Abstract
BACKGROUND
Inflammatory bowel disease (IBD) comprises two distinct diseases, Crohn’s disease (CD) and ulcerative colitis (UC), both of which are chronic, relapsing inflammatory disorders of the gastrointestinal tract with a mostly unknown etiology. The incidence and prevalence of IBD are continually increasing, indicating the need for further studies to investigate the genetic determinants of these diseases. Since microRNAs (miRNAs) regulate protein translation via complementary binding to mRNA, discovering differentially expressed miRNAs (DE) in UC or CD patients could be important for diagnostic biomarker identification, assisting in the appropriate disease differentiation progressing the understanding of IBD pathogenesis.
AIM
To determine the miRNA expression profile in UC and CD patients and the potential pathophysiological contributions of differentially expressed miRNA.
METHODS
A total of 20 formalin-fixed paraffin-embedded colonic samples were collected from the Pathology Department of Botucatu Medical School at São Paulo State University (Unesp). The diagnosis of UC or CD was based on clinical, endoscopic, radiologic, and histological criteria and confirmed by histopathological analysis at the time of selection. The TaqMan™ Array Human MicroRNA A+B Cards Set v3.0 (Applied Biosystems™) platform was used to analyze 754 miRNAs. Targets of DE-miRNAs were predicted using miRNA Data Integration Portal (mirDIP) and the miRNA Target Interaction database (MiRTarBase). All statistical analyses were conducted using GraphPad Prism software. Parametric and nonparametric data were analyzed using t-tests and Mann-Whitney U tests, respectively.
RESULTS
The results showed that of the 754 miRNAs that were initially evaluated, 643 miRNAs were found to be expressed in at least five of the patients who were diagnosed with either CD or UC; the remaining 111 miRNAs were not considered to be expressed in these patients. The expression levels of 28 miRNAs were significantly different between the CD and UC patients (P ≤ 0.05); 13 miRNAs demonstrated a fold-change in expression level greater than 1. Five miRNAs with a downregulated expression were selected for enrichment analysis. The miRNAs whose expression levels were significantly lower in UC patients than in CD patients were enriched in certain signaling pathways that were mostly correlated with cancer-related processes and respective biomarkers.
CONCLUSION
MiRNAs could be used to differentiate UC from CD, and differently expressed miRNAs could help explain the distinct pathophysiology of each disease.
Keywords: Crohn’s disease, Ulcerative colitis, Inflammatory bowel disease, miRNA, Differential diagnosis, Biomarker
Core Tip: This study identified 27 microRNAs (miRNAs) with significantly different expression levels between Crohn’s disease (CD) and ulcerative colitis (UC) patients. Five miRNAs whose expression levels were significantly lower in the UC patients than in the CD patients were selected for enrichment analysis, which revealed enrichment in certain signaling pathways that were mostly associated with cancer-related processes. The characterization and comparison of the differentially expressed miRNAs in this study could lead to novel diagnostic biomarkers to differentiate UC from CD. These markers might also predict prognosis, help elucidate the distinct pathophysiology of each disease, and lead to novel therapeutic target identification.
INTRODUCTION
Crohn’s disease (CD) and ulcerative colitis (UC), the two main phenotypes of inflammatory bowel disease (IBD), are chronic, relapsing inflammatory disorders of the gastrointestinal (GI) tract whose etiologies remain unknown[1,2]. Although its pathophysiological mechanisms are unclear, IBD has been linked to an inappropriate and exacerbated immune response to commensal bacteria in genetically susceptible hosts, as well as with environmental factors, defects of the epithelial barrier, and dysregulation of the innate and adaptive immune responses at the level of the intestinal mucosa[1,2].
IBD affects 6 to 8 million people worldwide, with a prevalence of 84.3 people (79.2-89.9) per 100000 in 2017 and a mortality rate of 0.51[3]. The incremental increase in the incidence and prevalence of IBD globally is indicative of the need for further population-based genetic studies of those affected by these diseases. Differences in miRNA expression between these two diseases could lead to the identification and validation of diagnostic biomarkers, facilitating differential diagnosis and improving the understanding of IBD pathogenesis[4].
MiRNAs are a class of small (18-25 nucleotides in length), endogenous, non-coding, single-stranded RNA molecules that can negatively regulate target gene expression at the post-transcriptional level through binding to the 3’untranslated regions of the target mRNAs and promoting mRNA degradation or translational repression[4]. Overall, there is evidence that miRNAs contribute to the regulation of at least one-third of all protein-coding mRNAs, including those involved in the development, metabolism and cell cycle control[5]. Although studies have shown that miRNA plays an important role in both CD and UC and that they are differentially expressed in these disease states, there may have been potential confounding factors that were not considered. For example, the patients in these studies were treated with various medication classes, such as immunosuppressants, corticosteroids, and/or aminosalicylates[5-8]. Therefore, the aim of the present study was to investigate miRNA expression patterns in the tissues of treatment-naïve CD and UC patients to assess changes in miRNAs without the confounding influence of pharmacological agents.
MATERIALS AND METHODS
Ethics statement
The study was approved by the Botucatu Medical School Research Ethics Committee (protocol No. 71379417.2.0000.5411).
Sample collection
A total of twenty formalin-fixed paraffin-embedded (FFPE) colonic samples were collected from patients in the Pathology Department of Botucatu Medical School at São Paulo State University (Unesp). IBD diagnosis was based on clinical, endoscopic, radiologic, and histological criteria[9]. After sample selection, the diagnoses of UC or CD were confirmed by histopathological analysis of the biopsied colonic samples collected from each patient during a diagnostic colonoscopy; therefore, none of the patients received any medication at the time of the examination.
RNA isolation, reverse transcription, pre-amplification, and qRT-PCR analyses
Total RNA from each colon biopsy was isolated using MagMAX™ FFPE DNA/RNA Ultra Kit (Thermo Fisher Scientific, Massachusetts, EUA). For reverse transcription, Megaplex™ Reverse Transcription Primer Pools and Taqman miRNA reverse transcription kits (Thermo Fisher Scientific) were used. Briefly, 3 μL of RNA (1-350 ng) were reverse-transcribed by combining the Megaplex RT Primer (Thermo Fisher Scientific) with the TaqMan® miRNA Reverse Transcription Kit (Thermo Fisher Scientific). A volume of 2.5 μL of the reverse transcription product was used for pre-amplification with Megaplex™ PreAmp Primers (Thermo Fisher Scientific) and TaqMan® PreAmp Master Mix (Thermo Fisher Scientific). For qRT-PCR, 9 μL of the pre-amplified product was mixed with 441 μL of H2O and 450 μL of TaqMan® Universal PCR Master Mix II without uracil-N-glycosylase (Thermo Fisher Scientific). Subsequently, 100 μL of each sample was loaded into each fill reservoir of the TaqMan microfluidic array cards. Real-time-PCR analysis was performed using a QuantStudio™ Real-Time PCR System (with TaqMan® Array Block) (Thermo Fisher Scientific) with universal cycling conditions [95°C/10 min, then (95°C/15 s, 60°C/60 s) for 40 cycles]. The TaqMan miRNA array output data (.sds files) were uploaded to the ThermoFisher Cloud App (https://www.thermofisher.com/mysso/LoginDisplay) and analyzed via the relative quantification (ΔΔCt) method using defined threshold settings for each miRNA. The geometric means of the threshold cycle (Ct) values of three small nucleolar RNAs (snRNAs), RNU44, RNU48, and U6, were used as the endogenous controls. Briefly, the change in quantification cycle (ΔCq) value was calculated as:
Cq (miRNA of interest) - mean Cq (endogenous control).
The ΔΔCq was subsequently calculated as:
ΔCq (miRNA of interest) - mean of ΔCq (miRNA of interest in the reference group - CD).
The relative quantification (Rq or gene expression fold-change) was calculated as 2-(ΔΔCq). Subsequently, the FC was calculated using the Rq and the FC values, and P values were log2- and log10-transformed, respectively, and plotted as a volcano plot, displaying the -log 10(P value) vs the log2 (FC) for each target in the UC group relative to that in the CD group. The volcano plot was generated using an online server (https://paolo.shinyapps.io/ShinyVolcanoPlot/) after defining the statistical cutoffs as a log2 fold-change > 1 and a P value < 0.05. A heat map was created using a web tool (http://www.heatmapper.ca/expression/).
Target prediction and gene enrichment analysis
The targets of the DE miRNAs were predicted using miRNA Data Integration Portal (mirDIP) (http://ophid.utoronto.ca/mirDIP/index.jsp)[10,11] and the miRNA Target Interaction database (MirTarBase) (http://miRTarBase.mbc.nctu.edu.tw/)[12]. The combined gene list of each miRNA was uploaded to the Enrichr database (http://amp.pharm.mssm.edu/Enrichr/), a web server for comprehensive gene set enrichment analysis[13,14]. Duplicate susceptibility genes were excluded prior to analysis. The CD and UC susceptibility genes were identified from the National Human Genome Research Institute-European Bioinformatics Institute (NHGRI-EBI) Catalog of human genome-wide association studies (GWAS)[15].
Statistical analysis
The statistical methods of this study were reviewed by Ana E. V. Quaglio from the Laboratory of Phytomedicines, Pharmacology, and Biotechnology (PhytoPharmaTec), Department of Biophysics and Pharmacology, São Paulo State University (Unesp). All statistical analyses were conducted using GraphPad Prism (GraphPad Software, San Diego, CA, United States). To determine if data is from a Gaussian distribution were performed D’Agostino-Person normality test. Parametric and non-parametric data were analyzed using unpaired and two-tailed t-tests and Mann-Whitney U tests, respectively. Statistical significance was considered for P ≤ 0.05.
RESULTS
This study included 20 patients with IBD (10 with UC and 10 with CD), with a mean age at the time of diagnosis of 36.1 and 31.6 years for those with UC and CD, respectively. In terms of sex, 40% of the UC and 30% of the CD patients were male, and 60% and 70% of the UC and CD patients were female, respectively. Other characteristics of the patients are presented in Table 1.
Table 1.
Demographic characteristics and clinical features of inflammatory bowel disease patients
|
|
Ulcerative colitis
|
Crohn’s disease
|
| Number of patients | 10 | 10 |
| Age at onset (yr, mean) | 36.1 ± 18.31 | 31.6 ± 14.6 |
| Sex, (%) | ||
| Male | 40 | 30 |
| Female | 60 | 70 |
| Race, (%) | ||
| Caucasian | 100 | 90 |
| Non-caucasian | 0 | 10 |
| Alcoholism, (%) | 10 | 20 |
| Smoking, (%) | 20 | 10 |
| Family history of IBD, (%) | 10 | 0 |
| Site of UC, (%) | ||
| Proctitis | 20 | - |
| Left-sided | 30 | - |
| Extensive | 50 | - |
| Site of CD, (%) | ||
| Ileal | - | 20 |
| Colonic | - | 20 |
| Ileocolonic | - | 60 |
IBD: Inflammatory bowel disease; UC: Ulcerative colitis; CD: Crohn’s disease.
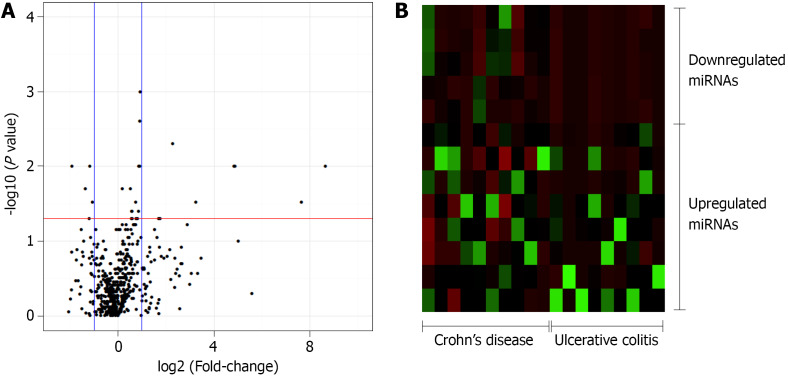
In this study, 377 miRNAs and four controls were analyzed in each plate, totalizing 754 miRNAs of interest. Of these, 643 miRNAs were expressed in at least five patients diagnosed with UC or CD, whereas 111 miRNAs were not considered to be expressed in these patients. The expression levels of 27 miRNAs significantly differed between the CD and UC patients. Relative to the expression levels in CD patients, the five miRNAs that were downregulated in UC patients with a fold-change greater than 1 were selected for the subsequent enrichment analysis: miR-192-3p/5p, miR-378a-3p/5p, and miR-429 (Figure 1).
Figure 1.
Volcano plot and heat map of the patients. A: Volcano plot of the 754-microRNA (miRNA) analyzed in inflammatory bowel disease patients. Control group: Crohn’s disease (CD). Threshold x: fold change = 1; Threshold y: P ≤ 0.05; B: Heat map of the 13 significantly different miRNA between the CD and ulcerative colitis patients with a fold-change in expression level greater than 1 relatively to the expression levels in CD patients (8 upregulated and 5 downregulated). miRNA: MicroRNA.
Target prediction and gene enrichment analysis of miRNAs
To assess the potential functions of miRNAs in UC or CD, the targets of the differentially expressed miRNAs (Supplementary Table 1) were predicted using an online database. Relative to the expression levels in CD patients, the miRNAs that were decreased in UC patients were those that were enriched in signaling pathways, such as forkhead box protein O (FOXO), transforming growth factor-beta (TGF-β), and mitogen-activated protein kinase (MAPK), as well as in pathways associated with cancer, particularly colorectal cancer (CRC) (Table 2).
Table 2.
Gene enrichment analysis of decreased microRNAs in ulcerative colitis patients compared with Crohn’s disease patients
|
Pathway
|
P
value
|
FDR
|
| Proteoglycans in cancer | 2.27E-11 | 6.99E-09 |
| FoxO signaling pathway | 2.37E-11 | 3.64E-09 |
| Pathways in cancer | 7.49E-11 | 7.69E-09 |
| Colorectal cancer | 1.25E-07 | 5.51E-06 |
| TGF-β signaling pathway | 1.47E-07 | 4.53E-06 |
| Signaling pathways regulating pluripotency of stem cells | 4.72E-07 | 1.32E-05 |
| Autophagy | 5.58E-07 | 1.32E-05 |
| ErbB signaling pathway | 9.95E-07 | 2.19E-05 |
| mTOR signaling pathway | 3.70E-06 | 7.61E-05 |
| MAPK signaling pathway | 1.66E-05 | 2.22E-04 |
FoxO: Forkhead box protein O; TGF: Transforming growth factor-β; ErbB: Erythroblastic leukemia viral oncogene homolog; mTOR: Mammalian target of rapamycin; MAPK: Mitogen-activated protein kinase; FDR: False discovery rate-adjusted.
Several of the miRNA gene targets have been previously flagged as CD or UC susceptibility genes. Using the IBD susceptibility gene list from the 2019 GWAS[17], the miRNA targets that had already been linked to one of the two diseases were identified. There are 619 and 474 genes that have been proposed as CD and UC susceptibility genes, respectively. Of the predicted targets of the miRNAs that were downregulated, 24 and 11 overlapped with the proposed CD and UC susceptibility genes, respectively. A total of 328 genes were common to both diseases, and 54 were common to the two diseases and the predicted targets (Figure 2 and Supplementary Table 2).
Figure 2.
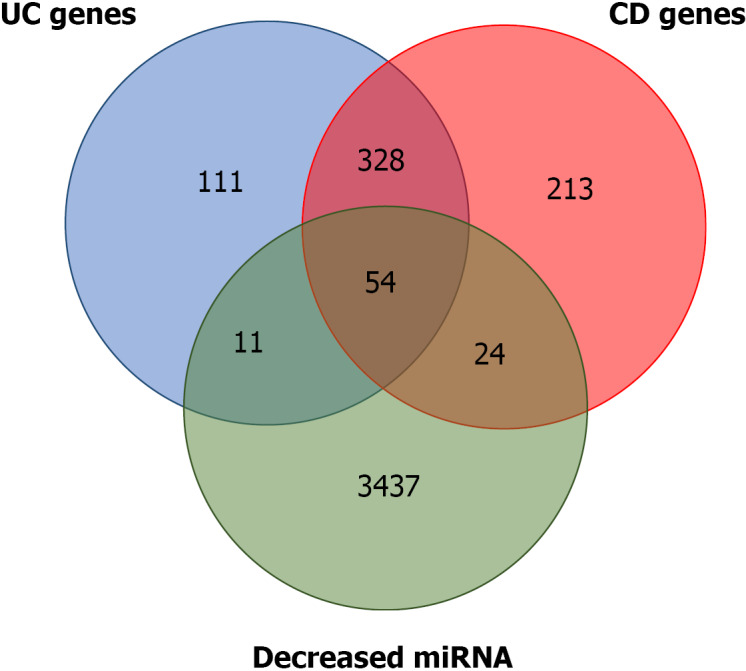
Overlap between predicted microRNA targets for microRNA differentially expressed and ulcerative colitis and Crohn’s disease susceptibility genes. Detailed information is provided in Supplementary Material 1. miRNA: MicroRNA; UC: Ulcerative colitis; CD: Crohn’s disease.
DISCUSSION
MiRNAs are short single-stranded, post-transcriptional regulatory RNAs that are involved in major cellular processes. The mature miRNA species may be derived from either the 5’ or 3’ arms of the precursor duplex and are referred to as the miRNA-5p and -3p species, respectively. Initially, it was believed that only one of these strands was functional and that the other strand, known as miRNA*, was destined for degradation[16]. However, recent reports have indicated that both the miRNA and miRNA* species often co-exist, and both are functional[16,17]. Based on this finding, the miRNA-5p and -3p nomenclature are used solely based on the 5’- or 3’-arm derivation of the miRNA species[16]. This study described, for the first time, the co-existence of miR-192-3p and miR-192-5p and, miR-378a-3p and miR-378a-5p in the colonic samples of IBD patients. In addition to miR-429, the expression levels of both miRNAs were decreased in UC compared with CD patients.
Most of the altered pathways identified following the enrichment analysis are related to cancer processes, alterations in the extracellular matrix (ECM), inflammatory mediators like TGF-β, members of the heat shock protein (HSP) family, and MAPKs. The decrease in the expression of miRNAs related to all these pathways leads to dysregulation. Proteoglycans are important components of the ECM that have multiple functions depending on both their protein and carbohydrate constituents[18]. Heparan sulphate proteoglycans on the cell membrane may play diverse roles in cancer-related processes, acting as either inhibitors or promoters of tumor progression depending on the tumor type and stage of progression[19]. In CRC, for example, the cell surface proteoglycan syndecan-2 (SDC2 gene) is upregulated, leading to increased cell migration[20]. Moreover, high SDC2 expression was shown to be related to tumorigenic behaviors mediated through the regulation of cell adhesion, proliferation, and migration[21]. Recently, chronic inflammatory hypoxia-mediated SD2 expression was reported to be correlated with CRC development[21]. In addition, acute inflammation could also induce SDC2 expression predominantly in the proximal colon, indicating its potential as a biomarker for acute colonic inflammation[22].
In the present study, SDC2 was predicted to be regulated by miR-429, which was proven to be related to cancer progression and metastasis, likely due to dysregulated SDC2 expression, increasing cell migration and proliferation. It has been reported that miR-429 becomes downregulated in esophageal squamous carcinoma cells, and its expression could predict poor prognosis for patients[23]. Moreover, they showed that miR-429 inhibited cellular proliferation through the nuclear factor kappa B (NF-κB) pathway and inhibited cell migration-mediated epithelial-mesenchymal transition (EMT) processes[23].
In nasopharyngeal carcinoma, miR-429 functions as a tumor suppressor by downregulating talin-1 (TLN1), a protein that enhances the migration and invasion of various carcinomas[24]. This miRNA is also related to an antimetastatic function, as it can regulate the metastasis of hepatocellular carcinoma by directly targeting Crk-like protein, which is involved in processes related to EMT, as well as the progression, development, invasion, metastasis, and apoptosis of a variety of cancers[25]. Most functional studies have reported that miR-429 plays an oncogenic role in CRC, and its expression is downregulated as the disease progresses[26,27]. In studies involving a dextran sodium sulphate-induced model of intestinal inflammation, miR-429 was downregulated[28], corroborating the findings of the present study. Others have reported that miR-429 modulates mucin secretion in human CRC cells and mouse colon tissues via upregulation of myristoylated alanine-rich protein kinase C substrate expression confirming that miR-429 is a candidate for intestinal anti-inflammatory therapy in human UC[28].
Another family of proteins with an important correlation with miRNA function is the HSP family. HSPs comprise several classes of constitutively active and/or stress-induced molecular chaperones that assist in proper polypeptide folding, the refolding of denatured proteins, protein transport, and stabilization of native protein structures, and numerous HSPs are overexpressed in inflamed tissues[29]. A recent review by Hoter & Naim (2019)[30] that analyzed the roles of HSPs in IBD found accumulating evidence linking the upregulation of HSP90 and/or HSP70 expression to the pathogenesis of IBD in the intestinal mucosa of patients with UC at the time of diagnosis, with the expression levels decreasing following the initiation of pharmacological therapy. Similar results were reported in a pre-clinical model of intestinal inflammation in which the colonic expression of HSP70 increased following the induction of intestinal inflammation[31]. Several authors have reported that HSPs are subject to post-transcriptional regulation by miRNAs[32]. The selected downregulated miRNAs found in the UC patients in the present study are predicted to regulate several members of the HSP70 family, such as HSPA1B and HSPA2, as well as HSP90B1, a member of the HSP90 family that plays critical roles in the folding of proteins such as Toll-like receptors and integrins[33], suggesting that the decrease in these miRNAs may have been related to the increased levels of the altered HSPs observed in the UC patients. Dysregulation on HSP70/90 Levels has also been related to intestinal inflammation associated with CRC. For example, Hoter & Naim (2019)[30] found that HSP90 was highly expressed and was implicated in the progression from UC to UC-associated CRC. Furthermore, HSP90 inhibition has been actively investigated as a treatment for gastrointestinal and CRC arising from IBD progression.
MiR-378a-3p/5p, two of the miRNAs correlated with HSP downregulation, have also been shown to be altered during intestinal inflammation[34]. The involvement of miR-378 in IBD patients has also been reported[35-37], indicating an upregulation in UC or a downregulation in CD. These differences may have resulted from the small number of patients included in these previous studies or the use of pharmacological agents by the participants, which could reflect treatment rather than disease effects. The patients in the present study were treatment naïve; therefore, the differences in expression resulted from the disease itself. A recent study conducted by Dubois-Camacho et al[38] in 2019, corroborates the present findings, as they showed that a decrease in miR-378a expression in UC patients was associated with higher levels of interleukin 33 and tumor necrosis factor-alpha (TNF-α), and that miR-378a expression levels increased following treatment with an anti-TNF-α monoclonal antibody[38].
In CRC, the overexpression of miR-378 inhibits the proliferation of colon cancer cells in vitro by inducing apoptosis and preventing migration and invasion[39]. For example, miR-378a also alleviated the malignant phenotypes of colon cancer cells by inhibiting the Wnt/β-catenin signaling pathway[39]. In addition, CRC patients with low miR-378a expression experienced a shorter survival time than those with high miR-378a expression, indicating that miR-378a may serve as an important diagnostic biomarker[40]. Collectively, it is possible to assume that the decrease in miR-378a-3p/5p levels during inflammation and the later progression to CRC can increase HSP90 Levels. Thus, increased HSP90 and decreased miR-378a-3p/5p levels could act as important biomarkers and potential targets of pharmacological interventions for UC and UC-related CRC.
Among the downregulated miRNAs observed in IBD patients, miR-192-3p/5p, which was first cloned in 2003[41], is a tumor-related miRNA, as its dysregulation has been reported in several types of cancer[42]. Overexpression of miR-192 has been shown to induce apoptosis in bladder cancer cells and arrest breast cancer cells' growth[42,43]. Moreover, in CRC, miR-192 regulates the enzyme dihydrofolate reductase and cellular proliferation through the p53 tumor suppressor network, decreasing cancer progression and metastases[42,43]. Decreased levels of miR-192 are also related to IBD. For example, Wu et al[6] reported a link between chronic IBD and the altered expression of certain miRNAs, demonstrating that miR-192 was downregulated in the colonic epithelial cells of active UC patients which corroborates the findings of the present study.
An inverse correlation exists between the expression of miR-192 and MPI-2α, an epithelial cell-expressed chemokine previously implicated in IBD[6]. In addition, miR-192 was shown to be induced by TGF-β, suggesting that miR-192 plays a key role in inflammatory, fibrotic processes[6]. Moreover, other putative miR-192 targets identified in the enrichment analysis include mediators of inflammation and fibrosis, such as nucleotide-binding oligomerization domain-containing protein 2, TNF receptor-associated factor-interacting protein 3, 4, and 5, as well as matrix metalloproteinase 16 and 20 (Supplementary Material 1). Changes in miR-192 expression have also been shown in a 2,4,6-trinitrobenzenesulfonic acid-induced intestinal inflammation model[44]. In that elegant study, the EMT was activated as a result of intestinal inflammation, along with a simultaneous increase in the early growth response protein 1 and fibroblast growth factor 2 expression levels (based on mesenchymal markers); on the other hand, miR-192 expression was decreased[44].
Conversely, elevated levels of miR-192 are associated with tumor suppression and cell proliferation[45-49]. Ji et al[45] demonstrated that miR-192 suppresses the growth of bladder cancer cells via targeting Yin Yang 1, a transcription factor that plays an important role in regulating development and cell proliferation. Similarly, Flammang et al[48], in a study involving pancreatic ductal adenocarcinoma, described miR-192 as a marker with prognostic value, as increased miRNA expression exerted a suppressive effect. In CRC, miR-192 also acts as a tumor suppressor, inhibiting CRC invasion[46,47]. Furthermore, Huang et al[49] found that certain metabolites of normal intestinal microflora were capable of upregulating miR-192 expression, suppressing the proliferation of colon cancer cells through a decrease in bone morphogenic protein type 2 receptor levels; thus, cell proliferation, migration, and invasion ability are diminished, and the rate of cellular apoptosis is improved[49]. Collectively, these data indicate a potential role of miR-192-3p/5p as a biomarker for UC activity and a target for pharmacological treatment.
These results provide information that may explain the differences in CRC and mortality rates between UC and CD patients. Two cohort studies from Olén et al[50,51] (2020a; 2020b) compared CRC mortality and incidence in UC and CD patients. In the first study, which evaluated UC patients, the authors found that UC patients had a 36% higher incidence of CRC than that of reference individuals [hazard ratio (HR) = 1.66][51]. Among CD patients, the incidence was 21.9% higher than that of the reference group (HR = 1.4)[50]. These findings demonstrated a tendency toward a higher probability of developing CRC in UC compared with CD, which was associated with higher mortality[50,51]. The lower CRC incidence in CD could be explained by the early removal of this portion of the intestine[52]. Nevertheless, there was differential expression of these selected miRNAs between those with CD and UC. The decreased expression levels of miR-378a-3p/5p, miR-429, and miR-192-3p/5p in UC patients could represent a possible mechanism responsible for increasing their likelihood of developing CRC.
Although the overlap between DE-miRNAs targets and IBD susceptibility genes (Figure 2; Supplementary Table 1) suggested some involvement with the disease onset, progression and future cancer development, the data are preliminary and the sample size is small, so further studies are required to better comprehension of miRNA role in IBD pathogenesis. Future studies should aim to address some of the limitations by increasing the number of patients and investigating the potential links between these miRNAs and pathological UC and UC-associated CRC mechanisms.
CONCLUSION
In conclusion, this study highlighted the potential involvement of miR-192-3p, miR-192-5p, miR-378a-3p, miR-378a-5p and miR-429 as players in IBD pathology, UC differentiation and UC-associated CRC, indicating the potential use of these miRNAs as specific biomarkers for UC. Moreover, these miRNAs could be also useful as biomarkers for UC-associated CRC. Since samples of treatment-naïve patients were used, the distinct expression found in this study was a result from the disease itself with no medication effect. This way, the DE-miRNAs may represent a new pharmacological target for UC treatment.
ARTICLE HIGHLIGHTS
Research background
Inflammatory bowel disease (IBD) is a chronic and relapsing disorder of the gastrointestinal tract including two distinct phenotypes, ulcerative colitis (UC) and Crohn’s disease (CD). IBD pathophysiological mechanisms are unclear.
Research motivation
IBD affects 6 to 8 million people worldwide and the incremental increase in the incidence and prevalence globally is indicative of the need for population-based genetic studies including microRNAs (miRNAs) expression profiles.
Research objectives
The present study aimed to investigate the miRNA expression patterns in tissue of treatment-naïve CD and UC patients and the potential pathophysiological contributions of differentially expressed (DE) miRNA in IBD.
Research methods
A total of 20 formalin-fixed paraffin embedded colonic samples were used in a TaqMan™ Array Human MicroRNA (Applied Biosystems™) platform aiming to analyze 754 miRNAs. After that, targets of DE-miRNAs were predicted using miRNa data integration portal (miRDIP) and the miRNA target interaction database (miRTarBase).
Research results
A total of 643 miRNAs were found to be expressed in both diseases but only 13 miRNAs were significantly different between the CD and UC patients (P ≤ 0.05; fold-change > 1). The miRNAs whose expression levels were significantly lower in UC patients than in CD patients (miR-192-3p/5p, miR-378a-3p/5p and miR-429) were enriched in signaling pathways that were mostly correlated with cancer-related processes and respective biomarkers.
Research conclusions
Ulcerative colitis and Crohn’s disease presented distinct patterns of miRNA expression that could be useful as new pharmacological targets besides acts as biomarkers for UC-associated CRC.
Research perspectives
New studies should be done with the DE-miRNAs using a large number of patients aiming to confirm the differences found in this pilot study. With this confirmation, the DE-miRNAs will be able to be used as pharmacological targets and differential markers for each disease.
ACKNOWLEDGEMENTS
The authors would like to acknowledge Professor dos Reis PP (PhD) for technical assistance.
Footnotes
Institutional review board statement: This study was approved by the Botucatu Medical School Research Ethics Committee, No. 71379417.2.0000.5411.
Conflict-of-interest statement: There is no conflict of interest.
ARRIVE guidelines statement: The authors have read the ARRIVE guidelines, and the manuscript was prepared and revised according to the ARRIVE guidelines.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review started: April 28, 2021
First decision: June 13, 2021
Article in press: November 20, 2021
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Brazil
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Corsello A S-Editor: Wu YXJ L-Editor: A P-Editor: Wu YXJ
Contributor Information
Ana Elisa Valencise Quaglio, Laboratory of Phytomedicines, Pharmacology and Biotechnology (PhytoPharmaTec), Department of Biophysics and Pharmacology, São Paulo State University (Unesp), Institute of Biosciences, Botucatu 18618-689, São Paulo, Brazil. anaequaglio@hotmail.com.
Felipe Jose Santaella, Department of Pathology, Botucatu Medical School, Sao Paulo State University (Unesp), Botucatu 18618-687, São Paulo, Brazil.
Maria Aparecida Marchesan Rodrigues, Department of Pathology, Botucatu Medical School, Sao Paulo State University (Unesp), Botucatu 18618-687, São Paulo, Brazil.
Ligia Yukie Sassaki, Department of Internal Medicine, Botucatu Medical School, São Paulo State University (Unesp), Botucatu 18618-687, São Paulo, Brazil.
Luiz Claudio Di Stasi, Laboratory of Phytomedicines, Pharmacology and Biotechnology (PhytoPharmaTec), Department of Biophysics and Pharmacology, São Paulo State University (Unesp), Institute of Biosciences, Botucatu 18618-689, São Paulo, Brazil.
Data sharing statement
No additional data are available.
References
- 1.Annese V. Genetics and epigenetics of IBD. Pharmacol Res. 2020;159:104892. doi: 10.1016/j.phrs.2020.104892. [DOI] [PubMed] [Google Scholar]
- 2.Khalili H, Chan SSM, Lochhead P, Ananthakrishnan AN, Hart AR, Chan AT. The role of diet in the aetiopathogenesis of inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2018;15:525–535. doi: 10.1038/s41575-018-0022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.GBD 2017 Inflammatory Bowel Disease Collaborators. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. 2020;5:17–30. doi: 10.1016/S2468-1253(19)30333-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pekow JR, Kwon JH. MicroRNAs in inflammatory bowel disease. Inflamm Bowel Dis. 2012;18:187–193. doi: 10.1002/ibd.21691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu F, Zhang S, Dassopoulos T, Harris ML, Bayless TM, Meltzer SJ, Brant SR, Kwon JH. Identification of microRNAs associated with ileal and colonic Crohn's disease. Inflamm Bowel Dis. 2010;16:1729–1738. doi: 10.1002/ibd.21267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, Brant SR, Chakravarti S, Kwon JH. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135:1624–1635.e24. doi: 10.1053/j.gastro.2008.07.068. [DOI] [PubMed] [Google Scholar]
- 7.Zahm AM, Hand NJ, Tsoucas DM, Le Guen CL, Baldassano RN, Friedman JR. Rectal microRNAs are perturbed in pediatric inflammatory bowel disease of the colon. J Crohns Colitis. 2014;8:1108–1117. doi: 10.1016/j.crohns.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Zhang S, Yu Q, Yang G, Guo J, Li M, Zeng Z, He Y, Chen B, Chen M. Circulating MicroRNA223 is a New Biomarker for Inflammatory Bowel Disease. Medicine (Baltimore) 2016;95:e2703. doi: 10.1097/MD.0000000000002703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Morain C, Tobin A, Leen E, Suzuki Y, O'Riordan T. Criteria of case definition in Crohn's disease and ulcerative colitis. Scand J Gastroenterol Suppl. 1989;170:7–11; discussion 16-19. doi: 10.3109/00365528909091340. [DOI] [PubMed] [Google Scholar]
- 10.Shirdel EA, Xie W, Mak TW, Jurisica I. NAViGaTing the micronome--using multiple microRNA prediction databases to identify signalling pathway-associated microRNAs. PLoS One. 2011;6:e17429. doi: 10.1371/journal.pone.0017429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tokar T, Pastrello C, Rossos AEM, Abovsky M, Hauschild AC, Tsay M, Lu R, Jurisica I. mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res. 2018;46:D360–D370. doi: 10.1093/nar/gkx1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, Chien CH, Wu MC, Huang CY, Tsou AP, Huang HD. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39:D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM, Jeon M, Ma'ayan A. Gene Set Knowledge Discovery with Enrichr. Curr Protoc. 2021;1(3) doi: 10.1002/cpz1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, Suveges D, Vrousgou O, Whetzel PL, Amode R, Guillen JA, Riat HS, Trevanion SJ, Hall P, Junkins H, Flicek P, Burdett T, Hindorff LA, Cunningham F, Parkinson H. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choo KB, Soon YL, Nguyen PN, Hiew MS, Huang CJ. MicroRNA-5p and -3p co-expression and cross-targeting in colon cancer cells. J Biomed Sci. 2014;21:95. doi: 10.1186/s12929-014-0095-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuchenbauer F, Mah SM, Heuser M, McPherson A, Rüschmann J, Rouhi A, Berg T, Bullinger L, Argiropoulos B, Morin RD, Lai D, Starczynowski DT, Karsan A, Eaves CJ, Watahiki A, Wang Y, Aparicio SA, Ganser A, Krauter J, Döhner H, Döhner K, Marra MA, Camargo FD, Palmqvist L, Buske C, Humphries RK. Comprehensive analysis of mammalian miRNA* species and their role in myeloid cells. Blood. 2011;118:3350–3358. doi: 10.1182/blood-2010-10-312454. [DOI] [PubMed] [Google Scholar]
- 18.Hardinghan T. Proteoglycans and Glycosaminoglycans. 2006. Dynamics of Bone and Cartilage Metabolism. 2nd ed. Seibel M, Robins S, Bilezikian J, editors; pp. 85–98. [Google Scholar]
- 19.Iozzo RV, Sanderson RD. Proteoglycans in cancer biology, tumour microenvironment and angiogenesis. J Cell Mol Med. 2011;15:1013–1031. doi: 10.1111/j.1582-4934.2010.01236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vicente CM, da Silva DA, Sartorio PV, Silva TD, Saad SS, Nader HB, Forones NM, Toma L. Heparan Sulfate Proteoglycans in Human Colorectal Cancer. Anal Cell Pathol (Amst) 2018;2018:8389595. doi: 10.1155/2018/8389595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi S, Chung H, Hong H, Kim SY, Kim SE, Seoh JY, Moon CM, Yang EG, Oh ES. Inflammatory hypoxia induces syndecan-2 expression through IL-1β-mediated FOXO3a activation in colonic epithelia. FASEB J. 2017;31:1516–1530. doi: 10.1096/fj.201601098R. [DOI] [PubMed] [Google Scholar]
- 22.Hong H, Song HK, Hwang ES, Lee AR, Han DS, Kim SE, Oh ES. Up-regulation of syndecan-2 in proximal colon correlates with acute inflammation. FASEB J. 2019;33:11381–11395. doi: 10.1096/fj.201900561R. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Yu XJ, Zhou W, Chu YX. MicroRNA-429 inhibits the proliferation and migration of esophageal squamous cell carcinoma cells by targeting RAB23 through the NF-κB pathway. Eur Rev Med Pharmacol Sci. 2020;24:1202–1210. doi: 10.26355/eurrev_202002_20172. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z, Zhu Z, Lin Z, Luo Y, Liang Z, Zhang C, Chen J, Peng P. miR-429 suppresses cell proliferation, migration and invasion in nasopharyngeal carcinoma by downregulation of TLN1. Cancer Cell Int. 2019;19:115. doi: 10.1186/s12935-019-0831-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo C, Zhao D, Zhang Q, Liu S, Sun MZ. miR-429 suppresses tumor migration and invasion by targeting CRKL in hepatocellular carcinoma via inhibiting Raf/MEK/ERK pathway and epithelial-mesenchymal transition. Sci Rep. 2018;8:2375. doi: 10.1038/s41598-018-20258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Du L, Yang Y, Wang C, Liu H, Wang L, Zhang X, Li W, Zheng G, Dong Z. MiR-429 is an independent prognostic factor in colorectal cancer and exerts its anti-apoptotic function by targeting SOX2. Cancer Lett. 2013;329:84–90. doi: 10.1016/j.canlet.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 27.Sun Y, Shen S, Tang H, Xiang J, Peng Y, Tang A, Li N, Zhou W, Wang Z, Zhang D, Xiang B, Ge J, Li G, Wu M, Li X. miR-429 identified by dynamic transcriptome analysis is a new candidate biomarker for colorectal cancer prognosis. OMICS. 2014;18:54–64. doi: 10.1089/omi.2012.0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mo JS, Alam KJ, Kim HS, Lee YM, Yun KJ, Chae SC. MicroRNA 429 Regulates Mucin Gene Expression and Secretion in Murine Model of Colitis. J Crohns Colitis. 2016;10:837–849. doi: 10.1093/ecco-jcc/jjw033. [DOI] [PubMed] [Google Scholar]
- 29.Tukaj S. Heat Shock Protein 70 as a Double Agent Acting Inside and Outside the Cell: Insights into Autoimmunity. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21155298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoter A, Naim HY. The Functions and Therapeutic Potential of Heat Shock Proteins in Inflammatory Bowel Disease-An Update. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20215331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quaglio AE, Castilho AC, Di Stasi LC. Experimental evidence of heparanase, Hsp70 and NF-κB gene expression on the response of anti-inflammatory drugs in TNBS-induced colonic inflammation. Life Sci. 2015;141:179–187. doi: 10.1016/j.lfs.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 32.Tranter M, Helsley RN, Paulding WR, McGuinness M, Brokamp C, Haar L, Liu Y, Ren X, Jones WK. Coordinated post-transcriptional regulation of Hsp70.3 gene expression by microRNA and alternative polyadenylation. J Biol Chem. 2011;286:29828–29837. doi: 10.1074/jbc.M111.221796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y, Liu B, Dai J, Srivastava PK, Zammit DJ, Lefrançois L, Li Z. Heat shock protein gp96 is a master chaperone for toll-like receptors and is important in the innate function of macrophages. Immunity. 2007;26:215–226. doi: 10.1016/j.immuni.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krist B, Florczyk U, Pietraszek-Gremplewicz K, Józkowicz A, Dulak J. The Role of miR-378a in Metabolism, Angiogenesis, and Muscle Biology. Int J Endocrinol. 2015;2015:281756. doi: 10.1155/2015/281756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duttagupta R, DiRienzo S, Jiang R, Bowers J, Gollub J, Kao J, Kearney K, Rudolph D, Dawany NB, Showe MK, Stamato T, Getts RC, Jones KW. Genome-wide maps of circulating miRNA biomarkers for ulcerative colitis. PLoS One. 2012;7:e31241. doi: 10.1371/journal.pone.0031241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaefer JS. MicroRNAs: how many in inflammatory bowel disease? Curr Opin Gastroenterol. 2016;32:258–266. doi: 10.1097/MOG.0000000000000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wohnhaas CT, Schmid R, Rolser M, Kaaru E, Langgartner D, Rieber K, Strobel B, Eisele C, Wiech F, Jakob I, Gantner F, Herichova I, Vinisko R, Böcher WO, Visvanathan S, Shen F, Panzenbeck M, Raymond E, Reber SO, Delić D, Baum P. Fecal MicroRNAs Show Promise as Noninvasive Crohn's Disease Biomarkers. Crohns Colitis 360. 2020;2:otaa003. doi: 10.1093/crocol/otaa003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dubois-Camacho K, Diaz-Jimenez D, De la Fuente M, Quera R, Simian D, Martínez M, Landskron G, Olivares-Morales M, Cidlowski JA, Xu X, Gao G, Xie J, Chnaiderman J, Soto-Rifo R, González MJ, Calixto A, Hermoso MA. Inhibition of miR-378a-3p by Inflammation Enhances IL-33 Levels: A Novel Mechanism of Alarmin Modulation in Ulcerative Colitis. Front Immunol. 2019;10:2449. doi: 10.3389/fimmu.2019.02449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng M, Zhu L, Li L, Kang C. miR-378 suppresses the proliferation, migration and invasion of colon cancer cells by inhibiting SDAD1. Cell Mol Biol Lett. 2017;22:12. doi: 10.1186/s11658-017-0041-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui Z, Liu QL, Sun SQ, Jiao K, Liu DR, Zhou XC, Huang L. MiR-378a-5p inhibits angiogenesis of oral squamous cell carcinoma by targeting KLK4. Neoplasma. 2020;67:85–92. doi: 10.4149/neo_2019_190306N191. [DOI] [PubMed] [Google Scholar]
- 41.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao Q, Kang Y, Wang HY, Guan WJ, Li XC, Jiang L, He XH, Pu YB, Han JL, Ma YH, Zhao QJ. Expression profiling and functional characterization of miR-192 throughout sheep skeletal muscle development. Sci Rep. 2016;6:30281. doi: 10.1038/srep30281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shang G, Mi Y, Mei Y, Wang G, Wang Y, Li X, Li Y, Zhao G. MicroRNA-192 inhibits the proliferation, migration and invasion of osteosarcoma cells and promotes apoptosis by targeting matrix metalloproteinase-11. Oncol Lett. 2018;15:7265–7272. doi: 10.3892/ol.2018.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boros É, Csatári M, Varga C, Bálint B, Nagy I. Specific Gene- and MicroRNA-Expression Pattern Contributes to the Epithelial to Mesenchymal Transition in a Rat Model of Experimental Colitis. Mediators Inflamm. 2017;2017:5257378. doi: 10.1155/2017/5257378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ji D, Jiang L, Li Y. MiR-192-5p suppresses the growth of bladder cancer cells via targeting Yin Yang 1. Hum Cell. 2018;31:210–219. doi: 10.1007/s13577-018-0201-6. [DOI] [PubMed] [Google Scholar]
- 46.Ast V, Kordaß T, Oswald M, Kolte A, Eisel D, Osen W, Eichmüller SB, Berndt A, König R. MiR-192, miR-200c and miR-17 are fibroblast-mediated inhibitors of colorectal cancer invasion. Oncotarget. 2018;9:35559–35580. doi: 10.18632/oncotarget.26263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao H, Chen J, Kong X, Zhu H, Zhang Y, Dong H, Wang J, Ren Q, Wang Q, Chen S, Deng Z, Chen Z, Cui Q, Zheng J, Lu J, Wang S, Tan J. miR-192/215-5p act as tumor suppressors and link Crohn's disease and colorectal cancer by targeting common metabolic pathways: An integrated informatics analysis and experimental study. J Cell Physiol. 2019;234:21060–21075. doi: 10.1002/jcp.28709. [DOI] [PubMed] [Google Scholar]
- 48.Flammang I, Reese M, Yang Z, Eble JA, Dhayat SA. Tumor-Suppressive miR-192-5p Has Prognostic Value in Pancreatic Ductal Adenocarcinoma. Cancers (Basel) 2020;12 doi: 10.3390/cancers12061693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang YL, Li XH, Ma H, Yue HY, Hu XY. Metabolites of intestinal microflora upregulate miR-192-5p to suppress proliferation of colon cancer cells via RhoA-ROCK-LIMK2 pathway. Eur Rev Med Pharmacol Sci. 2020;24:1794–1806. doi: 10.26355/eurrev_202002_20357. [DOI] [PubMed] [Google Scholar]
- 50.Olén O, Erichsen R, Sachs MC, Pedersen L, Halfvarson J, Askling J, Ekbom A, Sørensen HT, Ludvigsson JF. Colorectal cancer in Crohn's disease: a Scandinavian population-based cohort study. Lancet Gastroenterol Hepatol. 2020;5:475–484. doi: 10.1016/S2468-1253(20)30005-4. [DOI] [PubMed] [Google Scholar]
- 51.Olén O, Erichsen R, Sachs MC, Pedersen L, Halfvarson J, Askling J, Ekbom A, Sørensen HT, Ludvigsson JF. Colorectal cancer in ulcerative colitis: a Scandinavian population-based cohort study. Lancet. 2020;395:123–131. doi: 10.1016/S0140-6736(19)32545-0. [DOI] [PubMed] [Google Scholar]
- 52.Gillen CD, Walmsley RS, Prior P, Andrews HA, Allan RN. Ulcerative colitis and Crohn's disease: a comparison of the colorectal cancer risk in extensive colitis. Gut. 1994;35:1590–1592. doi: 10.1136/gut.35.11.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No additional data are available.