Abstract
Despite significant improvements in the way breast cancer is managed and treated, it continues to persist as a leading cause of death worldwide. If detected and diagnosed early, when tumours are small and localised, there is a considerably higher chance of survival. However, current methods for detection and diagnosis lack the required sensitivity and specificity for identifying breast cancer at the asymptomatic or very early stages. Thus, there is a need to develop more rapid and reliable methods, capable of detecting disease earlier, for improved disease management and patient outcome. Raman spectroscopy is a non-destructive analytical technique that can rapidly provide highly specific information on the biochemical composition and molecular structure of samples. In cancer, it has the capacity to probe very early biochemical changes that accompany malignant transformation, even prior to the onset of morphological changes, to produce a fingerprint of disease. This review explores the application of Raman spectroscopy in breast cancer, including discussion on its capabilities in analysing both ex-vivo tissue and liquid biopsy samples, and its potential in vivo applications. The review also addresses current challenges and potential future uses of this technology in cancer research and translational clinical application.
Subject terms: Cancer, Breast cancer, Microscopy
Introduction
Female breast cancer (BC) is the most commonly diagnosed malignancy and one of the leading causes of cancer-related deaths, worldwide; an estimated 2.26 million new cases and 680,000 deaths were recorded in 2020 alone [1]. Although significant advances have been made over the past few decades in the prevention, diagnosis and treatment of BC [2], it still remains a major global health burden. Distant (metastatic) recurrence is a significant clinical issue and is responsible for the majority of BC deaths [3]. Five-year survival of patients with localised BC is 99%; decreasing to 27% when diagnosed with late-stage disease [4]. Early detection and diagnosis, when the tumour is still localised [5], is imperative for improved patient outcome.
Various clinical methods exist for BC detection and diagnosis. Mammography is the gold-standard method for early BC detection and forms the basis of Mammographic Screening Programmes, introduced globally to detect small malignant tumours before symptoms develop. Although existing evidence suggests that mammography can reduce risk of mortality [6, 7], there has been considerable debate regarding the overall efficacy of mammography [8] both as a screening and diagnostic tool. Mammography is far less sensitive in women with dense breast tissue and there is also debate surrounding the appropriate age for screening [7, 9]. Moreover, overdiagnosis and subsequent over-treatment is now recognised as a major issue surrounding screening mammography [10]. However, imaging cannot be used solely for the diagnosis of BC; currently the only way to make a definitive diagnosis is through histopathological analysis of the patients’ tissue. Obtaining tissue involves invasive procedures such as surgical excision or needle core biopsy. Approximately 80% of biopsies are negative for BC thus rendering these invasive procedures unnecessary in most cases. Moreover, the morphological and molecular heterogeneity of breast tissue also presents challenges and may lead to interpretive disparity amongst pathologists. These limitations provide the motivation to develop new techniques capable of rapidly and accurately detecting and diagnosing BC in its early stages. Spectroscopic methods are emerging as powerful tools within biomedical research [11, 12] as they are non-invasive and have a high real-time spatial resolution. Of the many spectroscopic techniques available, Raman spectroscopy (RS) is particularly appealing as it has demonstrated the potential to rapidly, non-destructively and objectively, provide clinically relevant diagnostic information in a variety of specialities [13–15]. This review will explore the application of RS in BC diagnosis.
Raman spectroscopy for biological applications
RS is an optical spectroscopic technique that can probe the vibrational modes associated with chemical bonds in a sample; as different samples have distinct chemical compositions, a sample-specific spectral fingerprint, or ‘Raman spectrum’ can be obtained. This spectrum contains numerous bands at various frequencies, which are characteristic of the structural features and functional groups of a particular molecule. A more detailed description of the fundamental principles of RS, is in Supplementary File 1 and Supplementary Fig. S1.
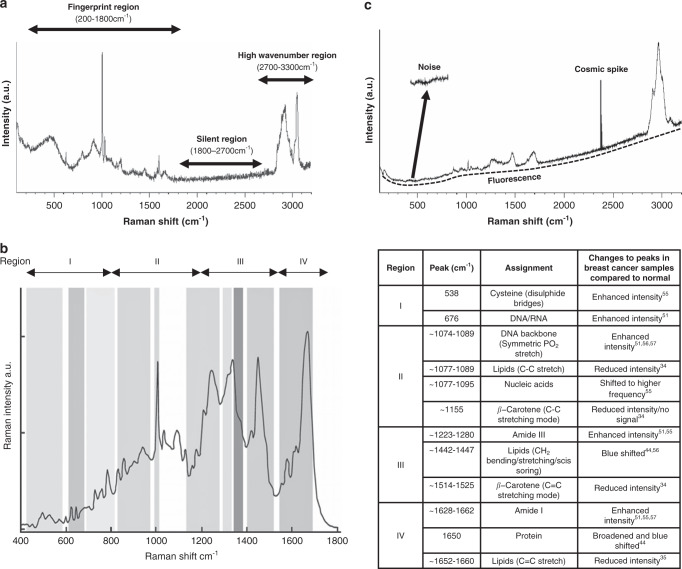
Different cells and tissues are made up of distinct combinations of proteins, carbohydrates, lipids and nucleic acids; each of which have a number of associated vibrational modes that can be probed by RS to reveal information on their structure and composition. Accordingly, RS can provide details on the current state and activity of cells and tissues. Fig. 1a displays an exemplar of a Raman spectrum recorded from MCF-7 BC cells, highlighting the three main regions of the spectrum: fingerprint (200–1800 cm−1), silent (1800–2700 cm−1) and high wavenumber (2700–3300 cm−1). The high wavenumber region is dominated by C–H, CH2, O–H, N–H vibrations of lipids, proteins and water whereas the ‘silent region’ is mostly free of bands from biological material; cellular Raman vibrations arising from triple bond functional groups like the alkynes are an exception [16]. Most peaks for biological samples can be found within the ‘fingerprint’ region where the richest molecular vibrational modes of proteins, lipids, carbohydrates and nucleic acids are found. A typical biological spectrum of this region is shown (Fig. 1b), with examples of peak assignments from studies centred around the detection of BC based on specific Raman signatures.
Fig. 1. Examples of Raman spectra and peak assignments.
a Extended scan from the Luminal A breast cancer cell line, MCF7 depicting the three main regions of the Raman spectrum: fingerprint, silent and high wavenumber regions are illustrated. Whilst the majority of biologically relevant molecular vibrations exist within the fingerprint, both the silent and high wavenumber regions may also contain molecular vibrations from a limited number of biomolecules. b Representation of the fingerprint region of a cellular Raman spectrum (left) with a variety of peaks that correspond to molecular vibrations of amino acids, proteins, lipids, nucleic acids and carbohydrates. Highlighted are examples of peak assignments from studies centred around the detection of breast cancer based on changes to specific Raman signatures (right). Image kindly provided by Renishaw. c Raman spectrum obtained from normal breast tissue displaying sources of spectral interference including noise, fluorescence and cosmic spikes (arrows).
Biological samples are highly complex and heterogeneous; they contain an extensive number of Raman-active vibrational modes, particularly within this fingerprint. Thus, extracting relevant information from such data-rich spectra is challenging. The many discrepancies, within the literature, associated with characterising spectral peak position reflect this [17]. Raman spectra of biological samples contain wide peaks that represent a combination of different molecules and neighbouring molecular interactions may cause a shift in peak position from the isolated functional group. Furthermore, most molecules within biological samples that contain Raman active vibrational modes, also exhibit fluorescence upon excitation; cellular autofluorescence is several orders of magnitude more intense than the Raman signal and thus can hinder spectral interpretation. Additionally, noise sources, either generated internally within the Raman system itself or exteriorly from other sources, include cosmic rays; dark current; shot noise and readout noise which further contaminate the spectra and complicate subsequent data processing [18]. Therefore, computational data pre-processing, to remove undesirable background noise (Fig. 1c), should be carried out prior to analysis. Computational spectral denoising techniques are beyond the scope of this review and reviewed elsewhere [19]. Moreover, if spectra, of the same sample, are not obtained simultaneously, under identical experimental parameters, then they may be subject to a shift in peak intensity. Normalisation is applied to address this disparity and a number of different approaches are available [19]. Prior to spectral analyses, it is important to identify any outliers within the dataset that could adversely affect interpretation and classification. Principal component analysis (PCA) and Signal to noise ratio based thresholding methods are two examples of outlier-detection algorithms [20].
Following pre-processing, corrected Raman spectra are subject to analysis to recognise vibrational signatures. Univariate methods can be used to analyse biological samples, but these are somewhat restricted to small datasets, for example, in studies on rare diseases or where samples are obtained from surgical resection with only a few suitable samples for Raman analysis. Moreover, univariate methods only make use of a limited number of variables like relative band positional or intensity changes from the whole spectrum hence resulting in a massive loss of information. Typically, Raman datasets in biological studies are large and multifaceted, and require more efficient, multivariate approaches, such as chemometrics, for a truly exploratory and comprehensive analysis. These methods involve an in-depth analysis of many spectra, simultaneously, to allow useful trends and patterns to emerge from the data, which may otherwise have been missed if analysed, only, at the single level. Moreover, such patterns can be modelled and subsequently used as a predictor for similar newly acquired data.
Multivariate methods can be classified into either unsupervised or supervised methods, depending on the objective of the analysis and any priori-knowledge of the sample [20]. Unsupervised methods, including: PCA [21], k-means clustering [22] and Hierarchical clustering analysis [23] do not require any previous knowledge of the sample and are thus exploratory in nature. They can assist in identifying patterns and trends within the dataset and the objective creation of groupings. Conversely, supervised methods, including: partial least squares (PLS) [24] linear discriminant analysis (LDA) [25] and multiple linear regression [26] rely on pre-existing class labels within the dataset, for example, the histopathological diagnosis, and thus are concerned with pattern recognition.
Raman variants in breast cancer studies
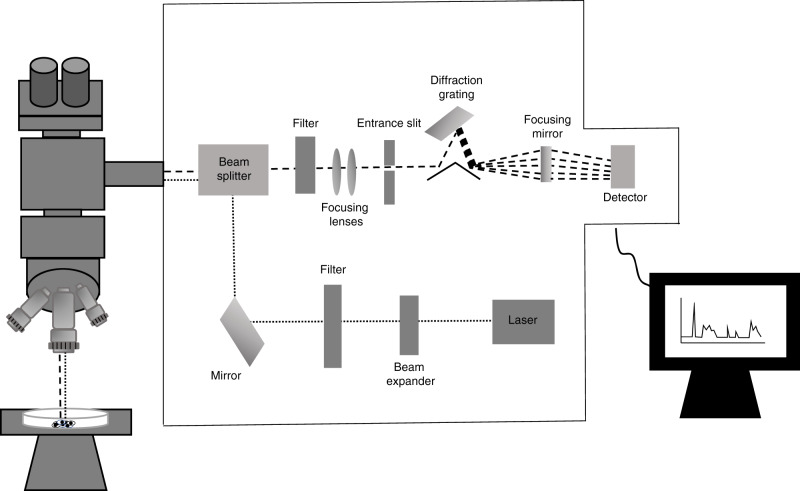
RS, in its simplest form, implies the phenomenon of Spontaneous Raman scattering (Fig. 2). Spontaneous Raman scattering has seen increasing popularity in biomedical research, but the Raman effect is inherently weak and is essentially competing with stronger signals from Rayleigh scattering and tissue autofluorescence. Experimentally, the use of higher-powered lasers and longer signal acquisition times can improve the Raman signal and photobleaching can quench the autofluorescence. However, the potential damage that can be induced following application of these experimental approaches poses significant challenges both to biological and clinical applications. Consequently, numerous Raman-based techniques have been developed to enhance the weak Raman signal and overcome issues with fluorescence interference, whilst minimising photodegradation (Table 1). Furthermore, the development of techniques such as Spatially offset RS and Transmission RS make it possible to probe biomolecules in deep tissue layers for potential non-invasive in vivo measurements. The basic principles and examples of where these variants have been applied in BC detection and diagnosis are described in Table 1.
Fig. 2. Schematic representation of the main components found within a typical spontaneous Raman scattering micro-spectroscopy system.
Laser light is guided through a beam expander onto a series of mirrors that focus the light onto the sample through a microscope objective lens. Scattered light is collected with this same objective lens in a 180° backscatter sampling geometry. Rayleigh scattered light is reduced through the use of edge filters and then the Raman scattered light is focused through an entrance slit and dispersed by a diffraction grating onto the detector. Variations in this general setup typically exist between manufacturers.
Table 1.
Examples of Raman variants with applications in breast cancer.
| Raman variants | Basic principles | Advantages | Examples of applications in breast cancer |
|---|---|---|---|
| Surface enhanced Raman spectroscopy (SERS) | This describes the large Raman signal enhancement provided when molecules are adsorbed onto a roughened metallic surface- typically silver or gold nanoparticles. | This technique provides an increase, in orders of magnitude, of the Raman signal intensity; molecules present only at a low concentration can still be detected. It is also capable of multiplex detection due to its molecularly narrow-band spectra. | Detection and diagnosis through analysing liquid biopsy [29, 30, 102, 117]; tissue samples [132]; exosomal miRNAs [133]; and in vivo imaging [134, 135]. |
| Spatially offset Raman spectroscopy (SORS) [136] | Raman scattered light is collected along the sample from areas that are laterally offset from the point of laser excitation. Differences in the chemical composition as a function of depth can be delineated by comparing spectra obtained with no offset (surface) and those collected with an offset (subsurface). As a result of photon diffusion mechanisms, the spectra obtained with offset have different contributions from various depths within the sample. | Raman signals can be retrieved from numerous individual layers within a sample and surface fluorescence signals are reduced, making in vivo measurements possible. | Detection and characterisation of breast microcalcifications buried in tissue at depths up to 10 mm [64]. |
| Transmission Raman spectroscopy (TRS) | The illumination and collection optics are found on opposites sides of the sample hence the generated spectra are representative of its whole volume. | This technique is insensitive to depth; it is possible to probe structures deep within a particular area without interference from surface Raman and fluorescence signals. TRS thus has potential for non-invasive in vivo applications. | Detection and characterisation of breast microcalcification buried in tissue at depths of 16–40 mm [93, 95, 96]. |
| Kerr-gated Raman spectroscopy | Exploits the differential time dependence of Raman signals (femtoseconds or picoseconds) and fluorescence (hundreds of picoseconds to nanoseconds) on short-repeated laser pulses. Kerr-gating can separate these distinct time domains by using excitation with a pico-second pulsed laser combined with ultra-fast gating of the Raman scattered light collected at various time delays. | Surface Raman and fluorescence signals tend to be far more prominent than Raman signals emanating deep within tissue, posing difficulty for depth profiling. The Kerr-gating technique functions to reject the temporally longer fluorescence signal and thus produce enhancement to improve the spectra collected from biological tissue for depth profiling. | Probing the biochemical composition of microcalcifications for early breast cancer detection [63]. |
| Near-infrared Fourier transform Raman spectroscopy (NIR FT-RS) | A non-dispersive Raman system that uses a Nd:YAG laser emitting at 1064 nm excitation. FT-Raman spectroscopy involves the coupling of the Raman sampling module with a FT-IR instrument, that contains an interferometer, in order to reduce the fluorescence typical of near-infrared excitation. | The use of pre-resonant excitation, where most biomolecules do not have electronic absorption bands, reduces the likelihood of fluorescence interference as well as minimising photodegradation. FT-RS also simultaneously measures the intensity of scattered light at many frequencies thus improving spectral resolution. | Diagnosis of breast cancer through in vivo and ex vivo measurements. [55, 137–139]. |
| Shell-isolated nanoparticle-enhanced Raman spectroscopy (SHINERS) | A monolayer of gold nanoparticle cores, coated with either silica or alumina, are applied to the surface of the sample to be probed. The protective shell prevents direct contact between the plasmonic nanoparticles and the probed surface, which if not present, could lead to structural changes of the biomolecules. | The plasmonic cores induce significant increase of the intensity of the electric field thus leading to a large enhancement of the Raman signal. It can do so without the requirement of a specialised substrate (SERS) and it is also not limited to only studying molecules with a large Raman cross-section (Tip-Enhanced Raman Spectroscopy). | Detection of breast pathology [40, 56, 57]; and microcalcification status [57, 69]. |
| Shifted excitation Raman difference spectroscopy (SERDS) | A laser employing two slightly different emission wavelengths is used to record two spectra for fluorescence rejection. A slight change to the wavelength of the incident radiation causes a shift in the Raman spectrum, but the fluorescence signal will remain unchanged. The different spectrum, obtained following subtraction of the two raw spectra, is free of fluorescence interference. | This technique is useful for probing samples that exhibit strong autofluorescence. Furthermore, this technique does not require complex sample preparation or experimental setup. As this approach eliminates fluorescence by experimental means rather than computational, the Raman features are not altered. | Classification of normal, benign and cancerous ex vivo breast tissue samples [74]. |
| Stimulated Raman spectroscopy (SRS) | Two synchronous pulsed lasers (pump beam and Stokes beam) are focused onto a sample. When the frequency difference between the lasers matches a molecular vibrational frequency, stimulated excitation of the vibrational transition occurs. During this excitation, energy of the pump photon is transferred to the chemical bond and thus the emitted Stokes photon has a lower energy. A loss of an incident photon at the pump frequency is paralleled by the generation of a new photon at the Stokes frequency. | SRS is much more efficient than spontaneous Raman as its signal strength is several orders of magnitude greater and it is unaffected by sample fluorescence. | Discrimination of type II microcalcifications [68, 70]; breast cancer diagnosis and surgical guidance [140]. |
| Coherent anti-Stokes Raman spectroscopy (CARS) | CARS involves the simultaneous interaction of two incident photons, at pump and Stokes frequencies, with the scattering material. This interaction can coherently excite a large proportion of the chemical bonds to excited vibrational states. The excited chemical bonds can exchange energy with a second pump photon leading to the coherent emission of a higher energy anti-stokes photon. | CARS allows detection even in the presence of a strong fluorescent background due to the anti-stokes shift; deep penetration into tissues as there is minimal scattering of NIR excitation beams and limited photodamage due to low absorption of NIR excitation beams. Moreover, CARS methods require a very short time to acquire spectra, which is imperative for in vivo applications. | Characterisation of ex vivo breast tissue [141, 142]; and type I and II microcalcifications buried at a 2 mm depth [94]. |
Raman spectroscopy in breast cancer detection and diagnosis
Morphological and functional changes that occur during the malignant transformation of cells and tissue are accompanied by significant biochemical changes. For example, unchecked cellular proliferation, a hallmark of cancer [27], increases the production of DNA, RNA and proteins and disrupts lipid metabolism. These changes, at the biochemical level, occur much earlier than the onset of clinical symptoms. As such, Raman-based methods can be used to probe and quantify these altered molecular signatures, with spectra serving as biomarker profiles for early disease classification and tumour grading.
Moreover, this information can be obtained without destroying the material since RS does not require extensive sample preparation or labelling. Consequently, additional processing and analyses are possible following the Raman acquisition, which in the case of diagnosing cancer, is essential to build a complete picture of individual tumours. Its non-destructive nature, in addition to its ability to be adapted to optical fibre techniques, demonstrates its potential for in-vivo intra-operative or bed side applications. Moreover, samples in a solid state are not a requisite for RS hence there has been increasing interest in analysing circulating markers in different body fluids [28–32] for early cancer detection and diagnosis. The following includes discussion on ex-vivo tissue, in vivo and liquid biopsy Raman diagnostic applications in BC.
Ex-vivo applications
Early Raman studies on breast tissue focused on the qualitative analysis of spectra; identifying specific features associated with the appearance of benign or malignant breast changes. The first observations made on human breast tissue demonstrated the ability of Fourier-Transform (FT) RS, with Near-Infrared (NIR) excitation (1064 nm), to discriminate normal and abnormal tissue [33]. Since then, considerable effort has focused on discerning the biochemical changes that accompany these spectral changes associated with breast pathology.
The most prominent biochemical differences between normal and abnormal breast tissue concern lipids, carotenoids and proteins. Spectra obtained from normal, benign and malignant breast tissue using visible excitation revealed marked reduction in the intensity of peaks corresponding to vibrational modes of β-carotene and lipids in abnormal versus normal breast tissue [34]. This same group demonstrated this reduction in peak intensity of lipids again in diseased breast tissue using NIR excitation [35]. They also found similarities between the spectra of human collagens and that of infiltrating ductal carcinoma specimen. The relevance of the vibrational modes of lipids, carotenoids and proteins as potential spectral biomarkers for distinguishing pre-cancerous/cancerous and noncancerous breast tissue has been further corroborated [36–43].
A common observation is that spectral differences between benign and malignant breast tissue are less obvious. For RS to be applied as a diagnostic tool, its ability to distinguish benign and malignant breast lesions, with a high sensitivity and specificity, is imperative. Indeed, benign and malignant breast tissue could be distinguished, for the first time, after performing PCA and logistic regression on spectra [44]. This diagnostic algorithm allowed correct classification of normal, benign and malignant lesions with 93%, 87% and 100% accuracy, respectively [44]. The coupling of multivariate statistical methods with RS has since proven to be indispensable for discriminating benign and malignant breast lesions [45–54].
Breast tissue abnormalities are inherently heterogeneous, both at the morphological and molecular level. This is reflected by the variety of benign and pre-cancerous/cancerous conditions that can affect the breast. Indeed, there has been considerable focus on distinguishing the wide variety of breast pathologies using RS. Efficacy of FT RS in differentiating benign, pre-invasive and malignant breast tissue has been demonstrated [55]. Through qualitative spectral analysis, a number of key differences were observed, including intensity changes of bands corresponding to amino acids, nucleic acids, proteins, carbohydrates and lipids. These differences could form the basis of spectral biomarkers for classifying different breast tissue states. Similarly, spontaneous RS and shell-isolated nanoparticle-enhanced Raman spectroscopy (SHINERS) have been used to distinguish normal, benign, precancerous, and pre-invasive and invasive malignant breast tissue [56, 57]. In keeping with other studies, they found that the main spectral features of normal breast tissue were associated with lipid vibrational modes, whereas the diseased tissues showed stronger peaks corresponding to that of protein and nucleic acids. They observed both a gradual increase in protein and nucleic acid concentration and a decrease in lipid concentration, as the breast tissue became more malignant. PLS-discriminant analysis demonstrated a high level of classification accuracy for five different tissue types. In addition to its ability to distinguish the morphological subtypes of BC, RS has also been able to classify the different molecular subtypes- Luminal A, Luminal B, HER2+ and triple negative [58]. Four hundred Raman spectra were obtained from breast tissue microarrays of various morphological classifications but stratified into their molecular subtypes, using a dispersive micro-Raman system operating at a 532 nm wavelength. They observed differences, between the subtypes, in the intensity, position and shape of certain peaks; including, the 1583 cm−1 peak of tryptophan and the 1667 cm−1 Amide I peak. LDA was able to correctly classify the BC subtypes with a specificity of 70%, 100%, 90% and 96.7% for luminal A, luminal B, HER2+ and triple negative BC, respectively.
Microcalcifications are small calcium deposits commonly observed on mammograms, as a result of benign cystic [59], or cancerous/early precancerous breast changes. Mammographic microcalcification classification relies on morphology and distribution rather than chemical composition and thus represents a major source of unnecessary biopsies in benign cases. Microcalcifications can be classed as either Type I or Type II. Type I microcalcifications are composed of calcium oxalate and are most often associated with benign lesions while Type II; are composed of calcium phosphate, mainly hydroxyapatite [60, 61] and are associated with both benign and malignant lesions. Calcium oxalate and calcium hydroxyapatite scatter photons effectively and have different spectral signatures, thus RS is powerful for differentiating type I and II microcalcifications [62–65]. The utility of RS in BC diagnoses, based on microcalcification- status, is also dependent on its ability to distinguish benign and malignant type II calcifications, where subtle chemical differences between both types of type II microcalcification were reported; malignant breast ducts contained lower levels of carbonate and higher amounts of protein compared to benign ducts [65]. The significance of carbonate as a spectroscopic marker for differentiating type II microcalcifications has been further demonstrated [66]. More recently, a Raman mapping approach was used to study the whole area of each microcalcification for a more intuitive description of all components within the lesion [61]. In addition to hydroxyapatite, whitlockite and amorphous carbonate were found in some benign type II microcalcifications. Raman profiles from microcalcifications could be correlated to their respective diagnostic categories with high sensitivity and specificity. In a pilot study, type II microcalcifications were also identified in benign and malignant canine mammary tumours with good discrimination between benign and malignant lesions supporting its use in the clinical diagnosis and management of human and canine BC [67].
Yang, et al. [68] combined spectral and morphological features in their analysis of type II microcalcification for improved discrimination of benign and malignant cases [68]. They extracted the relative abundance of the main chemical constituents of the microcalcifications- hydroxyapatite, carbonate and protein- using spontaneous and stimulated Raman spectroscopy (SRS) and found correlations between their abundance and tumour malignancy, similar to previous reports. Next, using SRS microscopy, they extracted features related to geometry (circularity, area, perimeter and Fourier descriptor) and texture from each calcification, and found there to be statistically significant differences between the benign and malignant cases. They then selected the most informative spectral and morphological features and analysed them with a support vector machine-based classification algorithm. Interestingly, it was the combination of extracted spectral and geometric features, as opposed to either pure spectroscopy or imaging-based methods, that yielded the best accuracy (99.05%) and precision (98.21%) for diagnosing tumour malignancy.
As mentioned previously, breast abnormalities are highly heterogenous and thus a binary classification system of either benign or malignant type II microcalcifications does not consider the broad spectrum of existing breast pathologies. This is necessary to better define calcification types. SHINERS, in combination with PCA, was used to accurately distinguish type II microcalcifications between fibroadenoma, atypical ductal hyperplasia and ductal carcinoma in situ [69]. More recently, hyperspectral SRS was combined with Second harmonic generation imaging to separate specific microcalcification diagnostic categories, such as: fibroadenoma, atypical ductal hyperplasia, ductal carcinoma in situ and invasive ductal carcinoma [70].
A tissue microarray study of 79 normal and 499 malignant breast tumours confirmed RS was a powerful technique in distinguishing normal from malignant mammary tissues with a sensitivity and specificity of 90% and 78%, respectively [71]. Lipid, particularly fatty acid spectra, dominated the normal tissue whereas protein spectra characterised the cancerous samples. While several previous studies used fresh and frozen breast samples to preserve the chemical composition of tissue, this study confirmed that RS can be easily used on paraffin wax embedded tissues thus widening its applicability.
A recent, albeit small study, of 36 frozen and paraffin embedded breast lesions, including normal, in situ carcinoma and invasive cancer, confirmed the reliability of the technique in differentiating benign from cancerous tissue in paraffin sections, despite some alteration of the chemical composition of the tissues due to dewaxing [72]. Nodal assessment, traditionally done by ultrasound imaging, with or without nodal sampling, is an integral component of the triple assessment of BC. Using frozen sections of axillary lymph nodes and comparison with the histological findings on paraffin sections, Raman imaging has provided promising potential in establishing nodal metastasis and the chemical composition of the metastatic lymph nodes in comparison with the primary BC [73].
In-vivo applications
Several proof-of-concept studies using portable, hand-held Raman probes for classifying ex vivo cancerous and non-cancerous breast tissue [22, 35, 47, 50, 74–78]; and for axillary lymph node assessment [79, 80] have been reported. This demonstrates the translational potential for possible in vivo applications for diagnosing BC intraoperatively; delineating tumour/normal tissue margins; eliminating residual tumour and intraoperative sentinel lymph node assessment.
The first in vivo Raman spectra from breast tissue were obtained from patients undergoing a partial mastectomy. Using a clinical Raman system and optical fibre Raman probe, surgical margins of the remaining tumour cavity following excision were assessed to investigate its feasibility for real-time, intraoperative margin assessment [81]. When comparing results to those from traditional histopathological assessment, Raman achieved an overall accuracy of 93%. Subsequently, spectra were acquired using a dispersive Raman spectrometer (785 nm) coupled to a fibre-optic probe, from breast tumour bearing Sprague-Dawley rats [82]. Probing eight regions of the tumoral mammary gland, a continuous decrease in band intensity, mainly of the band 1446 cm−1, which most likely corresponds to vibrations of CH2 [83], was noted as they moved from the histologically normal tumour marginal region to the actual lesion itself. A hand-held spectroscopic device, termed the ‘SpectroPen’, that can record both fluorescence and Raman signals has been developed [84]. This device, in combination with the fluorescent and SERS contrast agents, was able to detect cancer in 4T1 tumour bearing mice in vivo and pre/intra operatively. Moreover, this could evaluate, in real-time, the positive and negative tumour margins of the remaining tumour cavity. The same approach has been successfully applied to human BC for the intraoperative margin assessment in breast conserving surgery [85]. This is a promising application that could optimise patient management and reduce the need for further surgical procedures to achieve clear margins.
A strategy for image-guided surgical resection of murine breast tumours and intra-operative eradication of residual microtumours using a nanoprobe that combined photoacoustic imaging with SERS detection and photo-dermal therapy has been reported [86]. This achieved complete ablation of microtumours without local recurrence in a breast-tumour induced mouse model. If extended to human subjects, this could drastically improve patient outcomes, as residual tumour cells drive disease relapse.
Also in mouse models, the feasibility of obtaining spectra from the breast, transcutaneously, has been demonstrated [87–90] with 99% efficiency for the classification of transcutaneous normal and transcutaneous breast tumour tissue reported [90]. In vivo transcutaneous spectra were acquired using a commercial Raman spectrometer coupled to a fibre-optic probe. Although mouse models were used, spectra were comparable to that obtained from both ex vivo and intraoperative in vivo human breast spectra [90].
As mentioned previously, the detection of microcalcifications in mammograms can be an early sign of BC. The capability of RS in characterising type I and II microcalcifications in ex vivo tissue samples has been reviewed in the previous section. However, analysis in this way requires an invasive biopsy, which in patients who only harbour non-cancerous microcalcifications, is unnecessary as only a small proportion of microcalcifications detected with mammography are malignant [91]. This, therefore, necessitates the development of new techniques capable of characterising microcalcifications in vivo and in real time. Several groups have applied fibre-optic Raman sampling to probe the elemental constituents of microcalcifications. A portable clinical Raman system, delivering NIR excitation was used to examine freshly excised tissue [62]. A logistic regression diagnostic algorithm detected microcalcifications with a sensitivity of 86% and specificity of 96% and was able to characterise type I and II microcalcifications based on the presence or absence of the 912 cm−1 and 1477 cm−1 (calcium oxalate) or 960 cm−1 (calcium hydroxyapatite) bands. Using the same clinical Raman system, this group later examined biopsy samples (normal and lesions with/without microcalcifications) and developed a single-step algorithm to determine both microcalcification status and overall clinical diagnosis [92]. Their diagnostic algorithm yielded a sensitivity and specificity of 62.5% and 100% for diagnosing BC and an overall accuracy of 82.2% for classifying normal, benign and malignant breast samples.
A non-invasive Raman method for breast microcalcification characterisation would involve the use of instrumentation that can probe these calcifications, at a considerable depth, through different skin layers. However, the Raman signal weakens as the tissue sampling depth increases because the superficial Raman and fluorescence signals overwhelm the system. Thus, the use of Raman methods, capable of removing this fluorescence and enhancing the deep Raman signals, will overcome this and potentially have a role within the detection and evaluation of microcalcifications in the clinic. The application of Kerr-gated RS for depth profiling of microcalcification beneath the surface of chicken breast and fatty tissues as well as normal and cancerous breast tissue has been investigated [63]. Spectra were obtained from type I and II calcification standards through the different types of breast sections at depths of 0.9 mm. This same group also demonstrated the efficacy of Spatially offset Raman spectroscopy and transmission Raman Spectroscopy (TRS) to detect and characterise the chemical composition of calcified material through a 2–10 mm and 16 mm thick block of chicken breast tissue [64, 93]. Coherent anti-Stokes Raman micro-spectroscopy has also been applied to successfully image and distinguish type I and II calcifications buried at a depth of 2 mm between chicken tissue [94]. Using a breast phantom, constructed from porcine skin, adipose and muscular tissue at a clinically relevant depth of 27 mm, an improvement on the original TRS approach, it was possible to detect and identify the composition of calcification standards [95]. More recently, an advanced TRS instrument capable of detecting calcifications similar to those seen in patient samples, in soft tissue at a 40 mm tissue depth has been developed [96].
These studies have established that deep Raman can identify and distinguish both type I and II microcalcifications through up to 40 mm of tissue. However, to have utility in a clinical setting, it must also be able to distinguish benign and malignant type II microcalcifications. Transmission Raman methods can differentiate benign and malignant type II microcalcifications based on carbonate content [97]. The level of carbonate substitution in type II microcalcifications is known to vary significantly, depending on whether these are found within benign or malignant tissues [65]. Calcification standards, with different percentages of carbonate substitution, were inserted into a cuvette and buried within porcine soft tissue samples (5.6 mm). The group were able to determine the level of carbonate substitution through the 5.6 mm sample, by monitoring both the band position and width of 960 cm−1 (phosphate), and thus differentiate and probe the composition of type II microcalcifications.
Biofluids
Breast tumours exhibit marked intra-tumoral heterogeneity [98]; tumour cells within one compartment are often molecularly and genetically distinct from those in another. It is difficult to capture the complete tumour landscape in a single tissue biopsy. Accordingly, there is growing interest in the use of liquid biopsies in the early detection and diagnosis of BC as this facilitates the rapid, real-time analysis of the tumour as it evolves.
The first proof-of-concept study for BC diagnosis through fluid biopsy analysis using RS was performed on serum [99]. Using conventional Raman methods combined with PCA and LDA, BC patients’ samples were discriminated from healthy controls with a sensitivity and specificity of 97 and 78%. Using PCA loading vectors, they were able to characterise relevant differences in band position between both groups in seven band ratios, corresponding to proteins, phospholipids and polysaccharides. Using a similar micro-Raman spectroscopy set-up, analysis of serum from healthy and ductal carcinoma BC patients identified bands that differentiated the two types of serum sample; K-means clustering was applied to investigate their significance [100].
Whilst conventional RS methods have demonstrated efficacy for serum-based BC detection, a recent study compared its performance to SERS [101]. Normal Raman and SERS spectra were acquired from each serum sample; samples included those obtained from BC patients of Stage II-IV disease and normal individuals. SERS afforded a better overall diagnostic performance with improvements in both sensitivity and specificity over conventional Raman. A number of additional studies have since been undertaken to demonstrate the efficacy of SERS to accurately classify and discriminate serum (and serum-albumin) from BC patients and healthy controls [24, 102, 103]. In addition to distinguishing normal and cancerous samples, SERS analysis was also able to differentiate patients at different stages of disease, which agrees with an earlier study that used SERS, in combination with multivariate analysis, to distinguish serum samples from localised and locally advanced Luminal A BC patients with a sensitivity and specificity of over 80% [104]. For serum Raman analysis to be adopted for BC screening regimes, serum from BC patients must have its own unique biochemical signature relative to other malignancies. Indeed, serum-based Raman analysis has been able to distinguish BC patients from other malignancies, including: colorectal, lung, ovarian, oral, liver, leukaemia and cervical [105–107].
The potential for Raman micro-spectroscopy to aid in the diagnosis and staging of BC based on blood plasma composition has also been explored [108]. When comparing normal and BC samples (stage I-IV), a number of differences between certain Raman bands, corresponding to vibrational modes of numerous biomolecules in the fingerprint, have been observed. Stage II and stage III samples appeared biochemically similar, whereas the spectra from stage IV patients were more distinct. Nonetheless, the group applied PCA-Factorial Discriminant Analysis to their data and were able to differentiate all stages (II, III and IV) of the BC samples from each other as well as from the normal controls with very high sensitivity and specificity. This ability to detect BC through analysis of blood plasma by RS has recently been corroborated [109].
RS has also shown efficacy in probing whole blood samples to distinguish BC patients from normal controls. A PLS regression multivariate model, based on the Raman spectra from BC positive and healthy participants, was not only able to identify potential spectral biomarkers (vibrational modes of lycopene, phosphatidylserine, quinoid ring, calcium oxalate and calcium hydroxyapatite) for BC detection but also differentiate the two groups with a 90% sensitivity and 75% specificity [31].
Saliva is known to host a variety of biomarkers in many conditions [110–113]; recently, there has been growing interest in salivary biomarkers for early BC detection [114]. The appeal of saliva sampling is strongly attributed to the fact that it is a simple, non-invasive and low-cost procedure. Indeed, several groups have applied SERS to overcome this issue and shown its potential as a medium for BC diagnosis. SERS spectra of purified salivary proteins from normal controls and patients with benign and malignant breast tumours [115]. identified six prominent peaks, corresponding to different bond vibrations of salivary proteins [115]. Most of these peaks’ intensity was significantly different between the three groups. Furthermore, PLS-discriminant analysis, combined with the leave-one-patient-out cross-validation method, allowed them to discriminate salivary proteins with high sensitivity and specificity in all groups.
Elevated salivary levels of sialic acid have been shown to correlate with the presence of BC [116]. The feasibility of quantifying sialic acid levels by applying SERS to healthy and BC saliva samples showed the median sialic acid concentration of the healthy samples (3.5 mg dL−1) was significantly lower than that observed within BC samples (18.5 mg dL−1) [117]. SERS may have utility in the future as a simple test for quantifying sialic acid concentrations for BC diagnosis.
Urine-based RS has also been explored in the context of BC diagnosis. Raman spectra, acquired from the urine (unprocessed and concentrated) of adenocarcinoma-bearing and normal Sprague-Dawley rats, using a fibre optic Raman microprobe (785 nm), revealed intensity changes of several Raman bands between the two groups, including those corresponding to bond vibrations of urea and creatine [118]. PCA and PC-LDA were applied to the spectra of both unprocessed and concentrated urine; these algorithms were able to classify unprocessed urine with a 72% sensitivity and 80% specificity and concentrated urine with a sensitivity of 91% and specificity of 80%. The study also explored the feasibility of urine-based Raman for the early detection of BC by obtaining spectra from rats, prior to any breast tumour development. Indeed, they were able to classify cancerous urine samples, in the very early stages, with 72.5% sensitivity and 83% specificity [118]. Subsequently, its feasibility for BC diagnoses in human subjects has been demonstrated [30, 119].
Lacrimal fluid (tears) is perhaps the most unexpected potential source of biomarker for BC detection. One might assume that changes to the composition of tears only accompanies ocular disorders, however, recently there has been growing interest in investigating tear fluid in the context of systemic diseases, including BC [120]. In comparison to other body fluids, tears are relatively simple in composition [121], hence will require highly sensitive methods to detect their low-abundance analytes. As such, the practicality of SERS-based analysis of human tears for the discrimination of BC patients and healthy controls has recently been demonstrated. Using a portable Raman system, and a leave-one-out cross-validation-assisted PC-LDA identification method, a clinical sensitivity and specificity of 92% and 100%, respectively for detecting the presence of BC was achieved [29].
Current challenges and future outlook for clinical implementation
Capabilities of RS hold promise for future applications in BC detection and diagnosis. However, there are several barriers that remain with regards to its widespread clinical translation.
Good general information is currently available for obtaining spectra from biological samples [122], but, there is a real lack of uniformity regarding protocols amongst studies. This includes: the way the samples are prepared; the substrate on which they are mounted; the spectrometer instrument settings and the computational pre-processing methods applied (Table 2). The use of different protocols for analysing the same samples can result in significantly different spectra. As spectral differences between cancerous and non-cancerous samples can be very subtle, experimental variability may be responsible for disparity amongst studies with regards to spectral biomarkers for the same disease [123] or even lead to the discovery of false biomarkers. Therefore, it is of paramount importance to optimise and standardise the experimental setup, as well as validate its robustness, for future analysis of biological samples.
Table 2.
Examples of published data applying Raman spectroscopy in breast cancer, showing lack of uniformity in sample preparation, Raman spectrometer parameters and spectral pre-processing technique.
| Reference | Sample | Instrument settings | Spectral pre-processing | |||||
|---|---|---|---|---|---|---|---|---|
| Tissue / cells | Preparation | Substrate | Laser intensity | Laser wavelength | Microscope | Other | ||
| [36] | IDC and adjacent normal tissue. Number nor specified | 6μm snap frozen sections | Not defined | 10 mW | 532 nm; spot size 650 nm | ×50 objective | N/A | Spectra were processed (background subtraction, cosmic ray removal, spectral de-mixing) using WITEC Project Plus software (Ulm, Germany). |
| [143] | Normal (n = 49); fibrocystic change (n = 16); ductal epithelial hyperplasia (n = 3); fibroadenoma (n = 30); infiltrating carcinoma (n = 31) | Snap frozen. Spectra acquired using thawed tissue stored in PBS | Not required | 100–150 mW | 830 nm; spot size 100μm | ×63, water immersion objective Numerical aperture 0.9 | Integration time 10–30 s Spectral resolution 8 cm-1 | Spectra were Raman shift frequency-calibrated using known spectral lines of toluene. Fluorescence background was removed by fitting the spectrum to a fifth-order polynomial and then subtracting this polynomial from the spectrum. Cosmic rays were removed using a derivative filter. |
| [65] | Ductal epithelial hyperplasia (n = 3); sclerosing adenosis (n = 19); fibrocystic disease (n = 43); fibroadenoma (n = 5), MCS (n = 4); DCIS (n = 16) | 6 μm FFPE sections | MgF2 slides | 100–150 mW | 830 nm; spot size 2 μm | ×63 objective Numerical aperture 0.9 | Integration time- 60 s Spectral resolution- 8 cm−1 | See ref. [143]. |
| [144] | MCF7; MDA-MB-436; MCF-10A cell lines | 20μm sections of cell pellets in agar plugs | Glass slides | 10 mW | 532 nm | ×50 objective | Spectrograph aperture; 50 μm pinhole Exposure time and accumulations; 50 s with 5 accumulations | Baseline corrections and Unit Vector Normalisation were performed on spectra prior to analysis. |
| [145] | HMEC; HMLE; HMLE-Twist; HMLE-Ras; BT-474; T47D; MDA-MB-231 cell lines | Unfixed or fixed in methanol/acetone (1:1) | MgF2 coverslips | 28 mW | 785 nm; spot size 6µm2 | ×50 objective. Numerical aperture 0.5 | Diffraction grating 1200 groves/mm Acquisition time: Live cells: 25 s Fixed cells: 15 s | Origin version-8.5 software (Origin Lab Corporation, Northampton, MA, USA) was used for pre-processing. 10-point and 3-point baseline correction methods were applied to spectra in the low and high wavenumber regions, respectively. Spectral noise was reduced using a Savitsky–Golay filter. Spectra were normalised by dividing each point by the norm of the whole spectrum. |
| [81] | No preparation required- in vivo measurement of breast tissue- normal, fibrocystic change, fibroadenoma and infiltrating carcinoma | 82–125 mW | 830 nm | Not applicable | 1 s exposure | Following spectral acquisition, calibration data (spectrum of 4-acetamidophenol) was collected for spectral corrections. Chromatic intensity variations were corrected by collecting the spectrum of a tungsten white light source diffusely scattered by a reflectance standard (BaSO4). Background signals, generated in the optical fibres, were removed by collecting the scattered excitation light from a roughened aluminium surface and then optimally subtracted from the data in an iterative loop by using a scaling factor related to the tissues’ optical properties. Tissue fluorescence background was modelled with a 6th order polynomial. | ||
| [146] | BT-474; MCF-10A cell lines | Cell pellets | Quartz microscope slide | 30 mW | 785 nm | ×50 objective Numerical aperture 0.75 | 30 s exposure and 2 accumulations Spectral resolution- 3 cm−1 | Background spectrum of the quartz slide was subtracted from sample spectrum using an automatic spectral subtraction function (GRAMS/AI spectroscopy software- Thermo Fisher Scientific Inc., Waltham, Massachusetts). Fluorescence background was subtracted using an automated modified polynomial fitting method (MATLAB, MathWorks, Natick, Massachusetts). Spectra were normalised to the mean intensity in the fingerprint range (700–1750 cm−1) |
| [147] | DCIS (n = 30); IDC (n = 30); normal (n = 7) | FFPE 20μm sections that were subsequently deparaffinised | Glass slides | Low- value not specified | 786 nm | Not specified | 4 exposures, 16 scans 4 cm−1 spectral resolution | Not specified |
| [45] | Normal (n = 105); fibroadenoma) (n = 52); malignant (n = 101) | Stored in saline | Not specified | 150 mW | 785 nm | Not specified | 30 s integration time, 20 accumulations | Spectra were calibrated with a cubic fit to known frequencies of Tylenol. Baseline correction, smoothing, calibration, and normalization was carried out based on the ΔCH2 band using algorithms of Grams 32 (Galactic Industries Corporation, Salem, NH, USA). |
| [148] | 4T1 mouse breast cancer cell line | Mice were injected with a suspension of 4T1 cells (1 × 105) and developed tumour nodules at injection site. Mice were sacrificed and tumour, adjacent breast tissue, contralateral mammary gland and its adjacent lymph nodes were removed. | Not required | 50 mW | 785 nm | 20x objective | 3 exposures, 10 s exposure time 4 cm−1 spectral resolution | MATLAB (version 6.5) was used to pre-process data. A median filter was applied to the spectra to remove cosmic rays or spikes. Noise was filtered using wavelets. Background fluorescence was subtracted from the denoised spectra using a modified cubic spline algorithm. The spectrum was then normalised so that the minimum and maximum values of the spectrum were 0 and 1, respectively. |
| [71] | Tissue microarray of normal (n = 79) and breast cancer samples (n = 499) | 5um thick and 0.6 mm diameter | Not specified | 10 mW | 532 nm | ×50 long work distance objective | Not specified | Spectra were fluorescence-corrected by employing a six-polynomial algorithm (OmnicTM, Thermo Scientific, Waltham, MA, USA). Pre-processing (baseline correction; Savitzky-Golay smoothing and standard normal variate corrections) was performed using the Unscrambler X 10.2TM software (Camo software, Oslo, Norway) |
IDC invasive ductal cancer, PBS phosphate buffered saline, MCS Monckeberg medial calcific sclerosis, DCIS ductal carcinoma in situ, FFPE formalin-fixed paraffin-embedded.
As we have highlighted, RS has significant potential as a rapid in vivo tool to probe the biochemical changes that accompany malignancy. Over the past few decades, a variety of innovative fibre-optic based Raman probes have been developed to transform Raman from benchtop to bedside [123]. These can be inserted into the working channel of an endoscope for assessment of both hollow and solid organs. To date, there have been a number of large clinical studies conducted for skin [124], oral [125], GI [126], and cervical cancer [127]. However, research outcomes from studying BC, have relied on small sample sizes and thus validation in randomised large-scale clinical studies is warranted.
Moreover, analysing live tissues presents its own challenges. As discussed earlier, the Raman signal is inherently weak, which can be compromised by background emanating from the measurement device or the tissue itself. Most probes that have been developed have NIR excitation, which can limit autofluorescence interference. Furthermore, many of these probes are based on silica optical fibres, signals from which can overwhelm the tissue spectrum. Prolonging integration times or the use of higher laser powers are often used to improve signal to noise, when analysing resected tissue samples. However, this can be impractical and potentially unsafe for in vivo applications. Indeed, one of the most important considerations is the laser-tissue interaction. RS is generally considered non-destructive, however, the long-term effects of repeated irradiation on tissue architecture are unclear; rigorous safety testing is imperative, prior to clinical implementation. In addition to safety, its clinical performance value must also be demonstrated; either comparable with or outperforming the existing standard of care and if there would be economic value to health care systems in adopting such Raman devices into routine practice.
The coupling of RS with multivariate analysis provides a means to processing and analysing the rich information that is contained within the Raman spectra. However, RS is somewhat unknown to many clinicians and thus the complex and laborious task of analysing the large volume of data may serve as a hinderance and disrupt the standard workflow. Therefore, it is essential that fully automated spectral diagnostic frameworks are created using machine learning and/or artificial intelligence approaches that can be easily interpreted to help in clinical decision making. Not only is the ease of interpreting the output imperative for translation to the clinic but so too is obtaining this data and operating the instrument to ensure repeatability and reproducibility amongst centres. Moreover, it is imperative that clinicians are competent in interpreting this data in context. Every patient is different; some may have underlying fibrocystic breast disease and others may have had previous radiotherapy or surgery, leading to tissue damage and scarring. As such, to embrace the new era of personalised medicine, the clinician must be able to interpret the Raman results on an individual basis.
The COVID-19 pandemic has seen widespread disruption to multiple aspects of cancer care including the suspension of mammographic screening programmes and the deferral of routine diagnostics. Consequently, cancers that would otherwise have been detected early could now be allowed to progress and become more difficult to treat. Even before the pandemic, remote health monitoring, in the form of home and wearable optical technologies, has been rising in popularity. COVID-19 may have heightened their appeal, particularly in the medical space. Currently, these technologies use only a fraction of the physiological data that can be accessed by optical sensors, such as for heart rate and blood glucose monitoring. However, the potential exists to leverage portable and remote optical technologies for oncological applications [128] in the COVID and post-COVID landscape. Low-cost, handheld portable Raman spectrometers have already demonstrated capability for in vivo detection of skin cancers [129]. Moreover, a proof-of-concept study demonstrated the potential of leveraging a mobile phone camera and computing system to incorporate a Raman spectroscope into a phone [130]. Using a standard mobile phone camera as a detector for a Spatial-Heterodyne (miniature) Raman spectrometer (SHRS), spectra from ammonium nitrate and sodium sulphate could be obtained. The spectra obtained by this device were comparable to that obtained with a miniature SHRS with high quality optics and a CCD detector, although, as expected, with a lower SNR and signal intensity in the former. Refinements to this type of device could lead to the development of miniature, affordable, high-throughput Raman spectrometers that could eventually be trialled in the medical realm to transform at-home health care delivery. As highlighted recently, the management of BC has been modified during the pandemic, potentially impacting on delays to diagnosis [131]. Although not a replacement to conventional diagnosis, RS has potential to be used in easily accessible regions, like the breast, as a supplementary screening tool to identify, prioritise and streamline the most at-risk individuals for further testing.
Supplementary information
Acknowledgements
We are grateful to Friends of Anchor and the University of Aberdeen Development Trust for supporting this work and to Renishaw (Wotton-under-Edge, Gloucestershire, UK) for providing the original version of Fig. 1B. KH is in receipt of an Elphinstone Scholarship, University of Aberdeen. EK was a recipient of an Aberdeen Summer Research Scholarship. Special thanks to staff at Renishaw for helpful discussions.
Author contributions
Designed study RAE, DM and VS; wrote manuscript KH, EK and AMS; edited manuscript and approved final version all authors.
Funding
Friends of Anchor. The University of Aberdeen Development Trust. University of Aberdeen, Elphinstone Scholarship
Data availability
Not required.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Not required.
Consent to publish
The consent for publication is given by all authors.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-021-01659-5.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clinicians. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, et al. Breast cancer. Nat Rev Dis Prim. 2019;5:1–31. doi: 10.1038/s41572-018-0051-2. [DOI] [PubMed] [Google Scholar]
- 3.Dillekås H, Rogers MS, Straume O. Are 90% of deaths from cancer caused by metastases? Cancer Med. 2019;8:5574–6. doi: 10.1002/cam4.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: A Cancer J Clinicians. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 5.Milosevic M, Jankovic D, Milenkovic A, Stojanov D. Early diagnosis and detection of breast cancer. Technol Health Care. 2018;26:729–59. doi: 10.3233/THC-181277. [DOI] [PubMed] [Google Scholar]
- 6.Secretan BL, Ph D, Scoccianti C, Ph D, Loomis D, Ph D. Breast-cancer screening — viewpoint of the IARC working group. N. Engl J Med. 2015;372:2353–8. doi: 10.1056/NEJMsr1504363. [DOI] [PubMed] [Google Scholar]
- 7.Duffy SW, Vulkan D, Cuckle H, Parmar D, Sheikh S, Smith RA, et al. Effect of mammographic screening from age 40 years on breast cancer mortality (UK Age trial): final results of a randomised, controlled trial. Lancet Oncol. 2020;21:1165–72. doi: 10.1016/S1470-2045(20)30398-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esserman LJ, Investigators, the W. S. and A. The WISDOM Study: breaking the deadlock in the breast cancer screening debate. npj Breast Cancer. 2017;3:1–7. doi: 10.1038/s41523-017-0035-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolb TM, Lichy J, Newhouse JH. Comparison of the performance of screening mammography, physical examination, and breast US and evaluation of factors that influence them: an analysis of 27,825 patient evaluations. Radiology. 2002;225:165–75. doi: 10.1148/radiol.2251011667. [DOI] [PubMed] [Google Scholar]
- 10.Jacklyn G, McGeechan K, Houssami N, Bell K, Glasziou PP, Barratt A. Overdiagnosis due to screening mammography for women aged 40 years and over. Cochrane Database Syst Rev. 2018;2018:1–17. [Google Scholar]
- 11.Sahu RK, Mordechai S. Spectroscopic techniques in medicine: The future of diagnostics. Appl Spectrosc Rev. 2016;51:484–99. doi: 10.1080/05704928.2016.1157809. [DOI] [Google Scholar]
- 12.Kim JA, Wales DJ, Yang G-Z. Optical spectroscopy for in vivo medical diagnosis—a review of the state of the art and future perspectives. Prog Biomed Eng. 2020;2:042001. doi: 10.1088/2516-1091/abaaa3. [DOI] [Google Scholar]
- 13.Guevara E, Torres-Galván JC, Ramírez-Elías MG, Luevano-Contreras C, González FJ. Use of Raman spectroscopy to screen diabetes mellitus with machine learning tools. Biomed Opt Express. 2018;9:4998. doi: 10.1364/BOE.9.004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pavlou E, Zhang X, Wang J, Kourkoumelis N. Raman spectroscopy for the assessment of osteoarthritis. Ann Jt. 2018;3:83–83. doi: 10.21037/aoj.2018.09.10. [DOI] [Google Scholar]
- 15.Carlomagno C, Bertazioli D, Gualerzi A, Picciolini S, Banfi PI, Lax A, et al. COVID-19 salivary Raman fingerprint: innovative approach for the detection of current and past SARS-CoV-2 infections. Sci Rep. 2021;11:1–13. doi: 10.1038/s41598-021-84565-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shipp DW, Sinjab F, Notingher I. Raman spectroscopy: techniques and applications in the life sciences. Adv Opt Photonics. 2017;9:315–428. doi: 10.1364/AOP.9.000315. [DOI] [Google Scholar]
- 17.Stone N, Kendall C, Smith J, Crow P, Barr H. Raman spectroscopy for identification of epithelial cancers. Faraday Discuss. 2004;126:141–57. doi: 10.1039/b304992b. [DOI] [PubMed] [Google Scholar]
- 18.Barton SJ, Ward TE, Hennelly BM. Algorithm for optimal denoising of Raman spectra. Anal Methods. 2018;10:3759–69. doi: 10.1039/C8AY01089G. [DOI] [Google Scholar]
- 19.Ramírez-Elías, MG, González, FJ Raman Spectroscopy for In Vivo Medical Diagnosis. In: Gustavo MdN (ed). Raman Spectroscopy, 1st edn. (IntechOpen: London, UK, 2018) pp 293-311.
- 20.Gautam R, Vanga S, Ariese F, Umapathy S. Review of multidimensional data processing approaches for Raman and infrared spectroscopy. EPJ Tech Instrum. 2015;2:1–38. doi: 10.1140/epjti/s40485-015-0018-6. [DOI] [Google Scholar]
- 21.Marro M, Rodríguez-Rivero AM, Araujo-Andrade C, Fernández-Figueras MT, Pérez-Roca L, Castellà E, et al. Unravelling the encapsulation of dna and other biomolecules in hap microcalcifications of human breast cancer tissues by raman imaging. Cancers. 2021;13:2658. doi: 10.3390/cancers13112658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kothari R, Jones V, Mena D, Bermúdez Reyes V, Shon Y, Smith JP, et al. Raman spectroscopy and artificial intelligence to predict the Bayesian probability of breast cancer. Sci Rep. 2021;11:1–17. doi: 10.1038/s41598-021-85758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Depciuch J, Barnaś E, Skręt-Magierło J, Skręt A, Kaznowska E, Łach K, et al. Spectroscopic evaluation of carcinogenesis in endometrial cancer. Sci Rep. 2021;11:9079. doi: 10.1038/s41598-021-88640-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao N, Wang Q, Tang J, Yao S, Li H, Yue X, et al. Non-invasive SERS serum detection technology combined with multivariate statistical algorithm for simultaneous screening of cervical cancer and breast cancer. Anal Bioanal Chem. 2021;413:4775–84. doi: 10.1007/s00216-021-03431-3. [DOI] [PubMed] [Google Scholar]
- 25.Jeng M-J, Sharma M, Sharma L, Chao T-Y, Huang S-F, Chang L-B, et al. Raman Spectroscopy analysis for optical diagnosis of oral cancer detection. J Clin Med. 2019;8:1313. doi: 10.3390/jcm8091313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Yang T, Li CS, Song Y, Lou H, Guan D, et al. Surface enhanced Raman spectroscopy (SERS) for the multiplex detection of Braf, Kras, and Pik3ca mutations in plasma of colorectal cancer patients. Theranostics. 2018;8:1678–89. doi: 10.7150/thno.22502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 28.Qian K, Wang Y, Hua L, Chen A, Zhang Y. New method of lung cancer detection by saliva test using surface-enhanced Raman spectroscopy. Thorac Cancer. 2018;9:1556–61. doi: 10.1111/1759-7714.12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim S, Kim TG, Lee SH, Kim W, Bang A, Moon SW, et al. Label-free surface-enhanced Raman spectroscopy biosensor for on-site breast cancer detection using human tears. ACS Appl Mater Interfaces. 2020;12:7897–904. doi: 10.1021/acsami.9b19421. [DOI] [PubMed] [Google Scholar]
- 30.Moisoiu V, Socaciu A, Stefancu A, Iancu SD, Boros I, Alecsa CD, et al. Breast cancer diagnosis by surface-enhanced raman scattering (SERS) of urine. Appl Sci (Switz) 2019;9:806. doi: 10.3390/app9040806. [DOI] [Google Scholar]
- 31.Bilal M, Bilal M, Tabassum S, Saleem M, Mahmood H, Sarwar U, et al. Optical screening of female breast cancer from whole blood using Raman spectroscopy. Appl Spectrosc. 2017;71:1004–13. doi: 10.1177/0003702816667516. [DOI] [PubMed] [Google Scholar]
- 32.Zhang K, Liu X, Man B, Yang C, Zhang C, Liu M, et al. Label-free and stable serum analysis based on Ag-NPs/PSi surface-enhanced Raman scattering for noninvasive lung cancer detection. Biomed Opt Express. 2018;9:4345. doi: 10.1364/BOE.9.004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu CH, Alfano RR, Sha WL, Zhu HR, Akins DL, Cleary J, et al. Human breast tissues studied by IR Fourier-transform Raman spectroscopy. Conference on Lasers and Electro-Optics Vol 10 (1991).
- 34.Redd DCB, Feng ZC, Yue KT, Gansler TS. Raman spectroscopic characterization of human breast tissues: implications for breast cancer diagnosis. Appl Spectrosc. 1993;47:787–91. doi: 10.1366/0003702934067072. [DOI] [Google Scholar]
- 35.Frank CJ, McCreary RL, Redd DCB. Raman Spectroscopy of normal and diseased human breast tissues. Anal Chem. 1995;67:777–83. doi: 10.1021/ac00101a001. [DOI] [PubMed] [Google Scholar]
- 36.Brozek-Pluska B, Musial J, Kordek R, Bailo E, Dieing T, Abramczyk H. Raman spectroscopy and imaging: applications in human breast cancer diagnosis. Analyst. 2012;137:3773–80. doi: 10.1039/c2an16179f. [DOI] [PubMed] [Google Scholar]
- 37.Surmacki J, Brozek-Pluska B, Kordek R, Abramczyk H. The lipid-reactive oxygen species phenotype of breast cancer. Raman spectroscopy and mapping, PCA and PLSDA for invasive ductal carcinoma and invasive lobular carcinoma. Molecular tumorigenic mechanisms beyond Warburg effect. Analyst. 2015;140:2121–33. doi: 10.1039/C4AN01876A. [DOI] [PubMed] [Google Scholar]
- 38.Kopec M, Imiela A, Abramczyk H. Monitoring glycosylation metabolism in brain and breast cancer by Raman imaging. Sci Rep. 2019;9:1–13. doi: 10.1038/s41598-018-36622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Albayrak M, Senol O, Demirkaya-Miloglu F, Calik M, Kadioglu Y. Novel chemometrics-assisted spectroscopic methods for diagnosis and monitoring of invasive ductal carcinoma in breast tissue. Bratisl Med J. 2019;120:184–7. doi: 10.4149/BLL_2019_031. [DOI] [PubMed] [Google Scholar]
- 40.Zheng C, Jia HY, Liu LY, Wang Q, Jiang HC, Teng LS, et al. Molecular fingerprint of precancerous lesions in breast atypical hyperplasia. J Int Med Res. 2020;48:1–13. doi: 10.1177/0300060520931616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma D, Shang L, Tang J, Bao Y, Fu J, Yin J. Classifying breast cancer tissue by Raman spectroscopy with one-dimensional convolutional neural network. Spectrochimica Acta - Part A: Mol Biomolecular Spectrosc. 2021;256:119732. doi: 10.1016/j.saa.2021.119732. [DOI] [PubMed] [Google Scholar]
- 42.Abramczyk H, Brozek-Pluska B, Surmacki J, Jablonska J, Kordek R. The label-free Raman imaging of human breast cancer. J Mol Liq. 2011;164:123–31. doi: 10.1016/j.molliq.2011.04.021. [DOI] [Google Scholar]
- 43.Brozek-Pluska B, Jablonska-Gajewicz J, Kordek R, Abramczyk H. Phase transitions in oleic acid and in human breast tissue as studied by raman spectroscopy and raman imaging. J Medicinal Chem. 2011;54:3386–92. doi: 10.1021/jm200180f. [DOI] [PubMed] [Google Scholar]
- 44.Manoharan R, Shafer K, Perelman L, Wu J, Chen K, Deinum G, et al. Raman spectroscopy and fluorescence photon migration for breast cancer diagnosis and imaging. Photochemistry Photobiol. 1998;67:15–22. doi: 10.1111/j.1751-1097.1998.tb05160.x. [DOI] [PubMed] [Google Scholar]
- 45.Chowdary MVP, Kumar KK, Kurien J, Mathew S, Krishna CM. Discrimination of normal, benign, and malignant breast tissues by Raman spectroscopy. Biopolymers. 2006;83:556–69. doi: 10.1002/bip.20586. [DOI] [PubMed] [Google Scholar]
- 46.Abramczyk H, Placek I, Brozek-Płuska B, Kurczewski K, Morawiec Z, Tazbir M. Human breast tissue cancer diagnosis by Raman spectroscopy. Spectroscopy. 2008;22:113–21. doi: 10.1155/2008/842724. [DOI] [Google Scholar]
- 47.Zúñiga WC, Jones V, Anderson SM, Echevarria A, Miller NL, Stashko C, et al. Raman spectroscopy for rapid evaluation of surgical margins during breast cancer lumpectomy. Sci Rep. 2019;9:1–16. doi: 10.1038/s41598-019-51112-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lyng FM, Traynor D, Nguyen TNQ, Meade AD, Rakib F, Al-Saady R, et al. Discrimination of breast cancer from benign tumours using Raman spectroscopy. PLoS ONE. 2019;14:1–13. doi: 10.1371/journal.pone.0212376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu C, Wang J, Zheng C, Xu S, Zhang H, Liang Y, et al. Raman spectra exploring breast tissues: Comparison of principal component analysis and support vector machine-recursive feature elimination. Med Phys. 2013;40:063501. doi: 10.1118/1.4804054. [DOI] [PubMed] [Google Scholar]
- 50.Haka AS, Volynskaya Z, Gardecki JA, Nazemi J, Shenk R, Wang N, et al. Diagnosing breast cancer using Raman spectroscopy: prospective analysis. J Biomed Opt. 2009;14:054023. doi: 10.1117/1.3247154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu CH, Zhou Y, Sun Y, Li JY, Zhou LX, Boydston-White S, et al. Resonance Raman and Raman spectroscopy for breast cancer detection. Technol Cancer Res Treat. 2013;12:371–82. doi: 10.7785/tcrt.2012.500325. [DOI] [PubMed] [Google Scholar]
- 52.Thakur JS, Dai H, Serhatkulu GK, Naik R, Naik VM, Cao A, et al. Raman spectral signatures of mouse mammary tissue and associated lymph nodes: normal, tumor and mastitis. J Raman Spectrosc. 2007;38:127–34. doi: 10.1002/jrs.1565. [DOI] [Google Scholar]
- 53.Abramczyk H, Surmacki J, Brozek-Płuska B, Morawiec Z, Tazbir M. The hallmarks of breast cancer by Raman spectroscopy. J Mol Struct. 2009;924–926:175–82. doi: 10.1016/j.molstruc.2008.12.055. [DOI] [Google Scholar]
- 54.Li H, Ning T, Yu F, Chen Y, Zhang B, Wang S. Raman microspectroscopic investigation and classification of breast cancer pathological characteristics. Molecules. 2021;26:921. doi: 10.3390/molecules26040921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bitar RA, Martinho H, da S, Tierra-Criollo CJ, Zambelli Ramalho LN, Netto MM, et al. Biochemical analysis of human breast tissues using Fourier-transform Raman spectroscopy. J Biomed Opt. 2006;11:054001. doi: 10.1117/1.2363362. [DOI] [PubMed] [Google Scholar]
- 56.Zheng C, Liang L, Xu S, Zhang H, Hu C, Bi L, et al. The use of Au@SiO2 shell-isolated nanoparticle-enhanced Raman spectroscopy for human breast cancer detection. Anal Bioanal Chem. 2014;406:5425–32. doi: 10.1007/s00216-014-7967-5. [DOI] [PubMed] [Google Scholar]
- 57.Zheng C, Shao W, Paidi SK, Han B, Fu T, Wu D, et al. Pursuing shell-isolated nanoparticle-enhanced Raman spectroscopy (SHINERS) for concomitant detection of breast lesions and microcalcifications. Nanoscale. 2015;7:16960–8. doi: 10.1039/C5NR05319F. [DOI] [PubMed] [Google Scholar]
- 58.Talari ACS, Rehman S, Rehman IU. Advancing cancer diagnostics with artificial intelligence and spectroscopy: identifying chemical changes associated with breast cancer. Expert Rev Mol Diagnostics. 2019;19:929–40. doi: 10.1080/14737159.2019.1659727. [DOI] [PubMed] [Google Scholar]
- 59.Wilkinson L, Thomas V, Sharma N. Microcalcification on mammography: approaches to interpretation and biopsy. Br J Radiol. 2017;90:20160594. doi: 10.1259/bjr.20160594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Büsing CM, Keppler U, Menges V. Differences in microcalcification in breast tumors. Virchows Arch A Pathological Anat Histol. 1981;393:307–13. doi: 10.1007/BF00430830. [DOI] [Google Scholar]
- 61.Vanna R, Morasso C, Marcinno B, Piccotti F, Torti E, Altamura D, et al. Raman spectroscopy reveals that biochemical composition of breast microcalcifications correlates with histopathologic features. Cancer Res. 2020;80:1762–72. doi: 10.1158/0008-5472.CAN-19-3204. [DOI] [PubMed] [Google Scholar]
- 62.Saha A, Barman I, Dingari NC, McGee S, Volynskaya Z, Galindo LH, et al. Raman spectroscopy: a real-time tool for identifying microcalcifications during stereotactic breast core needle biopsies. Biomed Opt Express. 2011;2:2792. doi: 10.1364/BOE.2.002792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baker R, Matousek P, Ronayne KL, Parker AW, Rogers K, Stone N. Depth profiling of calcifications in breast tissue using picosecond Kerr-gated Raman spectroscopy. Analyst. 2007;132:48–53. doi: 10.1039/B614388A. [DOI] [PubMed] [Google Scholar]
- 64.Stone N, Baker R, Rogers K, Parker AW, Matousek P. Subsurface probing of calcifications with spatially offset Raman spectroscopy (SORS): Future possibilities for the diagnosis of breast cancer. Analyst. 2007;132:899–905. doi: 10.1039/b705029a. [DOI] [PubMed] [Google Scholar]
- 65.Haka AS, Shafer-Peltier KE, Fitzmaurice M, Crowe J, Dasari RR, Feld MS. Identifying microcalcifications in benign and malignant breast lesions by probing differences in their chemical composition using raman spectroscopy. Cancer Res. 2002;62:5375–80. [PubMed] [Google Scholar]
- 66.Sathyavathi R, Saha A, Soares JS, Spegazzini N, McGee S, Rao Dasari R, et al. Raman spectroscopic sensing of carbonate intercalation in breast microcalcifications at stereotactic biopsy. Sci Rep. 2015;5:1–12. doi: 10.1038/srep09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dantas D, Soares L, Novais S, Vilarinho R, Agostinho Moreira J, Silva S, et al. Discrimination of benign and malignant lesions in canine mammary tissue samples using Raman spectroscopy: a pilot study. Animals. 2020;10:1–16. doi: 10.3390/ani10091652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang Y, Yang Y, Liu Z, Guo L, Li S, Sun X, et al. Microcalcification-based tumor malignancy evaluation in fresh breast biopsies with hyperspectral stimulated Raman scattering. Anal Chem. 2021;93:6223–31. doi: 10.1021/acs.analchem.1c00522. [DOI] [PubMed] [Google Scholar]
- 69.Liang L, Zheng C, Zhang H, Xu S, Zhang Z, Hu C, et al. Exploring type II microcalcifications in benign and premalignant breast lesions by shell-isolated nanoparticle-enhanced Raman spectroscopy (SHINERS) Spectrochimica Acta - Part A: Mol Biomolecular Spectrosc. 2014;132:397–402. doi: 10.1016/j.saa.2014.04.147. [DOI] [PubMed] [Google Scholar]
- 70.Shin KS, Laohajaratsang M, Men S, Figueroa B, Dintzis SM, Fu D. Quantitative chemical imaging of breast calcifications in association with neoplastic processes. Theranostics. 2020;10:5865–78. doi: 10.7150/thno.43325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lazaro-Pacheco D, Shaaban AM, Titiloye NA, Rehman S, Rehman IU. Elucidating the chemical and structural composition of breast cancer using raman micro-spectroscopy. EXCLI J. 2021;20:1118–32. doi: 10.17179/excli2021-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ning T, Li H, Chen Y, Zhang B, Zhang F, Wang S. Raman spectroscopy based pathological analysis and discrimination of formalin fixed paraffin embedded breast cancer tissue. Vibrational Spectrosc. 2021;115:103260. doi: 10.1016/j.vibspec.2021.103260. [DOI] [Google Scholar]
- 73.Smith J, Kendall C, Sammon A, Christie-Brown J, Stone N. Raman spectral mapping in the assessment of axillary lymph nodes in breast cancer. Technol Cancer Res Treat. 2003;2:327–31. doi: 10.1177/153303460300200407. [DOI] [PubMed] [Google Scholar]
- 74.Gebrekidan MT, Erber R, Hartmann A, Fasching PA, Emons J, Beckmann MW, et al. Breast tumor analysis using shifted-excitation Raman difference spectroscopy (SERDS) Technol Cancer Res Treat. 2018;17:1–11. doi: 10.1177/1533033818782532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim S, Kim W, Bang A, Song J-Y, Shin J-H, Choi S. Label-free breast cancer detection using fiber probe-based Raman spectrochemical biomarker-dominated profiles extracted from a mixture analysis algorithm. Anal Methods. 2021;13:3249–55. doi: 10.1039/D1AY00491C. [DOI] [PubMed] [Google Scholar]
- 76.Kim, S, Bang, A, Choi, S. Multiple label-free detection of breast cancer biomarkers using Raman spectroscopy. Proc. SPIE 2019;11089:110892G.
- 77.Keller MD, Vargis E, de Matos Granja N, Wilson RH, Mycek M-A, Kelley MC, et al. Development of a spatially offset Raman spectroscopy probe for breast tumor surgical margin evaluation. J Biomed Opt. 2011;16:077006. doi: 10.1117/1.3600708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li Q, Li W, Zhang J, Xu Z. An improved: K-nearest neighbour method to diagnose breast cancer. Analyst. 2018;143:2807–11. doi: 10.1039/C8AN00189H. [DOI] [PubMed] [Google Scholar]
- 79.Horsnell J, Stonelake P, Christie-Brown J, Shetty G, Hutchings J, Kendall C, et al. Raman spectroscopy—a new method for the intra-operative assessment of axillary lymph nodes. Analyst. 2010;135:3042–7. doi: 10.1039/c0an00527d. [DOI] [PubMed] [Google Scholar]
- 80.Horsnell JD, Kendall C, Stone N. Towards the intra-operative use of Raman spectroscopy in breast cancer—overcoming the effects of theatre lighting. Lasers Med Sci. 2016;31:1143–9. doi: 10.1007/s10103-016-1959-y. [DOI] [PubMed] [Google Scholar]
- 81.Haka AS, Volynskaya Z, Gardecki JA, Nazemi J, Lyons J, Hicks D, et al. In vivo margin assessment during partial mastectomy breast surgery using Raman spectroscopy. Cancer Res. 2006;66:3317–22. doi: 10.1158/0008-5472.CAN-05-2815. [DOI] [PubMed] [Google Scholar]
- 82.Bitar R, Martins MA, Ribeiro D, Carvalho C, Santos EAP, Ramalho LNZ, et al. In vivo Raman spectroscopy for breast cancer: diagnosis in animal model. Biomed Optical Spectrosc. 2008;6853:68530O. doi: 10.1117/12.760089. [DOI] [Google Scholar]
- 83.Movasaghi Z, Rehman S, Rehman IU. Raman spectroscopy of biological tissues. Appl Spectrosc Rev. 2007;42:493–541. doi: 10.1080/05704920701551530. [DOI] [Google Scholar]
- 84.Mohs, AM, Mancini, MC, Singhal, S, Provenzale, JM, Wang, MD, Nie, S. NIH Public Access. 2012;82:1–17.
- 85.Hubbard T, Ferguson D, Shore A, Stone N. P122: Raman spectroscopy for intraoperative margin analysis in breast conserving surgery. Eur J Surgical Oncol. 2020;46:e43. doi: 10.1016/j.ejso.2020.03.161. [DOI] [Google Scholar]
- 86.Wen Y, Truong VX, Li M. Real-time intraoperative surface-enhanced Raman spectroscopy-guided thermosurgical eradication of residual microtumors in orthotopic breast cancer. Nano Lett. 2021;21:3066–74. doi: 10.1021/acs.nanolett.1c00204. [DOI] [PubMed] [Google Scholar]
- 87.Bhattacharjee T, Maru G, Ingle A, Krishna CM. Transcutaneous in vivo Raman spectroscopic studies in a mouse model: evaluation of changes in the breast associated with pregnancy and lactation. J Biomed Opt. 2013;18:047004. doi: 10.1117/1.JBO.18.4.047004. [DOI] [PubMed] [Google Scholar]
- 88.Bhattacharjee T, Maru G, Ingle A, Krishna CM. Transcutaneous in vivo Raman spectroscopy: detection of age-related changes in mouse breast. Vibrational Spectrosc. 2013;67:80–86. doi: 10.1016/j.vibspec.2013.04.004. [DOI] [Google Scholar]
- 89.Bhattacharjee T, Maru G, Ingle A, Murali Krishna C. Transcutaneous in vivo Raman spectroscopy of breast tumors and pretumors. J Raman Spectrosc. 2015;46:1053–61. doi: 10.1002/jrs.4739. [DOI] [Google Scholar]
- 90.Bhattacharjee T, Kumar P, Maru G, Ingle A, Krishna CM. Swiss bare mice: a suitable model for transcutaneous in vivo Raman spectroscopic studies of breast cancer. Lasers Med Sci. 2014;29:325–33. doi: 10.1007/s10103-013-1347-9. [DOI] [PubMed] [Google Scholar]
- 91.Matousek P, Stone N. Emerging concepts in deep Raman spectroscopy of biological tissue. Analyst. 2009;134:1058–66. doi: 10.1039/b821100k. [DOI] [PubMed] [Google Scholar]
- 92.Barman I, Dingari NC, Saha A, McGee S, Galindo LH, Liu W, et al. Application of raman spectroscopy to identify microcalcifications and underlying breast lesions at Stereotactic core needle biopsy. Cancer Res. 2013;73:3206–15. doi: 10.1158/0008-5472.CAN-12-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Matousek P, Stone N. Prospects for the diagnosis of breast cancer by noninvasive probing of calcifications using transmission Raman spectroscopy. J Biomed Opt. 2007;12:024008. doi: 10.1117/1.2718934. [DOI] [PubMed] [Google Scholar]
- 94.Petrov GI, Arora R, Yakovlev VV. Coherent anti-Stokes Raman scattering imaging of microcalcifications associated with breast cancer. Analyst. 2021;146:1253–9. doi: 10.1039/D0AN01962C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stone N, Matousek P. Advanced transmission Raman spectroscopy: A promising tool for breast disease diagnosis. Cancer Res. 2008;68:4424–30. doi: 10.1158/0008-5472.CAN-07-6557. [DOI] [PubMed] [Google Scholar]
- 96.Ghita A, Matousek P, Stone N. High sensitivity non-invasive detection of calcifications deep inside biological tissue using Transmission Raman Spectroscopy. J Biophotonics. 2018;11:1–8. doi: 10.1002/jbio.201600260. [DOI] [PubMed] [Google Scholar]
- 97.Kerssens MM, Matousek P, Rogers K, Stone N. Towards a safe non-invasive method for evaluating the carbonate substitution levels of hydroxyapatite (HAP) in micro-calcifications found in breast tissue. Analyst. 2010;135:3156–61. doi: 10.1039/c0an00565g. [DOI] [PubMed] [Google Scholar]
- 98.Turashvili G, Brogi E. Tumor heterogeneity in breast cancer. Front Med. 2017;4:227. doi: 10.3389/fmed.2017.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pichardo-Molina JL, Frausto-Reyes C, Barbosa-García O, Huerta-Franco R, González-Trujillo JL, Ramírez-Alvarado CA, et al. Raman spectroscopy and multivariate analysis of serum samples from breast cancer patients. Lasers Med Sci. 2007;22:229–36. doi: 10.1007/s10103-006-0432-8. [DOI] [PubMed] [Google Scholar]
- 100.Varghese S, Reveendran A, Senthil Kumar V, Karthikeyan TM, Ranganathan V. Micro raman spectroscopic analysis on blood serum samples of ductal carcinoma patients. Asian J Pharm Clin Res. 2018;11:176–8. doi: 10.22159/ajpcr.2018.v11i9.26806. [DOI] [Google Scholar]
- 101.Nargis HF, Nawaz H, Bhatti HN, Jilani K, Saleem M. Comparison of surface enhanced Raman spectroscopy and Raman spectroscopy for the detection of breast cancer based on serum samples. Spectrochimica Acta - Part A: Mol Biomolecular Spectrosc. 2021;246:119034. doi: 10.1016/j.saa.2020.119034. [DOI] [PubMed] [Google Scholar]
- 102.Lin D, Wang Y, Wang T, Zhu Y, Lin X, Lin Y, et al. Metabolite profiling of human blood by surface-enhanced Raman spectroscopy for surgery assessment and tumor screening in breast cancer. Anal Bioanal Chem. 2020;412:1611–8. doi: 10.1007/s00216-020-02391-4. [DOI] [PubMed] [Google Scholar]
- 103.Lin Y, Gao S, Zheng M, Tang S, Lin K, Xie S, et al. A microsphere nanoparticle based-serum albumin targeted adsorption coupled with surface-enhanced Raman scattering for breast cancer detection. Spectrochimica Acta - Part A: Mol Biomolecular Spectrosc. 2021;261:120039. doi: 10.1016/j.saa.2021.120039. [DOI] [PubMed] [Google Scholar]
- 104.Cervo S, Mansutti E, Mistro G, Del, Spizzo R, Colombatti A, et al. SERS analysis of serum for detection of early and locally advanced breast cancer. Anal Bioanal Chem. 2015;407:7503–9. doi: 10.1007/s00216-015-8923-8. [DOI] [PubMed] [Google Scholar]
- 105.Vlad M, Andrei S, Diana G, Radu B, Lorand M, Lajos R, et al. SERS-based differential diagnosis between multiple solid malignancies: Breast, colorectal, lung, ovarian and oral cancer. Int J Nanomed. 2019;14:6165–78. doi: 10.2147/IJN.S198684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gao S, Zheng M, Lin Y, Lin K, Zeng J, Xie S, et al. Surface-enhanced Raman scattering analysis of serum albumin via adsorption-exfoliation on hydroxyapatite nanoparticles for noninvasive cancers screening. J Biophotonics. 2020;13:1–8. doi: 10.1002/jbio.202000087. [DOI] [PubMed] [Google Scholar]
- 107.González-Solís JL. Discrimination of different cancer types clustering Raman spectra by a super paramagnetic stochastic network approach. PLoS ONE. 2019;14:1–15. doi: 10.1371/journal.pone.0213621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nargis HF, Nawaz H, Ditta A, Mahmood T, Majeed MI, Rashid N, et al. Raman spectroscopy of blood plasma samples from breast cancer patients at different stages. Spectrochimica Acta - Part A: Mol Biomolecular Spectrosc. 2019;222:117210. doi: 10.1016/j.saa.2019.117210. [DOI] [PubMed] [Google Scholar]
- 109.Știufiuc GF, Toma V, Buse M, Mărginean R, Morar-Bolba G, Culic B, et al. Solid plasmonic substrates for breast cancer detection by means of SERS analysis of blood plasma. Nanomaterials. 2020;10:1–15. doi: 10.3390/nano10061212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim JJ, Kim CJ, Camargo PM. Salivary biomarkers in the diagnosis of periodontal diseases. J Calif Dent Assoc. 2013;41:119–24. [PMC free article] [PubMed] [Google Scholar]
- 111.Abdul Rehman S, Khurshid Z, Hussain Niazi F, Naseem M, Al Waddani H, Sahibzada H, et al. Role of salivary biomarkers in detection of cardiovascular diseases (CVD) Proteomes. 2017;5:21. doi: 10.3390/proteomes5030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Janigro D, Kawata K, Silverman E, Marchi N, Diaz-Arrastia R. Is salivary S100B a biomarker of traumatic brain injury? A pilot study. Front Neurol. 2020;11:528. doi: 10.3389/fneur.2020.00528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee Y-H, Joshipura K, Vergara JL, Wong DT. Detection of type II diabetes mellitus using salivary transcriptomic biomarkers. Genomic Medicine. Biomark, Health Sci. 2012;4:7–11. [Google Scholar]
- 114.Abrao Nemeir I, Saab J, Hleihel W, Errachid A, Jafferzic-Renault N, Zine N. The advent of salivary breast cancer biomarker detection using affinity sensors. Sens (Basel, Switz) 2019;19:2373. doi: 10.3390/s19102373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Feng S, Huang S, Lin D, Chen G, Xu Y, Li Y, et al. Surface-enhanced Raman spectroscopy of saliva proteins for the noninvasive differentiation of benign and malignant breast tumors. Int J Nanomed. 2015;10:537–47. doi: 10.2147/IJN.S71811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ozturk LK, Emekli-Alturfan E, Kasikci E, Demir G, Yarat A. Salivary total sialic acid levels increase in breast cancer patients: a preliminary study. Medicinal Chem. 2011;7:443–7. doi: 10.2174/157340611796799230. [DOI] [PubMed] [Google Scholar]
- 117.Hernández-Arteaga A, de Jesús Zermeño Nava J, Kolosovas-Machuca ES, Velázquez-Salazar JJ, Vinogradova E, José-Yacamán M, et al. Diagnosis of breast cancer by analysis of sialic acid concentrations in human saliva by surface-enhanced Raman spectroscopy of silver nanoparticles. Nano Res. 2017;10:3662–70. doi: 10.1007/s12274-017-1576-5. [DOI] [Google Scholar]
- 118.Bhattacharjee T, Khan A, Maru G, Ingle A, Krishna CM. A preliminary Raman spectroscopic study of urine: Diagnosis of breast cancer in animal models. Analyst. 2015;140:456–66. doi: 10.1039/C4AN01703J. [DOI] [PubMed] [Google Scholar]
- 119.Lin X, Wang L, Lin H, Lin D, Lin J, Liu X, et al. A novel urine analysis technique combining affinity chromatography with Au nanoparticle based surface enhanced Raman spectroscopy for potential applications in non-invasive cancer screening. J Biophotonics. 2019;12:1–9. doi: 10.1002/jbio.201800327. [DOI] [PubMed] [Google Scholar]
- 120.Hagan S, Martin E, Enríquez-de-Salamanca A. Tear fluid biomarkers in ocular and systemic disease: potential use for predictive, preventive and personalised medicine. EPMA J. 2016;7:15. doi: 10.1186/s13167-016-0065-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Aass C, Norheim I, Eriksen EF, Thorsby PM, Pepaj M. Single unit filter-aided method for fast proteomic analysis of tear fluid. Anal Biochem. 2015;480:1–5. doi: 10.1016/j.ab.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 122.Butler HJ, Ashton L, Bird B, Cinque G, Curtis K, Dorney J, et al. Using Raman spectroscopy to characterize biological materials. Nat Protoc. 2016;11:664–87. doi: 10.1038/nprot.2016.036. [DOI] [PubMed] [Google Scholar]
- 123.Cordero E. In-vivo Raman spectroscopy: from basics to applications. J Biomed Opt. 2018;23:1. doi: 10.1117/1.JBO.23.7.071210. [DOI] [PubMed] [Google Scholar]
- 124.Schleusener J, Gluszczynska P, Reble C, Gersonde I, Helfmann J, Fluhr JW, et al. In vivo study for the discrimination of cancerous and normal skin using fibre probe-based Raman spectroscopy. Exp Dermatol. 2015;24:767–72. doi: 10.1111/exd.12768. [DOI] [PubMed] [Google Scholar]
- 125.Krishna H, Majumder SK, Chaturvedi P, Sidramesh M, Gupta PK. In vivo Raman spectroscopy for detection of oral neoplasia: A pilot clinical study. J Biophotonics. 2014;7:690–702. doi: 10.1002/jbio.201300030. [DOI] [PubMed] [Google Scholar]
- 126.Bergholt MS, Zheng W, Ho KY, Yeoh KG, Teh M, So JBY, et al. Real-time depth-resolved fiber optic Raman endoscopy for in vivo diagnosis of gastric precancer. SPIE BiOS. 2014;8939:89390M (2014).
- 127.Shaikh R, Dora TK, Chopra S, Maheshwari A, Kedar KD, Bharat R, et al. In vivo Raman spectroscopy of human uterine cervix: exploring the utility of vagina as an internal control. J Biomed Opt. 2014;19:087001. doi: 10.1117/1.JBO.19.8.087001. [DOI] [PubMed] [Google Scholar]
- 128.Teng F, Cormier T, Sauer-Budge A, Chaudhury R, Pera V, Istfan R, et al. Wearable near-infrared optical probe for continuous monitoring during breast cancer neoadjuvant chemotherapy infusions. J Biomed Opt. 2017;22:014001. doi: 10.1117/1.JBO.22.1.014001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lieber C, Mahadevan-Jansen A. Development of a handheld Raman microspectrometer for clinical dermatologic applications. Opt Express. 2007;15:11874. doi: 10.1364/OE.15.011874. [DOI] [PubMed] [Google Scholar]
- 130.Barnett PD, Angel SM. Miniature spatial heterodyne Raman spectrometer with a cell phone camera detector. Appl Spectrosc. 2017;71:988–95. doi: 10.1177/0003702816665127. [DOI] [PubMed] [Google Scholar]
- 131.Dave RV, Kim B, Courtney A, O’Connell R, Rattay T, Taxiarchi VP, et al. Breast cancer management pathways during the COVID-19 pandemic: outcomes from the UK ‘Alert Level 4’ phase of the B-MaP-C study. Br J Cancer. 2021;124:1785–94. doi: 10.1038/s41416-020-01234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li M, Wu J, Ma M, Feng Z, Mi Z, Rong P, et al. Alkyne-and nitrile-anchored gold nanoparticles for multiplex SERS imaging of biomarkers in cancer cells and tissues. Nanotheranostics. 2019;3:113–9. doi: 10.7150/ntno.30924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lee JU, Kim WH, Lee HS, Park KH, Sim SJ. Quantitative and specific detection of exosomal miRNAs for accurate diagnosis of breast cancer using a surface-enhanced Raman scattering sensor based on plasmonic head-flocked gold nanopillars. Small. 2019;15:1–10. doi: 10.1002/smll.201804968. [DOI] [PubMed] [Google Scholar]
- 134.Feng J, Chen L, Xia Y, Xing J, Li Z, Qian Q, et al. Bioconjugation of gold nanobipyramids for SERS detection and targeted photothermal therapy in breast cancer. ACS Biomater Sci Eng. 2017;3:608–18. doi: 10.1021/acsbiomaterials.7b00021. [DOI] [PubMed] [Google Scholar]
- 135.Ou YC, Webb JA, O’Brien CM, Pence IJ, Lin EC, Paul EP, et al. Diagnosis of immunomarkers: In vivo via multiplexed surface enhanced Raman spectroscopy with gold nanostars. Nanoscale. 2018;10:13092–105. doi: 10.1039/C8NR01478G. [DOI] [PubMed] [Google Scholar]
- 136.Mosca S, Conti C, Stone N, Matousek P. Spatially offset Raman spectroscopy. Nat Rev Methods Prim. 2021;1:22. doi: 10.1038/s43586-021-00025-2. [DOI] [Google Scholar]
- 137.García-Flores AF, Raniero L, Canevari RA, Jalkanen KJ, Bitar RA, Martinho HS, et al. High-wavenumber FT-Raman spectroscopy for in vivo and ex vivo measurements of breast cancer. Theor Chem Acc. 2011;130:1231–8. doi: 10.1007/s00214-011-0925-9. [DOI] [Google Scholar]
- 138.Bitar Carter RA, Martin AA, Netto MM, Soares FA. FT-Raman spectroscopy study of human breast tissue. Biomed Vib Spectrosc Biohazard Detect Technol. 2004;5321:190. [Google Scholar]
- 139.Moreno M, Raniero L, Arisawa EAL, do Espírito Santo AM, dos Santos EAP, Bitar RA, et al. Raman spectroscopy study of breast disease. Theor Chem Acc. 2010;125:329–34. doi: 10.1007/s00214-009-0698-6. [DOI] [Google Scholar]
- 140.Lu F-KF, Calligaris D, Suo Y, Santagata S, Golby AJ, Xie XS, et al. Label-free biomolecular characterization of human breast cancer tissue with stimulated Raman scattering (SRS) spectral imaging. Proc SPIE. 2017;10069:100690R (2017).
- 141.Yang Y, Gao L, Wang Z, Thrall MJ, Luo P, Wong KK, et al. Label-free imaging of human breast tissues using coherent anti-Stokes Raman scattering microscopy. SPIE BiOS. 2011;7903:79032G. [DOI] [PMC free article] [PubMed]
- 142.Huang X, Yuan Y, Bielecki TA, Mohapatra BC, Luan H, Silva-Lopez E, et al. Discrimination of tumor from normal tissues in a mouse model of breast cancer using CARS spectroscopy combined with PC-DFA methodology. J Raman Spectrosc. 2017;48:1166–70. doi: 10.1002/jrs.5201. [DOI] [Google Scholar]
- 143.Haka AS, Shafer-Peltier KE, Fitzmaurice M, Crowe J, Dasari RR, Feld MS. Diagnosing breast cancer by using Raman spectroscopy. Proc Natl Acad Sci USA. 2005;102:12371–6. doi: 10.1073/pnas.0501390102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Talari ACS, Evans CA, Holen I, Coleman RE, Rehman IU. Raman spectroscopic analysis differentiates between breast cancer cell lines. J Raman Spectrosc. 2015;46:421–7. doi: 10.1002/jrs.4676. [DOI] [Google Scholar]
- 145.Chaturvedi D, Balaji SA, Bn VK, Ariese F, Umapathy S, Rangarajan A. Different phases of breast cancer cells: Raman study of immortalized, transformed, and invasive cells. Biosensors. 2016;6:57. doi: 10.3390/bios6040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bi X, Rexer B, Arteaga CL, Guo M, Mahadevan-Jansen A. Evaluating HER2 amplification status and acquired drug resistance in breast cancer cells using Raman spectroscopy. J Biomed Opt. 2014;19:025001. doi: 10.1117/1.JBO.19.2.025001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rehman S, Movasaghi Z, Tucker AT, Joel SP, Darr JA, Ruban AV, et al. Raman spectroscopic analysis of breast cancer tissues: identifying differences between normal, invasive ductal carcinoma and ductal carcinomain situ of the breast tissue. J Raman Spectrosc. 2007;38:1345–51. doi: 10.1002/jrs.1774. [DOI] [Google Scholar]
- 148.Kast RE, Serhatkulu GK, Cao A, Pandya AK, Dai H, Thakur JS, et al. Raman spectroscopy can differentiate malignant tumors from normal breast tissue and detect early neoplastic changes in a mouse model. Biopolymers. 2008;89:235–41. doi: 10.1002/bip.20899. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not required.