Abstract
The Id subfamily of helix-loop-helix (HLH) proteins plays a fundamental role in the regulation of cellular proliferation and differentiation. The major mechanism by which Id proteins are thought to inhibit differentiation is through interaction with other HLH proteins and inhibition of their DNA-binding activity. However, Id proteins have also been shown to interact with other proteins involved in regulating cellular proliferation and differentiation, suggesting a more widespread regulatory function. In this study we demonstrate functional interactions between Id proteins and members of the Pax-2/-5/-8 subfamily of paired-domain transcription factors. Members of the Pax transcription factor family have key functions in regulating several developmental processes exemplified by B lymphopoiesis, in which Pax-5 plays an essential role. Id proteins bind to Pax proteins in vitro and in vivo. Binding occurs through the paired DNA-binding domain of the Pax proteins and results in the disruption of DNA-bound complexes containing Pax-2, Pax-5, and Pax-8. In vivo, Id proteins modulate the transcriptional activity mediated by Pax-5 complexes on the B-cell-specific mb-1 promoter. Our results therefore demonstrate a novel facet of Id function in regulating cellular differentiation by functionally antagonizing the action of members of the Pax transcription factor family.
Members of the Id subfamily of helix-loop-helix (HLH) proteins play important roles in promoting cell cycle entry, enhancing apoptosis, stimulating proliferation, and blocking cellular differentiation (reviewed in references 21, 28, and 30). The founding member of this subfamily, Idl, was originally identified as a protein that inhibits the DNA-binding activity of basic HLH (bHLH) proteins (4). Subsequently, three further genes that encode the related proteins Id2 (5, 41), Id3 (8, 11), and Id4 (35) were identified. Like Id1, the other Id proteins (Id2, Id3, and Id4) also inhibit DNA binding by bHLH proteins (reviewed in references 21, 28, and 30). Mechanistically, the Id proteins are thought to inhibit bHLH proteins by sequestering them in inactive heterodimers which are incapable of DNA binding due to the absence of the basic region in the Id proteins (4, 41; reviewed in references 21, 28, and 30). In addition to their association with bHLH transcription factors, Id proteins have also been shown to interact with several non-HLH proteins, including the retinoblastoma protein (pRB) and related pocket proteins (19, 22, 23), MIDA1 (20, 38), and, more recently, members of the TCF subfamily of ETS-domain transcription factors (48). Id proteins inhibit DNA binding by the TCF proteins through interaction with their ETS DNA-binding domains. This interaction also leads to the dissociation of TCFs from ternary TCF-SRF-SRE complexes and hence to the inhibition of c-fos promoter activity (48).
A subset of ETS-domain transcription factors, including Elk-1, can also form ternary complexes with the paired-domain transcription factor Pax-5 and the B-cell-specific mb-1 promoter (15). In this case, Pax-5, rather than SRF, serves to recruit the ETS-domain proteins to the promoter. Pax-5 is a member of a subfamily of Pax proteins which also contains Pax-2 and Pax-8 (reviewed in references 25 and 40). This subfamily is characterized by the presence of an octapeptide motif and a partial homeodomain in addition to the N-terminal paired DNA-binding domain. Pax-5 plays an important role in regulating B-cell development (reviewed in references 7 and 27). Several target genes have been identified, which are either up-regulated (mb-1, N-myc, and LEF-1) or down-regulated (PD-1) in keeping with the observation that Pax-5 can function as both a transcriptional activator and repressor. In the case of the mb-1 and LEF-1 genes, the paired domain of Pax-5 is sufficient to up-regulate their expression (31).
As Id proteins are also expressed during B-cell development and function as negative regulators of B lymphopoiesis (9, 41, 42, 44), we tested whether Id proteins could affect the activity of ETS-domain protein complexes that form on the mb-1 promoter. By analogy with the ternary complex that forms on the c-fos SRE, it was expected that Id-mediated dissociation of the ETS-domain protein component might be observed. However, DNA binding by Pax-5, in addition to that by Elk-1, is inhibited upon addition of Id proteins to ternary Pax-5–Elk-1–mb-1 complexes. Id proteins bind directly to Pax-5 in vitro and in vivo, and this leads to down-regulation of the activity of Pax-5–Elk-1–mb-1 complexes in vivo. Other members of the Pax-2/-5/-8 subfamily are also targets of the Id proteins. Collectively, our data reveal a novel facet of Id function in regulating the activity of an additional class of transcription factors, the Pax proteins.
MATERIALS AND METHODS
Plasmid constructs.
The following plasmids were used for expressing glutathione S-transferase (GST) fusion and C-terminal six-histidine-tagged proteins in Escherichia coli. pAS413 encodes full-length Elk-1 (amino acids 1 to 428) (24). pGEX-Id2 encodes GST-Id2 (Id2 amino acids 1 to 134 fused to GST) (12). pAS801 [encoding GST-Pax-5(1-149)] was constructed by PCR amplification from pREP4-BSAP (2; kindly provided by Ben Adams) using the oligonucleotide pair ADS578-ADS579, followed by ligation as a BglII-EcoRI fragment into the same sites of pGEX-2T (Pharmacia). pGEX-Id2ΔHLH was constructed by PCR and comprises the N-terminal 43 amino acids and C-terminal 69 amino acids from Id2 fused to GST.
The following plasmids were used in mammalian cell transfections. pmb-1-CAT (pAS1101) contains two copies of part of the mb-1 promoter (−95 to −58) upstream from the chloramphenicol acetyltransferase (CAT) gene and was constructed by ligating two copies of the annealed oligonucleotide pair ADS580 and ADS581 (5′-TCGACGAGTAAGGGCCACTGGAGCCCATCTCCGGCACGGC-3′ and 5′-TCGAGCCGTGCCGGAGATGGGCTCCAGTGGCCCTTACTCG-3′, respectively) into the SalI-XhoI sites of pBLCAT5 (pAS311) (6). pAS383 encodes full-length Flag epitope-tagged Elk-1 driven by a cytomegalovirus (CMV) promoter (46). pRSV.Elk-1-VP16 (pAS348) encodes full-length Elk-1 fused to the VP16 transcriptional activation domain (34). pAS1111 encodes full-length Pax-5 driven by a CMV promoter (26; kindly provided by Peter Gruss). pAS1120 (encoding CMV-driven Flag epitope-tagged full-length Pax-5) was constructed by ligating a HindIII fragment from pAS1106 into pcDNA3 cut with the same enzymes. pAS1106 (encoding T3-promoter driven Flag epitope-tagged full-length Pax-5) was constructed by lighting an XbaI-XhoI-cleaved PCR fragment (primer pair ADS582-ADS583 on the template pREP4-BSAP) into the same sites in pAS798 (43).
The following plasmids were used for in vitro transcription and translation and/or transfection purposes. pBluescript-NF-κB(p50) (encoding the p50 subunit of NF-κB) was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health, from Gary Nabel and Neil Perkins. pcDNA3Id1 (encoding full-length Id1; amino acids 1 to 148), pcDNA3Id2 (encoding full-length Id2; amino acids 1 to 134), pcDNA3Id3 (encoding full-length Id3; amino acids 1 to 119) (26), pAS68 [encoding MEF2A(1-86)] (39), pAS522 (encoding full-length MEF2A) (47), and pT7.SAP-1 (encoding full-length SAP-1; amino acids 1 to 431) (10) have been described previously.
pAS803 (encoding full-length Pax-5; amino acids 1 to 391) was constructed by ligating a HindIII-XbaI-cleaved PCR fragment (primers ADS448 and ADS449 and template pREP4-BSAP) into the same sites in pAS37 (36). pAS1106, pAS1107, pAS1113, and pAS1114 (encoding full-length Pax-5 [amino acids 1 to 391], Pax-5 amino acids 1 to 175, Pax-2 amino acids 1 to 175, and Pax-8 amino acids 1 to 175, respectively, fused to a C-terminal six-histidine tag and Flag tags) were constructed by ligating XbaI-XhoI-cleaved PCR fragments (primers ADS582-ADS583, ADS582-ADS584, ADS608-ADS609, and ADS610-ADS611 and templates pREP4-BSAP, pREP4-BSAP, pCMVPax2, and pCMVPax8, respectively) into the same sites in pAS798 (43). Pax-5 constructs used in this study were derived from human cDNAs, whereas Pax-2 and Pax-8 constructs were derived from mouse cDNAs.
Protein expression.
In vitro-translated proteins, GST fusion proteins, and six-histidine-tagged proteins were purified, and their concentrations were determined as described previously (48). Pax-5(1-175) was generated by in vitro transcription and translation using NaeI-linearized pAS803 as a template. For use in gel retardation assays, Pax-5(1-149) was cleaved from the GST moiety with thrombin while still attached to the reduced glutathione agarose beads (1).
In vitro protein-protein interaction assays.
Interactions between GST-Id2 and in vitro-translated Pax-2/-5/-8 derivatives or control proteins were investigated using pulldown assays as previously described (39).
Gel retardation assays.
Gel retardation assays were performed with 32P-labeled probes as described previously (37). The binding sites used include the mb-1 site for Pax-containing complexes (ADS576-ADS577 top strand, 5′-CGCGTGAGTAAGGGCCACTGGAGCCCATCTCCGGCACGG-3′) and the N10 site for MEF2A (36). DNA-protein complexes were formed at room temperature for 15 min using in vitro-translated or recombinant, bacterially expressed Pax-2/-5/-8 derivatives, in vitro-translated MEF2A(1-86), and bacterially expressed full-length Elk-1. In vitro-translated Id proteins were usually added at the same time as the Pax proteins or after the end of this incubation period in dissociation experiments. Reaction mixtures were normalized for reticulocyte lysate content. Protein-DNA complexes were analyzed on nondenaturing 5% polyacrylamide gels cast in 0.5× Tris-borate-EDTA buffer and visualized by autoradiography and phosphorimaging.
Cell culture, transfection, and reporter gene assays.
NIH 3T3 and Cos-7 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Gibco BRL) and 25 mM glucose. Transfection experiments were carried out using Superfect (Qiagen) as described previously (48) or by electroporation (PD31 cells). For reporter gene assays, mb-1 promoter-driven reporters were cotransfected alongside vectors encoding Pax-5, Elk-1-VP16, and Id2. DNA concentrations were normalized with appropriate empty vectors.
Extracts were prepared from transfected cells, and CAT-luciferase assays were carried out as previously described (24, 48). Results were normalized for equal concentrations of total protein. Data from CAT assays were quantified by phosphorimager analysis, and the data were presented graphically using Microsoft Excel software. Transfection efficiency was monitored by measuring the β-galactosidase activity from cotransfected pCH110 plasmid (0.5 μg) (Pharmacia KB Biotechnology Inc.), and β-galactosidase activities were determined as described previously (48).
Immunoprecipitation.
The antibody matrix was prepared by covalently coupling an Id3-specific rabbit polyclonal antibody (Santa Cruz Biotechnology Inc.) to protein A beads. Cos-7 cell extracts containing overexpressed Id3 and Flag-tagged Pax-5 or Elk-1 proteins were prepared from two 35-mm-diameter dishes in 400 μl of Triton lysis buffer (20 mM Tris [pH 7.4], 137 mM sodium chloride, 25 mM β-glycerophosphate, 50 mM sodium fluoride, 2 mM EDTA, 10% glycerol, 1% Triton X-100, 2 mM sodium pyrophosphate, 10 mM MgCl2) containing protease inhibitors (final concentrations: leupeptin, 2 μg/ml; pepstatin A, 1 μg/ml; phenylmethylsulfonyl fluoride, 100 μg/ml; and aprotinin, 2 μg/ml). A 25-μl sample of antibody matrix was incubated with the cell extract with rotation for 4 h at 4°C. Complexes were washed three times with Triton lysis buffer and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Western blot analysis. Immunoprecipitated Flag epitope-tagged proteins and Id3 were detected by immunoblot analysis with a mouse monoclonal anti-M2 Flag antibody (Kodak) and polyclonal Id3 antibody (Santa Cruz) followed by enhanced chemiluminescence (Amersham) as described previously (48). Endogenous Id3–Pax-5 complexes were coimmunoprecipitated from approximately 108 Nalm-6 pre-B cells after lysis in 20 ml of Triton lysis buffer (above) supplemented with 0.4 M NaCl. Immunoprecipitates were prepared by incubating the lysate with 10 μg of anti-Id3 antibody immobilized on protein A-Sepharose and then were subjected to Western analysis with Pax-5 antibody (sc-1974; Santa Cruz)
Figure generation.
Figures were generated electronically from scanned images of autoradiographic images by using Picture Publisher (Micrografix) and Powerpoint 7.0 (Microsoft) software. Results are representative of the original autoradiographic images.
RESULTS
Id3 inhibits the formation of binary and ternary DNA-bound Pax-5 complexes.
Id proteins have previously been demonstrated to cause the dissociation of the TCF component of the ternary TCF-SRF-SRE complex that forms on the c-fos promoter (48). A subset of the TCFs, among other ETS-domain proteins, form ternary complexes with Pax-5 on the B-cell-specific mb-1 promoter (15). We therefore investigated whether the Id proteins could cause the dissociation of the TCF Elk-1 from this ternary complex.
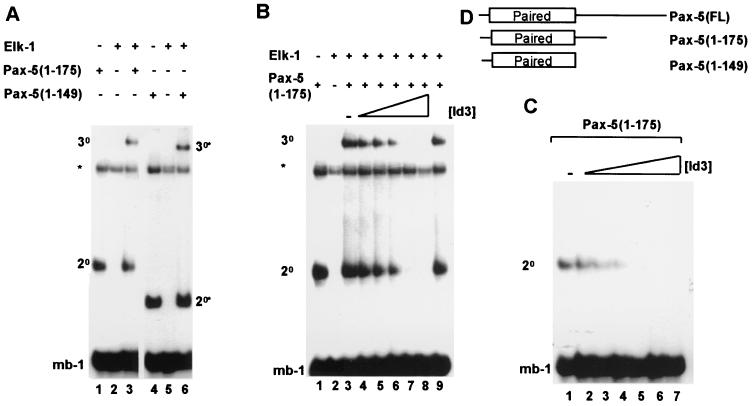
Elk-1 is unable to efficiently bind to the mb-1 site on its own (Fig. 1A, lane 2) but can be recruited by a C-terminally truncated derivative of Pax-5, Pax-5(1-175) (Fig. 1A, lane 3). Similarly, the truncated Pax-5 derivative Pax-5(1-149) is also able to recruit Elk-1 to the mb-1 site (Fig. 1A, lanes 4 to 6) (15). Pax-5(1-149) contains just the paired domain and lacks additional C-terminal sequences (Fig. 1D). In the results presented below, the experiments were subsequently carried out with in vitro-translated Pax-5(1-175). However, virtually identical results were produced using recombinant Pax-5(1-149).
FIG. 1.
Id3 inhibits Pax-5-mediated complex formation on the mb-1 promoter. Gel retardation analysis of complex formation by Pax-5 on the mb-1 promoter in the presence of Elk-1 and/or Id3. The locations of binary Pax-5–DNA (2°) and ternary Pax-5–Elk-1–DNA (3°) complexes are indicated. Asterisks indicate the position of a complex arising from the reticulocyte lysate. (A) Complex formation on the mb-1 site by in vitro-translated Pax-5(1-175) (lanes 1 and 3), recombinant Pax-5(1-149) (lanes 4 and 6), and full-length recombinant Elk-1 (lanes 2, 3, 5, and 6). (B) Ternary complex formation by Pax-5(1-175) and full-length Elk-1 and the mb-1 site in the presence of increasing amounts of in vitro-translated Id3 (shown schematically over lanes 3 to 8; relative molar ratios, 0, 1, 2, 4, 8, and 16, respectively). (C) Binary complex formation by Pax-5(1-175) in the presence of increasing amounts of in vitro-translated Id3 (shown schematically over lanes 1 to 7; relative molar ratios, 0, 1, 2, 4, 8, 12, and 16, respectively). (D) Diagrammatic representation of the Pax-5 derivatives used in this study. The location of the N-terminal paired-box DNA- binding domain is indicated.
To investigate a possible role of Id3 in regulating complex formation on the mb-1 site, increasing amounts of Id3 were added to DNA-binding reactions containing Elk-1 and Pax-5(1-175) (Fig. 1B, lanes 3 to 8). As the concentration of Id3 increased, the amount of the ternary Elk-1–Pax-5–mb-1 complex decreased to below detectable levels. However, unexpectedly, the amount of binary Pax-5–mb-1 complex was also reduced to a comparable degree by increasing amounts of Id3. This dose-dependent reduction in Pax-5 DNA binding by increasing concentrations of Id3 was also observed in the absence of Elk-1 (Fig. 1C, lanes 1 to 7).
Together, these results indicate that Id3 is able to inhibit the formation of ternary Elk-1–Pax-5–mb-1 complexes, and a major target for this inhibitory activity is the paired-box DNA- binding domain of Pax-5.
Specificity of Id-mediated inhibition of nucleoprotein complex formation by Pax-5.
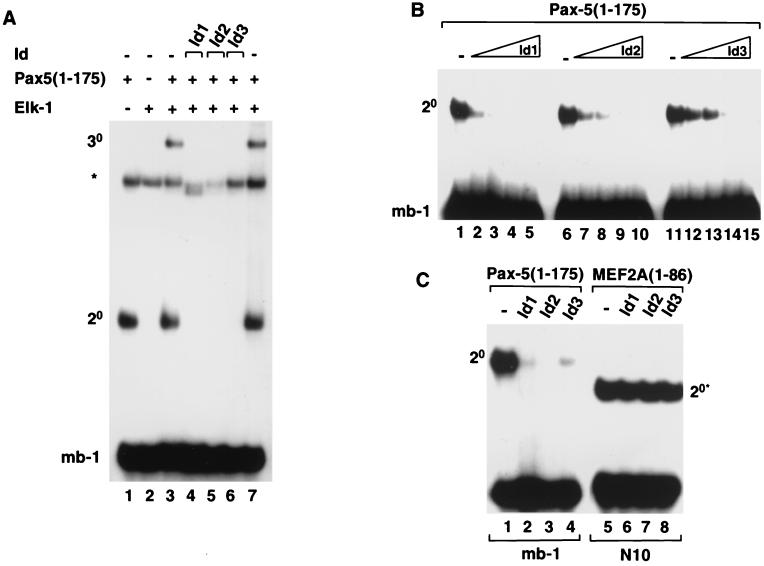
In addition to Id3, the related proteins Id1 and Id2 are also expressed in B cells (9) and might therefore have a role in regulating Pax-5 binding to the mb-1 promoter. We therefore tested whether these additional Id proteins inhibit DNA binding by binary Pax-5 and ternary Elk-1–Pax-5-containing complexes. As observed with Id3 (Fig. 1B and 2A, lane 6), both Id1 and Id2 inhibit the formation of both binary and ternary complexes containing Pax-5 (Fig. 2A, lanes 4 and 5, respectively). In order to compare the relative abilities of Id1, Id2, and Id3 to inhibit DNA binding by Pax-5, the amount of each Id protein added to the DNA-binding reactions was varied (Fig. 2B). Of these proteins, Id1 was the most potent (lanes 1 to 5), while Id3 (lanes 11 to 15) and Id2 (lanes 6 to 10) were both less efficient at inhibiting DNA binding by Pax-5. Subsequently, to test whether the inhibitory effect of the Id proteins was specific, we compared the ability of the Id's to inhibit DNA binding by Pax-5 and the MADS-box transcription factor MEF2A. In contrast to the effect of the Id proteins on DNA binding by Pax-5, no inhibition of DNA binding by MEF2A was observed upon addition of Id1, Id2, or Id3 (Fig. 2C).
FIG. 2.
Id1, Id2, and Id3 inhibit Pax-5-mediated complex formation on the mb-1 promoter. Gel retardation analysis of complex formation by Pax-5 on the mb-1 promoter in the presence of Elk-1 and/or Id1, Id2, or Id3. The locations of binary Pax-5–DNA (2°) and ternary Pax-5–Elk-1–DNA (3°) complexes are indicated. The asterisk indicates the position of a complex arising from the reticulocyte lysate. (A) Ternary complex formation by in vitro-translated Pax-5(1-175) and full-length recombinant Elk-1 and the mb-1 site in the absence (lanes 3 and 7) or presence of equimolar amounts (with respect to each Id protein) of each of the in vitro-translated Id proteins, Id1 (lane 4), Id2 (lane 5), and Id3 (lane 6). (B) Binary complex formation by Pax-5(1-175) in the presence of increasing amounts of in vitro-translated Id1 (lanes 1 to 5), Id2 (lanes 6 to 10), and Id3 (lanes 11 to 15). The relative molar ratios of Id's added (0, 1, 2, 4, and 8, respectively) are shown schematically above each set of lanes. (C) Id proteins do not inhibit DNA binding by MEF2A. Binary complex formation by equimolar amounts of in vitro-translated Pax-5(1-175) (lanes 1 to 4) and MEF2A(1-86) (lanes 5 to 8) and the mb-1 and N10 sites, respectively, in the absence (lanes 1 and 5) or presence of equimolar amounts (with respect to each Id protein) of each of the in vitro-translated Id proteins, Id1 (lanes 2 and 6), Id2 (lanes 3 and 7), and Id3 (lane 4 and 8). For all reactions, the volumes of reticulocyte lysate added to each lane were equalized by adding appropriate volumes of unprogrammed lysate.
Collectively, these experiments demonstrate that Id1, Id2, and Id3 inhibit DNA binding by Pax-5 with relative efficiencies of Id1 > Id2 > Id3 but do not affect the binding of a different transcription factor, MEF2A.
The Id proteins inhibit DNA binding by all members of the Pax-2/-5/-8 subfamily.
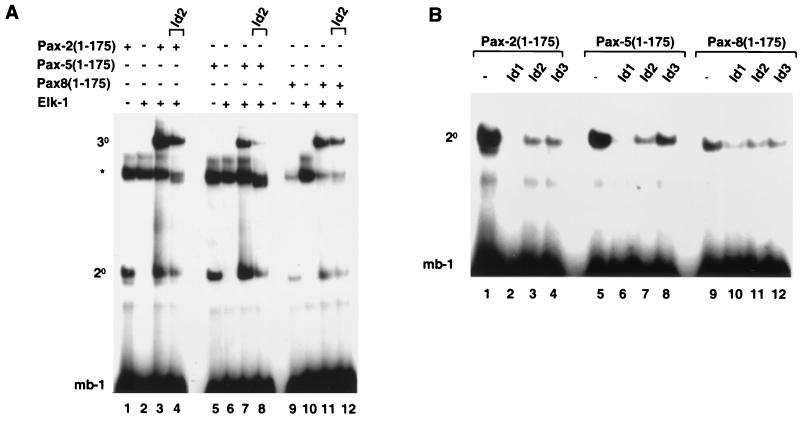
Two other proteins, Pax-2 and Pax-8, exhibit a high degree of sequence and functional similarity to Pax-5, which is especially pronounced within the paired DNA- binding domains (40). The ability of the Id proteins to inhibit nucleoprotein complex formation by Pax-2 and Pax-8 was therefore investigated. All three members of the Pax-2/-5/-8 subfamily can bind to the mb-1 site, albeit with reduced efficiency in the case of Pax-8 (Fig. 3A, lanes 1, 5, and 9). Similarly, all three Pax proteins can form ternary complexes with Elk-1 on this element (Fig. 3A, lanes 3, 7, and 11). Furthermore, upon addition of Id2, a reduction in the efficiency of binary and ternary complex formation involving each Pax protein could be observed (Fig. 3A, lanes 4, 8, and 12). Of the proteins tested, the efficiency of inhibition by Id2 increased, Pax-8 < Pax-2 < Pax-5, with Pax-8 complexes being inhibited the least.
FIG. 3.
Id proteins inhibit nucleoprotein complex formation by all members of the Pax-2/-5/-8 subfamily of Pax proteins. Shown is a gel retardation analysis of complex formation by Pax-2, Pax-5, and Pax-8 on the mb-1 promoter in the presence of Elk-1 and/or Id1, Id2, or Id3. The locations of binary Pax-DNA (2°) and ternary Pax–Elk-1–DNA (3°) complexes and the position of a complex arising from the reticulocyte lysate (asterisk) are indicated. (A) Complex formation by in vitro-translated Pax-2(1-175) (lanes 1 to 4), Pax-5(1-175) (lanes 5 to 8), and Pax-8(1-175) (lanes 9 to 12) and full-length recombinant Elk-1 (lanes 2, 3, 4, 6, 7, 8, 10, 11, and 12) and the mb-1 site. Equal molar ratios of each Pax protein were used. In vitro-translated Id2 was added to the binding reaction mixtures in lanes 4, 8, and 12. (B) Binary complex formation by Pax-2(1-175) (lanes 1 to 4), Pax-5(1-175) (lanes 5 to 8), and Pax-8(1-175) (lanes 9 to 12) in the presence of equimolar amounts (with respect to each Id protein) of in vitro-translated Id1 (lanes 2, 6, and 10), Id2 (lanes 3, 7, and 11), and Id3 (lanes 4, 8, and 12). For all reactions the volumes of reticulocyte lysate added to each lane were equalized by adding appropriate volumes of unprogrammed lysate.
The ability of the different Id proteins Id1, Id2, and Id3 to inhibit DNA binding by Pax-2, Pax-5, and Pax-8 was compared directly (Fig. 3B). All three Id proteins inhibit DNA binding by each Pax protein. However, differences in the efficiency of inhibition are apparent, with Pax-8 being inhibited to a lesser extent by each of the Id proteins. It is also noted that Id1 appears to have the greatest inhibitory effect and Id3 the least on all three Pax proteins. These results therefore demonstrate that DNA binding by all members of the Pax-2/-5/-8 subfamily is disrupted by the Id proteins and that all three Id proteins possess this inhibitory activity.
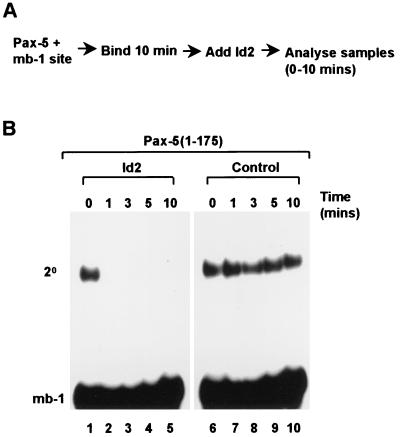
Id2 causes rapid dissociation of Pax-5–DNA complexes.
DNA binding by Pax-5 is inhibited upon prior incubation with Id proteins. To investigate whether Id proteins can dissociate preformed DNA-bound Pax-5 complexes, a time course experiment was performed in which Id proteins were added to binding reactions once they had reached equilibrium, and samples were subsequently analyzed over a 10-min time period (Fig. 4A). Rapid dissociation of the Pax-5–DNA complex was observed within 1 min of Id2 addition (Fig. 4B, lane 2), whereas no change in the intensity of the Pax-5–DNA complex could be observed in the absence of added Id2 over the same time period (Fig. 4B, lanes 7 to 10). Id2 is therefore capable of causing rapid dissociation of Pax-5–DNA complexes.
FIG. 4.
Id2 causes rapid disruption of Pax-5–DNA complexes. (A) Schematic representation of the experimental protocol used. (B) Gel retardation analysis of binary (2°) complex formation by Pax-5(1-175) and the mb-1 promoter in the presence (lanes 1–5) or absence (lanes 6–10) of added Id2. Equal amounts of rabbit reticulocyte lysate were added to each tube. The times at which samples were withdrawn following Id2 addition to the binding reaction mixture are indicated above each lane.
Id proteins interact with Pax-5 in vitro and in vivo.
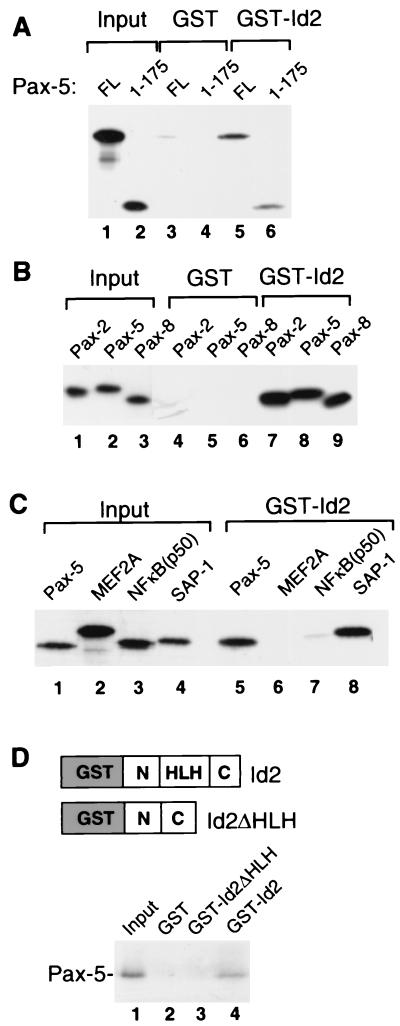
As the Id proteins inhibit DNA binding by Pax-5 and other members of the Pax-2/-5/-8 subgroup, it is likely that the two classes of protein interact directly. To test whether these proteins do indeed interact, the binding of in vitro-translated Pax-5 to a GST-Id2 fusion protein was investigated (Fig. 5A). Both full-length Pax-5 (lane 5) and Pax-5(1-175) (lane 6) bind to GST-Id2, indicating that the paired-box DNA-binding domain of Pax-5 is sufficient for binding to Id2. This is consistent with the DNA-binding studies, which demonstrate that the paired box is the target for the inhibitory activity of the Id proteins. Subsequently, the binding of Pax-2 and Pax-8 to GST-Id2 was investigated (Fig. 5B). As observed with Pax-5 (lane 8), both Pax-2 (lane 7) and Pax-8 (lane 9) also bind to Id2. In order to test whether the interaction of the Id proteins with Pax proteins was specific, we compared the ability of Id2 to bind to Pax-5, SAP-1 (a known interaction partner) (48), and two members of alternative transcription factor families, MEF2A and NF-κB(p50) (Fig. 5C). Both Pax-5 and SAP-1 bind efficiently to Id2 (lanes 5 and 8). However, in contrast, little binding of MEF2A and NF-κB(p50) was observed (lanes 6 and 7), indicating that Id2 interacts specifically with members of the Pax and ETS-domain transcription factor families. To further demonstrate the specificity of Id binding and to map the domain requirements, we tested the role of the HLH motif in Id2 for Pax-5 binding. In contract to wild-type Id2, no binding of Pax-5 to Id2ΔHLH was observed (Fig. 5D). Thus, although Id2ΔHLH retains part of its biological activity (16), this protein is unable to interact with Pax-5, thereby demonstrating a requirement for the HLH motif.
FIG. 5.
Pax-2/-5/-8 proteins interact directly with Id proteins in vitro. (A) GST pulldown analysis of full-length (FL) Pax-5 and Pax-5(1-175) with GST (lanes 3 and 4) and a GST-Id2 fusion protein (lanes 5 and 6); 20% of the input proteins are shown in lanes 1 and 2. (B) GST pulldown analysis of Pax-2(1-175), Pax-5(1-175), and Pax-8(1-175) with GST (lanes 4 to 6) and a GST-Id2 fusion protein (lanes 7 to 9); 10% of the input proteins are shown in lanes 1 to 3. (C) GST pulldown analysis of full-length Pax-5, MEF2A, NF-κB(p50), and SAP-1 with a GST-Id2 fusion protein (lanes 5 to 8); 10% of the input proteins are shown in lanes 1 to 4. (D) GST pulldown analysis of Pax-5(1-175) with the indicated GST-Id2 fusion proteins. The structures of these proteins are shown diagrammatically (above), and 10% of the input proteins are shown in lane 1.
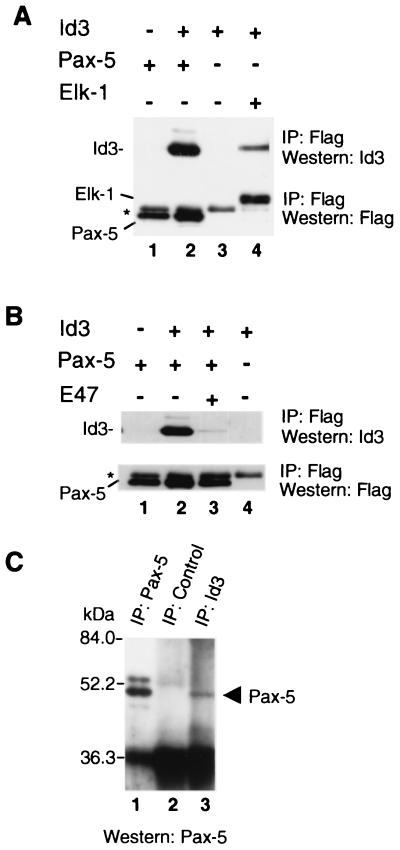
To investigate the interaction of Pax-5 and Id proteins in vivo, Cos-7 cells were cotransfected with Id3 and either Flag epitope-tagged Pax-5 or Elk-1 as a positive control. Subsequently, protein complexes were isolated by immunoprecipitation with anti-Flag antibody, followed by detection of the expressed Flag-tagged protein or coprecipitated protein by Western blotting (Fig. 6A). When either Pax-5 or Id3 was transfected alone, very little Id3 was precipitated with the anti-Flag antibody (lanes 1 and 3). However, when Pax-5 and Id3 were cotransfected, Id3 was coprecipitated (lane 2). Similarly, upon cotransfection of Elk-1 and Id3, Id3 could be coprecipitated by the anti-Flag antibody (lane 4) as observed previously with Elk-1 and Id2 (48). However, Id proteins have previously been shown to also interact with bHLH proteins (reviewed in references 21, 28, and 30). We therefore investigated whether the bHLH protein E47 could compete with Pax-5 for Id3 binding. Upon coexpression of E47, the amount of Id3 complexed with Pax-5 was significantly reduced (Fig. 6B; compare lanes 2 and 3). Thus, bHLH proteins and Pax proteins directly compete for Id protein binding (see Discussion). Finally, we used the pre-B cell line, Nalm-6, which expresses high levels of Id3 and Pax-5 proteins (T. Inoue and J. Norton, unpublished observations), to investigate whether endogenous Id3–Pax-5 complexes could be detected in a physiological context. Pax-5 protein was detectable in immunoprecipitates of Nalm-6 cells prepared using anti-Id3 antibody (Fig. 6C, lane 3). Cross-reactivity of anti-Pax-5 antibody with lower-molecular-weight immunoglobulin present in immunoprecipitates (as seen in Fig. 6C) prevented the reciprocal detection of Id3 protein in immune complexes prepared using anti-Pax-5 antibody (data not shown).
FIG. 6.
Pax-5 and Id3 interact in vivo. (A) Coimmunoprecipitation (IP) of Id3 with Pax-5 from Cos-7 cells. Cells were transfected with expression vectors encoding Flag-tagged Pax-5 alone (lane 1), Id3 and Flag-tagged Pax-5 (lane 2), Id3 alone (lane 3), or Id3 and Flag-tagged Elk-1 (lane 4). (B) bHLH proteins and Pax proteins compete for Id binding. Coimmunoprecipitation of Id3 with Flag-tagged Pax-5 in the presence or absence of exogenous E47 (indicated above each lane). Flag-tagged proteins were immunoprecipitated, followed by detection of the Flag-tagged protein and interacting Id3 proteins by Western blotting with the indicated antibodies. The asterisk indicates a band that cross-reacts with the Flag epitope antibody. (C) Coimmunoprecipitation of endogenous Id3 and Pax-5 from Nalm-6 pre-B cells. Samples of total cell extract (from 106 cells) and immunoprecipitates of Id3 complexes or of control serum (each from 108 cells) were analyzed by Western blotting using Pax-5 antibody.
Collectively, these results demonstrate that the Id proteins interact specifically with members of the Pax-2/-5/-8 subfamily of Pax proteins in vitro and in vivo. This interaction requires the Id HLH motif and occurs via the paired-box DNA-binding domain of the Pax proteins. The latter observation is consistent with the inhibitory effects of the Id proteins that also act on this minimal domain of Pax-5 (Fig. 1).
Id3 inhibits Pax-5-mediated transcriptional activation in vivo.
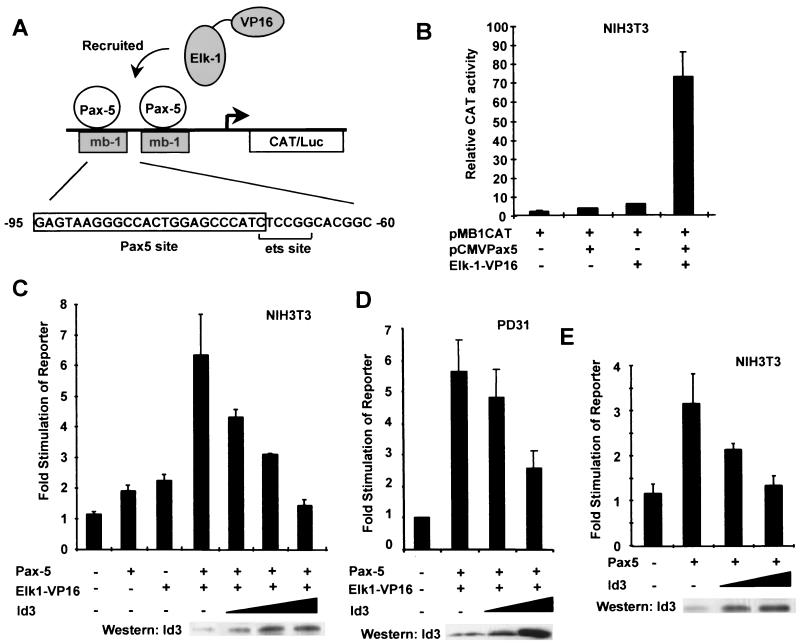
The mb-1 promoter is regulated by a ternary complex that is composed of the protein components Pax-5 and a subset of ETS-domain transcription factors (15). In order to generate a highly active ternary complex in vivo in the absence of additional regulatory cues, we used a fusion protein consisting of the ETS-domain transcription factor Elk-1 and the potent VP16 transcription activation domain. This has previously been shown to be recruited to the c-fos SRE by binding to SRF to allow high-level transcriptional activation in vivo (24, 34). In order to demonstrate that Pax-5 could act in a similar manner to recruit Elk-1–VP16 to the mb-1 site in vivo, cotransfection experiments were carried out in NIH 3T3 cells with vectors encoding Pax-5 and Elk-1–VP16 and a reporter vector containing two copies of the mb-1 site fused to a basal thymidine kinase promoter and a CAT gene (Fig. 7A). When either Pax-5 or Elk-1–VP16 was expressed alone, low levels of mb-1–CAT reporter gene induction were observed (twofold and fivefold, respectively, relative to induction by the reporter alone; Fig. 7B). In contrast, coexpression of the two proteins caused synergistic transcriptional activation (74-fold; Fig. 7B), demonstrating that Pax-5 does indeed recruit Elk-1–VP16 to the mb-1 promoter in vivo.
FIG. 7.
Id3 attenuates Pax-5-mediated activation of the mb-1 promoter. (A) Diagrammatic representation of the reporter system used, in which two copies of the indicated region from the mb-1 promoter drive the expression of a CAT or luciferase (Luc) gene. Pax-5 recruits ETS-domain proteins such as Elk-1 to this site (15). (B) Coexpression of Pax-5 and Elk-1–VP16 causes synergistic activation of the mb-1 promoter-regulated CAT reporter. NIH 3T3 cells were cotransfected with the mb-1–CAT reporter vector (1 μg) and with expression vectors encoding either Pax-5 (pAS1111; 0.25 μg) or Elk-1–VP16 (pAS348; 0.25 μg) alone or Pax-5 and Elk-1–VP16 together (0.25 μg each). (C) Id3 inhibits activation of the mb-1 reporter construct by Pax-5–Elk-1–VP16 complexes. NIH 3T3 cells were cotransfected with the mb-1–CAT reporter vector (1 μg), and expression vectors encoding either Pax-5 (pAS1111; 0.1 μg) or Elk-1–VP16 (pAS348; 0.1 μg) alone or Pax-5 and Elk-1–VP16 together (0.1 μg each). Where indicated, increasing amounts of the Id3 expression vector (pcDNA3Id3; 0.5, 2, and 4 μg) were cotransfected. (D) Id3 inhibits activation of the mb-1 reporter construct by Pax-5–Elk-1–VP16 complexes in B cells. PD31 cells were cotransfected with the mb-1–luc reporter vector and with expression vectors encoding Pax-5 (pAS1111; 5 μg) and Elk-1–VP16 (pAS348; 5 μg). Where indicated, increasing amounts of the Id3 expression vector (pcDNA3Id3; 5 and 20 μg) were cotransfected. (E) Id3 inhibits activation of the mb-1 reporter construct by Pax-5. NIH 3T3 cells were cotransfected with the mb-1–CAT reporter vector (1 μg), an expression vector encoding Pax-5 (pAS1111; 0.25 μg), and increasing amounts (0.5 and 4 μg) of an Id3 expression vector (pcDNA3Id3). Data are presented relative to those of the reporter plasmid alone (taken as 1). All values and standard errors were calculated from averages of duplicate (B, D, and E) or triplicate (C) samples and are representative of two or three independent experiments. In all cases, CAT-luciferase activity was measured 24 h after transfection. Gels (C to E) show Western blots of Id3 expression following transfection with increasing amounts of the Id3 expression vector.
The ability of Id3 to inhibit the activity of the ternary Pax-5–Elk-1–VP16–mb-1 complex in vivo was tested by coexpressing increasing amounts of Id3 with a constant amount of Pax-5 and Elk-1–VP16 in NIH 3T3 cells. A decrease in the activity of the mb-1-driven reporter gene was observed upon coexpression of Id3, which decreased back to basal levels at the highest concentration of Id3 expression vector used (Fig. 7C). In order to demonstrate that the same phenomenon could be observed in B cells, the experiment was repeated in PD31 cells. Again, increasing levels of Id3 led to inhibition of the mb-1-driven reporter construct (Fig. 7D). Id proteins can also inhibit the formation of binary Pax-5–DNA complexes in vitro (Fig. 1 to 4). Transfection of a vector encoding Pax-5 alone caused a modest activation of the mb-1-driven reporter gene (Fig. 7E). However, cotransfection of increasing amounts of an Id3 expression vector causes a decrease in the activity of the reporter back to basal levels (Fig. 7E). In all cases, Western blotting demonstrated a dose-dependent increase in Id3 expression upon transfection of higher amounts of expression vector (Fig. 7C to E, lower panels).
Collectively, these data demonstrate that Pax-5-mediated activation of the mb-1 promoter is inhibited by the Id proteins in vivo and is consistent with the direct interactions observed between these proteins and the inhibitory action of the Id proteins on DNA binding by Pax-5 in vitro.
DISCUSSION
The Id proteins act as key regulators of cell fate determination (reviewed in references 21, 28, and 30). Several molecular targets for Id proteins have been identified. In this study we demonstrate that Pax transcription factors represent novel targets for Id-mediated inhibition. Id proteins bind to the paired DNA-binding domain of members of the Pax-2/-5/-8 subfamily and disrupt their ability to bind DNA. Interaction of Id proteins with Pax proteins results in the inhibition of promoters regulated by Pax complexes in vivo. As Pax proteins themselves are also involved in regulating cellular proliferation and differentiation, this study provides novel insights into mechanisms by which these processes might be controlled at the molecular level.
Molecular targets of Id proteins.
Until recently, it was thought that Id proteins function by acting in a dominant-negative manner to sequester bHLH proteins away from their DNA targets (reviewed in references 21, 28, and 30). However, while this clearly represents a major mechanism of their action, recent studies have demonstrated that Id proteins can also bind to and alter the activities of a diverse set of regulatory proteins, including pRB and other pocket domain proteins (19, 22, 23), MIDA1 (20, 38), and the TCF subfamily of ETS-domain transcription factors (48). Pax-5 and the related proteins Pax-2 and Pax-8 can now be added to this list. However, the action of the Id proteins is not pleiotropic. Id proteins do not affect DNA binding by the MADS-box transcription factors SRF (48) or MEF2A (Fig. 2C). Furthermore, Id proteins do not efficiently interact with MEF2A or NF-κB(p50) (Fig. 5C). The latter case is substantiated by the observation that the activity of NF-κB-regulated promoters is not inhibited by Id proteins (44). However, it appears increasingly likely that additional molecular targets for the Id proteins might exist.
Id proteins act to cause dissociation of both Pax and ETS-domain proteins from DNA (Fig. 1 to 4) (48) and thus act in a manner conceptually similar to that observed with bHLH proteins. However, one key difference is that both Pax and ETS-domain proteins bind DNA as monomers. Therefore, removal of an obligatory dimerization partner, as occurs for inhibition of class A-class B bHLH protein heterodimers, is not a universal mode of Id action. Instead, a more likely mode of action in the antagonism of Pax and ETS-domain proteins would involve an allosteric affect on DNA binding. Indeed, in support of this hypothesis, Id proteins can cause the rapid dissociation of preformed complexes with both ETS-domain and Pax proteins (Fig. 4) (48), arguing that they are unlikely to act simply by sterically hindering DNA binding. It is, however, possible that in an equilibrium state, Id proteins block reassociation of free Pax proteins with DNA rather than stripping the proteins off the DNA. In this scenario, Id proteins might act either sterically or allosterically. A further key question arises: how do Id proteins interact with and regulate two apparently unrelated transcription factors? However, key features shared by ETS-domain and Pax proteins are that they both bind DNA as monomers and use helix-turn-helix (HTH) motifs to present recognition helices to the DNA major groove. Furthermore, these transcription factors have been shown to exhibit overlapping DNA-binding specificities (33). Finally, Pax proteins have been shown to form complexes with a subset of ETS-domain proteins (15; this study) and recent molecular modelling indicates that the ETS-domain can potentially interact with the C-terminal HTH motif of the paired domain while binding to an overlapping region of the DNA (45). Thus, Pax-ETS-domain protein complexes might be a common theme in the developmental regulation of gene expression. In this regard, it is interesting to note that in the case of the mb-1 promoter, both components of the ternary complex can be targeted by the Id proteins, leading to potent inhibition.
All three Id proteins inhibit the activities of Pax proteins. However, of the Pax-2/-5/-8 subfamily, Pax-8 is inhibited the least. A comparison of the different Id proteins reveals that Id1 is more potent than either Id2 or Id3 (Fig. 3). Interestingly, this relative order of potency differs from the situation observed with Elk-1, in which Id2 inhibits the most, followed by Id3, with Id1 being the weakest (48). Thus, by varying the composition and abundance of different Id proteins, distinct outcomes might be expected. It has also been shown that Pax-5 functions in a dose-dependent manner and that this dosage is in part controlled by a switch from monoallelic transcription in progenitor and mature B cells to biallelic transcription in immature B cells (32). Thus, the relative stoichiometries of Id proteins and Pax-5 in different B-cell populations are likely to play a key role in determining the transcriptional outcome on Pax-5-regulated genes.
Role of Id-Pax protein interactions in vivo.
Pax-5 plays a major role in regulating B-cell development (reviewed in references 7 and 27), whereas Pax-2 and Pax-8 are thought to be involved in kidney development (reviewed in reference 25). Significantly, Id proteins are also expressed in the B-cell lineage (9, 42, 44) and their expression levels vary during the differentiation program. Furthermore, overexpression of Id1 in transgenic mice impairs B-cell development (42). This effect was attributed to direct Id-mediated antagonism of bHLH proteins, although due to the abolition of Pax-5 expression in these transgenic mice, possible effects of Id proteins on the activity of Pax-5 during later stages of B-cell development could not be inferred. Our results indicate that as the relative stoichiometries of Pax-5 and Id proteins vary during B-cell development, then the inhibitory effects of the Id proteins are likely to also vary, thus providing a subtle mechanism for regulating the activity of Pax-5 in the B-cell differentiation program. Furthermore, as Id proteins also interact with bHLH proteins, pRb, and ETS-domain proteins (reviewed in reference 28), the relative abundance of each of these is likely to play an important role in fine-tuning the response to Id proteins. This may be particularly significant for cells expressing a functional excess of Id protein in which cellular pools of bHLH E proteins, with which Id proteins interact most avidly, exist in an Id-bHLH heterodimeric state. Further complexities arise due to fluctuating levels of Id proteins during the cell cycle (3, 8, 11). This is likely to manifest in differential effects of Id-mediated inhibition of Pax protein activity at different points during cell cycle progression. In addition, cell cycle-regulated Cdk-2-dependent phosphorylation of Id proteins is known to dramatically alter their affinities and specificities of interactions with bHLH proteins (12, 19). It is currently unknown whether the same applies to Id interactions with other classes of transcription factors. However, taken together, the available data are consistent with a dynamic situation in vivo, in which regulated exchange of Id partner proteins might represent a key regulatory step.
Similarly, the interaction of Id proteins with Pax-2 and Pax-8 in kidney cells is likely to affect their activity towards key target genes such as WT-1 (13, 14, 17) and hence also kidney development. Thus, Id-Pax protein interactions are likely to be of critical importance in regulating gene expression and the decision between proliferation and differentiation in several cell lineages and developmental programs in which these genes have been implicated (25, 28, 30).
In summary, we have demonstrated physical and functional interactions between Id proteins and Pax transcription factors. Interactions with Id proteins leads to a loss of DNA binding by Pax proteins and hence to down-regulation of their target promoters. A novel role of Id proteins has therefore been uncovered in regulating the activity of Pax transcription factors, which has implications for several important cellular processes.
ACKNOWLEDGMENTS
We thank Margaret Bell, Linda Shore, Kelly Warrington, and Luke Peterson for excellent technical assistance and Katherine Stewart for secretarial assistance. We also thank Bob Liddell for DNA sequencing and Ben Adams, Peter Gruss, Neil Perkins, and Richard Treisman for reagents. We are grateful to Julie Stinson and Shen-Hsi Yang for comments on the manuscript and members of our laboratory for helpful discussions.
This work was supported by the Wellcome Trust, the UK Cancer Research Campaign (CRC), and an MRC studentship to E.C.R. A.D.S. is a Research Fellow of the Lister Institute of Preventative Medicine.
REFERENCES
- 1.Abath F G, Simpson A J G. A simple method for the recovery of purified recombinant peptides cleaved from glutathione-S-transferase-fusion proteins. BioTechniques. 1991;10:178. [PubMed] [Google Scholar]
- 2.Adams B, Dorfler P, Aguzzi A, Kozmik Z, Urbanek P, Maurer-Fogy I, Busslinger M. Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes Dev. 1992;6:1589–1607. doi: 10.1101/gad.6.9.1589. [DOI] [PubMed] [Google Scholar]
- 3.Barone M V, Pepperkok R, Peverali F A, Philipson L. Id proteins control growth induction in mammalian cells. Proc Natl Acad Sci USA. 1994;91:4985–4988. doi: 10.1073/pnas.91.11.4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 5.Biggs J R, Murphy E V, Israel M A. A human Id-like HLH protein expressed during early development. Proc Natl Acad Sci USA. 1992;89:1512–1516. doi: 10.1073/pnas.89.4.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boshart M, Kluppel M, Schmidt A, Schutz G, Luckow B. Reporter constructs with low background activity utilizing the CAT gene. Gene. 1992;110:129–130. doi: 10.1016/0378-1119(92)90456-y. [DOI] [PubMed] [Google Scholar]
- 7.Busslinger M, Urbanek P. The role of BSAP (Pax-5) in B-cell development. Curr Opin Genet Dev. 1995;5:595–601. doi: 10.1016/0959-437x(95)80028-x. [DOI] [PubMed] [Google Scholar]
- 8.Christy B A, Sanders L K, Lau L F, Copeland N G, Jenkins N A, Nathans D. An Id-related helix-loop-helix protein encoded by a growth factor-inducible gene. Proc Natl Acad Sci USA. 1991;88:1815–1819. doi: 10.1073/pnas.88.5.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper C L, Brady G, Bilia F, Iscove N N, Quesenberry P J. Expression of the Id family helix-loop-helix regulators during growth and development in the hematopoietic system. Blood. 1997;89:3155–3165. [PubMed] [Google Scholar]
- 10.Dalton S, Treisman R. Characterisation of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 1992;68:597–612. doi: 10.1016/0092-8674(92)90194-h. [DOI] [PubMed] [Google Scholar]
- 11.Deed R W, Bianchi S M, Atherton G T, Johnston D, Santibanez-Akaref M, Murphy J J, Norton J D. An immediate early gene encodes an Id-like helix-loop-helix protein and is regulated by protein kinase C activation in diverse cell types. Oncogene. 1993;8:599–607. [PubMed] [Google Scholar]
- 12.Deed R W, Hara E, Atherton G T, Peters G, Norton J D. Regulation of Id3 cell cycle function by Cdk-2-dependent phosphorylation. Mol Cell Biol. 1997;17:6815–6821. doi: 10.1128/mcb.17.12.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dehbi M, Pelletier J. PAX8-mediated activation of the wt1 tumor suppressor gene. EMBO J. 1996;15:4297–4306. [PMC free article] [PubMed] [Google Scholar]
- 14.Dehbi M, Ghahremani M, Lechner M, Dressler G, Pelletier J. The paired-box transcription factor, PAX2, positively modulates expression of the Wilms' tumor suppressor gene (WT1) Oncogene. 1996;13:447–453. [PubMed] [Google Scholar]
- 15.Fitzsimmons D, Hodsdon W, Wheat W, Maira S M, Wasylyk B, Hagman J. Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter. Genes Dev. 1996;10:2198–2211. doi: 10.1101/gad.10.17.2198. [DOI] [PubMed] [Google Scholar]
- 16.Florio M, Hernandez M C, Yang H, Shu H K, Cleveland J L, Israel M A. Id2 promotes apoptosis by a novel mechanism independent of dimerization to basic helix-loop-helix factors. Mol Cell Biol. 1998;18:5435–5444. doi: 10.1128/mcb.18.9.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fraizer G C, Shimamura R, Zhang X, Saunders G F. PAX8 regulates human WT1 transcription through a novel DNA binding site. J Biol Chem. 1997;272:30678–30687. doi: 10.1074/jbc.272.49.30678. [DOI] [PubMed] [Google Scholar]
- 18.Hara E, Hall M, Peters G. Cdk2-dependent phosphorylation of Id2 modulates activity of E2A-related transcription factors. EMBO J. 1997;16:332–342. doi: 10.1093/emboj/16.2.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iavarone A, Garg P, Lasorella A, Hsu J, Israel M A. The helix-loop-helix protein Id-2 enhances cell proliferation and binds to the retinoblastoma protein. Genes Dev. 1994;8:1270–1284. doi: 10.1101/gad.8.11.1270. [DOI] [PubMed] [Google Scholar]
- 20.Inoue T, Shoji W, Obinata M. MIDA1 is a sequence-specific DNA binding protein with novel DNA binding properties. Genes Cells. 2000;5:699–709. doi: 10.1046/j.1365-2443.2000.00362.x. [DOI] [PubMed] [Google Scholar]
- 21.Israel M A, Hernandez M-C, Florio M, Andres-Barquin P J, Mantani A, Carter J H, Julin C M. Id gene expression as a key mediator of tumour cell biology. Cancer Res. 1999;59:1726–1730. [PubMed] [Google Scholar]
- 22.Lasorella A, Iavarone A, Israel M A. Id2 specifically alters regulation of the cell cycle by tumor suppressor proteins. Mol Cell Biol. 1996;16:2570–2578. doi: 10.1128/mcb.16.6.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lasorella A, Noseda M, Beyna M, Iavarone A. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature. 2000;407:592–598. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- 24.Ling Y, Lakey J, Roberts E C, Sharrocks A D. Molecular characterisation of the B-box protein-protein interaction motif of the ETS-domain transcription factors Elk-1. EMBO J. 1997;16:2431–2440. doi: 10.1093/emboj/16.9.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mansouri A, Hallonet M, Gruss P. Pax genes and their roles in cell differentiation and development. Curr Opin Cell Biol. 1996;8:851–857. doi: 10.1016/s0955-0674(96)80087-1. [DOI] [PubMed] [Google Scholar]
- 26.Maulbecker C C, Gruss P. The oncogenic potential of Pax genes. EMBO J. 1993;12:2361–2367. doi: 10.1002/j.1460-2075.1993.tb05890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michaelson J S, Singh M, Birshtein B K. B cell lineage-specific activator protein (BSAP). A player at multiple stages of B cell development. J Immunol. 1996;156:2349–2351. [PubMed] [Google Scholar]
- 28.Norton J D. Id helix-loop-helix proteins in cell growth, differentiation and tumourigenesis. J Cell Sci. 2000;113(Pt. 2):3897–3905. doi: 10.1242/jcs.113.22.3897. [DOI] [PubMed] [Google Scholar]
- 29.Norton J D, Atherton G T. Coupling of cell growth control and apoptosis functions of Id proteins. Mol Cell Biol. 1998;18:2371–2381. doi: 10.1128/mcb.18.4.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Norton J D, Deed R W, Craggs G, Sablitzky F. Id helix-loop-helix proteins in cell growth and differentiation. Trends Cell Biol. 1998;8:58–65. [PubMed] [Google Scholar]
- 31.Nutt S L, Morrison A M, Dorfler P, Rolink A, Busslinger M. Identification of BSAP (Pax-5) target genes in early B-cell development by loss- and gain-of-function experiments. EMBO J. 1998;17:2319–2333. doi: 10.1093/emboj/17.8.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nutt S L, Vambrie S, Steinlein P, Kozmik Z, Rolink A, Weith A, Busslinger M. Independent regulation of the two Pax5 alleles during B-cell development. Nat Genet. 1999;21:390–395. doi: 10.1038/7720. [DOI] [PubMed] [Google Scholar]
- 33.Plaza S, Grevin D, MacLeod K, Stehelin D, Saule S. Pax-QNR/Pax-6, a paired- and homeobox-containing protein, recognizes Ets binding sites and can alter the transactivating properties of Ets transcription factors. Gene Expr. 1994;4:43–52. [PMC free article] [PubMed] [Google Scholar]
- 34.Price M A, Rogers A E, Treisman R. Comparative analysis of the ternary complex factors Elk-1, SAP-1a and SAP-2 (ERP/NET) EMBO J. 1995;14:2589–2601. doi: 10.1002/j.1460-2075.1995.tb07257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riechmann V, van Cruchten I, Sablitzky F. The expression pattern of Id4, a novel dominant negative helix-loop-helix protein, is distinct from Id1, Id2 and Id3. Nucleic Acids Res. 1994;22:749–755. doi: 10.1093/nar/22.5.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharrocks A D, von Hesler F, Shaw P E. The identification of elements determining the different DNA binding specificities of the MADS box proteins p67SRF and RSRFC4. Nucleic Acids Res. 1993;21:215–221. doi: 10.1093/nar/21.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharrocks A D, Gille H, Shaw P E. Identification of the amino acids essential for DNA binding and dimerization in p67SRF: implications for a novel DNA-binding motif. Mol Cell Biol. 1993;13:123–132. doi: 10.1128/mcb.13.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shoji W, Inoue T, Yamamoto T, Obinata M. MIDA1, a protein associated with Id, regulates cell growth. J Biol Chem. 1995;270:24818–24825. doi: 10.1074/jbc.270.42.24818. [DOI] [PubMed] [Google Scholar]
- 39.Shore P, Sharrocks A D. The transcription factors Elk-1 and serum response factor interact by direct protein-protein contacts mediated by a short region of Elk-1. Mol Cell Biol. 1994;14:3283–3291. doi: 10.1128/mcb.14.5.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stuart E T, Kioussi C, Gruss P. Mammalian Pax genes. Annu Rev Genet. 1994;28:219–236. doi: 10.1146/annurev.ge.28.120194.001251. [DOI] [PubMed] [Google Scholar]
- 41.Sun X-H, Copeland N G, Jenkins N A, Baltimore D. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol. 1991;11:5603–5611. doi: 10.1128/mcb.11.11.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun X H. Constitutive expression of the Id1 gene impairs mouse B cell development. Cell. 1994;79:893–900. doi: 10.1016/0092-8674(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 43.West A G, Causier B, Davies B, Sharrocks A D. DNA binding and dimerisation determinants of Antirrhinum majus MADS-box transcription factors. Nucleic Acids Res. 1998;26:5277–5287. doi: 10.1093/nar/26.23.5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson R B, Kiledjian M, Shen C P, Benezra R, Zwollo P, Dymecki S M, Desiderio S V, Kadesch T. Repression of immunoglobulin enhancers by the helix-loop-helix protein Id: implications for B-lymphoid-cell development. Mol Cell Biol. 1991;11:6185–6191. doi: 10.1128/mcb.11.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu H E, Rould M A, Xu W, Epstein J A, Maas R L, Pabo C O. Crystal structure of the human Pax6 paired domain-DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding. Genes Dev. 1999;13:1263–1275. doi: 10.1101/gad.13.10.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang S-H, Whitmarsh A J, Davis R J, Sharrocks A D. The Elk-1ETS-domain transcription factor contains a MAP kinase targeting motif. Mol Cell Biol. 1998;18:710–720. doi: 10.1128/mcb.18.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang S-H, Galanis A, Sharrocks A D. Targeting of p38 MAPKs to MEF2 transcription factors. Mol Cell Biol. 1999;19:4028–4038. doi: 10.1128/mcb.19.6.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yates P R, Atherton G, Deed R W, Norton J D, Sharrocks A D. Id helix-loop-helix proteins inhibit nucleoprotein complex formation by the TCF ETS-domain transcription factors. EMBO J. 1999;18:968–976. doi: 10.1093/emboj/18.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]