Abstract
Background
Anti‐SARS‐CoV‐2 S antibodies prevent viral replication. Critically ill COVID‐19 patients show viral material in plasma, associated with a dysregulated host response. If these antibodies influence survival and viral dissemination in ICU‐COVID patients is unknown.
Patients/Methods
We studied the impact of anti‐SARS‐CoV‐2 S antibodies levels on survival, viral RNA‐load in plasma, and N‐antigenaemia in 92 COVID‐19 patients over ICU admission.
Results
Frequency of N‐antigenaemia was >2.5‐fold higher in absence of antibodies. Antibodies correlated inversely with viral RNA‐load in plasma, representing a protective factor against mortality (adjusted HR [CI 95%], p): (S IgM [AUC ≥ 60]: 0.44 [0.22; 0.88], 0.020); (S IgG [AUC ≥ 237]: 0.31 [0.16; 0.61], <0.001). Viral RNA‐load in plasma and N‐antigenaemia predicted increased mortality: (N1‐viral load [≥2.156 copies/ml]: 2.25 [1.16; 4.36], 0.016); (N‐antigenaemia: 2.45 [1.27; 4.69], 0.007).
Conclusions
Low anti‐SARS‐CoV‐2 S antibody levels predict mortality in critical COVID‐19. Our findings support that these antibodies contribute to prevent systemic dissemination of SARS‐CoV‐2.
Keywords: antibodies, antigenaemia, COVID‐19, RNA, mortality
Introduction
Anti‐SARS‐CoV‐2 spike (S) antibodies bind to multiple domains in this viral protein [1]. Host antibodies directed at the receptor binding domain (RBD) mediate inhibition of viral attachment to cell surface receptors [2]. Therefore, anti‐S antibodies could play a role in reducing viral replication during an ongoing acute infection by interfering with virus entry into a cell. Interestingly, whether levels of host‐produced endogenous antibodies against the S protein could influence mortality risk in severe COVID‐19 has not been sufficiently studied to the present date.
Recent works from our group and others have demonstrated the importance of SARS‐CoV‐2 RNAemia [3, 4, 5] and SARS‐CoV‐2 antigenaemia as markers of severity in COVID‐19 [6]. The presence of viral RNA and proteins in plasma could represent a surrogated marker of poor viral control. On top of this, viral RNAemia and antigenaemia associate with dysregulated host responses to the infection caused by SARS‐CoV‐2 [3, 7]. It is necessary to elucidate whether anti‐SARS‐CoV‐2 S antibodies could influence the dissemination of viral genomic material or viral proteins at the systemic level.
In this work, we profiled the concentration of anti‐SARS‐CoV‐2 S IgM and IgG antibodies in the plasma of patients with COVID‐19 in the first 24 h following admission to the ICU. We evaluated the association between antibody levels with the concentration of viral RNA and the presence of SARS‐CoV‐2 nucleoprotein (N) protein in plasma. In parallel, we assessed the impact of antibody levels, viral RNA load and antigenaemia on the mortality risk of these patients.
Materials and methods
Design
We recruited 92 critically ill adult patients with a positive nasopharyngeal swab polymerase chain reaction (PCR) test for SARS‐CoV‐2 from 16 March to 15 April 2020, during the first pandemic wave, at Hospital Universitario Río Hortega, Hospital Clínico Universitario (Valladolid); Hospital Gregorio Marañón, Hospital Príncipe de Asturias (Madrid), Spain. We obtained EDTA plasma in the first 24 h following admission to the ICU. The study was approved by the “Comite de Etica de la Investigacion con Medicamentos del Area de Salud de Salamanca”, code PI 2020 03 452. Informed consent was obtained orally when clinically possible. In the remaining cases, the informed consent waiver was authorized by the Ethics committee.
Laboratory assays
We quantified SARS‐CoV‐2 RNA in plasma using the Bio‐Rad SARS‐CoV‐2 droplet digital PCR kit (CA, USA) (File S1). Our team developed a specific immunoassay to quantify anti‐SARS‐CoV‐2 S IgG and IgM antibodies in plasma (File S1). We evaluated the presence/absence of N antigen of SARS‐CoV‐2 in plasma using the Panbio COVID‐19 Ag Rapid Test Device from Abbott (Chicago, IL, USA).
Statistical analysis
Statistical analysis was performed using IBM SPSS Statistics 25.0 (SPSS INC, Armonk, NY, USA). The level of significance was fixed at 0.05 (two‐tailed). Differences between independent groups were assessed using the chi‐square test or Fisher's exact test for categorical variables and the Mann–Whitney U test for continuous variables.
The Kaplan–Meier method was used to calculate survival probabilities and the log‐rank test to compare groups. We also used Cox proportional‐hazards models to estimate the risk of dying, adjusted by the significant covariates at baseline resulting from the comparison between survivors and non‐survivors (full description in File S1). The outcome was 30‐day mortality following ICU admission.
Results
Characteristics of the patients (Table 1): Patients who died were older than those who survived. In addition, patients who died had increased frequency of arterial hypertension and type‐2 diabetes, higher glucose and creatinine levels, decreased concentrations of platelets and monocytes, and higher APACHE and SOFA scores. Patients who died received more often beta‐interferon than those who survived.
Table 1.
Baseline characteristics of patients admitted to the intensive care unit. Statistics: Continuous variables are represented as median (interquartile range) and categorical variables as absolute count (%). p‐Values were calculated by Mann–Whitney U test for continuous variables and chi‐square tests or Fisher's exact test for categorical variables. Significant differences are shown in bold
| Characteristics | All | Alive by day 30 | Dead by day 30 | p‐Value |
|---|---|---|---|---|
| Number | 92 | 54 | 38 | |
| Epidemiology | ||||
| Age (years) | 66 (50; 71.5) | 60 (47; 67) | 70 (66; 75) | <0.001 |
| Male | 60 (65.2%) | 37 (68.5%) | 23 (60.5%) | 0.428 |
| Alcoholism | 1 (1.1%) | 0 (0%) | 1 (2.6%) | 0.413 |
| Smoking | 5 (5.4%) | 2 (3.7%) | 3 (7.9%) | 0.645 |
| Comorbidities | ||||
| Cardiac disease | 7 (7.6%) | 2 (3.7%) | 5 (13.2%) | 0.121 |
| Chronic vascular disease | 5 (5.4%) | 4 (7.4%) | 1 (2.6%) | 0.401 |
| COPD | 3 (3.3%) | 2 (3.7%) | 1 (2.6%) | 0.999 |
| Asthma | 2 (2.2%) | 1 (1.9%) | 1 (2.6%) | 0.999 |
| Obesity | 22 (23.9%) | 16 (29.6%) | 6 (15.8%) | 0.125 |
| Hypertension | 39 (42.4%) | 17 (31.5%) | 22 (57.9%) | 0.012 |
| Dyslipidemia | 30 (32.6%) | 17 (31.5%) | 13 (34.2%) | 0.783 |
| Chronic renal disease | 3 (3.3%) | 1 (1.9%) | 2 (5.3%) | 0.567 |
| Chronic hepatic disease | 3 (3.3%) | 1 (1.9%) | 2 (5.3%) | 0.567 |
| Type 1 diabetes | 3 (3.3%) | 3 (5.6%) | 0 (0%) | 0.265 |
| Type 2 diabetes | 19 (20.7%) | 7 (13%) | 12 (31.6%) | 0.031 |
| Measurements at ICU admission | ||||
| Temperature (°C) | 37 (36.5; 37.6) | 37 (36.6; 37.7) | 36.8 (36; 37.2) | 0.109 |
| Systolic pressure (mm Hg) | 120 (108.5; 130) | 120 (109; 132) | 115.6 (108; 125) | 0.274 |
| Oxygen saturation (%) | 92 (88; 94) | 92 (88; 94) | 91 (88; 95.5) | 0.851 |
| Bilateral pulmonary infiltrate | 86 (93.5%) | 51 (94.4%) | 35 (92.1%) | 0.688 |
| Glucose (mg/dl) | 160.5 (124.5; 208) | 152 (114; 173) | 174 (126; 267) | 0.027 |
| Creatinine (mg/dl) | 0.9 (0.7; 1.3) | 0.8 (0.7; 1) | 1.1 (0.7; 1.7) | 0.005 |
| Na (mEq/L) | 138.5 (135; 142) | 139 (136; 142) | 138 (134; 142) | 0.328 |
| K (mEq/L) | 3.9 (3.5; 4.4) | 3.9 (3.5; 4.4) | 4 (3.5; 4.4) | 0.763 |
| Platelets (cell x 103/ μl) | 206.5 (163.5; 289.5) | 262.5 (175; 309) | 179.5 (154; 216) | <0.001 |
| INR | 1.2 (1.1; 1.3) | 1.2 (1.1; 1.3) | 1.3 (1.1; 1.4) | 0.166 |
| D Dimer (ng/ml) | 6182 (1733; 51,407) | 5403 (1940; 50,079) | 6182 (976; 59,829) | 0.768 |
| LDH (UI/L) | 489 (358; 607.5) | 502 (330; 606) | 470.4 (362; 609) | 0.962 |
| GPT (UI/L) | 46.5 (25.5; 69.5) | 51.5 (29; 74) | 37.5 (23.4; 69) | 0.175 |
| Ferritin (ng/ml) | 885 (110; 1569) | 10295 (296; 1518) | 730 (75; 1621) | 0.134 |
| C‐reactive protein (mg/dl) | 97.6 (28; 182.8) | 74.5 (23.7; 144) | 117 (44.3; 227) | 0.211 |
| Hematocrit (%) | 37.7 (34.6; 40.7) | 38.4 (35.7; 41) | 35.8 (32.9; 39.6) | 0.116 |
| WBC (cells/mm3) | 9145 (6705; 13,095) | 9450 (7400; 13,720) | 8450 (6200; 11,900) | 0.216 |
| Lymphocytes (cells/mm3) | 525 (335; 800) | 595 (400; 830) | 465 (290; 670) | 0.073 |
| Neutrophils (cells/mm3) | 8310 (5865; 11,190) | 8395 (6630; 11,480) | 7950 (5300; 10,400) | 0.526 |
| Monocytes (cells/mm3) | 300 (170; 496.9) | 350 (200; 600) | 190 (120; 400) | <0.001 |
| Severity score | ||||
| APACHE | 15 (10.5; 19) | 13 (8; 16) | 18 (14; 23) | <0.001 |
| SOFA | 6 (4; 8) | 5.5 (4; 7) | 7 (5; 9) | 0.013 |
| Treatment during hospitalization | ||||
| Invasive ventilation | 88 (95.7%) | 50 (92.6%) | 38 (100%) | 0.140 |
| Non‐invasive ventilation | 32 (34.8%) | 19 (35.2%) | 13 (34.2%) | 0.923 |
| Hydroxychloroquine | 91 (98.9%) | 54 (100%) | 37 (97.4%) | 0.413 |
| Corticoids | 79 (85.9%) | 49 (90.7%) | 30 (78.9%) | 0.111 |
| Azithromycin | 76 (82.6%) | 44 (81.5%) | 32 (84.2%) | 0.732 |
| Remdesivir | 9 (9.8%) | 7 (13%) | 2 (5.3%) | 0.298 |
| Tocilizumab | 29 (31.5%) | 20 (37%) | 9 (23.7%) | 0.175 |
| Lopinavir/ritonavir | 89 (96.7%) | 52 (96.3%) | 37 (97.4%) | 0.999 |
| Beta Interferon | 51 (56%) | 25 (47.2%) | 26 (68.4%) | 0.044 |
| Time course and outcome | ||||
| Hospital stay (days) | 23.5 (17.5; 38) | 36 (21; 58) | 18 (14; 23) | <0.001 |
| ARDS | 88 (95.7%) | 50 (92.6%) | 38 (100%) | 0.140 |
| Sepsis | 51 (55.4%) | 29 (53.7%) | 22 (57.9%) | 0.690 |
| Septic shock | 43 (46.7%) | 24 (44.4%) | 19 (50%) | 0.599 |
Abbreviations: APACHE II, acute physiology and chronic health disease classification system II; ARDS, acute respiratory distress syndrome; COPD, chronic obstructive pulmonary disease; GPT, glutamic‐pyruvate transaminase; INR, international normalized ratio; LDH, lactic acid dehydrogenase; p‐value, level of significance; SOFA, sequential organ failure assessment; WBC, white blood cell.
Levels of anti‐SARS‐CoV‐2 S antibodies: Ten patients (of the total cohort) had no detectable levels in the plasma of anti‐SARS‐CoV‐2 S IgG, and 13 had no detectable levels of anti‐SARS‐CoV‐2 S IgM. Patients who died showed more often absence/lower levels of anti‐SARS‐CoV‐2 S IgM and IgG than those who survived by day 30 following ICU admission (Figure 1, panels 1 and 2). Concentrations of both anti‐SARS‐CoV‐2 S IgM and IgG showed a direct correlation with the number of days transcurred since the onset of COVID‐19 symptoms: IgM (0.40, 0.001); IgG (0.45, < 0.001) (Spearman correlation coefficient, p). The median time from the onset of the symptoms was 5 days for samples with no anti‐SARS‐CoV‐2 S IgM and 10 days for those with IgM (p = 0.008). In turn, the median time from the onset of the symptoms was 8 days for samples with no anti‐SARS‐CoV‐2 S IgG and 10 days for those with anti‐SARS‐CoV‐2 S IgG (p = 0.147).
Fig. 1.
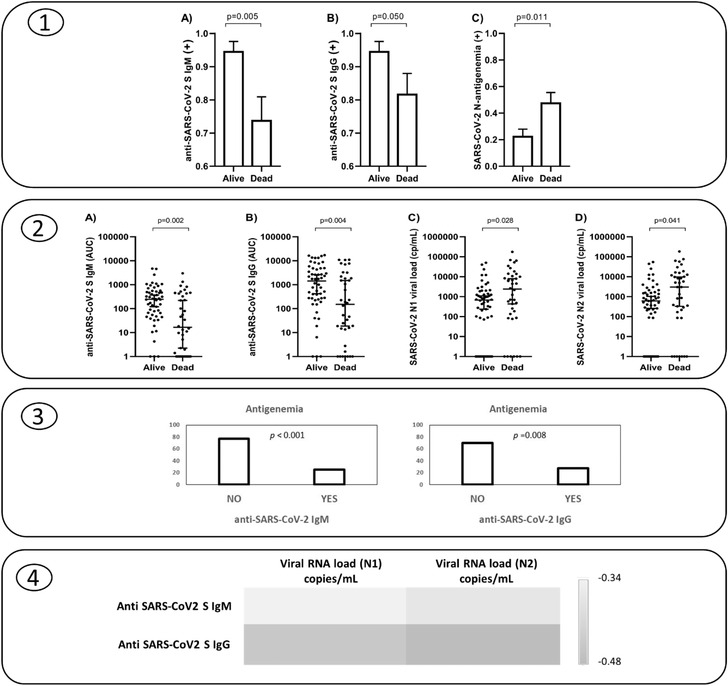
Panel 1: Frequencies of patients with positive SARS‐CoV‐2 S antibodies (IgM, IgG) and antigenaemia by survival status 30 days following ICU admission. Panel 2: Levels of SARS‐CoV‐2 S antibodies (IgM, IgG) and viral RNA load (N1, N2) in plasma following ICU admission by survival status at day 30. Panel 3: Frequency of N‐antigenaemia in those patients with absence or presence of anti SARS‐CoV‐2 S IgM or anti SARS‐CoV‐2 S IgG antibodies. Panel 4: Heat map showing the correlation coefficients between anti SARS‐CoV‐2 S antibodies and viral RNA load in plasma.
Prevalence of antigenaemia and viral RNA load: In contrast to that observed for antibodies, non‐survivors showed more frequently the presence of antigenaemia (Fig. 1, panel 1) along with higher viral RNA loads in plasma (Fig. 1, panel 2).
Correlate between antibody responses, antigenaemia and viral RNA load in plasma: Frequency of N‐antigenaemia was >2.5‐fold higher in patients with no anti‐SARS‐CoV‐2 S antibodies than in those patients who presented detectable antibodies (anti‐SARS‐CoV‐2 S IgM: 77%/25%; IgG: 70%/28%) (Fig. 1, panel 3). In turn, levels of anti‐S antibodies correlated inversely with viral RNA load in plasma: anti‐SARS‐CoV‐2 S IgM/N1 (copies/ml) (r = −0.34, p < 0.001); anti‐SARS‐CoV‐2 S IgM/N2 (copies/ml) (r = −0.37, p < 0.001) (Figure 1, panel 4); anti‐SARS‐CoV‐2 S IgG/N1 (copies/ml) (r = −0.45, p < 0.001); anti‐SARS‐CoV‐2 S IgG/N2 (copies/ml) (r = −0.48, p < 0.001) (Fig. 1, panel 4).
Impact of antibodies, antigenaemia and viral RNA load on 30‐day mortality: Multivariate analysis demonstrated that the absence/low levels of anti‐SARS‐CoV‐2 S IgM and IgG was an independent risk factor for mortality at day 30 following ICU admission (Fig. 2). In turn, the multivariate analysis evidenced that the presence of N‐antigenaemia and high viral RNA loads predicted also increased mortality (Fig. 2). When levels of anti‐SARS‐CoV‐2 S antibodies, viral RNA load and antigenaemia were simultaneously introduced in the multivariate analysis, anti‐S IgG, anti‐S IgM and viral RNA concentrations in plasma still predicted mortality (File S2). Kaplan–Meier analysis evidenced that low antibody levels, high viral RNA loads in plasma, or the presence of N‐antigenaemia translated into earlier mortality (Fig. 2).
Fig. 2.
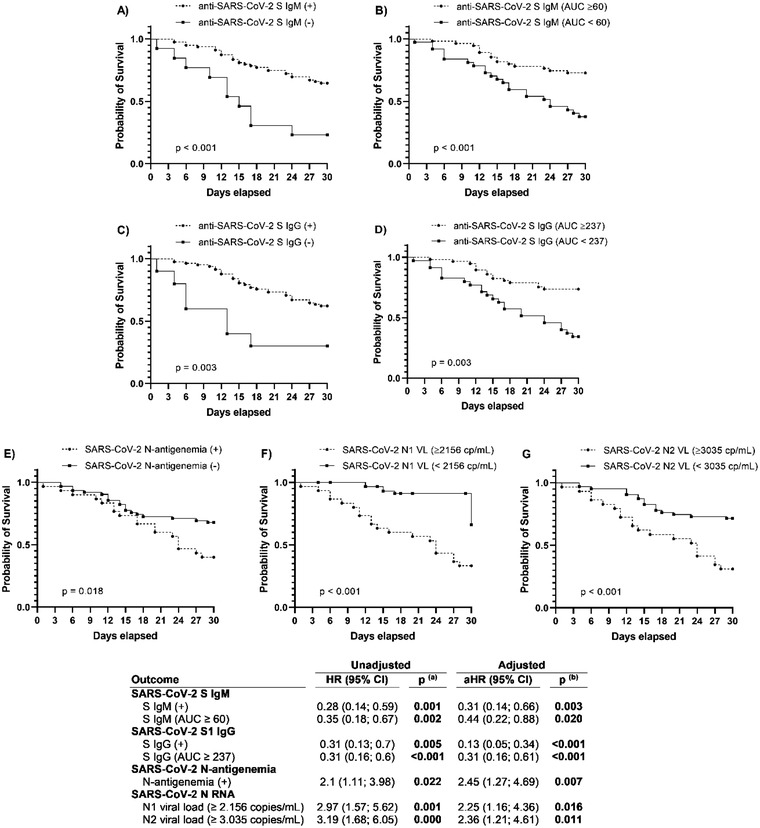
Upper panel: Kaplan–Meier curves to represent survival by day 30 following ICU admission depending on the presence or absence of SARS‐CoV‐2 S antibodies (IgM, IgG) (a and c), the presence or absence of antigenaemia (e), the presence of higher/lower levels of SARS‐CoV‐2 S antibodies (IgM, IgG) (b and d) or viral RNA load in plasma (N1,N2) (f and g). Lower panel: Cox regression analysis to assess risk of 30‐day mortality following ICU admission. Statistics: p‐values were calculated by univariate (a) and multivariate (b) analysis adjusted by the most relevant covariates (see Statistical Analysis section). Significant differences are shown in bold
Abbreviations: 95% CI, 95% confidence interval; aHR, adjusted HR; HR, hazard ratio; p‐value, level of significance.
Discussion
Our work evidenced an opposed impact of anti‐SARS‐CoV‐2 S antibodies and viral RNA load or antigenaemia in the mortality risk of critically ill COVID‐19 patients. While levels of both IgG and IgM anti‐SARS‐CoV‐2 S antibodies were positively associated with increased survival, the presence of viral components in plasma (RNA or N‐antigen) predicted a higher risk of death.
Our study is the largest one to the current date in profiling anti‐SARS‐CoV‐2 S antibodies in critically ill COVID‐19 patients, suggesting a protective role of endogenous anti‐S antibodies against mortality in these patients. Fourati et al., in a small study with 25 ICU patients, found that patients who were still alive at day 28 displayed significantly higher anti‐S1 IgA or IgG titers upon admission than those who had died [8]. Asif et al. reported that plasma concentrations of anti‐S1 IgA, IgG and IgM antibodies tended to be higher in COVID‐19 patients who survived, at both early and late timepoints following ICU admission [9]. Nonetheless, this work involved only 19 patients.
Previous studies have shown that critical COVID‐19 patients develop higher titers of SARS‐CoV‐2 antibodies than those with milder disease, suggesting that antibody response alone is insufficient to avoid severe disease [10]. Our results support nonetheless that critical COVID‐19 patients would need to mount a robust anti‐S antibody response to survive.
In turn, the connection between viral RNAemia and mortality in COVID‐19 has been demonstrated in a metanalysis from Tang et al., involving 2181 patients of different severity [11]. In ICU‐COVID‐19 patients, Gutmann et al. have recently reported that SARS‐CoV‐2 RNAemia is associated with higher 28‐day mortality [12]. Our work identifies the levels of viral RNA in plasma predicting fatal outcomes. As far as we know, this is the first study demonstrating that the presence of antigenaemia is also an independent predictor of mortality in critically ill COVID‐19 patients.
The inverse association found between levels of anti‐SARS‐CoV‐2 S antibodies with viral RNA load/antigenaemia suggests that these antibodies could be mediating their beneficial effects (at least in part) by contributing to the control of viral replication and preventing systemic dissemination of the virus or viral components. In this regard, whether the presence of viral RNA/antigens in plasma of severe COVID‐19 patients corresponds to the presence of infective viral particles is currently unknown, but spreading of viral material at the systemic level could by itself stimulate innate immunity through TLR or pattern recognition receptors leading to unabated pro‐inflammatory responses, which could contribute to the pathogenesis of multi‐organ failure in severe COVID‐19 [13]. In this regard, it has been described that viral RNA load in plasma and antigenaemia are associated with increased levels of cytokines or C‐reactive protein in COVID‐19 [3, 7]. The potential role of anti‐SARS‐CoV‐2 S antibodies in the control of viral replication/spreading is also suggested by the results from Röltgen et al., who found that the appearance of these antibodies correlated with a decrease in viral RNAemia along the course of COVID‐19 [14]. In turn, Li et al. found higher anti‐S IgG levels in those recovered patients who were SARS CoV‐2 RNA negative in respiratory samples than those who were RNA positive [15]. Lucas et al. reported that a delayed seroconversion correlates with impaired viral control and mortality in COVID‐19 [16]. In our cohort, patients with the lowest antibody titers were those more promptly admitted to the ICU since the onset of the symptoms, which suggests their inability to timely mount effective humoral responses. Our patients with no anti‐SARS‐CoV‐2 S antibodies at ICU admission showed more often antigenaemia, further supporting a protective role of these antibodies in preventing the dissemination of the virus or viral components. In a small study with 39 (critically and non‐critically ill) patients, Ogata et al. observed that seroconversion was followed by antigen clearance in plasma [6]. Also, recent findings from Hingrat et al. support the implication of anti‐SARS‐CoV‐2 antibodies in the clearance of antigenaemia [17].
Our findings pose several potential translational implications. (1) Profiling anti‐SARS‐CoV‐2 S antibodies following ICU admission could contribute to personalizing treatment with exogenous antibodies targeting the S protein of the virus and perhaps with convalescent serum [18, 19]. Results from the RECOVERY trial support this notion since only the patients with an absence of anti‐S antibodies seem to receive clinical benefit from exogenous monoclonal antibodies against the SARS‐CoV‐2 S‐protein [18]. (2) Quantification of viral RNA load in plasma could be helpful to identify those COVID‐19 patients at risk of suffering a fatal outcome. (3) Profiling antigenaemia using point of care tests could also contribute to early identify those patients at risk of clinical deterioration.
A limitation of our work was that we profiled antibodies, viral RNA load and antigenaemia on samples preserved from the first pandemic wave. The findings reported here should be confirmed with prospective studies with patients from subsequent waves. Another limitation is that we did not evaluate the neutralization activity of anti‐SARS‐CoV‐2 S antibodies. Further studies should clarify if the quantitative results provided here correlate with the functional ability of these antibodies to block viral replication.
Conclusion
Low anti‐SARS‐CoV‐2 S antibody levels predict mortality in critical COVID‐19. Our findings support that these antibodies contribute to prevent systemic dissemination of SARS‐CoV‐2.
Conflict of interest
The authors declare that they have no conflict of interest.
Funding information
This work was supported by awards from the Canadian Institutes of Health Research (CIHR OV2 – 170357), Research Nova Scotia, Atlantic Genome/Genome Canada, Li‐Ka Shing Foundation, Dalhousie Medical Research Foundation (David J. Kelvin), David J. Kelvin is a recipient of the Canada Research Chair in Translational Vaccinology and Inflammation, and the “Subvenciones de concesión directa para proyectos y programas de investigación del virus SARS‐CoV2, causante del COVID‐19”, FONDO ‐ COVID19, Instituto de Salud Carlos III (COV20/00110, CIBERES, 06/06/0028), (Antoni Torres) and finally by the “Convocatoria extraordinaria y urgente de la Gerencia Regional de Salud de Castilla y León, para la financiación de proyectos de investigación en enfermedad COVID‐19” (GRS COVID 53/A/20) (CA). Ana P. Tedim was funded by the Sara Borrell Research Grant CD018/0123 funded by Instituto de Salud Carlos III and co‐financed by the European Development Regional Fund (A Way to Achieve Europe programme). The funding sources did not play any role in the design of the study and collection, analysis, interpretation of data or writing the manuscript.
Author contributions
Salvador Resino, Raquel Almansa, Antoni Torres, Isidoro Martínez, Jesús F. Bermejo‐Martin, Ricard Ferrer, Ferrán Barbé and David J. Kelvin designed the study. Salvador Resino, Jesús F. Bermejo‐Martin, Isidoro Martínez and Amanda de la Fuente analysed the data and drafted the manuscript. Elena Bustamante, Luis Tamayo, César Aldecoa, José Manuel Gómez, Gloria Renedo, Jose Ángel Berezo, Jamil Antonio Cedeño, Nuria Mamolar, Pablo García Olivares, Rubén Herrán‐Monge, Ramón Cicuendez, Pedro Enríquez, Juan Bustamante‐Munguira, Milagros González‐Rivera, Carolina Puertas, Felipe Pérez‐García, Jesús Rico‐Feijoo, Silvia Martín, Anna Motos, Laia Fernandez‐Barat and Wysali Trapiello contributed with patient recruitment and data acquisition. María Martin‐Vicente, María José Muñoz‐Gómez, Isidoro Martínez, Vicente Más and Mónica Vázquez developed the antibody assay and profiled antibodies levels in plasma. Ana P. Tedim, Cristina Doncel, Alicia Ortega, Noelia Jorge, Jose María Eiros and Marta Dominguez‐Gil set up the viral RNA quantification assays and profiled viral RNA load in plasma. All the authors critically reviewed the article and provided final approval of the version submitted for publication. All the authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Supporting information
Supporting information
Supporting information
Acknowledgements
We would like to thank the IBSAL and CIBER for administrative support to perform this study.
Martin‐Vicente M, Almansa R, Martínez I, Tedim AP, Bustamante E, Tamayo L, et al. Low anti‐SARS‐CoV‐2 S antibody levels predict increased mortality and dissemination of viral components in the blood of critical COVID‐19 patients. J Intern Med. 2022;291:232–240.
Contributor Information
Jesús F. Bermejo‐Martin, Email: jfbermejo@saludcastillayleon.es.
Antoni Torres, Email: ATORRES@clinic.cat.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Heffron AS, McIlwain SJ, Amjadi MF, Baker DA, Khullar S, Armbrust T, et al. The landscape of antibody binding in SARS‐CoV‐2 infection. PLoS Biol. 2021;19:e3001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Deb P, Molla MdMA, Rahman KMS‐U. An update to monoclonal antibody as therapeutic option against COVID‐19. Biosaf Health. 2021;3:87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bermejo‐Martin JF, González‐Rivera M, Almansa R, Micheloud D, Tedim AP, Domínguez‐Gil M, et al. Viral RNA load in plasma is associated with critical illness and a dysregulated host response in COVID‐19. Crit Care. 2020;24:691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Veyer D, Kernéis S, Poulet G, Wack M, Robillard N, Taly V, et al. Highly sensitive quantification of plasma SARS‐CoV‐2 RNA shelds light on its potential clinical value. Clin Infect Dis. 2021;73:e2890–97. 10.1093/cid/ciaa1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hagman K, Hedenstierna M, Gille‐Johnson P, Hammas B, Grabbe M, Dillner J, et al. SARS‐CoV‐2 RNA in serum as predictor of severe outcome in COVID‐19: a retrospective cohort study. Clin Infect Dis. 2021;73:e2995–e3001. 10.1093/cid/ciaa1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ogata AF, Maley AM, Wu C, Gilboa T, Norman M, Lazarovits R, et al. Ultra‐sensitive serial profiling of SARS‐CoV‐2 antigens and antibodies in plasma to understand disease progression in COVID‐19 patients with severe disease. Clin Chem. 2020;66(12):1562–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Perna F, Bruzzaniti S, Piemonte E, Maddaloni V, Atripaldi L, Sale S, et al. Serum levels of SARS‐CoV‐2 nucleocapsid antigen associate with inflammatory status and disease severity in COVID‐19 patients. Clinical Immunol. 2021;226:108720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fourati S, Hue S, Pawlotsky J‐M, Mekontso‐Dessap A, de Prost N. SARS‐CoV‐2 viral loads and serum IgA/IgG immune responses in critically ill COVID‐19 patients. Intensive Care Med. 2020;46:1781–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Asif S, Frithiof R, Lipcsey M, Kristensen B, Alving K, Hultström M. Weak anti‐SARS‐CoV‐2 antibody response is associated with mortality in a Swedish cohort of COVID‐19 patients in critical care. Critical Care. 2020;24:639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lafon E, Diem G, Witting C, Zaderer V, Bellmann‐Weiler RM, Reindl M, et al. Potent SARS‐CoV‐2‐specific T cell immunity and low anaphylatoxin levels correlate with mild disease progression in COVID‐19 patients. Front Immunol. 2021;12:684014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tang K, Wu L, Luo Y, Gong B. Quantitative assessment of SARS‐CoV‐2 RNAemia and outcome in patients with coronavirus disease 2019. J Med Virol. 2021;93:3165–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gutmann C, Takov K, Burnap SA, Singh B, Ali H, Theofilatos K, et al. SARS‐CoV‐2 RNAemia and proteomic trajectories inform prognostication in COVID‐19 patients admitted to intensive care. Nat Commun. 2021;12:3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Birra D, Benucci M, Landolfi L, Merchionda A, Loi G, Amato P, et al. COVID 19: a clue from innate immunity. Immunol Res. 2020;68:161–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Röltgen K, Powell AE, Wirz OF, Stevens BA, Hogan CA, Najeeb J, et al. Defining the features and duration of antibody responses to SARS‐CoV‐2 infection associated with disease severity and outcome. Sci Immunol. 2020;5:eabe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li K, Huang B, Wu M, Zhong A, Li L, Cai Y, et al. Dynamic changes in anti‐SARS‐CoV‐2 antibodies during SARS‐CoV‐2 infection and recovery from COVID‐19. Nat Commun. 2020;11:6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lucas C, Klein J, Sundaram ME, Liu F, Wong P, Silva J, et al. Delayed production of neutralizing antibodies correlates with fatal COVID‐19. Nat Med. 2021;27:1178–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hingrat QL, Visseaux B, Laouenan C, Tubiana S, Bouadma L, Yazdanpanah Y, et al. Detection of SARS‐CoV‐2 N‐antigen in blood during acute COVID‐19 provides a sensitive new marker and new testing alternatives. Clin Microbiol Infect. 2020;27:789.e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. RECOVERY group , Horby PW, Mafham M, Peto L, Campbell L, Pessoa‐Amorim G, et al. Casirivimab and imdevimab in patients admitted to hospital with COVID‐19 (RECOVERY): a randomised, controlled, open‐label, platform trial. medRxiv. 2021. 10.1101/2021.06.15.21258542 [DOI] [Google Scholar]
- 19. Ali S, Luxmi S, Anjum F, Muhaymin SM, Uddin SM, Ali A, et al. Hyperimmune anti‐COVID‐19 IVIG (C‐IVIG) therapy for passive immunization of severe and critically ill COVID‐19 patients: a structured summary of a study protocol for a randomised controlled trial. Trials 2020;21:905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Supporting information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


