Summary
The disruption of the tumor microenvironment (TME) is a promising anti-cancer strategy, but its effective targeting for solid tumors remains unknown. Here, we investigated the anti-cancer activity of the mitochondrial complex I inhibitor intervenolin (ITV), which modulates the TME independent of energy depletion. By modulating lactate metabolism, ITV induced the concomitant acidification of the intra- and extracellular environment, which synergistically suppressed S6K1 activity in cancer cells through protein phosphatase-2A-mediated dephosphorylation via G-protein-coupled receptor(s). Other complex I inhibitors including metformin and rotenone were also found to exert the same effect through an energy depletion-independent manner as ITV. In mouse and patient-derived xenograft models, ITV was found to suppress tumor growth and its mode of action was further confirmed. The TME is usually acidic owing to glycolytic cancer cell metabolism, and this condition is more susceptible to complex I inhibitors. Thus, we have demonstrated a potential treatment strategy for solid tumors.
Subject areas: Microenvironment, Cell biology, Cancer
Graphical abstract
Highlights
-
•
Stromal cells enhance anti-cancer activity of complex I inhibitors
-
•
Anti-cancer activity of complex I inhibitors is independent on energy depletion
-
•
Complex I inhibitors suppress S6K1 through the intra- and extracellular acidification
-
•
Acidic tumor microenvironment is more susceptible to complex I inhibitors
Microenvironment; Cell biology; Cancer
Introduction
Tumor tissues contain cancer cells and several stromal cells, such as macrophages (Hao et al., 2018; Mantovani et al., 2017), vascular endothelial cells (Hida et al., 2004; Huang et al., 2019), immune cells (Lambrechts et al., 2018; Lavin et al., 2017), and fibroblasts (Kalluri, 2016; Mizutani et al., 2019). Among stromal cells in tumor tissues, cancer-associated fibroblasts (CAFs) produce cytokines, growth factors, and various proteins that constitute the extracellular matrix to support the proliferation of cancer cells (Erez et al., 2010; Kawada et al., 2015; Tian et al., 2019; Zhang et al., 2018). Such a tumor-supportive environment is established by not only these proteins but also metabolites from CAFs that are induced by tumor-stromal cell interactions to support the metabolic energy production of cancer cells in response to lower nutrient conditions in the tumor microenvironment (TME). The oncogenic TME makes cancer cells reliant on specific energy metabolism or proliferative signaling pathways (Eckert et al., 2019; Sousa et al., 2016). Thus, disruption of the TME can lead to the suppression of cancer cells adapted to the TME. Therefore, the TME is a target for cancer therapy, and a more detailed understanding of TME modulation could provide effective strategies for tumor suppression.
Thus, we focused on the TME, especially cancer-stromal cell interactions, to identify anti-cancer drug targets and detected many compounds, such as bacterial and fungal metabolites, from natural sources with potential anti-cancer activity (Kawada et al., 2009a, 2009b, 2010a, 2010b). Recently, we discovered a novel natural compound called intervenolin (ITV) from the culture medium of Nocardia sp. ML96-86F2 and evaluated its anti-cancer activity in a co-culture system of cancer and stromal cells (Kawada et al., 2013). We found that ITV suppresses gastric cancer cell growth upon co-culture with gastric fibroblasts and exhibits higher suppressive activity than in monoculture conditions (Yoshida et al., 2018). In addition, the conditioned medium (CM) of fibroblasts treated with ITV exhibited anti-cancer activity. Thus, we proposed that ITV suppresses cancer cell growth via direct and indirect mechanisms, the latter involving fibroblasts.
Interestingly, in the present study, we found that ITV is a potent mitochondrial complex I inhibitor. Compounds with complex I inhibitor mechanisms have been evaluated as anti-cancer drugs (Ashton et al., 2018). For instance, the anti-cancer activity of metformin, a well-known biguanide clinically used as a first-line therapy for type 2 diabetes, has been extensively studied (Pernicova and Korbonits, 2014). Metformin activates adenosine 5′-adenosine monophosphate-activated protein kinase (AMPK) via ATP reduction through the inhibition of mitochondrial complex I activity, thus leading to the inhibition of the mammalian target of rapamycin complex 1 (mTORC1) signaling, an important cascade in cancer growth (Hardie, 2014; Zhou et al., 2019). In accordance with these findings, ASP4132, another complex I inhibitor, exhibits anti-cancer activity by activating AMPK (Kuramoto et al., 2020). However, recent studies have identified novel targets of metformin whose mode of action is independent of AMPK (Kalender et al., 2010). Therefore, the anti-cancer activity of complex I inhibitors remains to be fully elucidated.
Recently, treatment with IACS-010759, a clinical-grade small molecule inhibitor of OXPHOS via mitochondrial complex I inhibition, was found to deplete the energy and suppress the growth of glioblastoma and leukemia cells, whose growth depends on oxidative phosphorylation (OXPHOS) for energy production (Molina et al., 2018). Currently, a phase I trial of IACS-010759 for patients with advanced solid tumors, relapsed/refractory acute myeloid leukemia, and lymphomas is underway. Thus far, IACS-010759 has a favorable safety profile (NCT02882321 and NCT03291938). Accordingly, OXPHOS has emerged as a target for treating these OXPHOS-dependent cancers (Ashton et al., 2018).
Therefore, to elucidate the direct and indirect mechanisms underlying the anti-cancer activity of ITV and mitochondrial complex I inhibitors, we analyzed the changes in the extracellular environment following the inhibition of complex I. We employed metabolic analyses of the extra- and intracellular environments and analyzed the cancer-suppressive signaling pathways related to the metabolic shift in cancer cell lines and studied mouse and patient-derived xenograft (PDX) models to determine the anti-cancer mechanism of mitochondrial complex I inhibitors.
Results
Anti-tumor activity of ITV is potentiated in co-culture conditions
First, we investigated the effect of ITV on gastric cancer cells co-cultured with gastric stromal cells. We confirmed that ITV (Figure 1A) significantly suppresses the growth of gastric cancer cell lines MKN-B and MKN-74 upon co-culture with gastric stromal cells (Hs738) at lower concentrations than in monoculture conditions (Figures 1B and S1A). As ITV also exhibited stronger growth inhibitory activity against cancer cells in non-contact co-culture experiments than in monoculture (Yoshida et al., 2018), we speculated that ITV exerts growth suppression on cancer cells by modulating extracellular conditions. Indeed, MKN-1, MKN-B, and MKN-74 cell growth was suppressed by the CM of stromal cells pre-treated with ITV (ITV CM) (Figures 1C, 1D, and S1B), thus indicating indirect ITV activity.
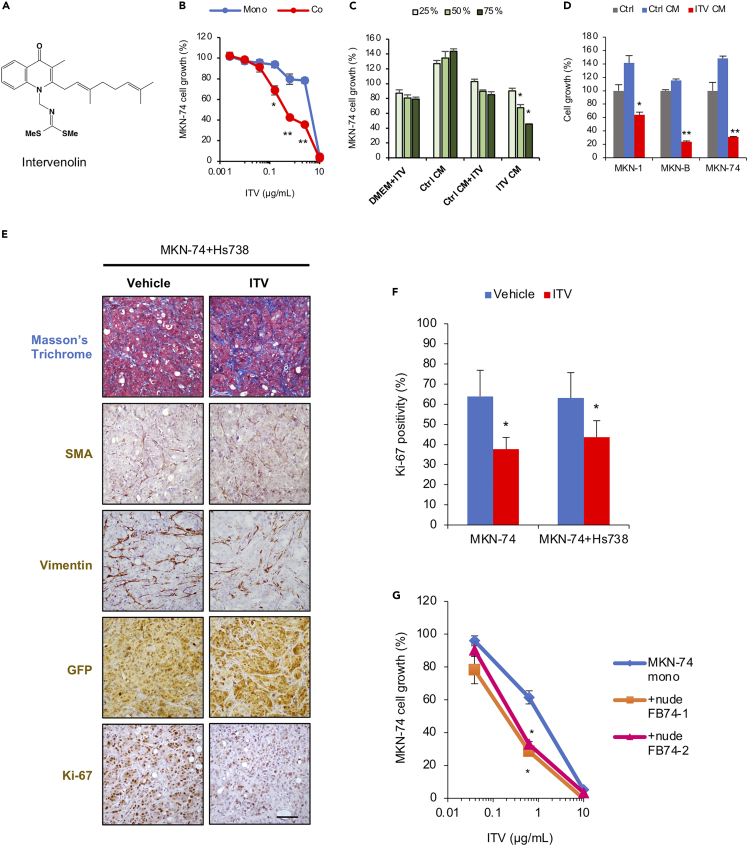
Figure 1.
Intervenolin (ITV) exhibits anti-cancer activity that is potentiated in co-culture conditions
(A) Structure of ITV.
(B) Growth of MKN-74 cells cultured alone (Mono) or with Hs738 (Co) for 3 days at the indicated ITV concentrations were measured using GFP fluorescence intensity and expressed as percentage relative to that of cells without ITV treatment (n = 3; ∗p < 0.005, ∗∗p < 0.001 versus mono).
(C) Growth of MKN-74 cells upon treatment with ITV CM, Ctrl CM, and their respective controls (DMEM + ITV and Ctrl CM + ITV) for 3 days (∗p < 0.001 versus Ctrl CM+ ITV). For each condition, 1 μg/mL ITV was used.
(D) Growth of cancer cell lines MKN-1, -B, and −74 (n = 3).
(E) Formalin-fixed, paraffin-embedded tumor sections from mice inoculated with MKN-74 and Hs738 cells treated with the vehicle or 12.5 mg/kg ITV were stained with Masson's trichrome stain and analyzed using immunohistochemistry with antibodies against the indicated proteins. Scale bar, 100 μm (n = 5).
(F) Percentages of Ki-67-positive cells in the high-power field of tumor sections (n = 5; ∗p < 0.01).
(G) MKN-74 cells were cultured alone (Mono) or with fibroblast cells isolated from mice tumor tissues (+nude FB74-1,2) (Co) for 3 days in the presence of ITV at the indicated concentrations (n = 3; ∗p < 0.0005 versus mono).
Data are presented as the mean ± SD and were analyzed using two-sided Student's t test.
The enhancement of ITV anti-cancer activity upon co-culture was further validated in several cancer cell lines from other organs, including the colon, lung, prostate, and pancreas, paired with stromal cells originating from the same organs. We found that ITV exhibited growth inhibitory activity against not only gastric cancer cell lines but also other cancer cell lines from different organs (Figure S1C) and that this inhibitory activity is significantly potentiated in co-culture conditions. These results suggest that ITV exhibits anti-cancer activity by regulating extracellular conditions.
ITV suppresses tumor growth in a mouse xenograft model
We further evaluated the anti-tumor activity of ITV against MKN-74 cells inoculated with or without Hs738 cells to determine whether stromal cells can enhance anti-tumor activity in vivo. Upon ITV treatment, mice bearing MKN-74 and Hs738 cells had significantly reduced tumor growth, tumor weight, and Ki-67 positivity without body weight loss at day 21 (Figures 1E, 1F, S2B, and S2C). Tumor growth was also suppressed by ITV treatment at the same extent in mice bearing MKN-74 alone (Figures 1F, S2A, and S2D).
Histochemical and immunohistochemical analysis results showed that fibroblast-like stromal cells are abundant in the tumor tissues regardless of co-inoculation with Hs738 cells (Figures 1E, and S2A). Fibroblast-like cells in tumors from mice bearing MKN-74 cells alone are predicted to be derived from host mice. Consequently, MKN-74 cell growth was significantly inhibited by ITV treatment upon co-culture with isolated mouse fibroblasts compared with that in monoculture conditions (Figure 1G). These results indicate that ITV suppresses cancer cell growth by modulating the TME in vivo regardless of the origin of stromal cells.
Mode of action of ITV
Using a panel of 39 human cancer cell lines (termed JFCR39), we predicted the mechanism of action of several compounds by comparing the fingerprint of GI50 (half growth inhibition) against JFCR39 with 250 reference compounds (Dan et al., 2002; Paull et al., 1989; Yamori, 2003). Some biguanide compounds showed high correlation with ITV in the JFCR39 system (buformin: r = 0.737; phenformin: r = 0.696; metformin: r = 0.586) (Figure S2E). Buformin and phenformin are derivatives of metformin, which is the first alternative drug for type 2 diabetes. Biguanides and metformin are also anti-cancer reagents, which function primarily by inhibiting mitochondrial complex I.
Next, we examined whether complex I inhibition is essential for the anti-cancer activity of ITV and found that all complex I inhibitors, including buformin, metformin, arctigenin, and rotenone (Bridges et al., 2014; Ravanel et al., 1984; Thomas et al., 2013), exhibit significantly stronger growth inhibitory activities against gastric cancer cells upon co-culture with Hs738 than in monoculture, similar to ITV (Figure S2F). This suggests that ITV is a possible complex I inhibitor and that mitochondrial complex I inhibition is required for its enhanced anti-cancer activity in the presence of stromal cells.
ITV inhibits mitochondrial complex I activity
We evaluated the potential of ITV as a complex I inhibitor by measuring oxidative consumption rate (OCR) and found that ITV significantly suppresses OCR and reactivity against FCCP, an uncoupler, in a dose-dependent manner (Figures 2A and S3A). Although glucose supplementation upregulated OCR of gastric cancer cells in glucose- and pyruvate-depleted control, ITV or rotenone (positive control as a complex I inhibitor) maintained low OCR levels even in the presence of sufficient glucose, but this inhibition was reversed by succinate supplementation (Figures S3B and S3C). Succinate is a substrate of complex II and can partially bypass the function of complex I (Ehinger et al., 2016).
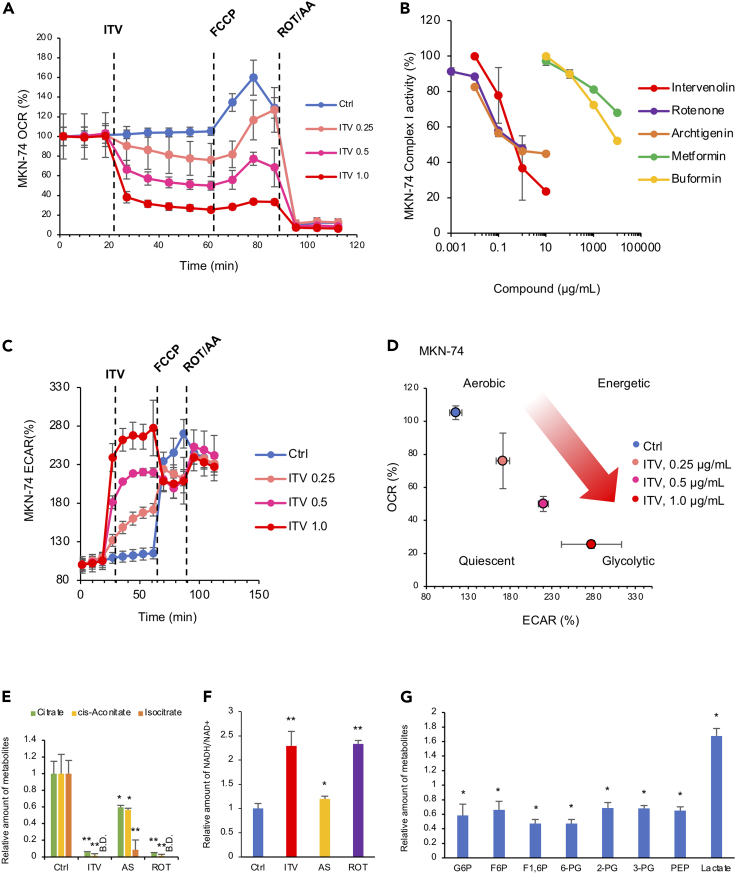
Figure 2.
Intervenolin (ITV) inhibits mitochondrial complex I activity and induces metabolic shift in cancer cells
(A) Results of seahorse flux analyzer assays showing the OCR of MKN-74 cells. Assay buffer (Ctrl) or the indicated concentrations of ITV were introduced at 24 min; FCCP, 64 min; and rotenone (ROT) and antimycin A (AA), 88 min.
(B) Activity of the mitochondrial complex I as measured using an NADH assay. Reduction rate of NADH that normalized by non-treated value was shown as complex I activity. The activity of mitochondria without compounds was defined as 100%.
(C) Seahorse flux analyzer assay results showing the extracellular acidification rate (ECAR) in MKN-74 cells as in (A).
(D) Energy map of MKN-74 cells based on OCR and ECAR values. The status of the metabolic pathways is classified as aerobic, glycolytic, energetic, or quiescent.
(E–G). Intracellular metabolite concentrations were determined using mass spectrometric analysis (E and F). Relative amounts of the indicated metabolites in MKN-74 cells treated by 1 μg/mL ITV, 1 μg/mL AS-1936 (AS), or 0.01 μg/mL rotenone (ROT) for 3 h are shown. B.D., below detection limit (n = 3; ∗p < 0.05 versus Ctrl, ∗∗p < 0.005 versus Ctrl).
(G) Relative amounts of the indicated metabolites in MKN-74 cells treated by ITV for 3 h are shown (n = 3; ∗p < 0.05 versus non-treated cells). Data are presented as the mean ± SD (n = 3) and normalized according to the baseline and analyzed using two-sided Student's t -test.
We further evaluated ITV inhibitory activity against the enzymatic activity of complex I using isolated mitochondria and observed that ITV more strongly inhibits complex I activity than do biguanides, such as buformin and metformin (Figures 2B, S3D, and S3E). Results of enzymatic analysis of complexes II, II/III, IV, and V showed that ITV does not inhibit other components of the electron transport chain (ETC) and OXPHOS (Figure S3F) up to 1 μg/mL.
Next, ITV binding to a target molecule was examined using pull-down assays; as AS-1936 is an ITV derivative and its inhibition of cancer cell growth is not potentiated in co-culture conditions, it was used as a control with a significantly lower activity (Figure S4A). NDUFB8, a subunit of complex I, coprecipitated with ITV, which was inhibited by excess ITV (Figure S4B), indicating that ITV binds and inhibits mitochondrial complex I. Because ITV (1 μg/mL), metformin (1,000 μg/mL), and rotenone (0.01 μg/mL) suppressed the OCR of gastric cancer cells to the same extent, the OCR values were well correlated with their inhibitory activity in co-culture conditions (Figures S4C and S4D).
Complex I inhibitory activity against MKN-B, MKN-74, and Hs738 cells showed no significant difference; thus, we speculated that target cells of ITV in the co-culture environment are both stromal and gastric cancer cells (Figures 2B, S3D, and S3E).
ITV increases extracellular lactate levels via metabolic shift
We determined the factor(s) present in the ITV-treated cell culture medium and found that ITV upregulates the extracellular acidification rate (ECAR) of cancer cells. According to the OCR and ECAR energy map of ITV-treated gastric cancer cells, energy production shifted to glycolysis to compensate for OXPHOS inhibition (Figures 2C, 2D, S4E, and S4F). Therefore, we speculated that metabolite(s) are involved in ITV CM activity.
We thus performed metabolomic analysis to clarify the metabolomic changes caused by ITV and other complex I inhibitors. We focused on glycolysis and the tricarboxylic acid (TCA) cycle because metabolites involved in these pathways were significantly altered (Figure S5). ITV or rotenone reduced the levels of the components of the TCA cycle, such as citrate, cis-aconitate, and isocitrate, to a lesser extent than did AS-1936 (Figure 2E). As the NADH/NAD+ ratio in MKN-74 cells treated with ITV or rotenone was significantly upregulated, we attributed the downregulation of the TCA cycle to the suppressed consumption of NADH due to complex I inhibition (Figure 2F). Levels of the intermediate metabolites of glycolysis, such as G6P, F6P, F1, 6P, 6-PG, 2-PG, 3-PG, and PEP, were reduced by ITV, but those of its end product, lactate, were elevated (Figure 2G). The decrease in intermediate metabolite levels compensated for the ITV- or rotenone-induced OXPHOS inhibition, and the increase in lactate levels was due to the suppression of the TCA cycle.
In accordance with these findings, the extracellular lactate levels in co-culture conditions were significantly greater (>10 mM) than in monocultures (3–5 mM at 0.6 μg/mL ITV) (Figure S6A). Moreover, ITV, metformin, and rotenone significantly increased lactate levels and decreased glucose levels in the CM of Hs738 cells compared with MKN-B and MKN-74 cells (Figures S6B–S6D). Thus, these results suggest that lactate is a candidate factor involved in the mechanism of ITV anti-cancer activity. Furthermore, the reduction in both intra- and extracellular nutrient levels caused by metabolic changes cannot be a critical factor for the anti-cancer activity of ITV in co-culture conditions as nutrient supplementation, such as with glutamine, pyruvate, and glucose, did not attenuate ITV CM activity (Figure S6E).
S6K1 is synergistically downregulated by extracellular acidification and complex I inhibition
We next evaluated whether the addition of lactate with ITV suppresses cancer cell growth. However, we observed that sodium lactate does not inhibit gastric cancer cell growth even in the presence of ITV, metformin, or rotenone (Figures S6F–S6I). We then investigated the environmental factors influencing the anti-cancer property of ITV. Interestingly, ITV CM had a lower pH (pH 6.9) than the control medium (pH 7.4) or control CM (pH 7.4) from Hs738. Therefore, we evaluated whether acidic conditions potentiate the anti-cancer activity of ITV and found that gastric cancer cell growth is inhibited at low pH even with ITV treatment (pH/ITV) at the same extent as with ITV CM (Figure 3A). However, pH neutralization suppressed the growth inhibitory activity of pH/ITV and ITV CM against cancer cells (Figure 3A). Hence, extracellular acidification caused by elevated lactate levels enhances the anti-cancer activity of ITV.
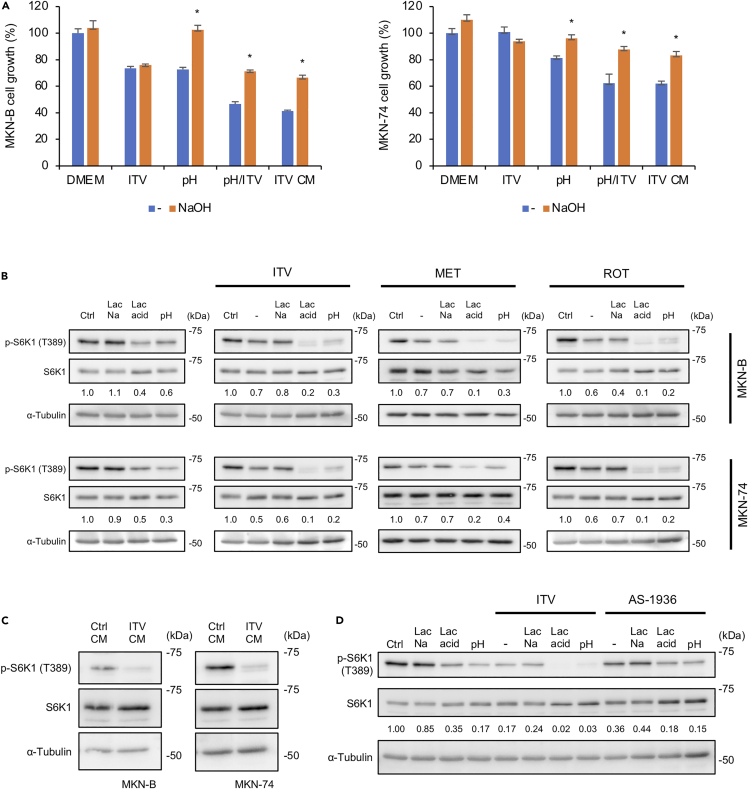
Figure 3.
Complex I inhibition and low extracellular pH synergistically suppress S6K1
(A) MTT assay results showing the growth of MKN-B and MKN-74 cells cultured with intervenolin (ITV; 1 μg/mL), low pH medium (pH), low pH medium with ITV (1 μg/mL) (pH/ITV), or ITV CM. Cell viability grown at DMEM without NaOH was defined as 100% (n = 3; ∗p < 0.05).
(B–D) Western blot analysis showing the phosphorylation levels of S6K1 (T389) in cancer cells with the following culture conditions: (B and D) MKN-B and MKN-74 cells treated with ITV (1 μg/mL), metformin (MET, 1 mg/mL), rotenone (ROT, 0.01 μg/mL), or AS-1936 (1 μg/mL) in DMEM supplemented 1% D-FBS (−), 10 mM sodium lactate (Lac Na), 10 mM lactic acid (Lac acid), or low pH medium (pH) for 3 h. Relative intensities of phospho-S6K1/total S6K1 ratio (versus ctrl) were analyzed by ImageJ and shown. (C) MKN-B and MKN-74 cells cultured in Ctrl CM or ITV CM for 3 h. Data were analyzed using two-sided Student's t test.
Next, we performed an anti-phospho-kinase antibody array to determine the intracellular signals involved in ITV-induced cell growth inhibition. Some kinases such as Akt (S473), ERK1/2 (T202/Y204), AMPKα1 (T172), and S6K1 (T389) were determined as suppressed targets of ITV CM (Figures S7A, S7B, and Table S1). Glucose levels in the ITV CM was significantly lower than in the control CM (Figures S6C and S6D). Immunoblot analysis showed that excess glucose recovered the phosphorylation status of Akt, ERK1/2, and AMPKα1 but not S6K1 (Figure S7B) and did not suppress the growth inhibitory activity of ITV CM (Figure S7C). Furthermore, ITV, but not AS-1936, suppressed S6K1 phosphorylation in gastric cancer cells cultured under acidic conditions (10 mM lactate supplementation and low pH medium) but not in 10 mM sodium lactate (Figures 3B and 3D). Similar results were observed using metformin and rotenone (Figure 3B). S6K1 was significantly suppressed in the ITV CM (Figure 3C). Therefore, potentiated ITV (or complex I inhibitors) action under acidic conditions due to elevated lactate levels suppresses S6K1, inhibiting tumor cell growth.
Suppression of S6K1 phosphorylation is protein phosphatase-2A dependent
S6K1 activity is necessary for protein synthesis, and the AMPK/mTORC1 pathway is a well-known S6K1-suppressive signal cascade (Hardie, 2014; Saxton and Sabatini, 2017). However, our results showed that AMPK is not involved in S6K1 suppression in gastric cancer cells treated with ITV CM (Figure S7B). Thus, we examined mTORC1 activation to investigate the pathway involved in S6K1 suppression by ITV under acidic conditions. As the localization of mTORC1 on the lysosome is a key event for its activation (Son et al., 2019), we analyzed the co-localization of mTOR and LAMP1, a lysosome surface marker, in gastric cancer cells treated with ITV under acidic conditions, ITV/acidic conditions, and ITV CM. Signal overlap of mTOR and LAMP1 was found at ITV, low pH, pH/acidic, and ITV CM except for EBSS, indicating that the suppression of S6K1 phosphorylation is mTORC1 activation independent (Figure S7D).
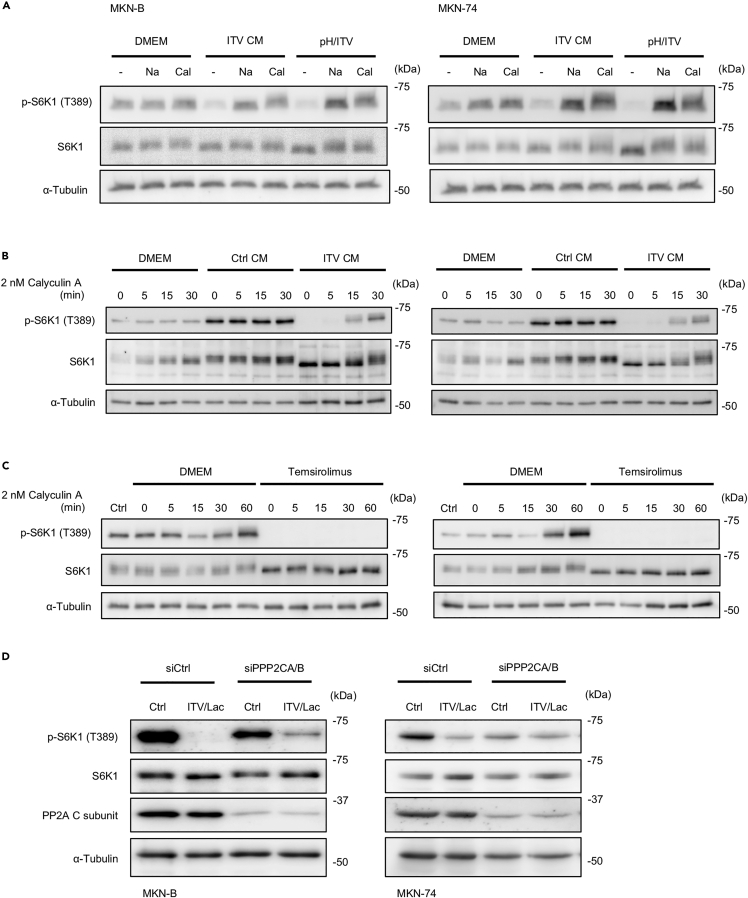
Sacco et al. (2016) reported that metformin treatment induces protein phosphatase-2A (PP2A)-dependent S6K1 inactivation via undetermined signaling. Thus, we further investigated the effect of PP2A on S6K1 phosphorylation in gastric cancer cells treated with acidic-potentiated ITV. Suppression of S6K1 phosphorylation by ITV CM or pH/ITV treatment was suppressed by calyculin A, an inhibitor of PP2A, and pH neutralization (Figure 4A). In addition, the complete suppression of S6K1 phosphorylation in gastric cancer cells due to ITV CM was recovered by calyculin A as early as 30 min (Figure 4B). In contrast, calyculin A treatment did not restore S6K1 phosphorylated levels after temsirolimus, an mTOR inhibitor, completely suppressed S6K1 phosphorylation (Figure 4C). Moreover, silencing both PPP2CA and PPP2CB (PPP2CA/B), genes encoding for the catalytic subunits of PP2A, in gastric cancer cells attenuated the suppression of S6K1 phosphorylation by ITV at low pH (Figure 4D). These results suggest that the suppression of S6K1 phosphorylation by ITV at low pH is caused by PP2A dephosphorylation.
Figure 4.
Suppression of S6K1 by intervenolin (ITV) under acidic conditions is dependent on PP2A
(A–D) Western blot analysis showing the phosphorylation levels of S6K1 (T389) in cancer cells with the following culture conditions: (A) MKN-B and MKN-74 cells cultured in DMEM supplemented with 1% D-FBS (DMEM), ITV CM, or low-pH medium containing ITV (1 μg/mL) (pH/ITV) for 3 h and added with NaOH (Na, 0.1 mM) or calyculin A (Cal, 2 nM) for further 30 min incubation; (B and C) MKN-B and MKN-74 cells cultured in DMEM supplemented 1% D-FBS (DMEM), Ctrl CM, ITV CM, or temsirolimus (25 μM) and added with calyculin A (2 nM) for the indicated time; (D) MKN-B and MKN-74 cells treated with non-target siRNA (siCtrl) or siRNA for PPP2CA and PPP2CB (PPP2CA/B) and cultured in DMEM supplemented 1% D-FBS (Ctrl) or the medium containing 10 mM lactate and ITV (1 μg/mL; ITV/Lac) for 3 h.
Proton-sensing G protein-coupled receptor regulates S6K1 suppression by ITV under acidic conditions
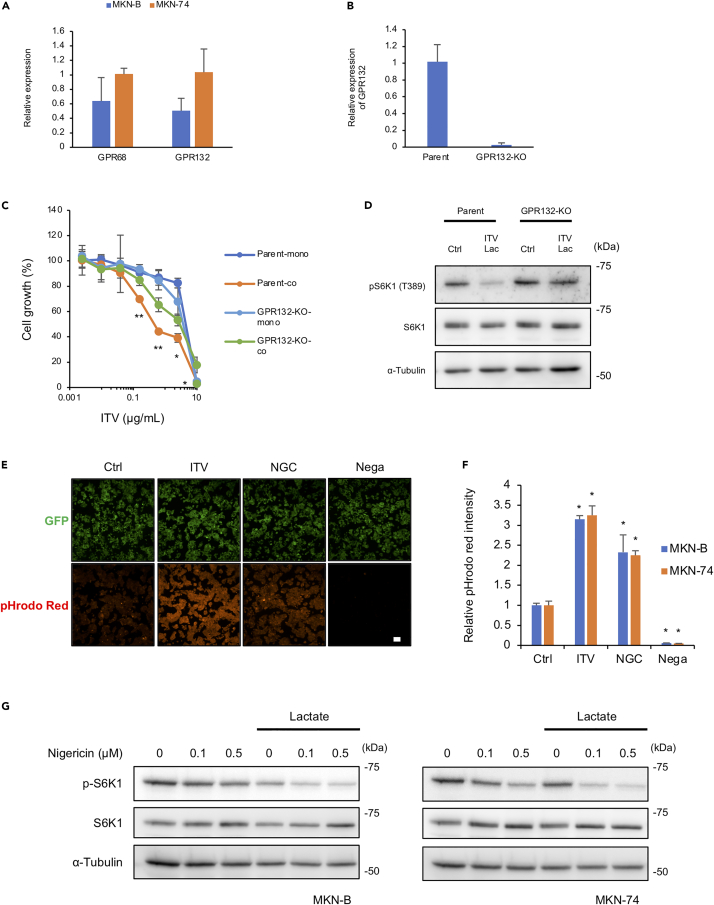
GPR4, GPR65 (TDAG8), GPR68 (OGR-1), and GPR132 (G2A) are proton-sensing G protein-coupled receptors (GPCRs) activated by extracellular acidic pH (Chen et al., 2011; Huang et al., 2016; Ihara et al., 2010; Murakami et al., 2004; Saxena et al., 2012). Therefore, we examined whether these GPCRs are involved in S6K1 suppression by complex I inhibitors under acidic conditions. We detected the expression of GPR68 and GPR132, but not GPR4 and GPR65, in MKN-B and MKN-74 cells (Figure 5A). Although ITV significantly inhibited parental cell growth when co-cultured with gastric stromal cells, its inhibitory effect was canceled in GPR132-knockout cells (Figures 5B and 5C). Consistent with this, S6K1 phosphorylation in GPR132-knockout cells was not suppressed by ITV under acidic conditions induced by lactate (Figure 5D).
Figure 5.
GPR132 is important for intervenolin (ITV) activity, and complex I inhibition leads to intracellular acidification
(A) Relative mRNA expression levels of GPR68 and GPR132 in MKN-B and MKN-74 (versus MKN-74) as measured using qPCR (n = 3).
(B) Relative mRNA expression levels of GPR132 in MKN-74 parent cells (parent) and GPR132-knockout cells (GPR132-KO).
(C) Parent and GPR132-KO cells were cultured alone (Mono) or with Hs738 (Co) for 3 days with the indicated concentrations of ITV. Cell growth was determined by measuring GFP fluorescence intensity (n = 3; ∗p < 0.05, ∗∗p < 0.005 versus mono).
(D) Parent and GPR132-KO cells were cultured in the medium containing 8 mM lactate with the indicated concentrations of ITV for 1 h. Western blot analysis shows the phosphorylation levels of S6K1 (T389) in cancer cells.
(E and F) Intracellular pH levels of cancer cells measured using pHrodo Red fluorescence intensity normalized to that of GFP. MKN-B and MKN-74 cells were treated with 1 μg/mL ITV or 0.1 μg/mL nigericin (NGC) for 3 h. Cells not treated with pHrodo Red served as the negative control group (Nega). Scale bar, 100 μm (n = 3; ∗p < 0.005 versus ctrl).
(G) MKN-B and MKN-74 cells cultured in the medium containing 10 mM lactate with the indicated concentrations of nigericin for 3 h. Western blot analysis shows phosphorylation levels of S6K1 (T389) in cancer cells.
Data are presented as the mean ± SD and were analyzed using two-sided Student's t test.
Proton-sensing GPCRs are fully activated at pH levels between 6.4 and 6.8 (Ludwig et al., 2003). Here, the pH values used in co-culture experiments were from 6.8 to 6.9. Thus, the results indicate that ITV treatment under acidic conditions suppresses gastric cancer cell growth through GPCRs, particularly GPR132.
Intracellular acidification is crucial for the suppressive activity of ITV under acidic conditions
Because PP2A dephosphorylated S6K1 in gastric cancer cells treated with ITV under acidic conditions, we next explored the related events induced by ITV that act synergistically with low extracellular pH to suppress S6K1. As large amounts of lactate are produced by complex I inhibition, we hypothesized that lactate also accumulates in the cytosol of cancer cells, altering intracellular pH. Following treatment with ITV or nigercin, an H+ and K+ ionophore that lowers intracellular pH, the intracellular pH of MKN-B and MKN-74 cells significantly decreased compared with that of control cells (Figures 5E and 5F). Furthermore, S6K1 phosphorylation in gastric cancer cells was suppressed by nigericin under acidic conditions induced by 10 mM lactate like complex I inhibitors (Figure 5G). These results suggest that reduced intracellular pH due to lactate accumulation acts synergistically with low extracellular pH to suppress S6K1.
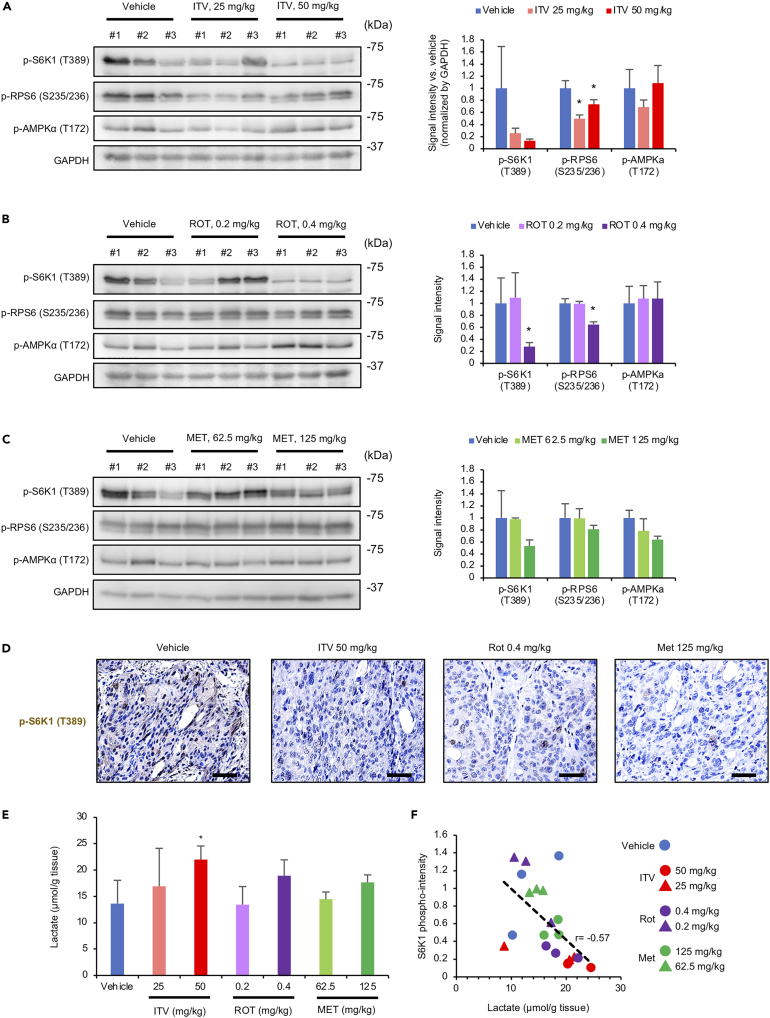
Complex I inhibitors suppress S6K1 activity in a mouse xenograft model
To confirm the mode of action of ITV in vivo, we performed immunohistochemistry using tumor tissues from MKN-74-bearing mice. We evaluated the phosphorylation of S6K1 and ribosomal protein S6 (RPS6), a substrate of S6K1. As expected, S6K1 and RPS6 phosphorylation levels were decreased by ITV (Figures 6A and 6D). Rotenone and metformin (albeit weakly) similarly suppressed S6K1 and RPS6 phosphorylation (Figures 6B–6D). Although rotenone (0.4 mg/kg) exhibited stronger activity than did ITV, this concentration was toxic for mice. Meanwhile, AMPK phosphorylation levels in tumor tissues remained unchanged after ITV, rotenone, and metformin treatment (Figures 6A–6C).
Figure 6.
Intervenolin (ITV) suppresses intratumor S6K1 phosphorylation and elevates intratumor lactate levels in MKN-74-inoculated mouse xenograft models
(A–C) Western blot analysis showing the phosphorylation levels of S6K1 (T389), RPS6 (S235/236), and AMPK (T172) in tumor tissues from mice treated with ITV, rotenone (ROT), or metformin (MET) at the indicated doses. Signal intensities of the bands are shown in the bar graph (n = 3; ∗p < 0.005 versus vehicle).
(D) Formalin-fixed, paraffin-embedded tumor sections from mice treated with the vehicle, ITV, ROT, or MET were analyzed by immunohistochemistry using antibodies against phospho-S6K1 (T389). Scale bar, 100 μm (n = 3).
(E) Lactate levels in tumor tissues from mice treated with the vehicle, ITV, ROT, or MET (n = 3; ∗p < 0.005 versus vehicle).
(F) S6K1 phosphorylation and lactate levels in tumor tissues from mice treated with the vehicle, ITV, ROT, or MET are significantly inversely correlated. r represents Pearson's correlation coefficient.
Data are presented as the mean ± SD and were analyzed using two-sided Student's t test.
In addition, ITV, rotenone, and metformin increased lactate levels in tumor tissues (Figure 6E). Of importance, phospho-S6K1 intensity and lactate level in tumor tissues were inversely correlated (r = −0.57; Figure 6F). These results indicate that the suppression of S6K1 phosphorylation in tumor tissues occurs in the TME under high lactate concentrations.
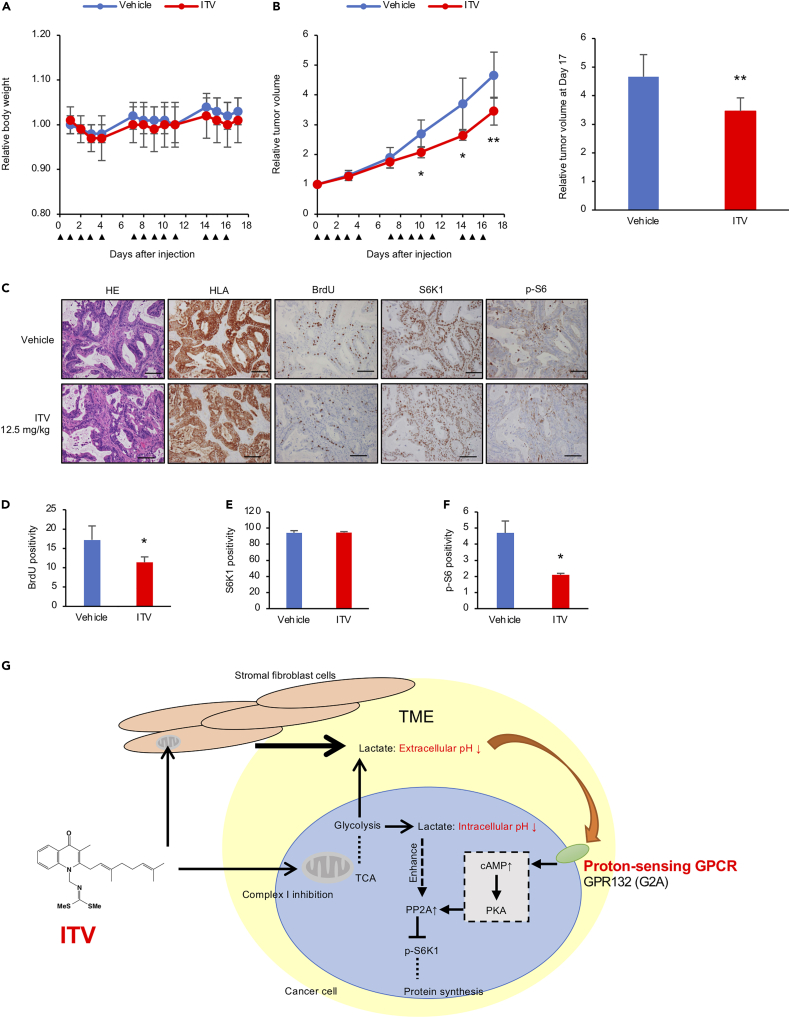
ITV exhibits anti-tumor activity in a human pancreatic cancer patient-derived xenograft mouse model
Pancreatic tumor tissues include a significant number of stromal components (von Ahrens et al., 2017). We next examined the anti-cancer activity of ITV and ASP4132, a complex I inhibitor, against pancreatic cancer cells. Both ITV and ASP4132 significantly suppressed the growth of pancreatic cancer cells co-cultured with pancreatic normal stellate cells HPaSteC at lower concentrations than in monocultures (Figure S8A). In vitro evaluation showed that ASP4132 more strongly inhibited complex I activity and gastric cancer cell growth upon co-culture than did ITV (Figures S8B and S8C). As expected, both ASP4132-treated CM from Hs738 cells and ITV-treated CM suppressed S6K1 phosphorylation (Figure S8D). In addition, ASP4132 significantly inhibited BxPC-3 xenograft growth without any body weight loss and reduced RSP6 phosphorylation in tumor tissues (Figures S9A and S9B).
Finally, we examined the anti-tumor activity of ITV using a human pancreatic cancer PDX model. ITV-treated mice showed significantly inhibited tumor growth on days 10 (P = 0.016), 14 (P = 0.023), and 17 (P = 0.009) compared with vehicle-treated mice without any body weight loss (Figures 7A and 7B). Moreover, ITV significantly reduced phosphorylated RSP6 levels and BrdU-positive cell number (Figures 7C–7F). These results indicate that ITV shows potent anti-tumor activity against human pancreatic cancer PDX models and exhibits the same mode of action in vivo.
Figure 7.
Intervenolin (ITV) suppresses tumor growth in mouse PDX models
(A) Mice treated with ITV (12.5 mg/kg; n = 6) did not show any weight loss compared with those with the vehicle (n = 7).
(B) Relative tumor volumes (∗p < 0.005, ∗∗p < 0.01 versus vehicle) at 17 days from ITV injection. ITV was injected on the days indicated by black triangles.
(C) Immunohistochemistry analysis of tumor sections from mice treated with the vehicle or ITV. Scale bar, 100 μm.
(D–F) Percentages of BrdU-, S6K1-, and p-S6-positive cells in the tumor sections (∗p < 0.05).
(G) Proposed mechanism of ITV anti-cancer activity.
Data are presented as the mean ± SD and were analyzed using two-sided Student's t test.
Discussion
In normal cells, OXPHOS is the main metabolic pathway that generates energy, producing ATP through the ETC, which comprises mitochondrial complexes I, II, III, IV, and ATP-synthase (complex V). Energy produced via OXPHOS is driven by the electron donors NADH and FADH2 generated by the TCA cycle and ATP produced in complex V via the proton gradient between the inner membrane space and mitochondrial matrix. As the first step of the ETC, the proton gradient is generated via the proton pump function of complexes I and II, but mainly complex I. Therefore, complex I is a target molecule for inhibiting OXPHOS in chemotherapy.
In cancer cells, glycolysis is facilitated by the Warburg effect, which is a hallmark feature of all cancers, and OXPHOS is suppressed (Balsa et al., 2020; Koppenol et al., 2011; Zhang et al., 2020). However, several human tumors such as glioblastoma and leukemia mainly depend on OXPHOS and not glycolysis for energy generation, leading to the formation of a hypoxic TME, which can be exploited for cancer therapy (DeBerardinis and Chandel, 2020; Hensley et al., 2016; Kennedy et al., 2013). Therefore, energy depletion induced by OXPHOS inhibition is a promising strategy for the treatment of such cancers.
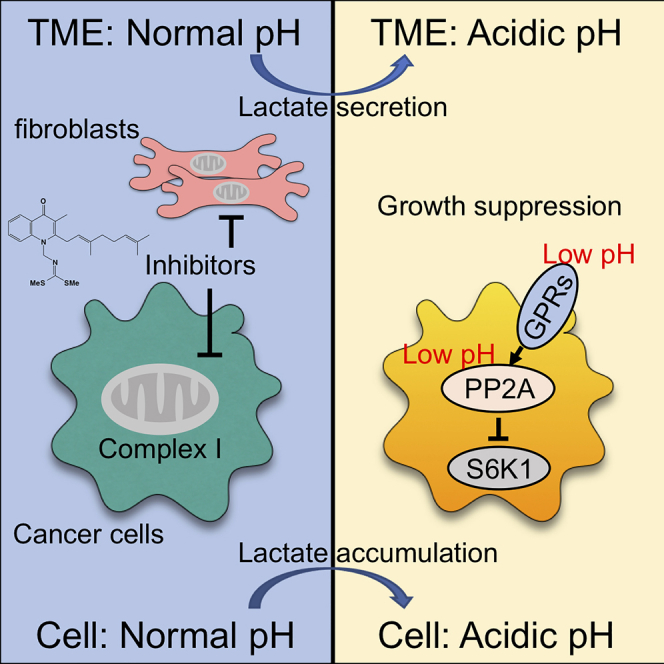
Here, we analyzed the mechanism of action of the natural compound ITV and revealed anti-tumor strategy that is independent of energy depletion. Complex I inhibitors shifted the metabolism from OXPHOS to glycolysis, resulting in the production of lactate in cancer cells, similar to the Warburg effect. Extracellular pH decreases due to excess lactate, and intracellular pH also decreases due to the accumulation of intracellular lactate. Intra- and extracellular acidification works synergistically to suppress S6K1 phosphorylation through PP2A. Proton-sensing GPCRs are fully active at a pH of ∼6.8, which is consistent with the pH of the ITV CM and the acidic conditions used in this study. The reduction in pH was sensed by proton-sensing GPCRs, whereas the growth suppressive activity of ITV under acidic conditions was partially attenuated in GPR132-knockout cells. Hence, GPR132 plays an important role in pH sensing to suppress S6K1 through PP2A (Figure 7G). The regulation of PP2A through activated GPCRs is functionally regulated by protein kinase A by increasing intracellular cAMP levels (Faubert et al., 2017; Leslie and Nairn, 2019). The regulation of the substrate specificity of PP2A is complicated. PP2A is a heterotrimer consisting of a scaffolding subunit A, catalytic subunit C, and regulatory subunit B; because each subunit has numerous isoforms (subunit A: 2, subunit C: 2, subunit B: at least 16 types in combination), many isoform combinations of PP2A exist, and the composition of each subunit determines substrate specificity (Ahn et al., 2007; DeGrande et al., 2013). Although the complete suppression of S6K1 requires intracellular acidification in addition to pH sensing by PP2A, the mechanism of PP2A regulation in response to acidification remains unclear and requires further investigation.
The pH inside solid tumors is extremely low (∼pH 6) owing to increased lactic acid produced by the Warburg effect. In our experiments, we found that complex I inhibition can further enhance glycolysis even inside cancer cells and that lactate secretion can be also further enhanced even in tumor tissues. Although the extracellular pH in tumor tissues is lower than in normal tissue, the intracellular pH in both tumor tissues and normal tissues was maintained (Webb et al., 2011). Although the extent of its activity could be possibly altered depending on expression levels of proton pumps and monocarboxylate transporters in cancer cells, anti-tumor activity via the concomitant acidification of both the intra- and extracellular environment through the inhibition of complex I is expected to be effective against solid tumors. Moreover, TMEs originally exhibiting a low pH are good targets for our proposed strategy using complex I inhibitors.
Unlike cancer cells, the energy production in stromal cells, such as Hs738 cells, depends on OXPHOS; thus, the magnitude of TME acidification induced by complex I inhibition in the presence of stromal cells markedly exceeded that of cancer cells alone. Therefore, the strategy of concomitant intra- and extra acidification induced by complex I inhibitors is more effective against solid tumors harboring abundant stromal cells. A strong stromal reaction is a hallmark of pancreatic ductal adenocarcinoma (PDAC). PDAC is the most common pancreatic cancer type and has the lowest overall survival rate of approximately 8% (Siegel et al., 2019). Patients with PDAC show poor response to chemotherapy and immune checkpoint inhibition therapy (Kabacaoglu et al., 2018). Therefore, the development of a more effective PDAC therapy is urgently needed. We found that ITV suppressed gastric cancer cell growth in both mouse xenograft models and human pancreatic cancer PDX models. Because ITV treatment under acidic conditions induced the death of gastric cancer cells (Figure S9C), the concomitant intra- and extra acidification strategy using complex I inhibitors is a promising therapeutic strategy for intractable cancers. This mechanism of action of complex I inhibitors against solid tumors can be used as a new anti-cancer strategy and for drug development.
In our in vitro experiments, rotenone, a well-known complex I inhibitor, strongly exhibited adverse toxicity, whereas metformin had a weak effect on tumor growth. Rotenone also inhibits myotube formation, which can have neurotoxic effects (Liu et al., 2005; Zhang et al., 2019). The limitations of metformin for cancer treatment, including inadequate potency and transport-mediated accumulation, have been previously reported (Wang et al., 2002). Although rotenone and metformin have similar in vitro inhibitory activity as ITV, we demonstrated that ITV is the best anti-cancer agent among these compounds. Indeed, ITV-treated mice did not show any weight loss even at the tumor-suppressive dose. Currently, we are synthesizing several derivatives of ITV that exhibit anti-tumor effects via complex I activity inhibition (Figure S9D, Table S2). Overall, our findings help contribute to the development of efficacious anti-cancer drugs.
Limitations of the study
Our current study demonstrated that anti-tumor activity via the concomitant acidification of both the intra- and extracellular environment through the inhibition of complex I could be applied to solid tumors. Since many types of transporters and proton pumps are involved in pH maintenance of cancer cells, it can be considered that the expression levels and genetic mutations of these factors could possibly alter the anti-tumor activity of complex I inhibitors. The extracellular acidification induced by complex I inhibitor is dependent on the activity of lactate transporters. Even if cancer cells have lower lactate transporter activity, cancer cells surrounding stromal cells could secrete lactate and induce extracellular acidification by complex I inhibitors. On the other hand, the intracellular acidification can be modified due to the activity of intracellular pH-maintenance factors.
We confirmed the anti-tumor activity of ITV through S6K1 suppression using a PDX model reproducing the heterogeneity of cancer cells of a patient origin, but it should be noted that we used only one PDX model derived from a single patient.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-phospho-(T389)-p70S6 kinase | Cell signaling Technology | Cat#9234; RRID: AB_2269803 |
| Anti-phospho-(S235/236)-S6 Ribosomal Protein | Cell signaling Technology | Cat#2211; RRID: AB_331679 |
| Anti-phospho-(T172)-AMPK alpha | Cell signaling Technology | Cat#2535; RRID: AB_331250 |
| Anti-AMPK alpha1 | Cell signaling Technology | Cat#2795; RRID: AB_560856 |
| Anti-PP2A C subunit | Cell signaling Technology | Cat#2259; RRID: AB_561239 |
| Anti-GAPDH | Cell signaling Technology | Cat#5174; RRID: AB_10622025 |
| Anti-phospho-(S473)-Akt | Cell signaling Technology | Cat#4060; RRID: AB_2315049 |
| Anti-p70S6 kinase | Santa Cruz Biotechnology | Cat#SC-230; RRID: AB_632156 |
| Anti-phospho-(T202/Y204)-ERK1/2 | Santa Cruz Biotechnology | Cat#SC-7383; RRID: AB_627545 |
| Anti-NDUFB8 | Abcam | Cat#ab110242; RRID: AB_10859122 |
| Anti-α-tubulin | Sigma-Aldrich | Cat#T5168; RRID: AB_477579 |
| Anti-rabbit IgG peroxidase-linked whole antibodies from donkey | GE Healthcare | Cat#NA 934; RRID: AB_772206 |
| Anti-mouse IgG peroxidase-linked whole antibodies from donkey | GE Healthcare | Cat#NA 931; RRID: AB_772210 |
| Anti-LAMP1 | Abcam | Cat#ab25630; RRID: AB_470708 |
| anti-mTOR | Cell Signaling Technology | Cat#2983; RRID: AB_2105622 |
| Alexa Fluor 546 anti-Mouse IgG | Thermo Fisher Scientific | Cat#A-21123; RRID: AB_2535765 |
| Alexa Fluor 488 anti-Rabbit IgG | Thermo Fisher Scientific | Cat#A-21206; RRID: AB_2535792 |
| Anti-a-SMA | Abcam | Cat#ab7817; RRID: AB_262054 |
| Anti-Vimentin | Santa Cruz Biotechnology | Cat#sc-6260; RRID: AB_628437 |
| Anti-GFP | Cell Signaling Technology | Cat#2955; RRID: AB_1196614 |
| Anti-Ki-67 | Abcam | Cat#ab16667; RRID: AB_302459 |
| Anti-Phospho-p70 S6 Kinase (Thr 389) | Aviva Systems Biology | Cat#OAAF07416; RRID: AB_2630782 |
| Anti-HLA | Hokudo | Cat#AB-46; RRID: AB_1962147 |
| Anti-BrdU | Dako | Cat#M0744; RRID: AB_10013660 |
| Anti-S6K | Santa Cruz Biotechnology | Cat#sc-8418; RRID: AB_628094 |
| Biological samples | ||
| Mouse tumor tissue | This manuscript | N/A |
| Tissue samples from PDAC patients and PDX mouse | JFCR | approval number 2012-1001 |
| Chemicals, peptides, and recombinant proteins | ||
| Metformin | Sigma-Aldrich | D150959 |
| Rotenone | Sigma-Aldrich | R8875 |
| Arctigenin | Sigma-Aldrich | SMB00075 |
| Buformin hydrocjloride | Santa Cruz Biotechnology | sc-207383 |
| NADH | Sigma-Aldrich | N8129 |
| Ubiquinone | Sigma-Aldrich | C7956 |
| Sodium L-lactate | Sigma-Aldrich | 71718 |
| L-(+)-Lactic acid | Sigma-Aldrich | L1750 |
| Calyculin A | Cell Signaling Technology | 9902 |
| Temsirolimus | Sigma-Aldrich | PZ0020 |
| Nigericin | Sigma-Aldrich | N7143 |
| TTFA (2-thenoyltrifluoroacetone | Sigma-Aldrich | T27006 |
| KCN (potassium cyanide | Sigma-Aldrich | 60178 |
| Antimycin A | Sigma-Aldrich | A8674 |
| Oligomycin | Sigma-Aldrich | O4876 |
| ASP4132 | Synthesized | (Kuramoto et al., 2020) |
| MTT | DOJINDO | 341-01823 |
| Matrigel | BD Biosciences | 354230 |
| ITV- and AS-1936-immobilized beads | This manuscript | N/A |
| Guide-it Recombinant Cas9 protein | Z2641N | TaKaRa Bio |
| pHrodo Red AM | Thermo Fisher Scientific | P35372 |
| Masson’s Trichrome staining | Sigma-Aldrich | HT15 |
| Critical commercial assays | ||
| XF mitostress kit | Agilent Technologies | 103-015-100 |
| MitoXpress | Agilent Technologies | MX-200-4 |
| Mitochondria Isolation Kit | Abcam | ab110171 |
| MitoCheck Complex Activity Assay Kit | Cayman Chemical | #700939, #700940, #700950, #700990, #701000 |
| Lactate Assay Kit II | BioVision | K627-100 |
| Lactate-Glo Assay | Promega | J5021 |
| LabAssay Glucose | Wako | 298065701 |
| Proteome Profiler Human Phospho-Kinase Array Kit | R&D systems | ARY003C |
| ChemMate EnVision kit | Dako | K5007 |
| Envision + Dual Link System-HRP | Dako | K4063 |
| Liquid DAB + Substrate Chromogen System | Dako | K3468 |
| BrdU solution | BD Biosciences | 550891 |
| PI/RNase Staining Buffer | BD Biosciences | 550825 |
| Experimental models: Cell lines | ||
| MKN-1 | JCRB | JCRB0252 |
| MKN-B | JCRB | JCRB1025 |
| MKN-74 | JCRB | JCRB0255 |
| DLD-1 | ATCC | CCL-221 |
| HCT-15 | ATCC | CCL-225 |
| HCT-116 | ATCC | CCL-247 |
| A549 | ATCC | CCL-185 |
| NCI-H23 | ATCC | CRL-5800 |
| DMS 273 | ECACC | 95062830 |
| LNCaP | ATCC | CRL-1740 |
| DU-145 | ATCC | HTB-81 |
| PC-3 | ATCC | CRL-1435 |
| Capan-1 | ATCC | HTB-79 |
| MIA PaCa-2 | ATCC | CRL-1420 |
| BxPC-3 | ATCC | CRL-1687 |
| CCD-18Co | ATCC | CRL-1459 |
| NHLF | Lonza | CC-2512 |
| PrSC | Lonza | CC-2508 |
| HPaSteC | ScienCell Research Laboratories | #5301 |
| Hs738 | ATCC | CRL-7869 |
| Experimental models: Organisms/strains | ||
| Mouse: CAnN.Cg-Foxn1nu/CrlCrlj (BALB/c nude) | Charles River Laboratories | https://www.crj.co.jp/product/rm/detail/balbcnu |
| Mouse: NOD.CB17-Prkdcscid/J (NOD-SCID) | Charles River Laboratories | https://www.crj.co.jp/product/rm/detail/nodscid |
| Mouse: Nod.Cg-PrkdcscidIL2rgtm1Wjl/SzJ (NSG) | Charles River Laboratories | https://www.crj.co.jp/product/rm/detail/nsg |
| PDX model | JFCR | approval number 2012-1001 |
| Oligonucleotides | ||
| ON-TARGETplus siRNA-SMARTpool, Non-targeting Pool | Dharmacon | D-001810-10-15 |
| ON-TARGETplus siRNA-SMARTpool, PPP2CA | Dharmacon | L-003598-01-0005 |
| ON-TARGETplus siRNA-SMARTpool, PPP2CB | Dharmacon | L-003599-00-0005 |
| crRNA, GPR132 | UAAGUGGCCCGUUGAUUCGU and UGCUGGUGAGGGGUCCGUAC | |
| tracrRNA | Sigma-Aldrich | TRACRRNA05N-5NMOL |
| GPR4 primers for qPCR | TaKaRa Bio | HA104269 |
| GPR65 primers for qPCR | TaKaRa Bio | HA159373 |
| GPR68 primers for qPCR | TaKaRa Bio | HA274097 |
| GPR132 primers for qPCR | TaKaRa Bio | HA274259 |
| GAPDH primers for qPCR | TaKaRa Bio | HA067812 |
| Software and algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| FACSuite software (version 1.2.1) | BD Biosciences | https://www.bdbiosciences.com/ja-jp/products/software/instrument-software/bd-facsuite-application#Overview |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Manabu Kawada (kawadam@bikaken.or.jp)
Materials availability
ITV-and AS-1936-immobilized beads are available by the lead contact upon request.
Experimental model and subject details
Animals
Mouse xenograft model
All animal studies were approved by the Institutional Committee for Animal Experiments of the Institute of Microbial Chemistry and performed in accordance with the relevant guidelines and regulations to minimize animal suffering. The anti-tumor activity of ITV was evaluated in vivo as previously mentioned (Kawada et al., 2013). Female BALB/c nude mice (5-week-old, n = 5) were purchased from Charles River Laboratories (Kanagawa, Japan) and used at 8 weeks of age.
Mouse PDX model
Male NOD.CB17-Prkdcscid/J (NOD-SCID) mice and female Nod.Cg-PrkdcscidIL2rgtm1Wjl/SzJ (NSG) mice were purchased from Charles River Laboratories Japan (Kanagawa, Japan). All animal procedures were performed according to protocols approved by the Japanese Foundation for Cancer Research Animal Care and Use Committee (JFCR; approval number 18-01-3) and were in accordance with the relevant guidelines and regulations. Mice were housed in specific pathogen-free conditions. Tissue samples were obtained from PDAC patients in accordance with the guidelines of the JFCR (approval number 2012-1001). Informed consent was obtained from all patients.
Tissue for the PDX was derived from a CA19-9 and MUC-1 positive moderately differentiated PDAC tumor (KRAS mutated; Gly12Arg) obtained from a patient (female, aged 43) who underwent surgical resection at the Cancer Institute Hospital of JFCR. Tumor samples were processed within 24 h of biopsy. Then, tumor tissues were subcutaneously implanted into a 5–6-week-old NOD-SCID male mouse, and PDX were expanded in 5–6-week-old NSG female mice; excess tissues were frozen in CELLBANKER.
Cell culture
Human gastric cancer cell lines MKN-1, MKN-B, and MKN-74; human colorectal cancer cell lines DLD-1, HCT-15, and HCT-116; human lung cancer cell lines A549, NCI-H23, and DMS 273; human prostate cancer cell lines LNCaP-CR, DU-145, and PC-3; human pancreatic cancer cell lines Capan-1, MIA PaCa-2, and BxPC-3; and stromal cells CCD-18Co, NHLF, PrSC, and HPaSteC were obtained as previously described (Kawada et al., 2015; Nishiya et al., 2021). These cancer cell lines stably express a transfected GFP vector as previously described (12). Cancer cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM; 05919, Nissui, Tokyo, Japan) supplemented with 10% fetal bovine serum (FBS, Sigma-Aldrich, St. Louis, MO), 100 U/mL penicillin G, 100 μg/mL streptomycin, and 0.292 mg/mL glutamine (10378-016, Thermo Fisher Scientific, Walthum, MA) at 37°C with 5% CO2. Hs738.St/Int (Hs738) human gastric stromal cells (CRL-7869) were obtained from the American Type Culture Collection. Stromal cells were maintained in DMEM supplemented with 10% FBS, 100 U/mL penicillin G, 100 μg/mL streptomycin, 0.292 mg/mL glutamine, insulin/transferrin/hydrocortisone (ITH: 5 μg/mL insulin, 5 μg/mL transferrin, and 1.4 μM hydrocortisone), and 5 ng/mL basic FGF (PeproTech, Rocky Hill, NJ) at 37°C with 5% CO2 (12). The preparation of the CM from stromal cells and the synthesis of ITV and its derivatives have been previously described (Abe et al., 2013a, 2013b).
Method details
Reagents
The reagents used and synthesized in this study were as follows: metformin (1,1-dimethylbiguanide hydrochloride: D150959, Sigma-Aldrich), rotenone (R8875, Sigma-Aldrich), arctigenin (SMB00075, Sigma-Aldrich), buformin (buformin hydrocjloride: sc-207383, Santa Cruz Biotechnology, Dallas, TX), NADH (N8129, Sigma-Aldrich), ubiquinone (coenzymeQ1: C7956, Sigma-Aldrich), sodium lactate (sodium L-lactate, 71718, Sigma-Aldrich), lactate (L-(+)-Lactic acid, L1750, Sigma-Aldrich), calyculin A (#9902, Cell Signaling Technology, Danvers, MA), temsirolimus (PZ0020, Sigma-Aldrich), nigericin (N7143, Sigma-Aldrich), TTFA (2-thenoyltrifluoroacetone, T27006, Sigma-Aldrich), KCN (potassium cyanide, 60178, Sigma-Aldrich), antimycin A (A8674, Sigma-Aldrich), and oligomycin (O4876, Sigma-Aldrich). ASP4132 was synthesized as previously described (Kuramoto et al., 2020).
Cell viability assays
Cells were inoculated into 96-well plates at 5 × 103 cells per well in 0.1 mL of DMEM supplemented with 1% dialyzed FBS (D-FBS) and ITH. After culture for the indicated number of days, cell growth was determined using 3-(4,5-dimethyl-2-lythiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT; 341-01823, DOJINDO, Kumamoto, Japan) assay (12). Alternatively, cells were lysed in 10 mm Tris–HCl at pH 7.4 containing 150 mm NaCl, 0.9 mm CaCl2, and 1% Triton X-100, and cell growth was measured based on GFP fluorescence intensity (excitation wavelength at 485 nm and emission wavelength at 538 nm) and assessed using a Cytation 5 plate reader (BioTek, Winooski, VT). Co-culture experiments and the evaluation of mode of action of the compounds using the JFCR 39 cell-line panel were performed as previously described (Kawada et al., 2015; Nakatsu et al., 2007).
Mouse xenograft models
Cancer cells (8 × 106) were suspended with or without the indicated stromal cells (8 × 106) in 0.3 mL of 10% FBS-DMEM and mixed with 0.5 mL of BD Matrigel Matrix Growth Factor Reduced (BD Biosciences Systems, Franklin Lakes, NJ), 100 μL of which (1 × 106) was subcutaneously injected into the left lateral flank of BALB/c nude mice. The vehicle (saline supplemented with 10% DMSO and 1% Tween 80) or the indicated samples were intravenously (for ITV) or intraperitoneally (for ASP4132) administered 5 days a week to mice. Tumor volume was estimated using the following formula: tumor volume (mm) = (length × width2)/2. Mice were euthanized on day 17 or 21 after cell implantation by cervical dislocation, and tumor weights were determined.
To evaluate the mechanism of action (Figure 6), three mice were used for each experimental set. After 24 h fasting, the vehicle or the indicated samples were intravenously administered into mice at 14 and 15 days after MKN-74 cell implantation. After 4 h, the mice were anesthetized with an intraperitoneal injection of mixed anesthesia containing medetomidine hydrochloride (0.75 mg/kg, Kyoritsu Seiyaku, Tokyo, Japan), midazolam (4 mg/kg, Sandoz, Tokyo, Japan), and butorphanol tartrate (5 mg/kg, Meiji Seika Pharma, Tokyo, Japan).
Isolation of mouse fibroblasts
Tumors were excised from euthanized mice, cut into small pieces (approximately 1 mm3), and cultured on a 35 mm dish with a drop of FBS. After air-drying, the tumor pieces were cultured in DMEM containing 1% D-FBS and ITH. After fibroblast expansion, they were trypsinized and pooled for further use.
OCR and extracellular acidification
Metabolic analysis was performed using Seahorse XFe24 analyzer (Agilent Technologies, Santa Clara, CA). Gastric cancer cells were cultured in 24-well plates for XFe24, and MKN-B cells (5 × 105) and the growth medium (2 × 105 cells/250 μL) were added. After equilibration in non-CO2 incubator for 1 h, the medium was replaced with 450 μL of the assay medium (XF DMEM, 103575-100, Agilent Technologies; containing 10 mm glucose, 1 mm pyruvate, and 2 mm glutamine). Mitostress test was performed with ITV (final concentration: 0.25, 0.5, and 1 μg/mL), FCCP (1 μM), and rotenone/antimycin A (0.5 μM).
For glucose-depleted conditions, the assay medium without glucose and pyruvate was used. Analysis was performed 1 h after incubation and after treatment with ITV (1 μg/mL), rotenone (0.01 μg/mL), glucose (16.7 mM), and succinate (30 mM). FCCP, rotenone, and antimycin A were purchased from Agilent Technologies (XF mitostress kit, 103015-100). MitoXpress (MX-200-4, Agilent) assay was performed according to manufacturers instructions. Cells (8 × 104 cells/well) were seeded in 96-well plates with the growth medium that was replaced with fresh medium containing 1% D-FBS with the MitoXpress reagent. The fluorescence signal of MitoXpress reagent was immediately read using a Cytation 5 plate reader after the addition of test compounds.
Enzymatic activity of complex I
Mitochondria were isolated from culture cells using a Mitochondria Isolation Kit (ab110171, Abcam, Cambridge, UK) according to manufacturer's instruction. Mitochondria extracts were frozen and thawed for three cycles. Protein concentration was determined using a protein assay (5000006JA, Bio-Rad Laboratories, Hercules, CA). For complex I activity assay, test compounds (20 μL) were added to 96-well plates, and mitochondria extract (5 μg) in 165 μL of assay buffer (containing 3.6 mg/mL fatty acid free BSA, 2.4 mg/mL antimycin A, 0.12 mM NADH, and 0.36 mM KCN in 60 mM potassium phosphate buffer at pH 7.5) was added to each well. After pre-heating the plate to 37°C and the addition of 15 μL of ubiquinone (1 mM), OD340 was immediately measured using Cytation 5 plate reader. The amount of reduced NADH was obtained by subtracting the value at 10 min from that at 5 min. The method for complex I activity assay was modified from a previous report (Spinazzi et al., 2012). To determine the activity of mitochondrial complexes I–V (Figure S3F), MitoCheck Complex Activity Assay Kit (#700939, #700940, #700950, #700990, #701000, Cayman Chemical, Ann Arbor, MI) was used according to manufacturer's instruction.
Pull-down assay
ITV- and AS-1936-immobilized beads were prepared (Kanoh et al., 2005; Suvarna et al., 2020). Mitochondrial fractions from Hs738 cells were resuspended in the lysis buffer used for western blotting and incubated with ITV-conjugated beads for 3 h at 4°C. After washing the beads with 50 mM HEPES-KOH (at pH 7.5 containing 150 mM NaCl, 1 mM EDTA, and 2.5 mM EGTA), the beads were denatured in SDS sample buffer by boiling and subjected to western blot analysis.
Measurement of lactate and glucose levels
Lactate Assay Kit II (K627-100, BioVision, Milpitas, CA) was used to determine lactate levels in co-cultured CM and monocultured cells according to the manufacturer’s instruction. Co-cultured CM was collected from the 96-well plates of co-culture experiment 3 days after the inoculation of cancer cells, whereas monocultured cells were seeded in 96-well plates (5 × 103 cells/well) in DMEM containing 1% D-FBS and ITH for 3 days. The supernatant of centrifuged CM (1,000 × g, 10 min at 25°C) and tissue samples from western blot analysis were used to measure lactate levels using Lactate-Glo Assay (J5021, Promega, Madison, WI). Extracellular glucose concentration of the CM from monocultured cells was measured using LabAssay Glucose (298065701, Wako, Tokyo, Japan).
Metabolite extraction and metabolomic analysis
Cells were seeded in 10 cm dishes (3 × 106 cells/dish) in DMEM with 10% FBS, and the medium was replaced with DMEM containing 1% D-FBS. Cells were incubated with the test compounds for 3 h while the control groups were treated with DMSO at equal concentrations. Then, cells were washed with mannitol solution and treated with 800 μL methanol for 30 s. The cell extract was added with 550 μL Milli-Q water containing internal standards (H3304-1002, Human Metabolome Technologies [HMT], Tsuruoka, Yamagata, Japan) for another 30 s. The obtained extract was centrifuged at 2,300 × g and 4°C for 5 min, after which 800 μL of the supernatant was centrifugally filtered at 9,100 × g and 4°C through a Millipore 5 kDa cutoff filter (Ultrafree MC-PLHCC, HMT). Metabolomic analysis was conducted with the C-SCOPE package of HMT using capillary electrophoresis time-of-flight mass spectrometry (CE-TOFMS) for cation analysis and CE-tandem mass spectrometry (CE-MS/MS) for anion analysis as previously described (Junker et al., 2006; Ohashi et al., 2008; Ooga et al., 2011; Sugimoto et al., 2010).
Western blot analysis
Cell lysates were prepared from cultured cells and subjected to western blotting with the following primary antibodies: anti-phospho-(T389)-p70S6 kinase (#9234), anti-phospho-(S235/236)-S6 Ribosomal Protein (#2211), anti-phospho-(T172)-AMPK alpha (#2535), anti-AMPK alpha1 (#2795), anti-PP2A C subunit (#2259), anti-GAPDH (#5174), and anti-phospho-(S473)-Akt (#4060), all from Cell Signaling Technology, anti-p70S6 kinase (SC-230, Santa Cruz Biotechnology), anti-phospho-(T202/Y204)-ERK1/2 (SC-7383, Santa Cruz Biotechnology), and NDUFB8 (ab110242, Abcam) (20). The following secondary antibodies were used: mouse monoclonal anti-α-tubulin (1:1,000 dilution; T5168; Sigma-Aldrich), anti-rabbit IgG peroxidase-linked whole antibodies from donkey (NA 934, GE Healthcare, Chicago, IL), and anti-mouse IgG peroxidase-linked whole antibodies from donkey (NA 931, GE Healthcare). Next, anti-phospho kinase antibody array was performed using a Proteome Profiler Human Phospho-Kinase Array Kit (ARY003C, R&D systems, Minneapolis, MN). MKN-74 cells were seeded in 6-well plates (3 × 104 cells/well) in DMEM supplemented with 1% D-FBS and cultured for 3 h, and the medium was replaced with the Ctrl CM or ITV CM.
siRNA transfection
For gene knockdown experiments, ON-TARGETplus siRNA-SMARTpool was used (Human PPP2CA: L-003598-01-0005, Human PPP2CB: L-003599-00-0005, Non-targeting Pool: D-001810-10-15, Dharmacon, Lafayette, CO). Lipofectamine RNAiMAX Reagent (13778, Thermo Fisher Scientific) was used for siRNA.
Immunofluorescence
Cells (1 × 105 cells/mL) were cultured on glass cover slips in 6-well plates in 2 mL of DMEM with 1% D-FBS, and the medium was replaced with the indicated prepared medium. Earle's balanced salt solution (EBSS) was purchased from Thermo Fisher Scientific (24010-043). Cells were washed with ice cold PBS (−) and fixed with 4% paraformaldehyde in PBS (−) for 10 min at 25°C. After washing with PBST (0.1% Tween-20 in PBS), cells were permeabilized with PBST-BSA (0.1% Tween-20, 0.1% BSA in PBS) for 45 min at 25°C and stained with DAPI, anti-LAMP1 (ab25630, Abcam), anti-mTOR (2983, Cell Signaling Technology) primary antibodies, and Alexa Fluor 546 anti-Mouse IgG (A-21123, Thermo Fisher Scientific) and Alexa Fluor 488 anti-Rabbit IgG (A-21206, Thermo Fisher Scientific) second antibodies. Stained cells were visualized under an Axiovert 200M microscope using the LSM5 LIVE scanning module of the LSM LIVE DUO software (ZEISS, Oberkochen, Germany).
Gene knockout
MKN-74 cells were seeded in 6-well plates (1 × 105 cells/well) in 2 mL DMEM with 10% FBS. Two CRISPR RNAs (crRNAs) for each gene were designed by CRISPRdirect (70) and purchased from Sigma-Aldrich (crRNA sequence, GPR132: UAAGUGGCCCGUUGAUUCGU and UGCUGGUGAGGGGUCCGUAC). The ribonucleoprotein (RNP) was constructed using two crRNAs (60 pmol), 60 pmol SygRNA Synthetic tracrRNA (TRACRRNA05N-5NMOL, Sigma-Aldrich), and 3 μg Guide-it Recombinant Cas9 protein (Z2641N, TaKaRa Bio, Shiga, Japan). Next, RNP was mixed with 10 mm Tris buffer at pH 7.5 (T2319, Sigma-Aldrich) and incubated on ice for 30 min. For transfection, 5 μL TransIT-CRISPR (T1706, Sigma-Aldrich) in 250 μL OPTI-MEM (31985062, Thermo Fisher Scientific) was added to the RNP solution and cell culture medium after incubation for 20 min.
Quantitative PCR
RNA was isolated using an RNeasy kit (74004, QIAGEN, Hilden, Germany), and cDNA was synthesized using a Reverse Transcription System (A3500, Promega). Quantitative PCR was performed using GoTaq qPCR Master Mix (A6001, Promega) on a Thermal Cycler Dice Real Time System III (TaKaRa Bio). The primer sets were purchased from TaKaRa Bio (GPR4: HA104269, GPR65: HA159373, GPR68: HA274097, GPR132: HA274259, GAPDH: HA067812). Relative expression levels were calculated by the 2 (-Delta Delta C(T)) method (Livak and Schmittgen, 2001), and GAPDH was used as the reference gene.
Intracellular pH
Measurement of intracellular pH using pHrodo Red AM (P35372, Thermo Fisher Scientific) was performed. Cells were seeded into 96-well plates (3 × 104 cells/well) in 0.1 mL DMEM supplemented with 1% D-FBS, and the medium was replaced with 0.1 mL HBSS/HEPES buffer (1.2 mM CaCl2, 0.8 mM MgSO4, 5.4 mm KCl, 0.4 mM KH2PO4, 4.2 mM NaHCO3, 137 mM NaCl, 0.3 mM Na2HPO4, 5.5 mM glucose, and 52 mM HEPES at pH 7.4) containing pHrodo Red and the test compound. After 3 h incubation in CO2 incubator, the buffer was replaced with fresh buffer, and fluorescence intensity was determined using Cytation 5 plate reader.
Sample preparation from mouse tumor tissues
Tumor tissues were washed with ice cold PBS (without calcium and magnesium), and their weight was measured. Tumor tissues were homogenized in a Dounce homogenizer with RIPA buffer (150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, 10 mM NaF, and 50 mM Tris-HCl at pH 7.4), PhosSTOP (4906845001, Sigma-Aldrich), and 25 μg/mL each of antipain, leupeptin, and pepstatin. The collected supernatants from a half volume of lysate that were centrifuged at 15,000 rpm for 15 min at 4°C were subjected to western blot analysis. Next, 0.125 mL 0.6 N HCl was added to the other half volume of lysate and mixed by vortex. After the addition of 0.125 mL Trizma base (0.121 g/mL; Sigma-Aldrich) and vortexing at 15,000 rpm for 15 min at 4°C, samples were centrifuged, and then the supernatants were used for the lactate assay.
Histology and immunohistochemistry
Immunohistochemical analysis for mouse xenograft model was performed as previously described (Ohishi et al., 2018). Briefly, paraffin-embedded tissues were cut and mounted on slides. To deplete paraffin, the sections were washed with xylene, ethanol, and methanol hydrogen peroxide. The deparaffinized sections were boiled in 0.01 M buffered sodium citrate solution (pH 6.0) for 10 min and subjected to Masson's Trichrome staining (Sigma-Aldrich) overnight or immunohistochemical staining with the following antibodies for 30 min: anti-a-SMA (1:2000, ab7817, Abcam), anti-Vimentin (1:400, sc-6260, Santa Cruz Biotechnology), anti-GFP (1:500, #2955, Cell Signaling Technology), anti-Ki-67 (1:200, ab16667, Abcam), anti-Phospho-p70 S6 Kinase (Thr 389) (1:50, OAAF07416, Aviva Systems Biology, San Diego, CA), and horseradish peroxidase-linked secondary antibodies. Tissues were stained with 3,3′-diaminobenzidine (DAB) using a ChemMate EnVision kit (Dako, Carpinteria, CA), counterstained with hematoxylin, visualized under a Nikon Biophot microscope (Tokyo, Japan), and photographed using a digital camera (Nikon Digital Sight DS- Ri1).
For the PDX model, tumors were fixed with 4% paraformaldehyde solution, embedded in paraffin, cut into 2 to 4 μm-thick sections, de-paraffinized with xylene, re-hydrated using graded ethanol, and subjected to hematoxylin and eosin (HE) staining or the subsequent immunohistochemistry. Antigen retrieval was performed using microwave heating for 10 min in sodium citrate-hydrochloric acid buffer (10 mM, pH 6.0). After rinsing in PBS, the sections were incubated with the primary antibodies overnight at 4°C. Thereafter, immunohistochemical staining was performed using a DAKO Envision + Dual Link System-HRP (K4063, Dako) with Liquid DAB + Substrate Chromogen System (K3468, Dako) and counterstained with hematoxylin. The following antibodies were used: anti-HLA (AB-46, Hokudo, Hokkaido, Japan), anti-BrdU (M0744, Dako), anti-S6K (sc-8418, Santa Cruz Biotechnology), and anti-phospho-S6RP(Ser235/236) (#2211, Cell Signaling Technology).
BrdU labeling assay
5-Bromo-2′-deoxyuridine (BrdU) labeling assay was conducted to evaluate tumor cell proliferation. At the end of experiments, animals were intraperitoneally injected with 0.2 mL of 10 mg/mL BrdU solution (550891, BD Biosciences) 90 min before euthanization. Paraffin-embedded tissue sections were immunostained using monoclonal antibodies against BrdU (M0744, Dako) and imaged using an ECLIPSE TE2000-U microscope (Nikon).
PDX model
When the tumor volume reached 200 mm3, mice were randomly divided into two groups administered with ITV (12.5 mg/kg in 100 μL saline containing 10% DMSO and 1% Tween 80) or the vehicle via the tail vain every 5 days except during weekends for a total of 2.5 weeks. Tumor size was assessed twice weekly using a caliper [(length × width2)/2]. Immunodeficient mice were then euthanized 18 days after ITV treatment. The areas containing the transplanted tumor tissues of PDX were measured and weighed.
Flow cytometry
Cultured cells were collected by trypsinization and fixed with cold 70% ethanol. After 2 h incubation in ice, cells were washed with PBS and stained with propidium iodide (PI/RNase Staining Buffer, 550825, BD Biosciences) for 15 min according to manufacturer's instruction. PI intensity was analyzed using BD FACSLyric flow cytometer (BD Biosciences). Data analysis was performed using the BD FACSuite software (version 1.2.1, BD Biosciences).
Quantification and statistical analysis
Statistical analysis was performed using two-tailed Student's t-test. Data are representative of at least three independent experiments. All data are presented as the mean ± standard deviation (s.d.). The number of replicates (n) and p values are indicated in each figure legend. A p value of <0.05 was considered to be statistically significant.
Acknowledgments
The authors thank Dr. Isao Momose, Dr. Daisuke Tatsuda, Dr. Shun-ichi Wada, Dr. Tomoko Nakanishi, and Ms. Akiko Harakawa (BIKAKEN); Dr. Ken Matsumoto, Dr. Yoko Yashiroda, and Dr. Minoru Yoshida (RIKEN); and Ms. Kanami Yamazaki, Dr. Tetsuo Mashima, and Dr. Akihiro Tomida (JFCR) for their fruitful suggestions and technical support. This work was supported in part by the Molecular Profiling Committee; Grant-in-Aid for Scientific Research on Innovative Areas “Advanced Animal Model Support (AdAMS)” from The Ministry of Education, Culture, Sports, Science and Technology, Japan (MEXT) (JSPS KAKENHI grant number JP 16H06276); the Project for Cancer Research and Therapeutic Evolution (P-CREATE) of the Japan Agency for Medical Research and Development (grant numbers JP20cm010623h to M.K. and JP20cm0106002h to Y.K.); Grant-in-Aid for Scientific Research (grant number 18K05366); and a Grant-in-Aid for Scientific Research on Innovative Areas (grant number 18H05503 to Y.K.) from MEXT.
Author contributions
J.Y. designed and performed most in vitro experiments, analyzed the data, and wrote the manuscript. T.O., S.O., and H.I. performed in vivo experiments with mouse xenograft models. H.A., R.O., C.S., and T.W. synthesized ITV and its derivatives. I.U. and M.A. performed some of the co-culture experiments using ITV derivatives and provided technical assistance. S.D. predicted the mode of action of ITV using JFCR39. M. Sugawara., T.K., J.U., Y.A., A.I., M.T., G.A., and T.N. established the PDX models. Y.K., K.H., and H.O. Prepared the compound-conjugate beads. T.S. and M. Shibasaki. supervised and edited the manuscript. M.K. supervised the work and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: December 17, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103497.
Supplemental information
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abe H., Kawada M., Inoue H., Ohba S., Nomoto A., Watanabe T., Shibasaki M. Synthesis of intervenolin, an antitumor natural quinolone with unusual substituents. Org. Lett. 2013;15:2124–2127. doi: 10.1021/ol400587a. [DOI] [PubMed] [Google Scholar]
- Abe H., Kawada M., Inoue H., Ohba S., Masuda T., Hayashi C., Igarashi M., Nomoto A., Watanabe T., Shibasaki M. Structure-activity relationship study of intervenolin derivatives: synthesis, antitumor, and anti-Helicobacter pylori activities. Tetrahedron. 2013;69:7608–7617. [Google Scholar]
- Ahn J.H., McAvoy T., Rakhilin S.V., Nishi A., Greengard P., Nairn A.C. Protein kinase A activates protein phosphatase 2A by phosphorylation of the B56delta subunit. Proc. Natl. Acad. Sci. U S A. 2007;104:2979–2984. doi: 10.1073/pnas.0611532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton T.M., McKenna W.G., Kunz-Schughart L.A., Higgins G.S. Oxidative phosphorylation as an emerging target in cancer therapy. Clin. Cancer Res. 2018;24:2482–2490. doi: 10.1158/1078-0432.CCR-17-3070. [DOI] [PubMed] [Google Scholar]
- Balsa E., Perry E.A., Bennett C.F., Jedrychowski M., Gygi S.P., Doench J.G., Puigserver P. Defective NADPH production in mitochondrial disease complex I causes inflammation and cell death. Nat. Commun. 2020;11:2714. doi: 10.1038/s41467-020-16423-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges H.R., Jones A.J., Pollak M.N., Hirst J. Effects of metformin and other biguanides on oxidative phosphorylation in mitochondria. Biochem. J. 2014;462:475–487. doi: 10.1042/BJ20140620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A., Dong L., Leffler N.R., Asch A.S., Witte O.N., Yang L.V. Activation of GPR4 by acidosis increases endothelial cell adhesion through the cAMP/Epac pathway. PLoS One. 2011;6:e27586. doi: 10.1371/journal.pone.0027586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan S., Tsunoda T., Kitahara O., Yanagawa R., Zembutsu H., Katagiri T., Yamazaki K., Nakamura Y., Yamori T. An integrated database of chemosensitivity to 55 anticancer drugs and gene expression profiles of 39 human cancer cell lines. Cancer Res. 2002;62:1139–1147. [PubMed] [Google Scholar]
- DeBerardinis R.J., Chandel N.S. We need to talk about the Warburg effect. Nat. Metab. 2020;2:127–129. doi: 10.1038/s42255-020-0172-2. [DOI] [PubMed] [Google Scholar]
- DeGrande S.T., Little S.C., Nixon D.J., Wright P., Snyder J., Dun W., Murphy N., Kilic A., Higgins R., Binkley P.F., et al. Molecular mechanisms underlying cardiac protein phosphatase 2A regulation in heart. J. Biol. Chem. 2013;288:1032–1046. doi: 10.1074/jbc.M112.426957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert M.A., Coscia F., Chryplewicz A., Chang J.W., Hernandez K.M., Pan S., Tienda S.M., Nahotko D.A., Li G., Blaženović I., et al. Proteomics reveals NNMT as a master metabolic regulator of cancer-associated fibroblasts. Nature. 2019;569:723–728. doi: 10.1038/s41586-019-1173-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehinger J.K., Piel S., Ford R., Karlsson M., Sjövall F., Frostner E., Morota S., Taylor R.W., Turnbull D.M., Cornell C., et al. Cell-permeable succinate prodrugs bypass mitochondrial complex I deficiency. Nat. Commun. 2016;7:12317. doi: 10.1038/ncomms12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erez N., Truitt M., Olson P., Arron S.T., Hanahan D. Cancer-associated fibroblasts are activated in incipient neoplasia to orchestrate tumor-promoting inflammation in an NF-kappaB-Dependent manner. Cancer Cell. 2010;17:135–147. doi: 10.1016/j.ccr.2009.12.041. [DOI] [PubMed] [Google Scholar]
- Faubert B., Li K.Y., Cai L., Hensley C.T., Kim J., Zacharias L.G., Yang C., Do Q.N., Doucette S., Burguete D., et al. Lactate metabolism in human lung tumors. Cell. 2017;171:358–371.e359. doi: 10.1016/j.cell.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J., Yan F., Zhang Y., Triplett A., Zhang Y., Schultz D.A., Sun Y., Zeng J., Silverstein K.A.T., Zheng Q., et al. Expression of adipocyte/macrophage fatty acid-binding protein in tumor-associated macrophages promotes breast cancer progression. Cancer Res. 2018;78:2343–2355. doi: 10.1158/0008-5472.CAN-17-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie D.G. AMPK--sensing energy while talking to other signaling pathways. Cell Metab. 2014;20:939–952. doi: 10.1016/j.cmet.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley C.T., Faubert B., Yuan Q., Lev-Cohain N., Jin E., Kim J., Jiang L., Ko B., Skelton R., Loudat L., et al. Metabolic heterogeneity in human lung tumors. Cell. 2016;164:681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hida K., Hida Y., Amin D.N., Flint A.F., Panigrahy D., Morton C.C., Klagsbrun M. Tumor-associated endothelial cells with cytogenetic abnormalities. Cancer Res. 2004;64:8249–8255. doi: 10.1158/0008-5472.CAN-04-1567. [DOI] [PubMed] [Google Scholar]
- Huang Y.H., Su Y.S., Chang C.J., Sun W.H. Heteromerization of G2A and OGR1 enhances proton sensitivity and proton-induced calcium signals. J. Receptor Signal Transduction Res. 2016;36:633–644. doi: 10.3109/10799893.2016.1155064. [DOI] [PubMed] [Google Scholar]
- Huang Z., Zhang M., Chen G., Wang W., Zhang P., Yue Y., Guan Z., Wang X., Fan J. Bladder cancer cells interact with vascular endothelial cells triggering EGFR signals to promote tumor progression. Int. J. Oncol. 2019;54:1555–1566. doi: 10.3892/ijo.2019.4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara Y., Kihara Y., Hamano F., Yanagida K., Morishita Y., Kunita A., Yamori T., Fukayama M., Aburatani H., Shimizu T., et al. The G protein-coupled receptor T-cell death-associated gene 8 (TDAG8) facilitates tumor development by serving as an extracellular pH sensor. Proc. Natl. Acad. Sci. U S A. 2010;107:17309–17314. doi: 10.1073/pnas.1001165107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker B.H., Klukas C., Schreiber F. VANTED: a system for advanced data analysis and visualization in the context of biological networks. BMC Bioinformatics. 2006;7:109. doi: 10.1186/1471-2105-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabacaoglu D., Ciecielski K.J., Ruess D.A., Algül H. Immune checkpoint inhibition for pancreatic ductal adenocarcinoma: current limitations and future options. Front. Immunol. 2018;9:1878. doi: 10.3389/fimmu.2018.01878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalender A., Selvaraj A., Kim S.Y., Gulati P., Brûlé S., Viollet B., Kemp B.E., Bardeesy N., Dennis P., Schlager J.J., et al. Metformin, independent of AMPK, inhibits mTORC1 in a rag GTPase-dependent manner. Cell Metab. 2010;11:390–401. doi: 10.1016/j.cmet.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer. 2016;16:582–598. doi: 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- Kanoh N., Honda K., Simizu S., Muroi M., Osada H. Photo-cross-linked small-molecule affinity matrix for facilitating forward and reverse chemical genetics. Angew. Chem. Int. Ed. Engl. 2005;44:3559–3562. doi: 10.1002/anie.200462370. [DOI] [PubMed] [Google Scholar]
- Kawada M., Inoue H., Ohba S., Hatano M., Amemiya M., Hayashi C., Usami I., Abe H., Watanabe T., Kinoshita N., et al. Intervenolin, a new antitumor compound with anti-Helicobacter pylori activity, from Nocardia sp. ML96-86F2. J. Antibiot. 2013;66:543–548. doi: 10.1038/ja.2013.42. [DOI] [PubMed] [Google Scholar]
- Kawada M., Inoue H., Ohba S., Masuda T., Momose I., Ikeda D. Leucinostatin A inhibits prostate cancer growth through reduction of insulin-like growth factor-I expression in prostate stromal cells. Int. J. Cancer. 2010;126:810–818. doi: 10.1002/ijc.24915. [DOI] [PubMed] [Google Scholar]
- Kawada M., Inoue H., Ohba S., Yoshida J., Masuda T., Yamasaki M., Usami I., Sakamoto S., Abe H., Watanabe T., et al. Stromal cells positively and negatively modulate the growth of cancer cells: stimulation via the PGE2-TNFα-IL-6 pathway and inhibition via secreted GAPDH-E-cadherin interaction. PLoS One. 2015;10:e0119415. doi: 10.1371/journal.pone.0119415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawada M., Inoue H., Usami I., Ikeda D. Phthoxazolin A inhibits prostate cancer growth by modulating tumor-stromal cell interactions. Cancer Sci. 2009;100:150–157. doi: 10.1111/j.1349-7006.2008.00996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawada M., Momose I., Someno T., Tsujiuchi G., Ikeda D. New atpenins, NBRI23477 A and B, inhibit the growth of human prostate cancer cells. J. Antibiot. 2009;62:243–246. doi: 10.1038/ja.2009.20. [DOI] [PubMed] [Google Scholar]
- Kawada M., Someno T., Inoue H., Ohba S., Masuda T., Kato T., Ikeda D. NBRI16716A, a new antitumor compound against human prostate cancer cells, produced by Perisporiopsis melioloides Mer-f16716. J. Antibiot. 2010;63:319–323. doi: 10.1038/ja.2010.42. [DOI] [PubMed] [Google Scholar]
- Kennedy K.M., Scarbrough P.M., Ribeiro A., Richardson R., Yuan H., Sonveaux P., Landon C.D., Chi J.T., Pizzo S., Schroeder T., et al. Catabolism of exogenous lactate reveals it as a legitimate metabolic substrate in breast cancer. PLoS One. 2013;8:e75154. doi: 10.1371/journal.pone.0075154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppenol W.H., Bounds P.L., Dang C.V. Otto Warburg's contributions to current concepts of cancer metabolism. Nat. Rev. Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- Kuramoto K., Yamada H., Shin T., Sawada Y., Azami H., Yamada T., Nagashima T., Ohnuki K. Development of a potent and orally active activator of adenosine monophosphate-activated protein kinase (AMPK), ASP4132, as a clinical candidate for the treatment of human cancer. Bioorg. Med. Chem. 2020;28:115307. doi: 10.1016/j.bmc.2020.115307. [DOI] [PubMed] [Google Scholar]
- Lambrechts D., Wauters E., Boeckx B., Aibar S., Nittner D., Burton O., Bassez A., Decaluwé H., Pircher A., Van den Eynde K., et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 2018;24:1277–1289. doi: 10.1038/s41591-018-0096-5. [DOI] [PubMed] [Google Scholar]
- Lavin Y., Kobayashi S., Leader A., Amir E.D., Elefant N., Bigenwald C., Remark R., Sweeney R., Becker C.D., Levine J.H., et al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell. 2017;169:750–765.e717. doi: 10.1016/j.cell.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie S.N., Nairn A.C. cAMP regulation of protein phosphatases PP1 and PP2A in brain. Biochim. Biophys. Acta Mol. Cel. Res. 2019;1866:64–73. doi: 10.1016/j.bbamcr.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.Q., Zhu X.Z., Weng E.Q. Intracellular dopamine oxidation mediates rotenone-induced apoptosis in PC12 cells. Acta Pharmacol. Sin. 2005;26:17–26. doi: 10.1111/j.1745-7254.2005.00003.x. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ludwig M.G., Vanek M., Guerini D., Gasser J.A., Jones C.E., Junker U., Hofstetter H., Wolf R.M., Seuwen K. Proton-sensing G-protein-coupled receptors. Nature. 2003;425:93–98. doi: 10.1038/nature01905. [DOI] [PubMed] [Google Scholar]
- Mantovani A., Marchesi F., Malesci A., Laghi L., Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 2017;14:399–416. doi: 10.1038/nrclinonc.2016.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani Y., Kobayashi H., Iida T., Asai N., Masamune A., Hara A., Esaki N., Ushida K., Mii S., Shiraki Y., et al. Meflin-positive cancer-associated fibroblasts inhibit pancreatic carcinogenesis. Cancer Res. 2019;79:5367–5381. doi: 10.1158/0008-5472.CAN-19-0454. [DOI] [PubMed] [Google Scholar]
- Molina J.R., Sun Y., Protopopova M., Gera S., Bandi M., Bristow C., McAfoos T., Morlacchi P., Ackroyd J., Agip A.A., et al. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat. Med. 2018;24:1036–1046. doi: 10.1038/s41591-018-0052-4. [DOI] [PubMed] [Google Scholar]
- Murakami N., Yokomizo T., Okuno T., Shimizu T. G2A is a proton-sensing G-protein-coupled receptor antagonized by lysophosphatidylcholine. J. Biol. Chem. 2004;279:42484–42491. doi: 10.1074/jbc.M406561200. [DOI] [PubMed] [Google Scholar]
- Nakatsu N., Nakamura T., Yamazaki K., Sadahiro S., Makuuchi H., Kanno J., Yamori T. Evaluation of action mechanisms of toxic chemicals using JFCR39, a panel of human cancer cell lines. Mol. Pharmacol. 2007;72:1171–1180. doi: 10.1124/mol.107.038836. [DOI] [PubMed] [Google Scholar]
- Nishiya N., Oku Y., Ishikawa C., Fukuda T., Dan S., Mashima T., Ushijima M., Furukawa Y., Sasaki Y., Otsu K., et al. Lamellarin 14, a derivative of marine alkaloids, inhibits the T790M/C797S mutant epidermal growth factor receptor. Cancer Sci. 2021;112:1963–1974. doi: 10.1111/cas.14839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y., Hirayama A., Ishikawa T., Nakamura S., Shimizu K., Ueno Y., Tomita M., Soga T. Depiction of metabolome changes in histidine-starved Escherichia coli by CE-TOFMS. Mol. bioSystems. 2008;4:135–147. doi: 10.1039/b714176a. [DOI] [PubMed] [Google Scholar]
- Ohishi T., Masuda T., Abe H., Hayashi C., Adachi H., Ohba S.I., Igarashi M., Watanabe T., Mimuro H., Amalia E., et al. Monotherapy with a novel intervenolin derivative, AS-1934, is an effective treatment for Helicobacter pylori infection. Helicobacter. 2018;23:e12470. doi: 10.1111/hel.12470. [DOI] [PubMed] [Google Scholar]
- Ooga T., Sato H., Nagashima A., Sasaki K., Tomita M., Soga T., Ohashi Y. Metabolomic anatomy of an animal model revealing homeostatic imbalances in dyslipidaemia. Mol. bioSystems. 2011;7:1217–1223. doi: 10.1039/c0mb00141d. [DOI] [PubMed] [Google Scholar]
- Paull K.D., Shoemaker R.H., Hodes L., Monks A., Scudiero D.A., Rubinstein L., Plowman J., Boyd M.R. Display and analysis of patterns of differential activity of drugs against human tumor cell lines: development of mean graph and COMPARE algorithm. J. Natl. Cancer Inst. 1989;81:1088–1092. doi: 10.1093/jnci/81.14.1088. [DOI] [PubMed] [Google Scholar]
- Pernicova I., Korbonits M. Metformin--mode of action and clinical implications for diabetes and cancer. Nat. Rev. Endocrinol. 2014;10:143–156. doi: 10.1038/nrendo.2013.256. [DOI] [PubMed] [Google Scholar]
- Ravanel P., Tissut M., Douce R. Effects of rotenoids on isolated plant mitochondria. Plant Physiol. 1984;75:414–420. doi: 10.1104/pp.75.2.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco F., Silvestri A., Posca D., Pirrò S., Gherardini P.F., Castagnoli L., Mann M., Cesareni G. Deep proteomics of breast cancer cells reveals that metformin rewires signaling networks away from a pro-growth state. Cell Syst. 2016;2:159–171. doi: 10.1016/j.cels.2016.02.005. [DOI] [PubMed] [Google Scholar]
- Saxena H., Deshpande D.A., Tiegs B.C., Yan H., Battafarano R.J., Burrows W.M., Damera G., Panettieri R.A., Dubose T.D., Jr., An S.S., et al. The GPCR OGR1 (GPR68) mediates diverse signalling and contraction of airway smooth muscle in response to small reductions in extracellular pH. Br. J. Pharmacol. 2012;166:981–990. doi: 10.1111/j.1476-5381.2011.01807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;169:361–371. doi: 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA: A Cancer J. Clinicians. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- Son S.M., Park S.J., Lee H., Siddiqi F., Lee J.E., Menzies F.M., Rubinsztein D.C. Leucine signals to mTORC1 via its metabolite acetyl-coenzyme A. Cell Metab. 2019;29:192–201.e197. doi: 10.1016/j.cmet.2018.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa C.M., Biancur D.E., Wang X., Halbrook C.J., Sherman M.H., Zhang L., Kremer D., Hwang R.F., Witkiewicz A.K., Ying H., et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536:479–483. doi: 10.1038/nature19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinazzi M., Casarin A., Pertegato V., Salviati L., Angelini C. Assessment of mitochondrial respiratory chain enzymatic activities on tissues and cultured cells. Nat. Protoc. 2012;7:1235–1246. doi: 10.1038/nprot.2012.058. [DOI] [PubMed] [Google Scholar]
- Sugimoto M., Wong D.T., Hirayama A., Soga T., Tomita M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010;6:78–95. doi: 10.1007/s11306-009-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvarna K., Honda K., Muroi M., Kondoh Y., Watanabe N., Osada H. Identification of target protein for bio-active small molecule using photo-cross linked beads and MALDI-TOF mass spectrometry. Bio-protocol. 2020;10:e3517. doi: 10.21769/BioProtoc.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S., Sharma N., Gonzalez R., Pao P.W., Hofman F.M., Chen T.C., Louie S.G., Pirrung M.C., Schonthal A.H. Repositioning of Verrucosidin, a purported inhibitor of chaperone protein GRP78, as an inhibitor of mitochondrial electron transport chain complex I. PLoS One. 2013;8:e65695. doi: 10.1371/journal.pone.0065695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C., Clauser K.R., Öhlund D., Rickelt S., Huang Y., Gupta M., Mani D.R., Carr S.A., Tuveson D.A., Hynes R.O. Proteomic analyses of ECM during pancreatic ductal adenocarcinoma progression reveal different contributions by tumor and stromal cells. Proc. Natl. Acad. Sci. U S A. 2019;116:19609–19618. doi: 10.1073/pnas.1908626116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Ahrens D., Bhagat T.D., Nagrath D., Maitra A., Verma A. The role of stromal cancer-associated fibroblasts in pancreatic cancer. J. Hematol. Oncol. 2017;10:76. doi: 10.1186/s13045-017-0448-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.S., Jonker J.W., Kato Y., Kusuhara H., Schinkel A.H., Sugiyama Y. Involvement of organic cation transporter 1 in hepatic and intestinal distribution of metformin. J. Pharmacol. Exp. Ther. 2002;302:510–515. doi: 10.1124/jpet.102.034140. [DOI] [PubMed] [Google Scholar]
- Webb B.A., Chimenti M., Jacobson M.P., Barber D.L. Dysregulated pH: a perfect storm for cancer progression. Nat. Rev. Cancer. 2011;11:671–677. doi: 10.1038/nrc3110. [DOI] [PubMed] [Google Scholar]
- Yamori T. Panel of human cancer cell lines provides valuable database for drug discovery and bioinformatics. Cancer Chemother. Pharmacol. 2003;52:S74–S79. doi: 10.1007/s00280-003-0649-1. [DOI] [PubMed] [Google Scholar]
- Yoshida J., Abe H., Watanabe T., Kawada M. Intervenolin suppresses gastric cancer cell growth through the induction of TSP-1 secretion from fibroblast-like stromal cells. Oncol. Lett. 2018;16:6777–6785. doi: 10.3892/ol.2018.9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Li L., Jiang H., Li Q., Wang-Gillam A., Yu J., Head R., Liu J., Ruzinova M.B., Lim K.H. Tumor-stroma IL1β-IRAK4 feedforward circuitry drives tumor fibrosis, chemoresistance, and poor prognosis in pancreatic cancer. Cancer Res. 2018;78:1700–1712. doi: 10.1158/0008-5472.CAN-17-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Reljić B., Liang C., Kerouanton B., Francisco J.C., Peh J.H., Mary C., Jagannathan N.S., Olexiouk V., Tang C., et al. Mitochondrial peptide BRAWNIN is essential for vertebrate respiratory complex III assembly. Nat. Commun. 2020;11:1312. doi: 10.1038/s41467-020-14999-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Guo H., Guo X., Ge D., Shi Y., Lu X., Lu J., Chen J., Ding F., Zhang Q. Involvement of Akt/mTOR in the neurotoxicity of rotenone-induced Parkinson's disease models. Int. J. Environ. Res. Public Health. 2019;16:3811. doi: 10.3390/ijerph16203811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Kuang Y., Liang S., Wang L. Metformin inhibits cell proliferation in SKM-1 cells via AMPK-mediated cell cycle arrest. J. Pharmacol. Sci. 2019;141:146–152. doi: 10.1016/j.jphs.2019.10.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.