Abstract
Exosomes are extracellular vesicles with unique membrane markers and components that participate in cellular communication. The contents of exosomes, including growth factors, microRNAs, long noncoding RNAs, and circular RNAs (circRNAs), have been recognized as prognostic biomarkers and promote cancer progression through cancer cell growth, metastasis, angiogenesis, and cancer development. One of the components of exosomes, circRNAs, are covalently closed and prevented from degrading, which results in their continually accumulating in exosomes. Evidence suggests that exosomal circRNAs are abundant and stable in body fluids and have been implicated in many diseases. In this article we summarize the biogenesis and function of circRNAs and explore the expressions of exosomal circRNAs in cancer, emphasizing the fact that exosomal circRNAs are a novel diagnostic biomarker in the early stages of cancer and/or a therapeutic target in further cancer treatment.
Keywords: Exosomes, exosomal circRNAs, cancers, biomarkers
Introduction
Exosomes were first found in sheep reticulocytes in 1983 and named in 1987 by Pan and Johnstone [1,2]. They are the smallest extracellular vesicles (30-150 nm in diameter), with a phospholipid bilayer structure, specific surface molecules (CD9 and CD63), and inner components, and they can be secreted by most eukaryotic cells in physiological and pathological conditions [3]. Growing evidence suggests that exosomes represent a novel type of cellular communication in the context of cancer through their bioactive cargos, including proteins [4-6], lipids [7], mRNAs [5], miRNAs [5], and circRNAs [8].
CircRNAs, a novel class of endogenous non-coding RNAs, were discovered in 1993 and are considered by-products of linear RNA erroneously splicing [9]. However, through functional research and the help of deep sequencing and transcriptome technologies, circRNAs show a widespread expression and functional pattern in mammalian species. With the function of miRNA sponging, protein interaction, transcriptional regulation, and translation regulation, circRNAs play an essential role in cancer. Through the vehicle of the exosome, circRNAs are transported to cells in the tumor microenvironment, affecting tumorigenesis, progression, invasion, metastasis, the epithelial-mesenchymal transition, and apoptosis in various mechanisms. Due to the covalently closed-loop structure generated through back-splicing without 5’caps and 3’tails [10,11], circRNAs are resistant to ribonuclease R (RNase R) and abundantly stable in the body fluid with a long half-life [12,13]. Emerging evidence suggests that the content of circRNAs is about ten times richer than the corresponding linear RNAs [14,15], and a large number of circRNAs exist in exosomes [8]. In addition, the exosomal circRNAs expression patterns are consistent with the corresponding tissues and cells, and they can serve a biological markers in various cancers diagnoses [16,17].
In this review, we summarize the functions of exosomal circRNAs in various cancers, raise the question of exosomal circRNAs translocation to nuclear, clarify the biomarker function in cancer detection and the underlying mechanism of exosomal circRNAs in promoting invasion, migration, metastasis, and drug resistance, and discuss clinical translation relevance-especially the potential of using liquid biopsies in the future.
Biogenesis of circRNAs
Hsu and Coca-Prados examined the circular structure of RNA in the cytoplasms of eukaryotic cells using electron microscopy in 1979 [18]. Traditionally, circRNAs are transcribed by RNA polymerase II and derived from precursor messenger RNAs (pre-mRNAs) through back-splicing (Figure 1) [19]. Alternative splicing and alternative circulation can produce tissue-specific circRNAs in the different cell cycles [15,20,21]. According to the structural components and the circulation mechanisms, there are five categories: exonic circRNAs (ecircRNAs), intronic circRNAs (ciRNAs), exonic-intronic circRNAs (EIciRNAs), tRNAs (tricRNAs), and intergenic circRNAs or fusion circRNAs (f-circRNAs) [22-26]. The term “circRNAs” mainly refers to the ecircRNAs located in the cytoplasm [27]. Intronic circRNAs and exonic-intronic circRNAs predominantly gather in the eukaryotic cell nuclei that can interact with U1 snRNP and promote the transcription of their parental genes [28].
Figure 1.
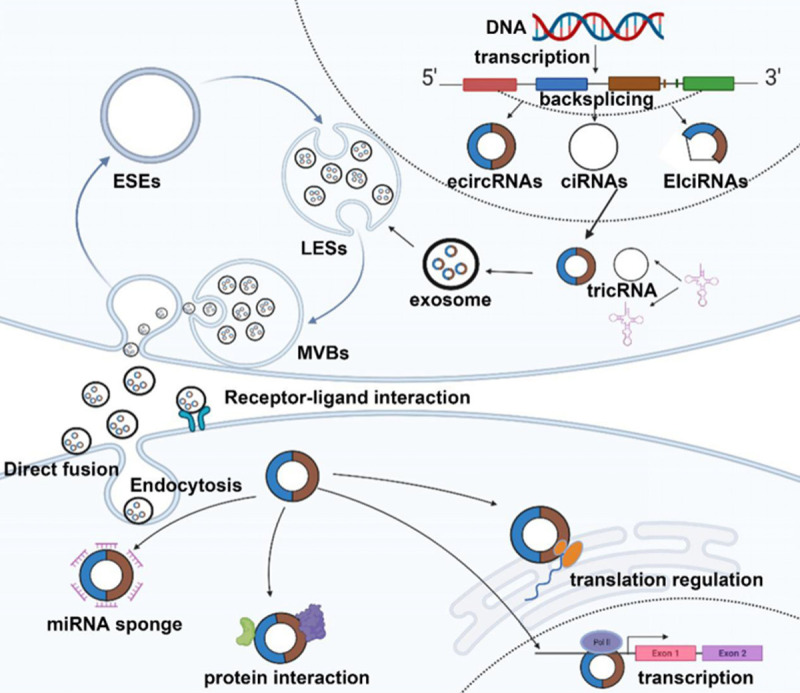
Biogenesis and delivery of exosomal circRNAs. CircRNAs are generated from pre-mRNA by back-splicing. According to the structural component, circRNAs are mainly divided into three types, including ecircRNAs, ciRNAs, and EIciRNAs. EcircRNAs are located in the cytoplasm and exert a regulatory function at the post-transcriptional level, while ciRNAs and EIciRNAs gather in the nuclei and interact with U1 snRNP to dominate the transcription. Exosomes are nanoscale extracellular vesicles of endocytic origin. The early endosome dissociates in the cytoplasm by folding the plasma membrane. With the help of the Golgi apparatus, the early endosome develops into the late endosome through the folding of the endosomal membrane. Vesicles accumulate in the late endosome to generate MVBs. MVBs are released in vitro by fusing with the plasma membrane to form exosomes. In this process, proteins, lipids, miRNA, mRNA, and non-coding RNAs are packaged into vesicles. Exosomes transfer circRNAs to recipient cells through direct fusion, receptor-ligand interaction, and endocytosis to perform the miRNA sponging, protein interaction, transcription, and translation regulation functions.
The biogenic process of circRNAs is ambiguous. However, three relatively mature models that permit the back-splicing procession have been broadly acknowledged: intron-pairing-driven circularization [15], lariat-driven circularization (also called exon-skipping) [29,30], and RNA-binding proteins (RBPs) that induce circularization [19,31,32]. Zhang discovered that the ALU repeat sequence can promote the base pairing of the intron to induce circularization [27,33,34]. Supposing that there are some particular nucleic acid sequences in introns, such as the seven nucleotides GU-rich motif near the 5’splice site and the 11 nucleotides C-rich motif near the branching point, the introns will escape the activity of the debranching enzyme and form circRNAs [24]. Two intronic circRNA fragments (ICFs) are bound by the GT-AG splicing signals, forming intergenic circRNA [35]. Another subtype of circRNA named tricRNAs comes from the precursor tRNA [26,36]. Also, the head and tail genes of the exons from different genes fuse using chromosomal translocations, thus giving rise to f-circRNAs [23]. The origination of the circRNAs is mainly caused by back-splicing that brings the downstream splice donor into proximity to the upstream splice acceptor, which is controlled by cis and trans-acting elements [13,37].
Functions of circRNA
A sponge of miRNA
MiRNA, an endogenous non-coding RNA with a length of 20-25 nucleotides, can bind to the miRNA response elements (MREs) sequence of mRNA and the suppression or activation target genes at the post-transcriptional level [38,39]. CircRNAs can act as competitive endogenous RNAs (ceRNAs) to regulate the expression of miRNA in various tumors [40-42] through absorbing miRNA with MREs and influence the target gene through its tumor suppressor or oncoprotein functions [43]. For example, MiR-7 is a tumor-suppressive miRNA in many malignant tumors. The circRNA, Cdr1as, harbors a total of 73 conserved miR-7 interaction sites [44], which keeps miR-7 stable and reverses the miR-7 target gene expression, activating the downstream signaling pathways to promote tumor proliferation, differentiation, migration, and invasion [29,45]. CircRNA-0000442 is down-regulated in breast cancer tissues compared to adjacent normal tissues. It restrains cancer growth by serving as a sponge for miR-148b-3p, contributing to PTEN inhibition and further PI3K/Akt activation [46]. In addition to the linear function model, circRNA and miRNA are network interplays for the multi-miRNA response elements found in single circRNA and multi-circRNA sponging for the same miRNA family [19,47]. A class of non-coding RNA established the network of circRNA, miRNA, and mRNA, for instance, the circUBE2D2/miR-512-3p/CDCA3 axis, the circRNF20/miR-487a/HIF-1α axis, and the circKDM4C/miR-548p/PBLD axis [48]; however, the precise regulatory mechanism remains unclarified.
Protein interaction
CircRNAs also have many binding sites for RNA-binding proteins (RBPs), which are also known as “protein decoys” with a scaffolding function [10]. Circ-Foxo3, generated from the forkhead box three genes, can interact with cyclin-dependent kinase inhibitors 1(p21) and cyclin-dependent kinase 2(CDK2) to form a circ-Foxo3-p21-CDK2 tertiary circRNAs protein complex to retard cell cycle progression [49,50]. CircMbl originates from the second exon of the splicing factor muscleblind (Mbl) with many Mbl binding sites [19]: the over-expression of Mbl, binding to the flanking introns, can promote the formation of circMbl and reduce the linear Mbl formed by conventional splicing [51]. Therefore, the circMbl levels are negatively correlated with mature linear mRNA [19]. CircACC1 can bind directly to the regulatory β and γ subunits, promoting AMPK holoenzyme activity [52]. In summary, circRNAs can directly or indirectly bind to the protein in an RNA-protein binding manner to activate or restrain the functions of those proteins.
Transcriptions regulation
CircRNAs regulate gene transcription and expression mainly through circRNA translocation to nuclear and binding to cis-regulatory elements (CREs). Previous research has shown that ciRNAs and EIciRNAs exist in the nuclei and regulate RNA polymerase II transcriptional activity (Pol II), thus controlling the transcription of the parental genes [28]. Ci-ankrd52 and Ci-sirt7 can interact with pol II to improve the expression of the parental gene [24]. In addition, Li Z et al. found that two exon-intron circRNAs, circEIF3J, and circPAIP2 can interact with U1 snRNP to form an RNA-protein complex, further connecting with RNA polymerase II (pol II) in the promoter region of the host gene to enhance transcription [28]. In general, the lines of evidence suggest that circRNA can control the expressions of the parent genes at the transcription and post-transcription levels [53]. However, the exosomal derived circRNA indirect nuclear transcription regulation remains unclear. MicroRNA and siRNA can translocate to nuclear through the importin and Argonaute mechanisms [54]. There is still a lack of clear conclusive evidence that exosomal circRNA are translocated to recipient cells’ nuclei even after the Argonaute binding possibility was found [44,55]. This fundamental evidence of RNA biology provides a possibility that exosomal derived circRNA can also participate in transcription regulation in recipient cells potentially.
Translation regulation
It was considered that circRNAs were short of 5’-3’ polarity and polyadenylated tails and lacked internal ribosome entry sites, because circRNAs could not translate into proteins [56]. However, Chang-you Chen and Peter Sarnow demonstrated that the synthesized circRNAs containing internal ribosome entry sites could be translated into short proteins or peptides in 1995 [57]. After that, it was proved that more and more circRNAs could be translated into proteins or peptides [58]. The translation mechanisms of circRNAs are divided into two types: dependent internal ribosome entry sites and independent internal ribosome entry sites [59,60]. For example, circSHPRH contains a small open reading frame driven by an internal ribosome entry site, translated into a 17kDa SHPRH-146aa protein in the internal ribosome entry site-dependent machinery, acting as a tumor suppressor in Glioblastomas [61]. While some circRNAs are translated into peptides through internal ribosome entry sites in an independent manner. Those circRNAs recruit YTHDF3 through the m6A modification and then combine with translation initiation factor eIF4G2 to initiate translation [62,63]. These findings show that circRNAs can take place in protein translation by IRES or m6A, but the roles of those peptides are still unknown.
Immunity moderation
CircRNAs have recently been implicated in the differentiation of immune cells and regulate the immune response. Macrophages express Circ-RasGEF1B upregulated under the stimulation of lipopolysaccharide (LPS) and positively regulate the expression of intercellular adhesion molecule 1 (ICAM-1) through the LPS/toll-like receptor 4 (TLR4) signaling pathway [64]. ICAM-1 is an adhesion molecule expressed by endothelial cells and is crucial for promoting lymphocyte homing and inflammation [65,66]. By regulating the stability of mature ICAM-1 mRNA, the endothelial and immune cells increase the cell-cell and cell-matrix interactions to promote antigen presentation and signal transduction [67]. Upon viral infection, the 2’-5’-oligoadenylate synthetase (OAS) is activated, which stimulates endonuclease RNase L to degrade the endogenous circRNAs, releasing PKR to bind with the virus dsRNA to inhibit replication [68]. In addition, those exogenous circRNAs without m6A modification are recognized by the immune receptor RIG-I, which triggers the immune response. However, the mechanism by which RIG-I recognizes the exogenous circRNAs is still not clear [69].
The formation and function of exosomal circRNAs in cancer
Exosomes are intraluminal vesicles (ILVs) generated within the payload that sprout inward containing components (nucleic acids, proteins, lipids, etc.) (Figure 1). Due to the invagination of the plasma membrane, the early endosomes dissociate in the cytoplasm [2]. The ILVs are accumulated in the late endosomes to generate multivesicular bodies (MVBs) [70]. Then the MVBs fuse with the cell membrane and release ILVs (exosomes) into the external environment [71]. As components of the exosome, the circRNAs exist stably in exosomes, resulting in highly abundant exosomes. However, different or tissue-specific circRNAs are found in the exosomes originating from the different cells, indicating that the sorting of circRNAs is selective and not random [72]. For example, hnRNPA2B1 can transport miRNA and lncARSR into vesicles by combining with exosome motifs [6,73]. It is speculated that exosomes may be with characteristic motifs that can combine with circRNAs to transfer them into vesicles. Li Y et al. introduced the ectopic expression of miR-7 into cells to down-regulate CDR1as/cirS-7 in exosomes, so the miRNA levels in the production cells can affect the sorting of circRNAs [74]. In addition, cooperation between the endosomal sorting complex required for transport (ESCRT) -0, -I, -II, and -III protein complexes promote the sorting of cargoes, the accumulation of ILVs, and the release of MVBs [75,76]. Other ESCRTs such as Alix and vps4A are expressed on the membrane of MVBs and contribute to the sorting of miRNA [77,78].
Accumulating evidence has shown that circRNAs play a critical role in the development of nervous system diseases [79], cardiovascular diseases [80], and cancer [23], especially in the processes of tumorigenesis [81], invasion [82], metastasis [83], angiogenesis [83], immune modulation [84] and drug resistance through protein-coding, miRNA sponging, and the cancer-related signaling pathways (Figure 2) [56]. Hsa_circ_0006528 [43], Hsa_circ_0001982 [85], and circ_ABCB10 [86] are upregulated in breast cancer to promote tumor growth and inhibit apoptosis. The knockdown of circDENND4C suppresses invasion and migration in breast cancer under hypoxia by increasing miR-200b and miR-200c [87]. CircPRRC2A promotes angiogenesis and metastasis through EMT and upregulates TRPM3 in renal cell carcinoma [88]. On the other hand, circ_0000442 [46], circ_Foxo3 [49], and circ_VRK1 [89] are down-regulated in breast cancer and decrease the sensitivity of cancer cells to chemotherapeutic drugs, playing tumor inhibitory roles.
Figure 2.
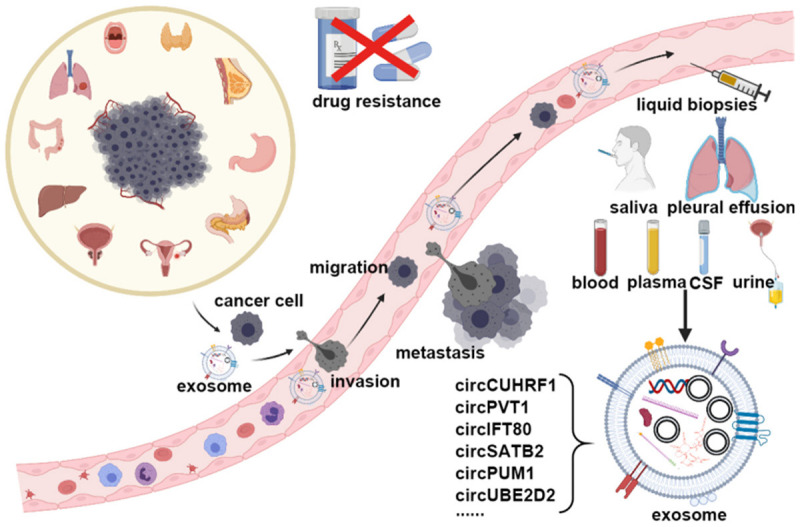
The pathway and detection of the exosomes. Cancer cells originating from different tissues can invade blood vessels and migrate to a particular destination with the help of exosomes and circRNAs. Then the cancer cells can settle in a new place, becoming a metastatic lesion. During those processes, exosomal circRNAs can be detected in the bodily fluids, such as the blood, saliva, pleural effusions, plasma, cerebrospinal fluid, and urine. Through liquid biopsies, the expressions of the circRNAs, for example, circCUHRF1, circPVT1, circIFT80, circSATB2, circPUM1, circUBE2D2 et al. can reflect cancer progression.
Exosomal circRNAs as a novel biomarker in cancer diagnosis and treatment
Cancer is a high-mortality disease characterized by dysregulation gene expression, genetic alternation, heterogeneity, invasiveness, and atypical clinical manifestations. Finding biomarkers with high sensitivity, high specificity, minimal invasiveness, and low cost is the basic requirement of liquid biopsies, especially in detecting cancer and finding the other risk factors contributing to cancer progression. Numerous works have shown that certain circRNAs are ectopically expressed in cancer cells and in the tumor microenvironment, promoting tumorigenesis and progression. Exosomal circRNAs exist stably in bodily fluids (Figure 2), and their phospholipid bilayer can protect circRNAs from degradation and exert their biological function through plasma membrane fusion, so the detection of exosomal circRNAs can be used as a new, minimally invasive biological marker in various cancers. CircRNA can be potentially employed in tumorigenesis detection in situ and in progression (Table 1).
Table 1.
Summary of the exosomal circRNAs in cancers
| Cancer type | Exo-circRNAS | Expression | Mechanisms | Sample type | lsolation method | Ref. |
|---|---|---|---|---|---|---|
| HCC | circ-DB | UP | circ-DB/miR34a/Usp7 | Medium and plasma | gradient centrifugation | [90] |
| circCUHRF1 | circCUHRF1/miR-449c-5p/TIM-3 | Serum and medium | ExoQuick | [92] | ||
| circTMEM45A | circTMEM45A/miR-665/IGF2 | plasma | ultracentrifugation | [95] | ||
| circ-100338 | mTOR signaling pathway | medium and plasma | ultracentrifugation | [83] | ||
| circ-0051443 | Down | Circ-0051443/miR-331-3p/BAKI | plasma | ExoQuick | [127] | |
| GC | ciRS-133 | UP | ciRS-133/miR-133/PRDM16 | medium and plasma | ultracentrifugation | [97] |
| circ-PVT1 | circ-PVT1/miR-30a-5p/YAP1 | serum | ExoQuick | [100] | ||
| CC | circFMN2 | UP | circFMN2/miR-1182/hTERT | serum | ultracentrifugation | [101] |
| circIFT80 | circIFT80/miR-1236-3p/HOXB7 | plasma | ultracentrifugation | [102] | ||
| ciRS-122 | ciRS-122/miR-122/PKM2 | medium and serum | ultracentrifugation | [105] | ||
| has_circ_0000338 | / | cell culture medium | QIAGEN exoRNeasy Midi Kit, ultracentrifugation | [106] | ||
| has_circ_0004771 | / | serum and medium | InvitrogenTM Total Exosome Isolation Kits | [104] | ||
| circPACRGL | circPACRGL/miR-142-3p/miR-506-3p/TGF- | plasma | ultracentrifugation | [4] | ||
| PC | circ-PDE8A | UP | circ-PDE8A/miR-338/MACC1/MET/AKT or ERK | plasma | ultracentrifugation | [107] |
| circ-IARS | circ-IARS/miR-122/RhoA | plasma | Total Exosome Isolation Kit | [108] | ||
| LC | circSATB2 | UP | circSATB2/miR-326/FSCN1 | cell culture medium | ultracentrifugation | [109] |
| has_circ_0002130 | has_circ_0002130/miR-498/GLIT1, HK2 and LDHA | Serum and medium | ultracentrifugation | [110] | ||
| has_circ_0001492 | / | plasma | Total Exosome Isolation Kit | [111] | ||
| has_circ_0001346 | / | plasma | Total Exosome Isolation Kit | [111] | ||
| OC | circWHSC1 | Up | circWHSC1/miR-145/MUC1 | cell culture medium | ultracentrifugation | [112] |
| circPUM1 | circPUM1/miR-6753-5p/MMP2; circPUM1/miR-615-5p/NF-B | cell culture medium | ultracentrifugation | [117] | ||
| BC | circ-UBE2D2 | Up | circ-UBE2D2/miR-200a-3p | cell culture media | ultracentrifugation | [128] |
| has_circRNA_0088088 | hsa-circRNA-0088088/let-7a-2-3p | cell culture media | Total Exosome Isolation Kit | [119] | ||
| circASS1 | circASS1/miR-443/Hippo signaling pathway | serum | / | [119] | ||
| has_circRNA_00005795 | Down | hsa-circRNA-0005795/hsa-miR-1304-3p or hsa-miR-3154 | cell culture media | Total Exosome Isolation Kit | [119] | |
| OSCC | circGDI2 | Down | circGDI2/miR-424-5p/SCAI | cell culture media | ExoQuick | [121] |
| PC | circ_0044516 | Up | circ_0044516/miR-29a-3p | / | / | [124] |
| TPC | has_circ_007293 | Up | / | serum | GSTM Exosome Isolation Reagent | [126] |
| has_circ_031752 | / | serum | GSTM Exosome Isolation Reagent | [126] | ||
| has_circ_020135 | / | serum | GSTM Exosome Isolation Reagent | [126] |
HCC, hepatocellular carcinoma, GC, gastric cancer, CC, colorectal cancer, PC, pancreatic cancer, LC, lung cancer, OC, ovarian cancer, BC, breast cancer, OSCC, oral squamous cell carcinoma, PC, prostate cancer, TPC, thyroid papillary carcinoma.
Hepatocellular carcinoma
Exosomal circ-deubiquitination (circ-DB) establishes a relationship between adipose tissue and hepatocellular carcinoma. Zhang et al. found that exosomal circ-DB secreted from adipose tissue can promote the progression of hepatocellular carcinoma and reduce DNA damage by reducing the miR-34a mediated USP7 suppression [90]. Also, the overexpression of circ-DB activates the USP7/cyclin-2 signal pathway, which promotes the synthesis of DNA in the S phase and transmits the cell cycle from the G2 phase to the M phase [91]. The knockout of exosomal circ-DB secreted by adipose tissue can inhibit the development of HCC. Reducing adipocytes may be an effective way to prevent HCC.
The serum exosomal circCUHRF1 expression is consistent with the HCC tumor load level. When the tumor is resected, the serum exosomal circCUHRF1 decreases [92]. In addition, there is a negative correlation between the serum exosomal circCUHRF1 level and the proportion of peripheral blood NK cells [93]. Exosomal circCUHRF1 acts as a sponge of miR-449c-5p to upregulate the expression of downstream TIM-3, which can reduce the anti-tumor immunity ability and the IFN-γ and TNF-α secretion functions of the NK cells to induce immune therapy failure [94].
Zhang et al. found that exosomal circTMEM45A plays a role in the progression of HCC through the circTMEM45A/miR-665/IGF2 axis [95]. Huang et al. had indicated that exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis by enhancing invasiveness and angiogenesis [83]. Sun et al. found the upregulation of hsa_circ_0004001, hsa_circ_0004123, and hsa_circ_0075792 in the plasma of hepatocellular carcinoma patients, which can be utilized as a diagnostic biomarker [96].
Gastric cancer
Serum exosomal ciRS-133 is significantly increased in patients with gastric cancer [97]. Luciferase assays show that the function of miR-133 is inhibited by being exposed to exosomal ciRS-133, which contains more than ten miR-133 binding sites. Exosomal ciRS-13 is transported into preadipocytes to induce energy consumption and weight loss through the miR-133/PRDM16 axis [98]. PRDM16, upregulated after the loss of miR-133, can induce the conversion of white fat to brown fat, which speeds up the body’s metabolism and energy expenditure [99]. The increased expression of PRDM16 leads to the browning of the white fat and the activation of UCP1 to promote the consumption of oxygen and glucose in brown fat and to energy metabolism disorders, leading to cachexia [97].
Exosomal circ-PVT1 promotes the progression, invasion, metastasis, and drug resistance of gastric cancer to cisplatin (DDP). Down-regulated exosomal circ-PVT1 can upregulate the expression of miR-30a-5p, negatively regulates the expression of its downstream target gene YAP1, and alleviates YAP1-mediated DDP resistance, reducing cell proliferation in gastric cancer [100].
Colorectal cancer
In colorectal cancer, the circRNA levels in the serum exosomes are related to the clinicopathological characteristics like tumor size, stage, and distant metastasis. The overexpression of exosomal hsa-circ-0005100 (circFMN2) and hsa_circ_0067835 (circIFT80) can promote the cell cycle from G0 to G1 and EMT through the circFMN2/miR-1182/hTERT axis and the circIFT80/miR-1236-3p/HOXB7 axis, leading to the progression of colorectal cancer [101,102]. Knockout circIFT80 can also induce cell apoptosis [103].
Several studies on colorectal cancer indicate that tumor-driven circRNA serves as a potential biomarker for auxiliary diagnosis. Pan B showed that tumor-derived exosomal hsa-circ-0004771 is used as a diagnostic marker for colorectal cancer [104]. Colorectal cancer-derived exosomal circPACRGL is significantly upregulated and promotes tumor growth, metastasis, and neutrophil N1-N2 differentiation through the miR-142-3p/miR-506-3p TGF-β1 axis [4]. Oxaliplatin-resistant cells transmit ciRS-122 to drug-sensitive cells and can promote drug resistance through the miR-122/PKM2 axis. PKM2 increases the consumption of ATP and glucose and then generates lactic acid. At the same time, the ectopic expression of si-ciRS-122 in the exosomes down-regulating ciRS-122 can reverse the occurrence of drug resistance [105]. Hon KW et al. showed that HCT116 drug resistant cells selectively transduce hsa-circ-0000338 into the parent cells via exosomes, reducing the sensitivity of parent cells to drugs and prolonging the cell activity, so high exosomal hsa-circ-0000338 levels indicate chemoresistance in colorectal cancer [106].
Pancreatic cancer
Pancreatic ductal adenocarcinoma (PDAC) was typically considered to have a poor prognosis, with a one-year survival rate of 25% and a five-year survival rate of 5%. Finding a PDAC biomarker for early detection is especially important for those with disease risk. Li Z et al. explained the intercellular communication of exosomes among PDAC cells using immunofluorescence. An RFP-tagged CD63 lentivirus was used to label the exosomes in Hs 766T cells, which were added to GFP-labeled HS 766T cells, finding a dot-like RFP signal in the cytoplasm of GFP-labeled Hs 766T cells. The phenomenon indicates that exosomes can transfer contents between PDCA cells. High expressions of exosomal circ-PDE8A in PDCA are associated with high tumor TNM stages, invasion, metastasis propensity, and poor prognoses. One multivariate analysis shows that exosomal circ-PDE8A is also negatively associated with the overall survival rate. Exosomal circ-PDE8A, acting as a competitive endogenous RNA, activates MACC1/MET/AKT or the ERK pathway by restraining miR-338 [107]. In addition, the circ-IARS levels in the serum exosomes also accelerate through the adsorption of miR-122 to upregulate the RhoA level, increasing the endothelial monolayer permeability to improve PDAC tumor invasion [108].
Lung cancer
Lung cancer is the most common cancer-related cause of death in humans. The manu subtype of lung cancer is defined according to its histological type. The expression of exosomal circSATB2 in non-small cell lung cancer is significantly higher than it is in normal bronchial epithelial cells. Exosomal circSATB2 can directly bind to miR-326, a tumor suppressor gene related to the progression of non-small cell lung cancer, and positively regulates the expression of FSCN1 to promote the proliferation, invasion, and metastasis of non-small cell lung cancer and the proliferation of the normal bronchial epithelia. One Pearson correlation analysis shows that exosomal circSATB2 is positively correlated with FSCN1, and both are negatively correlated with miR-326. An ROC curve analysis shows that exosomal circSATB2 have high sensitivity and specificity as a serum marker for diagnosing lung cancer. Serum exosomal circSATB2 is associated with the lymphatic metastasis of lung cancer, so it is helpful for judging the clinical stage and prognosis of lung cancer [109].
Exosomal hsa_circ_0002130 regulates GLUT1, HK2, and LDHA proteins by targeting miR-498 in Osimertinib-resistant NSCLC cells, affecting glycolysis and affecting tumor growth and progression [110]. The effects of upregulated exosomal hsa_circ_0002130 can be alleviated by the ectopic expression of miR-498. Chen et al. established a circular RNA expression profile of early-stage lung adenocarcinoma (LUAD) serum exosomes and found that 182 circRNAs expressed differently (105 upregulated and 77 downregulated) compared with the normal controls. Among those, hsa_circ_0001492 and hsa_circ_0001346 were significantly upregulated in plasma exosomes and potentially served as new markers for the early diagnosis of LUAD [111].
Ovarian cancer
CircRNAs in ovarian cancer tissue or cells can induce the endothelial-mesenchymal transformation of recipient cells and promote the progression and metastasis of ovarian cancer through the circWHSC1/miR-145/MUC1 axis. In vitro, exosomal circWHSC1 extracted from ovarian cancer was used to incubate the HMrSV5 cells, then the MMT (mesothelial-to-mesenchymal transition) process had happened. The morphology of the cells changed to fiber-like, and the E-adhesion decreased while the N-adhesion and MUC1 increased, resulting in cell loss tight junctions [112]. MUC1 is a membrane-bound mucin, and it transforms peritoneal mesothelial cells to malignancy through intracellular communication and provides a tumor micro-environment for peritoneal implantation and diffusion [113-115]. Exosomal circWHSC1 positively regulates the expression of MUC1 and promotes peritoneal diffusion and adhesion, so it is beneficial to the metastasis of ovarian cancer to the abdominal cavity. In addition, Cho et al. found that detecting the expression of MUC1 in ascites can distinguish metastatic adenocarcinoma cells from normal mesothelial cells [116].
Guan X et al. extracted exosomal circPUM1 from the circPUM1 overexpressed CAOV3 ovarian cancer cell line, which they used to incubate HMrSV5 cells. CircPUM1 upregulated NF-κB and MMP2 proteins by inhibiting miR-615-5p and miR-6753-5p, resulting in the occurrence of MMT and the metastasis of the cancer cells [117].
Breast cancer
In a recent study, qRT-PCR showed that the expression of circ-UBE2D2 in MCF-7/TAM-R-Exo was more than 20 fold than that in MCF-7/Par-Exo. Further, the CCK-8 and Transwell assays showed that MCF-7/TAM-R-Exo can enhance cell viability, the IC50 values, and the invasion and migration abilities. Those experiments demonstrated that the transduction of circ-UBE2D2 through exosomes in vitro can enhance tamoxifen resistance in breast cancer. Dual-luciferase reporter and RNA immunoprecipitation assays showed that MiR-200a-3p is the target of circ-UBE2D2 [118].
Yang et al. extracted serum exosomes from breast cancer patients and healthy volunteers. Their bioinformatics analysis and RT-qPCR found that hsa-circRNA-00005795 and hsa-circRNA-0088088 act as competitive endogenous RNA in breast cancer [119]. Exosomal circASS1 increases in breast cancer, which promotes growth and metastasis by inhibiting miR-443 or activating the Hippo signaling pathway [119]. In the 2020 ASCO Annual Meeting (May 29-June 2, 2020), researchers reported that the bioinformatics analysis and RT-qPCR further pointed out that circHSDL2 is upregulated in triple-negative breast cancer patients and adsorbed let-7a-2-3p to promote the proliferation and invasion of tumors [120]. Few studies exist on exosome-mediated circRNAs in breast cancer, so a large number of studies is needed to verify whether exosome-mediated circRNAs can be used as a new biological marker and therapeutic target for the prediction and treatment of breast cancer.
Oral squamous cell carcinoma
The upregulation of exosomal circGDI2 can inhibit the proliferation, invasion, metastasis, and glycolysis of oral squamous cell carcinoma [121]. Oral squamous cell carcinoma cells transmit circGDI2 to neighboring cancer cells through exosomes. Then circGDI2 plays an inhibitory role in oral squamous cell carcinoma progression through the circGDI2/miR-424-5P/SCAI axis [122,123]. The expression of serum exosomal circGDI2 is consistent with the expressions of the corresponding donor cells [122]. Therefore, exosomal circGDI2 can be used as a new non-invasive biological marker of cancer and a treatment target for inhibiting oral squamous cell carcinoma.
Other cancers
The serum exosomal circ-0044516 of prostate cancer patients is significantly upregulated, inhibiting the activity of miR-29a-3p and promoting the proliferation of cancer cells. MiR-29a-3p is a tumor suppressor gene, which has been proved to be involved in the formation of various tumors [124]. For example, the down-regulation of miR-29a-3p in gastric cancer cells can promote cancer metastasis [125]. The increased expression of exosomal Hsa-circ-007293, Hsa-circ-031752, and Hsa-circ-020135 can be used as a diagnostic marker for thyroid papillary carcinoma [126].
Clinical suggestions and prospects
In the past decade, significant advances in the screening, diagnosis, treatment, and prognostic evaluation of cancer have been made. Due to cancer heterogeneity, pre-existing drug resistant cells and the continuous self-differentiation of cancer cells leads to increased mortality and drug resistance. Cancer-related exosomes are secreted by cancer cells and cells in the tumor microenvironment, which play a regulatory role in the progression, migration, metastasis, invasion, and deterioration of cancers. CircRNAs are a covalently closed circular structure, lacking 5’-3’ends and poly-A tail, with the characteristics of stability, conservation, a long half-life, tissue-specific expression, and easy detection in bodily fluids. The circRNA research field has revealed the essential role of circRNA in tumor biology. CircRNA can act as biomarkers for early cancer detection, diagnosis, prognosis, and therapeutic efficacy evaluation. We can develop a standard for biopsy detection for various cancer types and cancer progression.
Although many studies clarify the molecular mechanism of circRNA biogenesis and functions in the context of tumors, more efforts are still needed to elucidate the exosomal circRNA detail functions of circRNA besides the sponging for miRNA and the clinical reality for exosomal circRNA communication between cells. Functioning as a novel therapeutic target, circRNAs can be divided into tumor suppressors and oncogenes. Oncogenic circRNAs are highly expressed in cancer tissues and act as competitive endogenous RNA to retain miRNA activity by adsorbing miRNA. We can inhibit the formation of circRNAs and degrade circRNAs using enzymes or circRNA inhibitors (blocking the combination of circRNAs and miRNA) to delay cancer growth. Tumor-suppressing circRNAs should be increased in tissue to inhibit tumor progression and migration. Exosomes are vesicles secreted by many cell types, including miRNA, mRNA, circRNA, proteins, and other cargos. Many previous studies have shown that cancer cells can transfer circRNAs to the surrounding cells through exosomes to promote carcinogenesis. Therefore, the synthetic tumor-suppressing circRNAs can be transferred into exosomes, which are transported to specific target cells to inhibit tumor metastasis and invasion. For example, Chen et al. injected circ-0051443-Eoxes into the HuH7 and Hep3b cell lines, stopping the cell cycle at the G0/G1 phase through the circ-0051443/miR-331-3p/BAKI axis, which inhibited the progression of hepatocellular cancer [127]. Exosomal circRNAs can be used as an effective therapeutic target in cancer.
Liquid biopsy provides a non-invasive method for early detection to monitor cancer progression, evaluate genetic alternation and adopt the individual approach in cancer treatment. As extracellular vesicles in liquid biopsy, exosomes can be isolated or detected via micro NMR devices, nanoplasmonic chips, a magneto-electrochemical sensor, and the like. Exosomal circRNA has a built-in advantage during exosomal liquid biopsy elevation in its relative stability over miRNA and lncRNA and inconvenience over lipids or proteins. However, more clinical randomized controlled trials are needed to evaluate the effectiveness and safety of the diagnosis and treatment using exosomal circRNAs before pre-market approval. The application of exosomal circRNAs in the diagnosis and treatment of cancer needs to be further explored.
Conclusion
In this review, we summarized the biogenesis and functions of circRNAs, especially in exosome-mediated cellular communication. Scientists have mainly studied the sponging of circRNAs, which act as a competitive endogenous RNA and inhibit miRNA activity by binding to miRNA, thus affecting the downstream signaling pathway. The circRNAs/miRNA/mRNA axis plays an essential role in cancer progression and has become a research hotspot. With the development of high-throughput technology and bioinformatics analysis, many exosomal circRNAs have been found in bodily fluids which potentially serve as a biomarker for detecting tumorigenesis, progression, and drug resistance. Exosomal circRNA detection in liquid biopsies is an emerging application in cancer.
Acknowledgements
We thank Dan-dan Wang and Da-he Zhang for their help in the revision of the present paper. This work was supported by the National Key Research and Development Program of China (2016YFC0905900) and the National Natural Science Foundation of China (No. 81872365).
Disclosure of conflict of interest
None.
References
- 1.Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33:967–978. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- 2.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262:9412–9420. [PubMed] [Google Scholar]
- 3.van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 4.Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, Chen C, Chang W, Ping Y, Ji P, Wu J, Quan W, Yao Y, Zhou Y, Sun Z, Li D. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-β1 axis. Mol Cancer. 2020;19:117. doi: 10.1186/s12943-020-01235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 6.Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y, Chen W, Liu F, Sun W, Li XF, Wang X, Wang Y, Xu ZY, Gao L, Yang Q, Xu B, Li YM, Fang ZY, Xu ZP, Bao Y, Wu DS, Miao X, Sun HY, Sun YH, Wang HY, Wang LH. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29:653–668. doi: 10.1016/j.ccell.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Skotland T, Sandvig K, Llorente A. Lipids in exosomes: current knowledge and the way forward. Prog Lipid Res. 2017;66:30–41. doi: 10.1016/j.plipres.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng DL, Xiang YY, Ji LJ, Lu XJ. Competing endogenous RNA interplay in cancer: mechanism, methodology, and perspectives. Tumour Biol. 2015;36:479–488. doi: 10.1007/s13277-015-3093-z. [DOI] [PubMed] [Google Scholar]
- 10.Hentze M, Preiss T. Circular RNAs: splicing’s enigma variations. EMBO J. 2013;32:923–925. doi: 10.1038/emboj.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18:6. doi: 10.1186/s12943-018-0934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeck W, Sorrentino J, Wang K, Slevin M, Burd C, Liu J, Marzluff W, Sharpless N. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao D, Zhang X, Liu B, Meng D, Fang K, Guo Z, Li L. Screening circular RNA related to chemotherapeutic resistance in breast cancer. Epigenomics. 2017;9:1175–1188. doi: 10.2217/epi-2017-0055. [DOI] [PubMed] [Google Scholar]
- 17.Lü L, Sun J, Shi P, Kong W, Xu K, He B, Zhang S, Wang J. Identification of circular RNAs as a promising new class of diagnostic biomarkers for human breast cancer. Oncotarget. 2017;8:44096–44107. doi: 10.18632/oncotarget.17307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu M, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 19.Ashwal-Fluss R, Meyer M, Pamudurti N, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 20.Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Öhman M, Refojo D, Kadener S, Rajewsky N. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 21.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Artemaki PI, Scorilas A, Kontos CK. Circular RNAs: a new piece in the colorectal cancer puzzle. Cancers. 2020;12:2464. doi: 10.3390/cancers12092464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165:289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 25.Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 26.Lu Z, Filonov G, Noto J, Schmidt C, Hatkevich T, Wen Y, Jaffrey S, Matera A. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA. 2015;21:1554–1565. doi: 10.1261/rna.052944.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 28.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 29.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 30.Kelly S, Greenman C, Cook PR, Papantonis A. Exon skipping is correlated with exon circularization. J Mol Biol. 2015;427:2414–2417. doi: 10.1016/j.jmb.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 31.Conn S, Pillman K, Toubia J, Conn V, Salmanidis M, Phillips C, Roslan S, Schreiber A, Gregory P, Goodall G. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 32.Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, Rajewsky N. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 33.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11:98. doi: 10.1186/s13045-018-0643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt C, Noto J, Filonov G, Matera A. A method for expressing and imaging abundant, stable, circular RNAs in vivo using tRNA splicing. Methods Enzymol. 2016;572:215–236. doi: 10.1016/bs.mie.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang JL, Yang L, Chen LL. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15:611–624. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 38.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 39.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang X, Li Z, Zhang Q, Wang W, Li B, Wang L, Xu Z, Zeng A, Zhang X, Zhang X, He Z, Li Q, Sun G, Wang S, Li Q, Wang L, Zhang L, Xu H, Xu Z. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer. 2019;18:71. doi: 10.1186/s12943-019-0969-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y, Zheng X, Xu B, Chen L, Wang Q, Deng H, Jiang J. Circular RNA hsa_circ_0004015 regulates the proliferation, invasion, and TKI drug resistance of non-small cell lung cancer by miR-1183/PDPK1 signaling pathway. Biochem Biophys Res Commun. 2019;508:527–535. doi: 10.1016/j.bbrc.2018.11.157. [DOI] [PubMed] [Google Scholar]
- 42.Zhu KP, Ma XL, Zhang CL. Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int J Biol Sci. 2018;14:321–330. doi: 10.7150/ijbs.24360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao D, Qi X, Zhang X, Fang K, Guo Z, Li L. hsa_circRNA_0006528 as a competing endogenous RNA promotes human breast cancer progression by sponging miR-7-5p and activating the MAPK/ERK signaling pathway. Mol Carcinog. 2019;58:554–564. doi: 10.1002/mc.22950. [DOI] [PubMed] [Google Scholar]
- 44.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 45.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 46.Zhang X, Mao L. Circular RNA Circ_0000442 acts as a sponge of MiR-148b-3p to suppress breast cancer via PTEN/PI3K/Akt signaling pathway. Gene. 2021;766:145113. doi: 10.1016/j.gene.2020.145113. [DOI] [PubMed] [Google Scholar]
- 47.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abdollahzadeh R, Daraei A, Mansoori Y, Sepahvand M, Amoli MM, Tavakkoly-Bazzaz J. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: a new look at hallmarks of breast cancer. J Cell Physiol. 2019;234:10080–10100. doi: 10.1002/jcp.27941. [DOI] [PubMed] [Google Scholar]
- 49.Du W, Yang W, Liu E, Yang Z, Dhaliwal P, Yang B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 2005;65:3980–3985. doi: 10.1158/0008-5472.CAN-04-3995. [DOI] [PubMed] [Google Scholar]
- 51.Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Q, Wang Y, Wu S, Zhou Z, Ding X, Shi R, Thorne RF, Zhang XD, Hu W, Wu M. CircACC1 regulates assembly and activation of AMPK complex under metabolic stress. Cell Metab. 2019;30:157–173. e157. doi: 10.1016/j.cmet.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 53.Zhang T, Zhang L, Han D, Tursun K, Lu X. Circular RNA hsa_Circ_101141 as a competing endogenous RNA facilitates tumorigenesis of hepatocellular carcinoma by regulating miR-1297/ROCK1 pathway. Cell Transplant. 2020;29:963689720948016. doi: 10.1177/0963689720948016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Wei Y, Li L, Wang D, Zhang CY, Zen K. Importin 8 regulates the transport of mature microRNAs into the cell nucleus. J Biol Chem. 2014;289:10270–10275. doi: 10.1074/jbc.C113.541417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 2013;4:283. doi: 10.3389/fgene.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang X, Fang L. Advances in circular RNAs and their roles in breast cancer. J Exp Clin Cancer Res. 2018;37:206. doi: 10.1186/s13046-018-0870-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen C, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 58.Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, Shenzis S, Samson M, Dittmar G, Landthaler M, Chekulaeva M, Rajewsky N, Kadener S. Translation of CircRNAs. Mol Cell. 2017;66:9–21. e27. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 60.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, Chen W, Gao X, Zhao K, Zhou H, Li Z, Ming L, Xie B, Zhang N. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 62.Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 5’ UTR m(6)A promotes cap-independent translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, Wong CC, Xiao X, Wang Z. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ng W, Marinov G, Liau E, Lam Y, Lim Y, Ea C. Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway. RNA Biol. 2016;13:861–871. doi: 10.1080/15476286.2016.1207036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hubbard AK, Rothlein R. Intercellular adhesion molecule-1 (ICAM-1) expression and cell signaling cascades. Free Radic Biol Med. 2000;28:1379–1386. doi: 10.1016/s0891-5849(00)00223-9. [DOI] [PubMed] [Google Scholar]
- 66.Ng WL, Marinov GK, Chin YM, Lim YY, Ea CK. Transcriptomic analysis of the role of RasGEF1B circular RNA in the TLR4/LPS pathway. Sci Rep. 2017;7:12227. doi: 10.1038/s41598-017-12550-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rubtsov Y, Goryunov К, Romanov А, Suzdaltseva Y, Sharonov G, Tkachuk V. Molecular mechanisms of immunomodulation properties of mesenchymal stromal cells: a new insight into the role of ICAM-1. Stem Cells Int. 2017;2017:6516854. doi: 10.1155/2017/6516854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu C, Li X, Nan F, Jiang S, Gao X, Guo S, Xue W, Cui Y, Dong K, Ding H, Qu B, Zhou Z, Shen N, Yang L, Chen L. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177:865–880. e821. doi: 10.1016/j.cell.2019.03.046. [DOI] [PubMed] [Google Scholar]
- 69.Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, Broughton JP, Kim J, Cadena C, Pulendran B, Hur S, Chang HY. N6-methyladenosine modification controls circular RNA immunity. Mol Cell. 2019;76:96–109. e109. doi: 10.1016/j.molcel.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stoorvogel W, Strous GJ, Geuze HJ, Oorschot V, Schwartz AL. Late endosomes derive from early endosomes by maturation. Cell. 1991;65:417–427. doi: 10.1016/0092-8674(91)90459-c. [DOI] [PubMed] [Google Scholar]
- 71.Mathieu M, Martin-Jaular L, Lavieu G, Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nature cell biology. 2019;21:9–17. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- 72.Hinger SA, Cha DJ, Franklin JL, Higginbotham JN, Dou Y, Ping J, Shu L, Prasad N, Levy S, Zhang B, Liu Q, Weaver AM, Coffey RJ, Patton JG. Diverse long RNAs are differentially sorted into extracellular vesicles secreted by colorectal cancer cells. Cell Rep. 2018;25:715–725. e714. doi: 10.1016/j.celrep.2018.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sánchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang M, Yu F, Li P, Wang K. Emerging function and clinical significance of exosomal circRNAs in cancer. Mol Ther Nucleic Acids. 2020;21:367–383. doi: 10.1016/j.omtn.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hurley J. ESCRTs are everywhere. EMBO J. 2015;34:2398–2407. doi: 10.15252/embj.201592484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Iavello A, Frech VS, Gai C, Deregibus MC, Quesenberry PJ, Camussi G. Role of Alix in miRNA packaging during extracellular vesicle biogenesis. Int J Mol Med. 2016;37:958–966. doi: 10.3892/ijmm.2016.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wei JX, Lv LH, Wan YL, Cao Y, Li GL, Lin HM, Zhou R, Shang CZ, Cao J, He H, Han QF, Liu PQ, Zhou G, Min J. Vps4A functions as a tumor suppressor by regulating the secretion and uptake of exosomal microRNAs in human hepatoma cells. Hepatology. 2015;61:1284–1294. doi: 10.1002/hep.27660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lukiw WJ. Circular RNA (circRNA) in Alzheimer’s disease (AD) Front Genet. 2013;4:307. doi: 10.3389/fgene.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, Gong Y, Liu J, Dong YH, Li N, Li PF. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 81.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maia J, Caja S, Strano Moraes MC, Couto N, Costa-Silva B. Exosome-based cell-cell communication in the tumor microenvironment. Front Cell Dev Biol. 2018;6:18. doi: 10.3389/fcell.2018.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang XY, Huang ZL, Huang J, Xu B, Huang XY, Xu YH, Zhou J, Tang ZY. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res. 2020;39:20. doi: 10.1186/s13046-020-1529-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang L, Yu D. Exosomes in cancer development, metastasis, and immunity. Biochim Biophys Acta Rev Cancer. 2019;1871:455–468. doi: 10.1016/j.bbcan.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tang YY, Zhao P, Zou TN, Duan JJ, Zhi R, Yang SY, Yang DC, Wang XL. Circular RNA hsa_circ_0001982 promotes breast cancer cell carcinogenesis through decreasing miR-143. DNA Cell Biol. 2017;36:901–908. doi: 10.1089/dna.2017.3862. [DOI] [PubMed] [Google Scholar]
- 86.Yang W, Gong P, Yang Y, Yang C, Yang B, Ren L. Circ-ABCB10 contributes to paclitaxel resistance in breast cancer through Let-7a-5p/DUSP7 axis. Cancer Manag Res. 2020;12:2327–2337. doi: 10.2147/CMAR.S238513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ren S, Liu J, Feng Y, Li Z, He L, Li L, Cao X, Wang Z, Zhang Y. Knockdown of circDENND4C inhibits glycolysis, migration and invasion by up-regulating miR-200b/c in breast cancer under hypoxia. J Exp Clin Cancer Res. 2019;38:388. doi: 10.1186/s13046-019-1398-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li W, Yang FQ, Sun CM, Huang JH, Zhang HM, Li X, Wang GC, Zhang N, Che JP, Zhang WT, Yan Y, Yao XD, Peng B, Zheng JH, Liu M. circPRRC2A promotes angiogenesis and metastasis through epithelial-mesenchymal transition and upregulates TRPM3 in renal cell carcinoma. Theranostics. 2020;10:4395–4409. doi: 10.7150/thno.43239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yan N, Xu H, Zhang J, Xu L, Zhang Y, Zhang L, Xu Y, Zhang F. Circular RNA profile indicates circular RNA VRK1 is negatively related with breast cancer stem cells. Oncotarget. 2017;8:95704–95718. doi: 10.18632/oncotarget.21183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, Fan Q, Li J, Ning T, Tian F, Li H, Sun W, Ying G, Ba Y. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38:2844–2859. doi: 10.1038/s41388-018-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 91.Wang Q, Ma S, Song N, Li X, Liu L, Yang S, Ding X, Shan L, Zhou X, Su D, Wang Y, Zhang Q, Liu X, Yu N, Zhang K, Shang Y, Yao Z, Shi L. Stabilization of histone demethylase PHF8 by USP7 promotes breast carcinogenesis. J Clin Invest. 2016;126:2205–2220. doi: 10.1172/JCI85747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, Cai JB, Ke AW. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer. 2020;19:110. doi: 10.1186/s12943-020-01222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ju Y, Hou N, Meng J, Wang X, Zhang X, Zhao D, Liu Y, Zhu F, Zhang L, Sun W, Liang X, Gao L, Ma C. T cell immunoglobulin- and mucin-domain-containing molecule-3 (Tim-3) mediates natural killer cell suppression in chronic hepatitis B. J Hepatol. 2010;52:322–329. doi: 10.1016/j.jhep.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 94.Zheng Y, Li Y, Lian J, Yang H, Li F, Zhao S, Qi Y, Zhang Y, Huang L. TNF-α-induced Tim-3 expression marks the dysfunction of infiltrating natural killer cells in human esophageal cancer. J Transl Med. 2019;17:165. doi: 10.1186/s12967-019-1917-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang T, Jing B, Bai Y, Zhang Y, Yu H. Circular RNA circTMEM45A acts as the sponge of MicroRNA-665 to promote hepatocellular carcinoma progression. Mol Ther Nucleic Acids. 2020;22:285–297. doi: 10.1016/j.omtn.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sun XH, Wang YT, Li GF, Zhang N, Fan L. Serum-derived three-circRNA signature as a diagnostic biomarker for hepatocellular carcinoma. Cancer Cell Int. 2020;20:226. doi: 10.1186/s12935-020-01302-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang H, Zhu L, Bai M, Liu Y, Zhan Y, Deng T, Yang H, Sun W, Wang X, Zhu K, Fan Q, Li J, Ying G, Ba Y. Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR-133/PRDM16 pathway. Int J Cancer. 2019;144:2501–2515. doi: 10.1002/ijc.31977. [DOI] [PubMed] [Google Scholar]
- 98.Liu W, Kuang S. miR-133 links to energy balance through targeting Prdm16. J Mol Cell Biol. 2013;5:432–434. doi: 10.1093/jmcb/mjt036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scimè A, Devarakonda S, Conroe HM, Erdjument-Bromage H, Tempst P, Rudnicki MA, Beier DR, Spiegelman BM. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yao W, Guo P, Mu Q, Wang Y. Exosome-derived Circ-PVT1 contributes to cisplatin resistance by regulating autophagy, invasion, and apoptosis via miR-30a-5p/YAP1 axis in gastric cancer cells. Cancer Biother Radiopharm. 2020;36:347–359. doi: 10.1089/cbr.2020.3578. [DOI] [PubMed] [Google Scholar]
- 101.Li Y, Li C, Xu R, Wang Y, Li D, Zhang B. A novel circFMN2 promotes tumor proliferation in CRC by regulating the miR-1182/hTERT signaling pathways. Clin Sci (Lond) 2019;133:2463–2479. doi: 10.1042/CS20190715. [DOI] [PubMed] [Google Scholar]
- 102.Feng W, Gong H, Wang Y, Zhu G, Xue T, Wang Y, Cui G. circIFT80 functions as a ceRNA of miR-1236-3p to promote colorectal cancer progression. Mol Ther Nucleic Acids. 2019;18:375–387. doi: 10.1016/j.omtn.2019.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 103.Baassiri A, Nassar F, Mukherji D, Shamseddine A, Nasr R, Temraz S. Exosomal non coding RNA in LIQUID biopsies as a promising biomarker for colorectal cancer. Int J Mol Sci. 2020;21:1398. doi: 10.3390/ijms21041398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pan B, Qin J, Liu X, He B, Wang X, Pan Y, Sun H, Xu T, Xu M, Chen X, Xu X, Zeng K, Sun L, Wang S. Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet. 2019;10:1096. doi: 10.3389/fgene.2019.01096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang X, Zhang H, Yang H, Bai M, Ning T, Deng T, Liu R, Fan Q, Zhu K, Li J, Zhan Y, Ying G, Ba Y. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 2020;14:539–555. doi: 10.1002/1878-0261.12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hon KW, Ab-Mutalib NS, Abdullah N, Jamal R, Abu N. Extracellular Vesicle-derived circular RNAs confers chemoresistance in colorectal cancer. Sci Rep. 2019;9:16497. doi: 10.1038/s41598-019-53063-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li Z, Yanfang W, Li J, Jiang P, Peng T, Chen K, Zhao X, Zhang Y, Zhen P, Zhu J, Li X. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018;432:237–250. doi: 10.1016/j.canlet.2018.04.035. [DOI] [PubMed] [Google Scholar]
- 108.Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K, Liu H, Bi H, Liu X, Li X. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37:177. doi: 10.1186/s13046-018-0822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang N, Nan A, Chen L, Li X, Jia Y, Qiu M, Dai X, Zhou H, Zhu J, Zhang H, Jiang Y. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol Cancer. 2020;19:101. doi: 10.1186/s12943-020-01221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ma J, Qi G, Li L. A novel serum exosomes-based biomarker hsa_circ_0002130 facilitates osimertinib-resistance in non-small cell lung cancer by sponging miR-498. Onco Targets Ther. 2020;13:5293–5307. doi: 10.2147/OTT.S243214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen F, Huang C, Wu Q, Jiang L, Chen S, Chen L. Circular RNAs expression profiles in plasma exosomes from early-stage lung adenocarcinoma and the potential biomarkers. J Cell Biochem. 2020;121:2525–2533. doi: 10.1002/jcb.29475. [DOI] [PubMed] [Google Scholar]
- 112.Zong ZH, Du YP, Guan X, Chen S, Zhao Y. CircWHSC1 promotes ovarian cancer progression by regulating MUC1 and hTERT through sponging miR-145 and miR-1182. J Exp Clin Cancer Res. 2019;38:437. doi: 10.1186/s13046-019-1437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jonckheere N, Van Seuningen I. The membrane-bound mucins: how large O-glycoproteins play key roles in epithelial cancers and hold promise as biological tools for gene-based and immunotherapies. Crit Rev Oncog. 2008;14:177–196. doi: 10.1615/critrevoncog.v14.i2-3.30. [DOI] [PubMed] [Google Scholar]
- 114.Jonckheere N, Van Seuningen I. The membrane-bound mucins: from cell signalling to transcriptional regulation and expression in epithelial cancers. Biochimie. 2010;92:1–11. doi: 10.1016/j.biochi.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 115.Deng J, Wang L, Chen H, Li L, Ma Y, Ni J, Li Y. The role of tumour-associated MUC1 in epithelial ovarian cancer metastasis and progression. Cancer Metastasis Rev. 2013;32:535–551. doi: 10.1007/s10555-013-9423-y. [DOI] [PubMed] [Google Scholar]
- 116.Cho J, Kim G, Lee J, Lee J, Nam J, Choi C. Diagnostic usefulness of MUC1 and MUC4 for distinguishing between metastatic adenocarcinoma cells and reactive mesothelial cells in effusion cell blocks. Acta cytologica. 2013;57:377–383. doi: 10.1159/000348499. [DOI] [PubMed] [Google Scholar]
- 117.Guan X, Zong ZH, Liu Y, Chen S, Wang LL, Zhao Y. circPUM1 promotes tumorigenesis and progression of ovarian cancer by sponging miR-615-5p and miR-6753-5p. Mol Ther Nucleic Acids. 2019;18:882–892. doi: 10.1016/j.omtn.2019.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nie W, Wu G, Zhang J, Huang LL, Ding J, Jiang A, Zhang Y, Liu Y, Li J, Pu K, Xie HY. Responsive exosome nano-bioconjugates for synergistic cancer therapy. Angew Chem Int Ed Engl. 2020;59:2018–2022. doi: 10.1002/anie.201912524. [DOI] [PubMed] [Google Scholar]
- 119.Yang SJ, Wang DD, Zhou SY, Zhang Q, Wang JY, Zhong SL, Zhang HD, Wang XY, Xia X, Chen W, Yang SY, Hu JH, Zhao JH, Tang JH. Identification of circRNA-miRNA networks for exploring an underlying prognosis strategy for breast cancer. Epigenomics. 2020;12:101–125. doi: 10.2217/epi-2019-0058. [DOI] [PubMed] [Google Scholar]
- 120.Yang S, Tang J. Abstract P6-10-29: circular RNAs in exosomes derived from breast cancer: promising biomarkers for diagnosis and prognosis of triple negative breast cancer (TNBC) 2020 [Google Scholar]
- 121.Su W, Wang Y, Wang F, Sun S, Li M, Shen Y, Yang H. Hsa_circ_0005379 regulates malignant behavior of oral squamous cell carcinoma through the EGFR pathway. BMC Cancer. 2019;19:400. doi: 10.1186/s12885-019-5593-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 122.Zhang Y, Tang K, Chen L, Du M, Qu Z. Exosomal CircGDI2 suppresses oral squamous cell carcinoma progression through the regulation of MiR-424-5p/SCAI axis. Cancer Manag Res. 2020;12:7501–7514. doi: 10.2147/CMAR.S255687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, Wang G, Wu P, Wang H, Jiang L, Yuan W, Sun Z, Ming L. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18:116. doi: 10.1186/s12943-019-1041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li T, Sun X, Chen L. Exosome circ_0044516 promotes prostate cancer cell proliferation and metastasis as a potential biomarker. J Cell Biochem. 2020;121:2118–2126. doi: 10.1002/jcb.28239. [DOI] [PubMed] [Google Scholar]
- 125.Bai F, Jiu M, You Y, Feng Y, Xin R, Liu X, Mo L, Nie Y. miR-29a-3p represses proliferation and metastasis of gastric cancer cells via attenuating HAS3 levels. Mol Med Rep. 2018;17:8145–8152. doi: 10.3892/mmr.2018.8896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yang C, Wei Y, Yu L, Xiao Y. Identification of altered circular RNA expression in serum exosomes from patients with papillary thyroid carcinoma by high-throughput sequencing. Med Sci Monit. 2019;25:2785–2791. doi: 10.12659/MSM.915658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chen W, Quan Y, Fan S, Wang H, Liang J, Huang L, Chen L, Liu Q, He P, Ye Y. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020;475:119–128. doi: 10.1016/j.canlet.2020.01.022. [DOI] [PubMed] [Google Scholar]
- 128.Hu K, Liu X, Li Y, Li Q, Xu Y, Zeng W, Zhong G, Yu C. Exosomes mediated transfer of Circ_UBE2D2 enhances the resistance of breast cancer to tamoxifen by binding to MiR-200a-3p. Med Sci Monit. 2020;26:e922253. doi: 10.12659/MSM.922253. [DOI] [PMC free article] [PubMed] [Google Scholar]


