Abstract
In order to survive subzero temperatures, some plants undergo cold acclimation (CA) where low, nonfreezing temperatures, and/or shortened day lengths allow cold-hardening and survival during subsequent freeze events. Central to this response is the plasma membrane (PM), where low temperature is perceived and cellular homeostasis must be preserved by maintaining membrane integrity. Here, we present the first PM proteome of cold-acclimated Brachypodium distachyon, a model species for the study of monocot crops. A time-course experiment investigated CA-induced changes in the proteome following two-phase partitioning PM enrichment and label-free quantification by nano-liquid chromatography-mass spectrophotometry. Two days of CA were sufficient for membrane protection as well as an initial increase in sugar levels and coincided with a significant change in the abundance of 154 proteins. Prolonged CA resulted in further increases in soluble sugars and abundance changes in more than 680 proteins, suggesting both a necessary early response to low-temperature treatment, as well as a sustained CA response elicited over several days. A meta-analysis revealed that the identified PM proteins have known roles in low-temperature tolerance, metabolism, transport, and pathogen defense as well as drought, osmotic stress, and salt resistance suggesting crosstalk between stress responses, such that CA may prime plants for other abiotic and biotic stresses. The PM proteins identified here present keys to an understanding of cold tolerance in monocot crops and the hope of addressing economic losses associated with modern climate-mediated increases in frost events.
Keywords: plasma membrane, Brachypodium distachyon, cold acclimation, freezing tolerance, nano-liquid chromatography-mass spectrometry
Introduction
Changing climatic conditions are associated with unpredictable weather patterns that can have devastating consequences on crop success (Raza et al. 2019). Higher average temperaturesand an increased frequency of winter freeze-thaw events present major challenges in temperate regions and can result in delayed bud-burst and freeze-induced injury, with acute exposure to temperatures below a thermal optimum generating chilling stress (Aroca et al. 2012; Tedla et al. 2020). Low-temperature effects include lower rates of biochemical and metabolic reactions, a loss in membrane fluidity, increased water viscosity, decreased water uptake in roots, the attenuated activity of numerous proteins and enzymes, as well as delayed energy dissipation associated with reduced photosynthesis and cellular respiration (Aroca et al. 2012). As temperatures lower further, there is the added challenge of freezing stress, resulting from the growth of extracellular ice crystals that physically damage plasma membranes (PMs), as well as cellular dehydration, which in turn results in the generation of reactive oxygens that together can ultimately lead to the collapse of membrane structures (Pearce 2001). Given the importance of cereal crops for food security, it is crucial to identify proteins associated with freeze protection in any effort to improve freezing resistance.
Prior to anthropogenic-induced climate change, plants in their native range would presumably only rarely be exposed to atypically acute and thus fatal exposure to freezing. Instead, cold acclimation (CA)—a process induced by low, nonfreezing temperatures, and/or shortened day lengths and involving epigenetic, biochemical, metabolic, and physiological changes—results in cold-hardening (Thomashow 1999, 2010; Fürtauer et al. 2019). Cold-hardened plants often accumulate unsaturated fatty acids, produce cryoprotective metabolites including soluble sugars and amino acids to mitigate osmotic stress, and elevated expression of molecular chaperons and reactive oxygen species scavengers (Suzuki and Mittler 2006). Many plants undergo CA, but the degree of their subsequent freezing tolerance is primarily determined by geographic origin, with some cold-hardy plants withstanding temperatures as low as −30°C (Lee et al. 2012; Zuther et al. 2012; Colton-Gagnon et al. 2014). Many of these species upregulate the expression of cryoprotective proteins including cold-responsive (COR) proteins, dehydrins, and ice-binding proteins (Bredow and Walker 2017; Liu et al. 2017).
Low-temperature sensing is initiated at the PM, in part by increased membrane rigidity and activation of mechanosensitive Ca2+ channels that initiate downstream signaling events key to cold tolerance (Mori et al. 2018; Yuan et al. 2018). In rice, Oryza sativa, low temperature is perceived by a PM G-protein signaling receptor, COLD1, required for Ca2+ channel activation (Ma et al. 2015). Analyses of the PM-associated proteome of thale cress, Arabidopsis thaliana, hereinafter Arabidopsis (Kawamura and Uemura 2002; Miki et al. 2019; Li et al. 2020), winter rye, Secale cereale (Uemura and Yoshida 1984), and oat, Avena sativa (Takahashi et al. 2010) suggest that the PM is also important for initiating cold-induced changes in these plants. Once cold-induced signaling has commenced, there is an accumulation of membrane-stabilizing COR proteins, a remodeling of the PM, as well as an upregulation of osmolyte synthases that help protect against dehydration-induced cell lysis (Minami et al. 2009; Takahashi et al. 2018).
Although not a crop plant, purple false brome Brachypodium distachyon (hereinafter, Brachypodium) is a model monocot with physiological similarity and high synteny with agriculturally important grasses such as rice and wheats (Triticeae). Evidence shows that Brachypodium evolved in Mediterranean regions and some cultivars tolerate low temperatures (Colton-Gagnon et al. 2014; Ryu et al. 2014; Bredow et al. 2016). Following CA, these survive to −10°C (Colton-Gagnon et al. 2014) with a diurnal freezing treatment allowing tolerance down to −12°C (Mayer et al. 2020). Here, we examine the profile of compatible solutes accumulated after CA, quantify cold-induced membrane protection, and present the first proteomic analysis of cold-acclimated (CA) Brachypodium from PM-enriched microsomal fractions in order to further advance our understanding of freeze-tolerance and the low-temperature protection in this model monocot.
Materials and methods
Plant growth and maintenance
Brachypodium seeds (ecotype: Bd21) were sown in potting soil and grown in a temperature-controlled chamber on a 20-h light (∼100 µmol m−2 s−1; 22°C) and 4 h dark (22°C) light cycle. CA Brachypodium were grown under standard conditions for 3 weeks and transferred to a low-temperature chamber (2°C, 12-h light as indicated above; 12-h dark), for 2–8 days with experimental groupings for 2, 4, 6, and 8 days designated CA2, CA4, CA6, and CA8, respectively. Nonacclimated (NA) Brachypodium were 3 weeks old at the time of use and not incubated further.
Electrolyte leakage
In order to assess the level of membrane damage associated with freezing, electrolyte leakage assays were conducted using NA or CA plants as described previously (Bredow et al. 2016). Briefly, one leaf was cut from the base of each plant, placed in 100 μL of deionized water, and immersed in a programmable circulating ethylene glycol bath set to 0°C. After lowering the temperature to −1°C over 30 min, the sample was nucleated with a single deionized ice chip to initiate ice crystal growth. The temperature of the bath was then lowered by 1°C every 15 min to a final temperature of −2 to −10°C. Samples were allowed to recover overnight at 4°C, transferred into conical tubes containing 25 mL of deionized water and then gently agitated at 150 rpm for 18 h. Conductivity measurements were taken before (Ci) and after (Cf) autoclaving samples to account for the total leaf mass, using a direct reading conductivity meter (TwinCond, Horiba, Kyoto, Japan) and presented as a percentage (100 * Ci/Cf) with 10 individual plants for each independent line, with the procedure carried out with three biological replicates.
Compatible solute profiling
Leaves [∼400 mg fresh weight (FW)] were frozen in liquid nitrogen and ground with a plastic pestle in 0.25 mL of 80% ethanol with fucose as an internal standard. Homogenates were transferred into 1.5 mL microcentrifuge tubes and incubated at 80°C for 30 min with occasional mixing. Samples were then centrifuged at 12,000 × g for 5 min at room temperature, the supernatants collected and the pellets re-extracted twice with 80% ethanol at 80°C and then centrifuged as described above. The supernatants were combined and dried under a dried N2 gas stream at 80°C. These extracts were then dissolved in methanol, followed by the addition of chloroform and water (a final ratio of chloroform:methanol:water, 1:1:0.9, v/v/v) to remove leaf pigments. After centrifugation at 600 × g for 5 min, the upper aqueous phase was collected and dried as described above. Dried samples were dissolved in water, centrifuged at 12,000 × g for 5 min, and the supernatants filtered through a 0.2 μm membrane filter and subsequently analyzed by high-pressure liquid chromatography (HPLC) using a tandem-connected Sugar KS801 and KS802 column (Shodex, Tokyo, Japan) at 80°C with a refraction index detector. The samples (50 μL) were injected and eluted with ultrapure water at a flow rate of 0.4 mL min−1. Each sugar was identified by comparison of its retention time relative to that of authentic standard sugars and quantified as a ratio of the area of the sample peak relative to that of the internal standard. All assays were replicated four times.
Microsome isolation and PM enrichment
The extraction of microsomal fractions from plant tissue and subsequent enrichment of PMs was performed as detailed previously (Kamal et al. 2020). Briefly, ∼30 g of leaf tissue was cut into small pieces in 300 mL of prechilled homogenizing medium [0.5 M sorbitol, 50 mM MOPS-KOH (pH 7.6), 5 mM EGTA (pH 8.0), 5 mM EDTA (pH 8.0), 1.5% (w/v) PVP-40, 0.5% (w/v) BSA, 2.5 mM PMSF, 4 mM SHAM, 2.5 mM DTT]. Tissue was further homogenized using a Polytron generator (PT10SK, Kinematica Inc., Lucerne, Switzerland) and filtrates were collected by sieving the homogenate through cheesecloth and removing cellular debris by centrifugation (10,000 × g for 10 min). Microsomal membrane fractions were collected by centrifugation at 231,000 × g for 35 min and homogenized in 1 mL of microsome suspension buffer (MSB; 10 mM KH2PO4/K2HPO4 buffer, pH 7.8, 0.3 M sucrose) with a teflon-glass homogenizer. Samples were centrifuged again at 231,000 × g for 35 min and the pellet was suspended in MSB (2 mL) prior to homogenization using an electric teflon-glass homogenizer.
A two-phase partition medium was produced by adding 1.45 g of polyethylene glycol (3350) and 1.45 g of dextran to 9.3 mL of MSB and 7.3 mL of NaCl medium (1.17 g of NaCl in 200 mL of MSB). The MSB homogenate was placed in the two-phase partition medium and incubated on ice for 10 min, with periodic shaking. Samples were centrifuged (650 × g for 5 min at 4°C) and the upper phase was transferred to a new partition medium, with this procedure conducted a total of three times. The upper phase was transferred to PM-suspension medium [10 mM MOPS buffer-KOH (pH 7.3), 1 mM EGTA (pH 8.0), 0.3 M sucrose, and 2 mM DTT] and centrifuged at 231,000 × g for 35 min at 4°C. The pellets were then homogenized in PM-suspension medium before repeating the 231,000 × g centrifugation. The final pellets were then homogenized on ice in 500 μL of PM-suspension medium using an electric Teflon-glass homogenizer, flash-frozen, and kept at −80°C until use. Isolations were done four times on independent plant material at each time point.
Enzyme activity assays
Verification of cell fractionation and PM enrichment was confirmed by enzyme marker assays for the presence of PM (vanadate-stimulated H+ ATPase), tonoplast (nitrate stimulated H+ ATPase), mitochondria (cytochrome c oxidase), endoplasmic reticulum (NADH cytochrome c reductase), and Golgi apparatus (Triton X-100 stimulated UDPase) in the microsomal and PM-enriched fractions as previously described (Uemura et al. 1995). In addition, chlorophyll content was determined in the two fractions to estimate the content of the thylakoid membrane of chloroplasts.
Tryptic digest and nano-liquid chromatography-mass spectrometry
Samples were electrophoresed on an e-PAGEL HRmini gel (ATTO Products, Tokyo, Japan) to remove nonproteinaceous material and excised for in-gel tryptic digestion as described previously (Takahashi et al. 2012; Kamal et al. 2020). Tryptic peptides were subsequently desalted and concentrated using a solid-phase extraction C-TIP T-300 (Nikkyo Technos Co., Tokyo, Japan). Peptide solutions were first concentrated with a trap column (L-column Micro 0.3 × 5 mm; CERI, Japan) and then separated with a Magic C18 AQ nano column (0.1 × 150 mm; MICHROM Bioresources, Auburn, CA, USA) using a linear gradient of 5–45% acetonitrile at a flow rate of 500 nL min−1. After peptides were ionized by an ADVANCE UHPLC spray source (MICHROM Bioresources, Auburn, CA, USA) at 1.8 KV spray voltage, mass analysis was performed using an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) equipped with Xcalibur software (version 2.0.7, Thermo Fisher Scientific). Full-scan mass spectra were obtained in the range of 400–1800 m/z with a resolution of 30,000. Collision-induced fragmentation was applied to the five most intense ions at a threshold above 500. Experiments for each treatment were conducted at least four times.
For semi-quantitative analysis, raw files were analyzed using Progenesis liquid chromatography-mass spectrometry (MS) software (version 4.0, Nonlinear Dynamics, New Castle, UK). Peptides were assigned to proteins by MASCOT searching (version 2.3.02, Matrix Science, London, UK) against the Brachypodium genome (version 2.0). Finally, proteins of interest (those with significantly different abundance profiles as described below) were filtered with analysis of variance (ANOVA, P < 0.05) and fold-changes (>1.5) reported according to normalized peptide intensity.
Statistical analysis
ANOVA was performed on the raw abundance levels of proteins among the four separate trials of each condition, with P-values adjusted via the Benjamini–Hochberg method (Ferreira and Zwinderman 2006) to account for multiple testing. Proteins with an absolute fold-change value >1.5 (adjusted P < 0.05) were classified as differentially abundant. For each protein that differed significantly across treatments, Dunnett’s tests were performed to determine the experimental conditions in which the protein’s abundance differed significantly from the control (NA) condition (P < 0.05; |fold-change| >1.5). Heatmaps were generated for proteins with significantly different abundance from the NA control in at least one treatment, using log2-transformed fold-change values. Proteins absent from one or more experimental conditions were not included in heatmaps. All code and scripts used in this study can be found in Supplementary File S1.
Protein functional classification and subcellular localization predictions
Descriptions were manually predicted using UniProt (UniProt Consortium), RIKEN Brachypodium FLcDNA database (Mochida et al. 2013), BLAST, and through literature searches. PM localizations were predicted using UniProt, TMHMM Server (version 2.0) for transmembrane helices (Krogh et al. 2001), GPS-lipid for N-myristoylation/-palmitylation sites, DeepLoc-1.0 (http://www.cbs.dtu.dk/services/DeepLoc-1.0/) (Almagro Armenteros et al. 2019), BUSCA (http://busca.biocomp.unibo.it/) (Savojardo et al. 2018), WolF PSORT (https://wolfpsort.hgc.jp/) (Horton et al. 2007), and known localization of orthologous plant proteins. Localization to other compartments was predicted using Uniprot (UniProt Consortium) and SignalP 5.0 (http://www.cbs.dtu.dk/services/SignalP/) (Almagro Armenteros et al. 2019) for localization to the extracellular space, mitochondria, and chloroplasts. Proteins were classified based on the functional categories as described by Bevan et al. (1998) and Miki et al. (2019).
Comparisons between the Brachypodium CA dataset (this study) and a previously reported Arabidopsis CA dataset (Miki et al. 2019) were conducted by analyzing minimum and maximum fold-changes following 2 days of CA for both species as well as following 6 days of CA for Brachypodium and 7 days of CA for Arabidopsis. A pseudocount of one was added for proteins not detected in NA conditions. Arabidopsis proteins were selected based on an ANOVA P < 0.05 and max fold-change >1.5 or min fold-change <0.67 using data provided in Supplementary Table S1 from Miki et al. (2019). Orthologous Brachypodium and Arabidopsis proteins were identified as reported in the publicly available OMA Browser (Altenhoff et al. 2021).
Construction of networks
A list of protein accession identifications for all significantly increased and decreased proteins obtained by MS was assembled and used as inputs for STRING (version 11.0) to predict protein–protein interactions (Franceschini et al. 2016; Szklarczyk et al. 2019) for CA2 and CA6 timepoints. A predicted network was prepared and exported to Cytoscape (version 3.8.1) for further modification. Additional protein metadata was input into Cytoscape including corresponding log2 fold-change values that were assigned to node fill mapping.
To construct a stress response meta-analysis network, individual protein accession identifications were subjected to literature searches (performed on January 1, 2021) and annotated according to their protein descriptions and involvement in stress response pathways (Supplementary File S2). Proteins with no reported involvement in stress responses were omitted. The dataset was then input into Cytoscape and log2 fold-changes were again selected as node fill mapping as described previously. All networks were centered in the plot area and exported as Scalable Vector Graphics files where further modification was performed and legends added in Inkscape (version 0.92.2). Interactive versions of each network were additionally exported as full webpages for viewing in any modern web browser as HTML files with all metadata.
Results
Freezing tolerance and accumulation of sugars
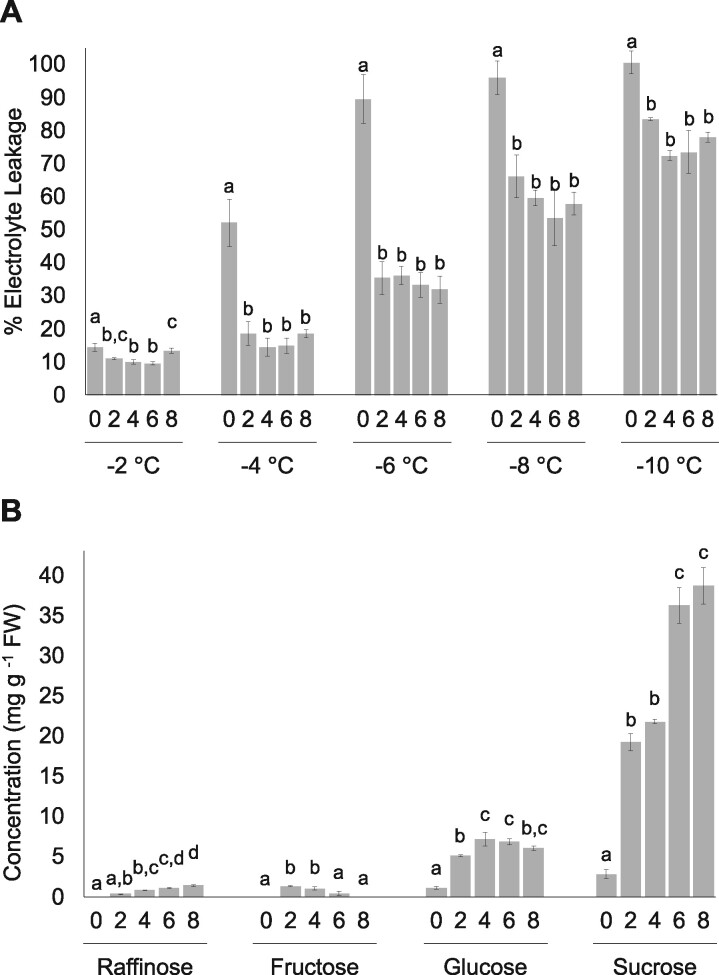
Electrolyte leakage assays were conducted to assess leaf membrane integrity of CA and NA Brachypodium following exposure to temperatures ranging from −2°C to −10°C (Figure 1A). Electrolyte leakage was significantly reduced compared to NA plants following two or more days of CA at temperatures between −4°C and −10°C. Increasing CA duration up to 28 days did not further reduce electrolyte leakage (Supplementary Figure S1). These results are consistent with earlier reports demonstrating that Brachypodium achieved peak freezing tolerance after 2 days of CA at 2°C (Colton-Gagnon et al. 2014; Bredow et al. 2016).
Figure 1.
Impact of duration of CA on B. distachyon freezing tolerance. (A) Electrolyte leakage assays conducted using leaf tissue from nonacclimated and cold-acclimated wildtype ecotype Bd21. Days of CA and temperatures are shown. Samples were subjected to the low temperatures indicated at the rate of 1°C every 15 min before assaying for electrolyte leakage (%). (B) Accumulation of soluble sugars in leaf tissue from nonacclimated and cold-acclimated wildtype ecotype Bd21. Days of CA and soluble sugar type are shown in four clusters of histograms and expressed as concentration in leaf fresh weights. Four biological replicates were conducted for all assays (n = 10) and ANOVA and post hoc Tukey’s tests were performed. Error bars represent the standard error of the mean. Letters above the histograms indicate statistically significant groups (P < 0.05) with separate analyses for each temperature and soluble sugar.
The accumulation of osmoprotectant sugars, or compatible solutes, mitigates the effects of freeze-induced cellular dehydration and is associated with increased freezing tolerance (Tarkowski and Van den Ende 2015). Here, the accumulation of sucrose increased from 2.8 mg g−1 in NA leaf tissue to 19.3 and 38.7 mg g−1 FW in CA2 and CA8, respectively (Figure 1B; Supplementary Figure S2). Glucose increased 4.6-fold to 5.1 mg g−1 in CA2 leaves and remained relatively stable for the duration of cold treatment. Raffinose showed a similar profile to sucrose with consistent increases in abundance throughout CA, reaching a maximum of 1.3 mg g−1 after 8 days. In contrast, fructose accumulation peaked in CA2 leaves (1.3 mg g−1 FW) and steadily declined in the remainder of the cold treatment groups. Thus, CA in Brachypodium is characterized by the early accumulation of glucose, raffinose, fructose, and sucrose, coincident with significantly reduced electrolyte leakage. Under longer acclimation regimes most soluble sugars increased in abundance, with the exception of fructose, and did not correspond to further changes in membrane integrity.
CA treatment time and quantifying recovered proteins
PM-associated proteins were analyzed in NA Brachypodium and following CA (CA2, CA4, CA6, and CA8). PM fractions were prepared by a method adapted for Arabidopsis (Kamal et al. 2020) with the effectiveness of fractionation and enrichment of PMs in Brachypodium validated using marker enzymes in NA and CA2 samples (Table 1). PM fractions of both NA and CA samples showed an overall decrease in mitochondria, endoplasmic reticulum, Golgi, and chloroplasts relative to the microsomal fractions. In contrast, vanadate-stimulated H+ ATPase activity, associated with the PM, showed an overall enrichment in PM fractions, with a small amount of contamination from the tonoplast, indicated by nitrogen-stimulated H+ ATPase activity (Table 1).
Table 1.
Specific activities of enzymes used as cellular compartment markers for the monitoring of the fractionation of tissue samples collected from B. distachyon either cold-acclimated (CA) for 2 days at 2°C, or non-CA (NA), with values not determined (ND) for microsomal fractions of the PM and tonoplasts; experiments were conducted four times with similar results, and as indicated by the standard deviation of the mean shown
| Compartment | Enzyme (activity units) | Microsomal fraction | PM-enriched fraction | |
|---|---|---|---|---|
| Mitochondria | Cytochrome c oxidase (μmol cyt c mg−1 min−1) | NA | 3.82 × 10−3 ± 3.27 × 10−4 | 2.64 × 10−3 ± 3.32 × 10−4 |
| CA | 4.38 × 10−3 ± 3.25 × 10−3 | 2.26 × 10−3 ± 2.58 × 10−4 | ||
| Endoplasmic reticulum | NADH cytochrome c reductase (μmol cyt c mg−1 min−1) | NA | 2.92 × 10−2 ± 1.34 × 10−2 | 1.03 × 10−3 ± 6.29 × 10−4 |
| CA | 4.17 × 10−2 ± 2.48 × 10−2 | 9.42 × 10−4 ± 5.41 × 10−4 | ||
| Golgi apparatus | Triton X-100 stimulated UDPase (μmol UDP mg−1 min−1) | NA | 4.15 × 10−1 ± 2.03 × 10−1 | 6.78 × 10−2 ± 5.93 × 10−2 |
| CA | 1.85 × 10−1 ± 3.20 × 10−2 | 5.06 × 10−2 ± 1.20 × 10−2 | ||
| Chlorophyll | Chlorophyll (mg mL−1) | NA | 1.40 ± 2.75 × 10−1 | 0.010 ± 1.16 × 10−1 |
| CA | 1.16 ± 1.20 × 10−2 | 0.0091 ± 6.00 × 10−3 | ||
| Plasma membrane | Vanadate-stimulated ɑ-H+ ATPase + Na3VO4 (μmol ATP mg−1 min−1) | NA | ND | 8.22 × 10−2 ± 3.00 × 10−2 |
| CA | ND | 5.12 × 10−2 ± 2.00 × 10−2 | ||
| Tonoplast | Nitrate stimulated ɑ-H+ ATPase + KNO3 (μmol ATP mg−1 min−1) | NA | ND | 2.52 × 10−2 ± 1.00 × 10−2 |
| CA | ND | 1.56 × 10−2 ± 2.00 × 10−2 | ||
PM-enriched fractions were subjected to nano-liquid chromatography-MS for the label-free quantification of proteins, resulting in the identification of a total of 1349 unique peptides corresponding to known proteins (Supplementary File S3). Many of these (848 or 63% of the total) showed a significant change in relative abundance (P < 0.05, |fold-change| >1.5) during cold treatment compared to NA plants. Of these proteins, 334 (39%) significantly increased >1.5-fold in relative abundance (Supplementary File S4) at one or more time points, with more (522; 62%) showing significant decreases >1.5-fold (Supplementary File S5). In line with the cold-induced membrane protection observed in electrolyte leakage assays (Figure 1), several protein families associated with cold stress were identified in our MS analysis including sugar transporters (sucrose transporter SUT1-like protein; Bradi1g73170.1; Tarkowski and Van den Ende 2015), dehydrins (dehydrin COR410-like; Bradi3g51200.1; Liu et al. 2017), and ROS scavengers (catalase; Bradi1g76330.1; Yousefi et al. 2018), and served to increase confidence in the analysis.
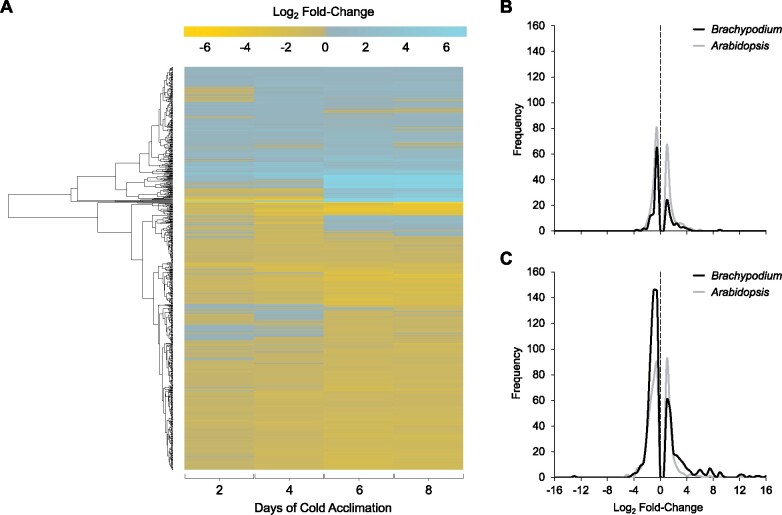
To better visualize changes in the proteome following CA, a heat map was generated using log2 transformed fold-changes at each treatment point for all proteins with significant changes in protein abundance (Figure 2A). Of the 334 proteins that showed relative increases in abundance after CA compared to NA plants, 57 (17%), 129 (39%), 224 (67%), and 205 (61%) were increased in CA2, CA4, CA6, and CA8 plants, respectively. The majority showed moderate increases over >1.5-fold compared to control levels, with some notable exceptions particularly after longer cold treatments. For example, after 2 days of CA, 7% and 3% of the proteins were one and two orders of magnitude more abundant, respectively, than in NA samples, but with higher levels achieved by more proteins at CA6. Overall, fold-changes for the 848 tracked proteins showed that the CA6 and CA8 treatment profiles were more alike than the similar CA2 and CA4 sample profiles. Using the CA2 proteome and the CA6 proteome as typifying the early and a sustained response, respectively, showed distinctive frequency distributions in the numbers of proteins that were relatively more or less abundant than proteins in the NA plants (Figure 2, B and C). Although as indicated, the relative abundance of more proteins was affected by the longer low-temperature treatment, in both CA2 and CA6 treatment groups there were about twice as many proteins that decreased vs increased in relative abundance (i.e., there were 52 and 97 proteins that increased and decreased in abundance, respectively in CA2, with corresponding numbers of 224 and 456 for CA6; Supplementary Figure S3).
Figure 2.
Global impact of CA on the B. distachyon PM proteome. (A) A heatmap showing the 848 proteins in the PM-enriched fractions with significant changes in relative abundance between nonacclimated and cold-acclimated samples following 2, 4, 6, or 8 days at 2°C. Data are mean fold-changes relative to nonacclimated plants based on four replicates and are presented as log2-transformed values. Hierarchical clustering analysis was used to group proteins displaying similar abundance proteins, and the results are presented by the dendrogram along the left-hand side of the figure. (B, C) Frequency distribution of log2 fold-changes shown for proteins with significant changes in relative abundance at (B) early CA (2 days for B. distachyon and 2 days for A. thaliana) and (C) sustained CA (6 days for B. distachyon and 7 days for A. thaliana). Log2 fold-change values of 0 are highlighted with a dotted line. Data were plotted using fold-change value bin widths of 0.5. Raw A. thaliana data were obtained from Miki et al. (2019) and reanalyzed to obtain fold-changes matching thresholds used in this manuscript of P < 0.05 and a fold-change >1.5 or <0.67 as opposed to >2.0 and <0.5 employed by Miki et al. (2019).
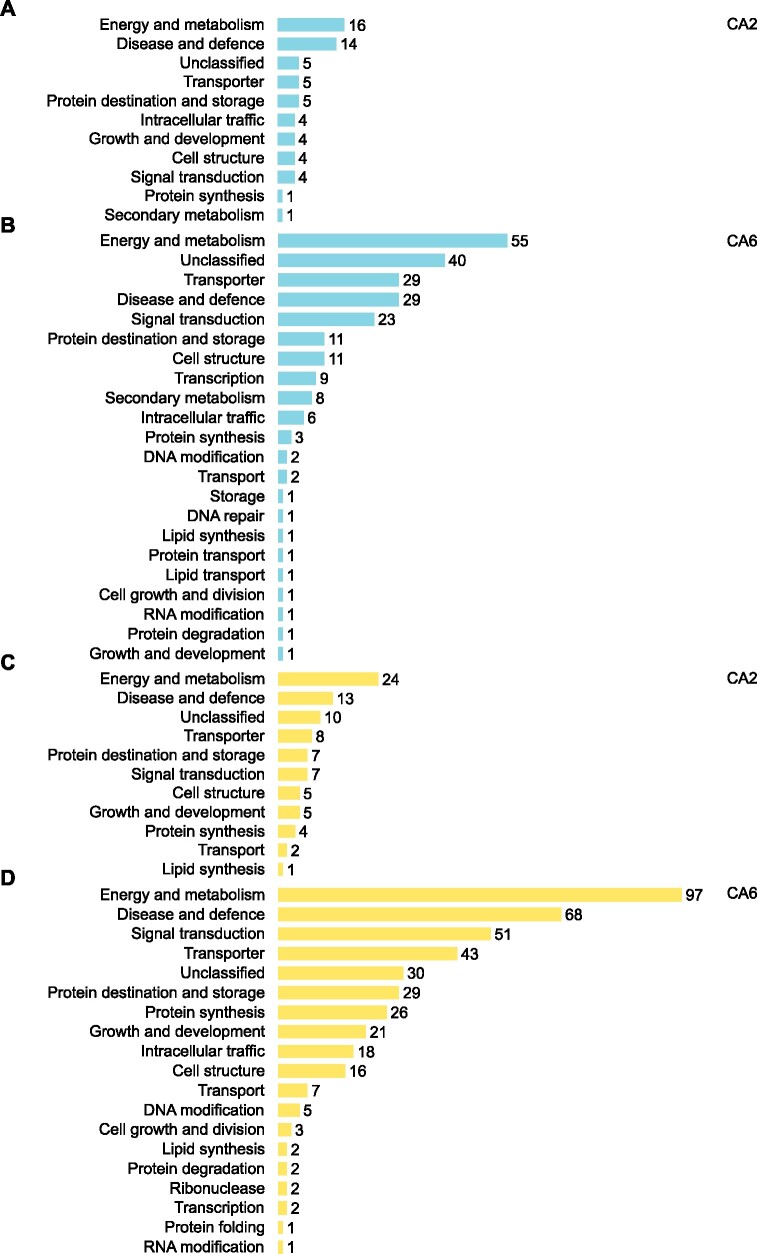
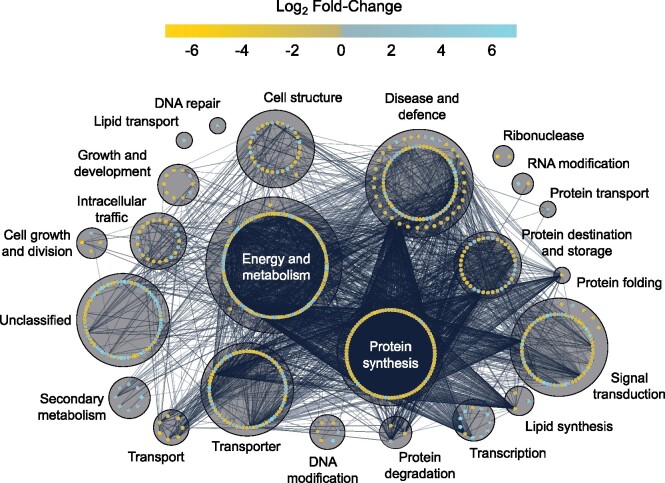
Proteins were functionally annotated and then categorized using UniProt (UniProt Consortium 2019), BLAST, and through the literature (Figures 3 and 4; Supplementary Files S6–S9). Many proteins that increased in relative abundance during CA were related to energy and metabolism (16 in CA2 and 55 in CA6 plants) such as PM ATPase (Bradi5g24690.1; Nguyen et al. 2018). They also represented disease and defense functions (14 in CA2 and 29 in CA6 plants) exemplified by dehydrin COR410-like (Bradi3g51200.1; Chiappetta et al. 2015), and transport and signal transduction affiliates (9 in CA2 and 25 in CA6 plants) such as heavy metal-associated isoprenylated plant protein 27 (Bradi1g70700.1; de Abreu-Neto et al. 2013). Proteins that increased in abundance with the greatest fold-changes are listed in Figure 3. Despite our division of an early (CA2 and CA4 plants) and a sustained response to low-temperature treatment (CA6 and CA8 plants), there were similarities in the cellular functions in both groups.
Figure 3.
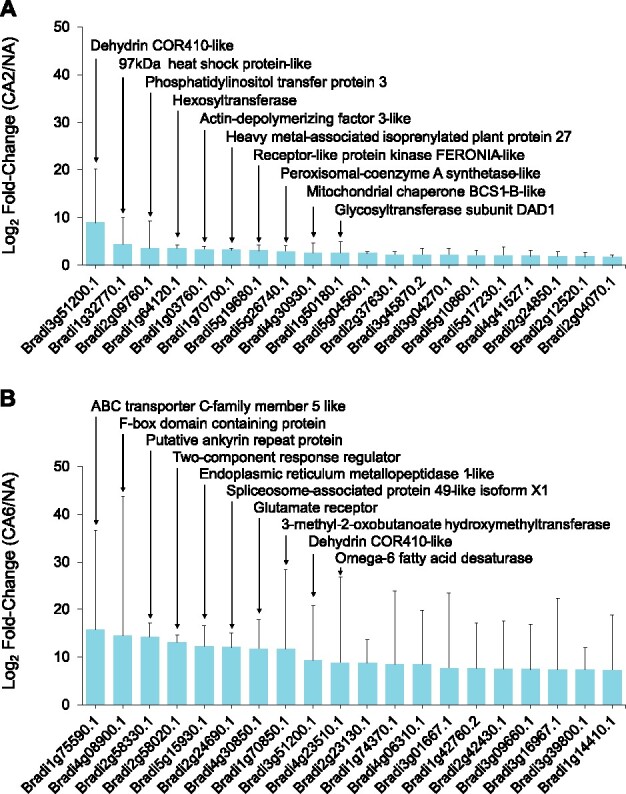
Brachypodium distachyon proteins undergoing the greatest fold increases in relative abundance during CA. Data are shown for (A) 2 days and (B) 6 days of CA. Values are the average of four replicate trials and protein annotations were predicted as previously described. Error bars represent the standard error of the mean. A pseudocount of one was added to the noncold-acclimated value for proteins that were not detected under that experimental condition. The 10 proteins with the greatest fold relative increases are labeled accordingly.
Figure 4.
Functional analysis of proteins whose relative abundance was impacted by 2 and 6 days of CA. Predicted functional categories of proteins that (A) increased in relative abundance at 2 days and (B) 6 days and (C) decreased in relative abundance at 2 days and (D) 6 days (|mean fold-change| > 1.5, P-value ≤ 0.05, based on four replicates) compared to nonacclimated controls. Functional descriptions were manually predicted using UniProt, RIKEN, Brachypodium FL cDNA database, BLAST, and literature searches. Scales are all equal.
The total of 522 proteins that decreased in relative abundance after CA when compared to NA plants showed a frequency profile that increased over time: 97 (19%), 332 (64%), 456 (87%), and 421 (81%) proteins showed significant relative decreases at CA2, CA4, CA6, and CA8, respectively (Figure 2; Supplementary Figure S3). For the most part (61%), the identified proteins showed relative decreases in abundance that were at, or just modestly greater than, 1.5-fold, but approximately 1% and <1% showed decreases that were one and two orders of magnitude greater, respectively. Proteins that decreased in relative abundance with the greatest fold-changes are listed (Supplementary Figure S4) and grouped according to functional categories (Figure 4). The most notable decreases in relative abundance were in proteins with likely functions in energy metabolism (24 in CA2 and 97 in CA6 plants), including cytochrome b5, Bradi2g62010.1 (Gou et al. 2019) and probable L-gulonolactone oxidase 4, Bradi3g13700.1 (Maruta et al. 2010). Proteins belonging to other functional classifications also decreased in relative abundance after low-temperature treatment. These included those associated with disease and defense (13 in CA2 and 68 in CA6 plants) as well signal transduction (7 in CA2 and 51 in CA6 plants). Not all of the identified proteins are known to localize to the PM, as discussed below.
Identification of proteins involved in the two-phase cold-induced response
As indicated, the numbers of significant changes in PM-associated protein relative abundance were highest at CA6, although electrolyte leakage assays indicated that CA2 plants were similarly protected from freezing damage (Figure 1A; Supplementary Figure S1). This suggests that early CA-mediated PM changes, including changes in the proteome, are sufficient for membrane protection. As indicated, the impact on the proteome could be grouped as an early (CA2 and CA4) and a sustained (CA6 and CA8) response and thus a categorical analysis of the annotated proteome focused on proteins increased in abundance in CA2 and CA6 plants to represent these two phases (Figures 2 and 4). In CA2 plants, the protein with the greatest relative increase (483-fold) was a predicted dehydrin COR410-like protein (Bradi3g51200.1), which increased an order of magnitude more than the second and third most abundantly increased proteins, a 97 kDa heat shock protein-like, Bradi1g32770.1 and a phosphatidylinositol transfer protein, Bradi2g09760.1, respectively (Figure 3A; Supplementary File S8). All of these proteins likely play important roles in ensuring membrane integrity. The accumulation of the COR410-like protein remained elevated with respect to NA plants in CA6 plants, however, eight other proteins showed greater relative fold-increases at the later time (Figure 3B). Thus, the early proteome response to low temperatures, coincident with CA-induced membrane protection, was followed by a later response that included a large number of additional proteins.
The three proteins with the greatest relative fold-increases in CA6 plants were an ABC transporter C-family 5 like protein (Bradi1g75590.1), an F-box domain-containing protein (Bradi4g08900.1), and a putative ankyrin repeat protein (Bradi2g58330.1) that showed a >19,000-fold increase in relative abundance over the level in NA plants (Figure 3B). Other proteins with large increases in relative abundance (>1.5-fold) following 6 days of CA included ROS scavengers (catalase, Bradi1g76330.1; phospholipid hydroperoxide glutathione peroxidase 1, Bradi1g47140.1), heat shock proteins or HSPs (97 kDa heat shock protein-like, Bradi1g32770.1; heat shock cognate 70 kDa protein-like, Bradi1g66590.1), sugar transporters (bidirectional sugar transporter SWEET1b-like, Bradi2g24850.1; sucrose transport protein SUT1-like, Bradi1g73170.1), dehydrins (the mentioned dehydrin COR410-like, Bradi3g51200.1; dehydrin COR410-like, Bradi5g10860.1), heavy metal-associated isoprenylated plant proteins or HIPPs (heavy metal-associated isoprenylated plant protein 27, Bradi1g70700.1; heavy metal-associated isoprenylated plant protein 20, Bradi3g04270.1), phospholipase D (Bradi4g36800.1), and a glutamate receptor (Bradi4g30850.1). As a group, these could function to support long-term survival under conditions of stress. In addition, there were five proteins that were only identified following CA, which are predicted to have roles in transcription (RRM domain-containing protein, Bradi2g24690.1; Lorković and Barta 2002), signal transduction (two-component response regulator, Bradi2g58020.1; Lohrmann and Harter 2002), defense signaling (putative ankyrin repeat protein, Bradi2g58330.1; Yang et al. 2012), and cold perception (glutamate receptor, Bradi4g30850.1; Gong et al. 2019), as well as an unclassified protein (ER metallopeptidase 1-like, Bradi5g15930.1; Marino and Funk 2012). The complete dataset of annotated CA6 increased proteins is available in Supplementary File S9.
Concomitant with the increased abundance of many PM proteins, those that decreased in relative abundance by 2 days of CA included enzymes likely required for nutrient uptake, PM H+ ATPases (PM ATPase, Bradi5g24690.1 and PM ATPase 1-like, Bradi1g54847.1; Morsomme and Boutry 2000). By CA6 some of these further decreased in relative abundance and were joined by additional PM H+ ATPases, consistent with reports from cold-stressed Arabidopsis (Muzi et al. 2016). Likewise, other PM ATPases decreased in relative abundance after 2 days (V-type ATPase, Bradi1g67960.1 and AAA+ ATPase, Bradi1g48010.1), and more after 6 days of CA (another AAA+ ATPase, Bradi1g36830.1; V-type proton ATPase subunit F, Bradi2g39610.1; and Obg-like ATPase, Bradi3g17680.1), further suggesting an early and sustained response to low-temperature exposure at least with regard to the numbers of impacted proteins. It is important to note that although many of the PM-associated proteins that decreased in relative abundance have been categorized to a single function, most notably in energy and metabolism (Figure 4), some may have other documented roles.
Protein localization algorithms (DeepLoc-1.0, BUSCA, WolF PSORT) predicted that 46% (26/57) of proteins at CA2 and 39% (88/224) of proteins at CA6 that increased in relative abundance were associated with the PM, which is not surprising given the technique designed to enrich for PM-associated proteins and highlighting the difficulty in the recovery of hydrophobic PM proteins. In contrast, 42% (41/97) of the proteins at CA2 and 38% (175/456) of those identified at CA6 that showed decreases in relative abundance were predicted to localize to the cytosol (Supplementary Files S6 and S7). Although highly abundant cytosolic proteins could have contaminated our PM preparations, some of these proteins may in fact loosely associate with the PM during normal growth and weaken as the PM is restructured in response to low temperature. Links to the PM are known to vary with reversible lipid interactions (Marmagne et al. 2004; Komatsu et al. 2007) although this remains to be experimentally determined for proteins identified in our analysis.
Crosstalk between stress response networks
Low temperatures can result in an increasing vulnerability to the effects of additional biotic and abiotic stresses in plants (Fujita et al. 2006; Rejeb et al. 2014), and thus CA regimes may result in abundance changes in seemingly unrelated stress response proteins to safeguard against the effects of combinatorial stresses. In order to gain a better understanding of the crosstalk between stress networks at the PM, a network map was constructed using stress-related proteins that increased or decreased in relative abundance compared to NA plants after 2 days of CA (Figure 5; see Supplementary File S10 for an interactive version of the network, and Supplementary File S2 for the underlying data). As expected, the cold stress node showed the largest number of interactions with other stress nodes at 41 proteins, including 20 interactions that were also associated with drought, 5 with osmotic stress, and 15 with salt stresses. These proteins included those with predicted functional categories of energy and metabolism (12), disease and defense (11), transporters (6), and signal transduction (4). A secondary cluster, the pathogen stress node with 39 proteins, showed interactions with other stress nodes, many of which overlapped with those seen with cold stress. Due to the significantly higher number of proteins, it was not practical to construct a corresponding network map for CA6 plants, but functional analysis of the proteins suggests that interactions between abiotic stresses would be retained.
Figure 5.
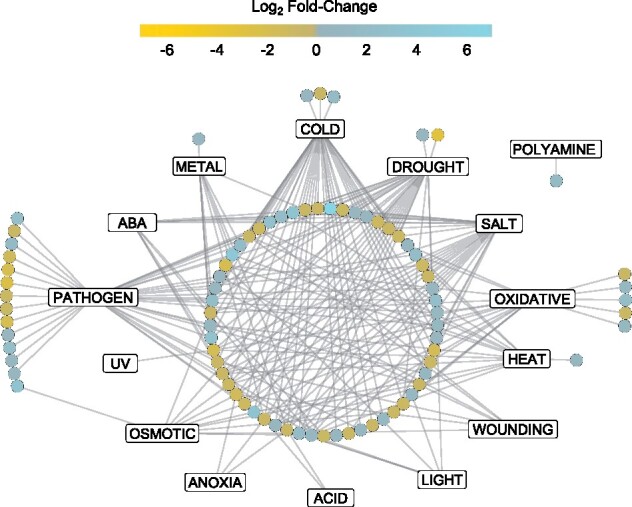
Network illustrating involved stress pathways and crosstalk between proteins significantly changed in relative abundance at 2 days of CA. Rectangle nodes represent functional protein stress response pathways while circular nodes represent individual proteins. Edges connecting protein nodes to stress response nodes represent validated functional associations from the literature. Protein function in the stress pathways was validated via Uniprot and the literature, and networks were built using Cytoscape version 3.8.1. Node color represents log2 fold-change of the protein in the cold-acclimated (2 days) vs nonacclimated PM proteomes. An interactive version of the network with encoded metadata is available in Supplementary File S10. Click on a node to see individual protein accession ID, protein description, log2 fold-change, predicted functional categories, and degrees of interaction. Click on an individual edge to see the literature for the specific protein–stress response relationship.
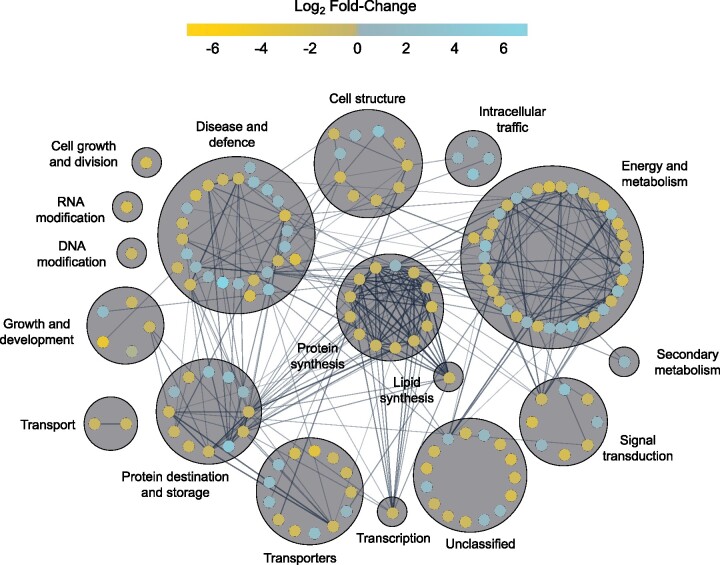
In order to better understand cellular physiology under cold stress conditions, protein–protein interaction networks were predicted in silico using STRING (Franceschini et al. 2016; Szklarczyk et al. 2019). Again, the proteome of CA2 and CA6 plants was chosen to represent early and sustained responses to low temperature. In CA2 plants, proteins largely associated with protein synthesis, with energy and metabolism and disease and defense, interacted together and with each other in interaction clusters, suggesting that low-temperature exposure initiates metabolic changes, which would not be unexpected (Figure 6; see Supplementary File S11 for an interactive version of the network with metadata). For example, a predicted subunit theta of T-complex protein 1, an ATP-dependent molecular chaperone protein that assists in protein folding (Bradi4g15450), showed 20 interactions with other proteins and these were primarily heat shock proteins, ribosomal subunits, and actins. Similar clusters were retained in the protein interaction network of CA6 plants, but involved many more protein–protein interactions, consistent with the increased number of differently abundant proteins at this later time (Figure 7; Supplementary File S12).
Figure 6.
Predicted protein–protein interaction network of all proteins at 2 days of CA. Nodes represent individual proteins (154) with node color representing the respective log2 fold-change of the protein in the cold-acclimated (2 days) vs nonacclimated PM proteomes. Edges between connected nodes represent predicted protein–protein interactions. Nodes are grouped based on predicted functional categories and labeled accordingly. Nodes on the outer circle of a main functional group have predicted functions in secondary categories. Interactions were predicted via STRING version 11 and the network was prepared using Cytoscape version 3.8.1. Darker and thicker edges between nodes represent stronger supporting data for the respective interaction. An interactive version of the network with encoded metadata is available in Supplementary File S11. Click on a node to see individual protein accession ID, protein description, log2 fold-change, functional categories, and degrees of interaction.
Figure 7.
Predicted protein–protein interaction network of all proteins at 6 days of CA. Nodes represent individual proteins (680) with fill color representing the respective log2 fold-change of the protein in the cold-acclimated (6 days) vs nonacclimated PM proteomes. Edges between connected nodes represent predicted protein–protein interactions between connected proteins. Nodes are grouped based on predicted functional categories and labeled accordingly. Nodes on the outer circle of a main functional group have predicted functions in secondary categories. Interactions were predicted via STRING version 11 and the network was prepared using Cytoscape version 3.8.1. Darker and thicker edges between nodes represent stronger supporting data for the respective interaction. An interactive version of the network with encoded metadata is available in Supplementary File S12. Click on a node to see individual protein accession ID, protein description, log2 fold-change, functional categories, and degrees of interaction.
STRING analysis predicted interactions between Suppressor of G-Two allele of SKP1 (SGT1, Bradi2g44030.1) with HSP90s (Bradi1g30130.1; Bradi1g32770.1) and HSP70s (Bradi1g66590.1; Bradi2g30660.1) at CA2 (Supplementary File S11). Interactions between SGT1, HSP90, and HSP70 regulate microbial disease resistance in a number of plant species including Arabidopsis (Takahashi et al. 2003; Azevedo et al. 2006; Noël et al. 2007; Spiechowicz et al. 2007) and barley (Azevedo et al. 2002; Shen et al. 2003; Hein et al. 2005; Chapman et al. 2021), suggesting that immune signaling pathways are upregulated in response to low-temperature treatment. In CA6 plants, such predicted interactions were conserved but also expanded (Supplementary File S12). These involved additional HSPs and other immune-related proteins, including stromal HSP70 (Bradi2g30560.1), HSP81-1s (Bradi3g39590; Bradi3g39620.1), activator of HSP90 (Bradi3g37790.1), respiratory burst oxidase homolog protein B-like (Bradi2g12790.2), HSP70-HSP90 organizing protein (Bradi3g50110.1), and two peptidyl-prolyl cis-trans isomerases (Bradi1g34750.1; Bradi2g39950.1) with known functions in immune response (Mokryakova et al. 2014). Interestingly, the respiratory burst oxidase homolog protein B-like, which is involved in pathogen resistance in Arabidopsis (Hawamda et al. 2020), shows additional predicted interactions with calcium-dependent protein kinase 13 (Bradi5g19430.1), mitogen-activated protein kinase (Bradi1g65810.1), catalase (Bradi1g76330.1), and an ammonium transporter (Bradi2g22750.1), further suggesting the activation of immune signaling pathways as a result of cold stress.
A single example of a protein with many interactions at CA6, allene oxide synthase 3 (Bradi3g08250.1), involved in the biosynthesis of jasmonic acid and plant defense (Farmer and Goossens 2019), showed 137 interactions with other proteins associated with functions in protein synthesis, as well as disease and defense . The correlation of time at low temperature and the number of protein interactions was also reflected in the 6.4-fold increase in the mean number of interactions shown by each protein depicted in the CA2 (3.3) and CA6 (21) networks. Overall, a short period of CA resulted in an interaction network containing 154 nodes as individual proteins, and 252 edges, representing predicted interactions. This increased to 680 nodes and 7152 edges in CA6 plants (Figures 6 and 7; Supplementary Files S11 and S12). The increasing complexity of protein–protein interactions and the increased number of crosstalk pathways as low-temperature exposure continues suggests that after the establishment of the early response, the cold resistance phenotype is supported by the proteins associated with the sustained response.
Discussion
Our motivation to understand PM proteome changes associated with CA in the model monocot Brachypodium was the need to address temperature-related challenges to food security linked to climate change. Our results suggest that freeze-tolerance in this species is a dynamic process, with an early frost-resistance response that can be achieved within 2 days of CA, but thereafter, additional changes to the PM occur four or more days later that presumably allow for a sustained response for low-temperature survival. The presence of these two phases of freezing tolerance is supported by multiple lines of evidence, most notably: (1) the acquisition of PM protection in CA2 plants, (2) the rapid accumulation of sucrose by CA2, followed by further sucrose accumulation after 6 days, and (3) changes in relative protein abundance, demonstrated by heatmaps, functional profiles, as well as network analysis generated from the MS protein discovery that could be divided into two groups: CA2–CA4 and CA6–CA8 (Figures 1–7).
The early response to CA
An early or rapid response to low temperature is sufficient to maintain PM integrity as assessed by electrolyte leakage. Of the 1349 PM-proteins identified by nano-liquid chromatography-MS, the relative abundance of 57 PM proteins increased >1.5-fold after 2 days of CA. These 57 proteins likely function in supportive roles to ensure membrane integrity at low temperatures. Consistent with this hypothesis, the vast majority (88%) of these proteins also increased in relative abundance in initial Brachypodium experiments that examined the NA and CA2 PM proteome using an acclimation temperature of 4°C (Supplementary File S13) demonstrating a conserved and repeatable proteomic response, coincident with membrane integrity. Functional annotation and categorization of the proteins suggested that the early response includes a global shift in proteins from those associated with energy and metabolism, as well as growth and development, identified in the NA PM proteome toward the accumulation of stress-related proteins after CA. Indeed, proteins identified in our two experimental CA Brachypodium PM proteomes were fairly consistent with those identified for the CA Arabidopsis PM proteome (Miki et al. 2019). For example, transporters and signal transduction proteins, including kinases, increased in relative abundance coincident with PM freeze protection in Arabidopsis following 2 days of CA. For Brachypodium proteins that significantly decreased and increased in relative abundance (27% and 13%, respectively), at least one ortholog was found to change in abundance in the same direction in the CA Arabidopsis PM proteome (Miki et al. 2019). When the reverse analysis was performed, 37% and 15% of the significantly decreased and increased proteins, respectively, from the CA Arabidopsis PM proteome had an ortholog in our corresponding dataset that changed in abundance in the same direction. This result suggests at least a partially conserved response to low temperature between Brachypodium and Arabidopsis at the PM. It is not surprising that they are not identical considering their evolutionary distance and the differences in freezing tolerance as determined by their capacity to survive low temperatures after CA, with Arabidopsis at −6°C to −11°C, depending on the accession, vs. Brachypodium at −12°C; (Kaplan et al. 2004; Hannah et al. 2006; Mayer et al. 2020).
Proteins with the highest increases in relative abundance are obvious candidates to assist in the protection of vulnerable PMs. A striking 483-fold elevation in relative abundance at CA2 was observed for COR410-like (Bradi3g51200.1), a dehydrin family cold-regulated protein. Transcripts encoding this protein have previously been shown to accumulate after low-temperature exposure in Brachypodium (Mayer et al. 2020) and orthologous COR47 (At1g20440.1), as well as the related COR78 (At5g52310.1), also showed relative increases in abundance after CA of Arabidopsis (Miki et al. 2019). It is thought that dehydrins, which are intrinsically disordered PM-associated proteins, act as chaperones to prevent protein denaturation during dehydration (Singh et al. 2019). These proteins might also interact with sucrose to change the glass transition temperature, decreasing the probability of ice crystal formation at subzero temperatures (Wolkers et al. 2001). Consistent with this finding, increasing levels of osmoprotectants including raffinose, glucose, and a 13-fold increase in sucrose were observed in CA2 plants (Figure 1). It should be recalled that under freeze conditions, most free water is bound to ice, thus a PM-dehydrin is likely critical to freezing tolerance and it is not surprising that the relative abundance would be increased to such a high level.
Other proteins that increased approximately twofold in relative abundance by CA2 also appear to be important for PM protection. For example, a protein closely related to 97 kDa HSP (Bradi1g32770.1) could act similarly to COR410 and serve as a chaperone for PM stabilization. Phosphatidylinositol transfer protein (Bradi2g09760.1) ferries phospholipids to the PM and would also play a key role in PM restructuring; indeed the abundance of glycosylphosphatidylinositol-anchored proteins in Arabidopsis has been reported to increase almost two-fold after CA (Takahashi et al. 2016).
At CA2, 97 proteins showed a significant decrease in relative abundance (Supplementary File S6). Notably, an estimated 42% of these proteins were annotated not as PM proteins but as cytoplasmic proteins. PM proteins can be reversibly associated with lipids or other proteins on the membrane (Marmagne et al. 2004). Thus it is possible that some of these proteins could be loosely associated with the PM but as low-temperature-induced restructuring of the PM commenced and continued, such associations could weaken or strengthen, resulting in their apparent decrease or increase in relative abundance, as has been reported for cold-stressed rice leaves (Komatsu et al. 2007). Proteins known to be associated with PMs under different conditions and that decreased in relative abundance were dominated by the so-called “master enzymes,” PM H+ ATPases (PM ATPase, Bradi5g24690.1; PM ATPase 1-like, Bradi1g54847.1) as well as other ATPases (V-type ATPase, Bradi1g67960.1; AAA+ ATPase, Bradi1g48010.1). Such PM ATPases are crucial for cell growth and maintain the transmembrane electrochemical gradient necessary for nutrient uptake (Morsomme and Boutry 2000). Thus, similar to other plants including Arabidopsis, there were relative abundance decreases in proteins annotated as having functions in cell growth, metabolism, and signal transduction (Huot et al. 2014; Fürtauer et al. 2019). Thus Brachypodium likely generally redirects energetics toward low-temperature protection, and away from growth as a general response during plant CA.
The sustained response to CA
As CA progressed over several days, dynamic changes in the PM-associated proteome were observed. Heatmaps for longer CA (CA6 and CA8) showed similar PM-proteome patterns that were distinct from the shorter exposure patterns (CA2 and CA4), again suggesting an early low-temperature response followed by a sustained response. By CA6, 224 PM proteins were increased in abundance, 167 more than at the earlier time point. Of these, 20% (45) and 9% (21) showed fold-changes upwards of two or three orders of magnitude, respectively. The protein showing the greatest relative increase (∼55,000-fold) was an ABC transporter C-family member 5 like protein (Bradi1g75590.1). Such transporters have been previously reported to function in abiotic and biotic stress responses through cellular detoxification, and through compound exchange of hormones, metabolites, and defense molecules (Kang et al. 2011; Hwang et al. 2016). After prolonged low temperatures, their role in detoxification is likely key to Brachypodium survival. Research with cold-stressed Arabidopsis has shown that similar ABC transporters have phosphorylation-level regulation within the membrane, further highlighting their importance (Kamal et al. 2020).
As noted, dehydrins interact with soluble carbohydrates, and after prolonged CA, sucrose reached concentrations in the leaves as high as 38 mg g−1. The time course of the increase in sugar concentrations in CA Brachypodium is similar to that reported for Arabidopsis as well as cereal crops (Plazek et al. 2003; Kamata and Uemura 2004; Klotke et al. 2004). Although this concentration is sufficient to lower the freezing point by a fraction of a degree, more importantly, sucrose has additional roles in directly stabilizing membranes (Strauss and Hauser 1986) and can scavenge ROS even more efficiently than dedicated scavenging enzymes (Nishizawa-Yokoi et al. 2008; Stoyanova et al. 2011; Tarkowski and Van den Ende 2015). Sucrose is also involved in signaling and sugar synthesis activation (Kooiker et al. 2013), and the “sweet immunity” that aids in pathogen recognition (Duran-Flores and Heil 2016). Coincident with the increase in sugar concentrations over the course of CA, a sugar transporter, (SWEET; Bradi2g24850.1), and a sucrose transporter SUT1-like protein (Bradi1g73170.1) increased in relative abundance, with two sucrose synthases (Bradi1g46670.1; Bradi1g60320.1) and two bidirectional SWEET sugar transporters (Bradi2g11920.1; Bradi2g56890.1) decreasing in relative abundance. Glycerol accumulation has also been shown to enhance abiotic stress resistance including cold resistance (Eastmond 2004). Although we did not assay for glycerol, we noted that glycerol kinase (Bradi5g23940.1) decreased in abundance after CA, suggesting reduced glycerol degradation allowing for accumulation of the polyol. After a week at low temperature, Arabidopsis also showed relative increases in monosaccharide and sucrose transporters (Miki et al. 2019) indicating that soluble carbohydrate regulation is a common strategy for cold tolerance in evolutionarily distinct plants.
By CA6, there were 456 proteins displaying significant decreases in abundance (>1.5-fold), approximately four-fold more than the number that decreased in abundance at CA2, including the PM ATPases (Bradi5g24690.1; Bradi1g54847.1; Bradi1g67960.1; Bradi1g48010.1). This observation is similar to reports from cold-stressed Arabidopsis (Muzi et al. 2016), but Brachypodium is notable for the large numbers of proteins that decreased in relative abundance during the sustained response, even with respect to the Arabidopsis dataset (Figure 2, B and C). This again underpins our contention that normal growth in CA plants is arrested in order to utilize resources for stress responses, and Brachypodium may be more efficient than the dicot in this regard.
Cross-resistance to abiotic and biotic stresses
A network map enabled the visualization of relationships between the PM proteins whose abundances were influenced by CA, with its construct partially motivated by the observation that some other proteins could have roles in stresses that could accompany low temperatures (Figure 5). Cold stress and pathogen stress represented the largest numbers of interactions (41 and 39, respectively) with proteins known to be associated with drought, osmotic, and salt stress clustering with the low-temperature responses, as might be expected from the known freeze-induced exclusion of solutes and cellular dehydration. The network indicates numerous instances of “crosstalk” among identified proteins, strongly suggesting that CA can “prime” Brachypodium for resistance to other abiotic and biotic stresses. STRING was used to interrogate protein–protein interactions in the CA-induced PM proteome, which led to the identification of protein-interaction clusters associated with protein synthesis, and metabolism and energy production (Figures 6 and 7). The translational apparatus showed numerous interactions, but overall, there were hundreds of proteins with reduced relative abundance after CA that appeared to reflect a change in metabolism and the redirection of energy away from growth. Translation reduction shown in cold-treated Arabidopsis triggers an intracellular increase in Ca2+ with the consequent regulation of COR genes (Guo et al. 2002; Zarka et al. 2003). Interactions in Brachypodium were generally conserved in CA2 and CA6, albeit with more interactions between the greater number of proteins identified after the longer cold exposure.
Cross-resistance to a variety of stresses is evolutionarily adaptive since plants are rarely exposed to a single stress and extensive crosstalk exists between biotic and abiotic stress response pathways (Fujita et al. 2006; Rejeb et al. 2014). Subsequent to CA, immune signaling involved a host of HSPs and chaperones as well as peptidyl-prolyl cis-trans isomerases, a respiratory burst oxidase homolog protein B-like involved in pathogen resistance, and calcium-dependent protein kinase 13, as detailed in Results. This strongly suggests the activation of immune signaling pathways as a result of cold stress.
A single example of a protein with many interactions at CA6, allene oxide synthase 3 (Bradi3g08250.1), involved in the biosynthesis of jasmonic acid and plant defense (Farmer and Goossens 2019), showed 137 interactions with other proteins associated with functions in protein synthesis, as well as disease and defense. Thus, the number and the identification of protein–protein interactions show an obvious link between low-temperature stress and defense. The incidence and severity of bacterial and fungal diseases are known to be influenced by abiotic stresses including drought, extreme temperatures, or salinity (Snapp 1992; Koga et al. 2004; Freeman and Beattie 2009; Moeder and Yoshioka 2009). With regards to low-temperature stress, wilting and necrosis caused by freezing damage can provide new entry points for pathogens, making plants susceptible at even low levels of infection. As well, psychrotolerant pathogens that thrive under the low ambient temperatures are more abundant, with some having the ability to initiate ice crystal formation at high subzero temperatures (Lindow et al. 1982; Wu et al. 2014). Indeed, CA winter cereals show higher disease resistance following low-temperature exposure (Kuwabara and Imai 2009; Wu et al. 2014; Pogány et al. 2016) and certain genes are similarly upregulated in response to both low temperature and exposure to pathogens (Yeh et al. 2000; Wu et al. 2014). As such, plants that are tolerant to one stress are likely to have elevated tolerance to another stress as is evidenced by our meta-analysis. It is possible that such cross-resistance could be exploited for the production of crops that are better suited to multiple abiotic stresses associated with our changing environment.
Conclusions
Taken together, we demonstrate the successful application of a two-phase partitioning system in combination with label-free quantification of proteins to achieve a PM-enriched proteome in the model monocot, Brachypodium. In addition to identifying promising new protein and gene targets for research into cold tolerance, we have provided large-scale datasets for future researchers to mine and analyze, and have importantly shown that changes in PM protein accumulation during days of CA are both dynamic and effective. We have also seen that monocots likely share certain freeze-resistance pathways and strategies with those that may be better known in dicots. Together our increased understanding of low-temperature responses represents an additional step toward enhanced cold tolerance in crops and a reduction in the sizable economic losses stemming from crop destruction due to increases in frost events attributed to our modern climate-mediated crisis.
Acknowledgments
M.B., T.N., and H.I. conducted the experiments. C.L.J., M.B., T.N., Y.K., and M.U. analyzed the data. C.L.J. prepared the figures. C.L.J., M.B., G.C.D., and V.K.W. wrote the paper, and all authors contributed to manuscript revision. M.U., G.C.D., and V.K.W. supervised the work. M.U. and V.K.W. secured funding for the work.
Funding
This research was funded by Kakenhi (#22120003 and #25650090) from the Japan Society for the Promotion of Science (JSPS) to M.U. and an NSERC (Canada) Discovery grant to V.K.W. M.B. was partially supported by a summer fellowship from the Japan Society for the Promotion of Science.
Data availability
All data described in this manuscript are original and produced by the authors. Supplementary files available at figshare https://doi.org/10.25387/g3.14462385. Supplementary File S1 contains all scripts and code used for the analysis of the Brachypodium cold-acclimated PM proteome. Supplementary File S2 contains the stress response network meta-analysis for the CA2 dataset of the Brachypodium PM proteome. Supplementary File S3 contains the total raw Brachypodium PM proteome dataset. Supplementary File S4 contains the significantly increased Brachypodium PM protein dataset. Supplementary File S5 contains the significantly decreased Brachypodium PM protein dataset. Supplementary File S6 contains the CA2 decreased annotated Brachypodium PM protein dataset. Supplementary File S7 contains the CA6 decreased annotated Brachypodium PM protein dataset. Supplementary File S8 contains the CA2 increased annotated Brachypodium PM protein dataset. Supplementary File S9 contains the CA6 increased annotated Brachypodium PM protein dataset. Supplementary File S10 contains the interactive network for the stress response meta-analysis of Brachypodium CA2 proteins. Supplementary File S11 contains the interactive network for the CA2 predicted protein–protein interactions. Supplementary File S12 contains the interactive network for the CA6 predicted protein–protein interactions. Supplementary File S13 contains the CA2 preliminary Brachypodium PM dataset.
Conflicts of interest
None declared.
Literature cited
- Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, et al. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 37:420–423. [DOI] [PubMed] [Google Scholar]
- Altenhoff AM, Train C-M, Gilbert KJ, Mediratta I, Mendes de Farias T, et al. 2021. OMA orthology in 2021: website overhaul, conserved isoforms, ancestral gene order and more. Nucleic Acids Res. 49:D373–D379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca R, Porcel R, Ruiz-Lozano JM.. 2012. Regulation of root water uptake under abiotic stress conditions. J Exp Bot. 63:43–57. [DOI] [PubMed] [Google Scholar]
- Azevedo C, Betsuyaku S, Peart J, Takahashi A, Noel L, et al. 2006. Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 25:2007–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, et al. 2002. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science. 295:2073–2076. [DOI] [PubMed] [Google Scholar]
- Bevan M, Bancroft I, Bent E, Love K, Goodman H, et al. 1998. Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature. 391:485–488. [DOI] [PubMed] [Google Scholar]
- Bredow M, Vanderbeld B, Walker VK.. 2016. Knockdown of ice-binding proteins in Brachypodium distachyon demonstrates their role in freeze protection. PLoS One. 11:e0167941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredow M, Walker VK.. 2017. Ice-binding proteins in plants. Front Plant Sci. 8:2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman AV, Hunt M, Surana P, Velásquez-Zapata V, Xu W, et al. 2021. Disruption of barley immunity to powdery mildew by an in-frame Lys-Leu deletion in the essential protein SGT1. Genetics. 217:iyaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappetta A, Muto A, Bruno L, Woloszynska M, Van Lijsebettens M, et al. 2015. A dehydrin gene isolated from feral olive enhances drought tolerance in Arabidopsis transgenic plants. Front Plant Sci. 6:392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colton-Gagnon K, Ali-Benali MA, Mayer BF, Dionne R, Bertrand A, et al. 2014. Comparative analysis of the cold acclimation and freezing tolerance capacities of seven diploid Brachypodium distachyon accessions. Ann Bot. 113:681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Abreu-Neto JB, Turchetto-Zolet AC, de Oliveira LFV, Zanettini MHB, Margis-Pinheiro M.. 2013. Heavy metal-associated isoprenylated plant protein (HIPP): characterization of a family of proteins exclusive to plants. FEBS J. 280:1604–1616. [DOI] [PubMed] [Google Scholar]
- Duran-Flores D, Heil M.. 2016. Sources of specificity in plant damaged-self recognition. Curr Opin Plant Biol. 32:77–87. [DOI] [PubMed] [Google Scholar]
- Eastmond PJ. 2004. Glycerol-insensitive Arabidopsis mutants: gli1 seedlings lack glycerol kinase, accumulate glycerol and are more resistant to abiotic stress. Plant J. 37:617–625. [DOI] [PubMed] [Google Scholar]
- Farmer EE, Goossens A.. 2019. Jasmonates: what ALLENE OXIDE SYNTHASE does for plants. J Exp Bot. 70:3373–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira JA, Zwinderman AH.. 2006. On the Benjamini–Hochberg method. Ann Statist. 34:1827–1849. [Google Scholar]
- Franceschini A, Lin J, von Mering C, Jensen LJ.. 2016. SVD-phy: improved prediction of protein functional associations through singular value decomposition of phylogenetic profiles. Bioinformatics. 32:1085–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman BC, Beattie GA.. 2009. Bacterial growth restriction during host resistance to Pseudomonas syringae is associated with leaf water loss and localized cessation of vascular activity in Arabidopsis thaliana. Mol Plant Microbe Interact. 22:857–867. [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, et al. 2006. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol. 9:436–442. [DOI] [PubMed] [Google Scholar]
- Fürtauer L, Weiszmann J, Weckwerth W, Nägele T.. 2019. Dynamics of plant metabolism during cold acclimation. Int J Mol Sci. 20:5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Liu J, Ronan EA, He F, Cai W, et al. 2019. A cold-sensing receptor encoded by a glutamate receptor gene. Cell. 178:1375–1386.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou M, Yang X, Zhao Y, Ran X, Song Y, et al. 2019. Cytochrome b5 is an obligate electron shuttle protein for syringyl lignin biosynthesis in Arabidopsis. Plant Cell. 31:1344–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xiong L, Ishitani M, Zhu J-K.. 2002. An Arabidopsis mutation in translation elongation factor 2 causes superinduction of CBF/DREB1 transcription factor genes but blocks the induction of their downstream targets under low temperatures. Proc Natl Acad Sci U S A. 99:7786–7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah MA, Wiese D, Freund S, Fiehn O, Heyer AG, et al. . 2006. Natural genetic variation of freezing tolerance in Arabidopsis. Plant Physiol. 142:98–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawamda AI, Zahoor A, Abbas A, Ali MA, Bohlmann H.. 2020. The Arabidopsis RboHB encoded by At1g09090 is important for resistance against nematodes. Int J Mol Sci. 21:5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein I, Barciszewska-Pacak M, Hrubikova K, Williamson S, Dinesen M, et al. 2005. Virus-induced gene silencing-based functional characterization of genes associated with powdery mildew resistance in barley. Plant Physiol. 138:2155–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park K-J, Obayashi T, Fujita N, Harada H, et al. 2007. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35:W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot B, Yao J, Montgomery BL, He SY.. 2014. Growth–defense tradeoffs in plants: a balancing act to optimize fitness. Mol Plant. 7:1267–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang JU, Song WY, Hong D, Ko D, Yamaoka Y, et al. 2016. Plant ABC transporters enable many unique aspects of a terrestrial plant’s lifestyle. Mol Plant. 9:338–355. [DOI] [PubMed] [Google Scholar]
- Kamal MM, Takahashi D, Nakayama T, Miki Y, Kawamura Y, et al. 2020. Proteomic approaches to identify cold-regulated plasma membrane proteins. Methods Mol Biol. 2156:171–186. [DOI] [PubMed] [Google Scholar]
- Kamata T, Uemura M.. 2004. Solute accumulation in heat seedlings during cold acclimation: contribution to increased freezing tolerance. Cryo Lett. 25:311–322. [PubMed] [Google Scholar]
- Kang J, Park J, Choi H, Burla B, Kretzschmar T, et al. 2011. Plant ABC transporters. Arabidopsis Book. 9:e0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, et al. . 2004. Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol. 136:4159–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Uemura M.. 2002. Changes in the plasma membrane from Arabidopsis thaliana within one week of cold acclimation. In:Li, P.H., Palva, E.T. (eds) Plant Cold Hardiness. Boston, MA: Springer. p. 181–194. [Google Scholar]
- Klotke J, Kopka J, Gatzke N, Heyer AG.. 2004. Impact of soluble sugar concentrations on the acquisition of freezing tolerance in accessions of Arabidopsis thaliana with contrasting cold adaptation - evidence for a role of raffinose in cold acclimation. Plant Cell Environ. 27:1395–1404. [Google Scholar]
- Koga H, Dohi K, Mori M.. 2004. Abscisic acid and low temperatures suppress the whole plant-specific resistance reaction of rice plants to the infection of Magnaporthe grisea. Physiol Mol Plant Pathol. 65:3–9. [Google Scholar]
- Komatsu S, Konishi H, Hashimoto M.. 2007. The proteomics of plant cell membranes. J Exp Bot. 58:103–112. [DOI] [PubMed] [Google Scholar]
- Kooiker M, Drenth J, Glassop D, McIntyre CL, Xue G-P.. 2013. TaMYB13-1, a R2R3 MYB transcription factor, regulates the fructan synthetic pathway and contributes to enhanced fructan accumulation in bread wheat. J Exp Bot. 64:3681–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL.. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305:567–580. [DOI] [PubMed] [Google Scholar]
- Kuwabara C, Imai R.. 2009. Molecular basis of disease resistance acquired through cold acclimation in overwintering plants. J Plant Biol. 52:19–26. [Google Scholar]
- Lee YP, Babakov A, de Boer B, Zuther E, Hincha DK.. 2012. Comparison of freezing tolerance, compatible solutes and polyamines in geographically diverse collections of Thellungiella sp. and Arabidopsis thaliana accessions. BMC Plant Biol. 12:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Takahashi D, Kawamura Y, Uemura M.. 2020. Plasma membrane proteome analyses of Arabidopsis thaliana suspension-cultured cells during cold or ABA treatment: relationship with freezing tolerance and growth phase. J Proteomics. 211:103528. [DOI] [PubMed] [Google Scholar]
- Lindow SE, Arny DC, Upper CD.. 1982. Bacterial ice nucleation: a factor in frost injury to plants. Plant Physiol. 70:1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Song Q, Li D, Yang X, Li D.. 2017. Multifunctional roles of plant dehydrins in response to environmental stresses. Front Plant Sci. 8:1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrmann J, Harter K.. 2002. Plant two-component signaling systems and the role of response regulators. Plant Physiol. 128:363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković ZJ, Barta A.. 2002. Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 30:623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Dai X, Xu Y, Luo W, Zheng X, et al. 2015. COLD1 confers chilling tolerance in rice. Cell. 160:1209–1221. [DOI] [PubMed] [Google Scholar]
- Marino G, Funk C.. 2012. Matrix metalloproteinases in plants: a brief overview. Physiol Plant. 145:196–202. [DOI] [PubMed] [Google Scholar]
- Marmagne A, Rouet M-A, Ferro M, Rolland N, Alcon C, et al. 2004. Identification of new intrinsic proteins in Arabidopsis plasma membrane proteome. Mol Cell Proteomics. 3:675–691. [DOI] [PubMed] [Google Scholar]
- Maruta T, Ichikawa Y, Mieda T, Takeda T, Tamoi M, et al. 2010. The contribution of Arabidopsis homologs of L-gulono-1,4-lactone oxidase to the biosynthesis of ascorbic acid. Biosci Biotechnol Biochem. 74:1494–1497. [DOI] [PubMed] [Google Scholar]
- Mayer BF, Bertrand A, Charron J-B.. 2020. Treatment analogous to seasonal change demonstrates the integration of cold responses in Brachypodium distachyon. Plant Physiol. 182:1022–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki Y, Takahashi D, Kawamura Y, Uemura M.. 2019. Temporal proteomics of Arabidopsis plasma membrane during cold- and de-acclimation. J Proteomics. 197:71–81. [DOI] [PubMed] [Google Scholar]
- Minami A, Kawamura Y, Yamazaki T, Furuto A, Uemura M.. 2009. Plasma membrane and plant freezing tolerance: possible involvement of plasma membrane microdomains in cold acclimation. In: Gusta L.V., Wisniewski M.E., Tanio K.K. (eds), Plant Cold Hardiness: From the Laboratory to the Field. Wallingford, U.K.: CAB International. p. 62–71. [Google Scholar]
- Mochida K, Uehara-Yamaguchi Y, Takahashi F, Yoshida T, Sakurai T, et al. 2013. Large-scale collection and analysis of full-length cDNAs from Brachypodium distachyon and integration with Pooideae sequence resources. PLoS One. 8:e75265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeder W, Yoshioka K.. 2009. Environmental sensitivity in pathogen resistant Arabidopsis mutants. In: Yoshioka, K., Shinozaki, K.(eds), Signal Crosstalk in Plant Stress Responses. Ames: Wiley-Blackwell. p. 113–135. [Google Scholar]
- Mokryakova MV, Pogorelko GV, Bruskin SA, Piruzian ES, Abdeeva IA.. 2014. The role of peptidyl-prolyl cis/trans isomerase genes of Arabidopsis thaliana in plant defense during the course of Xanthomonas campestris infection. Russ J Genet. 50:140–148. [PubMed] [Google Scholar]
- Mori K, Renhu N, Naito M, Nakamura A, Shiba H, et al. 2018. Ca 2+ -permeable mechanosensitive channels MCA1 and MCA2 mediate cold-induced cytosolic Ca2+ increase and cold tolerance in Arabidopsis. Sci Rep. 8:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsomme P, Boutry M.. 2000. The plant plasma membrane H(+)-ATPase: structure, function and regulation. Biochim Biophys Acta. 1465:1–16. [DOI] [PubMed] [Google Scholar]
- Muzi C, Camoni L, Visconti S, Aducci P.. 2016. Cold stress affects H-ATPase and phospholipase D activity in Arabidopsis. Plant Physiol Biochem. 108:328–336. [DOI] [PubMed] [Google Scholar]
- Nguyen TT, Sabat G, Sussman MR.. 2018. C ross-linking supports a head-to-tail mechanism for regulation of the plant plasma membrane P-type H-ATPase. J Biol Chem. 293:17095–17106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A, Yabuta Y, Shigeoka S.. 2008. The contribution of carbohydrates including raffinose family oligosaccharides and sugar alcohols to protection of plant cells from oxidative damage. Plant Signal Behav. 3:1016–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noël LD, Cagna G, Stuttmann J, Wirthmüller L, Betsuyaku S, et al. 2007. Interaction between SGT1 and cytosolic/nuclear HSC70 chaperones regulates Arabidopsis immune responses. Plant Cell. 19:4061–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce R. 2001. Plant freezing and damage. Ann Bot. 87:417–424. [Google Scholar]
- Plazek A, Hura K, Zur I, Niemczyk E.. 2003. Relationship between frost tolerance and cold-induced resistance of spring barley, meadow fescue and winter oilseed rape to fungal pathogens. J Agron Crop Sci. 189:333–340. [Google Scholar]
- Pogány M, Harrach B, Bozsó Z, Künstler A, Janda T, et al. 2016. Cold hardening protects cereals from oxidative stress and necrotrophic fungal pathogenesis. Open Life Sci. 11:78–85. [Google Scholar]
- Raza A, Razzaq A, Mehmood SS, Zou X, Zhang X, et al. 2019. Impact of climate change on crops adaptation and strategies to tackle its outcome: a review. Plants. 8:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejeb IB, Pastor V, Mauch-Mani B.. 2014. Plant responses to simultaneous biotic and abiotic stress: molecular mechanisms. Plants (Basel). 3:458–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JY, Hong S-Y, Jo S-H, Woo J-C, Lee S, et al. 2014. Molecular and functional characterization of cold-responsive C-repeat binding factors from Brachypodium distachyon. BMC Plant Biol. 14:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savojardo C, Martelli PL, Fariselli P, Profiti G, Casadio R.. 2018. BUSCA: an integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 46:W459–W466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QH, Zhou F, Bieri S, Haizel T, Shirasu K, et al. 2003. Recognition specificity and RAR1/SGT1 dependence in barley Mla disease resistance genes to the powdery mildew fungus. Plant Cell. 15:732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh PK, Srivastava D, Tiwari P, Tiwari M, Verma G, et al. 2019. Drought tolerance in plants: molecular mechanism and regulation of signaling molecules. In:Khan M.I.R., Reddy P.S., Ferrante A., Khan N.A. (eds) Plant Signaling Molecules. Cambridge, UK:: Woodhead Publishing. p. 105–123. [Google Scholar]
- Snapp SS. 1992. Salinity effects on root growth, root death and severity of infection by Phytophthora Parasitica Dast. in tomato (Lycopersicon esculentum Mill.) [Doctoral dissertation]. Davis, CA: University of California, Davis.
- Spiechowicz M, Zylicz A, Bieganowski P, Kuznicki J, Filipek A.. 2007. Hsp70 is a new target of Sgt1—an interaction modulated by S100A6. Biochem Biophys Res Commun. 357:1148–1153. [DOI] [PubMed] [Google Scholar]
- Stoyanova S, Geuns J, Hideg E, Van den Ende W.. 2011. The food additives inulin and stevioside counteract oxidative stress. Int J Food Sci Nutr. 62:207–214. [DOI] [PubMed] [Google Scholar]
- Strauss G, Hauser H.. 1986. Stabilization of lipid bilayer vesicles by sucrose during freezing. Proc Natl Acad Sci U S A. 83:2422–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Mittler R.. 2006. Reactive oxygen species and temperature stresses: a delicate balance between signaling and destruction. Physiol Plant. 126:45–51. [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, et al. 2019. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47:D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Casais C, Ichimura K, Shirasu K.. 2003. HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc Natl Acad Sci U S A. 100:11777–11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi D, Furuto A, Minami A, Uemura M.. 2010. Alterations of plasma membrane microdomains in oat and rye during cold acclimation. Cryobiology. 61:399–400. [Google Scholar]
- Takahashi D, Kawamura Y, Uemura M.. 2016. Cold acclimation is accompanied by complex responses of glycosylphosphatidylinositol (GPI)-anchored proteins in Arabidopsis. J Exp Bot. 67:5203–5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi D, Kawamura Y, Yamashita T, Uemura M.. 2012. Detergent-resistant plasma membrane proteome in oat and rye: similarities and dissimilarities between two monocotyledonous plants. J Proteome Res. 11:1654–1665. [DOI] [PubMed] [Google Scholar]
- Takahashi D, Uemura M, Kawamura Y.. 2018. Freezing tolerance of plant cells: from the aspect of plasma membrane and microdomain. Adv Exp Med Biol. 1081:61–79. [DOI] [PubMed] [Google Scholar]
- Tarkowski ŁP, Van den Ende W.. 2015. Cold tolerance triggered by soluble sugars: a multifaceted countermeasure. Front Plant Sci. 6:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedla B, Dang QL, Inoue S.. 2020. Freeze-thaw events delay spring budburst and leaf expansion while longer photoperiods have opposite effect under different [CO2] in white birch: advance it under elevated but delay it under ambient [CO2]. Environ Exp Bot. 173:103982. [Google Scholar]
- Thomashow MF. 1999. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 50:571–599. [DOI] [PubMed] [Google Scholar]
- Thomashow MF. 2010. Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol. 154:571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura M, Joseph RA, Steponkus PL.. 1995. Cold acclimation of Arabidopsis thaliana (effect on plasma membrane lipid composition and freeze-induced lesions). Plant Physiol. 109:15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura M, Yoshida S.. 1984. Involvement of plasma membrane alterations in cold acclimation of winter rye seedlings (Secale cereale L. cv Puma). Plant Physiol. 75:818–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium 2019. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47:D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkers WF, McCready S, Brandt WF, Lindsey GG, Hoekstra FA.. 2001. Isolation and characterization of a D-7 LEA protein from pollen that stabilizes glasses in vitro. Biochim Biophys Acta. 1544:196–206. [DOI] [PubMed] [Google Scholar]
- Wu J, Zhang Y, Yin L, Qu J, Lu J.. 2014. Linkage of cold acclimation and disease resistance through plant-pathogen interaction pathway in Vitis amurensis grapevine. Funct Integr Genomics. 14:741–755. [DOI] [PubMed] [Google Scholar]
- Yang Y, Zhang Y, Ding P, Johnson K, Li X, et al. 2012. The ankyrin-repeat transmembrane protein BDA1 functions downstream of the receptor-like protein SNC2 to regulate plant immunity. Plant Physiol. 159:1857–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh S, Moffatt BA, Griffith M, Xiong F, Yang DS, et al. 2000. Chitinase genes responsive to cold encode antifreeze proteins in winter cereals. Plant Physiol. 124:1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi V, Ahmadi J, Sadeghzadeh-Ahari D, Esfandiari E.. 2018. Influence of long-term cold stress on enzymatic antioxidative defense system in chickpea (Cicer arietinum L.). Acta Agrobot. 71:1–11. [Google Scholar]
- Yuan P, Yang T, Poovaiah BW.. 2018. Calcium signaling-mediated plant response to cold stress. Int J Mol Sci. 19:3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarka DG, Vogel JT, Cook D, Thomashow MF.. 2003. Cold induction of Arabidopsis CBF genes involves multiple ICE (Inducer of CBF Expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiol. 133:910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuther E, Schulz E, Childs LH, Hincha DK.. 2012. Clinal variation in the non-acclimated and cold-acclimated freezing tolerance of Arabidopsis thaliana accessions. Plant Cell Environ. 35:1860–1878. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data described in this manuscript are original and produced by the authors. Supplementary files available at figshare https://doi.org/10.25387/g3.14462385. Supplementary File S1 contains all scripts and code used for the analysis of the Brachypodium cold-acclimated PM proteome. Supplementary File S2 contains the stress response network meta-analysis for the CA2 dataset of the Brachypodium PM proteome. Supplementary File S3 contains the total raw Brachypodium PM proteome dataset. Supplementary File S4 contains the significantly increased Brachypodium PM protein dataset. Supplementary File S5 contains the significantly decreased Brachypodium PM protein dataset. Supplementary File S6 contains the CA2 decreased annotated Brachypodium PM protein dataset. Supplementary File S7 contains the CA6 decreased annotated Brachypodium PM protein dataset. Supplementary File S8 contains the CA2 increased annotated Brachypodium PM protein dataset. Supplementary File S9 contains the CA6 increased annotated Brachypodium PM protein dataset. Supplementary File S10 contains the interactive network for the stress response meta-analysis of Brachypodium CA2 proteins. Supplementary File S11 contains the interactive network for the CA2 predicted protein–protein interactions. Supplementary File S12 contains the interactive network for the CA6 predicted protein–protein interactions. Supplementary File S13 contains the CA2 preliminary Brachypodium PM dataset.