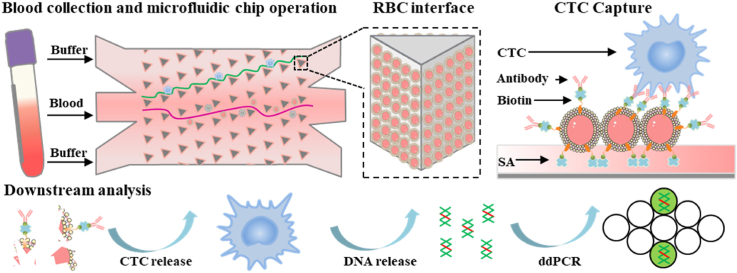
Abstract
Circulating tumor cells (CTCs), as important liquid biopsy target, can provide valuable information for cancer progress monitoring and individualized treatment. However, current isolation platforms incapable of balancing capture efficiency, specificity, cell viability, and gentle release have restricted the clinical applications of CTCs. Herein, inspired by the structure and functional merits of natural membrane interfaces, we established an antibody-engineered red blood cell (RBC-Ab) affinity interface on microfluidic chip for high-performance isolation and release of CTCs. The lateral fluidity, pliability, and anti-adhesion property of the RBC microfluidic interface enabled efficient CTCs capture (96.5%), high CTCs viability (96.1%), and high CTCs purity (average 4.2-log depletion of leukocytes). More importantly, selective lysis of RBCs by simply changing the salt concentration was utilized to destroy the affinity interface for efficient and gentle release of CTCs without nucleic acid contamination. Using this chip, CTCs were successfully detected in colon cancer samples with 90% sensitivity and 100% specificity (20 patients and 10 healthy individuals). After the release process, KRAS gene mutations of CTCs were identified from all the 5 cancer samples, which was consistent with the results of tissue biopsy. We expect this RBC interface strategy will inspire further biomimetic interface construction for rare cell analysis.
Keywords: Red blood cells, Circulating tumor cells, Colon cancer, Biomimetic interface
Graphical abstract
Highlights
-
(1)
Benefitting from the membrane fluidity, cellular softness, and anti-adsorption performance of the erythrocytes, CTCs were captured with high efficiency, high activity and high purity by microfluidic chip.
-
(2)
CTCs can be released gently without nucleic acid contamination by simple destruction of the erythrocytes interface by simply changing the salt concentration.
-
(3)
CTCs were successfully detected in colon cancer samples with 90% sensitivity and 100% specificity, which can used for cancer diagnosis and progress monitoring.
-
(4)
After CTCs release, KRAS gene mutations were identified from all the 5 cancer samples, which is important for individualized treatment.
1. Introduction
Circulating tumor cells (CTCs) shed from solid tumor tissues are considered to be important liquid biopsy targets [1]. CTCs enumeration can reflect the patient's tumor burden and facilitate diagnosis, therapy monitoring, and prognosis of cancers [[2], [3], [4]]. Moreover, in-depth molecular and functional analysis of CTCs can provide rich biological information for the precision treatment and biological research of cancers [[5], [6], [7], [8]]. However, the extremely rare CTCs are present in the complex whole blood matrix, with as low as several CTCs per mL of blood containing billions of blood cells [[9], [10], [11]]. Thus, efficient and specific CTCs isolation and gentle release of CTCs are urgently needed for clinical applications of CTCs and cancer study.
In recent years, the immunoaffinity interface combining magnetic nanoparticles [12,13], microarray structures [[14], [15], [16]], microfluidics [[17], [18], [19]], etc. has been widely used in the sorting and research of CTCs. These designs have improved CTCs analysis techniques from different perspectives, such as enrichment efficiency, cell viability, purity and throughput, etc. Existing interfaces are mainly fabricated of solid inorganic materials, such as monocrystalline, glass, and polydimethylsiloxane (PDMS), which was widely used in cell researches. In order to obtain better biocompatibility, higher softness and excellent anti-matrix adsorption of immunoaffinity interface, diverse biomimetic materials have been constructed for interface modification and functionalization [[20], [21], [22], [23], [24], [25]]. In particular, cell membrane interfaces have attracted considerable attention due to their natural antiadhesion properties [[26], [27], [28]]. For example, researchers have developed biomimetic magnetic nanoparticles by surface coating with cell membranes to achieve highly specific capture of CTCs [[29], [30], [31]]. Furthermore, cell membrane-based nanovesicles have been used to assemble microfluidic interfaces for higher capture efficiency [32]. Although promising, the interface assembly was complicated, requiring cell culture, purification of cellular membrane fractions, and preparation of nanovesicles. Alternatively, Pei's group constructed a biomimetic membrane interface by immobilizing intact cells on a substrate for simple construction of a cell membrane interface [33]. However, all these systems are still incapable of gentle and efficient release of CTCs. Aggregation of recognition molecules based on cell membrane fluidity was originally conducive to enhancing affinity recognition, but it may lead to steric hindrance in the competition or cleavage of recognition molecules, thus decreasing the release efficiency of CTCs. Additionally, nucleic acid contamination often occurred in membranes or nanovesicles derived from nucleated cells, leading to problems in downstream analysis.
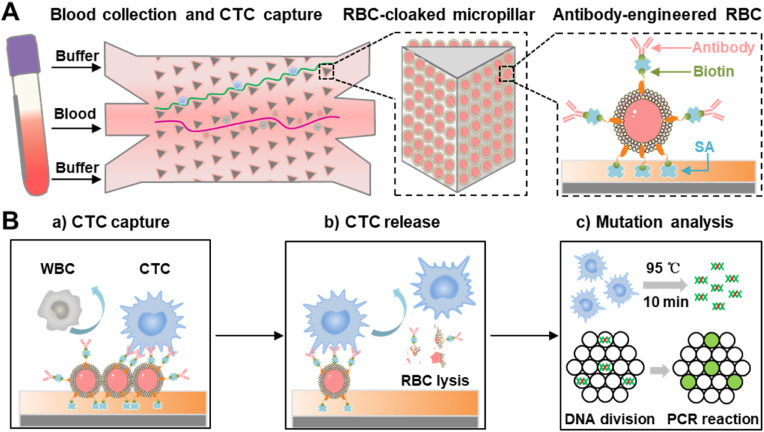
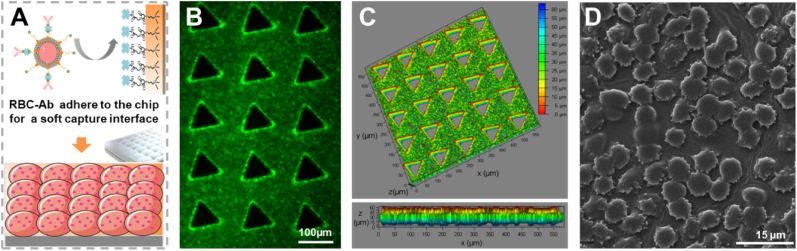
To avoid the constraints of cell-interface interactions mentioned above, we functionalized natural RBCs with antibodies, to provide improved biocompatibility and pliability [[34], [35], [36], [37], [38]], and used them for construction of an interface on a microfluidic chip (referred as RBC-Chip) for high-performance capture and release of CTCs. This RBC-Chip provides various benefits due to the combination of whole-cell RBCs interface and deterministic lateral displacement (DLD)-patterned interaction model [[39], [40], [41]]. First, placing a layer of RBCs on the capture substrate, which serves as a soft and flexible cushion, can maintain high CTCs viability during interfacial collision (Fig. 1A). In detail, although the substrate material of the microfluidic chip has good biocompatibility, the fluid velocity in the microfluidic chip is still large for the cells, and the cell-microcolumn collision at high flow rate can easily cause CTCs damage. Nano-scale biomimetic materials have been used to improve the activity of CTCs in chip [42], and micron-scale RBCs with soft properties might be a good choice. Second, benefitting from the anti-adsorption performance of the RBCs membrane, nonspecific capture of background cells can be effectively avoided (Fig. 1B–a). Third, the lateral fluidity of the RBCs membrane enable clustering of antibodies for highly efficient CTCs capture [43]. Once CTCs collide with the interface, antibodies and tumor biomarkers will cluster at the binding sites to form spatial multivalent recognition [35]. Most importantly, CTCs can be released gently without DNA contamination by simple destruction of the RBCs interface by treatment with RBCs lysis buffer (Fig. 1B–b) [44,45]. We also show for the first time, that the ligand-independent releasing strategy avoids the inefficient release of cells due to steric hindrance caused by the aggregation of recognition molecules at the binding site. In addition, the DLD-patterned microarray with size-dictated interaction selectively enhances the collision of large-sized cancer cells to improve the capture efficiency and purity of CTCs (Fig. 1A). To apply this strategy to clinical applications, we performed droplet digital PCR analysis (ddPCR) of the released CTCs based on KRAS gene mutations (Fig. 1B–c). Taken together, the RBC-Chip exhibited a variety of advantages of high immunoaffinity, high cell viability, low background interference and efficient cell recovery for isolation and downstream analysis of CTCs.
Fig. 1.
Schematic illustration of RBC-Chip. (A) Fabrication and sample loading of RBC-Chip. Antibody-engineered RBCs (RBC-Ab) are densely anchored on the inner surface of the solid-phase chip made of PDMS. CTCs tend to collide with the soft RBC-cloaked micropillars following the deterministic lateral displacement (DLD)-patterned interaction model. Detailed parameters of DLD-patterned interaction model can be seen in Fig. S1 (B) Working principle of CTCs capture and release on antibody-engineered RBCs interface. (a) CTCs are captured through fluidity-enhanced multivalent binding effect, while WBCs are not captured. (b) CTCs are released by the osmotic fragility of RBCs. (c) The recovered CTCs are applied for KRAS gene analysis by droplet digital PCR.
2. Results and discussion
2.1. Preparation of antibody-engineered red blood cells (RBC-Ab)
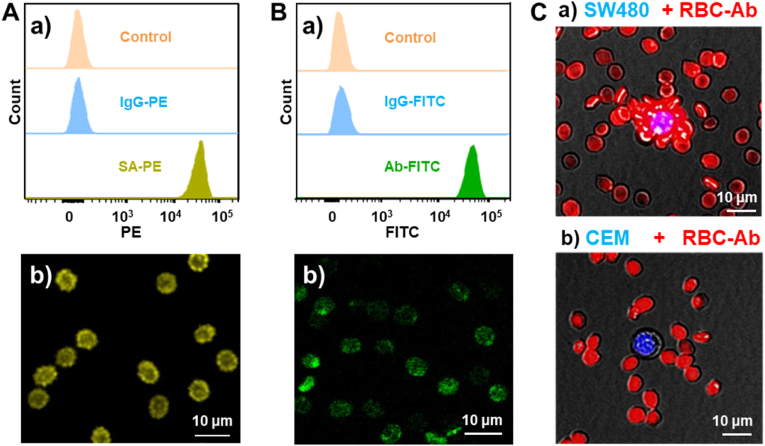
Epithelial cell adhesion molecule precursor (EpCAM), a major gastrointestinal tumor-associated protein, was used as a recognition target to participate in the capture of CTCs. To fabricate affinity RBCs as recognition elements for CTCs capture, EpCAM antibodies were modified on the RBCs surface (RBC-Ab) with dual-terminal functionalized DNA, 5′-Chol-DNA-Biotin-3′, as linkers. As shown in Fig. S2, 5′-end cholesterol modification enables the anchoring of DNA linkers to fabricate biotin-modified RBCs (RBC-Biotin) via hydrophobic interaction. Stability verification of RBC-Biotin in plasma can be seen in Fig. S3. 3′-End biotin modification allows streptavidin and biotinylated EpCAM antibodies to anchor on the RBCs. In order to ensure that streptavidin has remaining sites for biotin on the RBCs, the molar ratio of streptavidin to biotinylated EpCAM antibodies was set to 2:1 [43]. To characterize the fabrication, the RBC-Biotin was incubated with Streptavidin-Phycoerythrin (SA-PE), and RBC-Ab was incubated with FITC-modified detection antibody (Ab-FITC). The flow cytometry results and fluorescence images confirmed the successful construction of both RBC-Biotin and RBC-Ab (Fig. 2A–B).
Fig. 2.
Characterization of RBC-Ab. (A) Flow cytometric analysis (a) and fluorescence microscopic image (b) of RBC-biotin incubated with SA-PE. (B) Flow cytometric analysis (a) and fluorescence microscopic image (b) of RBC-Ab incubated with Ab-FITC. (C) Fluorescence images of target SW480 tumor cell (a) and control CCRF-CEM cell (b) incubated with RBC-Ab. The cell nuclei were labelled with DAPI (blue) and the RBC membrane was labelled with red fluorescent probe (red).
To evaluate the binding ability of RBC-Ab to tumor cells, EpCAM-positive model cells SW480 (human colorectal cancer cells) and EpCAM-negative model cells CCRF-CEM (human acute lymphoblastic leukemia T lymphocytes) were incubated with RBC-Ab. As shown in Fig. 2C, SW480 cells were bound by many RBC-Ab, while no binding was observed between CCRF-CEM cells and RBC-Ab. These results confirm the capability of RBC-Ab as recognition elements for EpCAM-positive tumor cells.
2.2. Verification of fluidity-enhanced and multivalent binding
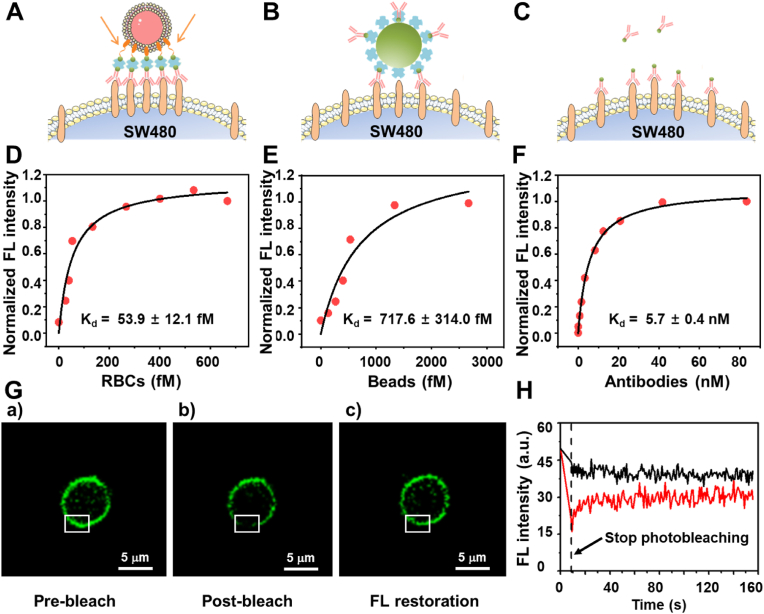
To better evaluate the attraction of CTCs to RBC-Ab, antibody-functionalized polystyrene beads (Bead-Ab) and RBC-Ab were employed to bind with SW480 in PBS. Although the antibody density and particle size are similar for Bead-Ab and RBC-Ab (calibrations of relevant conditions are shown in Fig. S4), antibodies inserted into the surface of the RBCs cytomembrane are able to laterally rearrange and accumulate at the binding site between RBC and SW480, resulting in enhancement of adhesion. In Fig. 3A–C, the binding diagrams of RBCs, beads, and free antibody binding to SW480 cells are shown in sequence, while Fig. 3D–F represents the corresponding dissociation constant (Kd) value. The dissociation constant (Kd) of RBC-Ab and SW480 was 53.9 ± 12.1 fM(Fig. 3D), while that between Bead-Ab and SW480 was 717.6 ± 314.0 fM, 13.3 times higher than the former (Fig. 3E). To verify multivalent binding, free antibodies were assessed (Fig. 3F), and the corresponding Kd was found to be 5.7 ± 0.4 nM, which was much poorer than the Kd of either RBC-Ab or Bead-Ab, indicating a significant binding affinity enhancement by the multivalent effect. Therefore, the excellent binding performance of RBC-Ab can be attributed to two effects. First, due to the modification of the RBCs surface with many antibodies, there is a multivalent effect in three-dimensional space, which can improve the binding affinity. Second, because of the excellent fluidity of the membrane's phospholipid bilayer, the EpCAM antibody modified on the RBCs surface can flow laterally and be continuously enriched at the RBC and CTC binding site, leading to improved affinity between cells. As shown in Fig. 3G–H, to demonstrate the membrane fludidity, the recognition molecules with fluorescent tags were evenly spread on the RBCs membrane. After strong light bleaching, the local fluorescence quenching appeared on the cell surface. However, the fluorescence was quickly restored in 30 s, which proved the mobility of the recognition molecules on the RBCs membrane.
Fig. 3.
Affinity assessment of multivalent binding and membrane fluidity effect. Diagrams of RBC-Ab (A), bead-Ab (B), and antibody (C) binding to SW480 cells. Characterization of the dissociation constants of RBC-Ab (D), Bead-Ab (E), and antibody (F) to SW480 cells. (G) Fluorescence images of RBC before blench (a), immediately after bleach with strong light irradiation (488 nm, 8.6 s) (b) and fluorescence restoration after 30s (c). The white box highlighted the change site. (H) Real-time fluorescence intensity of RBC membrane in the area of white box. Vertical dashed line indicates the time to stop the light irradiation.
2.3. Fabrication and characterization of RBC-Chip
The dense anchoring of RBC-Ab on the internal surface of the chip is an important step for the efficient capture of CTCs. Considering the excessive amount of biotin modified on the RBCs membrane, through the interaction between the biotin of RBC-Ab and the SA modified on chip, the RBC-Ab could be successfully captured on chip to form a supple interface (Fig. 4A). Step-by-step characterization of RBC-Chip was performed. First, the fluorescence signal from RBC-Ab stained with calcein reflects a dense cell anchoring (Fig. 4B). Then, considering the limitation of two-dimensional fluorescence imaging, we performed a confocal Z-Stack of Laser Scanning Confocal Microscopy (LSCM) on chip for three-dimensional perspective and comprehensive results. As shown in Fig. 4C, the color scale represents the height, showing that RBC-Ab was densely captured in a vertical distance of about 50 μm (blue to red). This result was consistent with the height of the chip (Fig. S5). Finally, scanning electron microscopy (SEM) was used to observe the structure of the RBC interface (Fig. 4D), showing that RBC-Abs were anchored on the PDMS substrate to form a cell interface with clean, intact cell microscopic morphology. Evaluation of the stability of the RBCs interface in the chip can be seen in Fig. S6.
Fig. 4.
Fabrication and characterization of RBC-Chip. (A) Schematic diagram of the RBC-Ab interface built on the PDMS substrate. (B) Fluorescence image of RBC-Ab on the chip stained with calcein. (C) Spatial distribution image of RBC-Ab stained with calcein. The color ruler indicated the height of the cell position. Fluorescence image showed that RBCs were modified on the chip at a high density (upper part), and the color change from blue to red, indicating that the cells have been successfully modified in the vertical direction of about 50 μm (lower part). (D) SEM image of the RBC-Ab on the PDMS substrate.
2.4. Performance of RBC-Chip
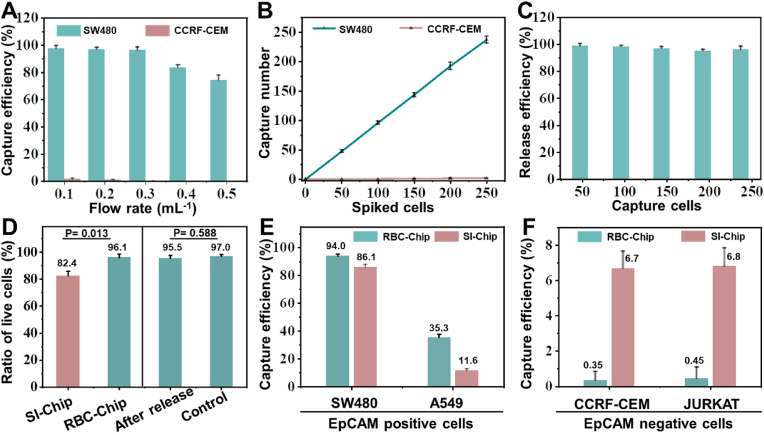
The performance evaluation of the chip included capture efficiency, capture specificity, capture stability, cell release efficiency, and cell viability. Preliminary evaluation was carried out in PBS buffer. As shown in Fig. 5A, both the cell capture efficiency and non-specific adsorption on chip gradually decreased as the flow rate increased. Therefore, to balance these two parameters, 0.3 mL h−1 was chosen for subsequent experiments, because at this flow rate, the capture efficiency of SW480 was as high as 96.5%, while the non-specific adsorption efficiency of CCRF-CEM (negative cells) was close to zero. To verify cell capture stability, as shown in Fig. 5B, different numbers of cells were introduced into the chip sequentially. The number of captured SW480 showed an excellent linear relationship with input cells, indicating good capture stability. The image of CTCs captured on the chip can be seen in Fig. S7, which provides the detailed positional relationship among the micropillars, RBC-Ab and CTCs.
Fig. 5.
Optimization and evaluation of RBC-Chip. (A) Cell capture efficiency at various flow rates in PBS. (B) The relationship between captured cell counts and spiked cell counts in PBS. (C) Release efficiency of SW480 cells by RBC lysis buffer treatment. (D) Cell viability of SW480 captured on different chips before release (left side), and the cell viability of SW480 after capture and release from RBC-Chip (right side, cells not treated with the chip as a control group). (E) Comparison of CTCs capture efficiency between RBC-Chip and SI-Chip in blood. (F) Comparison of non-specific adsorption of control cells between RBC-Chip and SI-Chip in blood. Plots and error bars represent mean values ± standard deviations obtained from three replicates.
Subsequently, release efficiency and viability of CTCs were investigated. As shown in Fig. 5C, after capturing different numbers of CTCs, RBC lysis buffer was injected into the chip for CTCs release. The data showed that for different numbers of input cells, the release efficiencies were about 96.9% with very small variation (images of CTCs before and after release can be seen in Fig. S8). Cell viability of released CTCs from RBC-Chip are shown in Fig. 5D (right part). The cell viability after release was as high as 95.5%, which was similar to the control group (cells in tube, 97.0%), indicating that neither capture nor release based on RBCs lysis caused CTCs damage. In order to further highlight the advantages of RBC-Chip as a soft capture interface, a comparison between RBC-Chip and solid interface based chip (SI-Chip) was performed to evaluate the captured cell viability (before release). As shown in Fig. 5D (left part), at the same flow rate, the cell viability of SI-Chip was 82.4%, significantly lower than that of RBC-Chip (96.1%). Therefore, these results showed that the biocompatibility and softness of RBCs can effectively prevent CTCs damage caused by collision [42] (detailed images revealing cell viability among different methods can be seen in Fig. S9). We also performed EpCAM protein characterization and cell reculture on the released tumor cells. The detailed information can be seen in Fig. S10- S11.
Before conducting clinical sample experiments, it was necessary to evaluate the performance of the chip with blood. A comparison was adopted for RBC-Chip and SI-Chip. As shown in Fig. 5E, when SW480 cells with high EpCAM expression were applied to analysis in blood, the capture efficiency of SW480 in RBC-Chip was 94.0%, slightly higher than that in SI-Chip (84.1%). Additionally, for A549 with low EpCAM expression, the RBC-Chip exhibited a much higher capture efficiency (35.3%) compared to SI-Chip (11.6%). The results showed that capture efficiency of RBC-Chip was higher than that of SI-Chip for each target cell line. More importantly, the non-specific adsorption of two control cells (CCRF-CEM and JURKAT) on RBC-Chip were much lower than that of SI-Chip (Fig. 5F). Movie S1 and Movie S2 demonstrate the capture of SW480 on RBC-Chip in PBS buffer and human blood, respectively.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bioactmat.2021.09.034.
The following are the supplementary data related to this article:
2.5. Clinical sample analysis
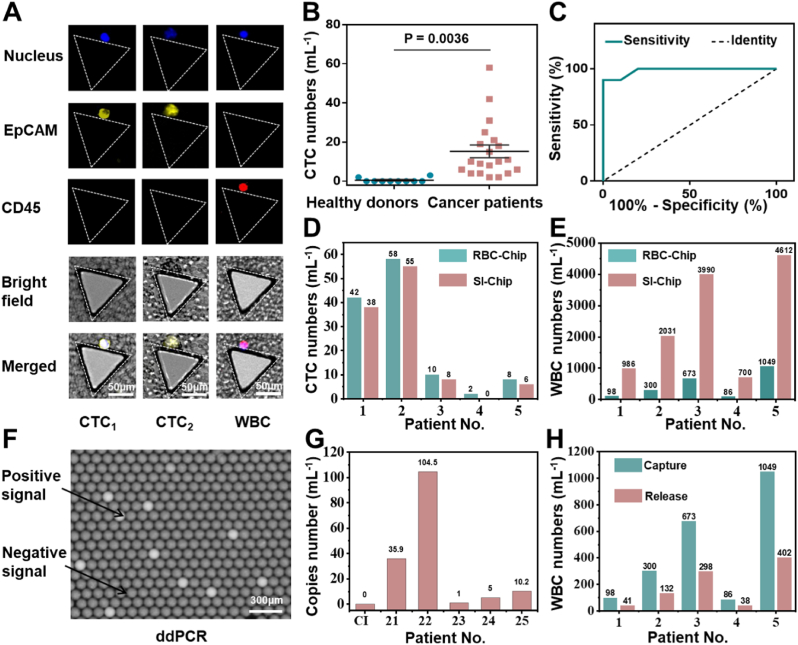
To illustrate the clinical application performance of RBC-Chip, the peripheral blood of colon cancer patients and healthy individuals were analyzed. After the cells were captured on chip, immunofluorescence staining was used to identify CTCs and WBCs. Generally, 2-(4-amidinophenyl)-6- indolecarbamidine dihydrochloride (DAPI) was used for nuclear staining to determine the presence of nucleated cells, mouse anti-human EpCAM antibody-PE (EpCAM-PE) was used to identify CTCs, and mouse anti-human CD45 antibody-APC (CD45-APC) was used to identify WBCs. As shown in Fig. 6A, DAPI+/EpCAM+/CD45- cells were identified as CTCs (CTC1, CTC2), and DAPI+/EpCAM−/CD45+ cells were identified as WBCs. Totally, CTCs in both peripheral blood of 20 patients and peripheral blood of 10 healthy individuals were identified and counted. As shown in Fig. 6B, the number of CTCs in the patient group (2–58 cells mL−1) was significantly higher than that in the healthy group (0–3 cells mL−1) (P = 0.0036), indicating that the chip was capable of distinguishing cancer patients from healthy individuals. From the ROC curve in Fig. 6C, AUC was calculated to be 0.985, indicating a reliable result (AUC: The area under the ROC curve and the coordinate axis. AUC closer to 1.0 represents higher reliability). When the threshold of the number of CTCs was set to 3.5 mL−1, the sensitivity of the chip for cancer detection was 90% and the specificity was 100% (Definition and calculation of sensitivity, specificity for clinical diagnosis were shown in Table 1.
Fig. 6.
Detecting CTCs in clinical samples from cancer patients with RBC-Chip. (A) Micrographs of representative CTCs and residual WBCs from patients' blood. (B) Enumeration of CTCs in 1 mL blood samples from 20 colon cancer patients and 10 healthy donors. (C) ROC analysis of CTC numbers between the cancer patients group and healthy donors group. (D) Numbers of CTCs captured by RBC-Chip and SI-Chip, respectively, from 1 mL of cancer patient blood. (E) Residual WBC numbers of RBC-Chip and SI-Chip, respectively, from 1 mL of cancer patient blood. (F) Image of ddPCR result. The bright droplets indicate the existence of the KRAS mutant gene, and the dark droplets indicate the absence of the KRAS mutant gene. (G) The KRAS mutation results of clinical samples or RBC lysis buffer (control group, Cl) by ddPCR. (H) Residual WBC numbers before and after CTCs release, respectively, from 1 mL of cancer patient blood.
Table 1.
Definition and calculation of sensitivity, specificity for clinical diagnosis.
| Patients with colorectal cancer (as confirmed on endoscopy and tissue biopsy) | ||||
|---|---|---|---|---|
| Condition positive | Condition negative | |||
| Peripheral Blood | Test positive | True positive (TP) = 18 | False positive (FP) = 0 | Positive predictive value = TP/(TP + FP) = 100% |
| Test negative | False negative (FN) = 2 | True negative (TN) 10 | Negative predictive value = TN/(FN + TN) = 83% | |
| Sensitivity = TP/(TP + FN) = 90% |
Specificity = TN/(FP + TN) = 100% |
|||
To further highlight the advantages of RBC-Chip in clinical application, we simultaneously counted the numbers of CTCs and WBCs of 5 patients detected by SI-Chip. As shown in Fig. 6D, the number of CTCs detected by the RBC-Chip in the same blood sample was slightly higher than that of the SI-Chip. In contrast, the average number of WBCs adsorbed by RBC-Chip was only 441 (4.2-log depletion of leukocytes, ranging from 86 to 1049), which is obviously superior to that of SI-Chip (average 2518, 3.4-log depletion of leukocytes, ranging from 700 to 4612) (Fig. 6E). In addition, after the release of CTCs based on RBC lysis, only a part of WBCs entered the recovered samples (average 182, 4.6-log depletion of leukocytes, ranging from 38 to 402), which further improved the CTCs purity and created favorable conditions for downstream analysis (Fig. 6H). Kirsten ratsarcoma viral oncogene homolog mutations (KRAS), as common point mutations between primary colorectal tumor and CTCs, are frequently analyzed in clinics for individualized and precision medicine [46]. Therefore, we performed ddPCR analysis of the released CTC samples based on the KRAS mutant gene. As shown in Fig. 6F–G, KRAS mutations were identified from all 5 patients by ddPCR, with 100% concordance with the primary tumour confirmed by tissue biopsy. Detailed information of patient or healthy samples can be found in Tables S2 and S6. These results indicated that the RBC-Chip interface has great potential in clinical sample analysis.
3. Conclusion
In summary, we have designed a natural, soft, and degradable microfluidics biomimetic interface based on intact RBCs (referred as RBC-Chip). Benefitting from the high surface area, membrane fluidity, cellular softness, biocompatibility of RBCs, this biomimetic interface has significant advantages in enhancing cell-interface interaction, reducing non-specific adsorption of blood cells, maintaining high activity and efficient release after cell isolation. In this work, RBC-Chip enabled efficient CTCs capture (96.5%), maintained high CTCs viability (96.1%) and high CTCs purity (average = 4.2-log depletion of WBCs). After selective lysis of RBCs, CTCs can be gently released from the chip with a release efficiency of about 96.9%, and high cell viability of about 95.5%. Simultaneously, the purity of CTCs have been further improved (average 4.6-log depletion of WBCs) after CTCs release. A comparison among different techniques for CTCs analysis was shown in Table S1. As for clinical application, CTCs were successfully detected in colon cancer samples with 90% sensitivity and 100% specificity, indicating superior achievement in clinical diagnosis and treatment evaluation. In addition, KRAS mutant genes from recovered CTCs were analyzed by ddPCR. Various KRAS mutant were identified from all five patients with 100% concordance with that of tissue biopsy, which was important for individualized medicine. Therefore, we expect that this RBC-Chip strategy will inspire further biomimetic interface construction for rare cell analysis.
4. Materials and methods
Reagents and Materials: SU8-3050 photoresist (MicroChem, USA), SU-8 Developer (propylene glycol monomethyl ether acetate), polydimethylsiloxane monomer (PDMS, RTV 615, Momentive) and its initiator reagent (PDMS, RTV 615, Momentive) were provided by CChip Technology Co., Ltd (Suzhou, China). Trichloro-(1H, 1H, 2H, 2H-perfluorooctyl) silane, 3-mercaptopropyl trimethoxysilane (MPTS), n-γ-maleimidobutyryl-oxysuccinimide ester (GMBS), streptavidin (SA, 85878), Tween-20, and bovine serum albumin (BSA) were provided by Sigma-Aldrich (St. Louis, MO, USA). 20 × PBS (phosphate buffer), red blood cell storage solution, and red blood cell lysis buffer were purchased from Solarbio Science and Technology Co., Ltd (Beijing, China). Biotinylated Goat anti-human EpCAM antibody was provided by the R&D System (BAF960, Minneapolis). 2-(4-Amidinophenyl)-6- indolecarbamidine dihydrochloride (DAPI, C1002), propidium iodide (PI, P8080), and DiD’ solid(1, 1′-dioctadecy l-3, 3, 3′, 3′-tetramethylindodicarbocyanine, 4-Chlorobenzenesulfonate Salt (a far-infrared plasma membrane fluorescent probe, C1039) were provided by Beyotime Biotechnology (Wuhan, China). Mouse anti-human EpCAM antibody-PE (EpCAM-PE, MA1-10198), streptavidin-PE (SA-PE, 12-4317-87) and calcein (AM, C3099) were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Mouse anti-human CD45 antibody-APC (CD45-APC, ab28106) and FITC-modified detection antibody (DA-FITC, A-11055) were provided by Abcam (Cambridge, MA, USA). RPMI 1640 Medium, DMEM Medium, fetal bovine serum (FBS), penicillin-streptomycin (P/S), and lymphocyte separation medium (LSM) were provided by Sigma-Aldrich (St. Louis, MO, USA). Cholesterol and Biotin two-terminal modified DNA chain (5′Chol-DNA-Biotin), KRAS mutation gene primers or probes were obtained from Sangon Biotech Co. Ltd. (Shanghai, China). 12 Sapphire Chips for the Naicatm Crystal Digital PCR System was purchased from STILLA technologles (F-94800 Villejuif, France).
Fabrication and modification of solid interface based chip (SI-Chip): The chip micromold was fabricated by a classical soft lithography technique [47]. Briefly, after spin-coating with SU-8 3050 and prebaking (65 °C for 5 min and 95 °C for 15 min), the silicon wafer covered with a photomask was quickly exposed to UV radiation with an energy of 100 mJ/cm2. After postbaking (65 °C for 5 min and 95 °C for 15 min), the silicon wafer was treated with photoresist developer to wash away the unexposed photoresist. Then, PDMS precursor (10:1) was poured onto the micromold and cured in a 135 °C oven for 7 min to replicate the SI-Chip. After punching holes to form inlets or outlets, the SI-Chip was bonded to the glass slide using a PDMS membrane by oxygen plasma for 1 min. As soon as a whole microchip was assembled completely, 4% (v/v) MPTS in absolute ethanol was poured into the chip for sulfhydryl modification for 1 h, and then the chip was washed with absolute ethanol and baked in a 100 °C oven for 1 h. Next, 0.5 mg mL−1 GMBS in absolute ethanol was poured into the chip for NHS ester modification for 30 min, and then washed with absolute ethanol and PBS. Next, 20 μg mL−1 SA in PBS was reacted with the chip for 1 h, and excess SA was washed away with PBS. Finally, 3% BSA in PBST (0.1% tween-20 in phosphate buffer) was used to block the chip at 4 °C overnight for further use.
Tumor cell culture and flow cytometry analysis: Tumor cell lines were cultured in RPMI 1640 or DMEM medium containing 10% fetal bovine serum in a 37 °C incubator with 5% CO2. CCRF-CEM (human acute lymphoblastic leukemia T lymphocytes) were used as model leukocytes, while SW480 (human colorectal cancer cells) were taken as model CTCs. To evaluate the EpCAM expression of CCRF-CEM and SW480, about 3 × 105 cells were incubated with 20 μL mouse anti-human EpCAM antibody (PE) in 200 μL PBS buffer at 25 °C for 30 min. SA-PE-stained cells were used as the negative control and non-stained cells were used as the blank. After twice washing with PBS buffer, cells were subjected to flow cytometry analysis by counting 1 × 104 events (BD FACS Verse).
Preparation of antibody-engineered red blood cells (RBC-Ab): Human blood came from one volunteer was put in a blood collection tube containing EDTA K2. First, after 1 : 1 dilution using DMEM medium, blood sample was carefully added onto the surface of LSM in a 15 mL centrifuge tube to form an obvious liquid–liquid interface. Then the bottom RBSs layer was extracted after centrifugation horizontally at 400 g at 18 °C for 30 min. Second, 50 μL of RBCs were diluted in 450 μL red blood cell storage solution and subsequently incubated with 10 μL (100 μM) 5′Chol-DNA-Biotin at 25 °C for 15 min, followed by centrifugation at 1300 rpm for 3 min and washing with PBS buffer three times, so that the RBCs labelled with biotin were formed (RBC-Biotin). Third, 10 μg SA and 1 μg biotinylated goat anti-human EpCAM antibody were mixed in 500 μL PBS for 30 min, before being added to the RBC-Biotin for 15 min. Finally, the solution was centrifuged at 1300 rpm for 3 min and washed with PBS buffer three times, so that the RBCs modified with antibody were formed (RBC-Ab).
To evaluate the biotinylation and antibody modification of RBCs, about 1.1 × 106 RBC-Biotin or RBC-Ab were incubated with1 μL SA-PE or 2 μL FITC-modified detection antibody (Ab-FITC) in 200 μL PBS buffer at 25 °C for 30 min, respectively. After twice washing with PBS buffer, cells were subjected to flow cytometry analysis by counting 1 × 104 events (BD FACS Verse). To verify the combination between RBC-Ab and SW480, 500 μL RBC-Ab (pretreated with 5 μM red plasma membrane fluorescent probe) and 100 μL SW480 (pretreated with 5 μg DAPI) were mixed together for 1 h of incubation at 25 °C. The results of the combination of RBC-Ab and SW480 were obtained using fluorescence microscopy.
Preparation and Characterization of antibody engineered red blood cell microfluidic chip (RBC-Chip): RBC-Ab (250 μL) was centrifuged at 1300 rpm for 3 min and resuspended in 25 μL PBS buffer. Then the concentrated RBC-Ab was injected into the SI-Chip for 3 min at a flow rate of 1 mL h−1, and the chip was flipped once during the RBC-Ab anchoring process. Finally, the chip was washed using PBS buffer for 3 min at a flow rate of 1 mL h−1. To verify the combination between RBC-Ab and SI-Chip, 2 μL calcein in 100 μL PBS buffer was used to dye RBC-Ab and the mixture was incubated for 30 min. Because two-dimensional imaging based on fluorescence microscopy was unable to reflect the modification in three-dimensions, a confocal Z-stack of the Laser Scanning Confocal Microscope (LSCM) was used to show more comprehensive results.
Performance of RBC-Chip in PBS buffer or peripheral blood: SW480 and CCRF-CEM were first pre-stained with DAPI and then spiked into PBS buffer or peripheral blood of healthy volunteers. For optimization of flow rate, about 100 pre-stained cells in PBS buffer were injected into the chip at a sample flow rate from 0.1 to 0.5 mL h−1, by cell counting at the sample inlet under a fluorescence microscope. To verify the consistency of capture efficiency at the optimal flow rate, various cell numbers from 50 to 250 were injected into the chip and the corresponding capture number was counted. For a more realistic assessment, two EpCAM positive cells (SW480, A549) and two EpCAM negative cells (CCRF-CEM, JURKAT) were spiked into peripheral blood to determine the capture efficiency of target cells and non-specific adsorption efficiency of control cells. SI-Chip was evaluated simultaneously to obtain a detailed comparation. After PBS washing, on-chip cells were imaged using a fluorescence microscope (Nikon Ti–U) and counted for calculation of the capture efficiency according to equation (1):
| (1) |
For release of the captured cells, red blood cell lysis buffer was injected into the chip and incubated for about 10 min, followed by elution with PBS at a flow rate of 1 mL h−1 for 3 min. Cells remaining on the micropillars of the chip were counted for calculation of the release efficiency according to equation (2):
| (2) |
The clearance rate of WBCs was used to reflect the purity of CTCs after capture and release. The higher the clearance rate, the higher the purity of CTCs. The WBCs depletion was calculated according to equation (3) [48]:
| (3) |
Clinical Sample collection from patients and ethical considerations: Sample collection was approved by the Human Research Ethics Committee at the First Affiliated Hospital of Xiamen University (Project number: KYX-2018-006). Informed consent was obtained from volunteers prior to blood collection. General information about peripheral blood donors is listed in Table S2. Peripheral blood was collected with a EDTA-modified vacuum tube, and analyzed within 12 h.
Isolation and detection of CTCs from Clinical Samples: The blood sample (1 mL) was pumped into RBC-Chip at the optimal flow rate of 0.3 mL h−1 for CTC capture. After PBS washing, 50 μL PBS containing 4 μL EpCAM-PE, 4 μL CD45-APC, 4 μL DAPI (20 μg mL−1) was injected into the chip at a flow rate of 1 mL h−1. After 1 h incubation in the dark, free fluorescent reagents were removed using PBS, the chip was scanned under a fluorescence microscope, and the captured cells were identified based on the fluorescent signals. Finally, CTCs were counted and then used for calculation of the sensitivity, specificity, positive predictive value, and negative predictive value according to Table 1:
CTCs recovery and KRAS mutation gene analysis: To release captured CTCs, 30 μL RBC lysis buffer was injected into the chip and incubated at 25 °C for 10 min. After washing with RBC lysis buffer, the released CTCs were collected. The recovered solution was incubated at 95 °C for 10 min, so that a large amount of RBCs debris will agglomerate and could be removed by centrifugation (2000 rpm, 3 min), and then was applied into droplet digital analysis. Detail reagent information, reaction information, and instrument parameter for KRAS mutation detection can be seen in Tables S3–S5.
Declaration of competing interest
All authors declare that they have no conflicts of interest and no competing interest.
CRediT authorship contribution statement
Haicong Shen: project design, experiment operation, formal analysis, manuscript writing. Rui Su: clinical sample acquisition, clinical data analysis. Jiao Peng: chip fabrication, chip modification. Lin Zhu: chip design, experimental assistance. Kunyue Deng: chip fabrication, material preparation. Qi Niu: experiment assistance, formal analysis. Yanling Song: experimental guidance, research design. Liu Yang: funding acquisition, experiment assistance. Lingling Wu: experimental guidance, research design, manuscript revision.. Zhi Zhu: funding acquisition, research design, supervised experiments, manuscript revision. Chaoyong Yang: funding acquisition, manuscript revision, All authors reviewed the manuscript.
Acknowledgements
We thank the National Natural Science Foundation of China (21775128, 21974113, 21735004, 21974112, and 21874089), National Key R&D Program of China (2019YFA0905800), Program for Chang Jiang Scholars and Innovative Research Teams in University (IRT13036), Medical and Health Program of Xiamen (3502Z20189005) and the National Science Fund for Fostering Talents in Basic Science (NFFTBS, J1310024) for their financial support.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2021.09.034.
Contributor Information
Lingling Wu, Email: linglingwu1990@126.com.
Zhi Zhu, Email: zhuzhi@xmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Yu L., Ng S.R., Xu Y., Dong H., Wang Y.J., Li C.M. Advances of lab-on-a-chip in isolation, detection and post-processing of circulating tumour cells. Lab Chip. 2013;13:3163–3182. doi: 10.1039/c3lc00052d. [DOI] [PubMed] [Google Scholar]
- 2.Maheswaran S., Sequist L.V., Nagrath S., Ulkus L., Brannigan B., Collura C.V., Inserra E., Diederichs S., Iafrate A.J., Bell D.W., Digumarthy S., Muzikansky A., Irimia D., Settleman J., Tompkins R.G., Lynch T.J., Toner M., Haber D.A. Detection of mutations in EGFR in circulating lung-cancer cells, N. Engl. J. Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wan Y., Winter M., Delalat B., Hardingham J.E., Grover P.K., Wrin J., Voelcker N.H., Price T.J., Thierry B. Nanostructured polystyrene well plates allow unbiased high-throughput characterization of circulating tumor cells. ACS Appl. Mater. Interfaces. 2014;6:20828–20836. doi: 10.1021/am505201s. [DOI] [PubMed] [Google Scholar]
- 4.Lin H.K., Zheng S., Williams A.J., Balic M., Groshen S., Scher H.I., Fleisher M., Stadler W., Datar R.H., Tai Y.-C., Cote R.J. Portable filter-based microdevice for detection and characterization of circulating tumor cells. Clin. Cancer Res. 2010;16:5011–5018. doi: 10.1158/1078-0432.CCR-10-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu M., Bardia A., Aceto N., Bersani F., Madden M.W., Donaldson M.C., Desai R., Zhu H., Comaills V., Zheng Z., Wittner B.S., Stojanov P., Brachtel E., Sgroi D., Kapur R., Shioda T., Ting D.T., Ramaswamy S., Getz G., Iafrate A.J., Benes C., Toner M., Maheswaran S., Haber D.A. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216–220. doi: 10.1126/science.1253533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aceto N., Bardia A., Miyamoto David T., Donaldson Maria C., Wittner Ben S., Spencer Joel A., Yu M., Pely A., Engstrom A., Zhu H., Brannigan Brian W., Kapur R., Stott Shannon L., Shioda T., Ramaswamy S., Ting David T., Lin Charles P., Toner M., Haber Daniel A., Maheswaran S. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baccelli I., Schneeweiss A., Riethdorf S., Stenzinger A., Schillert A., Vogel V., Klein C., Saini M., Bäuerle T., Wallwiener M., Holland-Letz T., Höfner T., Sprick M., Scharpff M., Marmé F., Sinn H.P., Pantel K., Weichert W., Trumpp A. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat. Biotechnol. 2013;31:539–544. doi: 10.1038/nbt.2576. [DOI] [PubMed] [Google Scholar]
- 8.Kalinich M., Bhan I., Kwan T.T., Miyamoto D.T., Javaid S., LiCausi J.A., Milner J.D., Hong X., Goyal L., Sil S., Choz M., Ho U., Kapur R., Muzikansky A., Zhang H., Weitz D.A., Sequist L.V., Ryan D.P., Chung R.T., Zhu A.X., Isselbacher K.J., Ting D.T., Toner M., Maheswaran S., Haber D.A. An RNA-based signature enables high specificity detection of circulating tumor cells in hepatocellular carcinoma. Proc. Natl. Acad. Sci. U.S.A. 2017;114:1123–1128. doi: 10.1073/pnas.1617032114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hao S.-J., Wan Y., Xia Y.-Q., Zou X., Zheng S.-Y. Size-based separation methods of circulating tumor cells. Adv. Drug Deliv. Rev. 2018;125:3–20. doi: 10.1016/j.addr.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Alix-Panabieres C., Pantel K. OPINION Challenges in circulating tumour cell research. Nat. Rev. Cancer. 2014;14:623–631. doi: 10.1038/nrc3820. [DOI] [PubMed] [Google Scholar]
- 11.Bankó P., Lee S.Y., Nagygyörgy V., Zrínyi M., Chae C.H., Cho D.H., Telekes A. Technologies for circulating tumor cell separation from whole blood. J. Hematol. Oncol. 2019;12 doi: 10.1186/s13045-019-0735-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allard W.J., Matera J., Miller M.C., Repollet M., Connelly M.C., Rao C., Stopeck A., Terstappen L. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with non-malignant diseases. J. Clin. Oncol. 2004;22 doi: 10.1158/1078-0432.CCR-04-0378. 847S-847S. [DOI] [PubMed] [Google Scholar]
- 13.Riethdorf S., Muller V., Coith C., Baack K., Janicke F., Fritsche H.A., Jr., Jackson S., Gornet T., Pantel M. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer - a multi-center validation study of the CellSearch (TM) system. Proc. Am. Assoc. Cancer Res. Annu. Meet. 2006;47 doi: 10.1158/1078-0432.CCR-06-1695. 1207-1207. [DOI] [PubMed] [Google Scholar]
- 14.Nagrath S., Sequist L.V., Maheswaran S., Bell D.W., Irimia D., Ulkus L., Smith M.R., Kwak E.L., Digumarthy S., Muzikansky A., Ryan P., Balis U.J., Tompkins R.G., Haber D.A., Toner M. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450 doi: 10.1038/nature06385. 1235-U10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang Y.-T., Hadlock T., Lo T.-W., Purcell E., Mutukuri A., Fouladdel S., Raguera M.D.S., Fairbairn H., Murlidhar V., Durham A., McLean S.A., Nagrath S. Dual-isolation and profiling of circulating tumor cells and cancer exosomes from blood samples with melanoma using immunoaffinity-based microfluidic interfaces. Advanced Science. 2020;7 doi: 10.1002/advs.202001581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L.X., Asghar W., Demirci U., Wan Y. Nanostructured substrates for isolation of circulating tumor cells. Nano Today. 2013;8:374–387. doi: 10.1016/j.nantod.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stott S.L., Hsu C.H., Tsukrov D.I., Yu M., Miyamoto D.T., Waltman B.A., Rothenberg S.M., Shah A.M., Smas M.E., Korir G.K., Floyd F.P., Gilman A.J., Lord J.B., Winokur D., Springer S., Irimia D., Nagrath S., Sequist L.V., Lee R.J., Isselbacher K.J., Maheswaran S., Haber D.A., Toner M. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glia A., Deliorman M., Sukumar P., Janahi F.K., Samara B., Brimmo A.T., Qasaimeh M.A. Herringbone microfluidic probe for multiplexed affinity-capture of prostate circulating tumor cells. Advanced Materials Technologies. 2021;6 [Google Scholar]
- 19.Zhang Q., Mao S.F., Li W.W., Huang Q.S., Feng S., Hong Z.Y., Lin J.M. Microfluidic adhesion analysis of single glioma cells for evaluating the effect of drugs. Sci. China Chem. 2020;63:865–870. [Google Scholar]
- 20.Zhang Q., Fang R.H., Gao W., Zhang L. A biomimetic nanoparticle to "lure and kill" phospholipase A2. Angew. Chem. Int. Ed. 2020;59:10461–10465. doi: 10.1002/anie.202002782. [DOI] [PubMed] [Google Scholar]
- 21.Monteiro N., Martins A., Reis R.L., Neves N.M. Liposomes in tissue engineering and regenerative medicine. J. R. Soc. Interface. 2014;11 doi: 10.1098/rsif.2014.0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao L., Bu L.-L., Cai B., Xu J.-H., Li A., Zhang W.-F., Sun Z.-J., Guo S.-S., Liu W., Wang T.-H., Zhao X.-Z. Cancer cell membrane-coated upconversion nanoprobes for highly specific tumor imaging. Adv. Mater. 2016;28:3460–3466. doi: 10.1002/adma.201506086. [DOI] [PubMed] [Google Scholar]
- 23.Fang R.H., Hu C.-M.J., Luk B.T., Gao W., Copp J.A., Tai Y., O'Connor D.E., Zhang L. Cancer cell membrane-coated nanoparticles for anticancer vaccination and drug delivery. Nano Lett. 2014;14:2181–2188. doi: 10.1021/nl500618u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rong Y., Zhang Z., He C.L., Chen X.S. Bioactive polypeptide hydrogels modified with RGD and N-cadherin mimetic peptide promote chondrogenic differentiation of bone marrow mesenchymal stem cells. Sci. China Chem. 2020;63:1100–1111. [Google Scholar]
- 25.Guo W.S., Liu L., Xiang C.Y., Chen J.Q., Liang X.J. Engineered nanoparticles circumvent the adaptive treatment tolerance to immune-checkpoint blockade therapy. Sci. China Chem. 2019;62:1557–1560. [Google Scholar]
- 26.Li R., He Y., Zhang S., Qin J., Wang J. Cell membrane-based nanoparticles: a new biomimetic platform for tumor diagnosis and treatment. Acta Pharm. Sin. B. 2018;8:14–22. doi: 10.1016/j.apsb.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang Z.-m., Zhou H., Yang C., Zhang R., You Q., Yan R., Li L., Ge M., Tang Y., Dong W.-f., Wang Z. Biomimetic immunomagnetic gold hybrid nanoparticles coupled with inductively coupled plasma mass spectrometry for the detection of circulating tumor cells. J. Mater. Chem. B. 2020;8:5019–5025. doi: 10.1039/d0tb00403k. [DOI] [PubMed] [Google Scholar]
- 28.Huang L.-L., Nie W., Zhang J., Xie H.-Y. Cell-membrane-based biomimetic systems with bioorthogonal functionalities. Acc. Chem. Res. 2020;53:276–287. doi: 10.1021/acs.accounts.9b00559. [DOI] [PubMed] [Google Scholar]
- 29.Xiong K., Wei W., Jin Y., Wang S., Zhao D., Wang S., Gao X., Qiao C., Yue H., Ma G., Xie H.-Y. Biomimetic immuno-magnetosomes for high-performance enrichment of circulating tumor cells. Adv. Mater. 2016;28:7929–7935. doi: 10.1002/adma.201601643. [DOI] [PubMed] [Google Scholar]
- 30.Meng Q.-F., Cheng Y.-X., Huang Q., Zan M., Xie W., Sun Y., Li R., Wei X., Guo S.-S., Zhao X.-Z., Rao L., Liu W. Biomimetic immunomagnetic nanoparticles with minimal nonspecific biomolecule adsorption for enhanced isolation of circulating tumor cells. ACS Appl. Mater. Interfaces. 2019;11:28732–28739. doi: 10.1021/acsami.9b10318. [DOI] [PubMed] [Google Scholar]
- 31.Zhang F., Wu L., Nie W., Huang L., Zhang J., Li F., Xie H.-Y. Biomimetic microfluidic system for fast and specific detection of circulating tumor cells. Anal. Chem. 2019;91:15726–15731. doi: 10.1021/acs.analchem.9b03920. [DOI] [PubMed] [Google Scholar]
- 32.Wu L.L., Ding H.M., Qu X., Shi X.N., Yang J.M., Huang M.J., Zhang J.L., Zhang H.M., Song J., Zhu L., Song Y.L., Ma Y.Q., Yang C.Y. Fluidic multivalent membrane nanointerface enables synergetic enrichment of circulating tumor cells with high efficiency and viability. J. Am. Chem. Soc. 2020;142:4800–4806. doi: 10.1021/jacs.9b13782. [DOI] [PubMed] [Google Scholar]
- 33.Ding P., Wang Z., Wu Z., Zhou Y., Sun N., Pei R. Natural biointerface based on cancer cell membranes for specific capture and release of circulating tumor cells. ACS Appl. Mater. Interfaces. 2020;12:20263–20270. doi: 10.1021/acsami.0c03355. [DOI] [PubMed] [Google Scholar]
- 34.Guo J.M., Yu Y.L., Zhu W., Serda R.E., Franco S., Wang L., Lei Q., Agola J.O., Noureddine A., Ploetz E., Wuttke S., Brinker C.J. Modular assembly of red blood cell superstructures from metal-organic framework nanoparticle-based building blocks. Adv. Funct. Mater. 2020;31 [Google Scholar]
- 35.Sun L.L., Shen F.Y., Xu J., Han X., Fan C.H., Liu Z. DNA-edited ligand positioning on red blood cells to enable optimized T cell activation for adoptive immunotherapy. Angew. Chem. Int. Ed. 2020;59:14842–14853. doi: 10.1002/anie.202003367. [DOI] [PubMed] [Google Scholar]
- 36.Koleva L., Bovt E., Ataullakhanov F., Sinauridze E. Erythrocytes as carriers: from drug delivery to biosensors. Pharmaceutics. 2020;12 doi: 10.3390/pharmaceutics12030276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Villa C.H., Anselmo A.C., Mitragotri S., Muzykantov V. Red blood cells: supercarriers for drugs, biologicals, and nanoparticles and inspiration for advanced delivery systems. Adv. Drug Deliv. Rev. 2016;106:88–103. doi: 10.1016/j.addr.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bogdanova A., Kaestner L. E. European red cell soc, early career scientists’ guide to the red blood cell - don’t panic! Front. Physiol. 2020;11 doi: 10.3389/fphys.2020.00588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGrath J., Jimenez M., Bridle H. Deterministic lateral displacement for particle separation: a review. Lab Chip. 2014;14:4139–4158. doi: 10.1039/c4lc00939h. [DOI] [PubMed] [Google Scholar]
- 40.Davis J.A., Inglis D.W., Morton K.J., Lawrence D.A., Huang L.R., Chou S.Y., Sturm J.C., Austin R.H. Deterministic hydrodynamics: taking blood apart. Proc. Natl. Acad. Sci. Unit. States Am. 2006;103:14779–14784. doi: 10.1073/pnas.0605967103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahmed M.G., Abate M.F., Song Y., Zhu Z., Yan F., Xu Y., Wang X., Li Q., Yang C. Isolation, detection, and antigen-based profiling of circulating tumor cells using a size-dictated immunocapture chip. Angew. Chem. Int. Ed. 2017;56:10681–10685. doi: 10.1002/anie.201702675. [DOI] [PubMed] [Google Scholar]
- 42.Wu L., Ding H., Qu X., Shi X., Yang J., Huang M., Zhang J., Zhang H., Song J., Zhu L., Song Y., Ma Y., Yang C. Fluidic multivalent membrane nanointerface enables synergetic enrichment of circulating tumor cells with high efficiency and viability. J. Am. Chem. Soc. 2020;142:4800–4806. doi: 10.1021/jacs.9b13782. [DOI] [PubMed] [Google Scholar]
- 43.Sun L., Shen F., Xu J., Han X., Fan C., Liu Z. DNA-edited ligand positioning on red blood cells to enable optimized. T Cell Activation for Adoptive Immunotherapy. 2020;59:14842–14853. doi: 10.1002/anie.202003367. [DOI] [PubMed] [Google Scholar]
- 44.Zhu D.-M., Wu L., Suo M., Gao S., Xie W., Zan M.-H., Liu A., Chen B., Wu W.-T., Ji L.-W., Chen L.-b., Huang H.-M., Guo S.-S., Zhang W.-F., Zhao X.-Z., Sun Z.-J., Liu W. Engineered red blood cells for capturing circulating tumor cells with high performance. Nanoscale. 2018;10:6014–6023. doi: 10.1039/c7nr08032h. [DOI] [PubMed] [Google Scholar]
- 45.Dong H., Xu X., Wang L.K., Mo R. Advances in living cell-based anticancer therapeutics. Biomater. Sci. 2020;8:2344–2365. doi: 10.1039/d0bm00036a. [DOI] [PubMed] [Google Scholar]
- 46.Krebs M.G., Metcalf R.L., Carter L., Brady G., Blackhall F.H., Dive C. Molecular analysis of circulating tumour cells-biology and biomarkers. Nat. Rev. Clin. Oncol. 2014;11:129–144. doi: 10.1038/nrclinonc.2013.253. [DOI] [PubMed] [Google Scholar]
- 47.Zhang J., Lin B., Wu L., Huang M., Li X., Zhang H., Song J., Wang W., Zhao G., Song Y., Yang C. DNA nanolithography enables a highly ordered recognition interface in a microfluidic chip for the efficient capture and release of circulating tumor cells. Angew. Chem. Int. Ed. 2020;59:14115–14119. doi: 10.1002/anie.202005974. [DOI] [PubMed] [Google Scholar]
- 48.Karabacak N.M., Spuhler P.S., Fachin F., Lim E.J., Pai V., Ozkumur E., Martel J.M., Kojic N., Smith K., Chen P.-i., Yang J., Hwang H., Morgan B., Trautwein J., Barber T.A., Stott S.L., Maheswaran S., Kapur R., Haber D.A., Toner M. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat. Protoc. 2014;9:694–710. doi: 10.1038/nprot.2014.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.