1 is regulated by Yak1-dependent phosphorylation of its N-terminal domain and that Zds1 is required for proper cytoplasmic localization of Bcy1 in carbon source-derepressed cells.
MATERIALS AND METHODS
Growth media, growth conditions, yeast strains, and plasmids.
Yeast media were prepared as described elsewhere (22). Cells were grown in synthetic complete (SC) medium supplemented with adenine, uracil, and amino acids as appropriate but lacking essential components to select for plasmids. For all of the experiments, cells were precultured in selective media and then inoculated in complete media. Carbon source-derepressed cells used for the experiments described in this study were grown on yeast extract - peptone - ethanol (YPE), YPA (acetate), or YPG (glycerol) until the cultures reached an A600 of 4 to 5. To obtain cells in stationary phase, these were cultured in YPD (dextrose) for at least 2 days.
Yeast strains used in this study are listed in Table 1. Deletion of YAK1 in strains GG101 and GG102 was achieved by transforming W303-1A and Wmsn2/4 with plasmid pGS136B (a gift of S. Garrett, New Jersey Medical School, Newark) after being digested by SmaI and HindIII. Leu+ transformants were checked for correct integration of the yak1::LEU2 deletion fragment using PCR. PA4036-2A and PA4036-2C were obtained by sporulation of DY4036 (a gift from D. Stillman, University of Utah Health Sciences Center, Salt Lake City) and subsequent isolation of spores (derived from a tetrad that yielded four viable spores) that contained zds1::LEU2 or zds2::TRP1, respectively.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| W303-1A | MATa ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1 | 30 |
| MR1 | W303-1A MATα bcy1::LEU2 | 12 |
| Wmsn2/4 | W303-1A MATa msn2::HIS3 msn4::TRP1 | 18 |
| GG101 | W303-1A MATa yak1::LEU2 | This study |
| GG102 | W303-1A MATa msn2::HIS3 msn4::TRP1 yak1::LEU2 | This study |
| DY 4036 | W303-1A/W303-1A ZDS1/zds1::LEU2 ZDS2/zds2::TRP1 | D. Stillman |
| PA4036-2A | W303-1A MATα zds1::LEU2 | This study |
| PA4036-2C | W303-1A MATa zds2::TRP1 | This study |
| PJ69-4A | MATa trp-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1- HIS3 met2::GAL7-lacZ | 15 |
All plasmids used in this study are listed in Table 2. Plasmids 313pBHB1–416, 313pBHB125–416, and 33AGHB1–124 are identical to plasmids 313HBwt, 313HBΔN2, and 33pAGHBΔC1, respectively, described earlier (12). 313pB(NLS)HB1–416 was created by introducing a double-stranded oligonucleotide obtained by annealing oligonucleotides 5′-C ATG CCA AAG AAG AAG AGA AAG GTC ATG CAT GC-3′ and 5′-CAT GGC ATG CAT GAC CTT TCT CTT CTT CTT TGG-3′, which encode the Simian virus 40 nuclear localization signal (SV40 NLS), in the NcoI site of 313pBHB1–416. Plasmid 33AGHB(1–124, Ser cluster II Ala) was created as follows. Two PCR fragments were generated by using a plasmid-borne copy of BCY1: one DNA fragment using forward primer 5′-GAC TGG ATC CAT GGT ATC TTC TTT GCC C-3′ (named BCY1-G-F) and reverse primer 5′-GCTA CTA GCT AGC TTG AGC TTG AGC TGC TTG AGG TCT GGA AAA TGA CTC CTC TGG-3′ (containing mutations that substitute Ser74, Ser77, and Ser79 by Ala and bearing an NheI site at the 5′ end [underlined]) and the second DNA fragment using forward primer 5′-CAC TAG TCT AGA GCC AGA GCC GCT GTT ATG TTC AAA TCC CCT TTT GTG AAC G-3′ (containing mutations that substitute Ser81, Ser83, and Ser84 by Ala and bearing a XbaI site at the 5′ end [underlined]) and reverse primer 5′-GAT GGA ATT CAT CGA TCT GTG TGG ATA GGG G-3′ (named BCY1-G-R). In several steps these were subsequently subcloned in 33pAGHBwt (12), using BamHI and EcoRI and with the NheI and XbaI sites of the two PCR-generated fragments fused, resulting in plasmid 33AGHB(Ser cluster II Ala) containing full-length BCY1 bearing cluster II Ser-to-Ala substitutions. Subsequently, this plasmid was used as a template for generating a DNA fragment by PCR using forward primer BCY1-G-F and reverse primer 5′-TTT CTG CAG TTA ATG CTG TTG TTC TTC CTG-3′ (named BΔC1) that was subcloned with PstI and BamHI in plasmid 33pAGHBwt (12).
TABLE 2.
Plasmids used in this study
| Plasmid | Description |
|---|---|
| pRS313 | CEN ARS HIS3 vector (26) |
| 313pBHB1–416 | Expresses a HA-BCY1 fusion, encoding HA-tagged wild-type Bcy1p, using the BCY1 promoter in vector pRS313; plasmid 313HBwt in Griffioen et al. (12) |
| 313pBHB125–416 | 313pBHB1–416 derivative encoding Bcy1 deleted for amino acids 1 to 124; plasmid 313HBΔN2 in Griffioen et al. (12) |
| 313pB(NLS)HBwt | 313pBHB1–416 derivative encoding Bcy1 fused with SV40 NLS |
| YCplac33 | CEN ARS URA3 vector (10) |
| 33AGHB1–124 | Expresses a HA-BCY1 fusion, encoding HA-tagged Bcy1 N terminus (amino acids 1 to 124) using the ADH1 promoter in vector YCplac33 |
| 33AGHB(1–124, Ser cluster I Ala) | 33AGHB1–124 derivative encoding Bcy1 bearing S3A, S4A, and S9A substitutions |
| 33AGHB(1–124, Ser cluster II Ala) | 33AGHB1–124 derivative encoding Bcy1 bearing S74A, S77A, S79A, S81A, S83A, and S84A substitutions |
| 33AGHB(1–124, Ser cluster I+II Ala) | 33AGHB1–124 derivative encoding Bcy1 bearing S3A, S4A, S9A, S74A, S77A, S79A, S81A, S83A, and S84A substitutions |
| 33pBGHBwt | Expresses a GFP-HA-BCY1 fusion, encoding wild-type Bcy1 fused with GFP-HA, using the BCY1 promoter in vector YCplac33 |
| 33pBGHB(Ser cluster I Ala) | 33pBGHBwt derivative encoding Bcy1 bearing S3A, S4A, and S9A substitutions |
| 33pBGHB(Ser cluster II Ala) | 33pBGHBwt derivative encoding Bcy1 bearing S74A, S77A, S79A, S81A, S83A, and S84A substitutions |
| 33pBGHB(Ser cluster I+II Ala) | 33pBGHBwt derivative encoding Bcy1 bearing S3A, S4A, S9A, S74A, S77A, S79A, S81A, S83A, and S84A substitutions |
| 195A2 | 2μm, ADE2 vector (this study) |
| 195A2-pBGHBwt | Expresses a GFP-HA-BCY1 fusion, encoding wild-type Bcy1p fused with GFP-HA, using the BCY1 promoter in vector 195ADE2 |
| 195A2-pBGHB(Ser cluster I Asp) | 195A2-pBGHBwt derivative encoding Bcy1 bearing S3D, S4D, and S9D substitutions |
| 195A2-BGHB(Ser cluster II Asp) | 195A2-pBGHBwt derivative encoding Bcy1 bearing S74D, S77D, S79D, S81D, S83D, and S84D substitutions |
| 195A2-pBGHB(Ser cluster I+II Asp) | 195A2-pBGHBwt derivative encoding Bcy1 bearing S3D, S4D, S9D, S74D, S77D, S79D, S81D, S83D, and S84D substitutions |
| pRS316 | CEN URA3 vector (10) |
| 316pADH1-ZDS1 | Expresses ZDS1, using the ADH1 promoter, in vector pRS316 |
| pGBT9 | Two-hybrid expression vector with GAL4 DNA-binding domain (DBD) (15) |
| T9-BCY1wt | BCY1 inserted in pGBT9; expresses Ga14-DBD fused Bcy1 |
| T9-BCY1125–416 | T9-BCY1wt derivative encoding Bcy1 deleted for amino acids 1 to 124 |
| T9-BCY11–179 | T9-BCY1wt derivative encoding Bcy1 deleted for amino acids 180 to 416 |
Plasmid 33AGHB(1–124, Ser cluster I Ala) and 33AGHB(1–124, Ser cluster I+II Ala) were constructed as follows. A PCR fragment was generated using a plasmid-borne copy of BCY1 and 33AGHB(Ser cluster II Ala), respectively, and forward primer 5′-CG CGC GGA TCC ATG GTA GCC GCC TTG CCC AAG GAA GCC CAA GCC GAA TTG CAA CTG-3′ (containing mutations that substitute Ser3, Ser4, and Ser9 by Ala; named MUT1-F) and reverse primer BΔC1. These PCR fragments were subcloned with BamHI and PstI in plasmid 33pAGHBwt (12). Plasmids 33AGHB(Ser cluster I Ala) and 33AGHB(Ser cluster I+II Ala) were generated as described above for plasmids 33AGHB(1–124, Ser cluster I Ala) and 33AGHB(1–124, Ser cluster I+II Ala) but using reverse primer BCY1-G-R instead of BΔC1. The PCR fragments were subcloned with BamHI and EcoRI.
33pBGHBwt was created by subcloning GFP-HA-BCY1 with EcoRI and SalI from plasmid 313GHBwt (12) in YCplac33 (10). 33pBGHB(Ser cluster I Ala), 33pBGHB(Ser cluster II Ala), and 33pBGHB(Ser cluster I+II Ala) were made by subcloning the corresponding BCY1 fragment from 33AGHB(Ser cluster I Ala), 33AGHB(Ser cluster II Ala), and 33AGHB(Ser cluster I+II Ala), respectively, in 33pBGHBwt using NotI and EcoRI.
Plasmid 195A2 was generated by replacing URA3 of YEplac195 (10) by ADE2.
195A2-pBGHB(Ser cluster II Asp) was created as follows. A PCR fragment was generated using a plasmid-borne copy of BCY1, forward primer 5′-GTT CTA TTT CCG GAA CCA GAG GAG TCA TTT TCC AGA CCT CAA GAC GCT CAA GAC CAA GAC AGA GAC AGA GAC GAC GTT ATG TTC AAA TCC CCC TTT GTG-3′ (containing mutations that substitute Ser74, Ser77, Ser79, Ser81, Ser83, and Ser84 by Asp) and reverse primer BCY1-G-R. It was cloned in 33pBGHBwt using BstEI and EcoRI. From the resulting plasmid [33pBGHB(Ser cluster II Asp)] a PvuII fragment containing GFP-HA-BCY1 under control of the BCY1 promoter was inserted in 195A2.
Plasmid 195A2-pBGHB(Ser cluster I Asp) and 195A2-pBGHB(Ser cluster I+II Asp) were constructed as follows. PCR fragments were generated using a plasmid-borne copy of BCY1 and 33pBGHB(Ser cluster II Asp), respectively, forward primer 5′-CG CGC GGA TCC ATG GTA GAC GAC TTG CCC AAG GAA GAC CAA GCC GAA TTG CAA CTG-3′ (containing mutations that substitute Ser3, Ser4, and Ser9 by Asp) and reverse primer BCY1-G-R. These PCR fragments were subcloned with BamHI and EcoRI in plasmid 33pAGHBwt. From the resulting plasmids GFP-HA-BCY1 was subcloned in 33pBGHBwt using EcoRI and NotI. Subsequently from the resulting plasmids, a PvuII fragment containing GFP-HA-BCY1 under control of the BCY1 promoter was inserted in 195A2.
T9-BCY1wt was created as follows. First, the NcoI-EcoRI fragment from 33pAGHBwt containing GFP-HA-BCY1 was subcloned correspondingly in pAS2-1 (Clontech). From the resulting plasmid the fragment encoding a fusion of Bcy1 with the Gal4 DNA-binding domain was inserted in pGBT9 (Clontech) using XhoI-EcoRI.
T9-BCY1125–416 and T9-BCY11–179 were made by subcloning the NotI-EcoRI fragments from plasmids 33pAGHBΔN2 and 33pAGHBΔC2 (12) in T9-BCY1wt.
316pADH1-ZDS1 was made as follows. A PCR fragment was generated using forward primer 5′-C CCA GAA TCC GGC GGC CGC GGA TCC ATG TCC AAT AGA GAT AAC GAG-3′ and reverse primer 5′-A AAA CTG CAG CTC GAG GTC GAC ATC TTC GTA CTAG-3′ and subcloned in a derivative of pADH1-MSN2-GFP in plasmid pRS316 (11) (containing a NotI site directly after the start codon) using NotI and SalI.
Western blot analysis.
Yeast cell cultures were grown at 30°C (see above). All subsequent steps were carried out at 4°C. Cells were harvested by centrifugation and washed in sterile water, and the pellets were resuspended in extraction buffer containing 10 mM Tris-HCl (pH 8.0), 150mM NaCl, 0.05% Tween 20, 10% glycerol, 5 mM EDTA, 5 mM NaF, 60 mM glycerol-2-phosphate, 1 mM dithiothreitol, 1 mM EGTA, and a mixture of protease inhibitors (Complete; Roche Molecular Biocemicals). Cells were disrupted by vortexing them for 5 min in the presence of glass beads. The resulting suspension was spun down in a microfuge at maximum speed, and this step was repeated once with the supernatant. Part of the resulting supernatant was taken up in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, separated by SDS-PAGE (16), and blotted onto nitrocellulose. Immunodetection of proteins was carried out using anti-hemagglutinin (HA) monoclonal antibody (12CA5 mouse immunoglobulin G [IgG], generous gift from T. Gartner). The secondary antibody used was anti-mouse IgG conjugated with horseradish peroxidase purchased from Amersham Pharmacia Biotech. Proteins were visualized using SuperSignal (Pierce) according to the manufacturer's instructions. Phosphatase treatment of protein extracts was carried out with λ phosphatase (New England Biolabs, Inc.) essentially according to the manufacturer's instructions. Then, 20 mM orthovanadate and 50 mM NaF were used for the inhibition of λ phosphatase activity. Cytoplasmic and nuclear fractions were obtained by differential centrifugation as previously described (1). The pellet and supernatant were used for Western analysis as nuclear and cytoplasmic fraction, respectively.
Green fluorescence protein (GFP) fluorescence microscopy.
Cells were used for fluorescence microscopy directly without fixation. Nuclei were stained by addition of 5 μg of DAPI (4′,6′-diamidino-2-phenylindole) per ml to the cell suspension. Cells were viewed using a Zeiss Axioplan 2 fluorescence microscope. Images were taken with a Quantix charge-coupled device camera using IP LAB software and then processed in Adobe Photoshop 4.0.
Two-hybrid screen.
T9-BCY11–416 was transformed into reporter strain PJ69-4A (15), and the resulting strain was transformed individually with each of three yeast genomic DNA fusion libraries, Y2HL-C1, Y2HL-C2, and Y2HL-C3 (15). Transformation mixtures were plated on medium lacking adenine, and positive transformants were tested for growth on SC-histidine medium supplemented with 2 mM 3-aminotriazole. From the remaining positives, plasmids containing the genomic DNA inserts were isolated and tested for their inability to interact with T9-BCY1125–416 (lacking the Bcy1 N-terminal domain). Finally, plasmids unable to confer growth were tested for interaction with T9-BCY11–179 (N-terminal domain of Bcy1) on SC-histidine medium supplemented with 10 mM 3-aminotriazole. For plasmids that also met this latter requirement, part of their genomic DNA inserts were sequenced to identify the corresponding gene (fragment). β-Galactosidase activity was assayed essentially as described previously (19).
RESULTS
Bcy1 is phosphorylated in its N-terminal targeting domain.
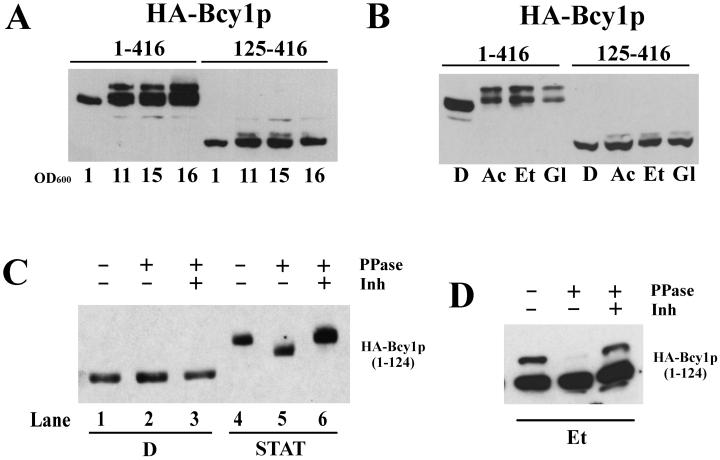
Recent studies have established that the localization of Bcy1 is carbon source dependent (12). In cells growing rapidly on glucose, Bcy1 is predominantly nuclear. In carbon source-derepressed cells it is distributed over the nucleus and cytoplasm. Previously, it had been shown that in stationary-phase cells Bcy1 was extensively modified (35). We investigated a possible functional link between the modification and localization of Bcy1. We studied these parameters both in ethanol-grown cells and in stationary-phase cells since these two physiological conditions appeared to have differential effects. Western blot analysis (Fig. 1A) of extracts of stationary-phase cells expressing full-length Bcy1 (HA-Bcy11–416) revealed the presence of several isoforms migrating more slowly than HA-Bcy11–416 detected in extracts of cells grown on glucose, a finding similar to what has been reported previously regarding Bcy1 modification in a different strain background (35). Also, in cells that were grown on various nonfermentable carbon sources, slower-migrating isoforms of Bcy1 were detected (Fig. 1B), although the nature of the modification appeared to be different compared to the situation in stationary-phase cells since fewer isoforms were observed. In cells expressing HA-Bcy1125–416 lacking its targeting domain, most of these isoforms were absent (Fig. 1A and B), demonstrating that the N-terminal domain is necessary for modification. We note that a minor isoform of HA-Bcy1125–416 is still detectable in carbon source-derepressed cells, most likely the result of autophosphorylation of Ser145 (35). To determine whether the N terminus is not only necessary but also sufficient for modification, we expressed the N-terminal part of Bcy1 in stationary-phase cells (Fig. 1C, lanes 4 to 6). In such cells the first 124 amino acids were sufficient to result in one or more isoform(s) migrating significantly slower than the form(s) detected in extracts from cells grown on glucose (Fig. 1C, lanes 1 to 3). This indicates that modification takes place near the Bcy1 N terminus. Upon phosphatase treatment of HA-Bcy11–124 from glucose grown cells, no change in migration was observed (Fig. 1C, lanes 1 to 3). The same fragment from stationary-phase cells migrated faster after phosphatase treatment (Fig. 1C, lanes 4 to 6), though migration was still slower than that observed for HA-Bcy11–124 detected in extracts from glucose-grown cells. This suggests that the slower migration of HA-Bcy11–124 is partly due to phosphorylation but presumably is also caused by a second, as-yet-unidentified type of modification. Also, in ethanol-grown cells expressing HA-Bcy11–124 the presence of at least one slower-migrating isoform was detected (Fig. 1D). After phosphatase treatment this isoform was almost undetectable, suggesting that also in ethanol-grown cells at least part of Bcy1 is phosphorylated. In contrast to the situation in stationary-phase cells, we note that in extracts of ethanol-grown cells no isoforms were observed that migrated significantly slower than HA-Bcy11–124 from glucose-grown cells after phosphatase treatment (data not shown).
FIG. 1.
Western analysis of extracts from yeast cells producing wild-type and mutant versions of HA-tagged Bcy1. (A) Extracts of strain MR1 transformed with 313pBHB1–416 and 313pBHB125–416 grown to stationary phase after growth on glucose. Samples were drawn at various cell densities as indicated by their optical densities at 600 nm. (B) Extracts isolated from the strains as referred to in Panel A grown on YP medium supplemented with glucose (D), acetate (Ac), ethanol (Et), or glycerol (Gl). (C) Extracts from strain W303-1A transformed with 33AGHB1–124. Samples were drawn from cultures grown logarithmically on YPD (D, lanes 1 to 3) and from stationary-phase cultures (STAT, lanes 4 to 6). Some extracts were treated with phosphatase in the presence or absence of inhibitors before loading on the gel. PPase, λ phosphatase; Inh, λ phosphatase inhibitors (20 mM vanadate and 50 mM NaF). (D) Phosphatase treatment of extracts isolated from the strains referred to in panel C grown on YP-ethanol (Et).
Bcy1 isoforms are differentially localized.
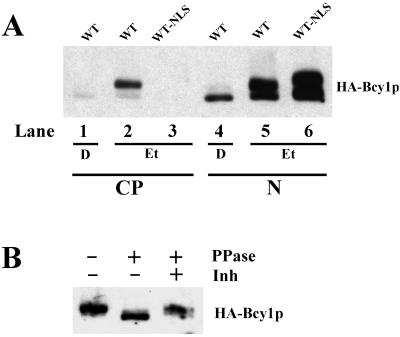
We reasoned that modification in the N-terminal domain of Bcy1 in carbon source-derepressed cells might be required for its cytoplasmic localization. To obtain evidence for this assumption, we performed fractionation experiments. Western analysis of cytoplasmic and nuclear fractions (Fig. 2A) showed that HA-Bcy1 is mostly found in the nuclear fraction of glucose-grown cells (lanes 1 and 4). In the cytoplasmic fraction of ethanol-grown cells (lane 2), a predominant slower-migrating isoform and very little of a faster-migrating form of HA-Bcy1 were detected. In the corresponding nuclear fraction, both slower- and faster-migrating isoforms were found (lane 5). Although this suggests that these are both nuclear, it cannot be completely ruled out in this case that detection of the slower-migrating isoform(s) is due to contamination of this fraction with the cytoplasmic fraction. It is evident, however, that unmodified Bcy1 is preferentially nuclear, irrespective of the carbon source used for growth, while virtually only modified Bcy1 is detected in the cytoplasm. To control the effectiveness of the fractionation procedure used, cells were tested expressing a fusion of HA-Bcy1 with the constitutive SV40 NLS. As expected, this fusion protein was detected exclusively in the nuclear fraction (lanes 3 and 6). Phosphatase treatment of HA-Bcy1 derived from the cytoplasmic fraction of ethanol-grown cells (sample from Fig. 2A, lane 2) resulted in an increase of migration (Fig. 2B), showing that cytoplasmic Bcy1 is phosphorylated.
FIG. 2.
Subcellular fractionation of cells grown on glucose (D)- and ethanol (Et)-based media. (A) Strain MR1 transformed with 313pBHBwt (WT) and 313pB(NLS)HBwt (WT-NLS). In lanes 1 to 3, the cytoplasmic fraction (CP) and in lanes 4 to 6, the nuclear fraction (N) were loaded. (B) Phosphatase treatment of the sample loaded in lane 2 of panel A.
Serine residues located in the N-terminal domain of Bcy1p are important for modification and proper subcellular localization.
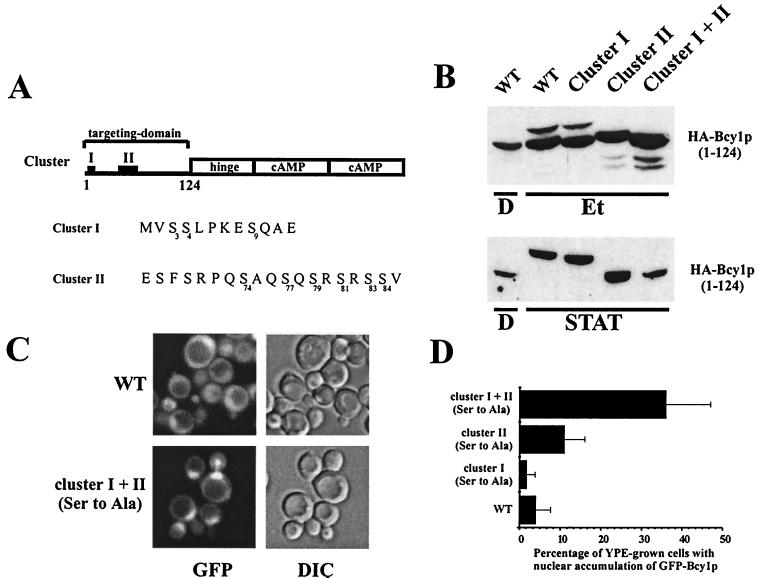
Since modification, partly through phosphorylation of the N-terminal domain, correlated well with localization, we wished to clarify whether phosphorylation of Bcy1 is necessary for cytoplasmic targeting. Inspection of the N-terminal domain revealed two clusters (I and II) which are particularly serine-rich. Cluster I is located near the N terminus, and cluster II is located between serines 68 and 84. We studied the effects on the modification and localization of serine-to-alanine substitutions in both clusters (affected amino acid residues are indicated in Fig. 3A). The wild-type N-terminal domain (Bcy11–124) from YPE-grown cells produced two major bands, representing at least two isoforms (Fig. 3B). The upper form (more slowly migrating) of these two isoforms disappeared when the serines in cluster II were mutated to alanines, while the lower form (more rapidly migrating) was unaffected. In contrast, substitution of the serines in cluster I to alanines had little effect on the upper isoform, while the migration of the lower isoform was somewhat increased. The combination of cluster I and cluster II serine-to-alanine substitutions caused an additive effect on observed isoforms, with the upper form disappearing and the lower form migrating more rapidly. These biochemical observations provide strong evidence that cluster II serines are phosphorylated but provide weaker evidence that cluster I is phosphorylated.
FIG. 3.
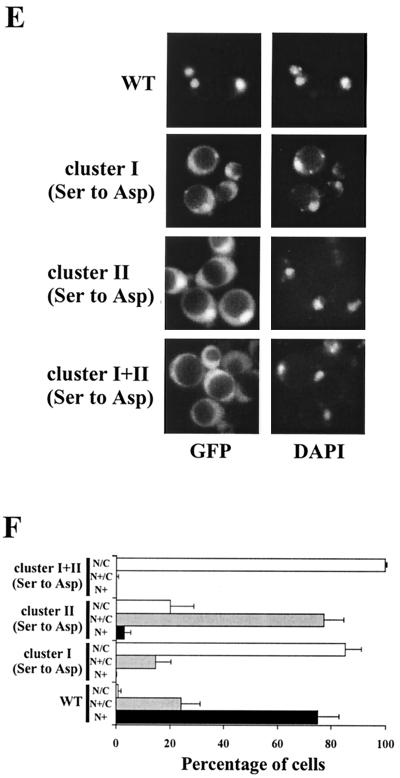
Characterization of cluster I and II substitution mutants. (A) Schematic representation of the domain structure of Bcy1. Residues 1 to 124 comprise the targeting domain necessary and sufficient for nutrient-controlled localization. The catalytic subunits associate with the hinge region; two cAMP-binding domains are present in the C-terminal region. “I” and “II” indicate the location of clusters I and II. The primary structure of the cluster I and II serine-rich regions is shown. The serines replaced in cluster I and cluster II are indicated with numbers in subscript. (B) Western analysis of extracts from yeast strain W303-1A transformed with 33AGHB1–124, 33AGHB(1–124, Ser cluster I Ala), 33AGHB(1–124, Ser cluster II Ala), and 33AGHB(1–124, Ser cluster I+II Ala) designated by WT, cluster I, cluster II, and cluster I+II, respectively. Cells were harvested from cultures growing on YP medium supplemented with glucose (D) or ethanol (Et) or grown on YPD to stationary phase (STAT). (C) Fluorescence microscopy of ethanol-grown MR1 cells transformed with 33pBGHBwt and 33pBGHB(Ser cluster I+II Ala) encoding GFP-Bcy1wt and GFP-Bcy1(Ser cluster I+II Ala), respectively. (D) Quantification of the localization pattern of MR1 cells transformed with 33pBGHBwt, 33pBGHB(Ser cluster I Ala), 33pBGHB(Ser cluster II Ala), and 33pBGHB(Ser cluster I+II Ala) encoding the corresponding versions of Bcy1 as indicated. The mean percentage of ethanol-grown cells with nuclear fluorescence stronger than cytoplasmic fluorescence was determined. Three independent transformants were assayed at least three times each (at least 100 cells were counted for each determination). The error bars indicate the standard deviation. (E) Fluorescence microscopy of glucose-grown W303-1A cells transformed with 195A2-pBGHBwt, 195A2-pBGHB(Ser cluster I Asp), 195A2-pBGHB(Ser cluster II Asp), and 195A2-pBGHB(Ser cluster I+II Asp) encoding the corresponding versions of GFP-Bcy1 as indicated. (F) Quantification of the localization pattern of W303-1A transformants shown in panel E. Three patterns of localization were distinguished. N+ (black bars), cells with nuclear accumulation of GFP-Bcy1 with no detectable cytoplasmic fluorescence; N+/C (gray bars), cells with nuclear accumulation of GFP-Bcy1 but with cytoplasmic fluorescence detectable; N/C (open bars), cells with fluorescence evenly distributed over the nucleus and cytoplasm. Three independent transformants were assayed at least three times each (at least 100 cells were counted for each determination). The error bars indicate the standard deviation.
In the stationary phase, as judged from the strong decrease in migration of the wild-type Bcy1 N-terminal domain, modification was more extensive than in the YPE-grown cells. However, also under these conditions Ala substitutions in cluster II resulted in a strongly increased migration, while cluster I substitutions caused a slight increase in migration. It is noteworthy that, although the migration shift in stationary phase was found to be only partly phosphatase sensitive (Fig. 1C), modification appeared to be completely abolished by a combination of Ala substitutions in both clusters I and II. This suggests that the serines substituted are important not only as targets of phosphorylation but also for mediating a putative additional modification.
Fluorescence microscopy of GFP-Bcy1 versions bearing Ser- to-Ala substitutions was carried out in both clusters to study their effects on localization (Fig. 3C and D). When cells were grown on YPE medium, GFP-Bcy1wt was found to be evenly distributed over the nucleus and cytoplasm in most cells as described previously (12). Substitution of Ser by Ala in either cluster I or cluster II alone [GFP-Bcy1(Ser cluster I Ala) and GFP-Bcy1(Ser cluster II Ala), respectively] did not have a strong impact on localization (Fig. 3D). However, GFP-Bcy1 mutated in both clusters [GFP-Bcy1(Ser cluster I+II Ala)] led to increased nuclear accumulation, suggesting that phosphorylation of cluster I and II serines is important for proper localization (Fig. 3C and D). In stationary-phase cells, all cluster substitution mutant versions of GFP-Bcy1 were distributed over the nucleus and cytoplasm, similar to GFP-Bcy1wt, indicating that mechanisms independent of the serines mutated are important for cytoplasmic localization in the stationary phase (data not shown).
To support the conclusion that the phosphorylation of clusters I and II serines is important for cytoplasmic localization, we studied the effects of Ser-to-Asp substitutions. Replacement of the serine residues by negatively charged amino acids supposedly mimics phosphorylated serines. In Fig. 3E and F, the effects of these substitutions on localization in cells growing on glucose are shown. Asp substitutions in cluster I led to a strong increase of cytoplasmic fluorescence compared to GFP-Bcy1wt. A similar observation was made when cluster II serines were substituted by aspartic residues, although the effect was somewhat less pronounced. Substitutions of Ser to Asp in both clusters led to an even more dramatic increase of cytoplasmic localization in glucose-grown cells and strongly resembles the localization pattern of wild-type Bcy1 in carbon source-derepressed cells. It should be noted that the Ser-to-Asp substitutions did not cause Bcy1 exclusion from the nucleus. This indicates that nuclear translocation per se is not blocked by the mutations. In conclusion, mimicking constitutive phosphorylation of the Bcy1 N-terminal domain by substitution of the serines in cluster I or II by aspartates is sufficient for cytoplasmic localization but does not exclude nuclear localization.
GFP-Bcy1 is accumulated in the nuclei of mutants with reduced modification of the Bcy1 N terminus.
The protein kinase Yak1 has previously been identified to be involved, directly or indirectly, in modifying Bcy1 (35). Furthermore, Yak1 kinase activity acts antagonistically to PKA (9). Its expression is dependent on Msn2 and Msn4, two transcription factors negatively regulated by PKA (27). Deletion of either YAK1 or MSN2 plus MSN4 can suppress the lethal phenotype of a simultaneous deletion of TPK1, TPK2, and TPK3, encoding different isoforms of PKA catalytic subunits (32).
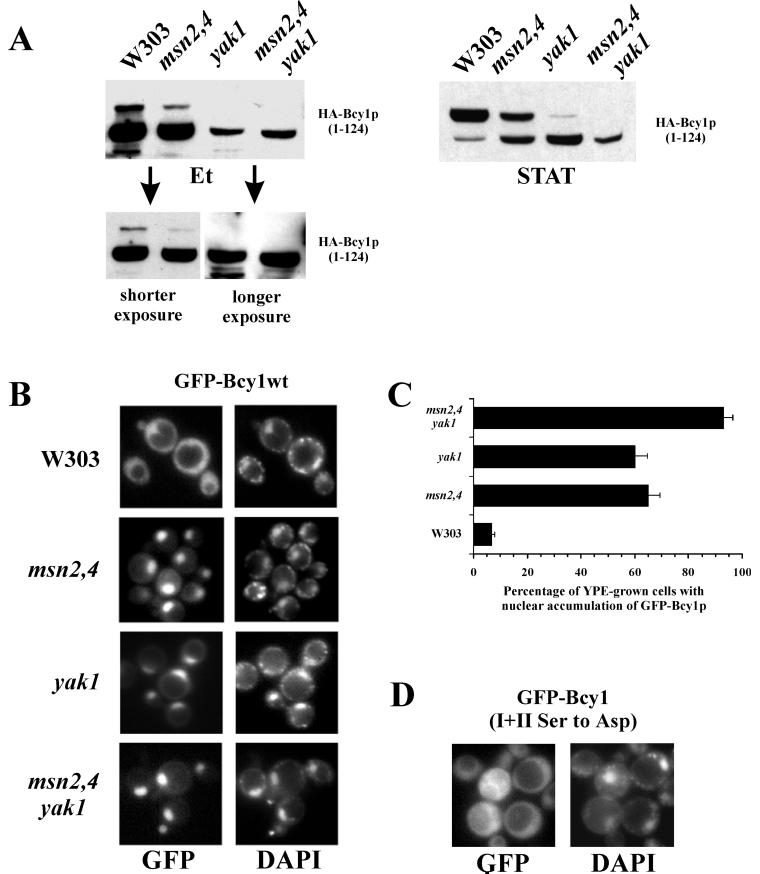
We wished to determine whether the Yak1-dependent modification of Bcy1 reported earlier is observed in the genetic background of our strains and under our experimental conditions. The results of Western analysis of extracts from the different mutants isolated from cells grown on ethanol or from stationary-phase expressing the N-terminal domain of Bcy1 are shown in Fig. 4A. As judged from the migration patterns, all of the mutant strains tested here, particularly the yak1 mutants, exhibit reduced modification compared to the wild type, both in ethanol-grown and in stationary-phase cells. Similar to what has been observed previously (35), we note that the levels of HA-Bcy11–124 in the mutant strains, especially in ethanol-grown cells, appear to be lowered. To rule out the possibility that slower-migrating isoforms are not detected because of the reduced overall levels of HA-Bcy11–124 (Fig. 4A, left upper panel), exposures are shown that exhibit the main HA-Bcy11–124 band at equal intensities (Fig. 4A, lower panel). It is obvious from these exposures that the relative levels of the slower-migrating isoform is significantly reduced or even not detectable in the mutants. Moreover, the migration of the main HA-Bcy11–124 band isolated from the mutant cells (especially from the msn2 msn4 yak1 cells) was slightly enhanced. The relative levels of slower-migrating isoforms are also significantly reduced in mutants grown to stationary phase (Fig. 4A, right panel). We investigated whether localization of Bcy1 is affected in strains with compromised Yak1 activity grown on ethanol (Fig. 4B and C). We observed in an msn2 msn4 deletion strain an increased nuclear localization of GFP-Bcy1wt. This effect appeared to be mainly dependent on Yak1 since a similar increase of nuclear accumulation was observed in a yak1 deletion strain. In a msn2 msn4 yak1 triple mutant, the extent of nuclear accumulation was further increased, suggesting that, apart from Yak1, other factors, whose expression is dependent on msn2 msn4, might be involved in controlling Bcy1 localization. In the stationary phase, however, these genomic deletions, as in the case of the Ser-to-Ala substitutions of clusters I and II, did not result in a different localization of GFP-Bcy1wt (data not shown), indicating that mechanisms independent of the activity of Msn2 and Msn4 or Yak1 are also involved in cytoplasmic localization. Importantly, in the msn2 msn4 yak1 strain grown on ethanol, GFP-Bcy1(Ser cluster I+II Asp) was evenly distributed over the nucleus and cytoplasm (Fig. 4D). This observation provides strong evidence that the nuclear concentration of GFP-Bcy1wt observed in Yak1-deficient strains is due to reduced modification of cluster I and/or cluster II serines and not to indirect effects caused by deletion of the genes.
FIG. 4.
Localization and modification of Bcy1 in different yeast mutants. (A) Western analysis of extracts isolated from mutant cells transformed with 33AGHB1–124. Prior to harvesting, cells were grown on ethanol (Et) or on YPD to stationary phase (STAT). Different exposures of the Western blot carried out with extracts of ethanol-grown cells are shown. (B) Fluorescence microscopy of ethanol-grown cells transformed with 195A2-pBGHBwt. Relevant genotypes of the different strains are indicated at the left. (C) Quantification of the localization pattern of the different strains shown in panel B. The percentage of ethanol-grown cells with stronger nuclear fluorescence compared to the cytoplasmic fluorescence was assayed. Three independent transformants were assayed at least three times each (at least 100 cells were counted for each determination). The error bars indicate the standard deviation. (D) Fluorescence microscopy of ethanol-grown msn2 msn4 yak1 mutant cells transformed with 195A2-pBGHB(Ser cluster I+II Asp).
Altogether, the data presented here show that modification of Bcy1 is strongly dependent on Yak1 and that for this reason Bcy1 is found preferentially in the nucleus of ethanol-grown cells in the absence of Yak1 kinase activity. Other mechanisms appear to play a role in stationary-phase cells.
Zds1 is required for cytoplasmic localization of GFP-Bcy1.
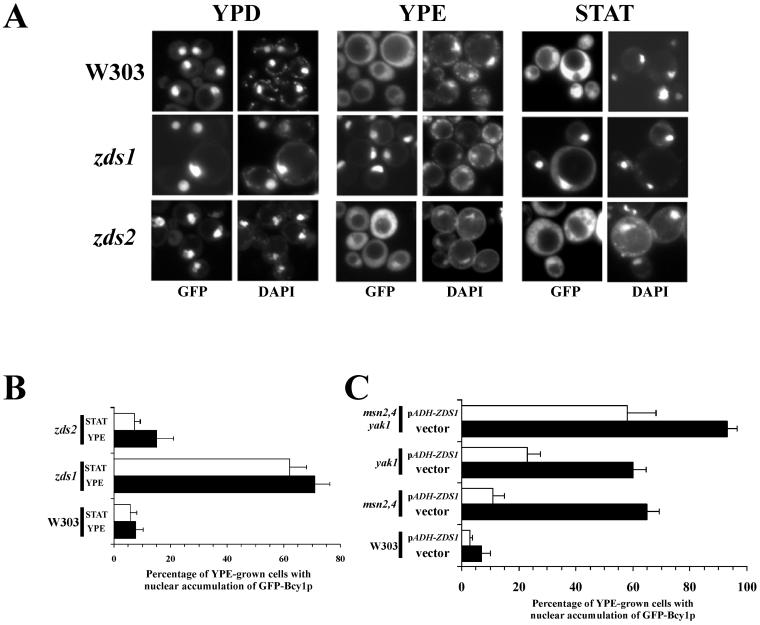
In mammalian cells, AKAPs have been identified that function as adaptor molecules directing RI or RII regulatory subunits to specific subcellular structures by interacting with their N-terminal domains (5). We hypothesized that functionally related proteins might exist that regulate the localization of Bcy1 in budding yeast. To identify proteins that interact with the N-terminal targeting region of Bcy1, we performed a two-hybrid screen. In this way we isolated a gene fragment encoding the C-terminal part (amino acids 781 to 942) of Zds2 (data not shown). ZDS2 has a close homolog, ZDS1. Especially the C-terminal part of Zds1 has very high homology to Zds2 (3, 36). To determine whether Zds1 or Zds2 affect the localization of Bcy1, we performed fluorescence microscopy with zds1 and zds2 mutants producing GFP-Bcy1wt (Fig. 5A and B). In glucose-grown cells, no effects of ZDS1 or ZDS2 deletion on the localization of GFP-Bcy1wt were observed. In ethanol-grown or in stationary-phase zds1 cells, however, GFP-Bcy1wt was found concentrated in the nucleus with no or low levels of cytoplasmic GFP-Bcy1wt present, indicating that Zds1 is involved in the cytoplasmic targeting of Bcy1. It should be pointed out that deletion of ZDS1 does not lead to a complete absence of cytoplasmic GFP-Bcy1. Quantification of the localization patterns of zds1 cells (Fig. 6B) revealed that in about 30% of the ethanol-grown or stationary-phase cells GFP-Bcy1 distribution was found to be unaffected, indicating that mechanisms independent of Zds1 also play a role in localization of GFP-Bcy1. In zds2 cells the effect on localization was found to be much weaker. Since zds1 zds2 double-deletion strains are unable to grow on nonfermentable carbon sources, we could not study the effect on localization of such a mutant in ethanol-grown or in stationary-phase cells. Since the absence of Zds1 results in increased nuclear accumulation of GFP-Bcy1wt in ethanol-grown cells, we tested whether Zds1 overproduction would result in the opposite effect. To this end, we ectopically expressed ZDS1, using the ADH1 promoter, in the mutant strains studied in the experiment illustrated in Fig. 4. The effects on GFP-Bcy1wt localization were determined (Fig. 5C). Overproduction of Zds1 led to an increase in cytoplasmically localized GFP-Bcy1wt, particularly in the msn2 msn4 strain (Fig. 5C).
FIG. 5.
Localization of GFP-Bcy1 as a function of Zds1 or Zds2. (A) Fluorescence microscopy of zds mutants transformed with 195A2-pBGHBwt. Fluorescence was determined from cells grown on YP plus glucose (YPD) or ethanol (YPE) and from cells in stationary phase (STAT). The relevant genotypes of the different strains are indicated. (B) Quantification of the localization pattern of the different strains shown in panel A. The percentage of ethanol-grown cells with stronger nuclear fluorescence compared to the cytoplasmic fluorescence was determined. Three independent transformants were assayed at least three times each (at least 100 cells were counted for each determination). The error bars indicate the standard deviation. (C) Quantification of the results of fluorescence microscopy with ethanol-grown cells transformed with 195A2-pBGHBwt. Relevant genotypes of the different strains are indicated at the left. Cells were cotransformed with either control plasmid pRS316 (indicated by vector, black bars) or plasmid 316pADH-ZDS1 (indicated by pADH-ZDS1, open bars), the latter encoding Zds1, whose production is regulated by the ADH1 promoter. Three independent transformants were assayed at least three times each (at least 100 cells were counted for each determination). The error bars indicate the standard deviation.
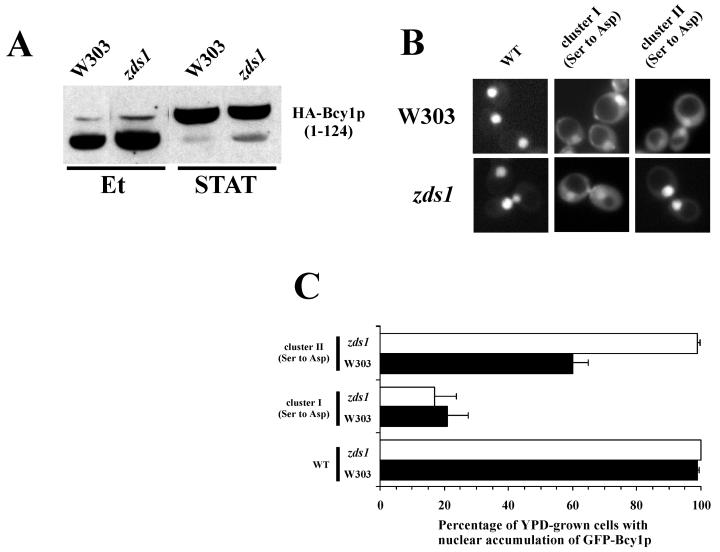
FIG. 6.
Functional interactions of Zds1 with Bcy1 versions mutated in their N-terminal domains. (A) Western analysis of extracts isolated from wild-type and zds1 cells transformed with 33AGHB1–124. Prior to harvesting, cells were grown on ethanol (Et) or YPD to stationary phase (STAT). (B) Fluorescence microscopy of glucose-grown W303-1A and zds1 cells transformed with 195A2-pBGHBwt, 195A2-pBGHB(Ser cluster I Asp), and 195A2-pBGHB(Ser cluster II Asp) encoding GFP fused to Bcy1, Bcy1(Ser cluster I Asp), and Bcy1(Ser cluster II Asp), respectively. (C) Quantification of the localization pattern shown in panel B. The percentage of glucose-grown W303-1A (black bars) and zds1 cells (open bars) with stronger nuclear fluorescence relative to cytoplasmic fluorescence was determined. Three independent transformants were assayed at least three times each (at least 100 cells were counted for each determination). The error bars indicate the standard deviation.
It is noteworthy that the efficiency of cytoplasmic localization of GFP-Bcy1 as a consequence of ZDS1 overexpression was found to be different in the mutants tested. The cytoplasmic localization of GFP-Bcy1 appears to be inversely correlated to the extent of affected HA-Bcy1 modification in the different mutant strains (Fig. 4A). This may suggest that phosphorylation of Bcy1 stimulates its Zds1-mediated cytoplasmic localization (see also below). Consistent with this hypothesis, in glucose-grown cells, a condition where Bcy1 is supposedly not modified, no significant effect of ZDS1 overexpression on GFP-Bcy1 localization was observed (data not shown).
Zds1-mediated cytoplasmic localization of GFP-Bcy1 is dependent on cluster II serines.
Above, we present evidence that modification of the Bcy1 N-terminal domain is an important requirement for cytoplasmic localization. Since Zds1 mediates the cytoplasmic recruitment of Bcy1, we tested whether modification of Bcy1 is affected by deletion of ZDS1. However, no migration differences of HA-tagged Bcy11–124 could be observed between wild type and a zds1 mutant in both ethanol-grown and stationary-phase cells (Fig. 6A), indicating that the Zds1 effect on GFP-Bcy1wt localization is not caused by a changed modification pattern. Furthermore, we investigated whether the cytoplasmic targeting of the GFP-Bcy1 alleles bearing cluster I or II Ser-to-Asp substitutions is affected by the deletion of zds1. We compared the localization patterns of GFP-Bcy1(Ser cluster I Asp) and GFP-Bcy1(Ser cluster II Asp) in glucose-grown wild-type and zds1 mutant cells (Fig. 6B and C). Cytoplasmic localization of GFP-Bcy1(Ser cluster I Asp) was found to be unaffected in zds1 mutant cells, but GFP- Bcy1(Ser cluster II Asp) displayed enhanced nuclear accumulation in zds1 mutant cells compared to wild-type cells. This result indicates that Zds1-mediated cytoplasmic localization of GFP-Bcy1wt is dependent on the phosphorylation of cluster II serines but not of cluster I serines.
The results of a two-hybrid analysis of an interaction between Bcy1 and Zds1 are consistent with these findings (data not shown). Altogether, the data obtained indicate that Zds1-mediated cytoplasmic localization of Bcy1 is regulated by cluster II serines. Cluster I-dependent localization appears to be independent of Zds1.
DISCUSSION
In previous studies, it has been demonstrated that the localization of the S. cerevisiae PKA subunit Bcy1 is carbon source regulated (12). Cytoplasmic localization of Bcy1 in carbon source-derepressed cells appears to be correlated with extensive modifications observed in such cells (35). In the present investigation, we have studied the nature of these modifications, whether modification and cytoplasmic localization are functionally linked, and which trans-acting factors are involved in control of Bcy1 localization.
Phosphorylation of the Bcy1 N terminus regulates its localization.
The following experimental data suggest a causal relationship between Bcy1 phosphorylation and its accumulation in the cytoplasm. (i) A major part of the phosphorylation observed in carbon source-derepressed cells can be localized to its N-terminal 124 amino acid residues, which are necessary and sufficient for proper nutrient-regulated localization (12). (ii) Fractionation experiments demonstrated that modified and unmodified Bcy1 are differentially localized. In the cytoplasmic fraction, phosphorylated Bcy1 is predominantly found, while unmodified Bcy1 is preferentially nuclear. (iii) Site-directed mutagenesis led us to identify serine residues, located in two different clusters, that are important for modification and subcellular distribution of Bcy1. While Ser-to-Ala mutations enhanced nuclear localization in ethanol-grown cells, Ser-to-Asp substitutions of cluster I and/or cluster II serines, presumably mimicking phosphorylated serine residues, were sufficient for cytoplasmic localization in glucose-grown cells.
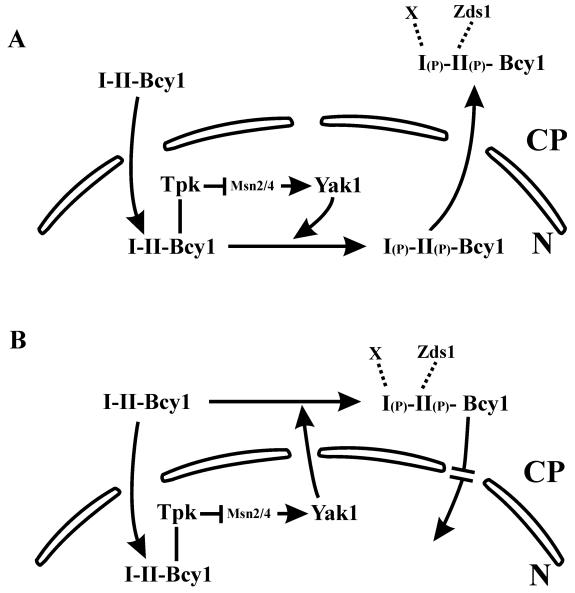
Based on these data we propose a working model (Fig. 7) that includes putative molecular mechanisms of regulated Bcy1 localization. Phosphorylation of Bcy1 may stimulate its nuclear export (Fig. 7A), but it does not cause its nuclear exclusion (Fig. 3E). Alternatively, phosphorylated Bcy1 synthesized de novo might be retained preferentially but not exclusively in the cytoplasm, or it may partly block its nuclear import (Fig. 7B).
FIG. 7.
Working model of localization of Bcy1 regulated by phosphorylation. Phosphorylation of cluster I or II serines is required for cytoplasmic localization of Bcy1 and is regulated by Yak1. Phosphorylation may stimulate export of nuclear Bcy1 (model A). Alternatively, phosphorylation of de novo-synthesized Bcy1 might trigger cytoplasmic retention or may inhibit its nuclear import (model B). Cluster II-mediated cytoplasmic localization of Bcy1 requires Zds1. A functional interaction between Bcy1 and Zds1 is dependent on phosphorylation of cluster II serines but not of cluster I serines. PKA (Bcy1-Tpk1) controls its regulatory subunit localization. Expression of Yak1 is activated by Msn2 and Msn4, two transcription factors negatively regulated by PKA. In the presence of glucose, PKA activity is high, resulting in low YAK1 expression. During growth on nonfermentable carbon sources, lower PKA activity allows enhanced expression of YAK1, presumably resulting in high Yak1 kinase activity, leading to Bcy1 phosphorylation. Abbreviations: CP, cytoplasm; N, nucleus; I, Bcy1 cluster I (amino acid residues 3 to 9); II, Bcy1 cluster II (amino acid residues 74 to 84); (P), phosphate; X, hypothetical factor interacting with phosphorylated cluster I.
PKA controls Yak1-dependent modification, and consequently localization, of Bcy1.
Bcy1 modification is dependent on a functional YAK1 gene (35). Here we obtained evidence that (i) YAK1-dependent modifications are localized in the N-terminal Bcy1 domain and are dependent on the PKA-inactivated transcription factors Msn2 and Msn4, which activate YAK1 expression (27), and (ii) the Bcy1 modification defect in msn2 msn4 and yak1 strains and in the respective triple mutant is correlated with a defect in cytoplasmic Bcy1 localization in ethanol-grown cells, whereas constitutive cytoplasmic localization of the cluster I plus cluster II S-D mutant version of Bcy1 is not impaired in such mutants.
Overall, these data lend support to the model depicted in Fig. 7, wherein PKA controls localization of its own regulatory subunit Bcy1 by negative regulation of Msn2 and Msn4: low PKA activity allows activation of Msn2 and Msn4, which will lead to enhanced Yak1 expression (27). Experiments with mutants with conditionally compromised PKA activity supported this model: a decreased cAMP level in adenylate cyclase mutants led to cytoplasmic localization of Bcy1 in glucose-grown cells (unpublished results). Importantly, this increase in cytoplasmic Bcy1 after lowering cAMP-levels was found to be dependent on Msn2 and Msn4. The underlying molecular mechanism of the function of Yak1 in this process remains to be established, but no evidence for the phosphorylation of the Bcy1 N-terminal domain in vitro has yet been obtained (unpublished results). Yak1 may activate protein kinases or downregulate phosphatases controlling the phosphorylation state of clusters I and II. Consistent with this notion, cluster I and II serines differ dramatically in their sequence context, suggesting that different kinases are responsible for the phosphorylation of serine residues in the two clusters.
Zds1 is required for the cytoplasmic localization of Bcy1 in carbon source-derepressed cells.
Our data provide evidence that Zds1 controls the subcellular localization of Bcy1. We observed that Bcy1 is largely absent from the cytoplasm of ethanol-grown zds1 mutant cells, in contrast to the situation in wild-type cells. Consistent with a regulatory role of Zds1 in the localization of Bcy1, the overproduction of Zds1 resulted in an increase of cytoplasmic Bcy1. The cytoplasmic localization of a Bcy1 version bearing S-D substitutions in cluster I appeared to be unaffected in a zds1 mutant, whereas the cytoplasmic localization of a cluster II S-D mutant version was found to be dependent on Zds1. Altogether, our results are consistent with a working model of Zds1-mediated recruitment of Bcy1 to the cytoplasm controlled by the phosphorylation of cluster II serines (Fig. 7). In this scenario, Zds1 may act as a cytoplasmic retention factor for Bcy1. Consistent with such a model, subcellular localization studies revealed that, apart from the concentration of Zds1 at the bud neck and bud tips reported earlier (3), Zds1 is also found in the cytoplasm (data not shown). In addition, two-hybrid analysis provided evidence for a weak Bcy1-Zds1 interaction (data not shown).
Comparison of mechanisms of PKA localization in yeast and in mammalian cells.
In budding yeast, mechanisms of PKA-regulatory subunit localization appear to be different from those acting in mammalian cells. Here we show that Bcy1 responds to extracellular nutritional signals that regulate the phosphorylation status of its N-terminal domain and consequently its subcellular distribution. This is a mechanism for controlling PKA-regulatory subunit localization not described for other eukaryotes. We identified two distinct regions in the Bcy1 N-terminal domain that control localization by different molecular mechanisms, whereas in mammalian RI or RII, principally one domain (the AKAP-binding region) is known to be involved in localization. Moreover, no AKAP homologues have been identified in yeast (5; unpublished results).
These differences seem to be reflected in the primary structure of the Bcy1 N-terminal domain. In RII, the first 23 residues form an AKAP-binding surface (21). Alignment studies of Bcy1 with RII revealed that the region in Bcy1 corresponding to the RII AKAP-binding domain is very poorly conserved, and critical residues involved in RII-AKAP interaction (14) are not present (unpublished observations). Despite the fact that no proteins have yet been identified that may interact with the extreme N-terminal domain of Bcy1 (factor X in Fig. 7), previous genetic studies revealed that residues 1 to 48 are (partially) sufficient for the nutrient-regulated localization of Bcy1 (12). In the present study we show that Ser residues at the extreme N-terminal domain (cluster I) are involved in this process. Therefore, functionally equivalent but structurally different analogue(s) of AKAPs might exist in yeast.
Our findings indicate fundamental differences between the molecular mechanism of AKAP-mediated RII localization and that of Zds1-dependent cytoplasmic localization of Bcy1. Zds1-mediated localization of Bcy1 is dependent on cluster II serines (residues 74 to 84) and not on its extreme N-terminal domain. In addition, a putative Zds1-Bcy1 interaction appears to be weak (or perhaps indirect) but dynamic and regulated by phosphorylation in response to extracellular nutritional signals, whereas RII-AKAP interaction is avid and considered to be constitutive (4, 5, 21). We therefore propose that Zds1 could represent a novel type of PKA-targeting protein.
Concluding remarks.
It has been described previously that mislocalization of Bcy1 results in reduced viability in the stationary phase, decreased sporulation efficiency, and delayed reproliferation of stationary-phase cells (12). The viability of cluster substitution mutants described in the present study was also found to be affected (data not shown), indicating that phosphorylation of the cluster serines is involved in a normal stationary-phase response. However, the underlying molecular mechanisms concerning the role of Bcy1 phosphorylation and localization and the function of Zds1 in these processes are not understood and require further studies.
Zds1 and its paralogue Zds2 have been identified in a wide variety of genetic screens, implying that these are multifunctional proteins involved in a large number of seemingly unrelated intracellular processes (3, 17, 20, 23–25, 34, 36). The role of Zds1 and Zds2 in transcriptional silencing might be the best-characterized phenotype at present. Zds1 and/or Zds2 were found to interact with Rap1, Sir2, Sir3 and Sir4 (24) (members of a nucleoprotein complex involved in transcriptional silencing [13]), and the deletion or overexpression of ZDS1 or ZDS2 affects transcriptional silencing (23). It remains to be seen whether Bcy1 phosphorylation and a putative interaction with Zds1 are connected mechanistically to transcriptional silencing or other Zds phenotypes. We note that, apart from a functional interaction of Zds1 and Zds2 with the Sir complex and Bcy1, Zds1 and Zds2 appear to interact also with protein kinases Snf1 and Pkc1, respectively (3, 33). This might be an interesting analogy with mammalian AKAPs, since some of these not only interact with RII but also appear to be multivalent platforms for a wide variety of signaling molecules. By way of analogy, Zds1 might be part of a multiprotein complex coordinating such cellular processes as signal transduction and transcription.
ACKNOWLEDGMENTS
We thank Andreas Hartig and Gustav Ammerer for critical reading of the manuscript and Mira Matz and Harald Nierlich for technical assistance.
This work was supported by the EU TMR network RYPLOS (contract number FMRX-CT96-0007), by grants P11303 and P13493 from the Fonds zur Förderung der wissenschaftlichen Forschung, Vienna, Austria, to H. R. and by a postdoctoral grant to J. N. from STINT (Stiftelsen för internationalisering av högre utbildning och forskning).
REFERENCES
- 1.Ausubel F M, Brent B, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Green Publishing Associates and Wiley Interscience; 1993. [Google Scholar]
- 2.Beebe S J. The cAMP-dependent protein kinases and cAMP signal transduction. Semin Cancer Biol. 1994;5:285–294. [PubMed] [Google Scholar]
- 3.Bi E, Pringle J R. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bregman D B, Bhattacharyya N, Rubin C S. High affinity binding protein for the regulatory subunit of cAMP-dependent protein kinase II-B. Cloning, characterization, and expression of cDNAs for rat brain P150. J Biol Chem. 1989;264:4648–4656. [PubMed] [Google Scholar]
- 5.Colledge M, Scott J D. AKAPs: from structure to function. Trends Cell Biol. 1999;9:216–221. doi: 10.1016/s0962-8924(99)01558-5. [DOI] [PubMed] [Google Scholar]
- 6.Edwards A S, Scott J D. A-kinase anchoring proteins: protein kinase A and beyond. Curr Opin Cell Biol. 2000;12:217–221. doi: 10.1016/s0955-0674(99)00085-x. [DOI] [PubMed] [Google Scholar]
- 7.Francis S H, Corbin J D. Structure and function of cyclic nucleotide-dependent protein kinases. Annu Rev Physiol. 1994;56:237–272. doi: 10.1146/annurev.ph.56.030194.001321. [DOI] [PubMed] [Google Scholar]
- 8.Francois J, Van Schaftingen E, Hers H G. The mechanism by which glucose increases fructose 2,6-bisphosphate concentration in Saccharomyces cerevisiae. A cyclic-AMP-dependent activation of phosphofructokinase 2. Eur J Biochem. 1984;145:187–193. doi: 10.1111/j.1432-1033.1984.tb08539.x. [DOI] [PubMed] [Google Scholar]
- 9.Garrett S, Broach J. Loss of Ras activity in Saccharomyces cerevisiae is suppressed by disruptions of a new kinase gene, YAK1, whose product may act downstream of the cAMP-dependent protein kinase. Genes Dev. 1989;3:1336–1348. doi: 10.1101/gad.3.9.1336. [DOI] [PubMed] [Google Scholar]
- 10.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 11.Görner W, Durchschlag E, Martinez-Pastor M T, Estruch F, Ammerer G, Hamilton B, Ruis H, Schüller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffioen G, Anghileri P, Imre E, Baroni M D, Ruis H. Nutritional control of nucleocytoplasmic localization of cAMP-dependent protein kinase catalytic and regulatory subunits in Saccharomyces cerevisiae. J Biol Chem. 2000;275:1449–1456. doi: 10.1074/jbc.275.2.1449. [DOI] [PubMed] [Google Scholar]
- 13.Grunstein M. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- 14.Hausken Z E, Coghlan V M, Hastings C A, Reimann E M, Scott J D. Type II regulatory subunit (RII) of the cAMP-dependent protein kinase interaction with A-kinase anchor proteins requires isoleucines 3 and 5. J Biol Chem. 1994;269:24245–24251. [PubMed] [Google Scholar]
- 15.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Ma X J, Lu Q, Grunstein M. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 1996;10:1327–1340. doi: 10.1101/gad.10.11.1327. [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Pastor M T, Marchler G, Schüller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 19.Miller J H. Experiments in molecular genetics Cold Spring Harbor Laboratory Press. N.Y: Cold Spring Harbor; 1972. [Google Scholar]
- 20.Mizunuma M, Hirata D, Miyahara K, Tsuchiya E, Miyakawa T. Role of calcineurin and Mpk1 in regulating the onset of mitosis in budding yeast. Nature. 1998;392:303–306. doi: 10.1038/32695. [DOI] [PubMed] [Google Scholar]
- 21.Newlon M G, Roy M, Morikis D, Hausken Z E, Coghlan V, Scott J D, Jennings P A. The molecular basis for protein kinase A anchoring revealed by solution NMR. Nat Struct Biol. 1999;6:222–227. doi: 10.1038/6663. [DOI] [PubMed] [Google Scholar]
- 22.Rose M, Winston F, Hieter P. Methods in yeast genetics, a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 23.Roy N, Runge K W. Two paralogs involved in transcriptional silencing that antagonistically control yeast life span. Curr Biol. 2000;10:111–114. doi: 10.1016/s0960-9822(00)00298-0. [DOI] [PubMed] [Google Scholar]
- 24.Roy N, Runge K W. The ZDS1 and ZDS2 proteins require the Sir3p component of yeast silent chromatin to enhance the stability of short linear centromeric plasmids. Chromosoma. 1999;108:146–161. doi: 10.1007/s004120050364. [DOI] [PubMed] [Google Scholar]
- 25.Schwer B, Linder P, Shuman S. Effects of deletion mutations in the yeast Ces1 protein on cell growth and morphology and on high copy suppression of mutations in mRNA capping enzyme and translation initiation factor 4A. Nucleic Acids Res. 1998;26:803–809. doi: 10.1093/nar/26.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith A, Ward M P, Garrett S. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 1998;17:3556–3564. doi: 10.1093/emboj/17.13.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor S S, Buechler J A, Yonemoto W. cAMP-dependent protein kinase: framework for a diverse family of regulatory enzymes. Annu Rev Biochem. 1990;59:971–1005. doi: 10.1146/annurev.bi.59.070190.004543. [DOI] [PubMed] [Google Scholar]
- 29.Thevelein J M, de Winde J H. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1999;33:904–918. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- 30.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 31.Toda T, Cameron S, Sass P, Zoller M, Scott J D, McMullen B, Hurwitz M, Krebs E G, Wigler M. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:1371–1377. doi: 10.1128/mcb.7.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toda T, Cameron S, Sass P, Zoller M, Wigler M. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell. 1987;50:277–287. doi: 10.1016/0092-8674(87)90223-6. [DOI] [PubMed] [Google Scholar]
- 33.Uetz P, Giot L, Cagney G, Mansfield T A, Judson R S, Knight J R, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg J M. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 34.Walowsky C, Fitzhugh D J, Castano I B, Ju J Y, Levin N A, Christman M F. The topoisomerase-related function gene TRF4 affects cellular sensitivity to the antitumor agent camptothecin. J Biol Chem. 1999;274:7302–7308. doi: 10.1074/jbc.274.11.7302. [DOI] [PubMed] [Google Scholar]
- 35.Werner-Washburne M, Brown D, Braun E. Bcy1, the regulatory subunit of cAMP-dependent protein kinase in yeast, is differentially modified in response to the physiological status of the cell. J Biol Chem. 1991;266:19704–19709. [PubMed] [Google Scholar]
- 36.Yu Y, Jiang Y W, Wellinger R J, Carlson K, Roberts J M, Stillman D J. Mutations in the homologous ZDS1 and ZDS2 genes affect cell cycle progression. Mol Cell Biol. 1996;16:5254–5263. doi: 10.1128/mcb.16.10.5254. [DOI] [PMC free article] [PubMed] [Google Scholar]