Abstract
Male sterility is one of the reproductive isolation systems in plants and quite useful for F1 seed production. We previously identified three independent quantitative trait loci (QTLs) for male sterility of cultivated strawberry, Here, we identified the specific subgenomes in which these QTLs are located by QTL-seq approach. QTLs qMS4.1, qMS4.2, and qMS4.3 were mapped separately in subgenomes Fvb4-4, Fvb4-3, and Fvb4-1, respectively, in ‘Camarosa’ genome assembly v. 1.0.a1. Candidate regions of qMS4.1 and qMS4.3 were clearly detected around 12–26 Mb in Fvb4-4 and 12–14 Mb in Fvb4-1, respectively; those of qMS4.2 were fragmented in Fvb4-3, which suggests that some scaffolds were incorrectly assembled in Fvb4-3. qMS4.3 was mapped to chr4X1 of ‘Reikou’ genome assembly r2.3, and qMS4.1 and qMS4.2 were both mapped to chr4Av, which indicates that differentiation of the subgenomes in which both QTLs are located was insufficient in ‘Reikou’ r2.3. Although ‘Camarosa’ genome assembly v. 1.0.a1 is an unphased map, which merges homologous chromosomes into one sequence, ‘Reikou’ genome assembly r2.3 is a phased map, which separates homologous chromosomes. QTL mapping to different reference genomes clearly showed the specific features of each reference genome, and that using different kinds of reference map could accelerate fine mapping and map-based cloning of certain genes of cultivated strawberry.
Keywords: ‘Camarosa’, mapping, QTL-seq, ‘Reikou’, strawberry
Introduction
Male sterility is defined as one of the reproductive isolation symptoms and frequently observed in many plants (Mohan and Kaul 1988). This feature not only attracts interest of researchers from the viewpoint of phylogenetic evolution but also is practically useful for F1 seed production of many commercial crops, because application of male sterility could reduce labor cost for artificial emasculation and pollination (Duvick 1959).
Cultivated strawberry (Fragaria × ananassa) is one of the most commercially important berry fruits in the world. Although genetic and phylogenetic analysis for male sterility had been conducted with many wild Fragaria species (Ashman et al. 2015, Goldberg et al. 2010, Govindarajulu et al. 2013, Spigler et al. 2008, 2010, Spigler and Ashman 2011, Tennessen et al. 2013, 2016, Wei et al. 2017), the male sterility of cultivated strawberry (Fragaria × ananassa) had not been reported until recently. Therefore, uncovering genetic regions for male sterility in cultivated strawberry was a critical issue to be solved from not only phylogenetic view point but also practical use for providing strawberry seedlings efficiently to local farmers.
Since the genome of cultivated strawberry is octoploid, alloploid, and highly heterozygous, it has been challenging to define its genome conformation and detect specific genomic regions related to agronomically important traits. However, recent advances in genome analysis and sequencing technology have made it possible to uncover the genome structure. Rousseau-Gueutin et al. (2008) conducted a phylogenetic analysis for the evolutionary history of current genome conformation of Fragaria species and supported the AA Aʹ Aʹ BB Bʹ Bʹ structure of cultivated strawberry proposed by Bringhurst (1990). Tennessen et al. (2014) studied the genome conformation and proposed the existence of Av, Bi, B1, and B2 subgenomes, of which one subgenome Av was derived from Fragaria vesca and the other three from Fragaria iinumae.
In our previous research, we performed QTL analysis using expressed sequence tag-simple sequence repeat (EST-SSR) markers (Isobe et al. 2013) and identified three independent quantitative traits loci (QTLs) for male sterility of cultivated strawberry (Wada et al. 2020). We also revealed that the accumulation of recessive alleles of the three QTLs induced male sterility (Wada et al. 2020) and all the primer sequences of the flanking SSR markers for the three QTLs were estimated to be derived from Fvb4 of the diploid F. vesca genome (Shulaev et al. 2011, Tennessen et al. 2013). Although our previous study was the first report of male sterility of cultivated strawberry, F. × ananassa ssp. ananassa. it did not detect specific genomic regions for the QTLs in the cultivated strawberry genome. As for the location of genetic regions for male sterility, Ashman et al. (2015) reported a QTLs for male sterility of F. vesca ssp. bracteata in Fvb4. Although Ashman et al. (2015) reported a different QTL for male sterility in Fvb6, the QTL in Fvb6 became functional only when the QTL in Fvb4 was not functional. These facts strongly suggest that genomic regions in Fvb4 are key regions for the control of male sterility in cultivated strawberry.
Recently, Edger et al. (2019b) reported a near-complete chromosome-scale assembly for cultivated strawberry using the American cultivar ‘Camarosa’ and proposed a novel phylogenetic theory for the origin of cultivated strawberry. According to their theory, cultivated strawberry originated from four different diploid ancestors: F. vesca, F. iinumae, F. viridis, and F. nipponica. Although their phylogenetic theory is debated (Edger et al. 2019a, Liston et al. 2019), the ‘Camarosa’ assembly they defined has been used as a reference genome for genomic studies of cultivated strawberry (Barbey et al. 2020, Negrini et al. 2020). A different chromosome-scale assembly for cultivated strawberry was developed using the Japanese cultivar ‘Reikou’ (https://www.biorxiv.org/content/10.1101/2021.04.23.441065v1). The ‘Camarosa’ map was an unphased map that merged homologous chromosomes into one sequence. In contrast, the ‘Reikou’ map was a phased map that differentiated homologous chromosomes. In order to detect chromosome conformation, find a linkage between a preferable allele of a certain gene and DNA marker accurately, and detect desirable haplotype, phased map is an ideal one (Browning and Browning 2011, Loh et al. 2016). But read length of recent next-generation sequencing technology was too short to define haplotype information of individual chromosomes (Leitwein et al. 2020). Therefore, constructing accurate phased map is challenging approach and utilizing unphased map is quite useful to map several QTLs efficiently. Since both maps have unique features, they will contribute in different ways to the progress of genomic research on cultivated strawberry.
The objective of this study was to map the three QTLs for male sterility that were identified by Wada et al. (2020) in two strawberry reference genome sequences on different reference maps (‘Camarosa’ and ‘Reikou’) of the cultivated strawberry genome (Edger et al. 2019b, https://www.biorxiv.org/content/10.1101/2021.04.23.441065v1).
Materials and Methods
Plant materials
The plant materials used in this study are listed in Table 1.
Table 1.
Plant materials used in this study
| Source | Generation | Target | Number of plantsa | |
|---|---|---|---|---|
| F_bulk | S_bulk | |||
| Fukuoka S6 | S2 | qMS4.1 | 20 | 20 |
| Fukuoka S6 | S1, S2 | qMS4.2 | 16 | 16 |
| Kaorino | S1 | qMS4.3 | 12 | 8 |
a ‘F_bulk’ and ‘S_bulk’ indicate a population which comprise of male-fertile and male-sterile plants, respectively.
(1) qMS4.1: The S2 population of ‘Fukuoka S6’ was used. As we revealed previously (Wada et al. 2020), ‘Fukuoka S6’ harbors two heterozygous alleles (in QTL regions qMS4.1 and qMS4.2) and a recessive homozygous allele (in QTL region qMS4.3). In the DNA marker-assisted selection (MAS) conducted here, we used the SSR marker FVES1264 for genotyping of qMS4.1 and FVES1356 for qMS4.2, both of which were developed by Isobe et al. (2013) and selected as flanking markers for the two QTLs (Wada et al. 2020). Before selecting S2 plants, we selected S1 plants derived from self-pollination of ‘Fukuoka S6’, which harbored a heterozygous allele in qMS4.1 and a recessive allele in qMS4.2, and conducted self-pollination of the relevant S1 plant to obtain S2 seeds. From the S2 population, we further selected 20 male-fertile and 20 male-sterile plants in the following manner: We grew 200 plants in a greenhouse and confirmed their male fertility or sterility at anthesis by anther color (Wada et al. 2020). From the male-fertile plants, we used MAS to select “perfect” male-fertile plants that carried dominant homozygote alleles and whose self-pollinated progeny did not include male-sterile plants. From the 200 plants, we also selected 20 male-sterile plants. We used these 40 plants in our QTL-seq analysis (Takagi et al. 2013).
(2) qMS4.2: We used a selection method similar to that for qMS4.1 to choose the S2 population of ‘Fukuoka S6’. However, we obtained only 6 perfect male-fertile plants and 4 male-sterile plants using MAS for use in our QTL-seq analysis.
(3) qMS4.3: We used a selection method similar to that for qMS4.1 to choose the S1 population of ‘Kaorino’. ‘Kaorino’ harbors one heterozygous allele in qMS4.3, and recessive homozygous alleles in qMS4.1 and qMS4.2 (Wada et al. 2020). We grew 50 S1 plants in a greenhouse, and used MAS to select 12 perfect male-fertile plants and 8 male-sterile plants for use in our QTL-seq analysis.
DNA extraction and genotyping for MAS
DNA extraction was performed with a DNeasy Plant Mini Kit (Qiagen Inc., Hilden, Germany) using young leaves from all of the selected plants. The polymerase chain reaction (PCR) was performed in an 8-μL reaction volume that included 1.0 ng of genomic DNA, 4.0 pmol of each primer (Supplemental Table 1), and 4 μL of 2× GoTaq Green master mix (Promega, Fitchburg, Wisconsin, USA). The amplification profile was as follows: an initial 5 min at 95°C, followed by 30 cycles of 30 s at 95°C, 30 s at 55°C, and 45 s at 72°C, and then 5 min at 72°C for final extension. PCR was done in a Biometra Tadvanced 384 thermal cycler (Analytik Jena AG, Jena, Germany). Amplified products were electrophoresed in non-denatured 12.0% polyacrylamide gels and the gels were stained with GelRed solution (Biotium, Fremont, CA, USA) according to the manufacturer’s protocol. The electropherogram was observed with a TP-20MP UV transilluminator (Atto Co., Ltd., Tokyo, Japan).
Sample preparation and whole-genome sequencing of bulked DNA for QTL-seq
For all three QTLs (qMS4.1, qMS4.2, and qMS4.3), we performed QTL-seq to detect candidate chromosomes and genomic regions. QTL-seq is an effective way to identify a candidate genomic region, which is responsible for a specific agronomic trait, and was proposed for the first time by Takagi et al. (2013). First, we prepared a bulked DNA pool for construction of the DNA library. As an example, for qMS4.1, we adjusted the DNA concentration of 20 perfect male-fertile plants to 50 ng/μL, and mixed 20 μL of each DNA solution to obtain one bulked DNA sample for DNA of the 20 (F_bulk). We prepared a bulked DNA sample from DNA of the 20 male-sterile plants (S_bulk) in the same manner, and used samples of both bulked DNA pools (F_bulk and S_bulk) for construction of the DNA library. Second, we constructed the DNA library and performed adapter ligation using the TruSeq DNA PCR-Free Library Prep Kit and TruSeq DNA Single Indexes (Illumina Inc., San Diego, CA, USA). Finally, we performed paired-end 150 bp sequencing using both DNA libraries on a HiSeq X sequencing platform (Illumina). Any short reads with a phred quality score of <30 (Q30) were excluded from the subsequent analysis.
Cleaning raw reads, mapping, and variant calls
The fastq files that contained the obtained raw reads were trimmed and cleaned with Trimmomatic software v. 0.39 (Bolger et al. 2014). Sequences were mapped to the reference sequences in BWA software (Li and Durbin 2009) separately using ‘Camarosa’ genome assembly v. 1.0.a1 (Edger et al. 2019b) and ‘Reikou’ genome assembly r2.3 (https://www.biorxiv.org/content/10.1101/2021.04.23.441065v1). The constructed sequence alignment map (sam) files were converted and sorted to produce binary alignment map (bam) files, then the duplicated reads generated as PCR duplication artifacts were excluded from the sorted bam files by the Markduplicates function of Picard tools (http://broadinstitute.github.io/picard). Variant calls were performed using the bam files for F_bulk and S_bulk in samtools software (https://github.com/samtools) and bcftools software (https://github.com/samtools/bcftools) to generate variant call format (vcf) files, then the generated vcf files were converted to tidy data to obtain comma-separated value (csv) files in the vcfR package for R software (Knaus and Grünwald 2017). The alternate allele frequency (Alt_AF) for each single-nucleotide polymorphism (SNP) position in F_bulk and S_bulk was calculated with the dplyr package for R software (Wickham et al. 2019), then we calculated a moving average of Alt_AF with the simpleSmoothTs command of the latticeExtra package (Sarkar and Andrews 2016) using Alt_AF values of the 1,000 front and 1,000 back SNPs. The regions where the difference in Alt_AF values between the two bulk populations was greater than the threshold value (P < 0.01) were designated as candidate genomic regions for the relevant QTL.
Validation of the QTL-seq results using amplified sequences of the flanking DNA markers
In our previous study, we selected the following SSR markers that were located near the male-sterility QTLs: FVES1264_196 for qMS4.1 (where the later numerical part represents the PCR amplicon base pair size), FVES1356_195 for qMS4.2, and FAES0001_269 for qMS4.3. We then conducted a BLAST search with ncbi-blast 2.8.1+ (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/) using the amplicon sequence (Supplemental Table 1) of those SSR markers in each reference map, and tried to detect the subgenome in which the SSR markers were located with the highest confidence.
Results
Sequence and mapping information from QTL-seq
Table 2 summarizes the sequence and mapping results. The largest amount of sequence for the three QTLs was 96,661 Mbp (F_bulk of qMS4.3), and the shortest sequence was 37 604 Mbp (S_bulk of qMS4.3). Since the genome size of cultivated strawberry was predicted to be 698 Mb (Hirakawa et al. 2014) and 805 Mbp (Edger et al. 2019b), the read depth of sequence in this study was estimated at 53.9× to 138.5× coverage of the cultivated strawberry genome.
Table 2.
Sequence and mapping information of QTL-seq
| Target | Population | Reference | Total reads (M) | Total bases (Mbp) | Q30a Reads (Mbp) | Q30 (%) |
Reads mapped (M) | Reads unmapped (M) | Mapping rate (%) |
|---|---|---|---|---|---|---|---|---|---|
| qMS4.1 | F_bulk | Camarosa | 522 | 78,865 | 75,334 | 95.5 | 503.0 | 10.8 | 97.9 |
| Reikou | 508.3 | 5.5 | 98.9 | ||||||
| S_bulk | Camarosa | 487 | 73,558 | 70,130 | 95.3 | 469.1 | 9.6 | 98.0 | |
| Reikou | 474.0 | 4.7 | 99.0 | ||||||
| qMS4.2 | F_bulk | Camarosa | 538 | 81,268 | 75,868 | 93.4 | 505.0 | 8.0 | 98.4 |
| Reikou | 424.6 | 3.6 | 99.2 | ||||||
| S_bulk | Camarosa | 496 | 74,946 | 70,029 | 93.4 | 466.3 | 7.3 | 98.5 | |
| Reikou | 394.4 | 3.3 | 99.2 | ||||||
| qMS4.3 | F_bulk | Camarosa | 640 | 96,661 | 90,479 | 93.6 | 612.1 | 23.8 | 96.3 |
| Reikou | 620.7 | 15.3 | 97.6 | ||||||
| S_bulk | Camarosa | 249 | 37,604 | 35,155 | 93.5 | 238.3 | 9.1 | 96.3 | |
| Reikou | 241.5 | 5.8 | 97.6 |
a Q30 indicates the proportion that met the criterion of phred quality <30.
(1) qMS4.1: Total reads, total bases, Q30 reads, and mapped reads were higher in F_bulk than in S_bulk. However, the proportion of Q30 reads (i.e., the proportion that met the criterion of phred quality <30) of F_bulk (95.5%) was almost equal to that of S_bulk (95.3%). Comparing the results of mapping to both reference maps, the number of reads mapped to ‘Camarosa’ genome assembly v. 1.0 (503.0 million of F_bulk and 469.1 million of S_bulk) was almost equal to that for ‘Reikou’ genome assembly r2.3 (508.3 million of F_bulk and 474.0 million of S_bulk). Therefore, the proportions of mapping to ‘Camarosa’ (97.9%, 98.0%) were slightly lower than those to ‘Reikou’ (98.9%, 99.0%).
(2) qMS4.2: Total reads, total bases, Q30 reads, and mapped reads were higher in F_bulk than in S_bulk, as in qMS4.1. Similarly, the proportion of Q30 reads of F_bulk (93.4%) was equal to that of S_bulk (93.4%). But the mapping results were the opposite of the qMS4.1 results; the number of reads mapped to ‘Camarosa’ was higher (505.0 million of F_bulk and 466.3 million of S_bulk) than the number mapped to ‘Reikou’ (424.6 million of F_bulk and 394.4 million of S_bulk). Therefore, the proportions of mapping to ‘Camarosa’ (98.4%, 98.5%) were slightly lower than those to ‘Reikou’ (99.2%, 99.2%).
(3) qMS4.3: Total reads, total bases, Q30 reads, and mapped reads were much higher in F_bulk than in S_bulk. Although the difference between the bulk populations was large, the trend was the same as in the qMS4.1 and qMS4.2 results. However, the proportion of Q30 reads of F_bulk (93.6%) was equal to that of S_bulk (93.5%), as in the qMS4.1 and qMS4.2 results. Mapped reads to ‘Camarosa’ (612.1 million of F_bulk and 238.3 million of S_bulk) were almost equal to those to ‘Reikou’ (620.7 million of F_bulk and 241.5 million of S_bulk). Therefore, the proportions of mapping to ‘Camarosa’ (96.3%, 96.3%) were also slightly lower than those to ‘Reikou’ (97.6%, 97.6%).
Mapping of QTLs for male sterility to the ‘Camarosa’ reference genome
(1) qMS4.1: Supplemental Fig. 1 shows the shift of the moving average of Alt_AF throughout the chromosomes. Significant differences of Alt_AF were detected only around the 12 to 26 Mb regions of Fvb4-4 (Fig. 1). Although the top hit chromosome region of the FVES1264_196 amplicon sequence (Supplemental Table 1) was detected in Fvb4-2 (Supplemental Table 2), the region in Fvb4-4 was the third hit.
Fig. 1.
Candidate genomic regions of qMS4.1 on Fvb4-4 of ‘Camarosa’ genome assembly v. 1.0.a1 as a reference genome. Vertical and horizontal axis indicates frequency of alternate allele (Alt_AF) reads out of total reads in each bulk population and physical position of each subgenome, respectively. Each line graph was drawn based on the moving average value calculated based on each Alt_AF of front and back 1000 SNP positions. Gray area indicates 99% confidence intervals of the difference of Alt_AF.
(2) qMS4.2: Supplemental Fig. 2 shows the shift of the moving average of Alt_AF throughout the chromosomes. Significant differences of Alt_AF were detected only around the 2 to 28 Mb regions of Fvb4-3 (Fig. 2), but the significant regions were dispersed discontinuously throughout this region. The top hit chromosome region of the FVES1356_195 amplicon sequence was detected in Fvb4-3 (Supplemental Table 2), which also matched the results of the QTL-seq.
Fig. 2.
Candidate genomic regions of qMS4.2 on Fvb4-3 of ‘Camarosa’ genome assembly v. 1.0.a1 as a reference genome. Other footnotes were the same as Fig. 1.
(3) qMS4.3: Supplemental Fig. 3 shows the shift of the moving average of Alt_AF throughout the chromosomes. Significant differences of Alt_AF were detected only around the 12 to 14 Mb regions of Fvb4-1 (Fig. 3), but the significant regions were divided into two parts. The top hit chromosome region of the FAES0001_269 amplicon sequence was in Fvb4-1 (Supplemental Table 2), which also matched the results of the QTL-seq. But the matched region (6.6 Mb) of FAES0001_269 amplicon was different from the candidate region (12 to14 Mb) in Fvb4-1.
Fig. 3.
Candidate genomic regions of qMS4.3 on Fvb4-1 of ‘Camarosa’ genome assembly v. 1.0.a1 as a reference genome. Other footnotes were the same as Fig. 1.
Mapping of QTLs for male sterility to the ‘Reikou’ reference genome
(1) qMS4.1: Supplemental Fig. 4 shows the shift of the moving average of Alt_AF throughout the chromosomes. Significant differences of Alt_AF were detected around the 7 to 14 Mb regions of chr4Ava (Fig. 4A) and the 3 to 7 Mb regions of chr4Avb (Fig. 4B). Compared with the mapping results for ‘Camarosa’ (Fig. 1), the difference in Alt_AF of the two bulk populations was less clear. Furthermore, the top hit region of the FVES1264_196 amplicon sequence was ch0, implying that the specific subgenome involved with FVES1264_196 was unclear (Supplemental Table 3).
Fig. 4.
Candidate genomic regions of qMS4.1 on chr4Ava (A) and chr4Avb (B) of ‘Reikou’ genome assembly r2.3 as a reference genome. Other footnotes were the same as Fig. 1.
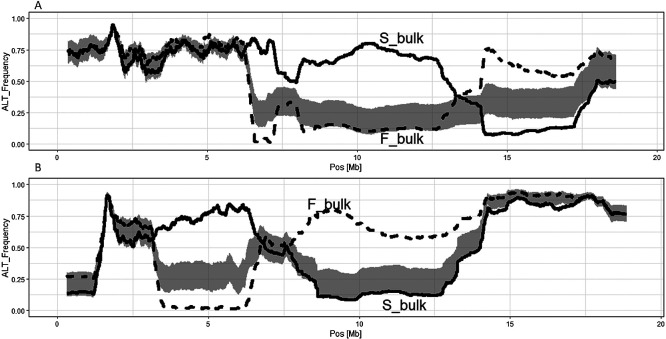
(2) qMS4.2: Supplemental Fig. 5 shows the shift of the moving average of Alt_AF throughout the chromosomes. Significant differences of Alt_AF were detected only around the 6.5 to 17 Mb regions of chr4Ava (Fig. 5A) and the 3 to 14 Mb regions of chr4Avb (Fig. 5B). The top hit chromosome region of the FVES1356_195 amplicon sequence was chr4Ava, and the second highest was chr4Avb (Supplemental Table 3), which also matched the results of the QTL-seq.
Fig. 5.
Candidate genomic regions of qMS4.2 on chr4Ava (A) and chr4Avb (B) of ‘Reikou’ genome assembly r2.3 as a reference genome. Other footnotes were the same as Fig. 1.
(3) qMS4.3: Supplemental Fig. 6 shows the shift of the moving average of Alt_AF throughout the chromosomes. Significant differences of Alt_AF were detected between the 6 to 15 Mb regions of chr4X1a and between the 4 to 13 Mb and 17 to 22 Mb regions of chr4X1b (Fig. 6). Significant regions on chr4X1b were divided into two regions of this subgenome. The top hit chromosome region of the FAES0001_269 amplicon sequence was chr4X1a, and the third highest was chr4X1b, which almost matched the results of the QTL-seq.
Fig. 6.
Candidate genomic regions of qMS4.3 on chr4X1a (A) and chr4X1b (B) of ‘Reikou’ genome assembly r2.3 as a reference genome. Other footnotes were the same as Fig. 1.
Discussion
Locations of QTLs for male sterility based on the reference genomes
In this study we detected candidate subgenomes in which the QTLs for male sterility were located, and successfully mapped qMS4.1, qMS4.2, and qMS4.3 in Fvb4-4, Fvb4-3, and Fvb4-1, respectively, of the ‘Camarosa’ genome assembly v. 1.0.a1 (Edger et al. 2019b).
We previously detected three independent male-sterility QTLs in the ‘Fukuoka S6’ and ‘Kaorino’ genetic backgrounds of cultivated strawberry, and hypothesized that these QTLs were located in the homoeologous regions of Fvb4 of wild strawberry, F. vesca (Wada et al. 2020). The present results strongly support those findings. In addition to that previous work, numerous studies have been conducted to reveal genetic regions responsible for male sterility of strawberry. Candidate chromosomes were Fvb4 (Ashman et al. 2015, Tennessen et al. 2013) and Fvb6 (Ashman et al. 2015, Govindarajulu et al. 2013, Spigler et al. 2008, 2010, Spigler and Ashman 2011) in F. vesca, and Fvb6 (Wei et al. 2017) in F. virginiana. From the viewpoint of allele effects, most of the male sterility regions harbor dominant alleles, except in the study of Ashman et al. (2015), which suggested that the QTL in Fvb6 has a recessive effect for male sterility only when the QTL in Fvb4 becomes non-functional. Taking those previous studies into account, our QTLs appear to be both novel QTLs for male sterility of cultivated strawberry and novel recessive QTLs in Fvb4.
It will be interesting to compare the map positions of the regions responsible for male sterility between our study and previous research. Edger et al. (2019b) successfully developed a first pseudomolecule for cultivated strawberry. However, we found no other studies that compared genetic regions of wild and cultivated strawberry except for our previous study (Wada et al. 2020). We hope that future studies will target the genetic regions responsible for male sterility.
Effectiveness and characteristics of QTL-seq approach
We used the QTL-seq approach (Takagi et al. 2013) to detect candidate chromosome regions for male sterility using male-fertile (F_bulk) and male-sterile (S_bulk) bulk populations (Table 2). The improvement of next-generation sequence technology contributed to the development of QTL-seq technique, and it has been applied for rapid detection of QTLs for many crops and vegetables. Lu et al. (2014) firstly identified a major QTL controlling early flowering in cucumber. Das et al. (2015) detected one major genomic region for seed weight of chickpea, and Wen et al. (2019) identified heat-tolerance QTLs for tomato. These previous studies and our study clearly demonstrated that QTL-seq approach was an effective and rapid detection method for detecting QTLs for agronomic traits.
In our QTL-seq result, the number of reads and bases for S_bulk were lower than those for F_bulk, especially in the analysis of qMS4.3. Although there are no previous reports which compared next-generation sequencer read length between male fertile plants and male sterile ones, the deletion of some genomic regions might have contributed to the loss of functions of some genes, which are responsible for the pollen fertility. Speaking of the difference of reference map, the mapping ratio for ‘Camarosa’ was slightly lower than that for ‘Reikou’ for all three QTLs. Since the materials used in this study (‘Fukuoka S6’ and ‘Kaorino’) are Japanese cultivars, the genetic distance of the American cultivar ‘Camarosa’ from these cultivars might be greater than that for the Japanese cultivar ‘Reikou’, and we would then expect more reads to be mapped to ‘Reikou’ than to ‘Camarosa’.
Comparative mapping of QTLs for male sterility using phased and unphased maps
As reference maps, we used ‘Camarosa’ genome assembly v. 1.0.a1 (Edger et al. 2019b) and ‘Reikou’ genome assembly r2.3 (https://www.biorxiv.org/content/10.1101/2021.04.23.441065v1). There are critical differences between these reference maps in addition to the differences between cultivars. In the ‘Camarosa’ map, subgenomes were designated as Fvb4-1, Fvb4-2, Fvb4-3, and Fvb4-4, which suggested that the haplotypes of homologous chromosomes were not differentiated and were unphased in this reference map. In contrast, the subgenomes were designated chr4Ava, chr4Avb, chr4Bia, chr4Bib, chr4X1a, chr4X1b, chr4X2a, and chr4X2b in the ‘Reikou’ map, which implied that haplotypes were differentiated and were phased in this map. Both maps were constructed using short reads from illumina HiSeq system and Chromium 10X Genomics data, which were initially assembled with DeNovo MAGIC (NRgene), but further assembly procedures were different from each other. In case of ‘Camarosa’ map, Dovetail HiC reads were used for HiRise (Putnam et al. 2016), and gapped regions were filled with PacBio reads. In contrast, linkage analysis of polymorphic markers was performed in case of constructing ‘Reikou’ map. The incorporation of linkage analysis of polymorphic markers could lead to the construction of phased map, and this difference of assembly could affect the results of subsequent comparative mapping of QTLs.
Although constructing a phased map is more difficult than constructing an unphased map, the ‘Reikou’ map is more desirable as a reference map than ‘Camarosa’ map because haplotype is clearly defined. In our study, the advantage of phased map was clearly indicated in the mapping of qMS4.3; qMS4.3 was mapped on the chr4X1 of ‘Reikou’ map, and Alt_AF of S_bulk was high in chr4X1a, low in chr4X1b (Fig. 6), implying that recessive allele of qMS4.3 was located on the haplotype of chr4X1b, not chr4X1a of ‘Reikou’ map. This linkage between the haplotype and the specific allele could not be detected in ‘Camarosa’ map.
We calculated the moving average of alternative allele frequency (Alt_AF) for both reference maps, which revealed the following two major differences. First, the moving average curves differ. These curves were commonly more notched (i.e., variable) in the ‘Camarosa’ map than in the ‘Reikou’ map. This difference could be due to the difference between unphased and phased maps; that is, the DNA sequences of the ‘Camarosa’ map represent the mixture of two homologous chromosomes, whereas those of the ‘Reikou’ map were separately assembled. This difference might be responsible for the notched form of the moving average for ‘Camarosa’.
Second, both maps show fragmentation of the candidate regions of the QTLs for male sterility. As Fig. 2 shows, the candidate regions for qMS4.2 in the ‘Camarosa’ map were fragmented into several regions of Fvb4-3. Additionally, FAES0001_269 amplicon, which is a neighboring SSR marker of qMS4.3, was mapped to the different region of Fvb4-1 from the significant region. “HiRise” used in the study of Edger et al. (2019b) is a software pipeline designed specifically to use proximity-ligation data from converting scaffold genome assemblies into chromosome-length pseudomolecules (Putnam et al. 2016). Although this method is a progressive way to construct pseudomolecules efficiently from short reads of next-generation sequencing data, it is based on statistical estimation, not real segregation data from actual sequences or DNA markers and has a limited power to reconstruct chromosomes for autopolyploid genomes, like cultivated strawberry (Zhang et al. 2019). Additionally, there were no heavily fragmentated regions on the results of ‘Reikou’ map. Therefore, incorporation of linkage analysis of polymorphic markers could contribute to the precise assemble of scaffolds and mis-assembly possibly occurred in the ‘Camarosa’ map. However, we believe that another possibility is likely: that the genetic distance between the American (‘Camarosa’) and Japanese (‘Reikou’, ‘Fukuoka S6’, and ‘Kaorino’) cultivars created this fragmentation, and that this should be taken into account in future research. Isobe et al. (2013) compared the genome sequence of F. × ananassa with that of F. vesca, and found that chromosome rearrangement had occurred between homeologous genomes of both species. Edger et al. (2019a) and Liston et al. (2019) called this event “homeologous exchange” and calculated the frequency of genome exchange to be 11.4% and 12.5%, respectively. Tennessen et al. (2018) demonstrated that repeated translocation of a gene cassette drives sex-chromosome turnover in octoploid strawberries. We previously performed cluster analysis of Japanese and world strawberry cultivars using SSR and cleaved amplified polymorphic sequence marker and revealed that USA strawberry cultivars (‘Aiko’, ‘Donner’, ‘Pajaro’, ‘Sequoia’, and ‘Tioga’) were classified into different clusters from modern Japanese cultivars (Wada et al. 2017). Therefore, the American cultivar might have experienced different chromosome exchanges from the Japanese cultivars.
qMS4.1 was mapped within the same subgenome, chr4Av, as qMS4.2 in the ‘Reikou’ reference map. Moreover, amplicons of SSR markers that are adjacent to the QTLs for male sterility were mapped only in ch0, chr4Av, and chr4X1, not only for qMS4.1 and qMS4.2 but also for qMS4.3. This suggests that scaffolds derived from different subgenomes were accidentally merged in the ‘Reikou’ map. Since filtering gaps with long reads (e.g. reads from PacBio) was not performed for constructing ‘Reikou’ map, scaffolds derived from different homoeologous chromosomes were merged incorrectly. We hope combining whole genome genotyping of segregation population which is derived from bi-parental cross with the assemble of scaffolds, which were filtered by long reads, could contribute to avoid incorrect merge of scaffolds.
Phylogenetic source of QTLs and candidate responsible genes for male sterility
Edger et al. (2019b) also mentioned wild strawberry species that were progenitors of cultivated strawberry. On this basis, we can propose hypothetical sources for the male sterility genes (Fig. 7). Fvb4-4 (qMS4.1), Fvb4-3 (qMS4.2), and Fvb4-1 (qMS4.3) were derived from F. iinumae, F. vesca, and F. viridis, respectively. Of course, since the origin of cultivated strawberry is currently controversial (Edger et al. 2019a, 2019b, Liston et al. 2019), it is difficult to define the progenitor species precisely. Outcrossing between different plants can increase genetic diversity, and Darwin (1877) mentioned females (male sterile plants) were generally developed from hermaphrodites, which finally lead to the occurrence of dioecious plants. Darwin (1877) also mentioned that females had been found in wild strawberry, such as, Fragaria vesca, virginiana, and chiloensis. Charlesworth and Charlesworth (1978) reported that females and hermaphrodites coexisted in the population of diploid strawberry, F. vesca ssp. bracteata. But Ashman (1999) mentioned that female plants and hermaphrodite plants evolved independently based on the evaluation of heritability of floral organs in F. virginiana population. These facts indicated that evolutionary procedure of females (male sterile plants) was a controversial topic, but they also suggested that it is likely that male sterile plants already existed in diploid species, and it is also likely that genetic regions for male sterility of different diploid species were incorporated independently into octoploid cultivated strawberry and contributed to the development of the male sterility and fertility system.
Fig. 7.
Location of QTLs for male sterility on reference map of cultivated strawberry and its inferred progenitor.
Although a genetic region for male sterility was detected in Fvb4 of F. vesca by Ashman et al. (2015), the allele effect of that region was dominant, not recessive. They also detected a region for male sterility in Fvb6, which was recessive and functional only when the region on Fvb4 was not functional. Therefore, it is possible that translocation from Fvb6 to Fvb4 might have occurred during the phylogenetic evolution of cultivated strawberry.
Tennessen et al. (2018) revealed that sex-determining regions (SDR) of wild North American octoploid strawberries were located on corresponding regions of Fvb6, and discovered only two genes in this region. One is GDP-mannose 3, 5-epimerase 2 (GMEW), and the other is 60S acidic ribosomal protein P0 (RPP0W). GMEW catalyzes the biosynthesis of ascorbic acid (vitamin C), which is involved in the antioxidant defence, photosynthesis, cell division, and growth regulation via hormone signaling pathways (Noctor and Foyer 1998, Pastori et al. 2003, Smirnoff 2000). Furthermore, ascorbic acid is also indicated to be involved in the formation of cell wall biosynthesis (Mounet-Gilbert et al. 2016). RPP0W is a kind of ribosomal protein and affects the development to stress response (McIntosh and Bonham-Smith 2006). Although Tennessen et al. (2013) proposed 57 genes included in the candidate genomic regions for male sterility of F. vesca, both genes are not found in this list. Furthermore, expression profiles of both genes in male-sterile strawberry still remained unknown. Paterson et al. (1988) presented a concept of ‘map-based cloning’ as a rapid gene isolation method based on the result of QTL analysis. Combining map-based cloning and expression analysis using our developed populations could lead to uncovering male sterility specific genes.
Future prospect for isolating responsible genes for male sterility
Currently, two different kinds of reference map are available to support genome and genetic analysis for cultivated strawberry: ‘Camarosa’ genome assembly v. 1.0.a1 (Edger et al. 2019b) and ‘Reikou’ genome assembly r2.3 (https://www.biorxiv.org/content/10.1101/2021.04.23.441065v1). In this study, we used both genomes to detect chromosome regions that contain QTLs for male sterility. When we apply this result to the development of F1 seed-type cultivars for use in practical breeding programs, restoration of female fertility will also be important, because most of the male-sterile plants were also female-sterile, and some male-sterile plants exhibited restoration of female fertility based on our experience (unpublished data). Extended improvement of those reference maps could contribute to future fine mapping of the genes responsible for both male sterility and other important agronomic traits, including the restoration of female fertility.
Author Contribution Statement
TW and SI designed the whole experiment, and respectively designed the field and molecular experiments. TW, HM, TS, and CH performed field experiment and evaluated male sterility. TW, HM, and CH conducted the genotyping experiment. TW, SI, and KS conducted the next-generation sequencing run and post-sequence analysis. MM, SN, and YT helped to design the analysis and statistical design for this study. SI and TW helped secure funding.
Supplementary Material
Acknowledgments
Funding: This study was partly supported by the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry (27015B) of the Ministry of Agriculture, Forestry and Fisheries of Japan, and grants from the Project of the NARO Bio-oriented Technology Research Advancement Institution (30020B, Research program on development of innovative technology).
We thank Prof. Masayoshi Shigyo of Yamaguchi University and Prof. Hiroki Takagi of Ishikawa Prefectural University for their critical and valuable comments, which accelerated our research. We also thank S. Sasamoto, T. Wada, C. Minami, H. Tsuruoka, M. Kato, K. Nannri, and A. Kurabayashi of the Kazusa DNA Research Institute for technical assistance. Sequencing of the bulk populations was performed by Eurofins Genomics, Co., Ltd. (Ohta, Tokyo, Japan). We also thank ELSS, Inc for English editing service of this manuscript.
Literature Cited
- Ashman, T.L. (1999) Quantitative genetics of floral traits in a gynodioecious wild strawberry Fragaria virginiana: implications for the independent evolution of female and hermaphrodite floral phenotypes. Heredity (Edinb) 83: 733–741. [DOI] [PubMed] [Google Scholar]
- Ashman, T.-L., Tennessen J.A., Dalton R.M., Govindarajulu R., Koski M.H. and Liston A. (2015) Multilocus sex determination revealed in two populations of gynodioecious wild strawberry, Fragaria vesca subsp. bracteate. G3 (Bethesda) 5: 2759–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbey, C., M. Hogshead, A.E. Schwartz, N. Mourad, S. Verma, S. Lee, V.M. Whitaker and K.M. Folta (2020) The genetics of differential gene expression related to fruit traits in strawberry (Fragaria × ananassa). Frontiers in Genetics 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger, A.M., Lohse M. and Usadel B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringhurst, R.S. (1990) Cytogenetics and evolution in American Fragaria. HortScience 25: 879–881. [Google Scholar]
- Browning, S.R. and Browning B.L. (2011) Haplotype phasing: existing methods and new developments. Nat. Rev. Genet. 12: 703–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B. and Charlesworth D. (1978) A model for the evolution of dioecy and gynodioecy. Am. Nat. 112: 975–997. [Google Scholar]
- Darwin, C. (1877) The different forms of flowers on plants of the same species. John Murray, London. [Google Scholar]
- Das, S., Upadhyaya H.D., Bajaj D., Kujur A., Badoni S., Laxmi, Kumar V., Tripathi S., Gowda C.L.L., Sharma S.et al. (2015) Deploying QTL-seq for rapid delineation of a potential candidate gene underlying major trait-associated QTL in chickpea. DNA Res. 22: 193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick, D.N. (1959) The use of cytoplasmic male-sterility in hybrid seed production. Econ. Bot. 13: 167–195. [Google Scholar]
- Edger, P.P., McKain M.R., Yocca A.E., Knapp S.J., Qiao Q. and Zhang T. (2019a) Reply to: Revisiting the origin of octoploid strawberry. Nat. Genet. 52: 5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger, P.P., Poorten T.J., VanBuren R., Hardigan M.A., Colle M., McKain M.R., Smith R.D., Teresi S.J., Nelson A.D.L., Wai C.M.et al. (2019b) Origin and evolution of the octoploid strawberry genome. Nat. Genet. 51: 541–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, M.T., Spigler R.B. and Ashman T.-L. (2010) Comparative genetic mapping points to different sex chromosomes in sibling species of wild strawberry (Fragaria). Genetics 186: 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajulu, R., Liston A. and Ashman T.-L. (2013) Sex-determining chromosomes and sexual dimorphism: insights from genetic mapping of sex expression in a natural hybrid Fragaria × ananassa subsp. cuneifolia. Heredity 110: 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa, H., Shirasawa K., Kosugi S., Tashiro K., Nakayama S., Yamada M., Kohara M., Watanabe A., Kishida Y., Fujishiro T.et al. (2014) Dissection of the octoploid strawberry genome by deep sequencing of the genomes of Fragaria species. DNA Res. 21: 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isobe, S.N., Hirakawa H., Sato S., Maeda F., Ishikawa M., Mori T., Yamamoto Y., Shirasawa K., Kimura M., Fukami M.et al. (2013) Construction of an integrated high density simple sequence repeat linkage map in cultivated strawberry (Fragaria × ananassa) and its applicability. DNA Res. 20: 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus, B.J. and Grünwald N.J. (2017) vcfr: a package to manipulate and visualize variant call format data in R. Mol. Ecol. Resour. 17: 44–53. [DOI] [PubMed] [Google Scholar]
- Leitwein, M., Duranton M., Rougemont Q., Gagnaire P.-A. and Bernatchez L. (2020) Using haplotype information for conservation genomics. Trends Ecol. Evol. (Amst.) 35: 245–258. [DOI] [PubMed] [Google Scholar]
- Li, H. and Durbin R. (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston, A., Wei N., Tennessen J.A., Li J., Dong M. and Ashman T.-L. (2019) Revisiting the origin of octoploid strawberry. Nat. Genet. 52: 2–4. [DOI] [PubMed] [Google Scholar]
- Loh, P.-R., Danecek P., Palamara P.F., Fuchsberger C., Reshef Y.A., Finucane H.K., Schoenherr S., Forer L., McCarthy S., Abecasis G.R.et al. (2016) Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 48: 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H., Lin T., Klein J., Wang S., Qi J., Zhou Q., Sun J., Zhang Z., Weng Y. and Huang S. (2014) QTL-seq identifies an early flowering QTL located near Flowering Locus T in cucumber. Theor. Appl. Genet. 127: 1491–1499. [DOI] [PubMed] [Google Scholar]
- McIntosh, K.B. and Bonham-Smith P.C. (2006) Ribosomal protein gene regulation: what about plants? Can. J. Bot. 84: 342–362. [Google Scholar]
- Mohan, L. and H. Kaul (1988) Male sterility in higher plants. Springer, Berlin. [Google Scholar]
- Mounet-Gilbert, L., Dumont M., Ferrand C., Bournonville C., Monier A., Jorly J., Lemaire-Chamley M., Mori K., Atienza I., Hernould M.et al. (2016) Two tomato GDP-D-mannose epimerase isoforms involved in ascorbate biosynthesis play specific roles in cell wall biosynthesis and development. J. Exp. Bot. 67: 4767–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini, F., O’Grady K., Hyvönen M., Folta K.M. and Baraldi E. (2020) Genomic structure and transcript analysis of the Rapid Alkalinization Factor (RALF) gene family during host-pathogen crosstalk in Fragaria vesca and Fragaria × ananassa strawberry. PLoS ONE 15: e0226448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor, G. and Foyer C.H. (1998) Ascorbate and glutathione: keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 249–279. [DOI] [PubMed] [Google Scholar]
- Pastori, G.M., Kiddle G., Antoniw J., Bernard S., Veljovic-Jovanovic S., Verrier P.J., Noctor G. and Foyer C.H. (2003) Leaf vitamin C contents modulate plant defense transcripts and regulate genes that control development through hormone signaling. Plant Cell 15: 939–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A.H., Lander E.S., Hewitt J.D., Peterson S., Lincoln S.E. and Tanksley S.D. (1988) Resolution of quantitative traits into Mendelian factors by using a complete linkage map of restriction fragment length polymorphisms. Nature 335: 721–726. [DOI] [PubMed] [Google Scholar]
- Putnam, N.H., O’Connell B.L., Stites J.C., Rice B.J., Blanchette M., Calef R., Troll C.J., Fields A., Hartley P.D., Sugnet C.W.et al. (2016) Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 26: 342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau-Gueutin, M., Lerceteau-Köhler E., Barrot L., Sargent D.J., Monfort A., Simpson D., Arús P., Guérin G. and Denoyes-Rothan B. (2008) Comparative genetic mapping between octoploid and diploid Fragaria species reveals a high level of colinearity between their genomes and the essentially disomic behavior of the cultivated octoploid strawberry. Genetics 179: 2045–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar, D. and F. Andrews (2016) latticeExtra: Extra Graphical Utilities Based on Lattice, version 6.28, Online manual. https://CRAN.R-project.org/package=latticeExtra.
- Shulaev, V., Sargent D.J., Crowhurst R.N., Mockler T.C., Folkerts O., Delcher A.L., Jaiswal P., Mockaitis K., Liston A., Mane S.P.et al. (2011) The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 43: 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnoff, N. (2000) Ascorbic acid: metabolism and functions of a multi-facetted molecule. Curr. Opin. Plant Biol. 3: 229–235. [PubMed] [Google Scholar]
- Spigler, R.B., Lewers K.S., Main D.S. and Ashman T.-L. (2008) Genetic mapping of sex determination in a wild strawberry, Fragaria virginiana, reveals earliest form of sex chromosome. Heredity 101: 507–517. [DOI] [PubMed] [Google Scholar]
- Spigler, R.B., Lewers K.S., Johnson A.L. and Ashman T.-L. (2010) Comparative mapping reveals autosomal origin of sex chromosome in octoploid Fragaria virginiana. J. Hered. 101: S107–S117. [DOI] [PubMed] [Google Scholar]
- Spigler, R.B. and Ashman T.-L. (2011) Sex ratio and subdioecy in Fragaria virginiana: the roles of plasticity and gene flow examined. New Phytol. 190: 1058–1068. [DOI] [PubMed] [Google Scholar]
- Takagi, H., Abe A., Yoshida K., Kosugi S., Natsume S., Mitsuoka C., Uemura A., Utsushi H., Tamiru M., Takuno S.et al. (2013) QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 74: 174–183. [DOI] [PubMed] [Google Scholar]
- Tennessen, J.A., Govindarajulu R., Liston A. and Ashman T.-L. (2013) Targeted sequence capture provides insight into genome structure and genetics of male sterility in a gynodioecious diploid strawberry, Fragaria vesca ssp. bracteata (Rosaceae). G3 (Bethesda) 3: 1341–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen, J.A., Govindarajulu R., Ashman T.-L. and Liston A. (2014) Evolutionary origins and dynamics of octoploid strawberry subgenomes revealed by dense targeted capture linkage maps. Genome Biol. Evol. 6: 3295–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen, J.A., Govindarajulu R., Liston A. and Ashman T.-L. (2016) Homomorphic ZW chromosomes in a wild strawberry show distinctive recombination heterogeneity but a small sex‐determining region. New Phytol. 211: 1412–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen, J.A., Wei N., Straub S.C.K., Govindarajulu R., Liston A. and Ashman T.-L. (2018) Repeated translocation of a gene cassette drives sex-chromosome turnover in strawberries. PLoS Biol. 16: e2006062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, T., Noguchi Y., Isobe S., Kunihisa M., Sueyoshi T. and Shimomura K. (2017) Development of a core collection of strawberry cultivars based on SSR and CAPS marker polymorphisms. Hort. J. 86: 365–378. [Google Scholar]
- Wada, T., Sueyoshi T., Hirata C., Takata K., Noguchi Y., Kataoka S., Isobe S., Mori M., Nagamatsu S., Tanaka Y.et al. (2020) Detection of chromosomal regions for male sterility in the cultivated strawberry Fragaria × ananassa Duch. Hort. J. 89: 147–160. [Google Scholar]
- Wei, N., Govindarajulu R., Tennessen J.A., Liston A. and Ashman T.-L. (2017) Genetic mapping and phylogenetic analysis reveal intraspecific variation in sex chromosomes of the Virginian strawberry. J. Hered. 108: 731–739. [DOI] [PubMed] [Google Scholar]
- Wen, J., Jiang F., Weng Y., Sun M., Shi X., Zhou Y., Yu L. and Wu Z. (2019) Identification of heat-tolerance QTLs and high-temperature stress-responsive genes through conventional QTL mapping, QTL-seq and RNA-seq in tomato. BMC Plant Biol. 19: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H., Averick M., Bryan J., Chang W., McGowan L., François R., Grolemund G., Hayes A., Henry L., Hester J.et al. (2019) Welcome to the tidyverse. J. Open Source Software 4: 1686. [Google Scholar]
- Zhang, X., Zhang S., Zhao Q., Ming R. and Tang H. (2019) Assembly of allele-aware, chromosomal-scale autopolyploid genomes based on Hi-C data. Nat. Plants 5: 833–845. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.