Abstract
Abstract
Inactivated and live attenuated vaccines have improved human life and significantly reduced morbidity and mortality of several human infectious diseases. However, these vaccines have faults, such as reactivity or suboptimal efficacy and expensive and time-consuming development and production. Additionally, despite the enormous efforts to develop vaccines against some infectious diseases, the traditional technologies have not been successful in achieving this. At the same time, the concerns about emerging and re-emerging diseases urge the need to develop technologies that can be rapidly applied to combat the new challenges. Within the last two decades, the research of vaccine technologies has taken several directions to achieve safe, efficient, and economic platforms or technologies for novel vaccines. This review will give a brief overview of the current state of the novel vaccine technologies, new vaccine candidates in clinical trial phases 1–3 (listed by European Medicines Agency (EMA) and Food and Drug Administration (FDA)), and vaccines based on the novel technologies which have already been commercially available (approved by EMA and FDA) with the special reference to pandemic COVID-19 vaccines.
Key points
• Vaccines of the new generation follow the minimalist strategy.
• Some infectious diseases remain a challenge for the vaccine development.
• The number of new vaccine candidates in the late phase clinical trials remains low.
Keywords: Vaccine, Immune response, Novel technologies, Infectious diseases, Clinical trials, Adjuvant
Introduction
Vaccination remains a very effective method of preventing infectious diseases and represents a relevant contribution to human health (Sallusto et al. 2010). The immense success of vaccines against polio, smallpox, measles, diphtheria, tetanus, and rabies demonstrates the potential of this phenomenal approach in reducing the global burden of infectious diseases and, in the case of smallpox, in completely eradicating it (Breman and Arita 1980; WHO 1980; Strassburg 1982). “Herd immunity” is a term describing threshold proportion of immune individuals which should cause a decline in incidence of infection in population (Fine et al. 2011). The coverage rate needed to stop the infection transmission depends on the basic reproduction number (Ro) which is defined as average number of transmissions expected from a single case introduced into a totally susceptible population. (Andre et al. 2008). Successful illustrations of community immunity which led in prevention of infection spreading or its complete elimination were identified in above-mentioned diseases, such as smallpox, measles, and poliovirus (Shelly et al. 2020). Infectious diseases with pandemic potential have regularly occurred and spread throughout history (plague, cholera, influenza, different corona viruses) (Piret and Boivin 2021). We are currently witnessing SARS-CoV-2 causing COVID-19 pandemic, which only proves that novel infectious agents with pandemic potential can emerge any time, any place. Still, every time it does emerge, it causes devastating consequences for human health and quality of life. Pandemic is the most dramatic form of emerging and spreading of infectious agents to which the effective vaccine cannot be prepared ahead. However, despite the remarkable successes of vaccines against some infectious diseases, others remain a major challenge, and there is an urgent need to develop vaccines against them. These are human pathogens causing a high rate of morbidity and mortality such as human immunodeficiency virus (HIV), Mycobacterium tuberculosis, Plasmodium falciparum, hepatitis C virus (HCV), respiratory syncytial virus (RSV), and others. Traditional approaches to vaccine development did not work for them. A long coexistence of host and pathogen has led to the evolution of the pathogen’s mechanisms enabling successful adaptation to the host and evasion of its immune response. Immune evasion encompasses a variety of complex mechanisms through which pathogen evades host anti-pathogen immunity and escapes recognition. The most common immune system evasion mechanisms are suppression of antigen presentation and T cell costimulatory pathways, induction of immunosuppressive cytokines, obstruction of interferon and chemokine functions, the establishment of persistency or latency, and high level of genetic variability of the pathogen (Arens 2012). Since the discovery of the HIV and beginning of the AIDS pandemic several decades ago, scientists have been making efforts to make an effective vaccine that would yield sterilizing neutralizing antibodies. However, due to the rapid establishment of latent HIV reservoir, heavy glycosylation of the envelope trimer thwarting neutralizing antibody attack, and extremely rapid HIV evolution, a vaccine is still not on the horizon (Haynes et al. 2019). A high mutational rate, structural plasticity of the immunodominant regions on the virion surface of diverse HCV genotypes, and constant amino acid substitutions on key structural components of envelope glycoproteins make the design of an effective HCV vaccine very challenging (Sevvana et al. 2021). Viruses evolved escape mechanisms by changing the antigenic regions at an extremely high rate. More than 250, 000 children under the age of five die from malaria every year (one child dies from malaria every two minutes), most of them in sub-Saharan Africa (https://www.who.int/publications/i/item/WHO-CDS-GMP-2018.05). Despite these catastrophic numbers, the malaria vaccine is still not available. A complex life cycle of the Plasmodium spp. and extensive polymorphism between strains from different geographical regions has only led to partial protection with some malaria vaccines in clinical trials (Bull et al. 1998; Sutherland 2007; Fowkes et al. 2010). The ultimate example of the failure to make an efficient vaccine by the traditional approach is tuberculosis (TB). TB is still a deadly disease, with an estimated 1.2 million TB deaths as a single infectious agent in 2019 and an additional 208, 000 deaths among HIV-positive patients (WHO 2020). The only licensed vaccine for the prevention of TB disease is the bacille Calmette-Guérin (BCG) vaccine developed a century ago. This vaccine prevents severe forms of TB in children and is widely used. There is currently no vaccine effective in preventing TB disease in adults, either before or after exposure to TB infection. The very complex physiology and pathogenesis of M. tuberculosis have so far led to unsuccessful vaccine candidates. M. tuberculosis has evolved different mechanisms to evade recognition by immune cells. It does not have a classical virulence factor like other bacterial pathogens either (Smith 2003). The lack of a validated immune correlate of protection, together with uncertainty as to which animal model, if any, best represents human disease, means vaccine development, and predicting which candidate vaccine might protect in humans is very challenging (Davenne and McShane 2016). New knowledge of M. tuberculosis genes and the proteins they encode should provide new bacterial targets that can be used to create new vaccines in combination with new vaccine technologies. These research breakthroughs are needed to rapidly reduce TB incidence worldwide to the levels already achieved in low-burden countries. (WHO 2020).
There is a number of infectious diseases that are awaiting efficacious and safe vaccines. However, improvement of the vaccines in use is also anticipated, e.g., for infections in adult age not only in childhood, for pregnant women, or older adults. Many vaccines, especially viral vaccines, have been developed within the last 70 years. Pathogen evolution, meanwhile, has continued with an emphasized selection of new pathogen variants under the influence of vaccine-driven evolutionary pressure what can lead to the failure of vaccine strategy. Mumps vaccine seems to be such an example. Despite the remarkable public health success of the mumps vaccine, evidence of virus escape from vaccine-induced immunity causing mumps resurgence is piling up. Vanning immunity, lack of natural boost, and a reduced capacity of vaccine-induced neutralizing antibodies to cross-neutralize circulating strains have been suggested as factors facilitating mumps virus to escape from vaccine-induced immunity (Santak et al. 2006; Ivancic-Jelecki et al. 2008; Cortese et al. 2011; Smits et al. 2013; Šantak et al. 2013; Šantak et al. 2015a, b; May et al. 2018; Ramanathan et al. 2018; Marshall and Plotkin 2019; Vermeire et al. 2019; Connell et al. 2020; Won et al. 2021) suggesting a new vaccine with better-matched epitopes will be needed soon. Hence, novel technologies for the development and production of vaccines are needed to effectively prevent and control infectious diseases in humans. The knowledge based on genomic analysis and systems biology and the novel vaccine technologies based on infectious pathogens can be applied to cancer vaccines and vice versa. This review will present significant and promising advances in cutting-edge vaccine technologies over the past decades.
A brief history of vaccine
The first preventive immunization was used to prevent smallpox. It was called variolation, and it was done by smearing skin tissue with smallpox pus. The description of variolation originates probably from China or India, and from there, it spreads to Ottoman Empire and later to Europe (Boylston 2012). The vaccine as we know it today dates back to the end of the eighteenth century. In 1796, an English physician Edward A. Jenner applied a common observation that milkmaids were immune to smallpox. He used a scraped pus from a cowpox blister from one milkmaid with cowpox (in Latin vacca means cow, hence the term vaccine) to inoculate a 13-year-old boy. Later on, he made a challenge with the variolous material to prove the effectiveness of the immunization process. From that point on, the smallpox vaccine was implemented worldwide, culminating in the smallpox eradication in 1980 (Breman and Arita 1980; WHO 1980; Strassburg 1982).
Ever since the door of the vaccine world has been ajar, there were many researches and trials. Some of them being a success, but many of them were not. After Jenner’s first trial, almost a hundred years passed until Louis Pasteur developed another successful vaccines: live attenuated chicken cholera vaccine, inactivated anthrax vaccine, and inactivated rabies vaccine in humans (in 1879, 1881 and 1885, respectively). As bacteriology developed over time vaccines against diphtheria, tetanus, anthrax, cholera, plague, typhoid, and tuberculosis rapidly followed (Aida et al. 2021).
The development of viral vaccines has awaited advances in modern technologies such as tissue culture and electron microscopy. Intensive research of new viral vaccines has been started since mid-twentieth century, and it lasts today. It is hard to single out vaccines with the most significant impact since each of them dramatically reduced the disease burden and decreased mortality (www.who.int/news-room/feature-stories/detail/counting-the-impact-of-vaccines). However, the development of polio and measles vaccines is worth mentioning since both diseases had high morbidity and mortality rates with severe sequelae prior introduction of vaccination. Vaccination has reduced the number of cases of illness, which gives hope that both of these diseases could be eradicated as smallpox was eradicated 40 years ago. Howe and Bodian (1941) observed that the virus which causes polio spreads by oral route. This observation led to the several vaccine candidates, all being oral vaccine candidates. One of them is Salk’s polio vaccine, a formaldehyde-inactivated (IPV) parenteral vaccine used from 1955 (Salk 1959). Another is Sabin’s live attenuated oral vaccine (OPV) used from the 1960s of the last century (Sabin et al. 1960). Although rarely, live attenuated polio vaccine was reported to cause vaccine-associated paralytic poliomyelitis. So today, IPV is preferred in most vaccination schedules (Wolbert and Higginbotham 2020). Edmonston strain of measles virus was isolated in tissue culture in 1954 (Enders and Peebles 1954). Most live attenuated measles vaccines used today (Edmonston-Zagreb, Schwartz, Moraten, and AIK-C) stem from this strain. Different regimes of virus attenuation in either chick embryo cells or human diploid cells are applied in different laboratories (Markowitz and Orenstein 1990). Attempts to make formaldehyde-inactivated measles vaccine failed because some individuals who received this vaccine developed atypical measles syndrome (Goodson and Seward 2015).
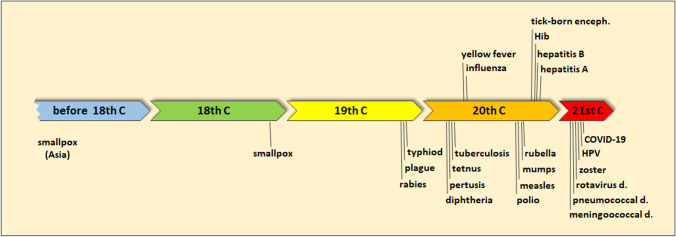
Vaccines for over twenty infectious diseases have been developed and successfully applied worldwide in the modern vaccinology era (Fig. 1). More than one vaccine type is available for some diseases, or better formulations replaced formulations with less efficacy and/or safety. In 20 years of this century, new vaccines against meningococcal disease, pneumococcal disease, rotavirus disease, herpes zoster, human papilloma virus, and COVID-19 have been approved (Fig. 1, Plotkin 2014). This shows that scientists and the pharmaceutical industry have recognized the potential and importance of vaccines as the human health improvement promoters.
Fig. 1.
History timeline of the most important vaccines
How should vaccines work?
The vaccination paradigm is the generation of a long-term immunity against the pathogen-specific antigen(s) by developing antibodies and cytotoxic T cells. To generate efficient vaccine-mediated protection requires activation of a complex network of cells and molecules of the immune system. Additionally, the vaccination should elicit a very similar immune response to one that develops upon resolving natural infection for those viruses that resolve on their own. Ideally, the immune response should be triggered at the site of the infection, e.g., a mucosa of the respiratory or gastrointestinal tract. Taken together, it is a complex challenge to achieve that everything cooperates as intended.
The first encounter of the body with the pathogen will result in adaptive response. However, this will take some time, and in the meantime, the pathogen will prevail, causing a disease. Vaccination prevents disease caused by the particular microorganism by enabling a fast response to infection and enhancing the immune response so that the microorganism cannot replicate at all (e.g., desired sterilizing immunity for HIV vaccine) or it will replicate, but at the level that will not cause a disease or at least it will cause only minor symptoms. In this way, the severe disease will not develop. Long-term protection requires a successful development of specific adaptive immune response and the persistence of vaccine antibodies above protective thresholds and/or the maintenance of memory immune cells capable of rapid and effective reactivation upon subsequent microbial exposure. Without many details, the series of events following the vaccination are (1) antigen-presenting cells (APC) (dendritic cells, monocytes, and neutrophils) are activated by the “danger signals”, (2) the activation changes the surface receptors on the APCs what initiate their migration to the local draining lymph nodes where (3) the activation of T and B lymphocytes will take place resulting in (4) specific B (antibody) and T (CD8 + T cell cytotoxicity) cell response and finally (5) development of memory cells (Siegrist 2008).
The very first requirement for effective response is for a vaccine to provide “danger signals” which will trigger reactions of the innate immune system. Antigen itself can be a trigger provider, but more often, it is the adjuvant. The primary role of the adjuvant is to enhance vaccine responses by inducing and modulating innate immunity, which will shape the adaptive responses. Most vaccines based on the live attenuated virus as an active substance do not require the addition of adjuvant because viral components and attenuated viral replication intensify and modulate immune response similar to natural infection, albeit to a slightly lower level (Markušić et al. 2014; Šantak et al. 2015a, b; Fleming 2016; Vasou et al. 2017; Nogales et al. 2018; Alsulaiman et al. 2020, reviewed in Griffin 2021 and Laksono et al. 2021). Other forms of vaccines follow minimalistic principles devoid of all components other than antigen. These forms of vaccines may require the action of adjuvant to induce effective immune response by vaccine design. This approach ensures the safety of the vaccine but is faced with poor immunogenicity. An essential point in the immune response to foreign infectious microorganisms and vaccines is distinguishing between “self” and “non-self”. This selectivity is achieved by the ability of the host to recognize (a) conserved products of microbial physiology and/or metabolism that are unique to microorganisms and are not produced by the host, (b) “markers of normal self”, or (c) “markers of abnormal self”. Conserved molecular patterns that are essential products of microbial physiology are called microbe-associated molecular patterns (MAMPs) and are recognized by receptors of the innate immune system cells called pattern recognition receptors (PRRs). The most common MAMPs are glycans, glycoconjugates, lipopolysaccharides, dsRNA, ssRNA, and unmethylated CpG motifs (Ramadan et al. 2017). The MAMPs are readily identified as “danger signals”, which lead to a series of events that develop adaptive immunity (Medzhitov and Janeway 2002). The use of adjuvants compensates for the lack of the natural “danger signal” omitted by the minimalistic vaccine design and may shape the type, duration, and intensity of the immune response (Siegrist 2008). An optimal adjuvant will stimulate innate immunity without inducing systemic inflammation that could elicit severe side effects (Teijaro and Farber 2021).
APC prime T cells (both CD4 + and CD8 +) what leads to a massive proliferation of antigen-specific T cells and the production of inflammatory and cytotoxic mediators. T helper (Th) cells primed by antigen-presenting DC encounter specific B cells that capture and process native antigen. The antigen-specific T-B interaction leads to a rapid expansion and differentiation of B cells into short-lived plasma cells producing unmutated IgM antibodies. T follicular helper (Tfh) cells and antigen trapped on follicular dendritic (FDC) networks drive proliferation, isotype switch, and affinity maturation of antigen-specific B cells. This proliferation leads to the generation of memory B cells and long-lived plasma cells that produce high affinity somatically mutated antibodies of switched isotypes (Allen et al. 2007). Upon infection, an immediate protection is mediated by neutralizing antibodies secreted by the long-lived plasma cells present in the bone marrow in an antigen-independent fashion and maintained at the constant level in serum and body fluids (Radbruch et al. 2006). Memory cells (B and T cells) confer immediate protection and generate secondary responses that are more rapid and of higher magnitude than primary responses. Memory T and B cells are the progeny of antigen-specific naive cells that have been clonally expanded in the course of an immune response and survive once antigen has been eliminated (Sallusto et al. 2010). Antigen-specific memory cells can take a few months to fully develop (Dogan et al. 2009).
Induction of solely neutralizing antibodies is very often considered as a correlate of the effective immune response. This may be true when the antibody is likely to be the main mechanism of protective immunity against bacteria and parasites with an exclusively extracellular lifestyle. But for viruses, bacteria, or parasites that spend most of their lives intracellularly, the induction of cytotoxic CD8 + T cells is a cornerstone, as CD8 + T cells are important for infection clearance (Siegrist 2008; Sallusto et al. 2010). Protection induced by vaccinations is mediated through a complex interplay between innate, humoral-, and cell-mediated immunity.
As mentioned earlier, live attenuated vaccines imitate natural infection, albeit at a level that disease symptoms are absent or less severe than in the natural infection. Hence, this type of vaccine will elicit an adequate immune response, but at the same time, it could pose a health risk due to the adverse effects upon vaccination. To avoid potential risk of serious adverse effects caused by live attenuated vaccines, individuals at risk are recommended not to be vaccinated. The risk is usually associated with allergy to any of the active substance or excipient, pregnancy, congenital or acquired immunodeficiency, and other underlying conditions. In very rare cases, the risk can be associated with the residual pathogenic potential of the vaccine virus which has been observed for some mumps vaccine strains and OPV. Symptomatic horizontal transmission of the vaccine virus has been recorded among previously healthy household members of the vaccinees after vaccination with L-Zagreb mumps vaccine strain (Atrasheuskaya et al. 2006, 2012; Kaic et al. 2008; Tesović et al. 2008). The use of the Sabin live attenuated OPV decreased the number of poliovirus infections by > 99%. However, in the past, the use of live poliovirus in the vaccine led to reversion of its wild-type virulence during production process resulting with rare cases of vaccine-associated poliomyelitis (VAP) (Cann et al. 1984). Ever since these events have been confirmed, strict measures have been undertaken during production and quality control procedures for OPV to maximally decrease the risk of VAP (Dörsam et al. 2000; Grachev et al. 2001). Also, a genetically divergent vaccine-derived poliovirus (VDPV) strains can emerge in vivo during vaccine use and spread in underimmunized populations, becoming circulating VDPV strain resulting in poliomyelitis outbreaks (Jorba et al. 2019).
In contrast to live attenuated vaccines, other types of vaccines will not jeopardize the health of the vaccines but are not able to completely activate the immune system. Inactivated vaccines are relatively simple and cheap to produce but induce B cell response primarily, while T cell response is mostly absent. In infections requiring a strong T cell-mediated response, this can be disadvantageous. Polysaccharide vaccines induce only a short-lived T cell-independent response, while polysaccharide-protein-conjugated vaccines show superior immunogenicity (Zimmermann and Curtis 2019). In the case of rapid spread of infections, which pose a risk to become pandemic threats, such as HIV, Ebola, Zika, and most recently COVID-19, fast development and distribution of vaccines are essential. Methods using live attenuated or inactivating pathogens require cultivation and propagation of a whole pathogen. Possible difficulties in cultivation of pathogens in vitro or biosafety requirements that specialized laboratories are obligatory to have might slow down vaccine production. (Rauch et al. 2018). A novel vaccine technologies, as will be described later in the text, are endeavor to overcome all these shortcomings.
One dose is rarely sufficient to provide effective and long-lasting protection. Hence, multiple dose regimes are recommended. Regardless of the type of the vaccine, there will always be a substantial variation between individuals in the immune response to vaccination, in both quantity and quality. As a result, there will be individuals who will receive a complete vaccination dose but remain susceptible to infection. Therefore, none of the vaccines will ever have 100% efficacy. Factors others than the vaccine itself that may affect the effectiveness of vaccination are intrinsic host factors (age, sex, genetics, comorbidities), perinatal host factor (gestational age, birth weight, breastfeeding, maternal antibodies, maternal infection during pregnancy, maternal malnutrition), extrinsic factors (infections, parasites, antibiotics, probiotics, prebiotics, microbiota, preexisting immunity), behavioral factors (smoking, alcohol consumption, exercise, acute physiological stress, chronic physiological stress, sleep), nutritional factors (body mass, nutritional status, micronutrients, enteropathy), environmental factors (rural vs. urban environment, geographic location, season, family size, toxins), vaccine factors (vaccine type and strain, adjuvants, dosage), and administration factors (schedule, vaccination site and route, co-administration with other vaccines or drugs) (reviewed in Zimmermann and Curtis 2019).
Despite the growing knowledge about how vaccines should work, the main challenge in the development of the new vaccines remains how to design the vaccine that will elicit adequate and effective immune response to confer a life-long immunity.
Novel vaccine technologies
Vaccines tailored by integrated approaches are urgently needed to address existing and new challenges in combat against human infectious diseases. A new era of vaccine technology started in the 1970s owing to the advent of recombinant DNA technology. The new technology enabled production of single protein or proteins constituting virus-like particles (VLP) potentially modified to achieve optimized vaccine. It also enabled the use of DNA or RNA encoding antigens as vaccines and the generation of modified vectors as carriers for antigens of different pathogens. Meanwhile, structural biology and systems biology have enabled the identification of key antigens and deciphered some aspects of the immune responses they generate. Rationally designed vaccines composed of antigens, delivery systems, and often adjuvants that elicit predictable immune responses against specific epitopes to protect against a particular pathogen are supposed to contain well-defined active substances and excipients. However, this is not always possible due to incomplete knowledge. There are no universally accepted strategies and tools to rationally design vaccines, and vaccine development is still generally a tedious and costly empiric process (Rueckert and Guzmán 2012).
In contrast to earlier generations of vaccines based on the whole microorganism (alive or inactivated) or toxoid, novel vaccine candidates are based on the minimalistic approaches to be devoid of the unwanted side effects. That, however, leads to suboptimal vaccine candidates in terms of immunogenicity or protection against different strains of the same pathogen (Table 1). The immunogenicity suffers due to the lack of the MAMPs in the vaccine formulation. To overcome this disadvantage of the novel vaccine technologies and achieve desirable and adequate efficacy, adjuvants are indispensable parts of most novel vaccine formulations.
Table 1.
Generalized comparisons of characteristics of novel vaccine technologies
| Protein vaccines | Synthetic peptide vaccines | Nucleic acid vaccines | Viral vector vaccines | Nanoparticles | |||
|---|---|---|---|---|---|---|---|
| DNA | RNA | Replicative | Non-replicative | ||||
|
Costa (USD/dose) |
35–200 | 20–50 | 6–10 | 20–40 | 100 | 5–10 | 20–40 |
|
Stability (low/high) |
Low | Low | High | Low | High | High | High |
| Immunogenicity: | |||||||
|
B cell (yes/no) |
Yes | Yes | Yes | Yes | Yes | Yes | Yes |
|
T cell (yes/no) |
Yes/no | Yes | Yes | Yes | Yes | Yes/no | Yes |
| Adjuvant (required/not required) | Required | Required | Required | Required | Not required | Not required | Not required |
| Number of doses (single/multiple) | Multiple | Multiple | Multiple | Multiple | Multiple | Multiple | Multiple |
|
Long-term protectionb (poor/good/excellent) |
Good | Poor | Good | Good | Excellent | Good | Good |
| Adverse effectsc: minor (frequency) | < 1/5 | < 1/3 | < 1/2 | < 1/2 | < 1/3 | < 1/2 | < 1/2 |
| Serious (frequency) | < 1/200 | None declared | < 1/250 | < 1/1000 | < 1/10000 | < 1/1000 | < 1/1000 |
|
Autoimmune response (yes/no) |
No | No | Yes | No | No | No | No |
|
Need for cold chain delivery (yes/no) |
Yes | No | No | Yes | Yes | Yes | Yes |
aEstimated cost is based on commercially available prices which are not always the reflection of real costs of the production; bpoor, protection up to 1 year; good, protection 1–5 years or a booster dose required for long-term protection; excellent, protection longer than 5 years; cbased on clinical trials data
Data are obtained either from vaccines in use or of those in final stage of clinical trial. Please note that some exceptions may contradict this generalized comparisons
In the next sections, a brief overview of the most commonly used technologies in the development of the new vaccine candidates will be presented, as well as vaccine candidates currently under the status of active clinical trial phases 1–3, and finally, the new generation vaccines which have been approved by the European Medicines Agency (EMA) and Food and Drug Administration (FDA) will be discussed.
Recombinant protein vaccines
Novel vaccine technology has been enabled by advances in recombinant DNA technology and genetic engineering, and the first trials in the 1980s focused on the recombinant structural proteins of specific pathogens (Lasky et al. 1986; Hollingdale et al. 1987; van Wyke Coelingh et al. 1987). The knowledge of the biology of the pathogen, the main proteins involved in pathogenesis and highly conserved ones, and the immune mechanisms that should be elicited for protection and identification of protective antigens has enabled a rational design of the recombinant protein vaccines. Recombinant protein vaccines are subunit vaccines that use only a small part of the pathogen and target immune response focused against few protective antigens. Thus, the composition of this type of vaccine can be well-defined and controlled. Since a single protein cannot replicate, the risk of having side effects is negligible (Nascimento and Leite 2012). That is the main advantage of using a single protein as a vaccine. However, there is a dark side to this concept. If applied alone, an antigen cannot provide an intrinsic “danger signal”, and the vaccine will not elicit an adequate and long-term immunity (Bomford 1998; Beverley 2002). To elicit a vigorous immune response, a recombinant protein vaccine requires adjuvants, and often multiple doses are needed as well (Hansson et al. 2000). So, making an efficient recombinant protein vaccine can be a challenge (Table 1).
Nowadays, there are various expression systems, each with its advantages and flaws, allowing the production of large-scale quantities of recombinant proteins. Which of the systems will be used depends on the desirable properties of the recombinant proteins as antigens: post-translational modifications, native or denatured/renatured protein, stability of the protein toward protease digestion, cost of the production etc. The most commonly used overexpression hosts used to produce recombinant proteins are bacteria (E. coli), yeast (S. cerevisiae, P. pastoris), insect, or mammalian cell lines. Transgenic plants as producers of recombinant vaccine protein hold promise as low-cost vaccine production systems as well (Mason et al. 1992; Kurup and Thomas 2020). Since each platform has been extensively reviewed many times, and they are not the main topic of this review, a brief comparison of each expression system will be given. The most popular platform is E. coli being easy to manipulate, genetically and physiologically well-defined, and yielding high expression levels (Rosano and Ceccarelli 2014). Yeast does not express proteins at the levels produced in E. coli, but the recombinant proteins have some post-translational modifications (Wang et al. 2016). Baculovirus-based expression in insect cells is relatively expensive, but the production level is higher than in yeast or mammalian cells and post-translational modification occurs, although not as in mammalian systems (Gopal and Schneemann 2018; Grose et al. 2021). A mammalian expression system would be an ideal choice because it will post-translationally modify expressed proteins in a human-like fashion. Still, the process involves the laborious and time-consuming generation of stable cell lines and the high cultivation costs, while the quantities of the expressed proteins are lowest compared to other expression systems (Gutiérrez-Granados et al. 2018; Tschorn et al. 2020).
Synthetic peptide vaccines
Synthetic peptide-based vaccine is technology using fragments of protein antigen sequences which are chemically synthesized and assembled into a single molecule with a potential to induce either T or B cells immune response, but preferably both (Moisa and Kolesanova 2011). Recombinant protein vaccine technology is tackling with certain problems such as the high price of protein overexpression, hosts cultivation and purification, possible induction of allergic and autoimmune responses, possible biological contaminants, and the need of special storage conditions, which should be overcome by using synthetic approach (Sobolev et al. 2005; Skwarczynski and Toth 2016). There are several crucial steps in synthetic peptide design. The first step is choosing an immunogen based on genome analysis and selection of potential targets of immune reaction. This step is followed by antigen mapping what means identification of antigenic determinants, testing the immune response and possible autoimmune reactions, and checking antigen determinants variability. Ideally, the vaccine should have both B and T cells epitope in order to stimulate B cell conversion and cytotoxic T cell immune response (Sobolev et al. 2005). B cell epitopes must retain native conformation so these epitopes must be significantly longer in order to achieve such conformation. Moreover, specific sequences have been used to flank or staple B cell epitopes. On the other hand, T cell epitopes can possess linear conformation and as such are presented to MHC-1 molecules (Skwarczynski and Toth 2016). The frequently used T cell epitope is universal epitope of tetanus toxin (Moisa and Kolesanova 2011). T helper epitope can induce both innate and adaptive immunity so presentation of T helper epitopes in synthetic peptide vaccine design was a great finding. (Skwarczynski and Toth 2016). Synthetic peptide vaccines also contain added adjuvants which help in vaccine delivery and activation of non-specific immune response (Moisa and Kolesanova 2011). Typically selected immunogen is linked to a carrier by chemical conjugation. Synthetic peptide approach is used in development of several vaccines for infectious diseases like Malaria, HCV, influenza virus, HIV and more, and certain peptide vaccine candidates are under clinical development (Malonis et al. 2020).
Nucleic acid vaccines
DNA vaccines
DNA vaccines consist of plasmid DNA (pDNA) containing the transgene encoding the antigen of interest under the control of a strong mammalian viral promoter. The transgene is expressed upon delivery of the pDNA to the cell, and an antigen-specific immune response could be elicited (Khatri et al. 2008). This very simple yet brilliant approach raised great interest. At the beginning of the research of the DNA vaccines, in the early 1990s, the scientific community had high expectations. Until 2010, the publication number showed constant growth, but then the growth stopped (Lee et al. 2018). There are three major limitations and/or disadvantages of DNA vaccines: (i) low level of intracellular/intranuclear transport of pDNA, which results in low immunogenicity (Hasson et al. 2015; Klimov 2019), (ii) safety issues regarding the possibility of integration of pDNA into the genomic DNA of the vaccinee and activation of oncogenes (Wurtele et al. 2003), and (iii) potential development of autoimmunity by elicitation of anti-DNA antibodies (Lilic and Ghosh 1994; Zafrir et al. 2012) (Table 1).
The main barriers for the successful delivery of pDNA acting as a vaccine are the stability of the pDNA, cellular uptake, endolysosomal escape, decomplexation from the carrier, and nuclear envelope translocation. Major efforts have been put toward the improvement of delivery methods of pDNA to the targeted cellular compartments. Electroporation, gene gun, and cationic lipid- or polymer-based nanoparticles were explored as more or less successful delivery technologies and showed promising results in eliciting both humoral and cell-mediated immune responses and generating protection against various pathogen challenges in small experimental animal models (Wolff et al. 1990; Fynan et al. 1993; Wang et al. 2008; reviewed in Lee et al. 2018). DNA vaccine may be taken up intracellularly by endocytosis within APCs (Dobrovolskaia and McNeil 2007) or interact with the TLR2 or TLR4 at the cell surface, which results in the expression of proinflammatory cytokines and chemokines (IL-8, IL-10, TNFa). Within the cell, pDNA can be randomly encapsulated by the nuclear envelope reforming post-mitosis or actively transported to the nucleus through nuclear localization signals (Xu et al. 2016).
DNA vaccines may contain unmethylated CpG sequences that function as MAMPs and can activate TLR9 in the endosome, eventually releasing IL-4 and interferon-alpha (IFN-a) (Kojima et al. 2002; Coban et al. 2005). Thus, DNA vaccine encodes an antigen of interest and contains elements that act as adjuvants. Different strategies have been developed to further enhance the elicitation and duration of immune protection by DNA vaccine. One of them is the co-administration of a cocktail of pDNA encoding an antigen and pDNA encoding immunomodulatory proteins (cytokines, chemokines, etc.) commonly named molecular adjuvants. The expression of molecular adjuvants can be optimized to tailor the immune response toward a desirable outcome without the concerns of a systemic cytokine storm. Many molecular adjuvants have been used in combination with DNA vaccines. To mention few most prominent ones which increased vaccine immunogenicity and induced sustained memory response: IL-2 (Kim et al. 1998, 2000), IL-12 (Chattergoon et al. 2004; Halwani et al. 2008; Naderi et al. 2013), and GM-CSF (Weiss et al. 1998; Ahlers et al. 2002; Yoon et al. 2006). For a more comprehensive review on molecular adjuvants, see Laddy and Weiner (2006), Abdulhaqq and Weiner (2008), and Suschak et al. (2017).
Furthermore, plasmid engineering and novel vector configurations can further enhance the effect of the DNA vaccine. Codon optimization adapts DNA sequences of transgene between species to augment its expression (Stachyra et al. 2016; Latanova et al. 2018). Bacterial regions that are not required for the transgene expression in mammalian cells can suppress the transgene’s promoter by heterochromatin formation. Removal of these bacterial regions generated mini circle vectors which have alleviated transgene silencing and show dramatically increased transgene’s expression (Chen et al. 2003). DNA vaccine production generally relies on a simple and inexpensive synthesis of the transgene in the plasmid vector, with reduced cost and time to manufacture. Plasmid DNA is also extremely stable at room temperature, canceling the need for a cold chain during transportation (Table 1). Given all this, after 30 years of intensive work on DNA vaccines, they have proven to be successful in small animal models. However, they appeared less successful in non-human primates and humans. Also, there are still some safety issues addressed, but without a final answer so far. As a result, there is no DNA vaccine approved for use in humans. Still, there are two prophylactic vaccines against infectious diseases for use in the veterinary vaccine: a vaccine against West Nile virus in horses and a vaccine against infectious hematopoietic necrosis virus in salmon (Bastos Pereira et al. 2014).
RNA vaccines
At the dawn of DNA vaccines in the early 1990s, also the concept of RNA vaccines appeared. RNA vaccines were reported to be effective for direct gene transfer for the first time by Woff et al. 1990. Initially, RNA vaccine technology was not extensively explored because of the concerns regarding their stability (Table 1) which was jeopardized by omnipresent RNases and the low production capacities. Recent technical advances have answered how to protect and stabilize RNA vaccine and how to produce it economically.
Two types of RNA molecules have been explored as vaccines against infectious pathogens: non-replicating mRNA vaccines and self-amplifying RNA vaccines.
Non-replicating mRNA molecules can be synthetically produced at a large scale through a cell-free enzymatic transcription reaction. The in vitro transcription reaction includes a linearized plasmid DNA encoding the mRNA vaccine as a template, a recombinant RNA polymerase, and nucleoside triphosphates as essential components. A cap structure is enzymatically added to the transcriptional product at the end of the reaction or as a synthetic cap analogue in a single-step procedure. Finally, a poly(A) tail will be provided to form a mature mRNA sequence (Zhang et al. 2019). To avoid digestion by RNases and higher expression of the heterogeneous antigen, various modification strategies can be applied to make mRNA more stable and highly translatable (Kariko et al. 2008, 2011; Thess et al. 2015). Synthetic cap analogues and capping enzymes (Martin et al. 1975; Stepinski et al. 2001) act to stabilize mRNA and increase protein translation via binding to eukaryotic translation initiation factor 4E (EIF4E). Regulatory elements in the 5′-untranslated region (UTR) and the 3′-UTR (Holtkamp et al. 2006; Trepotec et al. 2018) stabilize mRNA and increase protein translation. Poly(A) tail (Gallie 1991) stabilizes mRNA and increases protein translation. Modified nucleosides, such as pseudouridine (Ψ) and 5-methylcytidine (5-mC), decrease innate immune activation through innate immune sensors and improve translation (Kariko et al. 2008; Pardi and Weissman 2017; Meyer et al. 2018). Codon optimization additionally increases translation (Gustafsson et al. 2004).
Self-replicating RNA vaccines are based on a viral genome that encodes the viral proteins required for RNA replication (such as RNA-dependent RNA polymerase). A heterogeneous gene for the antigen of interest is also inserted. These vaccines carry out self-replication and generate multiple copies of the antigen-encoding mRNA (Geall et al. 2012; Ljungberg and Liljestrom 2015; Tews and Meyers 2017). Self-amplifying mRNA is mainly based on the genomes of positive-stranded RNA viruses Sindbis virus, Semliki Forest virus, Kunjin virus, alphaviruses and others (Zhou et al. 1994; Johanning et al. 1995; Anraku et al. 2002; Perri et al. 2003).
Non-replicating and self-replicating mRNA vaccines express high levels of the heterologous gene when they are introduced into the cytoplasm. The cell recognizes the newly expressed antigen as the antigen of intracellular pathogen that triggers both humoral and MHC-I-mediated CD8 + T cell responses (Ulmer and Geall 2016). Cell with the introduced RNA vaccine presents antigen on MHC-I and elicits CTL response by cross-priming to dendritic cells (Brito et al. 2015; Schnee et al. 2016; Chahal et al. 2017). Additionally, APCs engulf cells expressing vaccine antigens and present the antigen on MHC-II complexes what elicits a CD4 + T cell response (Aida et al. 2021). RNA vaccines are unique in their double function; they serve as immunogen (encoding the antigen) and as an adjuvant (through the intrinsic immunostimulatory properties of RNA). Single-stranded and double-stranded RNA function as MAMPs are immunosensed by various cellular innate sensors (TLR3, MDA5, RIG-I, NOD2, PKR) (Sabbah et al. 2009; Lester and Li 2014; Esser-Nobis et al. 2020; Fitzgerald and Kagan 2020; Rehwinkel and Gack 2020), resulting in cellular activation, and production of type I interferon and multiple inflammatory mediators (Pardi et al. 2018). Since self-replicating mRNA vaccines amplify the expression of the antigen through the transcription, they are more efficient than non-replicating mRNA vaccines if used at the same dosage. However, some studies indicated the need for adding an adjuvant to the mRNA vaccines. Besides using usual adjuvants such as MF59 and cationic emulsions, mRNAs that encode CD70, CD40 ligand, and TLR4 were designed within the name TriMix. These proteins act as activators of immune system. Also, the use of delivery systems (cationic lipid and protamine) appears to stimulate adjuvant outcome (Xu et al. 2020).
Delivery tools are proven highly important for the rapid uptake and effectiveness of mRNA vaccines. Similar to DNA vaccines, physical methods of gene gun and electroporation, cationic lipids, and polymers have been used to overcome the cytoplasmic membrane and transfer mRNA vaccines to the cytoplasm of the target cells (Guan and Rosenecker 2017; Tan and Sun 2018). Liposomal and polymer nanoparticles also shelter the nucleic acids and protect them from RNases.
The use of mRNA has several beneficial features over traditional vaccines and DNA vaccines. mRNAs have a better safety profile. They are non-infectious, and in contrast to DNA vaccines, mRNA vaccine exerts their function already in the cytoplasm and do not interact with the host’s genomic DNA, thus avoiding potential safety risk of integration into the genome and activating oncogenes.
Initial studies demonstrated good results of administrated non-replicating mRNA in animals but not so encouraging results in humans. Lipid- or polymer-based delivery systems or nucleoside-modified mRNA highly improved the efficacy of mRNA-based vaccines (reviewed in Pardi et al. 2018).
Recent technological advances have now largely overcome the issues which hindered the interest in mRNA vaccines. The most significant advantages of mRNA vaccines are high potency, low-cost manufacture, and safe administration. Multiple mRNA vaccine platforms against infectious diseases have demonstrated encouraging results in both animal models and humans. Recently, the mRNA vaccine has been proven a fast and straightforward solution to a desperate need for urgent vaccine demand in pandemic (see last chapter).
Recombinant viral vectors
The idea of viral vectors started in 1972 when lambda phage genes and galactose operon of E. coli were inserted in simian virus (SV) 40 DNA (Jackson et al. 1972). In 1982, foreign DNA was introduced in vaccinia virus DNA by homologous recombination, and in the same year, Panicali and Paoleti constructed vaccinia virus vector by adding herpes simplex gene in it (Mackett et al. 1982, Panicali and Paoletti 1982). From that point forward, numerous viruses are developed as experimental vaccine vectors. Recombinant viral vector platforms take advantage of the viral vector’s inherent ability to introduce genetic material of the desired antigen into the cell, which is subsequently expressed as a foreign protein, while the immune system reacts and treats it as such. This delivery system is often more efficient than other non-viral systems such as naked plasmid DNA and liposome nanoparticles. Another possible design of this technology is that viral vector carries a “ready-made” antigen on its particle and presents it to the immune system of the vaccine as its own protein. There are some significant advantages of the recombinant viral vectors compared to other technologies (Table 1). They are highly immunogenic due to the vector’s natural MAMPs, which eliminate the use of adjuvant in the vaccine formulation. The immune response is both cell-mediated, specifically CD8 + T cell response, and humoral immune response. As already mentioned above, they act as a very efficient delivery vehicle. Recombinant viral vectors can be designed so that they are replicative or non-replicative, depending on their ability to replicate in the organism of the vaccine upon vaccination. Generally, non-replicative viral vectors have a better safety profile than replicative vectors. An improved safety profile of the replicative viral vector can be achieved by selecting the favorable virus for a vector, which means they are either well-known attenuated human viruses or non-human viruses known as safe for humans. Another critical issue of the virus-vectored vaccines is viral immunodominance. A strong response against the virus epitopes can mask the response against the vaccinating antigen (Schöne et al. 2017).
Many viral vector platforms based on adenoviruses, lentiviruses, non-segmented negative-stranded RNA viruses (Sendai virus, measles virus, vesicular stomatitis virus, and others), poxviruses, herpes viruses, and baculoviruses have been explored as viral vectors for novel vaccines. Each of them will be described briefly.
Adenoviral vectors
Adenoviruses are non-enveloped viruses with a double-stranded DNA genome. Several vital properties make them desirable candidates for viral vectors: infection of dividing and non-dividing cells, they can be delivered both systemically or through mucosal surfaces, induction of both innate and adaptive immunity with strong Th1-polarized response, physical and genetic stability, easy to genetically engineer, with a packaging capacity of up to 34 kb they allow high levels of transgene expression and can grow to high titers in vitro (Tatsis and Ertl 2004; Gao et al. 2019). Adenovirus-based vaccines use both replicating- or non-replicating viruses as platforms to express the vaccinating antigen or epitope. The most commonly used adenoviruses used as viral vectors are human adenoviruses type 5 (Ad5) or type 26 (Ad26), and chimpanzee adenovirus (ChAdOX1). The antigen in the adenovirus vaccines can be present as a protein or as a transgene. Successful recombinant vaccines based on the use of replication-deficient adenoviruses have been developed. Although early vaccine candidates were replication-deficient due to the deletion of the E1 region, later versions of the vectors were improved by the deletion of E1 and E3 regions. Such vectors are stable, easy to manipulate, and can be produced at high titers, and they have shown safety and immunogenicity in clinical trials (Coughlan 2020). The major flaw of the replication-deficient adenovirus-based vaccines is the relatively high dose required to obtain immune protection. Adenovirus vaccines based on fully replicative viruses can expand antigen expression and trigger improved immunity. However, technical hurdles like pre-existing immunity to the adenoviral vector capsid reduce vaccination efficacy (Dull et al. 1998; Zhan et al. 2021).
Lentiviral vectors
Lentiviral vectors utilize the capacity of lentiviruses to mediate semi-random integration of DNA into the host cell genome to enable long-term transgene expression (Kalpana 1999). Thus, these vectors are rarely explored for the vaccines against infectious diseases but are very promising as gene therapy vectors of which some have already entered the clinical use (Naldini et al.1996; reviewed in Milone and O'Doherty 2018).
Non-segmented negative-stranded RNA viral vectors
Non-segmented negative-stranded RNA viruses (NNSV) have several features that make them attractive candidates for viral vectors (Bukreyev et al. 2006). Some of the most important are a well-established and relatively simple to manipulate a reverse genetic system that allows the recovery of the infectious virus entirely from cloned cDNA without the use of BACs or more complex technologies; replication in the cytoplasm without integration into the host genome, obviating concerns about cellular transformation; gene exchange involving NNSV has not been observed in nature; lack of recombination contributes to the stability of the inserted foreign gene; expression of few viral proteins decreases the problem of immunodominance; transcription of the NNSV genome has a polar gradient, such that promoter-proximal genes are expressed more efficiently than promoter-distal ones what enables fine-tuning of the expression of the transgene; and strong MAMPs which ensure a complete and good quality immune response (both cellular and humoral).
A relatively the low capacity of the genome for the transgene may be a drawback of NNSV vectors although recent studies show that multi-gene inserts are feasible (see later). RNA viruses possess the highest mutation rates in nature (Bordería et al. 2016). In spite of inherent lack of proof-reading capacity of the RNA-dependent RNA polymerase (RdRp) of NNSVs, they seem to be enough genetically stable when used as vaccine vectors. The plausible explanation for this seemingly paradoxical claim lies in genetic robustness of these RNA viruses. Genetic robustness is conferred if a virus has an ability to readily adapt in the face of a new environment without a change in phenotype. The genetic composition facilitates this (Barr and Fearns 2010). In case of recombinant NNSV vectors, the starting point of the virus are plasmid DNAs yielding a very homogene swarm of the initial genome variants. A limited number of passages at constant and controlled conditions should not pose a great challenge for genetic robustness of produced RNA vaccine vectors based on NNSVs. Thus, the variability of the produced virus batch can remain at low level. Although one would expect a higher mutation rate in inserted transgenes because there is no functional selection for them, they also seem to be relatively stable (reviewed in Willemsen and Zwart 2019). Importantly, extremely rare evidences of recombination of NNSV in nature prove this viruses to be safe and are not very likely that the risk of genetic exchange between vaccine virus and wild type virus would create a hazard exceeding.
As in adenoviral vectors, the antigen in the NNSV vectored vaccines can be present as a “ready-made” protein or as a transgene, depending on the design of the vaccine. Many NNSV has been explored for vectors (reviewed in Bukreyev et al. 2006). The use of Sendai virus (SeV), measles virus, and vesicular stomatitis virus (VSV) will be described in more details.
SeV is a member of the Respirovirus genus of the Paramyxoviridae family. It is particularly attractive as a human vaccine because it is a known pathogen of mice and has never caused a confirmed disease in humans (Karron and Collins 2007). SeV is not an attenuated human virus and is therefore not burdened with the concern that it will revert to its original pathogenic phenotype (Luongo et al. 2009; Schickli et al. 2012). It was established that SeV could accommodate foreign genes of 3 kb or even more. It can transiently infect human cells, allowing the cell to express antigens endogenously and with post-translational modifications matching target antigens and neutralizing epitopes (Henrickson et al. 1991). Endogenous expression of antigens also ensures robust activation of CD8 + T cells (York and Rock 1996). These cells provide a fail-safe mechanism; if antibodies do not eliminate all incoming infectious particles, the CD8 + T cells can kill infected mammalian cells to block virus amplification.
Measles virus is a member of the genus Morbillivirus in the family of Paramyxoviridae. Although the natural host of this virus is human, this virus is a very attractive vector for generating recombinant vaccines because there are several attenuated measles vaccine strains with proven efficacy, safety, and genetic stability. It is important to note that the measles vaccine is produced at a large scale in several countries. These production lines could be easily adapted to vectored vaccines in emergencies such as a pandemic. MV vector can take up foreign genes up to 6 kb in size and possibly more (Frantz et al. 2018). The capacity of measles virus to incorporate multiple transgenes has been recently shown by incorporating glycoprotein and nucleoprotein of Lassa fever virus into Schwarz measles vaccine vector (Mateo et al. 2021). Initial concern about the influence of the measles virus-preexisting immunity on the efficacy of the measles virus-vectored vaccines was unfounded as demonstrated by the immunization of previously immunized animals or humans (Frantz et al. 2018).
VSV is a member of the Rhabdoviridae family. This virus infects cattle, horses, and pigs. Human infection is very rare, and the seroprevalence in the human population is low. During the last decade, the recombinant VSV (rVSV) platform has been tested for multiple emerging and neglected viral diseases and therapeutic cancer vaccines in animal studies (Zemp et al. 2018). It was shown that this vector induces humoral and cellular immune responses upon vaccination (reviewed in Fathi et al. 2019). A VSV vaccine design strategy utilizes an rVSV vector lacking the VSV G protein (rVSVΔG) gene. This vector is thus modified to encode the glycoprotein (GP) of the pathogen of interest instead of VSV G protein and expresses the foreign GP on the viral membrane (Schnell et al. 1996; Garbutt et al. 2004; Publicover et al. 2004; Tani et al. 2012).
Vaccinia virus vectors
Vaccinia viruses (VVs) originated from highly efficacious vaccines for the eradication of smallpox (Ladnyĭ 1977), have been engineered to express foreign genes, serving as a highly appealing delivery system for heterologous viral antigens (Sutter and Staib 2003). A highly attenuated VV strain, the modified vaccinia virus Ankara (MVA) strain, is immunogenic and valuable for protecting against many infectious diseases (Carroll et al. 1997; Price et al. 2013). MVA is a well-studied vaccinia virus. It was used as a vaccine against smallpox with an established safety profile which allows scientists to use this virus under biosafety level 1 because it is deficient for factors required to replicate in mammalian cells productively. Despite replication deficiency, this vector still can induce an effective immune response upon immunization. MVA gained these features through serial passages in chicken embryo cells (Volz and Sutter 2017). Adaptation to a non-natural growth substrate led to deletions in the genome in regions indispensable for replication and in regions of many immunomodulatory genes that orthopoxviruses use to regulate the host cell environment (Perdiguero and Esteban 2009; Smith et al. 2013; Hernáez et al. 2018). Comparison with the genome maps of ancestor viruses revealed that the MVA genome harbors six major deletions and mutations, resulting in the loss of approx. 30 kb of genetic material (Meisinger-Henschel et al. 2007).
Except that this viral vector has an outstanding safety profile in humans, it has intrinsic adjuvant capacities that stimulate humoral and cellular immune responses (Sutter and Staib 2003; Gόmez et al. 2011). Next-generation VV vectors have been developed by specific deletion of gene products required for the replication and immune evasion in human cells and/or supplying the key factors in trans. The NYVAC is a highly attenuated vaccinia virus that is non-replicating in most human cells but can be grown in chicken embryo fibroblasts. It was generated by deleting 18 virulence-associated genes of a Copenhagen isolate (Tartaglia et al 1992). Sementis Copenhagen vector (SCV) was derived by deletion of the essential D13L gene of the Copenhagen strain. This vector is non-replicating in human cells and can only be grown in an engineered Chinese hamster ovary cell line which supplies the D13 protein and VV host range factor (CP77) in trans (Eldi et al. 2017).
Herpesvirus vectors
Herpesviruses (HVs) are enveloped double-stranded DNA viruses infecting vertebrate hosts with genome ranging from 150 to 240 kbp. Herpesviruses possess one of the largest DNA genomes within the mammalian viruses (McGeoch et al. 2006). BAC technology has facilitated the manipulation of HVs’ large DNA genome, and consequently, several herpes viruses BACs have been constructed (Zhou and Roizman 2005). A common feature of HVs is their lifelong latency of infected hosts, associated with maintaining their genome in specific host cells without lytic replication (Davison 2007). HV vaccine platforms are mostly based on the recombinant HV, which are attenuated by genetic interventions in the genome but can have limited replication upon vaccination. Alternatively, in trans complementation can be required for productive infection for a vector designed as non-replicative. A heterologous antigen is inserted in the genome, which is overexpressed during this short initial replication. The most common HV for vaccine vectors are CMV, EBV, and herpes simplex virus (HSV).
Baculovirus vectors
To overcome the effect of preexisting immunity, the researchers have employed viruses that do not infect mammals in nature. Such an alternative uses members of the family Baculoviridae, enveloped double-stranded DNA viruses, collectively named baculoviruses. Baculoviruses infect more than 600 insect species but are harmless to mammals (Lin et al. 2014; Kong et al. 2018). Of the many members of the Baculoviridae family, AcMNPV is the workhorse for producing recombinant proteins using insect cell lines (Felberbaum 2015; Van Oers et al. 2015). Virions of rAcMNPV could be modified as transduction vectors in mammalian cells (BacMam vectors) or to display antigenic proteins in their envelope (Kwang et al. 2016; Premanand et al. 2018). Baculovirus cloning capacity of a transgene is as large as 38 kb, thus allowing for the insertion of even multiple genes and regulatory elements (Cheshenko et al. 2001; Kost et al. 2005, 2010). The surface display system has been extensively used to develop pseudotyped baculoviruses as a vaccine delivery platform (reviewed in Lu et al. 2012). Baculovirus surface display system is using the transmembrane (TM) and C-terminal domain (CTD) of baculovirus gp64, or the expression of heterologous proteins is governed via the membrane anchor of VSV G protein, influenza virus neuraminidase, single chain antibody fragments, and Spodoptera exigua multiple nucleopolyhedrovirus F protein. Each of these systems leads to incorporation of the protein into baculovirus envelope. These strategies are the most widely adopted ones enhancing baculovirus transduction in vitro and in vivo (reviewed in Lu et al. 2012). Although baculovirus vectors do not replicate in mammalian cells what could lead to lower level of immune response, the effect of the baculovirus vector is enhanced by the immunogenic property of rAcMNPV by the existence of CpG-ODNs in its genome and their intracellular sensing by TLR9 molecules at APCs (Fragoso-Saavedra and Vega-López 2020).
Besides mentioned advantages, there are some weaknesses of viral vector-based vaccines which should be taken in concern. Mostly disadvantages are associated with virus manipulation and manufacturing such as possible integration into the genome, viral recombination during production, and unintentional introduction of microorganisms or contaminants (e.g., porcine cirovirus in rotavirus vaccine) (Rauch et al. 2018).
Nanoparticles
Nanoparticles (NP) are nanoscale particles (< 100 nm) that can serve as a delivery vehicle for vaccine antigens. Several types of NPs composed of gold, dendrimers, carbon polymers, and liposomes have been shown to improve vaccine efficacy, facilitate antigen uptake, and induce desired immunological responses (Lugade et al. 2013; Aida et al. 2021). NPs offer several advantages (Table 1): they can directly access lymphatic drainage systems for immune processing, can be modified to target specific subsets of immune cells, and can be delivered to specific intracellular compartments (Diaz-Arévalo and Zeng 2020; Aida et al. 2021). A comprehensive understanding of how NPs can optimize vaccine is still missing, and many experimental NP candidates are currently being explored in clinical trials (Diaz-Arévalo and Zeng 2020). In many cases, NPs have a multi-functional role in a vaccine formulation: they carry an antigen and stabilize it, and at the same time, they act as an adjuvant. Therefore, they gained much attention over the past two decades. The desirable characteristics of NPs depend on surface charge, hydrophobicity/hydrophilicity, and the steric effects of particle coating on particle size. Still, NP size has been confirmed as a key parameter that determines the potential of the nanoparticle to induce cytokine responses. The size of the NPs determines the cellular uptake mechanism, such as endocytosis, phagocytosis, and micropinocytosis. These mechanisms are responsible for the uptake and ingest of various particle sizes and critical parameters that affect the immunogenicity by promoting and inducing appropriate responses (Fifis et al. 2004; Mottram et al. 2007). Although nanoparticles strive to reach lymph nodes as their target tissue and APCs as their final goal, a certain proportion of nanoparticles will be wasted and cleared from the body. Clearance of nanoparticles could be achieved through degradation by the immune system or by renal (particles < 8 nm) or biliary (particles > 200 nm) clearance (Zhao et al. 2014). However, if they cannot be degraded or excreted from the body, they will accumulate at different locations in the body what may cause some adverse effects. Hence, to design a safe and highly efficacious vaccine based on nanoparticles, a fundamental understanding of how to improve delivery and immunostimulatory modes of nanoparticles, but also an understanding of in vivo distribution and metabolism, is required.
The interaction of immunogen and NPs is possible in three different ways: by covalent binding of the immunogen to the NP (conjugation), adsorption of the immunogen on the surface of the NP, and encapsulation of the immunogen within the NP. Which type of interaction will be applied depends on the nature of the NP and immunogen and specific application. Mostly used and explored NP building materials are virus-like particles (VLPs), liposomes, immunostimulatory complexes (ISCOMs), polymeric NPs, and emulsions (Kheirollahpour et al. 2020).
Virus-like particles (VLPs)
VLPs are self-assembling multi-protein nanostructures that are assembled from viral structural proteins and mostly mimic natural virus formation. VLP-forming proteins can be produced in any expression host system (e.g., E. coli, S. cerevisiae, baculovirus expression system, mammalian cell lines) depending on the desirable features of the protein(s). Following expression, the protein will self-assemble into the VLP, forming a repetitive geometric shape. These particles are devoid of any infectious materials and are incapable of replicating. The size of these particles (< 200 nm) facilitates direct drainage to the lymph nodes (Manolova et al. 2008; Mohsen et al. 2017a). The repetitive and stable structure is a potent pathogen-associated structural pattern (PASP) that facilitates cross-linking of BCRs. This causes effective uptake of VLPs by APCs, which activates adaptive immunity (Gomes et al. 2017). VLPs are also capable of binding natural IgM molecules efficiently and fixing C1q molecule, what leads to their deposition on follicular DCs (FDCs) (Link et al. 2012). The structure and the size of VLPs facilitate their cross-presentation of VLP-derived peptides on MHC class I molecules (Kovacsovics-Bankowski et al. 1993; Harding and Song 1994). Thus, VLPs are very efficient in activating B cell immunity and in stimulating the activation of CD8 + immune response (Mohsen et al. 2017a). Additionally, VLPs interior facets can package different DC-activating adjuvants, including dsRNA, ssRNA, and non-methylated CpGs resulting in effective stimulation of TLR3, TLR7/8, and TLR9, respectively (Storni et al. 2004; Ashley et al. 2011). VLP-based vaccine development is a rapidly growing field due to its simplicity, stability, uniformity, and efficacy in inducing humoral and cellular immunity and safety (Mohsen et al. 2017b). Also, VLP-based vaccines are the foremost commercialized class of NPs. There will be more on them in the later chapter.
Liposomes
Liposomes are spherical vesicles formed by self-assembled lipid molecules. Due to their dual nature (hydrophobic tail and hydrophilic head), lipid molecules form a lamellar bilayer which forms a spherical vesicle in water. Liposomes have shown significant clinical potential since they are biocompatible, stable within the body, and can be modified to display targeting moieties (Schwendener 2014). Liposomes can be tailored as carriers to incorporate hydrophilic molecules into the aqueous core or hydrophobic molecules within the phospholipid bilayers. In this way, they can incorporate virtually any immunogen (protein, nucleic acid, polysaccharide) or drug. The desired properties can be obtained by modulating the liposome composition (type of lipid molecules, charge, and size). Due to the chemical properties, water-soluble compounds can be localized within the aqueous inner space, whereas lipophilic compounds are immersed into the lipid bilayer. Also, some antigens may be attached to the outer surface either by adsorption or chemical cross-linking (Torchilin 2005; Watson et al. 2012).
Furthermore, liposomes can be modified to achieve immunostimulatory properties. For example, by modulating liposomes to present lectin binding mannose on their surface as well as to entrap monophosphoryl lipid A (MPLA) adjuvant, a novel nanoparticle was developed that was capable of targeting dendritic cells and facilitating enhanced antigen presentation to T cells against a model antigen (Wang et al. 2014). The size of the liposome may determine which arm of the immune system will dominate. Badiee et al. (2012) demonstrated that immunization with small liposomes (100 nm) favored a Th2 response, whereas large liposomes (≥ 400) favored a Th1 nm response, elevated interferon γ (IFNγ) levels, and immunoglobulin IgG2a/IgG1 ratios. The exceptional appeal of liposomes lies in their enormous versatility as carriers and adjuvant systems for vaccines.
Immune stimulating complex (ISCOM)
ISCOMs are self-assembled cage-like particles about 40 nm in size which can be used as delivery nanosystems for vaccines. They are produced by combining a protein antigen, saponin, cholesterol, or phospholipid held together by hydrophobic interactions. These spherical structures can be used for entrapping hydrophobic antigens (Morein et al. 1984; Alexyuk et al. 2019). ISCOMs-based vaccines have been shown to efficiently promote both humoral and cellular immune responses (Morein et al. 2004).
Polymeric NPs are of interest for vaccinology due to their biocompatibility, biodegradability, easy fabrication, solubility in water, non-toxic nature, and their ability to be easily modified into desired shapes and sizes (Smith et al. 2015). Alginate (Ekici et al. 2011), hyaluronic acid (Li et al. 2013), and chitosan (Akagi et al. 2012) are the natural polymers most widely used in medicals and pharmaceuticals. Synthetic polymers are also often explored as vaccine NPs. Among many of them, poly (d,l-lactic-coglycolic acid) (PLGA), poly (d,l-lactide-co-glycolide) (PLG) (Danhier et al. 2012), and polylactide (PLA) (Panyam and Labhasetwar 2003; Lou et al. 2019) are most often used. All polymers, both natural or synthetic, can self-assemble into different morphologies at nano- or microscale. The self-assembly is strongly dependent on the preparation method, the aqueous medium, and the other components of the formulation – nanoparticles, nanospheres, nanoemulsions, micelles, and (nano/hydro) gels (reviewed in Pippa et al. 2021).
Emulsions
Emulsions have been traditionally used as adjuvants in vaccine formulations for decades, and more recently, they are explored as vaccine delivery systems. Nanoscale emulsions consist of two immiscible liquid phases, emulsifier(s) and excipient(s). One is dispersed in the other to form a single phase. Complete and incomplete Freund’s adjuvants (CFA and IFA, respectively) are a water-in-oil (w/o) emulsions that are prepared from non-metabolized oils more than 70 years ago (Batista-Duharte et al. 2011). They are inflammation-inducing agents, and the stimulation of antibody-producing toward many antigens is greatly enhanced when these are administered with CFA or IFA. However, severe side effects of the CFA and IFA (Claassen et al. 1992) make them incompatible with practical human vaccinology. A novel, innocuous emulsions based on natural compounds have been prepared in the last two decades. A famous w/o squalene-based emulsion MF59® has been licensed and commonly used in vaccine development and has been proven to be a safe and potent adjuvant for use in influenza vaccines (O’Hagan 2007). Adjuvants AS03 and AF03, also squalene-based emulsions, are registered for administration with influenza vaccines (Nguyen-Contant et al. 2021). Squalene-based adjuvants modulate the innate immunity and the T cell component of adaptive immunity, leading to enhanced antibody response outcomes. Also approved, the saponin-based AS01 adjuvant system consists of two immunostimulants, monophosphoryl lipid A (MPL) and QS-21 saponin purified from the bark of the Quillaja saponaria Molina tree (Didierlaurent et al. 2017). QS-21 activates resident and migratory innate lymphoid cells, enhances the recruitment of neutrophils and monocytes, and activates the early interferon-gamma secretion (O'Hagan et al. 2020). Two more w/o emulsions are used in experimental immunizations, vaccine development, and clinical trials: Montanide ISA™ 51 and Montanide ISA™ 720. Montanide ISA™ 51 is a mix of mineral oil and surfactant from the mannide monooleate family. It renders w/o emulsion and is used in a ratio of 50 oil for 50 of the aqueous phase. Montanide ISA™ 720 is a mix of non-mineral oil of vegetable origin with the surfactant from the mannide monooleate family. It renders w/o emulsion and is used in a 70:30 oil to water ratio. It has been shown that the two mannide monooleate-based emulsions induce inflammation, stimulate the recruitment of APCs and favor the uptake of antigens by APCs, and stimulate the accumulation of lymphocytes in draining lymph nodes (Aucouturier et al. 2002).
Vaccine candidates in clinical trials
When a compound is developed to be a vaccine, it is tested in a non-clinical setting, including in vitro, ex vivo, and in vivo (animal models) methods. If non-clinical test results support further development, the candidate compound is tested in humans in phases 1–3 clinical trials as a pre-authorization process. A prevalent problem in vaccine translation is the time-consuming and costly transition from non-clinical to clinical development due to difficulties in predicting human immune responses. Although closer to humans, primate models are associated with ethical, logistic, and financial constraints (Rueckert and Guzmán 2012). National authority agencies have stringent regulations and guidelines that cover the design of clinical development programs for new vaccines intended to provide pre- and post-exposure prophylaxis against infectious diseases (EMA 2006). However, guidelines do not provide matters specific to each vaccine type. Therefore, planned protocols should specify in details methodologies to be used to evaluate immune responses to vaccination. The methods should be consistent across studies, validated and reproducible. When testing vaccines, the main focus is their immunogenicity. The testing results should provide relevant information on the quality and quantity of the immune response (both humoral and cell-mediated) and the safety of the tested compound. In developing any new vaccine, adequate data on immunogenicity should be assembled during the clinical development program (characterization of the immune response, investigation of an appropriate dose and primary schedule, assessment of the persistence of detectable immunity, and consideration of the need for and response to booster doses). Whenever possible, immune responses to vaccination should be compared to those seen due to natural infection. If a widely accepted immunological correlate of protection already exists, then the immune response’s characterization can be limited solely to those parameters. In some cases, it may be possible to generate only minimal data for new vaccines intended to prevent rare infections that carry considerable morbidity and mortality. The regulatory agencies also provide detailed guidance on post-authorization phase 4 clinical trials and pharmacovigilance, but they are not a subject of this review. From the idea to the vaccine authorization is a very long, laborious, and costly path with many pitfalls. Tables 2 and 3 show a list of ongoing initial clinical trials for novel prophylactic (P) and therapeutic (T) vaccines against human infectious diseases based on new technologies applied to the European Medicines Agency (EMA) and the Food and Drug Administration (FDA). Although EMA and FDA are independent regulatory agencies, they are continuously working on a harmonization of their regulatory processess. They have been rather successful in that as was shown by the analysis by Kashoki et al. (2020). The EMA and the FDA had high concordance (91–98%) in decisions on marketing approvals in period 2014–2016. Some differences were observed in the clinical data due to the difference in timing of submissions (more applications were submitted to the FDA before they were submitted to EMA). In this work, clinical trials of previously approved vaccines based on novel technologies that are in process of being approved for modified use (e.g., in combination with another vaccine or therapeutic indication for broader age group) are excluded from the lists. Despite many scientific publications describing experimental studies on novel vaccine technologies against infectious diseases in humans, the number of vaccine candidates in clinical trials is low (24 and 24, EMA and FDA, respectively). The numbers would be even lower if there were no COVID-19 pandemic. COVID-19 vaccine candidates represent 42% (10 out of 24, EMA) and 50% (12 out of 24, FDA) of the total number of vaccines based on the novel technologies in clinical trials (Tables 2 and 3). More than 40% of the applied clinical trials listed in Tables 2 and 3 are launched in early phases I or I/II (46% and 75%, EMA and FDA, respectively) (Fig. 2), indicating that results of the early phases often are not good enough to launch the next phase trial. Finally, there are only seven vaccine candidates (Tables 2 and 3, Fig. 2), three in the EMA’s list and four in the FDA’s list, in the last phase of the pre-authorization process. More than half of the phase III candidates (four out of seven) are intended against COVID-19. The current COVID-19 pandemic has forced regulatory agencies to treat marketing authorization applications for COVID-19 products, including vaccines, as an emergency. This has allowed the timeline for evaluation to be reduced, and the authorization for some COVID-19 vaccines was approved in an expedited manner (Fig. 3). In contrast, the non-emergency procedure usually takes much longer, on average 10 to 15 years. A comparative timeline of emergency and non-emergency procedures including complete development of the quadrivalent HPV vaccine Gardasil and the mRNA COVID-19 vaccine BNT126b (Comirnaty) is shown in Fig. 3.
Table 2.
Ongoing clinical trials for novel vaccine compounds against infectious diseases in humans in phases 1–3 listed by the EMA by the November 1, 2021 (https://www.clinicaltrialsregister.eu/)
| Medical condition | EudraCT Number | Start date | Vaccine type | Phase* | Sponsor |
|---|---|---|---|---|---|
| COVID-19 | 2020-002502-75 | 2020-11-26 | Multipeptide vaccine (P-pVAC-SARS-CoV-2) | I (P) | University Hospital Tuebingen |
| COVID-19 | 2021-000454-26 | 2021-03-25 | mRNA for Spike protein | I/II (P) | Leiden University Medical Centre |
| COVID-19 | 2020-003734-20 | 2021-02-03 | pDNA encoding Spike protein | I/II (P) | Takis S.r.l |
| COVID-19 | 2020-005997-82 | 2021-03-12 | Recombinant protein, RBD fused to Fc (SARS-CoV-2-RBD-Fc) | I/II (P) | UMCG |
| COVID-19 | 2021-000548-23 | 2021-07-07 | MVA encoding complete Spike protein | I/II (P) | University Medical Center Hamburg-Eppendorf |
| COVID-19 | 2020-005915-39 | 2021-02-19 | Gorilla-derived replication defective adenoviral vector encoding full-length Spike protein | II/III (P) | REITHERA SRL |
| COVID-19 | 2020-004272-17 | 2021-03-25 | Recombinant trimeric form of Spike protein | II/III (P) | Clover Biopharmaceuticals AUS Pty Ltd |
| COVID-19 | 2020-003998-22 | 2020-12-16 | Unmodified mRNA of Spike protein | II/III (P) | CureVac AG |
| COVID-19 | 2020-003370-41 | 2021-08-23 | Recombinant SARS-CoV-2 prefusion Spike delta TM protein | II/III (P) | Sanofi Pasteur Inc |
| COVID-19 | 2020-004123-16 | 2020-09-23 | Recombinant Spike protein | III (P) | Novavax, Inc |
| malaria | 2006-001743-66 | 2007-09-20 | pDNA encoding liver-stage-antigen-3 (PfLSA-3) | I/II (P) | Institute Pasteur |
| malaria | 2019-002872-14 | 2019-11-12 | MVA encoding P. vivax Duffy binding protein (PvDBPII) | I/II (P) | University of Oxford |
| RSV | 2018-000431-27 | 2018-12-18 | Chimpanzee-derived adenovector (ChAd155-RSV) encoding RSV proteins F, N and M2-1 | I/II (P) | GlaxoSmithKline Biologicals |
| RSV | 2020-000692-21 | 2020-12-03 | Recombinant subunit pre-fusion RSV antigen (RSVPreF3) | II (P) | GlaxoSmithKline Biologicals |
| RSV | 2020-003887-21 | 2020-11-03 | Adenoviral vector encoding preF | II (P) | hVIVO Services Ltd |
| RSV | 2020-000753-28 | 2021-05-11 | Recombinant subunit pre-fusion RSV antigen (RSVPreF3) | III (P) | GlaxoSmithKline Biologicals |
| HCV | 2010-022960-10 | 2011-03-30 | pDNA encoding HCVNS3/4a | II (P) | ChronTech Pharma AB |
| HIV | 2019-003102-26 | 2021-7-14 | MVA and chimpanzee adenoviral vector encoding envelope proteins | I/II (T) | OSPEDALE SAN RAFFAELE |
| HIV | 2005-003071-20 | 2005-09-20 | pDNA encoding multi HIV B clade fusion protein | II (T) | FIT Biotech Oyj Plc |
| HPV | 2019-001890-98 | 2020-05-29 | Chimpanzee adenovirus (ChAdOx1)-vectored multigenotype high risk human papillomavirus | I (T) | Vaccitech Ltd |
| influenza | 2015-001932-38 | 2016-07-22 | Peptide vaccine derived from conserved regions of internal proteins | II (P) | PepTcell |
| Klebsiella pneumoniae | 2020-005090-26 | 2021-06-21 | Tetravalent bioconjugate vaccine | I/II (P) | Limma Tech Biologicals AG |
| HBV | 2020-002118-42 | 2020-12-07 |
Nine synthetic peptides from highly conserved regions of HBV polymerase, core, and surface antigens formulated with IC31 |
II (T) | Altimmune, Inc |
| CMV | 2013-000903-18 | 2013-06-19 | pDNA encoding phosphoprotein 65 and glycoprotein B | III (T) | Astellas Pharma Global Development, Inc. (APGD) |
*Prophylactic vaccine (P); therapeutic vaccine (T)
Table 3.
Ongoing clinical trials for novel vaccine compounds against infectious diseases in humans in phases 1–3 listed by the FDA by the November 1, 2021 (https://www.clinicaltrials.gov/)
| Medical condition | Identifier | Start date | Vaccine type | Phase* | Sponsor |
|---|---|---|---|---|---|
| COVID-19 | NCT04568811 | 2020-09-26 | Adenovirus Ad5 encoding Spike protein | I (P) | Jiangsu Province Centers for Disease Control and Prevention |
| COVID-19 | NCT04405908 | 2020-06-19 | recombinant trimeric form of Spike protein | I (P) | Clover Biopharmaceuticals AUS Pty Ltd |
| COVID-19 | NCT04450004 | 2020-07-10 | Coronavirus-like particles | I (P) | Medicago |
| COVID-19 | NCT04760743 | 2020-12-17 | Recombinant Spike protein subunit vaccine | I (P) | SK bioscience Co., Ltd |
| COVID-19 | NCT04530656 | 2020-08-28 | Recombinant Spike protein produced in Sf9 cells | I (P) | Jiangsu Province Centers for Disease Control and Prevention |
| COVID-19 | NCT04368988 | 2020-05-25 | Recombinant Spike protein nanoparticles in baculovirus system | I/II (P) | Novavax |
| COVID-19 | NCT04713488 | 2021-01-15 | Adenovirus Ad26 encoding Spike protein | I/II (P) | Gamaleyeva Research Institute of Epidemiology and Microbiology |
| COVID-19 | NCT04537208 | 2020-09-03 | Recombinant Spike protein | I/II (P) | Sanofi Pasteur/GSK |
| COVID-19 | NCT04341389 | 2020-04-12 | Adenovirus Ad5 encoding Spike protein | II (P) | Institute of Biotechnology, China |
| COVID-19 | NCT04540419 | 2020-09-11 | Adenovirus Ad5 encoding Spike protein | III (P) | NPO Petrovax |
| COVID-19 | NCT04516746 | 2020-08-28 | Chimpanzee adenovirus Ad155 encoding Spike protein | III (P) | AstraZeneca |
| COVID-19 | NCT04780035 | 2020-11-18 | Chemically synthesized peptides of S protein bound to N protein carrier | III (P) | Federal Budgetary Research Institution State Research Center of Virology and Biotechnology "Vector" |
| RSV | NCT03636906 | 2019-04-08 | Chimpanzee adenovirus Ad155 encoding F, N and transcription antitermination proteins | I (P) | GlaxoSmithKline |
| RSV | NCT04752644 | 2021-02-22 | MVA encoding F and G glycoproteins, and N and M2 | II (P) | Bavarian Nordic |
| RSV | NCT04732871 | 2021-02-15 | Recombinant subunit pre-fusion RSV antigen (RSVPreF3) | III (P) | GlaxoSmithKline |
| HIV | NCT03856996 | 2019-05-23 | Recombinant gp120 protein produced in CHO cells | I (P) | National Institute of Allergy and Infectious Diseases (NIAID) |
| HIV | NCT03547245 | 2018-06-15 | Recombinant envelope outer domain nanoparticles in Expi293 cells | I (P) | International AIDS Vaccine Initiative |
| HIV | NCT03934541 | 2019-08-26 | MPER-656 gp41 peptide in liposomes | I (P) | National Institute of Allergy and Infectious Diseases (NIAID) |
| HIV | NCT02788045 | 2016-07 | Adenovirus Ad26 encoding gp140 | I/II (P) | Janssen Vaccines & Prevention |
| influenza | NCT04622592 | 2020-10-28 | Plant-derived recombinant quadrivalent VLP | I/II (P) | Medicago |
| tuberculosis | NCT04239313 | 2020-05-27 | A985b and fusion protein ESAT6-CFP10 combined with BCG | I (P) | Anhui Zhifei Longcom Biologic Pharmacy Co |
| tuberculosis | NCT02337270 | 2017-09-05 | Adenovirus Ad5 encoding Ag85A (Ad5AG85A) | I (P) | McMaster University |
| anthrax | NCT04148118 | 2020-01-08 | Recombinant PA protein | I/II (P) | BlueWillow Biologicals |
| HCV | NCT02772003 | 2016-06-04 | pDNA encoding NS3, NS4A, NS4B, NS5A | I (T) | National Cancer Institute (NCI) |
*Prophylactic vaccine (P); therapeutic vaccine (T)
Fig. 2.
Distribution of the clinical trial phases for new vaccines against infection diseases in humans based on novel technologies applied to the European Medicines Agency (EMA, a) or the Food and Drug Administration (FDA, b)
Fig. 3.
A comparative timelines of emergency and non-emergency procedures including a complete development and approval process of (a) HPV vaccine Gardasil (Merck Sharp and Dohme) and (b) the mRNA COVID-19 vaccine Comirnaty (BioNTech/Pfizer)
As mentioned in the “Introduction” section, despite the need for an efficient vaccine against HIV, tuberculosis, RSV, malaria, and HCV, they remain major challenges. There are four candidates for vaccine against RSV, two against malaria, six against HIV, and two against tuberculosis and HCV (Tables 2 and 3). Except one RSV vaccine which is in phase III, other are either in phase I, I/II, or II (Tables 2 or 3), so it will take some time until the decision is made and new vaccines will (maybe) be ready for use. The overlapping between the lists of ongoing clinical trials by the EMA and FDA is poor (Fig. 2). For COVID-19 candidate vaccine, there are two sponsors which are having trials at both regulatory agencies, namely Clover Biopharmaceuticals and Novavax. For conditions other than COVID-19, only GlaxoSmithKline Biologicals has filed a request for approval for RSV vaccine candidates by both agencies. The vast majority of sponsors applying to EMA are from EU/UK/Switzerland region (79%), while only 21% are non-European countries (USA (three requests), China (one request), and Japan (one request)) (Table 2). Within the FDA list of clinical trials for vaccines based on the novel technologies (Table 3), there is much more variety in geographical locations of sponsors: USA and EU/UK 25% each (6 requests), China 21% (5 requests), Canada and Russia 12,5% each (3 three requests each), and South Korea 4% (1 request).
Despite the low immunogenicity (no T cell immunity), a requirement of efficient adjuvant and no long-term protection (Table 1), recombinant protein technology prevails among the vaccine candidates listed in Tables 2 and 3 (Fig. 4). All vaccine candidates based on the recombinant proteins or synthetic peptides contain some kind of adjuvant to overcome the deficiencies. Twelve out of 48 novel vaccine candidates are based on recombinant adenoviral vectors (Fig. 4), indicating the attractiveness of this technology. Other new technologies are sporadically represented (Fig. 4).
Fig. 4.
Joint distribution of novel technologies used for new vaccines against infection diseases in humans under clinical trials applied to the European Medicines Agency (EMA) or the Food and Drug Administration (FDA)
Vaccines based on novel technologies in use
Today there is still a limited number of approved new generation vaccines based on novel technologies. As shown in Table 4, sixteen vaccines are on the market, four of them being approved in the last year during COVID-19 pandemic. Other twelve vaccines protect efficiently against hepatitis B (three vaccines), Ebola virus (three vaccines), HPV infection (three vaccines), varicella zoster, meningococcal meningitis caused by N. meningitides group B, and influenza viruses (one vaccine each) (Table 4).
Table 4.
Novel technologies-based vaccines against human infectious diseases approved by the EMA (www.ema.europa.eu) and FDA (https://www.fda.gov/vaccines-blood-biologics/vaccines/vaccines-licensed-use-united-states)
| Vaccine name | Active substance | Vaccination schedule | Adjuvant | Manufacturer (marketing authorization holder) |
|---|---|---|---|---|
| Engerix B | Recombinant hepatitis B virus surface antigen produced in S. cerevisiae | 0–1–6 months | AS04C, adsorbed on aluminum phosphate | GlaxoSmithKlein Biologicals |
| Recombivax HB1 | Recombinant hepatitis B virus surface antigen produced in S. cerevisiae | 0–1–6 months | Adsorbed on aluminum hydrixyphosphate sulfate | Merck Sharp and Dohme |
| HBVaxPro2 | Recombinant hepatitis B virus surface antigen produced in S. cerevisiae | 0–1–6 months | Adsorbed on aluminum hydrixyphosphate sulfate | Merck Sharp and Dohme |
| HEPLISAV | Recombinant hepatitis B virus surface antigen produced in Hansenula polymorpha | 0–1 month | CpG 1018 | Dynavax GmbH |
| Cervarix | Recombinant human papilloma virus types 16 and 18 L1 protein produced in baculovirus expression system | 0–1–6 months | AS04, adsorbed on aluminum phosphate | GlaxoSmithKlein Biologicals |
| Gardasil | Recombinant human papilloma virus types 6, 11, 16 and 18 L1 protein produced in S. cerevisiae | 0–2–6 months | Adsorbed on aluminum hydroxyphosphate sulfate | Merck Sharp and Dohme |
| Gardasil 9 | Recombinant human papilloma virus types 6, 11, 16, 18, 31, 33, 45, 52 and 58 L1 protein produced in S. cerevisiae |
0–6 to 12 months or 0–2–6 months |
Adsorbed on aluminum hydroxyphosphate sulfate | Merck Sharp and Dohme |
| Shingrix | Varicella zoster virus glycoprotein E produced in Chinese Hamster Ovary (CHO) cells | 0–2 months | AS01B | GlaxoSmithKlein Biologicals |
| Bexsero | Neisseria meningititdis group B NHBA fusion protein, NadA protein, fHbp fusion protein produced in E. coli | 0–1 month | Adsorbed on aluminum hydroxide | GSK Vaccines S.r.l |
| Trumenba | Neisseria meningititdis group B lipidated fHbp (factor H binding protein) of subfamily A and B produced in E. coli | 0–6 months or 0–1–6 months | Adsorbed on aluminum phophate | Pfizer |
| Flublok Quadrivalent1 | Recombinant influenza haemagglutinin proteins (A/XX (H1N1), A/XX (H3N2), B/XX, B/XX) produced in insect cell line Sf9 | Single dose | No adjuvant | Sanofi Pasteur |
| Supemtek2 | Recombinant influenza haemagglutinin proteins (A/XX (H1N1), A/XX (H3N2), B/XX, B/XX) produced in insect cell line Sf9 | Single dose | No adjuvant | Sanofi Pasteur |
| Ervebo | Recombinant vesicular stomatitis virus with a deletion of the envelope glycoprotein replaced with the Zaire Ebola Virus surface glycoprotein | Single dose | No adjuvant |
Burgwedel Biotech GmbH (Merck Sharp and Dohme) |
| Zabdeno3 | Adenovirus Ad26 encoding Zaire ebolavirus glycoprotein | 0 month | No adjuvant | Janssen Biologicals |
| Mvabea3 | MVA encoding glycoproteins of Zaire ebolavirus, Sudan ebolavirus and Marburg Marburgvirus, and nucleoprotein of Taï Forest ebolavirus | 8 weeks after Zabdeno | No adjuvant | Janssen Biologicals |
| Comirnaty | mRNA for SARS-CoV-2 Spike protein, nucleoside-modified | 0–21 day | Embedded in lipid nanoparticles | BioNTech/Pfizer |
| COVID-19 Vaccine Janssen | adenoviral vector (Ad26) encoding SARS-CoV-2 Spike protein | Single dose | No adjuvant | Janssen Biologicals |
| Spikevax (previously COVID-19 Vaccine Moderna) | mRNA for SARS-CoV-2 Spike protein, nucleoside-unmodified | 0–28 days | Embedded in SM-102 lipid nanoparticles |
Rovi Pharma Industrial Services (Moderna Biotech Spain) |
| Vaxzevria3 (previously COVID-19 Vaccine AstraZeneca) | Adenoviral vector (ChAdOX1) encoding SARS-CoV-2 Spike protein | 0–4 to 12 weeks | No adjuvant | AstraZeneca |
1Name of the anti-HBV vaccine in USA; 2name of the anti-HBV vaccine in EU; 3not approved by the FDA
The first recombinant technology-based vaccine was a vaccine against hepatitis B commercialized in 1986 and soon after it became widely administered for human use. These vaccines are based on a recombinant HBV surface antigen (HBsAg) produced in yeast that spontaneously self-assembles into 22 nm VLPs (Valenzuela et al. 1982; McAleer et al. 1984). Until then vaccine against HBV was produced as a highly purified HBsAg from the plasma of HBV-positive donors treated with formaldehyde for safety (Buynak et al 1976). This type of vaccine was associated with safety concerns because of the potential contamination of the vaccine with HBV. Additionally, the source of the HBV-positive plasma may be limited (McAleer et al. 1984). Today, there are three recombinant VLP-based HBV vaccines (Table 4). The main differences between them is the choice of adjuvant and the vaccination dose/schedule as indicated in Table 4. In spite of these differences, all three HBV vaccines have been shown as being highly effective.
Next VLP-based vaccines that have been approved and used successfully are vaccines against HPV infection. Almost 5% of all human cancers can be attributed to certain high risk HPV types (de Martel et al. 2017). This data alarmed for an urgent need for prophylactic vaccine against this virus. All HPV vaccine on the market today use HPV L1 capsid protein which self-assembles into VLPs (Yadav et al. 2019). The HPV vaccines in use include the bivalent (Cervarix), quadrivalent (Gardasil), and nonavalent (Gardasil 9) vaccines depending on the number of the HPV types they protect from. Bivalent vaccine was designed to contain L1 antigen of the most common high risk types HPV 16 and HPV 18, but it has been found to be cross protective for several years upon vaccination against HPV 31, 33, and 45, even with just one dose of the vaccine, in addition to reducing genital warts caused by HPV 6 and 11 (Tsang et al. 2020). Quadrivalent and nonavalent vaccines contain additional types of HPV (Table 4) inducing broader protection. Studies have demonstrated that three doses of the nonavalent vaccine led to adequate levels of antibodies against all nine genotypes (Manini and Montomoli 2018).
Vaccines based on recombinant proteins have also been proven as good vaccines against complications of the varicella zoster virus infection, against influenza and invasive disease caused by N. meningitides group B.
For prevention of herpes zoster (HZ) and post-herpetic neuralgia, there are two types of HZ vaccines currently in use: the live attenuated vaccine (Zostavax) and the recombinant protein vaccine (Shingrix). Both are indicated for use in adults 50 years of age or older, and Shingrix is also recommended for adults 18 years of age or older at increased risk of HZ. Although Shingrix requires two doses at intervals 0 and 2 months, while Zostavax requires only one shoot, Shingrix is safe to administer in individuals with primary and acquired immunodeficiency states, immunosuppressive therapy, active untreated tuberculosis, and during pregnancy, while Zostavax is contraindicated in these conditions (EMA, product information). Shingrix is also a rare example of the recombinant protein used as human drug produced in mammalian cell culture. Usually low levels of protein produced in such systems make them economically unprofitable. However, Haumont et al. (1996) successfully isolated a cell line expressing high levels of truncated anchorless varicella zoster glycoprotein E in the supernatant of the cell culture. The resulting protein was correctly processed, heavily glycosylated, and highly immunogenic.
Infection with N. meningitidis can cause severe life-threatening meningitis. There are six major serogroups of N. meningitides found worldwide, and all are associated with invasive meningococcal disease: A, B, C, X, Y, and W-135 (Garland 2020). Two effective quadrivalent polysaccharide conjugate meningococcal vaccines against serogroups A, C, W-135, and Y have been approved (Menveo and Menactra) in the first decade of the twenty-first century and are indicated for active immunization of children (from 2 years of age), adolescents, and adults at risk of exposure to N. meningitidis groups A, C, W-135, and Y, to prevent invasive disease (EMA, products information). Diversity of serogroup B and concerns about the potential autoimmunity of the serogroup B polysaccharide which resembles a human neural cell adhesion molecule made development of the vaccine against this serogroup more challenging (Garland 2020). In 2014, two vaccines indicated for active immunization of individuals 10 years and older to prevent invasive meningococcal disease caused by N. meningitidis serogroup B become available: Bexsero and Trumenba (EMA, product information). The specific composition of these vaccine and variations in the vaccination schedule is presented in Table 4. Although initially recommended as vaccines without need for boost, in 2020, the Advisory Committee on Immunization Practices (ACIP) made a recommendation (Mbaeyiet al. 2020) for individuals who remain at increased risk for invasive meningococcal disease. They should receive a booster dose of the same serogroup B vaccine 1 year after completing the primary vaccination and every 2–3 years thereafter. Additionally, ACIP recommended that during an outbreak, previously vaccinated individuals should receive a single booster dose if ≥ 1 year has passed since completion of the primary vaccination.
Today, various types of influenza vaccine are available: live attenuated or inactivated (whole virion, split virion, subunit vaccine), and also there is one recombinant protein-based vaccine (Šantak 2012). Traditional approach for influenza vaccine production is mostly based on inactivated influenza virus derived from embryonated hens’ eggs (reviewed in Šantak 2012). Rarely cell culture is used instead of embryonated eggs (Barberis et al. 2016; Bouvier 2018). The use of hens’ eggs can have some problematic features: it strongly relies on the timely supply of large number of eggs, adaptation of influenza virus to growth conditions in eggs may modify antigenic sites in the key viral proteins hemagglutinin (HA) and neuraminidase (NA) (Robertson et al. 1985, 1987), and also modifications in glycosylation patterns may occur which may be quantitatively and qualitatively different from those derived from viruses grown in human cells (An et al. 2019). A recombinant protein-based vaccine FluBlok Quadrivalent (approval name at the FDA)/Supemtek (approval name at the EMA) (Table 4) is advantageous over traditional approach because it is produced in insect cells Sf9 by baculovirus expression system. In this way, the problem with the egg supply is annulled, the vaccine production can begin as soon as the WHO recommends the strain composition of the influenza vaccine for the next season, and recommended HA and NA are expressed from the controlled system with no amino acid sequence modifications possible. Comparison of 28-day post-vaccination geometric mean titers (GMT) for Supemtek and comparator in adults 18–49 years of age show superior efficacy of the Supemtek except for B/Victoria lineage antigen (EMA, product information). Furthermore, Flublok vaccine exhibited superior performance in eliciting both CD4 T cell responses and HA-specific antibody responses compared to split and subunit vaccines (Richards et al. 2020). However, the issue of some differences in the N-glycosylation pattern in HA and NA produced in Sf9 cells remains (An et al. 2019).
The above-mentioned vaccines in use are based on the technology of the expression of recombinant proteins in different systems (E. coli, S. cerevisiae, H. polymorpha, baculovirus vectors, and insect Sf9 cells and CHO cells, Table 4). Recombinant proteins are rarely sufficiently immunogenic by themselves, and the use of adjuvant is required. All the vaccines discussed above have added adjuvant, mostly aluminum compounds and in some cases proprietary adjuvants. Interestingly, influenza vaccines Flublok/Supemtek consisted solely of the recombinant proteins as active substances, with no adjuvant added. The use of three times more HA (45 µg) of each lineage than is used in other inactivated vaccines (15 µg) may dismiss the use of adjuvant. But it is also important to emphasize here that seasonal inactivated vaccines against influenza based on the production in hens’ eggs have no adjuvant added either.
Recombinant viral vectors, as already discussed here in earlier chapters, seem as a very attractive platform due to the favorable features, especially in terms of immunogenicity and no need for an adjuvant. Recently approved vaccines against Ebola are additional proof of this. Multitude outbreaks of Ebola since its discovery in 1976 urged a development of the effective vaccines against this, very often lethal (> 40%), disease. Three vaccines have been approved by the EMA and one by the FDA (Table 4). The Ervebo vaccine, approved in 2019, is a live recombinant vaccine based on a VSV platform with the glycoprotein of the Zaire ebolavirus in place of the VSV envelope glycoprotein (Wolf et al. 2021). Another vaccine against Ebola consists of two heterologous components given 8 weeks apart (Pollard et al. 2020). For prime vaccination, the monovalent Zabdeno vaccine, based on the adenovirus Ad26, is used. For the boost, a multivalent vaccine Mvabea, based on the modified vaccinia Ankara Bavaria Nordic virus, is used (Table 4). This heterologous prime/boost vaccination regimen is approved by the EMA in 2020. None of the Ebola vaccines requires adjuvant to effectively induce protective immune response since they are based on the replicative or non-replicative viral vector platforms being an intrinsic adjuvant.
Vaccines against COVID-19
A previously unknown coronavirus was detected in December 2019 in Wuhan, China, associated with the severe acute respiratory syndrome (Wu et al. 2020). The virus was given the name SARS-CoV-2, being similar to the SARS virus identified as a causative agent of the severe acute respiratory syndrome in 2002/2003, and the disease was named COVID-19. Soon after detection, it struck the whole world. The COVID-19 pandemic have caused devastating world health and economic crisis. At the time of writing this text (November, 2021), there have been over 254 million confirmed cases of COVID-19, including more than 5 million deaths (https://covid19.who.int/). To strike back, many laboratories turned intensively to the research of this virus and the development of an effective vaccine. To response quickly to challenges of the spreading pandemic, new vaccine technologies that have been developed for other purposes come to the fore (Fig. 3b, Table 4). As a result, since the end of 2020, worldwide health regulatory agencies have approved several different types of COVID-19 vaccines under adjusted management of the clinical trials protocols for emergency approval. Until now, more than 7 billion vaccine doses have been administrated (https://covid19.who.int/).
Approved COVID-19 vaccines are either based on the traditional approaches or novel technologies such as mRNA and adenoviral vectors. A brief overview of COVID-19 vaccines based on the novel technologies will be here presented.
Two mRNA vaccines were approved worldwide: Comirnaty and Spikevax (Table 4). Both vaccines consist of the mRNA with modified nucleotides encoding full-length Spike protein of COVID-19 with modified amino acid sequence to stabilize antigenically preferred prefusion conformation (Wrapp et al. 2020; Vogel et al. 2021). The mRNA is formulated within lipid nanoparticles (Table 4), and both vaccines are proved to be able to elicit neutralizing antibodies (Fraley et al. 2021; Tenforde et al. 2021) and cellular immune response (Tarke et al. 2021). The dose schedule slightly differs between Comirnaty and Spikevax (Table 4). Clinical studies showed that the overall efficacy for Comirnaty 7 days after the second dose was 95% (95% CI), and for Spikevax 14 days after the second dose, the efficacy was 94.1% (95% CI) (https://www.ema.europa.eu/en/medicines/human/EPAR/comirnaty). Given the good results of efficacy and safety studies, Comirnaty was approved for use in December 2020 (both FDA (https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/pfizer-biontech-covid-19-vaccine) and EMA (https://www.ema.europa.eu/en/medicines/human/EPAR/comirnaty)) and Spikevax in December 2020 or January 2021 (FDA (https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/moderna-covid-19-vaccine) and EMA (https://www.ema.europa.eu/en/medicines/human/EPAR/spikevax-previously-covid-19-vaccine-moderna), respectively). Both vaccines were initially recommended for use in individuals 18 years and older, but after additional testing, the use was expanded to include adolescents 12 years and older. As millions of individuals received two doses of either of the mRNA vaccines against COVID-19, it became obvious that myocarditis and pericarditis coincided with the vaccination event (Gargano et al. 2021; Singh et al. 2021; Vidula et al. 2021; Watkins et al. 2021). These cases usually occurred around the second week after vaccination, more often after the second dose, and more often in younger men. On 9 July 2021, the COVID-19 subcommittee of the WHO Global Advisory Committee on Vaccine Safety (GACVS) published a statement regarding reports of myocarditis and pericarditis following COVID-19 mRNA vaccines which indicates a causal association between myocarditis and the mRNA vaccines. The estimated crude reporting rates of 40.6 cases per million second doses among males and 4.2 cases per million among females aged 12–29 years (Gargano et al. 2021). Although cases of post-vaccinal myocarditis and pericarditis are rare and generally mild and respond well to treatment, current evidence suggests a likely causal association between myocarditis and pericarditis and the application of COVID-19 mRNA vaccines. This has urged regulatory agencies to revise the approval fact sheets and some countries to limit the use of COVID-19 mRNA vaccines to individuals outside the risk groups.
The other two COVID-19 vaccines based on novel technologies and approved in many countries are Vaxzevria and COVID-19 Vaccine Janssen, both based on the replication-deficient adenoviral vectors (Table 4). The dynamic of the approval process varies in different countries or regions: Vaxzevria was approved by EMA in January 2021, while the approval process is still ongoing by the FDA; on the other hand, COVID-19 Vaccine Janssen was first approved by the FDA (February 2021) and then by EMA (March 2021). Both vaccines are indicated for immunization in individuals 18 years of age and older. The major difference between these two vaccines is the origin of the adenovirus: Vaxzevria is based on the chimpanzee adenovirus (ChAdOx1) vector and produced in HEK293 cells (Ramasamy et al. 2021), while COVID-19 Vaccine Janssen is based on the human adenovirus type 26 (Ad26) vector and produced in PER.C6 TetR cells (Bos et al. 2020). Both vectors carry the gene for the Spike protein of SARS-CoV-2, which is either unmodified (Vaxzevria) or modified to stabilize the protein in prefusion conformation (Bos et al. 2020). Although COVID-19 Vaccine Janssen is administered as a single dose, and the Vaxzevria vaccination course consists of two doses 4 to 12 weeks apart, the overall efficacy is very similar: 62,6% (95% CI) for Vaxzevria and 66,1% (95% CI) for COVID-19 Vaccine Janssen. As with the mRNA vaccines, the emergency use approval process did not identify severe side effects. After millions of doses have been administrated of both vaccines, a thrombosis with thrombocytopenia syndrome (TTS), accompanied by bleeding in some cases, has been observed. Venus thrombosis affected unusual sites such as cerebral venous sinus thrombosis and splanchnic vein thrombosis, as well as arterial thrombosis. Average incidence of TTS after vaccination with Vaxzevria was less than 8 cases per million doses and after vaccination with COVID-19 Vaccine Janssen was less than 0,5 per million doses (reviewed in Long et al. 2021). Most of these cases occurred within the first 3 weeks following vaccination, and women < 55 years of age were more commonly affected (https://www.ema.europa.eu/en/medicines/human/EPAR/vaxzevria-previously-covid-19-vaccine-astrazeneca; https://www.ema.europa.eu/en/medicines/human/EPAR/covid-19-vaccine-janssen; https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/janssen-covid-19-vaccine; Kantarcioglu et al. 2021; Long et al. 2021). The mortality rates for TTS were approximately 40% (Long et al. 2021). Thus, TTS is severe complication of adenovirus-based COVID-19 vaccines. The trigger for TTS is unclear. Vaccine impurities and the adenoviral vector are potential suspects. Also, the Spike protein itself is a very likely culprit because it has been observed by many case reports and other studies that COVID-19 patients often suffer from deep venous thrombosis (DVT) and pulmonary embolism (PE). The rate of DVT in the 573 non-COVID-19 patients, age 61 ± 17 years, male 44.9%, was 71/573 (12.4%) compared to 72/213 (33.8%) in COVID-19 patients, age 61 ± 16 years, male 61.0% (Marini et al. 2021). This indicates that SARS CoV-2 infection has tendency to initiate tromb formation. More studies are still required to gain a fundamental knowledge to be able to understand both the TTS following vaccination and DVT and PE during COVID-19.
All the four COVID-19 vaccines will be further monitored for efficacy and safety through phase 4 clinical trials and pharmacovigilance reporting.
Due to the massive transmission of the SARS-CoV-2 in the human population, new variants emerge and spread to new hosts. So far, several variants were designated as “variants of concern” (VOC) because they evolved with new, for virus advantageous, characteristics in terms of transmission, morbidity/mortality, and partial escape from neutralizing antibodies leading to reduced neutralization by the sera of convalescent or vaccinated individuals (Lazarevic et al. 2021). Therefore it is likely that in due course, the composition of COVID-19 vaccines, i.e., of the Spike protein, will have to be adjusted, similar as are for seasonal influenza vaccines each year, to evade immune evasion of the VOC. Novel technologies could enable the fast adjustment of effective tools to combat and control new VOC.
Conclusion
Despite the historical proof that vaccination can offer well-being to vaccine recipients and indirectly affects non-vaccinated individuals, the topic of vaccination in the high-income countries elicits different reactions: from unconditional acceptance through hesitancy to absolute refusal. One of the reasons for these conflicting views on the vaccination is that many diseases that once caused epidemics (smallpox, polio, measles, mumps, rubella, diphtheria, etc.) are now negligible, and the sense of security gives an impression that vaccination is no longer necessary. Also, the ratio of benefit-to-harm has changed in favor of risk of harm due to the low incidence, and therefore safer vaccines are wanted. Novel technologies offer new possibilities to answer the challenges to which traditional approaches failed (as an example, cervical cancer is today one of the most preventable cancers owing to novel technologies). On the other hand, limited data available about the safety of new technologies may also raise hesitancy and refusal. So vaccine developers may expect hard work in three fields: (1) to decide which strategy to use and how to design a new vaccine (and adjuvants), (2) to gather data from non-clinical and clinical trials demonstrating highly effective and safe new vaccine, but also (3) to develop a strategy based on the scientific results and proofs which will help to develop trust in health authorities, health professionals, and individuals to be vaccinated or their parents (if children are to be vaccinated) educating them from the position of unbiased resource of knowledge.
Acknowledgements
The authors thank Saša Kazazić for constructive comments and editing the manuscript.
Author contribution
ZM and MŠ searched the literature, integrated all contributions, and wrote the manuscript. All authors read and approved the manuscript.
Funding
This work was supported financially by the European Union through the European Regional Development Fund – the Competitiveness and Cohesion Operational Programme project “RAPTOVAX” (K.K.01.1.1.04.0099).
Declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zrinka Matić and Maja Šantak contributed equally to this work.
References
- Abdulhaqq SA, Weiner DB. DNA vaccines: developing new strategies to enhance immune responses. Immunol Res. 2008;42:219–232. doi: 10.1007/s12026-008-8076-3. [DOI] [PubMed] [Google Scholar]
- Ahlers JD, Belyakov IM, Terabe M, Koka R, Donaldson DD, Thomas EK, Berzofsky JA. A push-pull approach to maximize vaccine efficacy: abrogating suppression with an IL-13 inhibitor while augmenting help with granulocyte/macrophage colony-stimulating factor and CD40L. Proc Natl Acad Sci U S A. 2002;99:13020–13025. doi: 10.1073/pnas.192251199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida V, Pliasas VC, Neasham PJ, North F, McWhorter KL, Glover SR, Kyriakis CS. Novel vaccine technologies in veterinary medicine: a herald to human medicine vaccines. Front Vet Sci. 2021;8:654289. doi: 10.3389/fvets.2021.654289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T, Baba M, Akashi M. Biodegradable nanoparticles as vaccine adjuvants and delivery systems: regulation of immune responses by nanoparticle-based vaccine. In: Kunugi S, Yamaoka T, editors. Polymers in nanomedicine. Heidelberg: Springer-Verlag; 2012. pp. 31–64. [Google Scholar]
- Alexyuk PG, Bogoyavlenskiy AP, Alexyuk MS, Turmagambetova AS, Zaitseva IA, Omirtaeva ES, Berezin VE. Adjuvant activity of multimolecular complexes based on Glycyrrhiza glabra saponins, lipids, and influenza virus glycoproteins. Arch Virol. 2019;164:1793–1803. doi: 10.1007/s00705-019-04273-2. [DOI] [PubMed] [Google Scholar]
- Allen CD, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007;27:190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsulaiman JW, Khasawneh AI, Kheirallah KA. Could “trained immunity” be induced by live attenuated vaccines protect against COVID-19? Review of available evidence. J Infect Dev Ctries. 2020;14(9):957–962. doi: 10.3855/jidc.12805. [DOI] [PubMed] [Google Scholar]
- An Y, Parsons LM, Jankowska E, Melnyk D, Joshi M, Cipollo JF. N-Glycosylation of seasonal influenza vaccine hemagglutinins: implication for potency testing and immune processing. J Virol. 2019;93:e01693–e1718. doi: 10.1128/JVI.01693-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre FE, Booy R, Bock HL, Clemens J, Datta SK, John TJ, Lee BW, Lolekha S, Peltola H, Ruff HA, SantoshamM SHJ. Vaccination greatly reduces disease, disability, death and inequity worldwide. Bull World Health Organ. 2008;86:140–146. doi: 10.2471/BLT.07.040089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anraku I, Harvey TJ, Linedale R, Gardner J, Harrich D, Suhrbier A, Khromykh AA. Kunjin virus replicon vaccine vectors induce protective CD8+ T-cell immunity. J Virol. 2002;76:3791–3799. doi: 10.1128/JVI.76.8.3791-3799.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arens R. Rational design of vaccines: learning from immune evasion mechanisms of persistent viruses and tumors. Adv Immunol. 2012;114:217–243. doi: 10.1016/B978-0-12-396548-6.00009-3. [DOI] [PubMed] [Google Scholar]
- Ashley CE, Carnesc EC, Phillips GK, Durfee PN, Buley MD, Lino CA, Peabody DS. Cell-specific delivery of diverse cargos by bacteriophage MS2 viruslike particles. ACS Nano. 2011;5:5729–5745. doi: 10.1021/nn201397z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atrasheuskaya AV, Neverov AA, Rubin S, Ignatyev GM. Horizontal transmission of the Leningrad-3 live attenuated mumps vaccine virus. Vaccine. 2006;24(10):1530–1536. doi: 10.1016/j.vaccine.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Atrasheuskaya A, Kulak M, Fisenko EG, Karpov I, Ignatyev G, Atrasheuskaya A. Horizontal transmission of the Leningrad-Zagreb mumps vaccine strain: a report of six symptomatic cases of parotitis and one case of meningitis. Vaccine. 2012;30(36):5324–5326. doi: 10.1016/j.vaccine.2012.06.055. [DOI] [PubMed] [Google Scholar]
- Aucouturier J, Dupuis L, Deville S, Ascarateil S, Ganne V. Montanide ISA 720 and 51: a new generation of water in oil emulsions as adjuvants for human vaccines. Expert Rev Vaccines. 2002;1:111–118. doi: 10.1586/14760584.1.1.111. [DOI] [PubMed] [Google Scholar]
- Badiee A, Khamesipour A, Samiei A, Soroush D, Shargh V, Kheiri M. The role of liposome size on the type of immune response induced in BALB/C mice against leishmaniasis: rgp63 as a model antigen. Exp Parasitol. 2012;132:403–409. doi: 10.1016/j.exppara.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Barberis I, Myles P, Ault SK, Bragazzi NL, Martini M. History and evolution of influenza control through vaccination: from the first monovalent vaccine to universal vaccines. J Prev Med Hyg. 2016;57:E115–E120. [PMC free article] [PubMed] [Google Scholar]
- Barr JN, Fearns R. How RNA viruses maintain their genome integrity. J Gen Virol. 2010;91(Pt 6):1373–1387. doi: 10.1099/vir.0.020818-0. [DOI] [PubMed] [Google Scholar]
- Bastos Pereira V, Zurita-Turk M, Luerce Saraiva TD, Prósperi De Castro C, Mendes Souzac B, Mancha Agresti P, Alvarenga Lima F, Pfeiffer VN, Pacheco Azevedo MS, Santos Rocha C, Santos Pontes D, Azevedo V, Miyoshi A. DNA vaccines approach: from concepts to applications. World J Vaccines. 2014;4:50–71. doi: 10.4236/wjv.2014.42008. [DOI] [Google Scholar]
- Batista-Duharte A, Lindblad EB, Oviedo-Orta E. Progress in understanding adjuvant immunotoxicity mechanisms. Toxicol Lett. 2011;203(2):97–105. doi: 10.1016/j.toxlet.2011.03.001. [DOI] [PubMed] [Google Scholar]
- Beverley PCL. Immunology of vaccination. Br Med Bull. 2002;62:15–28. doi: 10.1093/bmb/62.1.15. [DOI] [PubMed] [Google Scholar]
- Bomford R. Will adjuvants be needed for vaccines of the future? Dev Biol Stand. 1998;92:13–17. [PubMed] [Google Scholar]
- Bordería AV, Rozen-Gagnon K, Vignuzzi M. Fidelity variants and RNA quasispecies. Curr Top Microbiol Immunol. 2016;392:303–322. doi: 10.1007/82_2015_483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos R, Rutten L, van der Lubbe JEM, Bakkers MJG, Hardenberg G, Wegmann F, Zuijdgeest D, de Wilde AH, Koornneef A, Verwilligen A, van Manen D, Kwaks T, Vogels R, Dalebout TJ, Myeni SK, Kikkert M, Snijder EJ, Li Z, Barouch DH, Vellinga J, Langedijk JPM, Zahn RC, Custers J, Schuitemaker H. Ad26 vector-based COVID-19 vaccine encoding a prefusion-stabilized SARS-CoV-2 Spike immunogen induces potent humoral and cellular immune responses. NPJ Vaccines. 2020;5:91. doi: 10.1038/s41541-020-00243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier NM. The future of influenza vaccines: a historical and clinical perspective. Vaccines (Basel) 2018;6:58. doi: 10.3390/vaccines6030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylston A. The origins of inoculation. J R Soc Med. 2012;105(7):309–313. doi: 10.1258/jrsm.2012.12k044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitburd F, Kirnbauer R, Hubbert NL, Nonnenmacher B, Trin-Dinh-Desmarquet C, Orth G, Schiller JT, Lowy DR. Immunization with viruslike particles from cottontail rabbit papillomavirus (CRPV) can protect against experimental CRPV infection. J Virol. 1995;69(6):3959–3963. doi: 10.1128/jvi.69.6.3959-3963.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breman JG, Arita I. The confirmation and maintenance of smallpox eradication. N Engl J Med. 1980;303:1263–1273. doi: 10.1056/NEJM198011273032204. [DOI] [PubMed] [Google Scholar]
- Brito LA, Kommareddy S, Maione D, Uematsu Y, Giovani C, Berlanda Scorza F, Otten GR, Yu D, Mandl CW, Mason PW, Dormitzer PR, Ulmer JB, Geall AJ. Self-amplifying mRNA vaccines. Adv Genet. 2015;89:179–233. doi: 10.1016/bs.adgen.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Bryan JT, Jansen KU, Lowe RS, Fife KH, McClowry T, Glass D, Brown DR. Human papillomavirus type 11 neutralization in the athymic mouse xenograft system: correlation with virus-like particle IgG concentration. J Med Virol. 1997;53(3):185–188. doi: 10.1002/(SICI)1096-9071(199711)53:3<185::AID-JMV1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Bryan JT, Buckland B, Hammond J, Jansen KU. Prevention of cervical cancer: journey to develop the first human papillomavirus virus-like particle vaccine and the next generation vaccine. Curr Opin Chem Biol. 2016;32:34–47. doi: 10.1016/j.cbpa.2016.03.001. [DOI] [PubMed] [Google Scholar]
- Bukreyev A, Skiadopoulos MH, Murphy BR, Collins PL. Nonsegmented negative-strand viruses as vaccine vectors. J Virol. 2006;80(21):10293–10306. doi: 10.1128/JVI.00919-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull PC, Lowe BS, Kortok M, Molyneux CS, Newbold CI, Marsh K. Parasite antigens on the infected red cell surface are targets for naturally acquired immunity to malaria. Nat Med. 1998;4:358–360. doi: 10.1038/nm0398-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buynak EB, Roehm RR, Tytell AA, Bertland AU, 2nd, Lampson GP, Hilleman MR. Vaccine against human hepatitis B. JAMA. 1976;235:2832–2834. doi: 10.1001/jama.1976.03260520026017. [DOI] [PubMed] [Google Scholar]
- Cann AJ, Stanway G, Hughes PJ, Minor PD, Evans DM, Schild GC, Almond JW. Reversion to neurovirulence of the live-attenuated Sabin type 3 oral poliovirus vaccine. Nucleic Acids Res. 1984;12(20):7787–7792. doi: 10.1093/nar/12.20.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll MW, Overwijk WW, Chamberlain RS, Rosenberg SA, Moss B, Restifo NP. Highly attenuated modified vaccinia virus Ankara (MVA) as an effective recombinant vector: a murine tumor model. Vaccine. 1997;15:387–394. doi: 10.1016/S0264-410X(96)00195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahal JS, Fang T, Woodham AW, Khan OF, Ling J, Anderson DG, Ploegh HL. An RNA nanoparticle vaccine against Zika virus elicits antibody and CD8+ T cell responses in a mouse model. Sci Rep. 2017;7:252. doi: 10.1038/s41598-017-00193-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattergoon MA, Saulino V, Shames JP, Stein J, Montaner LJ, Weiner DB. Co-immunization with plasmid IL-12 generates a strong T-cell memory response in mice. Vaccine. 2004;22:1744–1750. doi: 10.1016/j.vaccine.2004.01.036. [DOI] [PubMed] [Google Scholar]
- Chen ZY, He CY, Ehrhardt A, Kay MA. Minicircle DNA vectors devoid of bacterial DNA result in persistent and high-level transgene expression in vivo. Mol Ther. 2003;8:495–500. doi: 10.1016/S1525-0016(03)00168-0. [DOI] [PubMed] [Google Scholar]
- Cheshenko N, Krougliak N, Eisensmith RC, Krougliak VA. A novel system for the production of fully deleted adenovirus vectors that does not require helper adenovirus. Gene Ther. 2001;8:846–854. doi: 10.1038/sj.gt.3301459. [DOI] [PubMed] [Google Scholar]
- Claassen E, Leeuw W, Greeve P, Hendriksen C, Boersma W. Freund’s complete adjuvant: an effective but disagreeable formula. Res Immunol. 1992;143:478–483. doi: 10.1016/0923-2494(92)80057-R. [DOI] [PubMed] [Google Scholar]
- Coban C, Ishii KJ, Gursel M, Klinman DM, Kumar N. Effect of plasmid backbone modification by different human CpG motifs on the immunogenicity of DNA vaccine vectors. J Leukoc Biol. 2005;78:647–655. doi: 10.1189/jlb.1104627. [DOI] [PubMed] [Google Scholar]
- Connell AR, Connell J, Leahy TR, Hassan J. Mumps outbreaks in vaccinated populations-is it time to re-assess the clinical efficacy of vaccines? Front Immunol. 2020;11:2089. doi: 10.3389/fimmu.2020.02089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese MM, Barskey AE, Tegtmeier GE, Zhang C, Ngo L, Kyaw MH, Baughman AL, Menitove JE, Hickman CJ, Bellini WJ, Dayan GH, Hansen GR, Rubin S. Mumps antibody levels among students before a mumps outbreak: in search of a correlate of immunity. J Infect Dis. 2011;204(9):1413–1422. doi: 10.1093/infdis/jir526. [DOI] [PubMed] [Google Scholar]
- Coughlan L. Factors which contribute to the immunogenicity of non-replicating adenoviral vectored vaccines. Front Immunol. 2020;11:909. doi: 10.3389/fimmu.2020.00909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danhier F, Ansorena E, Silva JM, Coco R, Le Breton A, Préat V. PLGA-based nanoparticles: an overview of biomedical applications. J Control Release. 2012;161:505–522. doi: 10.1016/j.jconrel.2012.01.043. [DOI] [PubMed] [Google Scholar]
- Davenne T, McShane H. Why don’t we have an effective tuberculosis vaccine yet? Expert Rev Vaccines. 2016;15:1009–1013. doi: 10.1586/14760584.2016.1170599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison AJ. Overview of classification. In: Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, editors. Human herpesvirus: biology, therapy and immunoprophylaxis. Cambridge: Cambridge University Press; 2007. [PubMed] [Google Scholar]
- de Martel C, Plummer M, Vignat J, Franceschi S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int J Cancer. 2017;141:664–670. doi: 10.1002/ijc.30716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Arévalo D, Zeng M. Nanoparticle-based vaccines: opportunities and limitations. In: Shegokar R, editor. Nanopharmaceuticals. Amsterdam: Elsevier; 2020. pp. 135–150. [Google Scholar]
- Didierlaurent AM, Laupèze B, Pasquale AD, Hergli N, Collignon C, Garçon N. Adjuvant system AS01: helping to overcome the challenges of modern vaccines. Expert Rev Vaccines. 2017;16:55–63. doi: 10.1080/14760584.2016.1213632. [DOI] [PubMed] [Google Scholar]
- Dobrovolskaia MA, McNeil SE. Immunological properties of engineered nanomaterials. Nat Nanotechnol. 2007;2:469–478. doi: 10.1038/nnano.2007.223. [DOI] [PubMed] [Google Scholar]
- Dogan I, Bertocci B, Vilmont V, Delbos F, Mégret J, Storck S, Reynaud C-A, Weill J-C. Multiple layers of B cell memory with different effector functions. Nat Immunol. 2009;10:1292–1299. doi: 10.1038/ni.1814. [DOI] [PubMed] [Google Scholar]
- Dörsam V, Weimer T, Schmeel A, Hein B, Enssle K, Chumakov KM, Fibi MR. Increased safety level of serotype 3 Sabin oral poliomyelitis vaccine lots by improved seed virus, and tissue culture and virus infection conditions. Vaccine. 2000;18(22):2435–2443. doi: 10.1016/S0264-410X(99)00531-9. [DOI] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/JVI.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekici S, Ilgin P, Butun S, Sahiner N. Hyaluronic acid hydrogel particles with tunable charges as potential drug delivery devices. Carbohydr Polym. 2011;84:1306–1313. doi: 10.1016/j.carbpol.2011.01.028. [DOI] [Google Scholar]
- Eldi P, Cooper TH, Liu L, Prow NA, Diener KR, Howley PM, Suhrbier A, Hayball JD. Production of a Chikungunya vaccine using a CHO cell and attenuated viral-based platform technology. Mol Ther. 2017;25:2332–2344. doi: 10.1016/j.ymthe.2017.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EMA (2006) Guideline on clinical evaluation of new vaccines. https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-clinical-evaluation-new-vaccines_en.pdf. Accessed 18 Aug 2021
- Enders JF, Peebles TC. Propagation in tissue cultures of cytopathogenic agents from patients with measles. Proc Soc Exp Bioi Med. 1954;86:277. doi: 10.3181/00379727-86-21073. [DOI] [PubMed] [Google Scholar]
- Esser-Nobis K, Hatfield LD, Gale M., Jr Spatiotemporal dynamics of innate immune signaling via RIG-I-like receptors. Proc Natl Acad Sci USA. 2020;27:15778–15788. doi: 10.1073/pnas.1921861117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathi A, Dahlke C, Addo MM. Recombinant vesicular stomatitis virus vector vaccines for WHO blueprint priority pathogens. Hum Vaccin Immunother. 2019;15:2269–2285. doi: 10.1080/21645515.2019.1649532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felberbaum RS. The baculovirus expression vector system: a commercial manufacturing platform for viral vaccines and gene therapy vectors. Biotechnol J. 2015;10:702–714. doi: 10.1002/biot.201400438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fifis T, Gamvrellis A, Crimeen-Irwin B, Pietersz GA, Li J, Mottram PL, McKenzie IFC, Plebanski M. Size dependent immunogenicity: therapeutic and protective properties of nano-vaccines against tumors. J Immunol. 2004;173:3148–3154. doi: 10.4049/jimmunol.173.5.3148. [DOI] [PubMed] [Google Scholar]
- Fine P, Eames K, Heymann DL. Herd immunity: a rough guide. Clin Infect Dis. 2011;52:911–916. doi: 10.1093/cid/cir007. [DOI] [PubMed] [Google Scholar]
- Fitzgerald KA, Kagan JC. Toll-like receptors and the control of immunity. Cell. 2020;180:1044–1066. doi: 10.1016/j.cell.2020.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming SB. Viral inhibition of the IFN-induced JAK/STAT signalling pathway: development of live attenuated vaccines by mutation of viral-encoded IFN-antagonists. Vaccines (basel) 2016;4(3):23. doi: 10.3390/vaccines4030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowkes FJ, Richards JS, Simpson JA, Beeson JG. The relationship between anti-merozoite antibodies and incidence of plasmodium falciparum malaria: a systematic review and meta-analysis. PLoS Med. 2010;7:e1000218. doi: 10.1371/journal.pmed.1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso-Saavedra M, Vega-López MA. Induction of mucosal immunity against pathogens by using recombinant baculoviral vectors: Mechanisms, advantages, and limitations. J Leukoc Biol. 2020;108(3):835–850. doi: 10.1002/JLB.4MR0320-488R. [DOI] [PubMed] [Google Scholar]
- Fraley E, LeMaster C, Geanes E, Banerjee D, Khanal S, Grundberg E, Selvarangan R, Bradley T. Humoral immune responses during SARS-CoV-2 mRNA vaccine administration in seropositive and seronegative individuals. BMC Med. 2021;19:169. doi: 10.1186/s12916-021-02055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz PN, Teeravechyan S, Tangy F. Measles-derived vaccines to prevent emerging viral diseases. Microbes Infect. 2018;20(9–10):493–500. doi: 10.1016/j.micinf.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fynan EF, Webster RG, Fuller DH, Haynes JR, Santoro JC, Robinson HL. DNA vaccines: protective immunizations by parenteral, mucosal, and gene-gun inoculations. Proc Natl Acad Sci U S A. 1993;90:11478–11482. doi: 10.1073/pnas.90.24.11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- Gao J, Mese K, Bunz O, Ehrhardt A. State-of-the-art human adenovirus vectorology for therapeutic approaches. FEBS Lett. 2019;593:3609–3622. doi: 10.1002/1873-3468.13691. [DOI] [PubMed] [Google Scholar]
- Garbutt M, Liebscher R, Wahl V, Jones S, Möller P, Wagner R, Volchkov VE, Klenk H-D, Feldmann H, Ströher U. Properties of replication-competent vesicular stomatitis virus vectors expressing glycoproteins of filoviruses and arenaviruses. J Virol. 2004;78:5458–5465. doi: 10.1128/JVI.78.10.5458-5465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargano JW, Wallace M, Hadler SC, Langley G, Su JR, Oster ME, Broder KR, Gee J, Weintraub E, Shimabukuro T, Scobie HM, Moulia D, Markowitz LE, Wharton M, McNally VV, Romero JR, Talbot HK, Lee GM, Daley MF, Oliver SE. Use of mRNA COVID-19 vaccine after reports of myocarditis among vaccine recipients: update from the advisory committee on immunization practices - United States, June 2021. MMWR Morb Mortal Wkly Rep. 2021;70:977–982. doi: 10.15585/mmwr.mm7027e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland JM. An update on meningococcal vaccination. R I Med J. 2020;103:41–43. [PubMed] [Google Scholar]
- Geall AJ, Verma A, Otten GR, Shaw CA, Hekele A, Banerjee K, Cu Y, Beard CW, Brito LA, Krucker T, O'Hagan DT, Singh M, Mason PW, Valiante NM, Dormitzer PR, Barnett SW, Rappuoli R, Ulmer JB, Mandl CW. Nonviral delivery of self-amplifying RNA vaccines. Proc Natl Acad Sci USA. 2012;109:14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AC, Mohsen M, Bachmann MF. Harnessing nanoparticles for immunomodulation and vaccines. Vaccines (basel) 2017;5:6. doi: 10.3390/vaccines5010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gόmez CE, Najera JL, Krupa M, Perdiguero B, Esteban M. MVA and NYVAC as vaccines against emergent infectious diseases and cancer. Curr Gene Ther. 2011;11:189–217. doi: 10.2174/156652311795684731. [DOI] [PubMed] [Google Scholar]
- Goodson JL, Seward JF. Measles 50 years after use of measles vaccine. Infect Dis Clin North Am. 2015;29(4):725–743. doi: 10.1016/j.idc.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Gopal R, Schneemann A. Production and application of insect virus-based VLPs. Methods Mol Biol. 2018;1776:125–141. doi: 10.1007/978-1-4939-7808-3_8. [DOI] [PubMed] [Google Scholar]
- Grachev VP, Karganova GG, Rumyantsev AA, Ivanova OE, Eremeeva TP, Drozdov SG. Evaluation of the new control methods for oral poliomyelitis vaccine. Dev Biol (basel) 2001;105:211–217. [PubMed] [Google Scholar]
- Griffin DE. Measles immunity and immunosuppression. Curr Opin Virol. 2021;46:9–14. doi: 10.1016/j.coviro.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grose C, Putman Z, Esposito D. A review of alternative promoters for optimal recombinant protein expression in baculovirus-infected insect cells. Protein Expr Purif. 2021;186:105924. doi: 10.1016/j.pep.2021.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan S, Rosenecker J. Nanotechnologies in delivery of mRNA therapeutics using nonviral vector-based delivery systems. Gene Ther. 2017;24:133–143. doi: 10.1038/gt.2017.5. [DOI] [PubMed] [Google Scholar]
- Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Granados S, Cervera L, Kamen AA, Gòdia F. Advancements in mammalian cell transient gene expression (TGE) technology for accelerated production of biologics. Crit Rev Biotechnol. 2018;38:918–940. doi: 10.1080/07388551.2017.1419459. [DOI] [PubMed] [Google Scholar]
- Halwani R, Boyer JD, Yassine-Diab B, Haddad EK, Robinson TM, Kumar S, Parkinson R, Wu L, Sidhu MK, Phillipson-Weiner R, Pavlakis GN, Felber BK, Lewis MG, Shen A, Siliciano RF, Weiner DB, Sekaly RP. Therapeutic vaccination with simian immunodeficiency virus (SIV)-DNA + IL-12 or IL-15 induces distinct CD8 memory subsets in SIV-infected macaques. J Immunol. 2008;180:7969–7979. doi: 10.4049/jimmunol.180.12.7969. [DOI] [PubMed] [Google Scholar]
- Hansson M, Nygren PA, Ståhl S. Design and production of recombinant subunit vaccines. Biotechnol Appl Biochem. 2000;32(2):95–107. doi: 10.1042/BA20000034. [DOI] [PubMed] [Google Scholar]
- Harding CV, Song R. Phagocytic processing of exogenous particulate antigens by macrophages for presentation by class I MHC molecules. J Immunol. 1994;153:4925–4933. doi: 10.4049/jimmunol.153.11.4925. [DOI] [PubMed] [Google Scholar]
- Hasson SSAA, Al-Busaidi JKZ, Sallam TA. The past, current and future trends in DNA vaccine immunisations. Asian Pac J Trop Biomed. 2015;5:344–353. doi: 10.1016/S2221-1691(15)30366-X. [DOI] [Google Scholar]
- Haumont M, Jacquet A, Massaer M, Deleersnyder V, Mazzu P, Bollen A, Jacobs P. Purification, characterization and immunogenicity of recombinant varicella-zoster virus glycoprotein gE secreted by Chinese hamster ovary cells. Virus Res. 1996;40:199–204. doi: 10.1016/0168-1702(95)01270-2. [DOI] [PubMed] [Google Scholar]
- Haynes BF, Burton DR, Mascola JR. Multiple roles for HIV broadly neutralizing antibodies. Sci Transl Med. 2019;11:eaaz2686. doi: 10.1126/scitranslmed.aaz2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrickson KJ, Kingsbury DW, van Wyke Coelingh KL, Naeve CW, Portner A. Neutralizing epitopes of human parainfluenza virus type 3 are conformational and cannot be imitated by synthetic peptides. Vaccine. 1991;9:243–249. doi: 10.1016/0264-410X(91)90107-H. [DOI] [PubMed] [Google Scholar]
- Hernáez B, Alonso-Lobo JM, Montanuy I, Fischer C, Sauer S, Sigal L, Sevilla N, Alcamí A. A virus-encoded type I interferon decoy receptor enables evasion of host immunity through cell-surface binding. Nat Commun. 2018;9:1–14. doi: 10.1038/s41467-018-07772-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingdale MR, Ballou WR, Aley SB, Young JF, Pancake S, Miller LH, Hockmeyer WT. Plasmodium falciparum: elicitation by peptides and recombinant circumsporozoite proteins of circulating mouse antibodies inhibiting sporozoite invasion of hepatoma cells. Exp Parasitol. 1987;63:345–351. doi: 10.1016/0014-4894(87)90182-2. [DOI] [PubMed] [Google Scholar]
- Holtkamp S, Kreiter S, Selmi A, Simon P, Koslowski M, Huber C, Türeci O, Sahin U. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood. 2006;108:4009–4017. doi: 10.1182/blood-2006-04-015024. [DOI] [PubMed] [Google Scholar]
- Howe HA, Bodian D. Poliomyelitis in the chimpanzee: a clinical pathological study. Bull Johns Hopk Hosp. 1941;69:149–181. [Google Scholar]
- Ivancic-Jelecki J, Santak M, Forcic D. Variability of hemagglutinin-neuraminidase and nucleocapsid protein of vaccine and wild-type mumps virus strains. Infect Genet Evol. 2008;8(5):603–613. doi: 10.1016/j.meegid.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Symons RH, Berg P. Biochemical method for inserting new genetic information into DNA of Simian virus 40: Circular SV40 DNA molecules containing lambda phage genes and the galactose operon of Escherichia coli. Proc Nat Acad Sci. 1972;69:2904–2909. doi: 10.1073/pnas.69.10.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen KU, Rosolowsky M, Schultz LD, Markus HZ, Cook JC, Donnelly JJ, Martinez D, Ellis RW, Shaw AR. Vaccination with yeast-expressed cottontail rabbit papillomavirus (CRPV) virus-like particles protects rabbits from CRPV-induced papilloma formation. Vaccine. 1995;13(16):1509–1514. doi: 10.1016/0264-410X(95)00103-8. [DOI] [PubMed] [Google Scholar]
- Johanning FW, Conry RM, LoBuglio AF, Wright M, Sumerel LA, Pike MJ, Curiel DT. A Sindbis virus mRNA polynucleotide vector achieves prolonged and high level heterologous gene expression in vivo. Nucleic Acids Res. 1995;23:1495–1501. doi: 10.1093/nar/23.9.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorba J, Diop OM, Iber J, Henderson E, Zhao K, Quddus A, Sutter R, Vertefeuille JF, Wenger J, Wassilak SGF, Pallansch MA, Burns CC. Update on vaccine-derived poliovirus outbreaks - worldwide, January 2018-June 2019. MMWR Morb Mortal Wkly Rep. 2019;68(45):1024–1028. doi: 10.15585/mmwr.mm6845a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaic B, Gjenero-Margan I, Aleraj B, Ljubin-Sternak S, Vilibic-Cavlek T, Kilvain S, Pavic I, Stojanovic D, Ilic A. Transmission of the L-Zagreb mumps vaccine virus, Croatia, 2005–2008. Euro Surveill. 2008;13(16):18843. doi: 10.2807/ese.13.16.18843-en. [DOI] [PubMed] [Google Scholar]
- Kalpana GV. Retroviral vectors for liver-directed gene therapy. Semin Liver Dis. 1999;19(1):27–37. doi: 10.1055/s-2007-1007095. [DOI] [PubMed] [Google Scholar]
- Kantarcioglu B, Iqbal O, Walenga JM, Lewis B, Lewis J, Carter CA, Singh M, Lievano F, Tafur A, Ramacciotti E, Gerotziafas GT, Jeske W, Fareed J. An update on the pathogenesis of COVID-19 and the reportedly rare thrombotic events following vaccination. Clin Appl Thromb Hemost. 2021;27:10760296211021498. doi: 10.1177/10760296211021498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariko K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariko K, Muramatsu H, Ludwig J, Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karron RA, Collins PL. Parainfluenza viruses. In: Knipe DM, Howley PM, Griffin DE, editors. Fields Virology. 5. Philadelphia: Lippincott Williams and Wilkins; 2007. pp. 1497–1526. [Google Scholar]
- Kashoki M, Hanaizi Z, Yordanova S, Veselý R, Bouygues C, Llinares J, Kweder SL. A comparison of EMA and FDA decisions for new drug marketing applications 2014–2016: concordance, discordance, and why. Clin Pharmacol Ther. 2020;107(1):195–202. doi: 10.1002/cpt.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri K, Goyal AK, Gupta PN, Mishra N, Vyas SP. Plasmid DNA loaded chitosan nanoparticles for nasal mucosal immunization against hepatitis B. Int J Pharm. 2008;354:235–241. doi: 10.1016/j.ijpharm.2007.11.027. [DOI] [PubMed] [Google Scholar]
- Kheirollahpour M, Mehrabi M, Dounigh NM, Mohammadi M, Masoudi A. Nanopart Vaccine Deve Pharm Nanotechnol. 2020;8:6–21. doi: 10.2174/2211738507666191024162042. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Trivedi NN, Nottingham LK, Morrison L, Tsai A, Hu Y, Mahalingam S, Dang K, Ahn L, Doyle NK, Wilson DM, Chattergoon MA, Chalian AA, Boyer JD, Agadjanyan MG, Weiner DB. Modulation of amplitude and direction of in vivo immune responses by co-administration of cytokine gene expression cassettes with DNA immunogens. Eur J Immunol. 1998;28:1089–1103. doi: 10.1002/(SICI)1521-4141(199803)28:03<1089::AID-IMMU1089>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Yang JS, Montaner L, Lee DJ, Chalian AA, Weiner DB. Coimmunization with IFN-gamma or IL-2, but not IL-13 or IL-4 cDNA can enhance Th1-type DNA vaccine-induced immune responses in vivo. J Interferon Cytokine Res. 2000;20:311–319. doi: 10.1089/107999000312450. [DOI] [PubMed] [Google Scholar]
- Klimov VV. Vaccination. In: Klimov VV, editor. From basic to clinical immunology. New York: Springer; 2019. pp. 291–304. [Google Scholar]
- Kojima Y, Xin KQ, Ooki T, Hamajima K, Oikawa T, Shinoda K, Ozaki T, Hoshino Y, Jounai N, Nakazawa M, Klinman D, Okuda K. Adjuvant effect of multi-CpG motifs on an HIV-1 DNA vaccine. Vaccine. 2002;20:2857–2865. doi: 10.1016/S0264-410X(02)00238-4. [DOI] [PubMed] [Google Scholar]
- Kong M, Zuo H, Zhu F, Hu Z, Chen L, Yang Y, Lv P, Yao Q, Chen K. The interaction between baculoviruses and their insect hosts. Dev Comp Immunol. 2018;83:114–123. doi: 10.1016/j.dci.2018.01.019. [DOI] [PubMed] [Google Scholar]
- Kost TA, Condreay JP, Jarvis DL. Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nat Biotechnol. 2005;23:567–575. doi: 10.1038/nbt1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost TA, Condreay JP, Ames RS. Baculovirus gene delivery: a flexible assay development tool. Curr Gene Ther. 2010;10:168–173. doi: 10.2174/156652310791321224. [DOI] [PubMed] [Google Scholar]
- Kovacsovics-Bankowski M, Clark K, Benacerraf B, Rock KL. Efficient major histocompatibility complex class I presentation of exogenous antigen upon phagocytosis by macrophages. Proc Natl Acad Sci U S A. 1993;90:4942–4946. doi: 10.1073/pnas.90.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurup VM, Thomas J. Edible vaccines: promises and challenges. Mol Biotechnol. 2020;62:79–90. doi: 10.1007/s12033-019-00222-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwang TW, Zeng X, Wang S. Manufacturing of AcMNPV baculovirus vectors to enable gene therapy trials. Mol Ther - Methods Clin Dev. 2016;3:15050. doi: 10.1038/mtm.2015.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laddy DJ, Weiner DB. From plasmids to protection: a review of DNA vaccines against infectious diseases. Int Rev Immunol. 2006;25:99–123. doi: 10.1080/08830180600785827. [DOI] [PubMed] [Google Scholar]
- Ladnyĭ ID. Global program of smallpox eradication. 1. Smallpox in the world before acceptance of the program of its eradication by the World Health Organization. Zh Mikrobiol Epidemiol Immunobiol. 1977;3:98–105. [PubMed] [Google Scholar]
- Laksono BM, de Vries RD, Duprex WP, de Swart RL. Measles pathogenesis, immune suppression and animal models. Curr Opin Virol. 2021;41:31–37. doi: 10.1016/j.coviro.2020.03.002. [DOI] [PubMed] [Google Scholar]
- Lasky LA, Groopman JE, Fennie CW, Benz PM, Capon DJ, Dowbenko DJ, Nakamura GR, Nunes WM, Renz ME, Berman PW. Neutralization of the AIDS retrovirus by antibodies to a recombinant envelope glycoprotein. Science. 1986;233:209–212. doi: 10.1126/science.3014647. [DOI] [PubMed] [Google Scholar]
- Latanova AA, Petkov S, Kilpelainen A, Jansons J, Latyshev OE, Kuzmenko YV, Hinkula J, Abakumov MA, Valuev-Elliston VT, Gomelsky M, Karpov VL, Chiodi F, Wahren B, Logunov DY, Starodubova ES, Isaguliants MG. Codon optimization and improved delivery/immunization regimen enhance the immune response against wild-type and drug-resistant HIV-1 reverse transcriptase, preserving its Th2-polarity. Sci Rep. 2018;8:8078. doi: 10.1038/s41598-018-26281-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic I, Pravica V, Miljanovic D, Cupic M. Immune evasion of SARS-CoV-2 emerging variants: what have we learnt so far? Viruses. 2021;13:1192. doi: 10.3390/v13071192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Arun Kumar S, Jhan YY, Bishop CJ. Engineering DNA vaccines against infectious diseases. Acta Biomater. 2018;80:31–47. doi: 10.1016/j.actbio.2018.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester SN, Li K. Toll-like receptors in antiviral innate immunity. J Mol Biol. 2014;426:1246–1264. doi: 10.1016/j.jmb.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Luo Z, Liu P. Bioreducible alginate poly(ethylenimine) nanogels as an antigen-delivery system robustly enhance vaccine-elicited humoral and cellular immune responses. J Control Release. 2013;168:271–279. doi: 10.1016/j.jconrel.2013.03.025. [DOI] [PubMed] [Google Scholar]
- Lilic D, Ghosh SK. Liver dysfunction and DNA antibodies after hepatitis B vaccination. Lancet. 1994;344:1292–1293. doi: 10.1016/S0140-6736(94)90776-5. [DOI] [PubMed] [Google Scholar]
- Lin SY, Chung YC, Hu YC. Update on baculovirus as an expression and/or delivery vehicle for vaccine antigens. Expert Rev Vaccines. 2014;13(12):1501–1521. doi: 10.1586/14760584.2014.951637. [DOI] [PubMed] [Google Scholar]
- Link A, Zabel F, Schnetzler Y, Titz A, Brombacher F, Bachmann MF. Innate immunity mediates follicular transport of particulate but not soluble protein antigen. J Immunol. 2012;188:3724–3733. doi: 10.4049/jimmunol.1103312. [DOI] [PubMed] [Google Scholar]
- Ljungberg K, Liljestrom P. Self-replicating alphavirus RNA vaccines. Expert Rev Vaccines. 2015;14:177–194. doi: 10.1586/14760584.2015.965690. [DOI] [PubMed] [Google Scholar]
- Long B, Bridwell R, Gottlieb M. Thrombosis with thrombocytopenia syndrome associated with COVID-19 vaccines. Am J Emerg Med. 2021;49:58–61. doi: 10.1016/j.ajem.2021.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou B, De Beuckelaer A, Boonstra E, Li D, De Geest BG, De Koker S, Mastrobattista E, Hennink WE. Modular core-shell polymeric nanoparticles mimicking viral structures for vaccination. J Control Release. 2019;293:48–62. doi: 10.1016/j.jconrel.2018.11.006. [DOI] [PubMed] [Google Scholar]
- Lu HY, Chen YH, Liu HJ. Baculovirus as a Vaccine Vector Bioengineered. 2012;3:271–274. doi: 10.4161/bioe.20679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugade AA, Bharali DJ, Pradhan V, Elkin G, Mousa SA, Thanavala Y. Single low-dose un-adjuvanted HBsAg nanoparticle vaccine elicits robust, durable immunity. Nanomedicine. 2013;9:923–934. doi: 10.1016/j.nano.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Luongo C, Yang L, Winter CC, Spann KM, Murphy BR, Collins PL, Buchholz UJ. Codon stabilization analysis of the “248” temperature sensitive mutation for increased phenotypic stability of respiratory syncytial virus vaccine candidates. Vaccine. 2009;27:5667–5676. doi: 10.1016/j.vaccine.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackett M, Smith GL, Moss B. Vaccinia virus: a selectable eukaryotic cloning and expression vector. Proc Nat Acad Sci. 1982;79:7414–7419. doi: 10.1073/pnas.79.23.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malonis RJ, Lai JR, Vergnolle O. Peptide-based vaccines: current progress and future challenges. Chem Rev. 2020;120:3210–3229. doi: 10.1021/acs.chemrev.9b00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manini I, Montomoli E. Epidemiology and prevention of human papillomavirus. Ann Ig. 2018;30:28–32. doi: 10.7416/ai.2018.2231. [DOI] [PubMed] [Google Scholar]
- Manolova V, Flace A, Bauer M, Schwarz K, Saudan P, Bachmann MF. Nanoparticles target distinct dendritic cell populations according to their size. Eur J Immunol. 2008;38:1404–1413. doi: 10.1002/eji.200737984. [DOI] [PubMed] [Google Scholar]
- Marini CP, Lewis E, Petrone P, Zenilman A, Lu Z, Rivera A, McNelis J (2021) The incidence and the impact of deep vein thrombosis on the outcome of patients with COVID-19 infection. J Vasc Surg Venous Lymphat Disord S2213-333X(21)00521-7 [DOI] [PMC free article] [PubMed]
- Markowitz LE, Orenstein WA. Measles vaccines. Pediatr Clin N Am. 1990;37(3):603–625. doi: 10.1016/S0031-3955(16)36907-3. [DOI] [PubMed] [Google Scholar]
- Markušić M, Šantak M, Košutić-Gulija T, Jergović M, Jug R, Forčić D. Induction of IFN-α subtypes and their antiviral activity in mumps virus infection. Viral Immunol. 2014;27(10):497–505. doi: 10.1089/vim.2014.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall HS, Plotkin S. The changing epidemiology of mumps in a high vaccination era. Lancet Infect Dis. 2019;19:118–119. doi: 10.1016/S1473-3099(18)30541-3. [DOI] [PubMed] [Google Scholar]
- Martin SA, Paoletti E, Moss B. Purification of mRNA guanylyltransferase and mRNA (guanine-7-) methyltransferase from vaccinia virions. J Biol Chem. 1975;250:9322–9329. doi: 10.1016/S0021-9258(19)40646-7. [DOI] [PubMed] [Google Scholar]
- Mason HS, Lam DM, Arntzen CJ. Expression of hepatitis B surface antigen in transgenic plants. Proc Natl Acad Sci U S A. 1992;89:11745–11749. doi: 10.1073/pnas.89.24.11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo M, Reynard S, Journeaux A, Germain C, Hortion J, Carnec X, Picard C, Baillet N, Borges-Cardoso V, Merabet O, Vallve A, Barron S, Jourjon O, Lacroix O, Duthey A, Dirheimer M, Jouvion G, Moreau PH, Fellmann L, Carbonnelle C, Raoul H, Tangy F, Baize S. A single-shot Lassa vaccine induces long-term immunity and protects cynomolgus monkeys against heterologous strains. Sci Transl Med. 2021;13(597):eabf6348. doi: 10.1126/scitranslmed.abf6348. [DOI] [PubMed] [Google Scholar]
- May M, Rieder CA, Rowe RJ. Emergent lineages of mumps virus suggest the need for a polyvalent vaccine. Int J Infect Dis. 2018;66:1–4. doi: 10.1016/j.ijid.2017.09.024. [DOI] [PubMed] [Google Scholar]
- Mbaeyi SA, Bozio CH, Duffy J, Rubin LG, Hariri S, Stephens DS, MacNeil JR. Meningococcal vaccination: recommendations of the advisory committee on immunization practices, United States, 2020. MMWR Recomm Rep. 2020;69:1–41. doi: 10.15585/mmwr.rr6909a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAleer WJ, Buynak EB, Maigetter RZ, Wampler DE, Miller WJ, Hilleman MR. Human hepatitis B vaccine from recombinant yeast. Nature. 1984;307:178–180. doi: 10.1038/307178a0. [DOI] [PubMed] [Google Scholar]
- McGeoch DJ, Rixon FJ, Davison AJ. Topics in herpesvirus genomics and evolution. J Gen Virol. 2006;117:90–104. doi: 10.1016/j.virusres.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Janeway CA., Jr Decoding the patterns of self and nonself by the innate immune system. Science. 2002;296:298–300. doi: 10.1126/science.1068883. [DOI] [PubMed] [Google Scholar]
- Meisinger-Henschel C, Schmidt M, Lukassen S, Linke B, Krause L, Konietzny S, Goesmann A, Howley P, Chaplin P, Suter M, Hausmann J. Genomic sequence of chorioallantois vaccinia virus Ankara, the ancestor of modified vaccinia virus Ankara. J Gen Virol. 2007;88:3249–3259. doi: 10.1099/vir.0.83156-0. [DOI] [PubMed] [Google Scholar]
- Meyer M, Huang E, Yuzhakov O, Ramanathan P, Ciaramella G, Bukreyev A. Modified mRNA-based vaccines elicit robust immune responses and protect guinea pigs from ebola virus disease. J Infect Dis. 2018;217:451–455. doi: 10.1093/infdis/jix592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milone MC, O'Doherty U. Clinical use of lentiviral vectors. Leukemia. 2018;32(7):1529–1541. doi: 10.1038/s41375-018-0106-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohsen MO, Gomes AC, Cabral-Miranda G, Krueger CC, Leoratti FM, Stein JV, Bachmann MF. Delivering adjuvants and antigens in separate nanoparticles eliminates the need of physical linkage for effective vaccination. J Control Release. 2017;251:92–100. doi: 10.1016/j.jconrel.2017.02.031. [DOI] [PubMed] [Google Scholar]
- Mohsen MO, Zha L, Cabral-Miranda G, Bachmann MF. Major findings and recent advances in virus-like particle (VLP)-based vaccines. Semin Immunol. 2017;34:123–132. doi: 10.1016/j.smim.2017.08.014. [DOI] [PubMed] [Google Scholar]
- Moisa AA, Kolesanova EF. Synthetic Peptide Vaccines Biomed Khim. 2011;57:14–30. doi: 10.18097/pbmc20115701104. [DOI] [PubMed] [Google Scholar]
- Morein B, Sundquist B, Höglund S, Dalsgaard K, Osterhaus A. Iscom, a novel structure for antigenic presentation of membrane proteins from enveloped viruses. Nature. 1984;308:457–460. doi: 10.1038/308457a0. [DOI] [PubMed] [Google Scholar]
- Morein B, Hu KF, Abusugra I. Current status and potential application of ISCOMs in veterinary medicine. Adv Drug Deliv Rev. 2004;56:1367–1382. doi: 10.1016/j.addr.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Mottram PL, Leong D, Crimeen-Irwin B, Gloster S, Xiang SD, Meanger J, Ghildyal R, Vardaxis N, Plebanski M. Type 1 and 2 immunity following vaccination is influenced by nanoparticle size: formulation of a model vaccine for respiratory syncytial virus. Mol Pharm. 2007;4:73–84. doi: 10.1021/mp060096p. [DOI] [PubMed] [Google Scholar]
- Naderi M, Saeedi A, Moradi A, Kleshadi M, Zolfaghari MR, Gorji A, Ghaemi A. Interleukin-12 as a genetic adjuvant enhances hepatitis C virus NS3 DNA vaccine immunogenicity. Virol Sin. 2013;28:167–173. doi: 10.1007/s12250-013-3291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. In Vivo Gene Delivery and Stable Transduction of Nondividing Cells by a Lentiviral Vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- Nascimento IP, Leite LCC. Recombinant vaccines and the development of new vaccine strategies. Braz J Med Biol Res. 2012;45(12):1102–1111. doi: 10.1590/S0100-879X2012007500142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Contant P, Sangster MY, Topham DJ. Squalene-based influenza vaccine adjuvants and their impact on the hemagglutinin-specific B cell response. Pathogens. 2021;10:355. doi: 10.3390/pathogens10030355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales A, Martinez-Sobrido L, Topham DJ, DeDiego ML. Modulation of Innate Immune Responses by the Influenza A NS1 and PA-X Proteins. Viruses. 2018;10(12):708. doi: 10.3390/v10120708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hagan DT. MF59 is a safe and potent vaccine adjuvant that enhances protection against influenza virus infection. Expert Rev Vaccines. 2007;6:699–710. doi: 10.1586/14760584.6.5.699. [DOI] [PubMed] [Google Scholar]
- O'Hagan DT, Lodaya RN, Lofano G. The continued advance of vaccine adjuvants - 'we can work it out'. Semin Immunol. 2020;50:101426. doi: 10.1016/j.smim.2020.101426. [DOI] [PubMed] [Google Scholar]
- Panical D, Paoletti E. Construction of poxviruses as cloning vectors: Insertion of the thymidine kinase gene from herpes simplex virus into the DNA of infectious vaccinia virus. Proc Nat Acad Sci. 1982;79:4927–4931. doi: 10.1073/pnas.79.16.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panyam J, Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Adv Drug Deliv Rev. 2003;55:329–347. doi: 10.1016/S0169-409X(02)00228-4. [DOI] [PubMed] [Google Scholar]
- Pardi N, Weissman D. Nucleoside modified mRNA vaccines for infectious diseases. Methods Mol Biol. 2017;1499:109–121. doi: 10.1007/978-1-4939-6481-9_6. [DOI] [PubMed] [Google Scholar]
- Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines - a new era in vaccinology. Nat Rev Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdiguero B, Esteban M. The interferon system and vaccinia virus evasion mechanisms. J Interferon Cytokine Res. 2009;29:581–598. doi: 10.1089/jir.2009.0073. [DOI] [PubMed] [Google Scholar]
- Perri S, Greer CE, Thudium K, Doe B, Legg H, Liu H, Romero RE, Tang Z, Bin Q, Dubensky TW, Jr, Vajdy M, Otten GR, Polo JM. An alphavirus replicon particle chimera derived from venezuelan equine encephalitis and sindbis viruses is a potent gene-based vaccine delivery vector. J Viral. 2003;77:10394–10403. doi: 10.1128/JVI.77.19.10394-10403.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pippa N, Gazouli M, Pispas S. Recent advances and future perspectives in polymer-based nanovaccines. Vaccines (basel) 2021;9:558. doi: 10.3390/vaccines9060558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piret J, Boivin G. Pandemics throughout history. Front Microbiol. 2021;11:631736. doi: 10.3389/fmicb.2020.631736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin S. History of vaccination. Proc Natl Acad Sci U S A. 2014;111(34):12283–12287. doi: 10.1073/pnas.1400472111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard AJ, Launay O, Lelievre JD, Lacabaratz C, Grande S, Goldstein N, Robinson C, Gaddah A, Bockstal V, Wiedemann A, Leyssen M, Luhn K, Richert L, Bétard C, Gibani MM, Clutterbuck EA, Snape MD, Levy Y, Douoguih M, Thiebaut R, EBOVAC2 EBL2001 study group Safety and immunogenicity of a two-dose heterologous Ad26.ZEBOV and MVA-BN-Filo Ebola vaccine regimen in adults in Europe (EBOVAC2): a randomised, observer-blind, participant-blind, placebo-controlled, phase 2 trial. Lancet Infect Dis. 2020;21:493–506. doi: 10.1016/S1473-3099(20)30476-X. [DOI] [PubMed] [Google Scholar]
- Premanand B, Zhong Wee P, Prabakaran M. Baculovirus surface display of immunogenic proteins for vaccine development. Viruses. 2018;10:298. doi: 10.3390/v10060298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price PJ, Torres-Domínguez LE, Brandmüller C, Sutter G, Lehmann MH. Modified Vaccinia virus Ankara: innate immune activation and induction of cellular signalling. Vaccine. 2013;31(39):4231–4234. doi: 10.1016/j.vaccine.2013.03.017. [DOI] [PubMed] [Google Scholar]
- Publicover J, Ramsburg E, Rose JK. Characterization of nonpathogenic, live, viral vaccine vectors inducing potent cellular immune responses. J Virol. 2004;78:9317–9324. doi: 10.1128/JVI.78.17.9317-9324.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radbruch A, Muehlinghaus G, Luger EO, Inamine A, Smith KG, Dorner T, Hiepe F. Competence and competition: the challenge of becoming a long-lived plasma cell. Nat Rev Immunol. 2006;6:741–750. doi: 10.1038/nri1886. [DOI] [PubMed] [Google Scholar]
- Ramadan A, Land WG, Paczesny S. Editorial: danger signals triggering immune response and inflammation. Front Immunol. 2017;8:979–981. doi: 10.3389/fimmu.2017.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan R, Voigt EA, Kennedy RB, Poland GA. Knowledge gaps persist and hinder progress in eliminating mumps. Vaccine. 2018;36(26):3721–3726. doi: 10.1016/j.vaccine.2018.05.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy MN, Minassian AM, Ewer KJ, Flaxman AL, Folegatti PM, Owens DR, Voysey M, Aley PK, Angus B, Babbage G, Belij-Rammerstorfer S, Berry L, Bibi S, Bittaye M, Cathie K, Chappell H, Charlton S, Cicconi P, Clutterbuck EA, Colin-Jones R, Dold C, Emary KRW, Fedosyuk S, Fuskova M, Gbesemete D, Green C, Hallis B, Hou MM, Jenkin D, Joe CCD, Kelly EJ, Kerridge S, Lawrie AM, Lelliott A, Lwin MN, Makinson R, Marchevsky NG, Mujadidi Y, Munro APS, Pacurar M, Plested E, Rand J, Rawlinson T, Rhead S, Robinson H, Ritchie AJ, Ross-Russell AL, Saich S, Singh N, Smith CC, Snape MD, Song R, Tarrant R, Themistocleous Y, Thomas KM, Villafana TL, Warren SC, Watson MEE, Douglas AD, Hill AVS, Lambe T, Gilbert SC, Faust SN, Pollard AJ, Oxford COVID Vaccine Trial Group Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): a single-blind, randomised, controlled, phase 2/3 trial. Lancet. 2021;396:1979–1993. doi: 10.1016/S0140-6736(20)32466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch S, Jasny E, Schmidt KE, Petsch B. New vaccine technologies to combat outbreak situations. Front Immunol. 2018;9:1963. doi: 10.3389/fimmu.2018.01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Gack MU. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol. 2020;20:537–551. doi: 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards KA, Moritzky S, Shannon I, Fitzgerald T, Yang H, BrancheA TDJ, Treanor JJ, Nayak J, Sant AJ. Recombinant HA-based vaccine outperforms split and subunit vaccines in elicitation of influenza-specific CD4 T cells and CD4 T cell-dependent antibody responses in humans. NPJ Vaccines. 2020;5:77. doi: 10.1038/s41541-020-00227-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson JS, Naeve CW, Webster RG, Bootman JS, Newman R, Schild GC. Alterations in the hemagglutinin associated with adaptation of influenza B virus to growth in eggs. Virology. 1985;143:166–174. doi: 10.1016/0042-6822(85)90105-9. [DOI] [PubMed] [Google Scholar]
- Robertson JS, Bootman JS, Newman R, Oxford JS, Daniels RS, Webster RG, Schild GC. Structural changes in the haemagglutinin which accompany egg adaptation of an influenza A(H1N1) virus. Virology. 1987;160:31–37. doi: 10.1016/0042-6822(87)90040-7. [DOI] [PubMed] [Google Scholar]
- Rosano GL, Ceccarelli A. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol. 2014;5:172. doi: 10.3389/fmicb.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueckert C, Guzmán CA. Vaccines: from empirical development to rational design. PLoS Pathog. 2012;8(11):e1003001. doi: 10.1371/journal.ppat.1003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbah A, Chang TH, Harnack R, Frohlich V, Tominaga K, Dube PH, Xiang Y, Bose S. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10(10):1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin A, Ramos-Alvarez M, Alvarez-Amezquita J, Pelon W, Michaels RH, Spigland I, Koch MA, Barnes JM, Rhim JS. Live, orally given poliovirus vaccine. Effects of rapid mass immunization on population under conditions of massive enteric infection with other viruses. JAMA. 1960;173:1521–1526. doi: 10.1001/jama.1960.03020320001001. [DOI] [PubMed] [Google Scholar]
- Salk JE. Poliomyelitis vaccine preparation and administration; analysis of basic premises and current practices. JAMA. 1959;169:1829–1838. doi: 10.1001/jama.1959.03000330001001. [DOI] [PubMed] [Google Scholar]
- Sallusto F, Lanzavecchia A, Araki K, Ahmed R. From vaccines to memory and back. Immunity. 2010;33:451–463. doi: 10.1016/j.immuni.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šantak M. Old and new ways to combat human influenza virus. Period Biol. 2012;114:221–234. [Google Scholar]
- Santak M, Kosutić-Gulija T, Tesović G, Ljubin-Sternak S, Gjenero-Margan I, Betica-Radić L, Forcić D. Mumps virus strains isolated in Croatia in 1998 and 2005: genotyping and putative antigenic relatedness to vaccine strains. J Med Virol. 2006;78(5):638–643. doi: 10.1002/jmv.20587. [DOI] [PubMed] [Google Scholar]
- Šantak M, Lang-Balija M, Ivancic-Jelecki J, Košutić-Gulija T, Ljubin-Sternak S, Forcic D. Antigenic differences between vaccine and circulating wild-type mumps viruses decreases neutralization capacity of vaccine-induced antibodies. Epidemiol Infect. 2013;141(6):1298–1309. doi: 10.1017/S0950268812001896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šantak M, Markušić M, Balija ML, Kopač SK, Jug R, Örvell C, Tomac J, Forčić D. Accumulation of defective interfering viral particles in only a few passages in Vero cells attenuates mumps virus neurovirulence. Microbes Infect. 2015;17:228–236. doi: 10.1016/j.micinf.2014.11.006. [DOI] [PubMed] [Google Scholar]
- Šantak M, Örvell C, Gulija TK. Identification of conformational neutralization sites on the fusion protein of mumps virus. J Gen Virol. 2015;96:982–990. doi: 10.1099/vir.0.000059. [DOI] [PubMed] [Google Scholar]
- Schickli JH, Kaur J, Tang RS. Nonclinical phenotypic and genotypic analyses of a phase 1 pediatric respiratory syncytial virus vaccine candidate MEDI-559 (rA2cp248/404/1030DeltaSH) at permissive and non-permissive temperatures. Virus Res. 2012;169:38–47. doi: 10.1016/j.virusres.2012.06.027. [DOI] [PubMed] [Google Scholar]
- Schnee M, Vogel AB, Voss D, Petsch B, Baumhof P, Kramps T, Stitz L. An mRNA vaccine encoding rabies virus glycoprotein induces protection against lethal infection in mice and correlates of protection in adult and newborn pigs. PLoS Negl Trop Dis. 2016;10:e0004746. doi: 10.1371/journal.pntd.0004746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell MJ, Buonocore L, Kretzschmar E, Johnson E, Rose JK. Foreign glycoproteins expressed from recombinant vesicular stomatitis viruses are incorporated efficiently into virus particles. Proc Natl Acad Sci USA. 1996;93:11359–11365. doi: 10.1073/pnas.93.21.11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöne D, Hrycak CP, Windmann S, Lapuente D, Dittmer U, Tenbusch M. Immunodominance of adenovirus-derived CD8(+) T cell epitopes interferes with the induction of transgene-specific immunity in adenovirus-based immunization. J Virol. 2017;91:17. doi: 10.1128/JVI.01184-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwendener RA. Liposomes as vaccine delivery systems: a review of the recent advances. Ther Adv Vaccines. 2014;2:159–182. doi: 10.1177/2051013614541440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevvana M, Keck Z, Foung SK, Kuhn RJ. Structural perspectives on HCV humoral immune evasion mechanisms. Curr Opin Virol. 2021;49:92–101. doi: 10.1016/j.coviro.2021.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelly A, Gupta P, Ahuja R, Srichandan S, Meena J, Majumdar T. Impact of microbiota: a paradigm for evolving herd immunity against viral diseases. Viruses. 2020;12:1150. doi: 10.3390/v12101150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist CA (2008) Vaccine Immunology. Elsevier. https://www.who.int/immunization/documents/Elsevier_Vaccine_immunology.pdf. Accessed 15 Aug 2021
- Singh B, Kaur P, Cedeno L, Brahimi T, Patel P, Virk H, Shamoon F, Bikkina M. COVID-19 mRNA vaccine and myocarditis. Eur J Case Rep Intern Med. 2021;8:002681. doi: 10.12890/2021_002681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skwarczynski M, Toth I. Peptide-based synthetic vaccines. Chem Sci. 2016;7:842–854. doi: 10.1039/C5SC03892H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin Microbiol Rev. 2003;16(3):463–496. doi: 10.1128/CMR.16.3.463-496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GL, Benfield CTO, Maluquer de Motes C, Mazzon M, Ember SWJ, Ferguson BJ, Sumner RP. Vaccinia virus immune evasion: mechanisms, virulence and immunogenicity. J Gen Virol. 2013;94:2367–2392. doi: 10.1099/vir.0.055921-0. [DOI] [PubMed] [Google Scholar]
- Smith JD, Morton LD, Ulery BD. Nanoparticles as synthetic vaccines. Curr Opin Biotechnol. 2015;34:217–224. doi: 10.1016/j.copbio.2015.03.014. [DOI] [PubMed] [Google Scholar]
- Smits G, Mollema L, Hahné S, de Melker H, Tcherniaeva I, Waaijenborg S, van Binnendijk R, van der Klis F, Berbers G. Seroprevalence of mumps in the Netherlands: dynamics over a decade with high vaccination coverage and recent outbreaks. PLoS One. 2013;8(3):e58234. doi: 10.1371/journal.pone.0058234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolev BN, Olenina LV, Kolesanova EF, Poroikov VV, Archakov AI. Computer design of vaccines: approaches, software tools and informational resources. Curr Comput Aided Drug Des. 2005;1:207–222. doi: 10.2174/1573409053585681. [DOI] [Google Scholar]
- Stachyra A, Redkiewicz P, Kosson P, Protasiuk A, Góra-Sochacka A, Kudla G, Sirko A. Codon optimization of antigen coding sequences improves the immune potential of DNA vaccines against avian influenza virus H5N1 in mice and chickens. Virol J. 2016;13:143. doi: 10.1186/s12985-016-0599-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepinski J, Waddell C, Stolarski R, Darzynkiewicz E, Rhoads RE. Synthesis and properties of mRNAs containing the novel “anti-reverse” cap analogs 7-methyl(3′-O-methyl)GpppG and 7- methyl (3′-deoxy) GpppG. RNA. 2001;7:1486–1495. [PMC free article] [PubMed] [Google Scholar]
- Storni T, Ruedl C, Schwarz K, Schwendener RA, Renner WA, Bachmann MF. Nonmethylated CG motifs packaged into virus-like particles induce protective cytotoxic T cell responses in the absence of systemic side effects. J Immunol. 2004;172:1777–1785. doi: 10.4049/jimmunol.172.3.1777. [DOI] [PubMed] [Google Scholar]
- Strassburg MA. The global eradication of smallpox. Am J Infect Control. 1982;10(2):53–59. doi: 10.1016/0196-6553(82)90003-7. [DOI] [PubMed] [Google Scholar]
- Suschak JJ, Williams JA, Schmaljohn CS. Advancements in DNA vaccine vectors, non-mechanical delivery methods, and molecular adjuvants to increase immunogenicity. Hum Vaccin Immunother. 2017;13:2837–2848. doi: 10.1080/21645515.2017.1330236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland C. A challenge for the development of malaria vaccines: polymorphic target antigens. PLoS Med. 2007;4:e116. doi: 10.1371/journal.pmed.0040116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter G, Staib C. Vaccinia vectors as candidate vaccines: the development of modified vaccinia virus Ankara for antigen delivery. Curr Drug Targets Infect Disord. 2003;3:263–271. doi: 10.2174/1568005033481123. [DOI] [PubMed] [Google Scholar]
- Suzich JA, Ghim SJ, Palmer-Hill FJ, White WI, Tamura JK, Bell JA, Newsome JA, Jenson AB, Schlegel R. Systemic immunization with papillomavirus L1 protein completely prevents the development of viral mucosal papillomas. Proc Natl Acad Sci U S A. 1995;92(25):11553–11557. doi: 10.1073/pnas.92.25.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Sun X. Recent advances in mRNA vaccine delivery. Nano Res. 2018;11:5338–5354. doi: 10.1007/s12274-018-2091-z. [DOI] [Google Scholar]
- Tani H, Morikawa S, Matsuura Y. Development and applications of VSV vectors based on cell tropism. Front Microbiol. 2012;2:272. doi: 10.3389/fmicb.2011.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A, Sidney J, Methot N, Yu ED, Zhang Y, Dan JM, Goodwin B, Rubiro P, Sutherland A, Wang E, Frazier A, Ramirez SI, Rawlings SA, Smith DM, da Silva Antunes R, Peters B, Scheuermann RH, Weiskopf D, Crotty S, Grifoni A, Sette A. Impact of SARS-CoV-2 variants on the total CD4+ and CD8+ T cell reactivity in infected or vaccinated individuals. Cell Rep Med. 2021;2:100355. doi: 10.1016/j.xcrm.2021.100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia J, Perkus ME, Taylor J, Norton EK, Audonnet J-C, Cox WI, Paoletti E. NYVAC: A highly attenuated strain of vaccinia virus. Virology. 1992;188:217–232. doi: 10.1016/0042-6822(92)90752-B. [DOI] [PubMed] [Google Scholar]
- Tatsis N, Ertl HC. Adenoviruses as Vaccine Vectors. Mol Ther. 2004;10:616–629. doi: 10.1016/j.ymthe.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijaro JR, Farber DL. OVID-19 vaccines: modes of immune activation and future challenges. Nat Rev Immunol. 2021;21:195–197. doi: 10.1038/s41577-021-00526-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenforde MW, Patel MM, Ginde AA, Douin DJ, Talbot HK, Casey JD, Mohr NM, Zepeski A, Gaglani M, McNeal T, Ghamande S, Shapiro NI, Gibbs KW, Files DC, Hager DN, Shehu A, Prekker ME, Erickson HL, Exline MC, Gong MN, Mohamed A, Henning DJ, Steingrub JS, Peltan ID, Brown SM, Martin ET, Monto AS, Khan A, Hough CT, Busse L, Ten Lohuis CC, Duggal A, Wilson JG, Gordon AJ, Qadir N, Chang SY, Mallow C, Gershengorn HB, Babcock HM, Kwon JH, Halasa N, Chappell JD, Lauring AS, Grijalva CG, Rice TW, Jones ID, Stubblefield WB, Baughman A, Womack KN, Lindsell CJ, Hart KW, Zhu Y, Olson SM, Stephenson M, Schrag SJ, Kobayashi M, Verani JR, Self WH, Influenza and Other Viruses in the Acutely Ill (IVY) Network (2021) Effectiveness of SARS-CoV-2 mRNA vaccines for preventing Covid-19 hospitalizations in the United States. medRxiv 2021.07.08.21259776
- Tesović G, Poljak M, Lunar MM, Kocjan BJ, Seme K, Vukić BT, Sternak SL, Cajić V, Vince A. Horizontal transmission of the Leningrad-Zagreb mumps vaccine strain: a report of three cases. Vaccine. 2008;26(16):1922–1925. doi: 10.1016/j.vaccine.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Tews BA, Meyers G. Self-Replicating RNA. Methods Mol Biol. 2017;1499:15–35. doi: 10.1007/978-1-4939-6481-9_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thess A, Grund S, Mui BL, Hope MJ, Baumhof P, Fotin-Mleczek M, Schlake T. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol Ther. 2015;23:1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchilin VP. Recent advances with liposomes as pharmaceutical carriers. Nat Rev Drug Discov. 2005;4:145–160. doi: 10.1038/nrd1632. [DOI] [PubMed] [Google Scholar]
- Trepotec Z, Aneja M, Geiger J, Hasenpusch G, Plank C, Rudolph C. Maximizing the translational yield of mRNA therapeutics by minimizing 5'-UTRs. Tissue Eng Part A. 2018;25:69–79. doi: 10.1089/ten.tea.2017.0485. [DOI] [PubMed] [Google Scholar]
- Tsang SH, Sampson JN, Schussler J, Porras C, Wagner S, Boland J, Cortes B, Lowy DR, Schiller JT, Schiffman M, Kemp TJ, Rodriguez AC, Quint W, Gail MH, Pinto LA, Gonzalez P, Hildesheim A, Kreimer AR, Herrero R, Costa Rica HPV Vaccine Trial (CVT) Group Durability of cross-protection by different schedules of the bivalent HPV vaccine: the CVT trial. J Natl Cancer Inst. 2020;112:1030–1037. doi: 10.1093/jnci/djaa010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschorn N, Berg K, Stitz J. Transposon vector-mediated stable gene transfer for the accelerated establishment of recombinant mammalian cell pools allowing for high-yield production of biologics. Biotechnol Lett. 2020;42:1103–1112. doi: 10.1007/s10529-020-02889-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmer JB, Geall AJ. Recent innovations in mRNA vaccines. Curr Opin Immunol. 2016;41:18–22. doi: 10.1016/j.coi.2016.05.008. [DOI] [PubMed] [Google Scholar]
- Valenzuela P, Medina A, Rutter WJ, Ammerer G, Hall BD. Synthesis and assembly of hepatitis B virus surface antigen particles in yeast. Nature. 1982;298(5872):347–350. doi: 10.1038/298347a0. [DOI] [PubMed] [Google Scholar]
- Van Oers MM, Pijlman GP, Vlak JM. Thirty years of baculovirus-insect cell protein expression: from dark horse to mainstream technology. J Gen Virol. 2015;96:6–23. doi: 10.1099/vir.0.067108-0. [DOI] [PubMed] [Google Scholar]
- van Wyke Coelingh KL, Murphy BR, Collins PL, Lebacq-Verheyden AM, Battey JF. Expression of biologically active and antigenically authentic parainfluenza type 3 virus hemagglutinin-neuraminidase glycoprotein by a recombinant baculovirus. Virology. 1987;160:465–472. doi: 10.1016/0042-6822(87)90018-3. [DOI] [PubMed] [Google Scholar]
- Vasou A, Sultanoglu N, Goodbourn S, Randall RE, Kostrikis LG. Targeting pattern recognition receptors (PRR) for vaccine adjuvantation: from synthetic PRR agonists to the potential of defective interfering particles of viruses. Viruses. 2017;9(7):186. doi: 10.3390/v9070186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeire T, Barbezange C, Francart A, Hamouda A, Litzroth A, Hutse V, Martens L, Vandermarliere E, Van Gucht S. Sera from different age cohorts in Belgium show limited cross-neutralization between the mumps vaccine and outbreak strains. Clin Microbiol Infect. 2019;25(7):907. doi: 10.1016/j.cmi.2018.11.016. [DOI] [PubMed] [Google Scholar]
- Vidula MK, Ambrose M, Glassberg H, Chokshi N, Chen T, Ferrari VA, Han Y. Myocarditis and other cardiovascular complications of the mRNA-based COVID-19 vaccines. Cureus. 2021;13:e15576. doi: 10.7759/cureus.15576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel AB, Kanevsky I, Che Y, Swanson KA, Muik A, Vormehr M, Kranz LM, Walzer KC, Hein S, Güler A, Loschko J, Maddur MS, Ota-Setlik A, Tompkins K, Cole J, Lui BG, Ziegenhals T, Plaschke A, Eisel D, Dany SC, Fesser S, Erbar S, Bates F, Schneider D, Jesionek B, Sänger B, Wallisch AK, Feuchter Y, Junginger H, Krumm SA, Heinen AP, Adams-Quack P, Schlereth J, Schille S, Kröner C, de la Caridad Güimil Garcia R, Hiller T, Fischer L, Sellers RS, Choudhary S, Olga-Gonzalez O, Fulvia Vascotto F, Gutman MR, Fontenot JA, Hall-Ursone S, Brasky K, Griffor MC, Han S, Su AAH, Lees JA, Nedoma NL, Mashalidis EH, Sahasrabudhe PV, Tan CY, Pavliakova D, Singh G, Fontes-Garfias C, Pride M, Scully IL, Ciolino T, Obregon J, Gazi M, Carrion R, Jr, Alfson KJ, Kalina WV, Kaushal D, Shi PY, Klamp T, Rosenbaum C, Kuhn AN, Türeci Ö, Dormitzer PR, Jansen KU, Sahin U. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021;592:283–289. doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- Volz A, Sutter G. Modified Vaccinia Virus Ankara. Adv Virus Res. 2017;97:187–243. doi: 10.1016/bs.aivir.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Jiang S, Wang Y. Recent advances in the production of recombinant subunit vaccines in Pichia pastoris. Bioengineered. 2016;7:155–165. doi: 10.1080/21655979.2016.1191707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Wang T, Zhang M, Chen R, Niu R, Deng Y. Mannose derivative and lipid A dually decorated cationic liposomes as an effective cold chain free oral mucosal vaccine adjuvant-delivery system. Eur J Pharm Biopharm. 2014;88:194–206. doi: 10.1016/j.ejpb.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Wang S, Zhang C, Zhang L, Li J, Huang Z, Lu S. The relative immunogenicity of DNA vaccines delivered by the intramuscular needle injection, electroporation and gene gun methods. Vaccine. 2008;26:2100–2110. doi: 10.1016/j.vaccine.2008.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins K, Griffin G, Septaric K, Simon EL. Myocarditis after BNT162b2 vaccination in a healthy male. Am J Emerg Med. 2021;S0735–6757(21):00536–542. doi: 10.1016/j.ajem.2021.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson D, Endsley A, Huang L. Design considerations for liposomal vaccines: influence of formulation parameters on antibody and cell-mediated immune responses to liposome associated antigens. Vaccine. 2012;30:2256–2272. doi: 10.1016/j.vaccine.2012.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss WR, Ishii KJ, Hedstrom RC, Sedegah M, Ichino M, Barnhart K, Klinman DM, Hoffman SL. A plasmid encoding murine granulocyte-macrophage colony-stimulating factor increases protection conferred by a malaria DNA vaccine. J Immunol. 1998;161:2325–2332. doi: 10.4049/jimmunol.161.5.2325. [DOI] [PubMed] [Google Scholar]
- WHO Smallpox eradication. Wkly Epidemiol Rec. 1980;55:33–40. [Google Scholar]
- WHO (2020) Global tuberculosis report 2020
- Willemsen A, Zwart MP. On the stability of sequences inserted into viral genomes. Virus Evol. 2019;5(2):vez045. doi: 10.1093/ve/vez045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolbert JG, Higginbotham K (2020) Poliomyelitis. In: StatPearls [Internet]. StatPearls Publishing, Treasure Island
- Wolf J, Jannat R, Dubey S, Troth S, Onorato MT, Coller BA, Hanson ME, Simon JK. Development of pandemic vaccines: ERVEBO case study. Vaccines (Basel) 2021;9:190. doi: 10.3390/vaccines9030190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, Felgner PL. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- Won H, Kim AR, Yoo JS, Chung GT, Kang HJ, Kim SJ, Kim SS, Lee JW. Cross-neutralization between vaccine and circulating wild-type mumps viruses in Korea. Vaccine. 2021;39(13):1870–1876. doi: 10.1016/j.vaccine.2021.01.039. [DOI] [PubMed] [Google Scholar]
- Wrapp D, Wang N, Crbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtele H, Little KC, Chartrand P. Illegitimate DNA integration in mammalian cells. Gene Ther. 2003;10(21):1791–1799. doi: 10.1038/sj.gt.3302074. [DOI] [PubMed] [Google Scholar]
- Xu S, Yang K, Li R, Zhang L. mRNA vaccine era – mechanisms, drug platform and clinical prospection. Int J Mol Sci. 2020;21:6582. doi: 10.3390/ijms21186582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Liang W, Qiu Y, Cespi M, Palmieri GF, Mason AJ, Lam JK. Incorporation of a nuclear localization signal in pH responsive LAH4-L1 peptide enhances transfection and nuclear uptake of plasmid DNA. Mol Pharm. 2016;13:3141–3152. doi: 10.1021/acs.molpharmaceut.6b00338. [DOI] [PubMed] [Google Scholar]
- Yadav R, Zhai L, Tumban E. Virus-like Particle-Based L2 Vaccines against HPVs: Where Are We Today? Viruses. 2019;12:18. doi: 10.3390/v12010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HA, Aleyas AG, George JA, Park SO, Han YW, Lee JH, Cho JG, Eo SK. Cytokine GM-CSF genetic adjuvant facilitates prophylactic DNA vaccine against pseudorabies virus through enhanced immune responses. Microbiol Immunol. 2006;50:83–92. doi: 10.1111/j.1348-0421.2006.tb03773.x. [DOI] [PubMed] [Google Scholar]
- York IA, Rock KL. Antigen processing and presentation by the class I major histocompatibility complex. Annu Rev Immunol. 1996;14:369–396. doi: 10.1146/annurev.immunol.14.1.369. [DOI] [PubMed] [Google Scholar]
- Zafrir Y, Agmon-Levin N, Paz Z, Shilton T, Shoenfeld Y. Autoimmunity following hepatitis B vaccine as part of the spectrum of ’Autoimmune (Autoinflammatory) Syndrome induced by Adjuvants’ (ASIA): analysis of 93 cases. Lupus. 2012;21:146–152. doi: 10.1177/0961203311429318. [DOI] [PubMed] [Google Scholar]
- Zemp F, Rajwani J, Mahoney DJ. Rhabdoviruses as vaccine platforms for infectious disease and cancer. Biotechnol Genet Eng Rev. 2018;34:122–138. doi: 10.1080/02648725.2018.1474320. [DOI] [PubMed] [Google Scholar]
- Zhan W, Muhuri M, Tai PWL, Gao G. Vectored immunotherapeutics for infectious diseases: can rAAVs Be the game changers for fighting transmissible pathogens? Front Immunol. 2021;12:673699. doi: 10.3389/fimmu.2021.673699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Maruggi G, Shan H, Li J. Advances in mRNA vaccines for infectious diseases. Front Immunol. 2019;10:594. doi: 10.3389/fimmu.2019.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Seth A, Wibowo N, Zhao CX, Mitter N, Yu C, Middelberg AP. Nanoparticle vaccines. Vaccine. 2014;32(3):327–337. doi: 10.1016/j.vaccine.2013.11.069. [DOI] [PubMed] [Google Scholar]
- Zhou J, Sun XY, Stenzel DJ, Frazer IH. Expression of vaccinia recombinant HPV 16 L1 and L2 ORF proteins in epithelial cells is sufficient for assembly of HPV virion-like particles. Virology. 1991;185(1):251–257. doi: 10.1016/0042-6822(91)90772-4. [DOI] [PubMed] [Google Scholar]
- Zhou X, Berglund P, Rhodes G, Parker SE, Jondal M, Liljestrom P. Self-replicating semliki forest virus RNA as recombinant vaccine. Vaccine. 1994;12:1510–1514. doi: 10.1016/0264-410X(94)90074-4. [DOI] [PubMed] [Google Scholar]
- Zhou G, Roizman B. Characterization of a recombinant herpes simplex virus 1 designed to enter cells via the IL13Rα2 receptor of malignant glioma cells. J Virol. 2005;79:5272–5277. doi: 10.1128/JVI.79.9.5272-5277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Curtis N. Factors that influence the immune response to vaccination. Clin Microbiol Rev. 2019;32:e00084–e118. doi: 10.1128/CMR.00084-18. [DOI] [PMC free article] [PubMed] [Google Scholar]