Abstract
Objective
Access to palliative care (PC) is important for many patients with uncontrolled symptom burden from serious or complex illness. However, many patients who could benefit from PC do not receive it early enough or at all. We sought to address this problem by building a predictive model into a comprehensive clinical framework with the aims to (i) identify in-hospital patients likely to benefit from a PC consult, and (ii) intervene on such patients by contacting their care team.
Materials and Methods
Electronic health record data for 68 349 inpatient encounters in 2017 at a large hospital were used to train a model to predict the need for PC consult. This model was published as a web service, connected to institutional data pipelines, and consumed by a downstream display application monitored by the PC team. For those patients that the PC team deems appropriate, a team member then contacts the patient’s corresponding care team.
Results
Training performance AUC based on a 20% holdout validation set was 0.90. The most influential variables were previous palliative care, hospital unit, Albumin, Troponin, and metastatic cancer. The model has been successfully integrated into the clinical workflow making real-time predictions on hundreds of patients per day. The model had an “in-production” AUC of 0.91. A clinical trial is currently underway to assess the effect on clinical outcomes.
Conclusions
A machine learning model can effectively predict the need for an inpatient PC consult and has been successfully integrated into practice to refer new patients to PC.
Keywords: palliative care, machine learning, decision support systems, clinical, precision medicine
INTRODUCTION
A challenging issue in modern medicine is that of aligning care decisions with patients’ personal preferences. This alignment problem is particularly true for patients with progressed stages of illness who might prefer treatment courses focusing on comfort or on the potential for remaining in their home. Several recent studies have shown that many individuals prefer such care, however, there is a large gap between what they want and what they receive.1–3 According to the National Palliative Care (PC) registry for example, less than half of all hospital admissions that could benefit from PC actually receive it, despite a large recent growth in the availability of PC providers.4
The literature identifies several factors that contribute to this gap in care received versus preferred care, including a general lack of PC providers,4 the potential for physician overoptimism,5–7 and the time frame required for a patient to make a decision to engage in PC.8 Given these challenges and the wide gap in preferred versus received care, a better method for identifying patients for PC assessment presents a clear opportunity for improving standards of care.
Naturally, many attempts have been made to address this issue. Traditional predictive models based on point scales or classical statistics were common in early years in both hospital and ICU populations and are discussed in Section II of Avati et al.9 They take a deep learning approach to inpatient populations, building a binary classifier to predict 12-month mortality based on prior year electronic health record (EHR) data including demographics, diagnoses, procedures, medications, and encounters. Jung et al10 take a similar set of predictors but consider an outpatient population and build models based on regularized logistic regression and gradient-boosted trees. These recent approaches are strong from a methodological point of view, but they use mortality as a proxy for PC need. Additionally, to the best of our knowledge, none of these models are actually implemented into the clinical workflow at their respective institutions.
When considering related works, it is important to distinguish between PC and the closely related hospice care.8,11,12 PC focuses on integrating therapeutic regimens while concurrently considering social, emotional, and spiritual dynamics of both patients and their families. In contrast, hospice care, or end-of-life care, centers on symptom control rather than curative treatment and typically focuses on patients with a life expectancy of less than 6 months.
In this work we address the PC treatment gap by building a predictive model to identify patients likely to benefit from assessment by a PC team. This technical approach leverages 3 recent advances in modern medicine: the adoption of EHRs, a growing acceptance of the role of machine learning (ML) models in clinical care pathways, and an improved informatics approach to deploying such models in clinical environments. Unlike previous approaches, we directly predict PC consult rather than using mortality as a proxy. Most importantly, we effectively deliver the model results in a production setting and have integrated its predictions into the clinical workflow of our PC team. We are hopeful that other teams can use an approach similar to the 1 described here to develop and deploy PC consult models at their own institutions.
MATERIALS AND METHODS
Base data and standardization
Our IRB-exempt quality improvement initiative consisted of a base dataset of 68 349 encounter records of 50 143 adult (age 18 years or older) patients admitted to a Mayo Clinic hospital during the 1-year period 01/01/2017–12/31/2017. We included all adult patients with a hospitalization during this period, with repeat hospitalizations treated as (conditionally) independent events. Baseline data can be found in Table 1.
Table 1.
Cohort baseline properties
| Did not receive palliative care (N = 702 814) | Received palliative care (N = 2380) | Total (N = 705 194) | P value | |
|---|---|---|---|---|
| Age | <.001 | |||
| Mean (SD) | 61.072 (17.411) | 67.881 (15.790) | 61.095 (17.410) | |
| Gender | .048 | |||
| F | 311 571 (44.3%) | 1115 (46.8%) | 312 686 (44.3%) | |
| M | 391 242 (55.7%) | 1265 (53.2%) | 392 507 (55.7%) | |
| Community Status | <.001 | |||
| Community | 343 313 (48.8%) | 1389 (58.4%) | 344 702 (48.9%) | |
| Non-community | 359 501 (51.2%) | 991 (41.6%) | 360 492 (51.1%) | |
| Admission Source | <.001 | |||
| SNF/ICF | 11 969 (1.7%) | 111 (4.7%) | 12 080 (1.7%) | |
| Non-SNF/ICF | 690 845 (98.3%) | 2269 (95.3%) | 693 114 (98.3%) | |
| Hospitalizations in past 6 months | <.001 | |||
| Mean (SD) | 0.652 (1.258) | 1.203 (1.557) | 0.654 (1.259) | |
| Hospitalizations in past 12 months | <.001 | |||
| Mean (SD) | 0.940 (1.767) | 1.708 (2.214) | 0.943 (1.769) | |
| ICU in past 6 months | <.001 | |||
| Mean (SD) | 0.080 (0.366) | 0.171 (0.482) | 0.080 (0.366) | |
| ICU in past 12 months | <.001 | |||
| Mean (SD) | 0.118 (0.486) | 0.238 (0.616) | 0.118 (0.486) | |
| ICU Transfer Rate | <.001 | |||
| Mean (SD) | 0.167 (0.373) | 0.236 (0.425) | 0.167 (0.373) | |
| Troponin | .092 | |||
| N-Miss | 583 085 | 1674 | 584 759 | |
| Mean (SD) | 0.357 (1.225) | 0.435 (1.573) | 0.357 (1.228) | |
| Bilirubin | .004 | |||
| N-Miss | 333 127 | 640 | 333 767 | |
| Mean (SD) | 1.575 (3.973) | 1.851 (4.851) | 1.576 (3.977) | |
| Albumin | <.001 | |||
| N-Miss | 446 016 | 1155 | 447 171 | |
| Mean (SD) | 3.300 (0.715) | 3.040 (0.648) | 3.299 (0.715) | |
| Anion Gap | <.001 | |||
| N-Miss | 93 156 | 72 | 93 228 | |
| Mean (SD) | 13.955 (3.381) | 14.587 (3.803) | 13.957 (3.382) | |
| Neutrophil Count | <.001 | |||
| N-Miss | 148 989 | 195 | 149 184 | |
| Mean (SD) | 7.423 (6.510) | 8.512 (7.450) | 7.427 (6.514) | |
| Metastatic Cancer and Acute Leukemia (HCC) | <.001 | |||
| Mean (SD) | 0.056 (0.290) | 0.231 (0.614) | 0.057 (0.292) | |
| Lung and Other Severe Cancers (HCC) | <.001 | |||
| Mean (SD) | 0.071 (0.296) | 0.180 (0.473) | 0.071 (0.297) | |
| Pressure Pre-ulcer Skin Changes or Unspecified Stage (HCC) | <.001 | |||
| Mean (SD) | 0.026 (0.232) | 0.072 (0.435) | 0.026 (0.233) | |
| Septicemia Sepsis Systemic Inflammatory Response Syndrome Shock (HCC) | <.001 | |||
| Mean (SD) | 0.026 (0.175) | 0.051 (0.240) | 0.026 (0.175) | |
| Congestive Heart Failure (HCC) | <.001 | |||
| Mean (SD) | 0.149 (0.474) | 0.217 (0.616) | 0.150 (0.475) | |
| Previous Palliative Care (HCC) | <.001 | |||
| Mean (SD) | 0.009 (0.095) | 0.075 (0.264) | 0.009 (0.096) | |
| Previous Palliative Note | <.001 | |||
| Mean (SD) | 0.231 (1.769) | 2.203 (6.554) | 0.238 (1.810) | |
| Days since previous Palliative Care | <.001 | |||
| N-Miss | 674 857 | 1722 | 676 579 | |
| Mean (SD) | 128.516 (150.763) | 53.834 (99.959) | 126.799 (150.205) |
Abbreviations: HCC, hierarchical condition categories; ICF, intermediate care facility; SD, standard deviation; SNF, skilled nursing facility.
Predictors and response
Predictors (also called covariates or features) were drawn from 4 major categories: patient demographics, prior utilization, comorbidities, and time varying data, such as laboratory values and current stay length. Although known to be potentially informative, some variables, including medications and vital signs, were excluded as predictors until further validation and integration performance improvement could be done for future iterations of the algorithm. A complete list of predictors used is provided in Supplementary Table S1. Comorbidity predictors were established by normalizing ICD10 diagnosis codes to the hierarchical condition categories in the EHR data, then assigning a 1 or 0 based on whether or not the patient had been given a diagnosis code for a given comorbidity within the 2 years prior to admission. Prior utilization predictors were constructed by counting recent inpatient, ICU, and PC encounters prior to admission. The response variable was constructed using time from admission until a PC consult (right-censored at time of discharge). As described in the Temporal Component and Deployment Predictions sections below, we model the response as a heterogenous Poisson process, directly predicting the rate of PC intervention per unit time. For practical use in the clinic, we convert this rate into the 7-day probability of PC consult. When this probability exceeds a triggering threshold, the PC team is notified. This choice of outcome is a distinguishing feature of this work since the bulk of the PC prediction literature (eg, 9,10) predicts mortality rather than PC intervention. Mortality as a proxy is suboptimal as many terminal outcomes are not caused by conditions amenable to PC, and many patients can benefit from a PC consult even if their short-term mortality risk is low.
Temporal component
One of the major goals of this work was to produce a time-varying model that calculates updated predictions whenever new information becomes available; this process required the following steps. Any update in predictor values resulted in the creation of a new row in our data table, with unchanged values from previous measurements carried forward. For example, if a new laboratory value was recorded for a patient, a new row in our cohort was created with all predictors equal to their previous values other than the newly recorded laboratory value. Time from admission, recorded in days, was also directly included as a time-varying predictor. Thus, each row in the dataset included all predictors describing the state of the patient, including whether or not a PC consult occurred, as well as the time period (day) during which the patient was in that state. This carry-it-forward dataset construction approach resulted in training data containing 705 194 observations from 68 349 encounters of 50 143 unique patients. Implicit in this approach is the assumption that all predictor values remain constant between observations.
Modeling approach
We consider the PC consult as a time-to-event outcome that follows a heterogeneous Poisson process (that is, a Poisson process whose rate can vary over time) with rate equal to , where is a vector of all static predictors, and is a vector of all time-varying predictors. For practical purposes, a computational approximation is used that assumes is constant during certain windows of time. As described above, any change in values resulted in the creation of a new row in our data table, with unchanged values from previous measurements carried forward and treated as constant over the given window of time. During this period of constant , the likelihood of this model is Poisson.13 That is, the outcome of PC consult or not for each row is treated as Poisson with rate equal to , where , is the corresponding length of the window of time for that row (also called the exposure). Thus, any ML model that can use Poisson likelihood can be used to estimate . In particular, Gradient Boosting Machine (GBM)14,15 has an option to allow for a loss function equal to the minus log likelihood for Poisson, where
with an additive expansion of simple trees. Thus, we treat each row as Poisson with an offset term of log in a GBM to estimate the rate of the desired Poisson process.
The GBM estimation described above was carried out via the “gbm” package implemented by Ridgeway15 in R 3.4.2.16 This approach may be less familiar than the ubiquitous Cox proportional hazards model. However, GBM brings many advantages over Cox proportional hazards, such as a robust inclusion of both nonlinearities and interactions, as well as implicit and appropriate handling of missing data. As with all tree-based algorithms, GBM can handle missing values seamlessly as separate nodes in its variable splits. This is particularly important in the current work due to the natural sparsity and informative missingness found in laboratory values and diagnosis codes.
The data set was split into 80% training and 20% test. Hyperparameters for the GBM model (ie, number of trees, shrinkage, and interaction depth) were chosen via grid search in concert with 10-fold cross-validation17,18 on the training set. The optimal hyperparameters were chosen by selecting the values that produced the highest area under the curve (AUC) for the out-of-sample predictions.18,19 These values (n.trees = 4000, shrinkage = 0.025, depth = 2) were then used to train a model on the entire training data set. Predictions were then made on the 20% test sample and these test predictions and outcomes were used to produce the cross-validation results. Lastly, a final model was fit to all available data to be used in production.
Deployment predictions
Our model directly predicts the rate of PC intervention per unit time, conditional on the current values of a patient’s predictors. In order to make the score more interpretable, we convert this rate into a 7-day probability. Specifically, given a current patient state, we compute the 7-day probability of PC consult assuming all variables remain constant except for days in hospital. We also report a categorical “low,” “medium,” or “high.” The “high” threshold was calibrated to fall in line with the average capacity of the PC service (about 12 consults a day, including consults via the traditional pathway).
Model environment
Translating our predictive model for practical use in the hospital is 1 of the key contributions of this work. To do so, we leverage model publication concepts described by Murphree et al20 and expand significantly in terms of practical connections to institutional data sources as well as effective dissemination to clinical end users. Broadly speaking there are 3 main components to our deployment architecture: a) making the model available as a web service, b) ingesting and preprocessing input data from our informatics environment, and c) effectively communicating predictions from the model to the clinical team via a graphical user interface (GUI).
Model provision
As architected in,20 the model is published as a web service21 embedded in a docker container (www.docker.com). Publishing the model as a web service means that consumer applications can use it by constructing URLs that incorporate the predictor variables, most commonly via a JSON text file, and making a web request at a specific application programming interface (API) endpoint. We used the R package “jug” v0.1.722 to conveniently create the API. Embedding the model in a docker container means that we install our trained model and all of the dependencies needed to run it into a special lightweight, portable package that can be run on any machine with a kernel-compatible operating system. Our deployment is based on Docker v17.12.1-ce with custom base containers designed by our Information Technology team to meet institutional security standards.
This web service/docker approach affords us several advantages, including streamlined and flexible consumption by downstream display applications as well as seamless transition from sandbox to enterprise environments. By publishing our model as a web service we are able to effectively decouple model development from display development—either can change independently of the other as long as the API structure is maintained. By embedding our model in a docker container we can guarantee that it will function identically in our development environment and the enterprise-hardened production environment, as well as future versions of these environments. This directly improves the reproducibility and robustness of our work.
Data ingestion and preprocessing
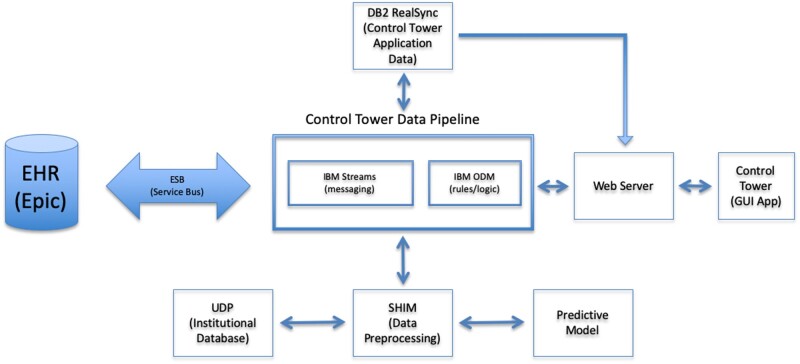
A full discussion of our hospital informatics environment is beyond the scope of this document. Briefly however, the relevant architecture consists of the following (Figure 1, glossary in Table 2). When any update to our institutional health record (www.epic.com) is made, the Epic system generates HL7 messages which are propagated institutionally across the Enterprise Service Bus (ESB). Messages on this bus are continuously monitored by the Control Tower Data Pipeline, which consists of an IBM Streams application (www.ibm.com) for reading and writing messages to the ESB and a rules manager based on IBM Operational Decision Manager (ODM) for applying clinical enrichment rules to messages. Output from ODM can consist of information important to the function and display of the Control Tower application. It can also kick off prediction processing if changes in relevant variables occur for patients being monitored. The primary information needed by the Control Tower GUI is stored in our institutional data store known as the Unified Data Platform (UDP) via FHIR APIs, with the enrichment data in an IBM DB2 database and patient state data for complex event processing in the ODM cache.
Figure 1.
Overall architecture schematic.
Table 2.
Glossary
| API | Application Programming Interface, a set of functions in our case allowing access to the predictive model |
| CT | Control Tower, a web browser application that displays information about patients, including output from the predictive model |
| docker | A docker container is a software package that includes an executable program as well as all dependencies necessary to run it |
| DP |
Control Tower data pipeline, the central actor in our informatics architecture. Reads and writes messages to the ESB via IBM Streams, applies application logic via ODM, calls the predictive model via the Model Broker (MB) |
| ESB | Enterprise Service Bus, the institution-wide pipeline for communication between applications |
| GUI | Graphical user interface |
| HL7 | Messaging standard for EHRs |
| JSON | JavaScript Object Notation, a human-readable text file format |
| ODM | Operational Decision Manager, an IBM product that applies logic or rules |
| MB | Model Broker, Data preprocessing and ingestion application. Feeds data to and from model |
| UDP | Unified Data Platform, our institution’s primary data resource |
| URL | Universal resource locator, or web address |
| web service | A piece of software made available over the internet |
When a prediction call is triggered, a predict request with patient identifier, admit date, and current time information is sent to the preprocessing environment or model broker. This Java Spring Boot application runs on a virtual server on the institution’s internal cloud. Using input from the data pipeline as well as any needed information collected from institutional data stores, the model broker assembles the data required by the model, calls the predictive model’s web service, and then returns the results to the ESB where they can be read and acted upon by the data pipeline.
All model input and output are also logged by the model broker so that production model input and output can be evaluated and compared to training results. It is also necessary to monitor the log data for drift and/or abrupt changes in time to the model predictor distribution or model performance. Currently, this is being done manually but should be automated and is a subject of further work.
Control tower application
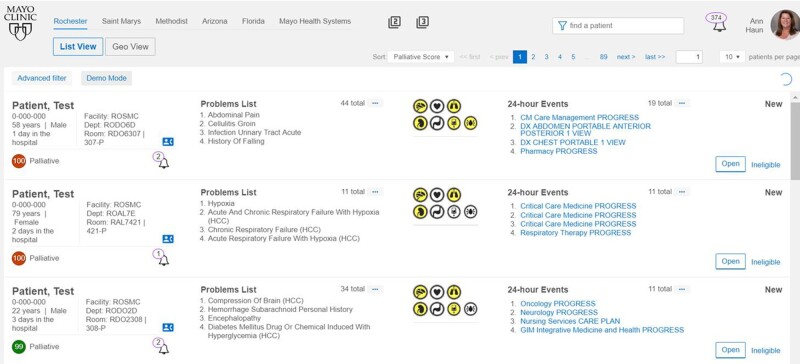
The browser-based monitoring application, known as Control Tower was developed using Angular 7. A prototypical screenshot can be seen in Figure 2. The algorithm is currently running on all inpatients in Mayo Clinic’s St. Mary’s and Methodist Hospitals in Rochester, Minnesota in an automated fashion and monitored by the PC team. Patients receive PC probability scores (0–100) from Control Tower and are subsequently ranked from highest to lowest need. In addition to the PC score, data on age, sex, problem list, hospitalization duration, vital signs, and laboratory results are available and presented to give the score some context.
Figure 2.
Screenshot of Control Tower application. This AngularJS browser-based application is monitored by a dedicated operator team. When a palliative care alert is triggered, the team considers the patient for forwarding to the PC team.
RESULTS
Predictive performance
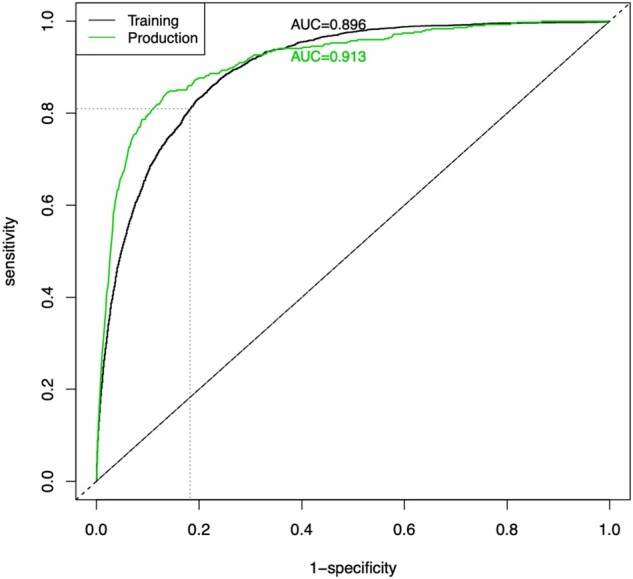
The model was evaluated using a 20% holdout set and achieves an AUC = 0.90; see Figure 3. In our clinical deployment we currently set the “high” threshold at a 7-day probability of PC equal to 0.08 (in order to match the capacity of our PC team as described above) which results in 82% specificity. At this threshold the positive predictive value is 0.19, however, that is based on historical PC consults where it was suspected that many patients who needed a PC consult did not receive 1. Out of the over 500 patients reviewed in production thus far from 11/14/19 to 01/13/20, the PC team has accepted 43% of the patients above this threshold and has rejected only 29% (28% deferred to next day).
Figure 3.
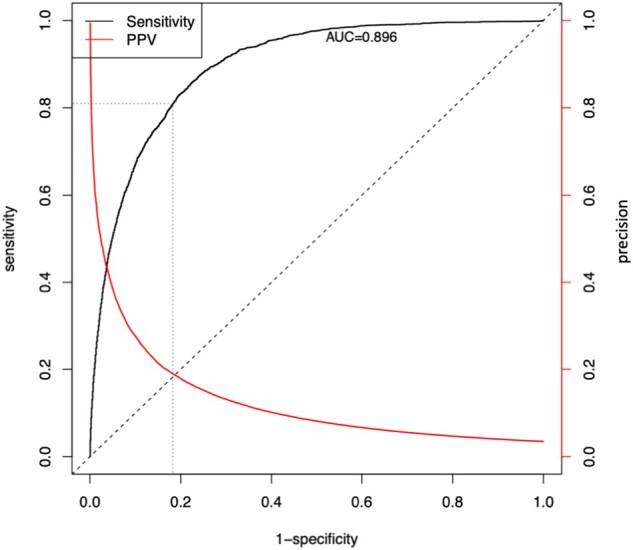
ROC curve and Positive Predictive Value (Precision) curve of the current model predictions with cross-validation. The threshold that is currently in clinical use are denoted by the fine dashed lines.
The time-varying aspect of modeling requires the receiver operating characteristic (ROC) plot in Figure 3 to be constructed nontraditionally. Recall that in a traditional ROC curve, the sensitivity and 1-specificity of a model are plotted at a series of different classification thresholds (cutoff values for turning a predicted probability into a predicted output class such as PC consult). For our model, probability of PC consult isn’t a single number but instead changes whenever a time-varying predictor changes. Furthermore, as described above in Deployment Predictions, the score that is calculated and displayed to the clinical team at any time point during an encounter is the “probability of receiving a PC consult in the next 7 days if all predictors were to remain in the current state.” Because of this, to produce the ROC plot in Figure 3 we take the maximum score a patient received during their encounter (prior to an event or discharge) and we use this maximum score as the probability needed to construct the curve. This performance assessment method is chosen over a time-dependent ROC curve or survival concordance in order to mirror use in actual practice (ie, once a patient’s score crosses a threshold at any point during their encounter, that patient will be brought to the PC team’s attention).
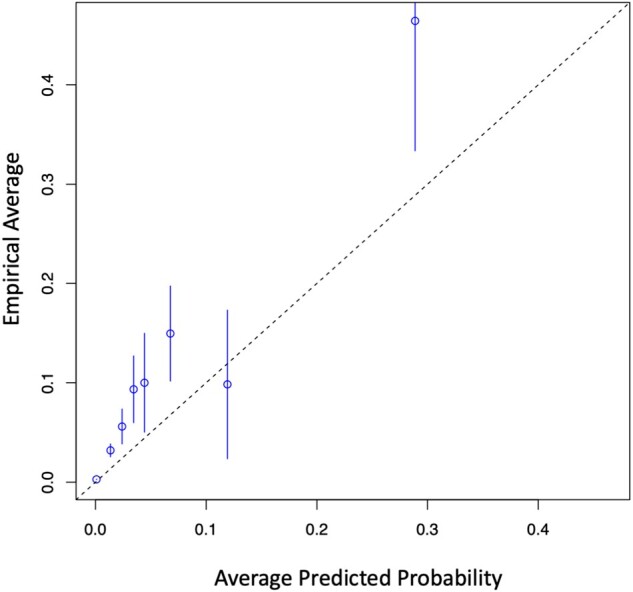
In addition to AUC, we evaluated the precision and calibration (Figure 4) of the model. Precision, also known as positive predictive value, is found on the right axis of Figure 3. One can tune the false positive rate and its inevitable effect on alert fatigue by selecting an appropriate threshold from this curve.
Figure 4.
Calibration curve of cross-validated model predictions from training.
Variable importance
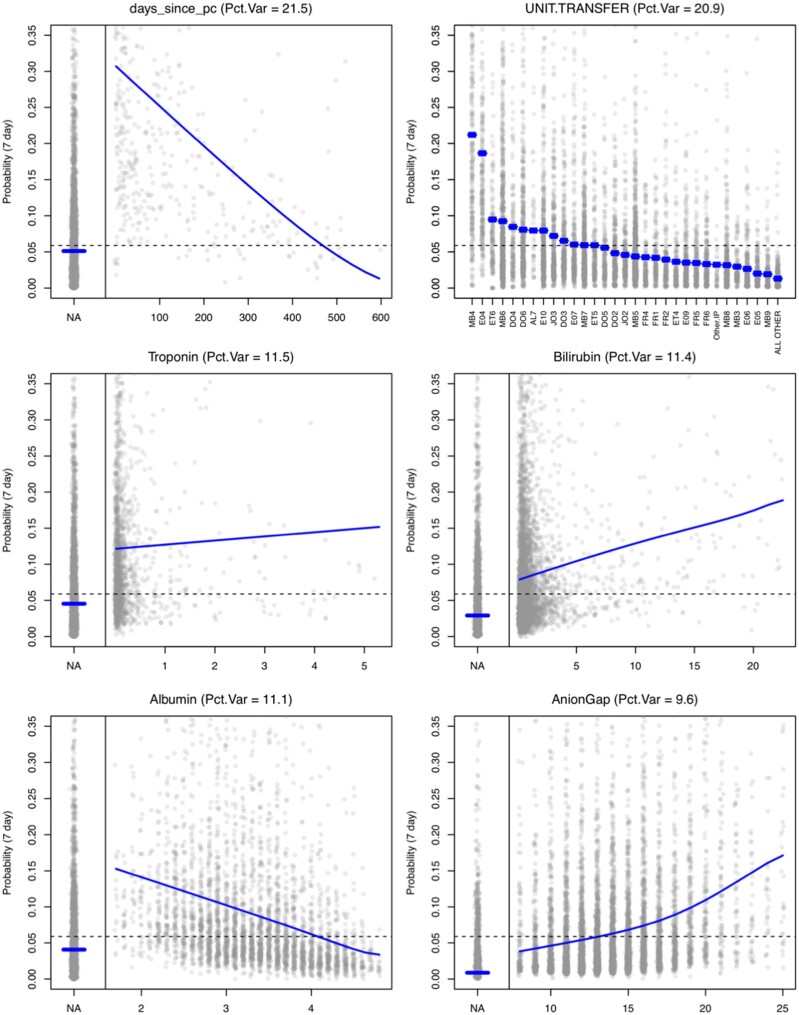
Figure 5 displays main effect plots for the 6 most informative variables. These plots are based on the predicted probability of getting a PC consult in the next 7 days. Grey points are (a sample of 10 000) model predictions displayed across the values of the respective predictor. The blue curve represents the average prediction across a particular predictor (ie, averaged over all other predictors). This main effect curve is a visual display of how important the respective predictor is (how much prediction changes across that variable). The dashed line is the population average prediction score. The proportion of variance explained by this predictor (Pct.Var at the top of each plot) is calculated as the main effect index23 of the predicted probabilities. This is the variance of the main effect curve across the marginal distribution of the respective predictor divided by the total variance of the predictions. This quantity can be interpreted as the proportion of the variability in the predictions that could be explained by this predictor alone. Supplementary Table S1 presents a list of every predictor used in the model and its variable type, along with the percentage of variance explained by each variable, sorted by contribution. The top 6 variables according to this measure are those presented in Figure 5. Supplementary Figure S1 presents the main effect plots for all predictors in the model for completeness. The most influential variable overall was days since palliative care consult. This variable is the number of days (prior to the admission date) of the most recent PC service note (it takes a value of “NA” if there is not such a note in the previous 2 years for this patient). A PC consult in the past couple weeks, followed by a hospital admission is a good indication that something may have escalated. The second variable is the hospital unit where the patient currently resides, which is a proxy for current medical need (ie, Oncology, Cardiac ICU, Transplant, etc). The next 4 are laboratory values (which are often missing), but we can see the influence that a missing value (a lab not being ordered) has on the prediction; generally, a missing lab is somewhat protective. If a clinician does not order a lab, it may indicate they are not worried about that particular lab value and its corresponding organ system.
Figure 5.
Main effect plots for the 6 most informative variables. These plots are based on the predicted probability of getting a PC consult in the next 7 days. Grey points are a sample of 10 000 model predictions displayed across the values of the respective predictor. The blue curve represents the average prediction across a particular predictor averaged over all other predictors.
The main effect curves can be used to assess individual influence on a particular prediction as well. Namely, we can simply use the difference between the overall average (dashed line) and the main effect curve for a particular value of a predictor presented by a patient. When this difference is large, then this predictor can be thought to “lift” the predicted probability higher. When a score is presented in the Control Tower GUI, a “hover over” feature also the variable influence score (ie, probability lift) for the 5 most influential variables. This can help the operator understand what the model is seeing as the reasons why this patient is a good candidate for a PC consult.
Clinical deployment
Our clinical deployment consists of a Control Tower operator who will interact with the inpatient PC consult service at Mayo Clinic’s St. Mary’s and Methodist hospitals in Rochester, Minnesota. The operator will monitor the Control Tower during weekday mornings and select daily a cohort of patients with the highest need who may benefit from PC review. In addition to assessing whether the patient is appropriate, the operator will exclude patients satisfying exclusion criteria (ie, patients who are or are about to be discharged or are currently in hospice care). The on service PC team will also assess the need for each patient; and for those patients which they agree could benefit, they will approach the attending clinical team (via a secure message, page, or phone) to suggest a PC referral.
Deployment model performance
Once the model was running in real time on production data, we inspected the model performance to ensure it was comparable to that achieved in cross-validation of the training data. Initially, production performance was poor (eg, AUC = 0.77) due to several issues, but the largest 2 problems were the following. (i) The patient population in production was different than that in training. The intended cohort was all inpatients, but the production system was providing predictions on all patients (ie, even those visiting for routine appointments or out-patient procedures, etc). (ii) Several labs had different units of measure, causing them to be inconsistent with training data. Once these problems were resolved, the production data and model results fell in line well with what was expected from the training results. Figure 6 shows the ROC curve comparison from training to production (0.90 vs 0.91).
Figure 6.
ROC curve (black) of cross-validated model predictions from training and the ROC curve (green) resulting from the model predictions on the production data.
DISCUSSION
We have built a machine learning (ML) model that can accurately identify patients likely to benefit from assessment by a PC team. Two major distinguishing features of this work are our choice of a more finely targeted outcome, namely time until PC intervention rather than a proxy, such as mortality, and our practical translation of the model to practice. Thanks to this translation, our work is already having direct impact on patient care. Although this current application targets PC, the broad objective of our program is to develop a framework for the hospital where predictive modeling, visualization tools, and alert-delivery mechanisms are able to assist clinicians in the identification of patients at risk of a variety of diagnostic delays and who may benefit from review with a specialist.
One of the largest challenges faced on this project stemmed from differences between the retrospectively collected training dataset and the streaming data the model received in production. We expect many applications of ML models to face similar challenges. This concern can be mitigated with collection of training data from exactly the same source as eventual production data.
One of the key elements in the success of this project was bringing together the necessary people and technologies required to operationalize ML models in clinical practice. Our broader team depended on a tight integration of clinical, research, informatics, and IT teams overseen by capable project management. For other teams interested in similar predictive model translation projects, it is critical to arrange buy-in from all potential project participants carefully ahead of time and plan a specific intervention and assessment.
As with any study, ours includes known weaknesses as well as areas for future improvement. One weakness is that the carry-it-forward assumption for time-varying predictors is restrictive (eg, it is clearly not the case that a patient’s hemoglobin value is constant for 3 days just because a lab was ordered on a Monday and not measured again until Thursday). In terms of future improvement, an important next step is to include vital signs and medications. Work in progress includes utilizing a patient’s history of laboratory measurements and vitals rather than just the single most recent value. Additionally, the ideal outcome of interest is whether this patient would benefit from a PC consult, but the model was trained on an outcome of historical PC consult which is not without error; namely, there are many people who could benefit from a PC consult that did not receive 1. This mismatch could potentially lead to missed detections by the algorithm, particularly if there is a systematic bias for PC consults under the current practice.
CONCLUSION
We have trained and effectively deployed into clinical workflow a ML model that can accurately and efficiently predict which adult hospital patients are likely to benefit from a PC consult. This enables providers to target patients most likely to benefit, lessening the gap between those who receive PC and those who could benefit from it. The strategy we have taken to translate our model from research to practice is both robust and effective and can serve as a blueprint for future efforts. Although the current application focuses on PC interventions, our deployment and monitoring framework is flexible and general, and the project’s ultimate goal is to have a portfolio of patient-centric predictive models to improve care.
FUNDING
DHM, PMW, SWA, GD, and CBS are supported by the Mayo Clinic Robert D. and Patricia E. Kern Center for the Science of Health Care Delivery.
AUTHOR CONTRIBUTIONS
VH and BWP conceived of the idea and, along with JS, provided critical clinical direction. CBS designed the modeling approach, led the statistical team and, with GD, performed revisions to the analysis. DHM performed the original analysis and contributed to model/data integration. PMW and SWA designed and performed model preprocessing, integration, and error control. PM, DJQ, and YL architected and developed data and pipelining infrastructure and oversaw the integration process to which DM and BW contributed. NC developed the Control Tower end user application. DHM drafted the initial manuscript, with all authors contributing to critical revision and results interpretation.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
None declared.
Supplementary Material
REFERENCES
- 1. Virdun C, Luckett T, Lorenz K, Davidson PM, Phillips J.. Dying in the hospital setting: a meta-synthesis identifying the elements of end-of-life care that patients and their families describe as being important. Palliat Med 2017; 31 (7): 587–601. [DOI] [PubMed] [Google Scholar]
- 2. Gott M, Ingleton C, Bennett MI, Gardiner C.. Transitions to palliative care in acute hospitals in England: qualitative study. BMJ Support Palliat Care 2011; 342 (1): d1773–8. [DOI] [PubMed] [Google Scholar]
- 3. Steinhauser KE, Christakis NA, Clipp EC, et al. Preparing for the end of life: preferences of patients, families, physicians, and other care providers. J Pain Symptom Manage 2001; 22 (3): 727–37. [DOI] [PubMed] [Google Scholar]
- 4. Dumanovsky T, Augustin R, Rogers M, Lettang K, Meier DE, Morrison RS.. The growth of palliative care in US hospitals: a status report. J Palliat Med 2016; 19 (1): 8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Christakis NA, Lamont EB.. Extent and determinants of error in doctors’ prognoses in terminally ill patients: prospective cohort study. BMJ 2000; 320 (7233): 469–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Selby D, Chakraborty A, Lilien T, Stacey E, Zhang L, Myers J.. Clinician accuracy when estimating survival duration: the role of the patient’s performance status and time-based prognostic categories. J Pain Symptom Manage 2011; 42 (4): 578–88. [DOI] [PubMed] [Google Scholar]
- 7. Vigano A, Dorgan M, Bruera E, Suarez-Almazor ME.. The relative accuracy of the clinical estimation of the duration of life for patients with end of life cancer. Cancer 1999; 86 (1): 170–6. [PubMed] [Google Scholar]
- 8. Morrison RS, Meier DE.. Clinical practice. Palliative care. N Engl J Med 2004; 350 (25): 2582–90. [DOI] [PubMed] [Google Scholar]
- 9. Avati A, Jung K, Harman S, Downing L, Ng A, Shah NH.. Improving palliative care with deep learning. BMC Med Inform Decis Mak 2018; 18 (S4): 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jung K, Sudat SEK, Kwon N, Stewart WF, Shah NH.. Predicting need for advanced illness or palliative care in a primary care population using electronic health record data. J Biomed Inform 2019; 92: 103115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buss MK, Rock LK, McCarthy EP.. Understanding palliative care and hospice: a review for primary care providers. Mayo Clin Proc 2017; 92 (2): 280–6. [DOI] [PubMed] [Google Scholar]
- 12. Rome RB, Luminais HH, Bourgeois DA, Blais CM.. The role of palliative care at the end of life. Ochsner J 2011; 11 (4): 348–52. [PMC free article] [PubMed] [Google Scholar]
- 13. Therneau TM, Grambsch PM.. Modeling Survival Data: Extending the Cox Model. New York: Springer; 2000. [Google Scholar]
- 14. Natekin A, Knoll A.. Gradient boosting machines, a tutorial. Front Neurorobot 2013; 7: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ridgeway G. gbm: generalized boosted regression models. R package version 2006; 1 (3): 55. [Google Scholar]
- 16.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 17. Kuhn M, Johnson K.. Applied Predictive Modeling. New York: Springer; 2013. [Google Scholar]
- 18. James G, Witten D, Hastie T, Tibshirani R.. An Introduction to Statistical Learning: With Applications in R. New York: Springer; 2013. [Google Scholar]
- 19. Kumar R, Indrayan A.. Receiver operating characteristic (ROC) curve for medical researchers. Indian Pediatr 2011; 48 (4): 277–87. [DOI] [PubMed] [Google Scholar]
- 20. Murphree DH, Quest DJ, Allen RM, Ngufor C, Storlie CB.. Deploying predictive models in a healthcare environment—an open source approach. Conf Proc IEEE Eng Med Biol Soc 2018; 2018: 6112–6. [DOI] [PubMed] [Google Scholar]
- 21. Barbaglia G, Murzilli S, Cudini S.. Definition of REST web services with JSON schema. Softw Pract Exper 2017; 47 (6): 907–20. [Google Scholar]
- 22. Smeets B. jug. Secondary jug 2017. https://cran.r-project.org/src/contrib/Archive/jug/jug_0.1.7.tar.gz Accessed May 25, 2018.
- 23. Storlie CB, Swiler LP, Helton JC, Sallaberry CJ.. Implementation and evaluation of nonparametric regression procedures for sensitivity analysis of computationally demanding models. Reliab Eng Syst Safe 2009; 94 (11): 1735–63. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.