Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is the cause of the ongoing coronavirus disease 2019 (COVID‐19) pandemic. The S protein is the key viral protein for associating with ACE2, the receptor for SARS‐CoV‐2. There are many kinds of posttranslational modifications in S protein. However, the detailed mechanism of palmitoylation of SARS‐CoV‐2 S remains to be elucidated. In our current study, we characterized the palmitoylation of SARS‐CoV‐2 S. Both the C15 and cytoplasmic tail of SARS‐CoV‐2 S were palmitoylated. Fatty acid synthase inhibitor C75 and zinc finger DHHC domain‐containing palmitoyltransferase (ZDHHC) inhibitor 2‐BP reduced the palmitoylation of S. Interestingly, palmitoylation of SARS‐CoV‐2 S was not required for plasma membrane targeting of S but was critical for S‐mediated syncytia formation and SARS‐CoV‐2 pseudovirus particle entry. Overexpression of ZDHHC2, ZDHHC3, ZDHHC4, ZDHHC5, ZDHHC8, ZDHHC9, ZDHHC11, ZDHHC14, ZDHHC16, ZDHHC19, and ZDHHC20 promoted the palmitoylation of S. Furthermore, those ZDHHCs were identified to associate with SARS‐CoV‐2 S. Our study not only reveals the mechanism of S palmitoylation but also will shed important light into the role of S palmitoylation in syncytia formation and virus entry.
Keywords: palmitoylation, SARS‐CoV‐2, spike, syncytia, ZDHHC
Highlights
SARS‐CoV‐2 S protein is palmitoylatedin both N terminal and C terminal cysteines.
Palmitoylation of S is critical for S mediated syncytia formation.
Palmitoylation of S promotes SARS‐CoV‐2pp entry.
Multiple ZDHHCs promoted palmitoylation of SARS‐CoV‐2 S.
1. INTRODUCTION
Coronavirus disease 2019 (COVID‐19) caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is a global pandemic. The first step of SARS‐CoV‐2 infection is virus entry, which is mediated by the association between virus‐encoded spike (S) protein and host receptor angiotensin‐converting enzyme 2 (ACE2). 1 To fulfill its function, SARS‐CoV‐2 S undergoes complex posttranslational modifications, including glycosylation, 2 , 3 , 4 phosphorylation, 5 and ubiquitination. 6 However, the mechanism of palmitoylation in S is not fully understood.
The addition of palmitic acid (PA), a 16‐carbon saturated fatty acid, to the thiol group of a cysteine residue of the substrate is so‐called S‐palmitoylation, which is generalized as “palmitoylation.” Palmitoylation is modified by the zinc finger Asp‐His‐His‐Cys (DHHC) domain‐containing (ZDHHC) palmitoyl acyltransferase (PAT) family proteins. 7 As an important form of lipid posttranslational modification, palmitoylation increases protein hydrophobicity, and affects the function of proteins by regulating their stability, transport, and localization. 7
S from SARS‐CoV, transmissible gastroenteritis coronavirus (TGEV) and mouse hepatitis virus (MHV) have been proved to be palmitoylated. 8 , 9 , 10 A previous proteomic study suggests that SARS‐CoV‐2 S interacts with ZDHHC5. 11 In the current study, we find that SARS‐CoV‐2 S is palmitoylated in both N terminal and C terminal cysteines. Palmitoylation of S is not essential for the plasma membrane localization of S but is critical for S‐mediated formation of syncytia and virus entry. We identified that multiple ZDHHCs associated with SARS‐CoV‐2 S and mediated its palmitoylation. Taken together, our findings identified the key enzymes for the palmitoylation of SARS‐CoV‐2 S and revealed that palmitoylation of S contributes to syncytia formation and virus entry.
2. METHODS
2.1. Plasmids
Plasmid transfection into 293T cells was performed using polyethylenimine (PEI). GFP‐tagged SARS‐CoV‐2 S was from Sino Biological. Constructs expressing S‐C1, S‐C9, and S‐C10 were generated by GENEWIZ. The plasmids expressing FLAG‐tagged ZDHHC1 (MG5A3618‐CF), ZDHHC2 (MG5A1611‐CF), ZDHHC3 (MG5A0430‐CF), ZDHHC4 (HG16190‐NF), ZDHHC5 (MG5A2307‐CF), ZDHHC6 (MG53691‐NF), ZDHHC7 (HG19166‐CF), ZDHHC8 (MG5A0792‐CF), ZDHHC9 (HG21413‐NF), ZDHHC11 (MG55410‐CF), ZDHHC12 (MG5A7057‐CF), ZDHHC13 (MG5A1809‐CF), ZDHHC14 (MG54007‐CF), ZDHHC15 (MG5A4866‐CF), ZDHHC16 (MG5A5172‐CF), ZDHHC17 (MG5A0647‐CF), ZDHHC18 (MG5A2533‐CF), ZDHHC19 (MG54666‐CF), ZDHHC20 (MG5A6992‐CF), ZDHHC21 (MG5A1530‐CF), ZDHHC22 (MG54364‐CF), ZDHHC23 (MG5A6405‐CF), and ZDHHC24 (MG5A6597‐CF) were from Sino Biological. All ZDHHC plasmids contained a cytomegalovirus (CMV) promoter and a kanamycin resistance cassette.
2.2. Antibodies
The mouse antibodies used in this study have been described previously and included anti‐FLAG (Sigma‐Aldrich, A2220), anti‐GFP (XHY, XHY038L), IgG control (MBL, M075‐3), and anti‐actin (Sigma‐Aldrich, A1978) antibodies. The rabbit antibodies used in this study are anti‐GFP (XHY, XHY026L), anti‐HIV‐1 p24 (Sino Biological, 11695‐R002) antibodies. The secondary antibodies included: horseradish peroxidase (HRP)‐conjugated electrochemiluminescence (ECL) goat anti‐rabbit IgG (Sigma‐Aldrich, A6154), HRP‐conjugated ECL goat anti‐mouse IgG (Sigma‐Aldrich, A4416).
2.3. Co‐Immunoprecipitation (Co‐IP)
Cells were washed three times with ice‐cold phosphate‐buffered saline (PBS) (Solarbio, P1010) and lysed in radioimmunoprecipitation assay lysis buffer (50 mM Tris [pH 7.4], 150 mM NaCl, and 1% Triton X‐100, 1% sodium deoxycholate, 1% sodium dodecyl sulfate [SDS], without inhibitors) (Beyotime, P0013K) supplemented with a protease inhibitor cocktail for mammalian cell and tissue extracts (100×) (Beyotime, P1010). Both the cell lysate and the antibody need to be bound to protein A + G agarose beads (Beyotime, P2055). The beads need to be equilibrated (washed three times with PBS stored at 4°C) before use to remove the preservatives in the bead storage solution. The cell lysate was incubated with the equilibrated protein A + G agarose beads slurry at 4°C for 30 min and centrifuged at 12 000 rpm at 4°C for 15 min. Then the cell lysate was incubated with protein A + G agarose beads bound to the antibody for 1.5 h and washed three times with PBS at 4°C followed by centrifugation at 500 g for 5 min. The proteins bound to the beads were boiled in two‐times loading buffer (Beyotime, P0015B) for 5 min and then subjected to sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) (10% separation gel in the lower layer, 5% concentrated gel in the upper layer).
2.4. Prediction of palmitoylation sites
Palmitoylation sites of S from SARS‐CoV‐2 were predicted in CSS‐palm 4.0 (http://csspalm.biocuckoo.org/online.php).
2.5. The acyl‐biotin exchange (ABE) assay
Detection of S palmitoylation by an ABE assay was performed based on a previous study. 12 The cell lysate buffer contained 10% glycerol, 50 mM Tris‐HCI buffer (pH 7.5) (Solarbio, T1140), IGEPAL CA‐630 (Sigma‐Aldrich, I3021), and 150 mM NaCl, protease inhibitor cocktail for mammalian cell and tissue extracts (Beyotime, P1010) and 0.1 mM phenylmethylsulfonyl fluoride (Beyotime, ST506). In general, ABE assay contains three steps: blocking the unmodified cysteine thiol groups by N‐ethylmaleimide (NEM) (Aladdin, E100553), cleaving and unmasking the palmitoylated cysteine's thiol group by hydroxylamine (HAM) (MACKLIN, H828371), and labeling the palmitoylated cysteine by a thiol‐reactive biotinylation reagent, 1‐biotinamido‐4‐[4ʹ‐(maleimidomethyl)cyclohexanecarboxamido]butane (biotin‐BMCC) (ProteoChem, B2112). HRP‐conjugated streptavidin was from GenScript (M00091).
2.6. Plasma membrane isolation
The plasma membrane was isolated using a plasma membrane protein isolation and cell fractionation kit (Bestbio, BB‐31161‐2), and the assay was performed according to the manufacturer's protocol.
2.7. Western blot analysis
The protein samples were separated by SDS‐PAGE, transferred to a polyvinylidene difluoride (PVDF) membrane. After transferring the membrane, we sealed it with a solution of 1×TBST (pH 8.0, CWBIO, CW0043S) and 5% skimmed milk powder, and then probed with appropriate primary and secondary antibodies. In the ABE experiment, 3% BSA (Beyotime, ST023) was used for blocking instead of 5% skimmed milk powder.
2.8. Syncytia formation assay
The 293T cells in 100 mm plates were transfected using PEI with 10 μg plasmids of S‐WT, S‐C1, S‐C9, S‐C10, and human ACE2, respectively. At 24 h posttransfection, cells were detached with trypsin (0.25%). Donor cells and recipient cells were evenly mixed in a ratio of 1:1 and seeded in 100 mm plates. After 12 h post culture, cells were incubated with a fresh medium at pH8.0. After 4 h of incubation, and images of syncytia formation were captured using a ×10 objective lens with OLYMPUS IX73P2F.
2.9. Reporter assay
SARS‐CoV‐2 pseudovirus particle (SARS‐CoV‐2pp) were produced in HEK293T cells by transfection with expression plasmids encoding HIV Gag/Pol (pLP1), HIV Rev (pLP2), pLenti6 encoding luciferase, and SARS‐CoV‐2 S. SARS‐CoV‐2pp‐infected 293T cells were assessed by monitoring the firefly luciferase activity (Promega, E4530). Cell viability was determined by measuring the cellular ATP level using a CellTiter‐Lumi™ Luminescent Cell Viability Assay Kit (Beyotime, C0065S) according to the manufacturer's protocol. The normalized luciferase activity was determined by dividing the luciferase activity by the ATP level. Statistically significant differences were assessed using a paired Student t‐test in GraphPad Prism, version 8 (GraphPad Software, Inc.). The data represent the means from at least three independent experiments ± standard deviations (SD). ****Means p < 0.0001, ns means no significance.
3. RESULTS
3.1. SARS‐CoV‐2 S protein is palmitoylated at multiple sites
Based on online software CSS‐palm 4.0 (http://csspalm.biocuckoo.org/online.php) prediction, the cysteines at sites C15, C1235, C1236, C1240, C1243, C1247, C1248, C1250, C1253, and C1254 in SARS‐CoV‐2 S (GenBank: QLJ57699.1) were found to be potential residues for S palmitoylation (Figure 1A). Subsequently, we performed homology analysis by WebLogo (http://weblogo.berkeley.edu) to compare the sequence of SARS‐CoV‐2 S with those of other coronaviruses (GenBank: BAC81404.1 for SARS‐CoV, QFQ59587 for MERS‐CoV, QDH43726.1 for HCoV‐OC43, AYN64561.1 for HCoV‐HKU1, AFV53148.1 for HCoV‐NL63, APT69890.1 for HCoV‐229E, QHR63300.2 for Bat coronavirus RaTG13, QIQ54048.1 for Pangolin coronavirus, AFD97607.1 for Murine hepatitis virus strain). We found that only S proteins from SARS‐CoV‐2, RaTG13, and HCoV‐NL63 contain a cysteine at 15th AA (Figure 1B). However, C terminal cysteine‐rich sequences are conserved (Figure 1C). We generated C15S (C1), C1235S/C1236S/C1240S/C1243S/C1247S/C1248S/C1250S/C1253S/C1254S (C9), and C15S/C1235S/C1236S/C1240S/C1243S/C1247S/C1248S/C1250S/C1253S/C1254S (C10) mutants of SARS‐CoV‐2 (Figure 1D) and investigated the palmitoylation of S by ABE assay. Upon mutating nine cysteines in the cytoplasmic tail of S, the palmitoylation signal is largely reduced (Figure 1E). As we did not perform the single point mutation in the cytoplasmic tail of S, we cannot draw the conclusion that all C in the cytoplasmic tail are required for palmitoylation of S. Nevertheless, some palmitoylation sites are located in C enriched region of the cytoplasmic tail. Interestingly, a single C15S mutant also reduced the palmitoylation level compared with that in wild‐type S (Figure 1E). When 10 predicted cysteines are all mutated to serine (S), the palmitoylation signal is even less than those in C1 or C9 mutant (Figure 1E). Thus, the cysteines at sites C15, and C terminal cytoplasmic tail are two key sites for SARS‐CoV‐2 S palmitoylation.
Figure 1.
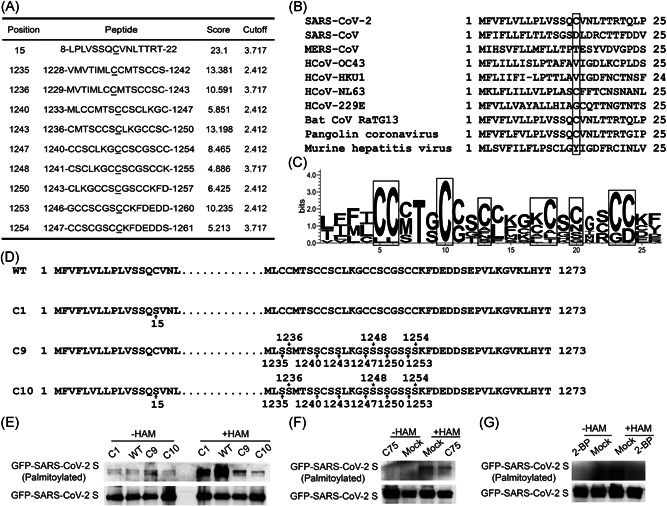
SARS‐CoV‐2 S protein is palmitoylated. (A) Prediction of potential palmitoylation sites on SARS‐CoV‐2 S by CSS‐palm 4.0. (B) Alignment of N terminal sequences of S from coronavirus (GenBank: QLJ57699.1 for SAR‐CoV‐2, BAC81404.1 for SARS‐CoV, QFQ59587 for MERS‐CoV, QDH43726.1 for HCoV‐OC43, AYN64561.1 for HCoV‐HKU1, AFV53148.1 for HCoV‐NL63, APT69890.1 for HCoV‐229E, QHR63300.2 for Bat coronavirus RaTG13, QIQ54048.1 for Pangolin coronavirus, and AFD97607.1 for Murine hepatitis virus strain). Box showed different residue at AA15. (C) WebLogo of C terminal sequences of S from coronavirus strains in Figure 1B. (D) Design of SARS‐CoV‐2 mutants. (E) ABE experiment showed SARS‐CoV‐2 S was modified by palmitoylation. (F) C75 reduced the palmitoylation of SARS‐CoV‐2 S. (G) 2‐BP inhibited S palmitoylation. GFP, green fluorescent protein; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
Fatty acid synthase generates PA, while protein palmitoylation is mediated by ZDHHC. To further confirm the palmitoylation of SARS‐CoV‐2, we applied fatty acid synthase inhibitor C75 and ZDHHC inhibitor 2‐BP. As shown in Figure 1F,G, both C75 and 2‐BP reduced the palmitoylation of S.
3.2. Palmitoylation of S is not required for plasma membrane targeting of S but important for S‐mediated syncytia formation and SARS‐CoV‐2 entry
As palmitoylation increases protein hydrophobicity and could facilitate protein trafficking to cellular membranes, next we investigated whether palmitoylation of S is important for plasma membrane targeting of S. Palmitoylation has no significant effect on the plasma membrane targeting of S as the level of S‐WT or C10 mutant in the plasma membrane portion are similar (Figure 2A). The expression of mutant S and wild‐type remained the same as shown in the input part of Figure 2A. Next, we investigated whether palmitoylation of S was important for SARS‐CoV‐2pp entry. We generated SARS‐CoV‐2 S/mutants based SARS‐CoV‐2pp in the HIV backbone and analyzed those pseudovirions with Western blot. As shown in Figure 2B, incorporation of SARS‐CoV‐2 S mutants into pseudovirions is less efficient compared with that of wild‐type S. Consistent with that, the entry abilities of all three mutant S based SARS‐CoV‐2pp were reduced, indicating that palmitoylation of S is critical for SARS‐CoV‐2pp entry (Figure 2C). Interestingly, the C1, C9, and C10 mutants of SARS‐CoV‐2 S‐mediated fewer syncytia formation, indicating less virus entry (Figure 2D). Thus, we concluded that the cysteines at sites C15, C1235, C1236, C1240, C1243, C1247, C1248, C1250, C1253, and C1254 of S are crucial for S‐mediated syncytia formation and SARS‐CoV‐2pp entry.
Figure 2.
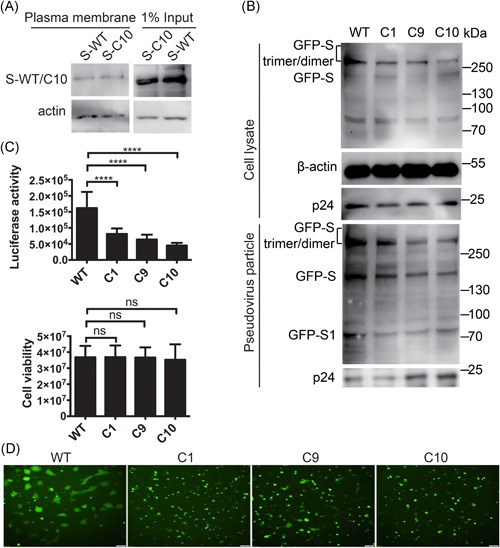
Palmitoylation of S is not required for plasma membrane targeting of S, but critical for S‐mediated syncytia formation and SARS‐CoV‐2pp entry. (A) The plasma membrane distribution of WT and C10 mutant S. (B) Incorporation of SARS‐CoV‐2 S and mutants proteins into pseudovirions. (C) Palmitoylation of S is important for SARS‐CoV‐2pp entry. Upper: luciferase assay showed palmitoylation sites mutant S alleviated entry of SARS‐CoV‐2pp. Bottom: cell viability was of no difference among groups. (D) Syncytia formation induced by transfection of GFP‐tagged S and S mutants. Scale bar: 100 μm. GFP, green fluorescent protein; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
3.3. Multiple ZDHHCs promoted SARS‐CoV‐2 S palmitoylation
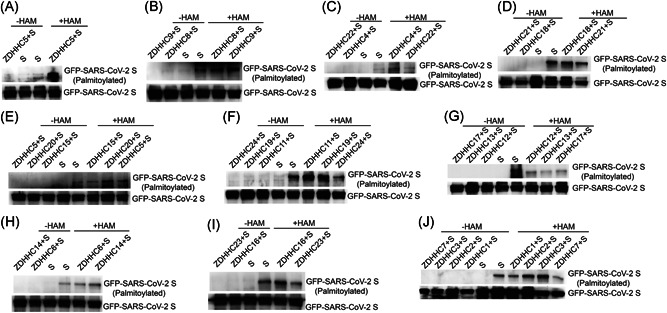
Protein palmitoylation is mediated mainly by ZDHHCs. Therefore, we screened for ZDHHCs promoted S palmitoylation. 293T cells were individually transfected with 23 FLAG‐tagged ZDHHCs and GFP‐tagged S, and the lysates were incubated with an anti‐FLAG antibody to perform Co‐IP experiments (Figure 4). ABE assays showed that the palmitoylation of S was enhanced after overexpression of ZDHHC5 (Figure 3A), ZDHHC8 (Figure 3B), ZDHHC9 (Figure 3B), ZDHHC4 (Figure 3C), ZDHHC20 (Figure 3E), ZDHHC11 (Figure 3F), ZDHHC19 (Figure 3F), ZDHHC14 (Figure 3H), ZDHHC16 (Figure 3I), ZDHHC2 (Figure 3J), or ZDHHC3 (Figure 3J). The ABE results are summarized in Table 1. These results suggested key roles for ZDHHC2, ZDHHC3, ZDHHC4, ZDHHC5, ZDHHC8, ZDHHC9, ZDHHC11, ZDHHC14, ZDHHC16, ZDHHC19, and ZDHHC20 in the palmitoylation of S.
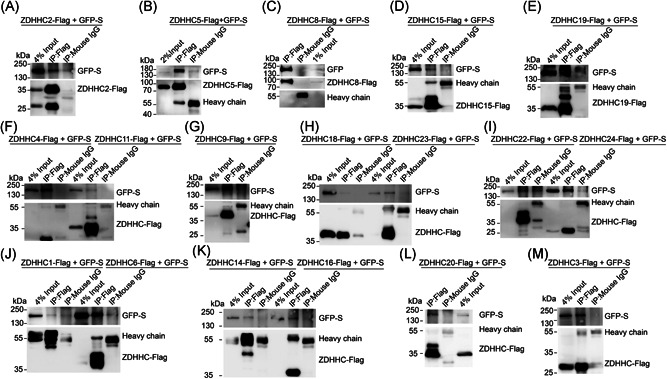
Figure 4.
Multiple ZDHHCs interacted with SARS‐CoV‐2 S. (A) ZDHHC2 associated with SARS‐CoV‐2. (B) ZDHHC5 associated with SARS‐CoV‐2 S. (C) ZDHHC8 associated with SARS‐CoV‐2 S. (D) ZDHHC15 associated with SARS‐CoV‐2 S. (E) ZDHHC19 associated with SARS‐CoV‐2 S. (F) ZDHHC4 and ZDHHC11 associated with SARS‐CoV‐2 S. (G) ZDHHC9 associated with SARS‐CoV‐2 S. (H) ZDHHC23 but not ZDHHC18 associated with SARS‐CoV‐2 S. (I) ZDHHC24 but not ZDHHC22 associated with SARS‐CoV‐2 S. (J) ZDHHC6 but not ZDHHC1 associated with SARS‐CoV‐2 S. (K) ZDHHC14 and ZDHHC16 associated with SARS‐CoV‐2 S. L. ZDHHC20 associated with SARS‐CoV‐2 S. (L) ZDHHC3 associated with SARS‐CoV‐2 S. SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; ZDHHCs, zinc finger DHHC domain‐containing palmitoyltransferases
Figure 3.
Identification of ZDHHCs which promoted the palmitoylation of SARS‐CoV‐2 S. 293T cells were individually transfected with GFP‐tagged S and ZDHHC5 (A), ZDHHC8 (B), ZDHHC9 (B), ZDHHC4 (C), ZDHHC22 (C), ZDHHC18 (D), ZDHHC21 (D), ZDHHC20 (E), ZDHHC11 (F), ZDHHC19 (F), ZDHHC24 (F), ZDHHC12 (G), ZDHHC13 (G), ZDHHC17 (G), ZDHHC6 (H), ZDHHC14 (H), ZDHHC16 (I), ZDHHC23 (I), ZDHHC1 (J), ZDHHC2 (J), ZDHHC3 (J), or ZDHHC7 (J). The lysates were incubated with an anti‐FLAG antibody to perform Co‐IP experiments and ABE assays. Co‐IP, coimmunoprecipitation; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; ZDHHCs, zinc finger DHHC domain‐containing palmitoyltransferases
Table 1.
Summary of ABE results and the interaction between SARS‐CoV‐2 S and ZDHHCs
| SARS‐CoV‐2 S | ZDHHCs | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Z1 | Z2 | Z3 | Z4 | Z5 | Z6 | Z7 | Z8 | Z9 | Z11 | Z12 | Z13 | Z14 | Z15 | Z16 | Z17 | Z18 | Z19 | Z20 | Z21 | Z22 | Z23 | Z24 | |
| ABE | − | + | + | + | ++ | − | − | + | + | + | − | − | + | − | + | − | − | + | ++ | − | − | − | − |
| Co‐IP | − | + | + | + | ++ | + | / | ++ | ++ | + | / | / | + | − | + | / | − | + | + | / | − | − | − |
Note: Z1–Z24, ZDHHC1–ZDHHC24. The interactions in the same row were scored relative to each other from not detectable (−) and strong to very strong (from + to ++). /, not tested.
Abbreviations: ABE, acyl‐biotinyl exchange; Co‐IP, co‐immunoprecipitation; ZDHHCs, zinc finger DHHC domain‐containing palmitoyltransferases.
3.4. Multiple ZDHHCs associated with SARS‐CoV‐2 S
As multiple ZDHHCs have been identified to promote S palmitoylation, we hypothesized that those ZDHHCs participated in the palmitoylation of S associated with S. To test this hypothesis, the plasmids expressing ZDHHC2, ZDHHC3, ZDHHC4, ZDHHC5, ZDHHC8, ZDHHC9, ZDHHC11, ZDHHC14, ZDHHC16, ZDHHC19, or ZDHHC20 were individually cotransfected with the plasmid expressing GFP‐S in 293T for 24 h (Figure 4). We used mouse IgG as a negative control to perform the co‐IP experiment. As shown in Figure 4, those ZDHHCs interacted with S. The results of interaction between S and ZDHHCs assayed by IP are summarized in Table 1.
4. DISCUSSION
Previous studies showed the palmitoylation of S from other coronaviruses including SARS‐CoV, MHV, and PEDV. 8 , 9 , 10 Consistent with our findings, two recent studies found that the palmitoylation of SARS‐CoV‐2 S protein is essential for viral infectivity. 13 , 14 However, they identified the key palmitoylation sites are restricted in the C terminal of S. In the current study, we have shown not only the C terminal cysteines of S but also the N‐terminal C15 is palmitoylated. The discrepancy between Wu's work and our study is due to the interpretation of their results. In Wu's paper, the palmitoylation signals of wild‐type S (S‐WT), C terminus (S‐ΔC‐Palm), and N terminus (S‐C15A) were 1.0, 0.1, and 0.9 using densitometric analysis with Image‐Pro Plus software. 14 However, the protein level of S‐WT was lower than that of S‐C15A. When the palmitoylation levels were normalized with protein levels, their results also indicated the palmitoylation in the N terminus of S protein.
A previous study only discovered ZDHHC5 as the major ZDHHC for SARS‐CoV‐2 S palmitoylation. 14 In the current study, we not only expanded the palmitoylation sites in SARS‐CoV‐2 S but also found that multiple ZDHHCs could palmitoylate SARS‐CoV‐2 S. As multiple ZDHHCs are involved in S palmitoylation, we proposed that the following three scenarios may occur. One possible scenario is that ZDHHCs are located in different cellular compartment palmitoylate S in different intracellular locations, such as Golgi or plasma membrane. The second scenario is that different ZDHHCs may contribute to different palmitoylation sites in S. The third scenario is that multiple ZDHHCs are redundant for the palmitoylation of S.
Palmitoylation of S from SARS‐CoV, MHV, or PEDV is not critical for plasma membrane localization. 8 , 9 , 10 For SARS‐CoV‐2, it is the same case. Plasma membrane isolation experiment showed that the protein level in the plasma membrane of palmitoylation deficient mutant of C10 was the same as that in wild‐type S. However, the syncytia formation was reduced by palmitoylation deficient mutation of S. Interestingly, palmitoylation of SARS‐CoV‐2 is critical for the incorporation of S to pseudovirus particles, which could explain the important role for S palmitoylation in SARS‐CoV‐2pp entry. The detailed mechanism of how palmitoylation affects S‐mediated fusion is unknown, which needs to be explored in the future.
In conclusion, we confirmed the palmitoylation of SARS‐CoV‐2 S and its function, proposing that palmitoylation of SARS‐CoV‐2 S is critical for S‐mediated syncytia formation and virus entry. C15 and C terminal multiple sites of cysteines are the key sites for palmitoylation. ZDHHC2, ZDHHC3, ZDHHC4, ZDHHC5, ZDHHC8, ZDHHC9, ZDHHC11, ZDHHC14, ZDHHC16, ZDHHC19, and ZDHHC20 are the major PATs for S.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Daoqun Li, Y Liu, and Y Lu performed the experiments. Shan Gao revised the manuscript. Leiliang Zhang conceived the work and wrote the manuscript. All authors read and approved the final manuscript.
ACKNOWLEDGMENTS
This study was supported by grants from the National Natural Science Foundation of China (82072270 and 81871663), the Shandong Provincial Natural Science Foundation (ZR2020QH273), and the Academic Promotion Programme of Shandong First Medical University (2019LJ001). We thank Drs. Ping Zhao and Zhaohui Qian for reagents.
Li D, Liu Y, Lu Y, Gao S, Zhang L. Palmitoylation of SARS‐CoV‐2 S protein is critical for S mediated syncytia formation and virus entry. J Med Virol. 2021;94:342‐348. 10.1002/jmv.27339
Daoqun Li, Yihan Liu, and Yue Lu are as Co‐first authors.
REFERENCES
- 1. Gao S, Zhang L. ACE2 partially dictates the host range and tropism of SARS‐CoV‐2. Comput Struct Biotechnol J. 2020;18:4040‐4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang Y, Zhao W, Mao Y, et al. Site‐specific N‐glycosylation characterization of recombinant SARS‐CoV‐2 spike proteins. Mol Cell Proteom. 2020;20:100058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boson B, Legros V, Zhou B, et al. The SARS‐CoV‐2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus‐like particles. J Biol Chem. 2021;296:100111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tian W, Li D, Zhang N, et al. O‐glycosylation pattern of the SARS‐CoV‐2 spike protein reveals an "O‐Follow‐N" rule. Cell Res. 2021:1‐3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bouhaddou M, Memon D, Meyer B, et al. The global phosphorylation landscape of SARS‐CoV‐2 infection. Cell. 2020;182(3):685‐712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lun CM, Waheed AA, Majadly A, Powell N, Freed EO. Mechanism of viral glycoprotein targeting by membrane‐associated RING‐CH proteins. mBio. 2021;12:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lemonidis K, Werno MW, Greaves J, et al. The ZDHHC family of S‐acyltransferases. Biochem Soc Trans. 2015;43(2):217‐221. [DOI] [PubMed] [Google Scholar]
- 8. Thorp EB, Boscarino JA, Logan HL, Goletz JT, Gallagher TM. Palmitoylations on murine coronavirus spike proteins are essential for virion assembly and infectivity. J Virol. 2006;80(3):1280‐1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Petit CM, Chouljenko VN, Iyer A, et al. Palmitoylation of the cysteine‐rich endodomain of the SARS‐coronavirus spike glycoprotein is important for spike‐mediated cell fusion. Virology. 2007;360(2):264‐274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gelhaus S, Thaa B, Eschke K, Veit M, Schwegmann‐Weßels C. Palmitoylation of the Alphacoronavirus TGEV spike protein S is essential for incorporation into virus‐like particles but dispensable for S‐M interaction. Virology. 2014;464‐465:397‐405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gordon DE, Jang GM, Bouhaddou M, et al. A SARS‐CoV‐2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459‐468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang N, Zhao H, Zhang L. Fatty acid synthase promotes the palmitoylation of chikungunya virus nsP1. J Virol. 2019;93:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nguyen HT, Zhang S, Wang Q, et al. Spike glycoprotein and host cell determinants of SARS‐CoV‐2 entry and cytopathic effects. J Virol. 2020;95:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu Z, Zhang Z, Wang X, et al. Palmitoylation of SARS‐CoV‐2 S protein is essential for viral infectivity. Signal Transduct Target Ther. 2021;6(1):231. [DOI] [PMC free article] [PubMed] [Google Scholar]


