Abstract
Several species belonging to the genus Burkholderia are clinically relevant, opportunistic pathogens that inhabit major environmental reservoirs. Consequently, the availability of means for adequate identification and epidemiological characterization of individual environmental or clinical isolates is mandatory. In the present communication we describe the use of the Riboprinter microbial characterization system (Qualicon, Warwick, United Kingdom) for automated ribotyping of 104 strains of Burkholderia species from diverse sources, including several publicly accessible collections. The main outcome of this analysis was that all strains were typeable and that strains of Burkholderia gladioli and of each species of the B. cepacia complex, including B. multivorans, B. stabilis, and B. vietnamiensis, were effectively discriminated. Furthermore, different ribotypes were discerned within each species. Ribotyping results were in general agreement with strain classification based on restriction fragment analysis of 16S ribosomal amplicons, but the resolution of ribotyping was much higher. This enabled automated molecular typing below the species level. Cluster analysis of the patterns obtained by ribotyping (riboprints) showed that within B. gladioli, B. multivorans, and B. cepacia genomovar VI, the different riboprints identified always clustered together. Riboprints of B. cepacia genomovars I and III, B. stabilis, and B. vietnamiensis did not show distinct clustering but rather exhibited the formation of loose assemblages within which several smaller, genomovar-specific clusters were delineated. Therefore, ribotyping proved useful for genomovar identification. Analysis of serial isolates from individual patients demonstrated that infection with a single ribotype had occurred, despite minor genetic differences that were detected by pulsed-field gel electrophoresis of DNA macrorestriction fragments. The automated approach allows very rapid and reliable identification and epidemiological characterization of strains and generates an easily manageable database suited for expansion with information on additional bacterial isolates.
Bacterial species belonging to the genus Burkholderia are renowned for their pathogenicity in both plants and people. Although the details of their pathogenic potential and their capacity to adapt to a potentially hostile host environment remain elusive to a large extent (9), research on their taxonomic classification is continuously ongoing. The taxonomic positions of two of the current Burkholderia species, B. cepacia and B. gladioli, have evolved dynamically over the past few decades (7, 44). Still, accurate species-level identification is often difficult, and, especially in relation to their pathogenicity in man, this may introduce uncertainties in the clinical relevance of strains belonging to the different species. The importance of B. gladioli is rising with the number of reports describing this bacterial species as an invasive human pathogen (2, 12, 17, 20, 21, 35). However, B. gladioli remains controversial as a putative pathogen, since mere colonization of the lungs of cystic fibrosis (CF) patients without apparent pathology has also been noted for this bacterial species (6).
Similarly, strains of B. cepacia, presently referred to as the B. cepacia complex, have been classified into five genomic species, called genomovars (labeled I to V), some of them with attributed names, namely B. multivorans (genomovar II) (39), B. stabilis (genomovar IV) (40), and B. vietnamiensis (genomovar V) (10). Recently, a sixth genomovar was identified (T. Coenye, J. J. LiPuma, D. Henry, B. Hoste, K. Vandemeulebroeke, M. Gillis, D. P. Speert, and P. Vandamme, submitted for publication). Distinguishing between genomovars relies on DNA-DNA hybridization, whole-cell protein pattern similarity, and phenotypic markers. Classification of B. cepacia-like strains in multiple species is stimulating the hypothesis that the genetic identities of the infective strains influence the clinical outcome. As a matter of fact, Burkholderia spp. pathogenicity can vary greatly, particularly in CF patients (19). Strains of different genomovars also greatly differ in their transmissibility (11), stressing the importance of rapid and reliable strain identification systems.
Given the dynamics and controversies mentioned above, novel diagnostic tests for B. cepacia, including its different genomovars, and B. gladioli have been reported. These assays include culture approaches (15, 16, 42), enzyme-linked immunosorbent assays (27), and, of course, PCR (3, 4, 23, 43). Some of these assays have the capacity to discriminate reliably among the various members of the B. cepacia complex (3, 4, 23). Here we present evidence that the automated Riboprinter microbial characterization system can be used to distinguish between isolates of the B. cepacia complex species and B. gladioli and at the same time provide information on subspecific genetic heterogeneity.
MATERIALS AND METHODS
Bacterial isolates.
The strains analyzed in the present study were gathered from diverse sources, including several publicly accessible reference collections (41). In addition to the collection described in reference 41, strains from the BCCM/LMG Bacterial Collection (Ghent, Belgium) and from the collection of the Medizinische Hochschule (Hannover, Germany) were included. A single strain of B. plantarii and three strains of Ralstonia pickettii (formerly Burkholderia pickettii [45]) were included for reasons of comparison and control. Serial isolates derived from individual patients were included. Table 1 describes the origins of the strains and provides information on the geographical and environmental or clinical sources of several of the isolates. For the strains included in reference 41, PCR-restriction fragment length polymorphism (RFLP) characterization profiles are also indicated. The four-letter codes presented in Table 1 identify the types as established by using the restriction enzymes AluI, CfoI, MspI, and DdeI.
TABLE 1.
Origin, characterization, and identification of the Burkholderia strains analyzed
| ENARE code | Other code | Source | Country | Ribogroup | 16S PCR-RFLP type | Species or genomovar as determined by:
|
|
|---|---|---|---|---|---|---|---|
| Protein profile analysis | Ribotyping analysis | ||||||
| 41C092 | Incub. water | Incubator water | ?a | 210-236-S-7 | AAAB | NDb | B. cepacia complex |
| 41C106 | Pat9 H118 | Patient 9 | Germany | ND | ND | ND | B. cepacia complex |
| 41C012 | 95-664 RIVM | Patient Ma | The Netherlands | 210-226-S-6 | BBEC | ND | B. gladioli |
| 41C011 | 95-665 RIVM | Patient Ma | The Netherlands | 210-226-S-6 | BBEC | ND | B. gladioli |
| 41C013 | 96-568 RIVM | Patient Ma | The Netherlands | 210-226-S-6 | BBEC | ND | B. gladioli |
| 41C014 | 96-580 RIVM | Patient Ma | The Netherlands | 210-226-S-6 | BBEC | ND | B. gladioli |
| 41C055 | ATCC 19302 | Onion bulb root | USAc | 210-226-S-8 | BBBC | B. gladioli | B. gladioli |
| 41C016 | CCUG2115, ATCC 19302 | Onion bulb root | USA | 210-226-S-8 | BBBC | B. gladioli | B. gladioli |
| 41C056 | ATCC 10248 | Gladiolus sp. | USA | 210-232-S-1 | BBBC | B. gladioli | B. gladioli |
| 41C038 | ATCC 10248, CEP0032 | Gladiolus sp. | USA | 210-232-S-1 | BBBC | B. gladioli | B. gladioli |
| 41C017 | CCUG1782, ATCC 10248 | Gladiolus sp. | USA | 210-232-S-1 | BBBC | B. gladioli | B. gladioli |
| 41C108 | CEP0025 | Sputum, CF patient | Canada | 210-232-S-1 | BBBC | B. gladioli | B. gladioli |
| 41C039 | CEP0029 | ? | Canada | 210-232-S-3 | BBEC | B. gladioli | B. gladioli |
| 41C069 | Pat10 H129 | ? | Germany | 210-236-S-1 | BBBC | ND | B. gladioli |
| 41C088 | Pat15 H176 | Patient 15 | Germany | 210-237-S-4 | BBBC | ND | B. gladioli |
| 41C079 | 601397 LVF | ? | The Netherlands | 210-216-S-1 | AAAA | ND | B. multivorans |
| 41C095 | 611881 LVF | ? | The Netherlands | 210-216-S-1 | AAAA | ND | B. multivorans |
| 41C020 | CCUG36978 | CF patient | Austria | 210-216-S-1 | AAAA | B. multivorans | B. multivorans |
| 41C086 | Pat4 H194 | Patient 4 | Germany | 210-216-S-1 | AAAA | ND | B. multivorans |
| 41C065 | Pat4 H115 | Patient 4 | Germany | 210-216-S-1 | AAAA | ND | B. multivorans |
| 41C064 | Pat4 H161 | Patient 4 | Germany | 210-216-S-1 | AAAA | ND | B. multivorans |
| 41C001 | Pat A-1 | Patient A | The Netherlands | 210-216-S-1 | AAAA | ND | B. multivorans |
| 41C005 | Pat A-2 | Patient A | The Netherlands | 210-216-S-1 | AAAA | ND | B. multivorans |
| 41C111 | Pat A-3 | Patient A | The Netherlands | 210-216-S-1 | AAAA | ND | B. multivorans |
| 41C007 | Pat A-4 | Patient A | The Netherlands | 210-216-S-1 | AAAA | ND | B. multivorans |
| 41C110 | Pat A-5 | Patient A | The Netherlands | 210-216-S-1 | AAAA | ND | B. multivorans |
| 41C109 | Pat A-6 | Patient A | The Netherlands | 210-216-S-1 | AAAA | ND | B. multivorans |
| 41C107 | Pat A-7 | Patient A | The Netherlands | 210-216-S-1 | AAAA | ND | B. multivorans |
| 41C004 | Pat C-1 | Patient C | The Netherlands | 210-216-S-1 | AAAA | ND | B. multivorans |
| 41C008 | Pat C-2 | Patient C | The Netherlands | 210-216-S-1 | AAAA | ND | B. multivorans |
| 41C009 | Pat C-3 | Patient C | The Netherlands | 210-216-S-1 | AAAA | ND | B. multivorans |
| 41C006 | Pat D-1 | Patient D | The Netherlands | 210-216-S-4 | AAAA | ND | B. multivorans |
| 41C051 | Pat1 H132 | Patient 1 | Germany | 210-216-S-5 | AAAA | ND | B. multivorans |
| 41C053 | Pat1 H59 | Patient 1 | Germany | 210-216-S-5 | AAAA | ND | B. multivorans |
| 41C052 | Pat1 H62 | Patient 1 | Germany | 210-216-S-5 | AAAA | ND | B. multivorans |
| 41C080 | Pat13 H174 | Patient 13 | Germany | 210-216-S-5 | AAAA | ND | B. multivorans |
| 41C050 | Pat2 H107 | Patient 2 | Germany | 210-216-S-5 | AAAA | ND | B. multivorans |
| 41C049 | Pat2 H119 | Patient 2 | Germany | 210-216-S-5 | AAAA | ND | B. multivorans |
| 41C048 | Pat2 H133 | Patient 2 | Germany | 210-216-S-5 | AAAA | ND | B. multivorans |
| 41C010 | 96-499 RIVM | Patient Ve | The Netherlands | 210-216-S-5 | AAAA | ND | B. multivorans |
| 41C043 | C5393 | Sputum, CF patient | Canada | 210-216-S-5 | AAAA | B. multivorans | B. multivorans |
| 41C054 | Pat G-1 | Patient G | The Netherlands | 210-219-S-2 | AAAA | B. multivorans | B. multivorans |
| 41C082 | Pat11 H158 | Patient 11 | Germany | 210-219-S-5 | AAAA | ND | B. multivorans |
| 41C063 | Pat5 H125 | Patient 5 | Germany | 210-219-S-5 | AAAA | ND | B. multivorans |
| 41C062 | Pat5 H126 | Patient 5 | Germany | 210-219-S-5 | AAAA | ND | B. multivorans |
| 41C061 | Pat5 H140 | Patient 5 | Germany | 210-219-S-5 | AAAA | ND | B. multivorans |
| 41C060 | Pat5 H141 | Patient 5 | Germany | 210-219-S-5 | AAAA | ND | B. multivorans |
| 41C059 | Pat5 H142 | Patient 5 | Germany | 210-219-S-5 | AAAA | B. multivorans | B. multivorans |
| 41C042 | CEP0144, ATCC 17616 | ? | ? | 210-232-S-7 | AAAA | B. multivorans | B. multivorans |
| 41C112 | 96-602 RIVM | ? | The Netherlands | 210-237-S-8 | AAAA | B. multivorans | B. multivorans |
| 41C115 | LMG13010 | CF patient | Belgium | 210-254-S-8 | ND | B. multivorans | B. multivorans |
| 41C120 | LMG17588, ATCC 17616 | ? | ? | 210-257-S-5 | ND | B. multivorans | B. multivorans |
| 41C015 | 96-673 RIVM | Patient Sy | The Netherlands | 210-226-S-7 | BBBC | B. plantarii | B. plantarii |
| 41C081 | Pat12 H162 | Patient 12 | Germany | 210-233-S-8 | ABBB | B. stabilis | B. stabilis |
| 41C083 | Pat14 H175 | Patient 14 | Germany | 210-233-S-8 | ABBB | B. stabilis | B. stabilis |
| 41C084 | Pat16 H177 | Patient 16 | Germany | 210-233-S-8 | ABBB | ND | B. stabilis |
| 41C085 | Pat17 H193 | Patient 17 | Germany | 210-233-S-8 | ABBB | ND | B. stabilis |
| 41C078 | Pat6 H134 | Patient 6 | Germany | 210-233-S-8 | ABBB | ND | B. stabilis |
| 41C077 | Pat6 H135 | Patient 6 | Germany | 210-233-S-8 | ABBB | ND | B. stabilis |
| 41C075 | Pat6 H154 | Patient 6 | Germany | 210-233-S-8 | ABBB | B. stabilis | B. stabilis |
| 41C122 | LMG18888 | Human blood | ? | 210-235-S-5 | ND | B. stabilis | B. stabilis |
| 41C074 | Pat7 H145 | Patient 7 | Germany | 210-235-S-5 | ABBB | ND | B. stabilis |
| 41C073 | Pat7 H146 | Patient 7 | Germany | 210-235-S-5 | ABBB | B. stabilis | B. stabilis |
| 41C087 | H177E | Water outlet | Germany | 210-235-S-5 | ABBB | ND | B. stabilis |
| 41C116 | LMG14294 | CF patient | Belgium | 210-254-S-2 | ND | B. stabilis | B. stabilis |
| 41C032 | SKMM | ? | The Netherlands | 210-225-S-2 | AAAB | B. vietnamiensis | B. vietnamiensis |
| 41C021 | CCUG9631 | Patient | Sweden | 210-236-S-8 | AAAB | B. vietnamiensis | B. vietnamiensis |
| 41C113 | LMG10929 | Soil | Vietnam | 210-257-S-6 | ND | B. vietnamiensis | B. vietnamiensis |
| 41C117 | LMG16232 | Sputum, CF patient | ? | 210-257-S-7 | ND | B. vietnamiensis | B. vietnamiensis |
| 41C036 | CCUG788, LMG6986 | ? | Sweden | 210-225-S-4 | AAAB | Genomovar I | Genomovar I |
| 41C040 | CEP0521 | CF patient | Australia | 210-225-S-6 | AAAB | Genomovar I | Genomovar I |
| 41C121 | LMG18821 | Sputum, CF patient | ? | 210-254-S-5 | ND | Genomovar I | Genomovar I |
| 41C037 | ATCC 25416 | Onion | ? | 210-216-S-7 | AAAB | Genomovar I | Genomovar I |
| 41C047 | ATCC 25416 | Onion | ? | 210-216-S-7 | AAAB | Genomovar I | Genomovar I |
| 41C022 | CCUG12691, ATCC 2541 | Onion | ? | 210-216-S-7 | AAAB | Genomovar I | Genomovar I |
| 41C041 | CEP0031, ATCC 25416 | Onion | ? | 210-216-S-7 | AAAB | Genomovar I | Genomovar I |
| 41C114 | LMG1222, ATCC 25416 | Onion | ? | 210-216-S-7 | AAAB | Genomovar I | Genomovar I |
| 41C057 | ATCC 25609, LMG6981 | ? | ? | 210-219-S-3 | AAAB | Genomovar I | Genomovar I |
| 41C058 | ATCC 17759 | ? | ? | 210-219-S-4 | AAAB | Genomovar I | Genomovar I |
| 41C003 | Pat B-2 | Patient B | The Netherlands | 210-216-S-2 | ND | ND | Genomovar III |
| 41C002 | Pat B-1 | Patient B | The Netherlands | 210-216-S-2 | ND | Genomovar III | Genomovar III |
| 41C035 | Pat E-1 | Patient E | The Netherlands | 210-216-S-6 | AAAA | ND | Genomovar III |
| 41C034 | Pat E-2 | Patient E | The Netherlands | 210-216-S-6 | AAAA | ND | Genomovar III |
| 41C033 | Pat E-3 | Patient E | The Netherlands | 210-216-S-6 | AAAA | ND | Genomovar III |
| 41C029 | Pat E-4 | Patient E | The Netherlands | 210-216-S-6 | AAAA | Genomovar III | Genomovar III |
| 41C068 | Pat3 H111 | Patient 3 | Germany | 210-219-S-7 | AAAB | ND | Genomovar III |
| 41C067 | Pat3 H116 | Patient 3 | Germany | 210-219-S-7 | AAAB | ND | Genomovar III |
| 41C066 | Pat3 H124 | Patient 3 | Germany | 210-219-S-7 | AAAB | Genomovar III | Genomovar III |
| 41C072 | Pat8 H149 | Patient 8 | Germany | 210-219-S-8 | AAAB | ND | Genomovar III |
| 41C071 | Pat8 H157 | Patient 8 | Germany | 210-219-S-8 | AAAB | ND | Genomovar III |
| 41C070 | Pat8 H169 | Patient 8 | Germany | 210-219-S-8 | AAAB | Genomovar III | Genomovar III |
| 41C044 | C6433 | Sputum, CF patient | Canada | 210-225-S-8 | AAAB | Genomovar III | Genomovar III |
| 41C045 | C5424 (ET12) | Sputum, CF patient | Canada | 210-226-S-1 | AAAB | Genomovar III | Genomovar III |
| 41C046 | C1257 | Sputum, CF patient | Canada | 210-226-S-2 | AAAB | Genomovar III | Genomovar III |
| 41C118 | LMG16654 | Sputum, CF patient | ? | 210-254-S-3 | ND | Genomovar III | Genomovar III |
| 41C119 | LMG16656 (ET12) | Sputum, CF patient | ? | 210-254-S-4 | ND | Genomovar III | Genomovar III |
| 41C123 | LMG18941 | Sputum, CF patient | ? | 210-254-S-7 | ND | Genomovar VI | Genomovar VI |
| 41C124 | LMG18942 | Sputum, CF patient | ? | 210-254-S-7 | ND | Genomovar VI | Genomovar VI |
| 41C019 | CCUG3314 | ? | ? | 210-232-S-6 | ND | R. pickettii | R. pickettii |
| 41C093 | Pat (41C093) | ? | ? | 210-237-S-5 | ND | ND | R. pickettii |
| 41C090 | Fish water | Fish water | ? | 210-237-S-5 | ND | ND | R. pickettii |
| 41C091 | Pat BK ICP | Patient | ? | 210-237-S-5 | ND | ND | R. pickettii |
| 41C018 | CCUG3318 | Patient | USA | 210-238-S-1 | ND | R. pickettii | R. pickettii |
?, not known.
ND, not determined.
USA, United States of America.
Automated ribotyping.
Strains were grown overnight on Brucella blood agar medium (bioMerieux, Marcy l'Étoile, France). Automated ribotyping was performed under the conditions recommended by the manufacturer of the Riboprinter microbial characterization system (Qualicon Europe Ltd., Warwick, United Kingdom) (34). Restriction enzymes that were validated with respect to their usefulness were EcoRI and PvuII. A large DNA probe harboring the genes for both the small-subunit rRNA and the large-subunit rRNA of Escherichia coli was employed. The ribotypes were categorized in different ribogroups by the Riboprinter. The banding patterns were compared by using the GelCompar software (Applied Maths, Ghent, Belgium). Clustering was performed by the unweighted pair group method with arithmetic averages (UPGMA) method based on the Pearson correlation coefficient, using an optimization coefficient of 1.2% (identical to that used by the Riboprinter analysis system).
Whole-cell protein electrophoresis.
Whole-cell protein electrophoresis was used to determine the species of representative strains of most ribogroups. Bacteriological purity was determined by plating and examining living cells by phase-contrast microscopy and by Gram staining. Strains were grown on nutrient agar (CM3; Oxoid, Haarlem, The Netherlands) supplemented with 0.04% (wt/vol) KH2PO4 and 0.24% (wt/vol) Na2HPO4 · 12H2O. Agar plates were incubated aerobically at 28°C. After 48 h, whole-cell protein extracts were prepared and sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed as described previously (39). The densitometric analysis, normalization, and interpolation of the protein profiles, including numerical analyses, were performed by using the Pearson product moment correlation coefficient and are expressed as percent similarity values. Data for referral have been presented on previous occasions (39, 40; Coenye et al., submitted). Only a representative set of isolates, described in Table 1, was analyzed in this way.
PFGE.
Strains from the Hannover region of Germany were analyzed by pulsed-field gel electrophoresis (PFGE) in accordance with experimental protocols described previously (32). Total SpeI digests of agarose-embedded genomic DNA were separated in 1% agarose gels in 0.5× TBE buffer (45 mM Tris, 45 mM boric acid, 1 mM EDTA [pH 8.3]) by using a Bio-Rad CHEF-DR III apparatus (two linear ramps [17 h, 3 to 35 s; 20 h, 5 to 80 s], 120° reorientation angle, 210 V). After electrophoresis, gels were stained with ethidium bromide and photographed by using Polaroid equipment.
Epidemic-marker PCR.
PCR amplification to test for the presence of the B. cepacia epidemic strain marker was performed as described elsewhere (26). Control experiments to ensure the quality of DNA were performed by amplifying part of the 16S RNA gene region, using primers BuRa-16-1 and BuRa-16-2 (4).
RESULTS
Technical issues.
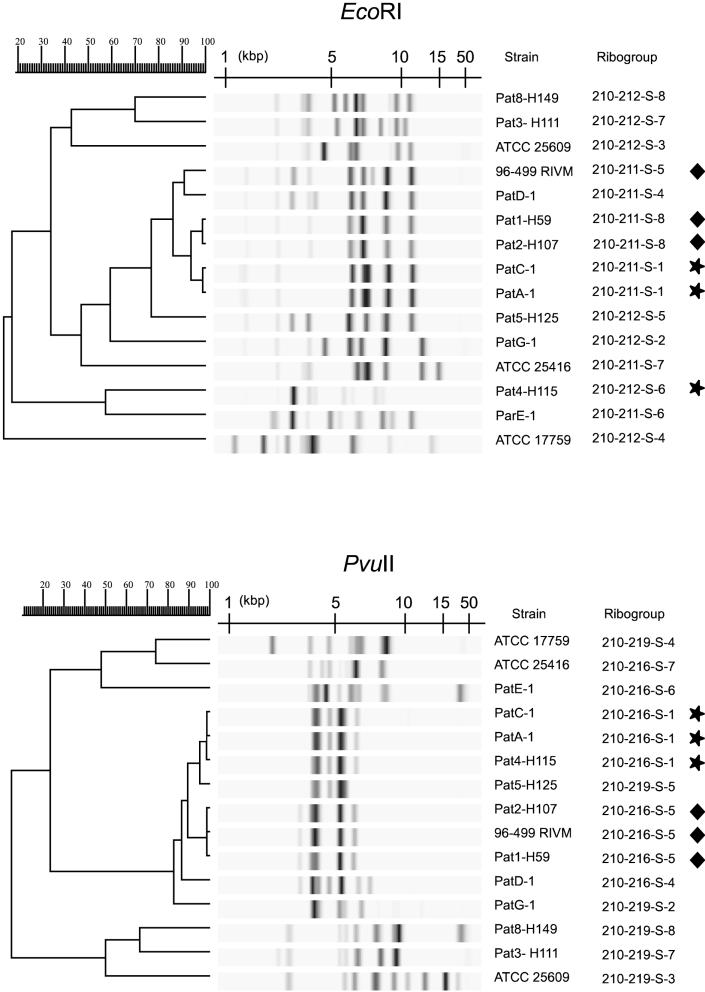
Pilot experiments performed with a limited number of strains revealed that EcoRI provides a higher level of resolution than PvuII (Fig. 1). Indeed, two groups of three strains each that were indistinguishable with PvuII were each separated into two different ribogroups with EcoRI. However, since the use of PvuII resulted in a sharper clustering of all of the B. multivorans strains tested, this enzyme appeared potentially more useful for genomovar identification. Therefore, the entire collection of strains was tested with PvuII. In total, the strains analyzed exhibited 39 distinct patterns, or ribotypes (Fig. 2). Adequate reproducibility of the tests was indicated by the fact that the duplicate reference strains generated identical patterns (CCUG12691 equals ATCC 25416, CCUG2115 equals ATCC 19302, and CCUG1782 equals ATCC 10248). The only exception was the duplicate ATCC 17616-LMG17588, which was categorized into two distinct ribogroups by the Riboprinter (Table 1). However, the two patterns were very similar (Fig. 2), and the difference could be attributed to a slightly incomplete digestion of the DNA of strain LMG17588. The pattern of this strain presented a number of faint bands as well as a high-molecular-weight band not evident when strain ATCC 17616 was analyzed. This result indicates that when strains are being typed, Riboprinter analysis results must be checked by visual inspection of the patterns, and differences in the high-molecular-weight range should be interpreted with caution. Importantly, the differences between the patterns of these two samples did not affect their clustering (Fig. 2).
FIG. 1.
Comparative analysis of the riboprints obtained, using either EcoRI or PvuII, for a subset of Burkholderia spp. strains. The two triplets of strains undistinguished using PvuII (labeled with stars and diamonds, respectively) were each separated into two groups by using EcoRI. Within each triplet, EcoRI distinction was in agreement with the geographic origin (Table 1). For cluster analysis, the UPGMA method was used, based on the matrix of Pearson correlation (see Materials and Methods). In the dendrogram scale, correlation levels were converted to percent similarity levels.
FIG. 2.
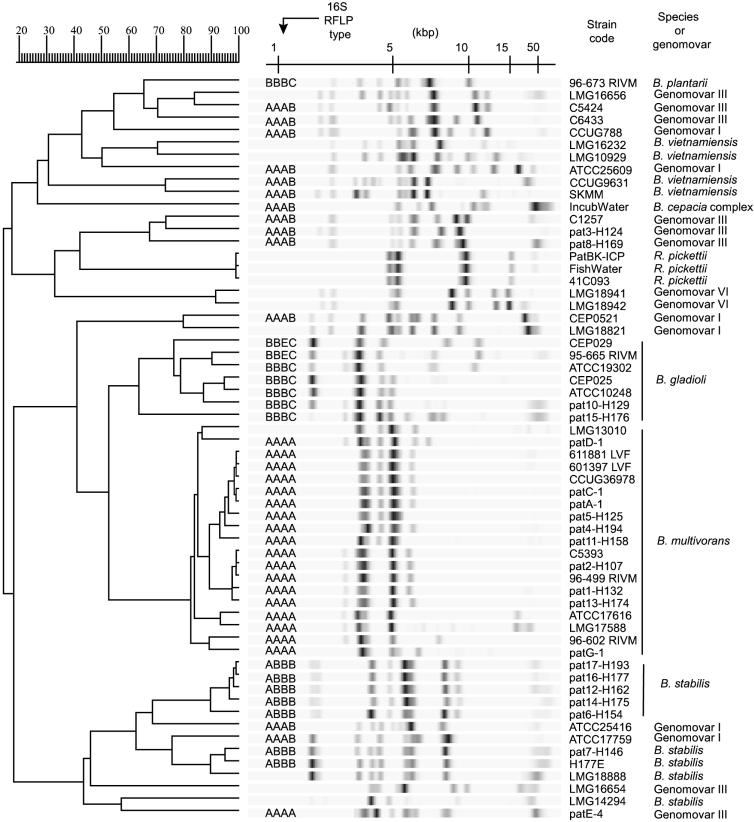
Overview of the PvuII riboprints obtained for Burkholderia spp. and R. pickettii strains, showing correspondence with 16S rRNA gene PCR-RFLP information gathered previously (41). Strain codes correspond to those listed in Table 1. Serial samples belonging to individual Dutch or German patients are indicated by lettering (patients A to G) and figures (patients 1 to 17), respectively. Only one isolate per patient was included. Note that not all strains for all patients are included. On the right are the species or genomovar names, as deduced from the ribotyping analysis and via characterization by whole-cell protein profiling of reference strains of each ribogroup (see Table 1). For cluster analysis, the UPGMA method was used, based on the matrix of Pearson correlation (see Materials and Methods). In the dendrogram scale, correlation levels were converted to percent similarity levels.
When the entire collection of strains was considered, the patterns of four strains (two B. cepacia strains from patient B and two R. pickettii strains) reproducibly showed only a single large DNA fragment (data not shown). Whether this is due to a scarcity of restriction sites or differences in restriction and modification systems is currently under investigation. Of the four discrepant strains, the DNAs of the one B. cepacia strain and the one R. pickettii strain tested with EcoRI were cut effectively, however (data not shown).
Concordance between genomovar classification and ribotype identification.
Figure 2 surveys all of the different ribotypes that were identified in the present set of strains. Only one isolate per patient or per duplicate strain is shown (except for ATCC 17616-LMG17588 [see above]). Table 1 shows the number of occurrences of each ribotype and indicates the number of samples tested for each patient and duplicate strain. One or several representative strains of most ribogroups were identified at the species level by whole-cell protein electrophoresis. These strains, considered as ribogroup-specific reference strains, are indicated in Table 1.
When the riboprints were clustered by the UPMGA method, several interesting subsets of strains were observed (Fig. 2). First, all B. gladioli strains fell into six ribogroups which clustered into a single branch of the dendrogram. Similarly, strains belonging to the species R. pickettii, as well as the single strain of B. plantarii, segregated separately. Second, distinctions between the genomovars were evident since strains belonging to different genomovars never showed the same riboprint pattern.
Several ribotypes were distinguished within each of the genomovars. For example, among nine strains in genomovar III, eight ribogroups were found. Moreover, within this genomovar, the two strains (C5424 and LMG16656) previously characterized by multilocus enzyme electrophoresis as belonging to electrophoretic type ET12 (39) had clearly distinct riboprints. Genomovar VI could not be separated into different ribogroups, but only two strains with this genomovar type were analyzed. These two isolates (LMG18941 and LMG18942) were clearly distinct from all other strains, in agreement with their recent distinction as a separate genomovar (Coenye et al., submitted).
With respect to the potential of ribotyping for genomovar identification, it was very interesting that all of the B. multivorans isolates (formerly genomovar II) clustered into one group. Moreover, B. multivorans strains could be distinguished from the other B. cepacia complex strains on the basis of their unique riboprints. Generally, these patterns contained only three or four bands restricted to a narrow range of molecular weights, whereas most of the other strains showed patterns with more hybridizing fragments and more variation in their size.
In contrast to the strains of B. multivorans and of genomovar VI, genomovar I, III, IV, and V strains did not cluster into separate branches (Fig. 2). However, a tendency of strains of a given genomovar to cluster was observed. For example, all strains of B. stabilis (genomovar IV) clustered in the lower branch of the dendrogram. More interestingly, however, smaller clusters specifically comprised several ribotypes of a single genomovar (e.g., strains LMG16656, C5424, and C6433 of genomovar III, CCUG9631 and SKMM of B. vietnamiensis, and CEP0521 and LMG18821 of genomovar I).
Concordance between 16S PCR-RFLP classification and ribotype identification.
The clustering obtained on the basis of ribotyping was in good agreement with 16S PCR-RFLP analysis data (Fig. 2). For example, most strains of the upper branch of the dendrogram showed the AAAB type, although they belonged to three distinct genomovars (I, III, and V). In genomovar III, the only analyzed strain (patE-4) that did not cluster with the other strains of that genomovar was also the only one with a different RFLP type (AAAA, versus AAAB in the others).
Obviously, the riboprints showed a higher degree of variability than RFLP typing. For instance, among the strains identified with PCR-RFLP code AAAA, eight different ribotypes were discerned. The increased heterogeneity among the riboprints was more obvious among the PCR-RFLP AAAB types; 11 different riboprint patterns were found. Finally, ribotyping distinguished between B. plantarii on the one hand and B. gladioli strains that shared the BBBC type with B. plantarii on the other hand. In no instance were different RFLP types found among the strains of a given ribogroup. In the B. cepacia complex, when considering only the 48 strains from different patients and from different environmental isolates, Simpson's index of diversity (used as an estimation of the discriminatory power [18]) for the PvuII characterization was 0.967. In comparison, RFLP discrimination among the 38 of such strains analyzed yielded a discriminatory power index of 0.647.
Analysis of correspondence between genomovar classification and RFLP type showed that except for the patient E strains, RFLP type AAAA was specific for genomovar II. Moreover, RFLP type ABBB was characteristic of B. stabilis (genomovar IV). Although genomovar I strains were scattered across the dendrogram, all showed the AAAB type, but this type was also found among genomovar III and V strains.
Serial samples from individual patients and geographic spread of clones.
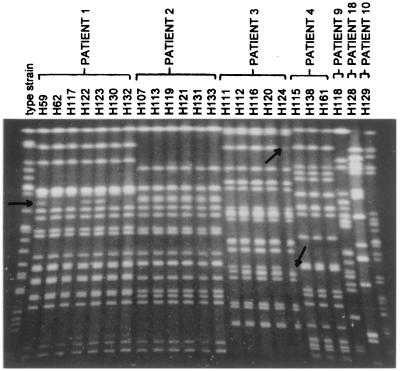
For several individual German or Dutch patients, three to seven isolates collected at different times were included in the analysis. In all instances, strains belonged to a single ribogroup when individual patients were considered. Consequently, persistent colonization by a single ribotype appears to be the most prevalent situation. However, using PFGE, minor differences could be detected between strains from the same patient (Fig. 3), suggesting evolution of the PFGE pattern in the course of the infection.
FIG. 3.
Comparison of the PFGE fingerprints obtained for serial isolates of B. cepacia from individual CF patients from the Hannover region (Germany). From left to right, the lanes contain SpeI-digested DNA derived from strains isolated from patients 1, 2, 3, 4, 9, 18, and 10. The strain isolated from the latter patient is a B. gladioli isolate and is included for comparative reasons. The H-prefixed numbers identify the individual isolates, whose characteristics are listed in Table 1. Arrows identify banding pattern polymorphism among clusters of strains derived from the same patient.
In several instances, interpatient transfer was suspected since strains belonging to the same ribogroup were encountered. This was the case for ribogroup 210-216-S-1 in Dutch patients A and C and in German patient 4, as well as for ribogroup 210-216-S-5 in German patients 1, 2, and 13 and in Dutch patient Ve (strain 96-499 RIVM) (Table 1). When investigated with EcoRI, however, these two sets of strains were separated according to their geographic origins (Fig. 1). Moreover, although the strains from German patients 1 and 2 were distinguished neither by PvuII nor by EcoRI, they proved to represent two different, although closely related, subtypes by PFGE (Fig. 3). Therefore, ribotype characterization with PvuII could not in itself be considered as an indication of an epidemiological link. However, epidemiological data suggested that nosocomial acquisition of B. cepacia occurred in three of the five patients who were harboring ribogroup 210-233-S-8, which belongs to genomovar IV (Table 1). Indeed, B. cepacia was initially detected in sputa from patients 14, 16, and 17 when they were seen for the first time in the Hannover clinic after returning from a stay at the same CF rehabilitation center. The B. cepacia isolates from these three patients exhibited identical SpeI restriction fragment patterns. However, interpreting typing data for strains of B. cepacia genomovar IV is not straightforward since genetic variability in this genomovar appears to be very limited (40).
To check for a correlation between ribogroups and the presence of a genetic marker recently shown to correlate with the epidemicity of B. cepacia strains (26), strains from each ribogroup and patient were screened by PCR. Only seven isolates (LMG16656, C5424, C6433, C1257, and the three strains from patient 8), representing five ribogroups, were positive, and all of them belonged to genomovar III. Among these seven were the two representatives of ET12. The possibly epidemic clone of B. stabilis identified in this study (ribogroup 210-233-S-8) did not harbor the epidemicity marker.
DISCUSSION
Identification of Burkholderia species.
Efforts to develop diagnostic tests for B. cepacia genomovars are presently intense (3, 4, 23, 33, 41). However, distinguishing between some genomovars, in particular genomovars I, III, and IV, has proven difficult or impossible, as was also illustrated for PCR-RFLP in the present study. Presently existing automated systems for bacterial identification, such as the Vitek and MicroScan systems, perform with varying but consistently insufficient accuracy for B. cepacia and B. gladioli identification, as was illustrated recently (41). Simple and reliable routine identification systems for Burkholderia species are currently lacking. In a previous study (41), PCR-RFLP analysis with four restriction enzymes was shown to be one of the best methods for Burkholderia species identification when done after screening by selective agar plate culture and performing API 20NE galleries to exclude all irrelevant bacterial species. Our results show that automated ribotyping has potential for identification of species of the B. cepacia complex.
Our results indicate that automated ribotyping with the Riboprinter has great potential for reliable distinction between strains of B. cepacia genomovars I, III, and VI, B. multivorans (genomovar II), B. stabilis (genomovar IV), B. vietnamiensis (genomovar V), and B. gladioli. Moreover, there is real potential for identification at the species level. First, strains belonging to different species or genomovars never showed the same ribotype. Therefore, the hypothesis that two strains belonging to the same ribogroup also belong to the same genomovar seems reasonable. Thus, determining the genomovar to which a strain belongs could be achieved by investigating whether its ribotype corresponds to one already present in the database. As with any library-based identification system, progressive expansion of the database of patterns with those of newly characterized strains will increase the probability of identifying a new strain. For this, any strain showing a new ribotype should be identified at the genomovar level by current methods. In addition, electronic communication between Riboprinters located in different laboratories or countries is now being initiated, making it possible to exploit fully the potential of library typing systems based on automated ribotyping.
Second, clustering analysis could also be useful. Our results show that when PvuII is used, B. multivorans, B. gladioli, and B. cepacia genomovar VI each correspond to a single cluster. In these cases, a new ribotype that falls within such a cluster could also be considered as belonging to this species. For example, although strains from patient D showed a unique ribotype, it clustered with B. multivorans patterns and therefore probably corresponds to B. multivorans. This approach could also be used for the smaller, species-specific clusters identified in genomovars I, II, and V.
As stated above, we were able to determine, based on the ribotyping analysis, the species of all strains in the study (Table 1) except the incubator water strain. This strain was not characterized by protein profiling and fell in a unique and unclustered ribogroup. As illustrated by this strain, two limitations to this approach exist. First, depending on the number of ribotypes existing in a given species, obtaining a comprehensive database including most of the representative ribotypes of this species may be difficult to achieve. Second, further research on more isolates will be needed to determine if any newly characterized strain of a given species will fall in its corresponding cluster. At the same time, it will have to be verified that no strain of any other species will by chance fall in this cluster. This risk is illustrated, for example, by the fact that strains ATCC 25416 and ATCC 17759 of genomovar I fell within the cluster containing all B. stabilis strains (Fig. 2).
Both limitations might be overcome by searching for restriction enzymes with less discriminatory power. Indeed, this not only would reduce the number of different ribotypes within a species but also would result in patterns comprising more-conserved characters, including diagnostic ones, and therefore in a better species-specific clustering. For these reasons, we opted to use PvuII rather than EcoRI, because of the lower discriminatory power of the former enzyme. However, a search for even less-discriminatory enzymes might be warranted. The identification potential of enzymes HincII and SmaI is presently under investigation, since they proved to yield interesting clustering results for a wide range of pseudomonads (5).
Characterization of Burkholderia spp. strains.
Many different procedures have proven useful for discrimination of Burkholderia spp. strains below the species level. The technologies used generally identify polymorphism at the protein level or screen chromosomes for variability in restriction sites or PCR primer annealing sites (14, 24, 25, 39). The PCR-RFLP approach is one that can be used for monitoring the spread of bacterial strains as well as the dynamics of patient colonization and infection (8, 36). It must be emphasized, however, that the resolution of PCR-RFLP is lower than that of PFGE. German patients 1 and 2, for instance, were carrying strains that were both of the PCR-RFLP AAAA type (Table 1) but were clearly differentiated by PFGE (Fig. 3).
Conventional ribotyping is not a novel approach for the genetic characterization of bacteria in general or for Burkholderia spp. in particular. Ribotyping has been instrumental in the analysis of nosocomial outbreaks of infection (28), in the deciphering of the spread of infectious strains in restricted locales such as summer camps for CF patients (29), and for the identification of clones capable of extensive geographic spread (31, 37). Over the years, ribotyping has acquired a central position among the technologies available to the microbial epidemiologist. Due to its automation, the Riboprinter microbial characterization system is suited for rapid and high-throughput typing of bacterial strains. This approach was shown to be successful for the epidemiological study of strains belonging to species such as Pseudomonas aeruginosa, Listeria monocytogenes, and E. coli (1, 13, 30). An important advantage of this automated system is its standardization and easily manageable database. Therefore, it can be developed as a library typing system by which strains characterized at different times and various locations can be reliably compared (38). Whether there is a link between pathogenicity or epidemicity potential and ribotype cannot be assessed on the basis of the present data set, but the determination of such a link will certainly be facilitated by this characterization system.
Using ribotyping, assessments of subspecific variation, suited for epidemiological and patient-related research, were also possible in this study. In agreement with the data of LiPuma et al. (22), long-term colonization with a single strain was documented in all patients investigated. Geographic dissemination was determined, and we showed that both local prevalence and extensive spread can be documented for different ribotypes. However, achieving the best discrimination between strains was not the goal of the present study, and PFGE typing proved to be more discriminatory than PvuII and EcoRI ribotyping, even on a limited test sample. However, due to its speed (8 h for characterization of strains), automated ribotyping might be useful for epidemiological investigations in a hierarchical approach combining a preliminary screening for ribotype differences with complementary, slower approaches when no discrimination is evidenced (30).
ACKNOWLEDGMENTS
We gratefully acknowledge Cindy van Pelt (Department of Medical Microbiology and Infectious Diseases, Erasmus University Medical Center Rotterdam, Rotterdam, The Netherlands) for maintaining the culture collection of Burkholderia spp. and for help with culture of the strains. Helke van Dessel and Karlijn Kusters are thanked for practical assistance in performing part of the riboprint analyses.
Peter Vandamme is a postdoctoral research fellow of the Fund for Scientific Research—Flanders (Belgium). Sylvain Brisse was supported by a European Communities TMR grant.
REFERENCES
- 1.Allerberger F, Fritschel S J. Use of automated ribotyping of Austrian Listeria monocytogenes isolates to support epidemiological typing. J Microbiol Methods. 1999;35:237–244. doi: 10.1016/s0167-7012(99)00025-1. [DOI] [PubMed] [Google Scholar]
- 2.Barker P M, Wood R E, Gilligan P H. Lung infection with Burkholderia gladioli in a child with cystic fibrosis: acute clinical and spirometric deterioration. Pediatr Pulmonol. 1997;23:123–125. doi: 10.1002/(sici)1099-0496(199702)23:2<123::aid-ppul9>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 3.Bauernfeind A, Schneider I, Jungwirth R, Roller C. Discrimination of Burkholderia gladioli from other Burkholderia species detectable in cystic fibrosis patients by PCR. J Clin Microbiol. 1998;36:2748–2751. doi: 10.1128/jcm.36.9.2748-2751.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauernfeind A, Schneider I, Jungwirth R, Roller C. Discrimination of Burkholderia multivorans and Burkholderia vietnamiensis from Burkholderia cepacia genomovars I, III, and IV by PCR. J Clin Microbiol. 1999;37:1335–1339. doi: 10.1128/jcm.37.5.1335-1339.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosch R, Lefrevre M, Grimont F, Grimont P. Taxonomic diversity of pseudomonads revealed by computer-interpretation of ribotyping data. Syst Appl Microbiol. 1996;19:541–555. [Google Scholar]
- 6.Christenson J C, Welch D F, Mukwaya G, Muszynski M J, Weaver R E, Brenner D J. Recovery of Pseudomonas gladioli from respiratory tract specimens of patients with cystic fibrosis. J Clin Microbiol. 1989;27:270–273. doi: 10.1128/jcm.27.2.270-273.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coenye T, Holmes B, Kersters K, Govan J R, Vandamme P. Burkholderia cocovenenans (van Damme et al. 1960) Gillis et al. 1995 and Burkholderia vandii Urakami et al. 1994 are junior synonyms of Burkholderia gladioli (Severini 1913) Yabuuchi et al. 1993 and Burkholderia plantarii (Azegami et al. 1987) Urakami et al. 1994, respectively. Int J Syst Bacteriol. 1999;49:37–42. doi: 10.1099/00207713-49-1-37. [DOI] [PubMed] [Google Scholar]
- 8.Dasen S E, LiPuma J J, Kostman J R, Stull T L. Characterization of PCR ribotyping for Burkholderia (Pseudomonas) cepacia. J Clin Microbiol. 1994;32:2422–2424. doi: 10.1128/jcm.32.10.2422-2424.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans E, Poxton I R, Govan J R. Lipopolysaccharide chemotypes of Burkholderia cepacia. J Med Microbiol. 1999;48:825–832. doi: 10.1099/00222615-48-9-825. [DOI] [PubMed] [Google Scholar]
- 10.Gillis M, Van Van T, Bardin R, Goor M, Hebbar P, Willems A, Segers P, Kersters K, Heulin T, Fernandez M P. Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int J Syst Bacteriol. 1995;45:274–289. [Google Scholar]
- 11.Govan J R W, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graves M, Robin T, Chipman A M, Wong J, Khashe S, Janda J M. Four additional cases of Burkholderia gladioli infection with microbiological correlates and review. Clin Infect Dis. 1997;25:838–842. doi: 10.1086/515551. [DOI] [PubMed] [Google Scholar]
- 13.Grif K, Karch H, Schneider C, Daschner F D, Beutin L, Cheasty T, Smith H, Rowe B, Dierich M P, Allerberger F. Comparative study of five different techniques for epidemiological typing of Escherichia coli O157. Diagn Microbiol Infect Dis. 1998;32:165–176. doi: 10.1016/s0732-8893(98)00103-5. [DOI] [PubMed] [Google Scholar]
- 14.Hales B A, Morgan J A W, Hart C A, Winstanley C. Variation in flagellin genes and proteins of Burkholderia cepacia. J Bacteriol. 1998;180:1110–1118. doi: 10.1128/jb.180.5.1110-1118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henry D, Campbell M, McGimpsey C, Clarke A, Louden L, Burns J L, Roe M H, Vandamme P, Speert D. Comparison of isolation media for recovery of Burkholderia cepacia complex from respiratory secretions of patients with cystic fibrosis. J Clin Microbiol. 1999;37:1004–1007. doi: 10.1128/jcm.37.4.1004-1007.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henry D A, Campbell M E, LiPuma J J, Speert D P. Identification of Burkholderia cepacia isolates from patients with cystic fibrosis and use of a simple new selective medium. J Clin Microbiol. 1997;35:614–619. doi: 10.1128/jcm.35.3.614-619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoare S, Cant A J. Chronic granulomatous disease presenting as severe sepsis due to Burkholderia gladioli. Clin Infect Dis. 1996;23:411. doi: 10.1093/clinids/23.2.411. [DOI] [PubMed] [Google Scholar]
- 18.Hunter P R, Gaston M A. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isles A, Maclusky I, Corey M, Gold R, Prober C, Fleming P, Levison H. Pseudomonas cepacia in cystic fibrosis: an emerging problem. J Pediatr. 1984;104:206–210. doi: 10.1016/s0022-3476(84)80993-2. [DOI] [PubMed] [Google Scholar]
- 20.Kanj S S, Tapson V, Davis R D, Madden J, Browning I. Infections in patients with cystic fibrosis following lung transplantation. Chest. 1997;112:924–930. doi: 10.1378/chest.112.4.924. [DOI] [PubMed] [Google Scholar]
- 21.Khan S U, Gordon S M, Stillwell P C, Kirby T J, Arroliga A C. Empyema and bloodstream infection caused by Burkholderia gladioli in a patient with cystic fibrosis after lung transplantation. Pediatr Infect Dis J. 1996;15:637–639. doi: 10.1097/00006454-199607000-00020. [DOI] [PubMed] [Google Scholar]
- 22.LiPuma J J, Fischer M C, Dasen S E, Mortensen J E, Terrence L S. Ribotype stability of serial pulmonary isolates of Pseudomonas cepacia. J Infect Dis. 1991;164:133–136. doi: 10.1093/infdis/164.1.133. [DOI] [PubMed] [Google Scholar]
- 23.LiPuma J J, Dulaney B J, McMenamin J D, Whitby P W, Stull T L, Coenye T, Vandamme P. Development of rRNA-based PCR assays for identification of Burkholderia cepacia complex isolates recovered from cystic fibrosis patients. J Clin Microbiol. 1999;37:3167–3170. doi: 10.1128/jcm.37.10.3167-3170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu P Y-F, Shi Z-Y, Lau Y-J, Hu B-S, Shyr J-M, Tsai W-S, Lin Y-H, Tseng C-Y. Comparison of different PCR approaches for characterization of Burkholderia (Pseudomonas) cepacia isolates. J Clin Microbiol. 1995;33:3304–3307. doi: 10.1128/jcm.33.12.3304-3307.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livesley M A, Baxter I A, Lambert P A, Govan J R, Weller P H, Lacey D E, Allison D G, Giwercman B, Hoiby N. Subspecific differentiation of Burkholderia cepacia isolates in cystic fibrosis. J Med Microbiol. 1998;47:999–1006. doi: 10.1099/00222615-47-11-999. [DOI] [PubMed] [Google Scholar]
- 26.Mahenthiralingam E, Simpson D A, Speert D P. Identification and characterization of a novel DNA marker associated with epidemic Burkholderia cepacia strains recovered from patients with cystic fibrosis. J Clin Microbiol. 1997;35:808–816. doi: 10.1128/jcm.35.4.808-816.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otterbein C K, Splettstoesser W D, Linde H J, Grunow R, Wolf H, Finke E J, Neubauer H. Development and characterization of a murine monoclonal antibody reactive with a 64 kDa somatic antigen of Burkholderia cepacia. Hybridoma. 1998;17:143–150. doi: 10.1089/hyb.1998.17.143. [DOI] [PubMed] [Google Scholar]
- 28.Ouchi K, Abe M, Karita M, Oguri T, Igari J, Nakazawa T. Analysis of strains of Burkholderia (Pseudomonas) cepacia isolated in a nosocomial outbreak by biochemical and genomic typing. J Clin Microbiol. 1995;33:2353–2357. doi: 10.1128/jcm.33.9.2353-2357.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pegues D A, Carson L A, Tablan O C, Fitzsimmons S C, Roman S B, Miller J M. Acquisition of Pseudomonas cepacia at summer camps for patients with cystic fibrosis. J Pediatr. 1994;124:694–702. doi: 10.1016/s0022-3476(05)81357-5. [DOI] [PubMed] [Google Scholar]
- 30.Pfaller M A, Wendt C, Hollis R J, Wenzel R P, Fritschel S J, Neubauer J J, Herwaldt L A. Comparative evaluation of an automated ribotyping system versus pulsed-field gel electrophoresis for epidemiological typing of clinical isolates of Escherichia coli and Pseudomonas aeruginosa from patients with recurrent gram-negative bacteremia. Diagn Microbiol Infect Dis. 1996;25:1–8. doi: 10.1016/0732-8893(96)00082-x. [DOI] [PubMed] [Google Scholar]
- 31.Pitt T L, Kaufmann M E, Patel P S, Benge L C, Gaskin S, Livermore D M. Type characterisation and antibiotic susceptibility of Burkholderia (Pseudomonas) cepacia isolates from patients with cystic fibrosis in the United Kingdom and the Republic of Ireland. J Med Microbiol. 1996;44:203–210. doi: 10.1099/00222615-44-3-203. [DOI] [PubMed] [Google Scholar]
- 32.Römling U T, Heuer T, Tümmler B. Bacterial genome analysis by pulsed field gel electrophoresis techniques. Adv Electrophoresis. 1994;7:355–406. [Google Scholar]
- 33.Segonds C, Heulin T, Marty N, Chabanon G. Differentiation of Burkholderia species by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene and application to cystic fibrosis isolates. J Clin Microbiol. 1999;37:2201–2208. doi: 10.1128/jcm.37.7.2201-2208.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sethi M R. Fully automated microbial characterization and identification for industrial microbiologists. Am Lab. 1997;5:31–35. [Google Scholar]
- 35.Shin J H, Kim S H, Shin M G, Suh S P, Ryang D W, Jeong M H. Bacteremia due to Burkholderia gladioli: case report. Clin Infect Dis. 1997;25:1264–1265. doi: 10.1086/516973. [DOI] [PubMed] [Google Scholar]
- 36.Shreve M R, Johnson S J, Milla C E, Wielinski C L, Regelmann W E. PCR ribotyping and endonuclease subtyping in the epidemiology of Burkholderia cepacia infection. Am J Respir Crit Care Med. 1997;155:984–989. doi: 10.1164/ajrccm.155.3.9117036. [DOI] [PubMed] [Google Scholar]
- 37.Steinbach S, Sun L, Jiang R Z, Flume P, Gilligan P, Egan T M, Goldstein R. Transmissibility of Pseudomonas cepacia infection in clinic patients and lung transplant recipients with cystic fibrosis. N Engl J Med. 1994;331:981–987. doi: 10.1056/NEJM199410133311504. [DOI] [PubMed] [Google Scholar]
- 38.Struelens M J, de Gheldre Y, Deplano A. Comparative and library epidemiological typing systems: outbreak investigations versus surveillance systems. Infect Control Hosp Epidemiol. 1998;19:565–569. doi: 10.1086/647874. [DOI] [PubMed] [Google Scholar]
- 39.Vandamme P, Holmes B, Vancanneyt M, Coenye T, Hoste B, Coopman R, Revets H, Lauwers S, Gillis M, Kersters K, Govan J R. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;47:1188–1200. doi: 10.1099/00207713-47-4-1188. [DOI] [PubMed] [Google Scholar]
- 40.Vandamme P, Mahenthiralingam E, Holmes B, Coenye T, Hoste B, de Vos P, Henry D, Speert D P. Identification and population structure of Burkholderia stabilis sp. nov. (formerly Burkholderia cepacia genomovar IV) J Clin Microbiol. 2000;38:1042–1047. doi: 10.1128/jcm.38.3.1042-1047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Pelt C, Verduin C M, Goessens W H F, Vos M C, Tümmler B, Segonds C, Reubsaet F, Verbrugh H, van Belkum A. Identification of Burkholderia spp. in the clinical microbiology laboratory: comparison of conventional and molecular methods. J Clin Microbiol. 1999;37:2158–2164. doi: 10.1128/jcm.37.7.2158-2164.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welch D F, Muszynski M J, Pai C H, Marcon M J, Hribar M M, Gilligan P H, Matsen J M, Ahlin P A, Hilman B C, Chartrand S A. Selective and differential medium for recovery of Pseudomonas cepacia from the respiratory tracts of patients with cystic fibrosis. J Clin Microbiol. 1987;25:1730–1734. doi: 10.1128/jcm.25.9.1730-1734.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitby P W, Dick H L N, Campbell III P W, Tullis D E, Matlow A, Stull T L. Comparison of culture and PCR for detection of Burkholderia cepacia in sputum samples of patients with cystic fibrosis. J Clin Microbiol. 1998;36:1642–1645. doi: 10.1128/jcm.36.6.1642-1645.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yabuuchi E, Kosako Y, Oyaizu H, Yano I, Hotta H, Hashimoto Y, Ezaki T, Arakawa M. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol Immunol. 1992;36:1251–1275. doi: 10.1111/j.1348-0421.1992.tb02129.x. [DOI] [PubMed] [Google Scholar]
- 45.Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuchi N. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia pickettii (Ralston, Palleromi and Doudoroff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol Immunol. 1995;39:897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x. [DOI] [PubMed] [Google Scholar]