Abstract
SARS‐CoV‐2 variants of concern (VOCs) have emerged worldwide and gained significant importance due to their high transmissibility and global spread, thus meriting close monitoring. In Pakistan, limited information is available on circulation of these variants as the alpha variant has been reported the main circulating lineage. The current study was designed to detect and explore the genomic diversity of SARS‐CoV‐2 lineages circulating during the third wave of the pandemic in the indigenous population. From May 01 to June 09, 2021, a total of 16 689 samples were tested using TaqPath™ COVID‐19 kit for the presence of SARS‐CoV‐2. Overall, 2562 samples (15.4%) were COVID‐19 positive. Out of these positive samples, 2124 (12.7%) did not show the spike gene amplification (spike gene target failure ([SGTF]), whereas 438 (2.6%) showed spike gene amplification (non‐SGTF). A subset (n = 58/438) of non‐SGTF samples were randomly selected for whole‐genome sequencing. Among VOCs, 45% (n = 26/58) were delta, 46% (n = 27/58) were beta, and one was gamma variant. The delta variant cases were reported mainly from Islamabad (n = 15; 58%) followed by Rawalpindi and Azad Kashmir (n = 1; 4% each). Beta variant cases originated mainly from Karachi (n = 8; 30%) and Islamabad (n = 11; 41%) and the gamma variant case was reported in a traveler from Italy. The delta, beta, and gamma variants possessed lineage‐specific spike mutations. Notably, two rare mutations (E484Q and L5F) were found in the delta variant. Furthermore, in the beta variant, two significant rare non‐synonymous spike mutations (A879S and K444R) were also reported. High prevalence of beta and delta variants in local population may increase the number of cases in the near future and provides an early warning to national health authorities to take timely decisions and devise suitable interventions to contain a possible fourth wave.
Keywords: alpha variant, beta variant, delta variant, gamma variant, SARS‐CoV‐2, variants of concern
1. INTRODUCTION
The COVID‐19 pandemic is still one of the leading cause of infectious mortality worldwide due to emergence of novel SARS‐CoV‐2 variants. As of July 31, 2021, the global total of SARS‐CoV‐2‐related infections has surpassed over 198 million, with 4.22 million deaths. In recent months, a diversification of SARS‐CoV‐2 has been observed globally because of evolution and adaptation processes. Some emerging mutations may confer a selective advantage to the virus, resulting in the selection of the “variants of concern” (VOCs) with significant epidemiological and pathogenic consequences. 1 , 2 Four specific viral lineages reflecting VOCs have emerged worldwide and warrant close monitoring: B.1.1.7 (alpha), B.1.351 (beta), P.1 (gamma), and B.1.617.2 (delta). 3 These VOCs have gained significant importance because of their contribution in sustained disease transmissibility in the upcoming waves of pandemic. Among the VOCs, alpha, beta, and delta variant has been recently the most important VOCs, which has contributed significantly to the upsurge of new waves worldwide.
The beta variant was first reported in South Africa in December, 2020 and is characterized by seven different lineage‐defining mutations in the spike protein, with three significant mutations (N501Y, E484K, and K417N) in the receptor‐binding domain (RBD). 4 The gamma variant was first detected in Manaus, Brazil in November, 2020 with the following lineage‐defining mutations: E484K, K417T, and N501Y. Interestingly, the gamma and beta lineages share three common mutations (K417N/T, E484K, and N501Y) in spike protein.
There is an evidence that strains with N501Y substitution have increased transmissibility due to enhanced binding affinity with human angiotensin‐converting enzyme 2 (ACE2) as determined by deep mutation scanning in a mouse model. 5 , 6 , 7 Additionally, the beta and gamma variants may provide an immune escape mechanism from antibodies due to E484K mutation in the spike protein. 8 It has been recently identified that these variants are capable of evading monoclonal and serum antibody responses. 9 , 10 The K417N/T, E484K, and N501Y mutations significantly decreased the neutralizing activity of convalescent and messenger RNA vaccine‐induced serum. 11
The delta variant was first detected in Maharashtra, India in October, 2020. Its sub‐lineage, B.1.617.1, is defined by the presence of a constellation of mutations, L452R, P681R, and E484Q in the spike region, whereas B.1.617.2 is characterized by following spike mutations: L452R, P681R, and T478K. The RBD mutations enhance infectivity due to the presence of L452R and T478K by increasing the spike protein's affinity for human ACE2 receptor. 12 , 13 Both mutations reduce the binding affinity of monoclonal antibodies, thereby impairing neutralizing ability. Additionally, structural analysis of mutations (L452R and E484Q) in RBD and furin cleavage site (P681R) revealed increased ACE2 binding and cleavage rate resulting in increased transmissibility. 14
The delta variant has now spread to 112 countries, with a global cumulative prevalence of 12% (n = 267 594) with the most notable prevalence of 44% (n = 14 938) in India. The beta variant has been spread to 107 countries with a global cumulative prevalence of 1% (n = 29 843) and the highest prevalence of 61% (n = 6121) in South Africa. The gamma variant has spread to 65 countries, with a global cumulative prevalence of 2% (n = 55 587) and highly prevalent (n = 15 011, 62%) in Brazil (https://outbreak.info, accessed on July 31, 2021). The prevalence of these variants around the globe and in the originating countries gives a clue about the emergence of possible new outbreaks/waves throughout the world.
As of July 31, 2021, there has been 1 029 811 positive cases of SARS‐CoV‐2 with 23 360 deaths in Pakistan. According to province‐wise data, the maximum number of cases were reported in Sindh (n = 380 093), Punjab (n = 356 211), Khyber Pakhtunkhwa (n = 143 673) and Islamabad (n = 87 304). 15 In comparison to the number of reported cases. the genome sequencing data submitted to global SARS‐CoV‐2 databases (GISAID and NCBI) from Pakistan is very limited (n = 614), resulting in limited assessment of the introduction, geographic spread, and community transmission of SARS‐CoV‐2 variants. Hence, the current study aimed to detect and explore the genomic diversity of different lineages circulating in Pakistan during the third wave.
2. MATERIALS AND METHODS
2.1. Sampling
Oropharyngeal samples (n = 16 689) were collected from COVID‐19 suspected patients and received at the Department of Virology, National Institute of Health, Islamabad.
2.2. RNA extraction and real‐time polymerase chain reaction (PCR)
RNA was extracted from the samples using a KingFisher™ Flex Purification System (Thermo Fisher Scientific). For the detection of SARS‐CoV‐2, TaqPath™ COVID‐19 CE‐IVD RT‐PCR kit (Thermo Fisher Scientific) that targets three genes (ORF1ab, N, and S) was used.
2.3. Sample selection criteria/strategy for whole genome sequencing
Previously, we had used the SGTF as a proxy for the detection of alpha variant in Pakistan using the TaqPath™ kit (Thermo Fisher Scientific). 13 This detection method can also be used the other way around for surveillance of other lineages. Based on this strategy, the non‐SGTF samples having low cycle threshold (C t) values (≤27) followed by selection based on their geographical location, that is, representing the whole country were selected for whole‐genome sequencing.
2.4. Complementary DNA (cDNA) synthesis, and amplification
The cDNA synthesis and amplification were performed according to the Primal‐Seq Nextera XT protocol (version 2) using SuperScript™ IV VILO™ Master Mix (Invitrogen) and Q5® High‐Fidelity 2X Master Mix (New England BioLabs), with the ARTIC nCoV‐2019 Panel V3 (Integrated DNA Technologies, Inc.). 16
2.5. Next generation sequencing
The paired‐end sequencing library (2 × 150 bp) was prepared from the generated amplicons using Illumina DNA Prep Kit (Illumina, Inc.) by following the standard protocol. The prepared libraries were pooled and subjected to sequencing on Illumina iSeq platform, using sequencing reagent, iSeq. 100 i1 Reagent v2 (300‐cycle) (Illumina, Inc.) at Department of Virology, National Institute of Health, Islamabad, Pakistan.
2.6. Data analysis
The FastQC tool (v0.11.9) was used to assess the read quality of sequenced files. 16 Trimmomatic (v0.39) 17 was employed to remove Illumina adapter sequences and low‐quality base calls with scores less than 30 to eliminate technical biases and artifacts. The filtered reads were assembled by aligning with the available reference genome of SARS‐CoV‐2 (Accession number: NC_045512.2) using the Burrows‐Wheeler Aligner's (BWA, v0.7.17) with default settings. 18 Picard Tools was used to remove PCR duplicates from aligned reads (v2.25.4). 19 The variants were called and consensus sequences of all the genomes were generated as per Centers for Diseases Control and Prevention (CDC) guidelines. 20 The assembled genomes were classified into PANGO lineages using the Pangolin v3.1.7 and pangoLEARN model dated 28‐07‐2021. 21
2.7. Phylogenetic analysis
Nextstrain's standard protocol for analyzing SARS‐CoV‐2 genomes was used for the phylogenetic analysis. To begin, BLAST search was conducted on the GISAID database against each of the current study's isolates (beta, gamma, and delta). This resulted in a total of 286 sequences including the current study's sequences. The sequences were clustered using Augur Nextstrain's phylodynamic pipeline. 22 Alignment of sequences to the Wuhan reference genome was performed using MAFFT v7.470. 23 The initial phylogenetic tree was constructed using IQTREE v1.6.12, 24 which utilizes generalized time‐reversible (GTR) model to generate the tree. The bootstrapping was performed to ensure a high degree of confidence in tree topology. Using reference genome (GISAID ID: EPI_ISL_402125) the raw tree was rooted. TreeTime v0.8.1 was used to further process the tree to generate a time‐resolved phylogeny based on maximum likelihood. 25 The resulting tree was visualized using Auspice.
3. RESULTS
From May 01, 2021 to June 09, 2021, a total of 16 689 samples were tested on real‐time PCR for the presence of SARS‐CoV‐2. Of the total samples tested, 15.4% (n = 2562) were COVID‐19 positive. Out of these positive samples, 2.6% (n = 438) isolates have shown the amplification of spike gene (non‐SGTF), whereas 12.7% (n = 2124) samples had the SGTF (Figure 1). A subset of 58 samples from non‐SGTF were selected for whole‐genome sequencing based on the criteria defined above. The clinical data of patients is summarized in Table 1. All the patients enrolled in the study didn't reported any noticeable sign and symptoms and recovery was uneventful. Some generable symptoms included fever, body ache, and sore throat. Only two patients reported respiratory illness (Table 1).
Figure 1.
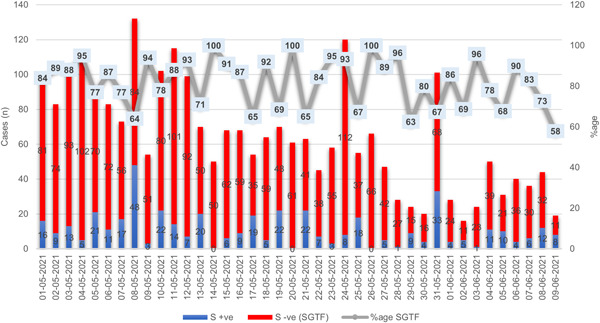
The distribution of COVID‐19 patients according to the Δ69–70 deletion during May and June 2021. The TaqPath real‐time PCR kit was used to detect SGTF in SARS‐CoV‐2 patients. The X‐axis indicates the months, while the Y‐axis indicates the number of cases. S +ve indicates the presence of a spike gene (non‐SGTF). SGTF indicates the absence of a spike gene
Table 1.
Clinical findings of patients enrolled in the study
| Parameter | Total number | Beta | Delta | Gamma |
|---|---|---|---|---|
| Number of patients | 58 (100%) | 27 (46.5%) | 26 (44.8%) | 1 (1.7%) |
| Male | 28 (48.2%) | 13 (48.1%) | 12 (46.1%) | |
| Female | 25 (43.1%) | 13 (48.1%) | 11 (42.3%) | 1 (100%) |
| Signs and Symptoms | ||||
| Fever | 27 (46.5%) | 13 (48.1%) | 14 (53.8%) | No |
| Cough | 8 (13.7%) | 3 (11.1%) | 3 (11.5%) | No |
| Sore throat | 15 (25.8%) | 5 (18.5%) | 8 (30.7%) | No |
| Body ache | 25 (43.1%) | 10 (37.0%) | 13 (50.0%) | No |
| Breathing difficulty | 2 (3.4%) | 2 (7.4%) | 0 (0.0%) | No |
| A‐symptomatic | 13 (22.4%) | 10 (37%) | 2 (7.7%) | 1 (100%) |
A total of six lineages, that is, B.1.1.448, B.1.36, B.1.1, B.1.351, P.1.1, and B.1.617.2 were identified from Pakistan. Among VOCs, delta variant turned out to be 45% (n = 26/58) followed by beta 46% (n = 27/58) and gamma variant (n = 1; 1%). The remaining 7% reported lineages were B.1.1.448 (n = 2), B.1.36 (n = 1), and B.1.1 (n = 1) (Figure 2). Among delta variant cases, Islamabad reported the highest numbers (n = 15; 58%) followed by Rawalpindi and Azad Jammu Kashmir (n = 1 each). Nine patients (35%) infected with delta variant had travel history with four from Afghanistan (GISAID IDs: EPI_ISL_2757757, EPI_ISL_2757758, EPI_ISL_2757759, EPI_ISL_2757756) and one each from Saudi Arabia (GISAID ID: EPI_ISL_2434174), Oman (GISAID ID: EPI_ISL_2757736), UAE (GISAID ID: EPI_ISL_2438666), and Bahrain (GISAID ID: EPI_ISL_2894982). Beta variant cases originated mainly from Karachi (n = 8; 30%) and Islamabad (n = 11; 41%), followed by Rawalpindi (n = 2; 7%), Azad Jammu Kashmir (AJK) (n = 1), Dera Ismail Khan (n = 1), Multan (n = 1), and Shangla (n = 1). Furthermore, two patients infected with the beta variant had travel history of the UAE (GISAID ID: EPI_ISL_2438665 and EPI_ISL_2438683). The one case of gamma variant (EPI_ISL_2894980) was having the travel history from Italy (Figure 3).
Figure 2.
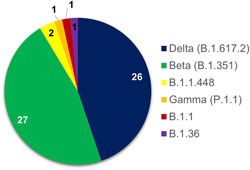
The distribution of variants of concerns reported in the current study. Delta and beta reported in the highest numbers (n = 54) followed by other lineages
Figure 3.
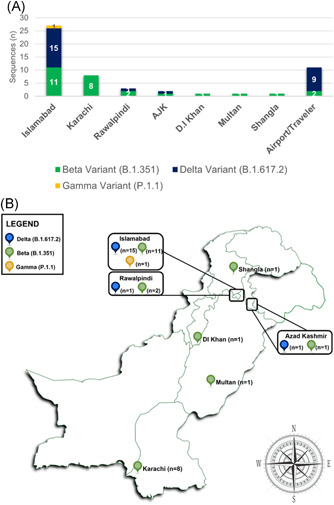
(A) Geography‐wise distribution of variants of concerns reported in the current study. X‐axis representing the geographical location and Y‐axis representing the number of beta (B.1.351), gamma (P.1.1), and delta (B.1.617.2) sequences. (B) Geographical distribution of beta and delta cases of SARS‐CoV‐2 in Pakistan from May 01 to June 9, 2021
The median age of patients infected with delta variant was 32.5 ranging from 20 to 53 years comprising 46% males and 42% females. The median age of patients infected with beta variant was 30 years ranging from 2 to 61 years. The female to male ratio was 1:1 with 48% of females and 48% of males.
Table 2 summarizes the mutations identified in all the 58 studied sequences. The delta variant isolates reported following significant mutations: S:L452R (22917 T>G), S:T478K (22995 C>A), S:P681R (23604 C>G), S:D950N (24410 G>A), ORF3a:S26L (25469 C>T), M:I82T (26767 T>C), ORF7a:V82A (27638 T>C), ORF7a:T120I (27752 C>T), N:D63G (28461 A>G), N:R203M (28881 G>T) and N:D377Y (29402 G>T). A highly significant less prevalent spike mutation, E484Q (23012 G>C), having a role in the immune escape was found in one 52‐year‐old male patient having a travel history of Bahrain (GISAID ID: EPI_ISL_2894982). Another distinct and rare spike mutation (L5F [21575 C>T]) was also reported in one 40‐year female patient (GISAID ID: EPI_ISL_2894977).
Table 2.
Genome‐wide amino acid mutations
| S. No | GASID ID | Lineage | ORF1ab | Spike | ORF3a | Nucleocapsid protein | Membrane | Envelope | ORF7a | ORF7b | ORF8 | ORF6 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | EPI_ISL_2757735 | B.1.351 | NSP2: T85I, K111E, A570V, E574DNSP3: S794L, K837N, A171VNSP5: K90RNSP12b: P314L, M809V | D80A, D215G, K417N, E484K, N501Y, D614G, A701V | Q57H, S171L | T205I, T362I | P71L | |||||
| 2 | EPI_ISL_2757739 | B.1.617.2 | NSP2:R27C, K111E, P129L NSP3: P822L, H1274Y NSP4: A446V NSP6: V149A NSP12b: P314L, G662S NSP13: V169FNSP15: E3K, H234Y | T19R,G142D, L452R, T478K, D614G, P681R, D950N | S26L | D63G, R203M, D377Y, R385K | I82T | V82A, L116F, T120I | ||||
| 3 | EPI_ISL_2757741 | B.1.617.2 | NSP2: K111ENSP3: A488S, P1228L, S1578GNSP4: V167L, T492I, NSP6: T77ANSP12b: P314L, G662S NSP13: Q194P, R392C NSP14:A394V | T19R,L452R, T478K, D614G, D950N | S26L, V112F | D63G, R203M, G215C, D377Y | I82T | T120I | T40I | |||
| 4 | EPI_ISL_2434174 | B.1.617.2 | NSP2: P129LNSP3: P822L, H1274Y, T1334INSP4: A446VNSP6: V149ANSP12b: P314L, G662S NSP13: P77LNSP15: H234Y | T19R,G142D,E156,L452R, T478K, D614G, D950N, S1061V | S26L | D63G, R203M, T362I, D377Y, R385K | I82T | V82A, T120I | A43V | D119 | ||
| 5 | EPI_ISL_2757743 | B.1.617.2 | NSP2: K111E, P129LNSP3: P822L, H1274Y NSP4: A446VNSP6: V149ANSP7: E50GNSP12b: P314L, G662S NSP13: P77LNSP15:H234Y | T19R,F106L, G142D, L452R, T478K, D614G, P681R, D950N | S26L, F79L | D63G, L139F, R203M, D377Y, R385K | V82A, L116F, T120I | F120L | ||||
| 6 | EPI_ISL_2757744 | B.1.617.2 | NSP2: K111E, P129LNSP3: P822L, H1274YNSP4: A446VNSP6: V149ANSP12b: P314L, G662S NSP13: P77LNSP15:H234Y | T19R,F106L, G142D, L452R, T478K, D614G, P681R, D950N | S26L | D63G, R203M, D377Y, R385K | I82T | V82A, L116F, T120I | ||||
| 7 | EPI_ISL_2757756 | B.1.617.2 | NSP1: Y68CNSP3: P822L, H1274Y I1723VNSP4: A446VNSP6: T181INSP12b: P314L, G662S NSP13: P77L | T19R,G142D, A222V, L452R, T478K, D614G, P681R, D950N | S26L | D63G, R203M, D377Y | I82T | V82A, T120I | ||||
| 8 | EPI_ISL_2757757 | B.1.617.2 | NSP1: Y68CNSP3: P822LNSP4: A446VNSP6: V149ANSP12b: G662SNSP13: P77L | T19R,G142D, L452R, T478K, D614G, P681R, D950N | D63G, D377Y | I82T | V82A, T120I | |||||
| 9 | EPI_ISL_2757758 | B.1.617.2 | NSP2: P129LNSP3: P822LNSP4: S163A, A446V NSP6: V149ANSP12b: P314L, G662S NSP13: P77LNSP14: P46LNSP16: K182N | T19R,G142D, L452R, T478K, D614G, P681R, D950N | S26L | D63G, D377Y | I82T | V82A, L116F, T120I | ||||
| 10 | EPI_ISL_2757759 | B.1.617.2 | NSP3: A488S, P1228L NSP4: V167L, T492INSP6: T77ANSP12b: P314L, G662S NSP13: P77L, Q194PNSP14: A394V | T19R,L452R, T478K, D614G, P681R, D950N | S26L | D63G, R203M, G215C, D377Y | I82T | V82A, T120I | ||||
| 11 | EPI_ISL_2757760 | B.1.617.2 | NSP3: A488S, P1228L S1578GNSP4: V167L, T492INSP6: T77ANSP8: T187INSP12b: P314L, K565N, G662SNSP13: P77LNSP14: A394V | T19R,L452R, T478K, D614G, P681R, D950N | S26L | D63G, R203M, G215C, A308G, D377Y | I82T | V82A, T120I | T40I | |||
| 12 | EPI_ISL_2757736 | B.1.617.2 | NSP3: A488S, P1228L P1469SNSP4: V167L, T492INSP6: T77ANSP12b: P314L, V345E, G662SNSP13: P77LNSP14: A394V | T19R,L452R, T478K, D614G, P681R, D950N | S26L | D63G, R203M, G215C, D377Y | I82T | V82A, T120I | T40I | |||
| 13 | EPI_ISL_2757761 | B.1.617.2 | NSP2: A375VNSP3: A375V L445V, P822LNSP4: A446VNSP6: V149ANSP12b: P314L, G662S NSP13: P77L | T19R,G142D, A222V, L452R, T478K, D614G, P681R, D950N | S26L | D63G, T141S, R203M, D377Y | I82T | V82A, T120I | ||||
| 14 | EPI_ISL_2757762 | B.1.617.2 | NSP3: S400G A488S, P1228LNSP4: V167L, T492INSP6: T77ANSP12b: P314L, G662S NSP13: P77LNSP14: A394V | T19R,L452R, T478K, D614G, P681R, D950N | S26L | D63G, R203M, G215C, D377Y | I82T | V82A, T120I | ||||
| 15 | EPI_ISL_2757737 | B.1.617.2 | NSP2: P129LNSP3: P822L H1274Y NSP4: A446VNSP6: V149ANSP12b: P314L, G662S NSP13: P77LNSP15: H234Y | T19R,G142D, L452R, T478K, D614G, P681R | S26L | D63G, R203M, D377Y, R385K | I82T | V82A, L116F, T120I | ||||
| 16 | EPI_ISL_2757738 | B.1.617.2 | NSP3: P822LNSP4: A446VNSP6: T181INSP12b: P314L, G662S NSP13: P77L | T19R, G142D, A222V, L452R, T478K, D614G, P681R | S26L | D63G, R203M, D377Y | I82T | V82A, T120I | ||||
| 17 | EPI_ISL_2434174 | B.1.617.2 | NSP2: P129LNSP3: P822L, H1274Y, T1334INSP4: A446VNSP6: V149ANSP12b: P314L, G662S NSP13: P77LNSP15:H234Y | T19R,G142D, L452R, T478K, D614G, P681R, D950N | S26L | D63G, R203M, T362I, D377Y, R385K | I82T | V82A, L116F, T120I | A43V | |||
| 18 | EPI_ISL_2438666 | B.1.617.2 | NSP3: P822LNSP4: A446VNSP6:V149A, T181I NSP12b: P314L, G662S NSP13: P77L | T19R,G142D, A222V, L452R, T478K, D614G, P681R, D950N | S26L | D63G, R203M, D377Y | I82T | V82A, T120I | ||||
| 19 | EPI_ISL_2434781 | B.1.617.2 | NSP2: P129LNSP3: R58, P822L, H1274Y, Y1695SNSP4: A446VNSP6: V149ANSP12b: P314L, G662S NSP13: P77LNSP15:H234Y | E156,T19R,G142D, L452R, T478K, D614G, P681R | S26L | D63G, R203M, D377Y, R385K | I82T | V82A, L116F, T120I | D119 | |||
| 20 | EPI_ISL_2434976 | B.1.351 | NSP2: P129LNSP3: H1274YNSP4: A446VNSP12b: P314L, E427, G662SNSP13: P77L NSP15:H234Y, S287L | T19R,G142D, L452R, T478K, D614G, P681R | S26L | D63G, R203M, D377Y, R385K | I82T | N52K, V82A, L116F, T120I | K42E | |||
| 21 | EPI_ISL_2434982 | B.1.617.2 | NSP2: P129LNSP3: P822L, H1274Y NSP4: A446VNSP6: V149ANSP12b: G219S, P314L, G662SNSP13: P77LNSP15:H234Y | E156,T19R,G142D, L452R, T478K, D614G, P681R, D950N | S26L | D63G, R203M, H300Y, D377Y, R385K | I82T | V82A, L116F, T120I | ||||
| 22 | EPI_ISL_2438547 | B.1.617.2 | NSP4: A446VNSP5: P96LNSP6: Q160KNSP8: A16V, Y138H NSP12b: P314L | V143,H49Y,F157S, N501T, D614G,P812S | S26L, I118T | D63G, R203M, D377Y, R385K | I82T | V82A, L116F, T120I | ||||
| 23 | EPI_ISL_2314809 | B.1.1.448 | NSP2: P129LNSP3: P822L, H1274Y NSP4: A446VNSP6: V149ANSP12b: P314L, G343, G662SNSP13: P77L, V169F, G439ENSP15:H234Y, S287L | N87,E156,T19R, G142D, L452R, T478K, D614G, P681R, D950N | I84V, RG203KR | F2S | ||||||
| 24 | EPI_ISL_2313082 | B.1.36 | NSP4: A446VNSP5: P96LNSP6: Q160KNSP8: A16V, Y138H | V143,H49Y,F157S, N501T, D614G,P812S | I84V, RG203KR | |||||||
| 25 | EPI_ISL_2438599 | B.1.1 | NSP2: E167DNSP3: K387NNSP4: A446VNSP5: P96LNSP6: Q160KNSP8: A16VNSP12b: P314L | S12F,H49Y,F157S, N501T, D614G | I84V, RG203KR | D35Y | F2S | |||||
| 26 | EPI_ISL_2438598 | B.1.36 | NSP3: D110GNSP12b: A7V, W153C, P314L | A67V,D614G | S60, Q57H, V77I, G172C | S194L, D377Y | ||||||
| 27 | EPI_ISL_2438683 | B.1.351 | NSP2: L97, T85INSP3: K387NNSP5: P252LNSP6: S106, Y80FNSP12b: P314L, L820F | A27S,L242,I726,D80A, G181V, D215G, K417N, E484K, N501Y, D614G, A701V | Q57H, S171L, T271I | T205I, K387N | P71L | |||||
| 28 | EPI_ISL_2894980 | P.1.1 | NSP3: K977QNSP6: S106NSP13: E341D | L18F,T20N,R19S,K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F | S235P | P80R, RG203KR | E92K | |||||
| 29 | EPI_ISL_2894979 | B.1.351 | NSP2: T85INSP3: S93F, K837NNSP5: T24I, K90RNSP6: S106NSP12b: A34V, P314L NSP13: T588INSP14: V161L | D80A, D215G, K417N, E484K, N501Y, D614G, A701V, A879S, D1163Y | Q57H, S171L | T205I | P71L | I121L | Y49H | |||
| 30 | EPI_ISL_2313084 | B.1.351 | NSP2: T85INSP3: K837NNSP5: K90RNSP6: S106NSP12b: P314LNSP13: T588I | D80A, D215G, K417N, E484K, N501Y, D614G, A701V, A879S, D1163Y | Q57H, S171L | T205I | P71L | |||||
| 31 | EPI_ISL_2313098 | B.1.351 | NSP2: T85INSP3: K837NNSP5: K90RNSP6: S106NSP12b: P314L | D80A, D215G,L242,K417N, E484K, N501Y, D614G, A701V | Q57H, S171L | P13S, T205I | P71L | |||||
| 32 | EPI_ISL_2313099 | B.1.351 | NSP2: T85INSP3: T217I, K837NNSP5: K90RNSP6: S106NSP8: Q24RNSP12b: P314L | L18F,D80A, D215G,L242,K417N, E484K, N501Y, D614G, A701V | Q57H, S171L | P13S, T205I | P71L | |||||
| 33 | EPI_ISL_2313112 | B.1.351 | NSP2: T85INSP3: T217I, K837NNSP5: K90RNSP6: S106NSP12b: P314L | L18F,D80A, D215G,L242,K417N, E484K, N501Y, D614G, A701V | Q57H, S171L | P13S, T205I | P71L | |||||
| 34 | EPI_ISL_2313114 | B.1.351 | NSP2: T85INSP3: T217I, K837NNSP5: K90RNSP6: S106NSP12b: P314L | L18F,D80A, D215G,L242,K417N, E484K, N501Y, D614G, A701V | Q57H, S171L | P13S, T205I | P71L | |||||
| 35 | EPI_ISL_2314185 | B.1.351 | NSP2: T85INSP3: T217I, K837NNSP5: K90RNSP6: S106NSP12b: P314L | L18F,D80A, D215G,L242,K417N, E484K, N501Y, D614G, A701V | Q57H, S171L | G30R, T205I | P71L | W29L | ||||
| 36 | EPI_ISL_2894975 | B.1.351 | NSP2: T85I, G88RNSP3: D178Y, S794L, K837NNSP5: K90RNSP6: S106NSP12b: P314L | D80A, D215G, K417N, E484K, N501Y, D614G, A701V | Q57H, S171L | G30R, T205I | P71L | W29L | ||||
| 37 | EPI_ISL_2894976 | B.1.351 | NSP3: S794L, K837NNSP5: K90RNSP6: S106NSP12b: P314L | D80A, D215G, K417N, E484K, N501Y, D614G, A701V | Q57H, S171L | G30R | P71L | W29L | ||||
| 38 | EPI_ISL_2313081 | B.1.351 | NSP2: T3I, T85INSP3: S794L, K837NNSP4: S336LNSP5: K90RNSP6: S106NSP7: L56F | D80A, D215G,L242,K417N, E484K, N501Y, D614G, A701V | Q57H, S171L | G30R, T205I | P71L | W29L | ||||
| 39 | EPI_ISL_2757745 | B.1.351 | NSP2: T85I, K111ENSP3: S794L, K837NNSP5: K90RNSP12b: P314L, V728F | D80A, D215G, K417N, E484K, N501Y, D614G, A701V | Q57H, S171L | G30R, T205I | P71L | |||||
| 40 | EPI_ISL_2757746 | B.1.351 | NSP2: T85I, K111ENSP3: S794L, K837N, K1155RNSP5: K90RNSP12b: P314LNSP14: Y154H | D80A, D215G, K417N, E484K, N501Y, D614G, A701V | Q57H | G30R, N192K, T205I, P365L | P71L | |||||
| 41 | EPI_ISL_2757747 | B.1.351 | NSP2: T85I, K111ENSP3: S794L, K837NNSP5: K90RNSP12b: P314LNSP15: A92V | D80A, D215G, K417N, E484K, N501Y, D614G, A701V | Q57H, S171L | G30R, T205I | P71L | |||||
| 42 | EPI_ISL_2757748 | B.1.351 | NSP2: T85I, K111ENSP3: V613I, S794L, K837NNSP5: K90RNSP12b: P314LNSP16: P80A | K77N,D80A, D215G, K417N, E484K, N501Y, D614G, A701V | Q57H, S171L | G30R, P151S, T205I | P71L | W29L | ||||
| 43 | EPI_ISL_2757749 | B.1.351 | NSP2: T85I, K111E, E442G NSP3: S794L, K837NNSP5: K90RNSP12b: P314LNSP15: G229C | D80A, D215G, K417N, K444R, E484K, N501Y, D614G, A701V | Q57H | G30R, T205I | P71L | W29L | ||||
| 44 | EPI_ISL_2757750 | B.1.351 | NSP2: T85I, A318VNSP3: S794L, K837N, S1578GNSP5: K90RNSP12b: P314L, K565N NSP13: Q194P | D80A, D215G, K417N, E484K, N501Y, D614G, A701V | Q57H, S171L | G30R, T205I | P71L | |||||
| 45 | EPI_ISL_2757751 | B.1.351 | NSP2: T85INSP3: S794L, K837N, S1578GNSP5: K90RNSP12b: P314LNSP13: Q194P | D80A, D215G, K417N, E484K, N501Y, D614G, A701V | Q57H, S171L | G30R, T205I | P71L | |||||
| 46 | EPI_ISL_2757752 | B.1.351 | NSP2: T85I, R121QNSP3: G307V, S794L, K837N, L1515PNSP4: F255SNSP5: K90RNSP12b: P314LNSP13: Q194P | D80A, D215G, K417N, E484K, N501Y, D614G, A701V | Q57H, W69L, S171L | G30R, T205I | P71L | |||||
| 47 | EPI_ISL_2757753 | B.1.351 | NSP2: T85INSP3: P395S, S794L, K837NNSP5: K90RNSP6: N40INSP12b: P314LNSP14: K34Q | D80A, D215G, K417N, E484K, N501Y, D614G, A701V | Q57H, S171L | G30R, T205I | P71L | |||||
| 48 | EPI_ISL_2757754 | B.1.351 | NSP2: T85INSP3: S794L, K837NNSP4: S432GNSP5: K90RNSP6: C221GNSP10: T102INSP12b: P314L | D80A, D215G, K417N, E484K, N501Y, D614G, A701V | Q57H, S171L | G30R, T205I | P71L | |||||
| 49 | EPI_ISL_2757755 | B.1.351 | NSP2: T85INSP3: S794L, K837N NSP5: K90RNSP12b: P314L | D80A, D215G, K417N, E484K, N501Y, D614G, A701V | Q57H, S171L | G30R, T205I | P71L | |||||
| 50 | EPI_ISL_2757740 | B.1.351 | NSP2: T85I, K111ENSP3: S794L, K837NNSP5: K90RNSP12b: P314L | D80A, D215G, K417N, E484K, N501Y, D614G, A701V | Q57H, S171L | G30R, T205I | P71L | |||||
| 51 | EPI_ISL_2438665 | B.1.351 | NSP2: T85INSP3: S794L, K837NNSP5: K90RNSP6: S106, A161V NSP12b: P314L | D80A, D215G,L242,K417N, E484K, N501Y, D614G, A701V | Q57H, S60, S171L | T205I, T362I | P71L | |||||
| 52 | EPI_ISL_2434247 | B.1.351 | NSP2: T85INSP3: S794L, K837NNSP5: K90RNSP6: S106NSP12b: P314L | D80A, D215G,L242,K417N, E484K, N501Y, D614G, A701V | Q57H, S171L | G30R, T205I | P71L | W29L | ||||
| 53 | EPI_ISL_2894974 | B.1.617.2 | NSP2: P129LNSP3: P822LNSP4: A446VNSP6: V149ANSP12b: P314L, G662S NSP13: V169F, G439E NSP15:H234Y | T19R,E156,L452R, T478K, D614G, P681R, D950N | S26L | D63G, R203M, D377Y, R385K | I82T | V82A, T120I | A43V | D119 | K42E | |
| 54 | EPI_ISL_2313086 | B.1.617.2 | NSP3: A488S, P1228L NSP4: V167L, T492INSP6: T77ANSP12b: P314L, G662S, K774NNSP13: P77LNSP14:A394V | T19R,L452R, T478K, D614G, P681R, D950N | S26L | D63G, R203M, G215C, D377Y | I82T | V82A, T120I | T40I | D120 | ||
| 55 | EPI_ISL_2894982 | B.1.617.2 | NSP2: P129L, V447FNSP3: P822LNSP4: A446VNSP6: V149ANSP12b: P314L, G662S NSP13: P77LNSP14:P46L | T19R,E156,G142D, L452R, T478K, E484Q, D614G, P681R, D950N | S26L | D63G, D377Y | I82T | V82A, L116F, T120I | D119 | |||
| 56 | EPI_ISL_2894983 | B.1.617.2 | NSP2: P129L, V447FNSP3: P822LNSP4: A446VNSP6: V149ANSP12b: P314L, G662S NSP13: P77LNSP14:P46L | T19R,E156,L452R, D614G, P681R | S26L | D63G, R203M, D377Y | I82T | V82A, L116F, T120I | D119 | |||
| 57 | EPI_ISL_2894977 | B.1.617.2 | NSP2: P129LNSP3: P822LNSP4: A446VNSP6: V149ANSP12b: G662SNSP13: P77L, V169F NSP15: H234Y | L5F,T19R,E156,L452R, T478K, D614G, P681R | S26L | D63G, R203M, D377Y, R385K | I82T | N52K, V82A, L116F | D119 | K42E | ||
| 58 | EPI_ISL_2757734 | B.1.617.2 | NSP2: K500NNSP3: A488S, P1228L, P1469SNSP4: V167L, T492INSP6: T77ANSP12b: P314L, G662S NSP13: P77LNSP14: A394V | T19R,F106L, G142D, L452R, T478K, D614G, P681R, D950N | S26L | D63G, R203M, M210I, G215C, D377Y | I82T | V82A, T120I | T40I |
The mutational analysis revealed that all the beta variant isolates reported significant and lineage defining mutations as: ORF1a:K1655N (NSP3: K837N) (5230 G>T), S:D80A (21801 A>C), S:D215G (22206 A>G), S:K417N (22813 G>T), S:E484K (23012 G>A), S:N501Y (23063 A>T), S:A701V (23664 C>T), E:P71L (26456 C>T) and N:T205I (28887 C>T). Corresponding to the person who traveled back to Pakistan from UAE reported several rare missense mutations in the spike region, A27S (21641 G>T), G181V (22104 G>T), ORF1b region, L820F (NSP12b: L820F) (15925 C>T), ORF3a region, T271I (26204 C>T) and N region, K387N (29434 G>T) (GISAID ID: EPI_ISL_2438683). Three rare mutations in the ORF1a region, T183I (NSP2: T3I) (813 C>T), S3099L (NSP4: S336L) (9561 C>T), and L3915F (NSP7: L56F) (12008 C>T) were observed in one of the virus isolates (GISAID ID: EPI_ISL_2313081). Furthermore, rare spike mutations, A879S (24197 G>T) and D1163Y (25049 G>T), were found in two patients (GISAID IDS: EPI_ISL_2894979 and EPI_ISL_2313084) and a unique mutation, K444R (22893 A>G), in a 26 years old male patient (GISAID ID: EPI_ISL_2757749). The gamma variants harbors non‐synonymous lineage defining mutations, ORF1a: K1795Q (NSP3: K977Q) (A5648C), ORF1a: del:11288:9 (NSP6: S106), S:T20N (C21621A), S:R19S (G22132T), S:K417T (A22812C), S:E484K (G23012A), S:N501Y (A23063T), S:H655Y (C23525T), S:T1027I (C24642T), ORF8:E92K (G28167A) and N:P80R (C28512G).
To infer the origin of Pakistani isolates, we built a maximum likelihood phylogenetic tree using 286 full‐length genomes of SARS‐CoV‐2. In phylogenetic tree, the VOCs detected in the current study showed close similarities with isolates originating from Asia, Europe, and North America. Two beta variant infected patients (GISAID ID: EPI_ISL_2438665 and EPI_ISL_2438683) having a travel history from UAE, clustered with Bangladesh, India, and United Kingdom viral isolates. The delta variant (GISAID ID: EPI_ISL_2434174, having travel history of Saudi Arabia) clustered with viruses from Bangladesh, Nepal and England. Moreover, delta variant isolates with travel history of UAE (GISAID ID: EPI_ISL_2438666) and Afghanistan (GISAID ID: EPI_ISL_2757757 and GISAID ID: EPI_ISL_2757756) showing close homology with strains from the United States, Nepal, and India. Similarly, the delta variant detected in a traveler from Oman (EPI_ISL_2894982) clustered with viral strains from the United Kingdom, India, and United States. The one case of gamma variant (GISAID ID: EPI_ISL_2894980) grouped with Italian viral isolates (Figure 4). These results suggest the probable introduction of VOCs through inbound travelers in Pakistan from different countries.
Figure 4.
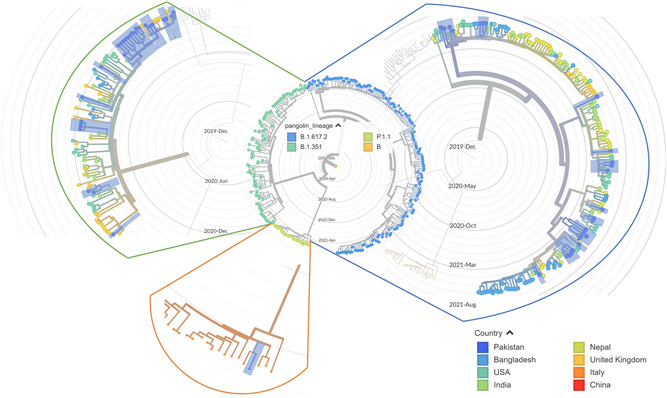
Phylogenetic distribution of beta, gamma, and delta lineages in 286 SARS‐CoV‐2 viral genomes around the world, including current study sequences from Pakistan with reference to Wuhan/Hu‐1/2019 (GISAID ID: EPI_ISL_402125). The maximum likelihood phylogenetic tree was constructed using Nextstrain's Augur tree implementation pipeline and the default parameters were used for IQ‐TREE. The time‐resolved phylogenetic tree with selected metadata information was constructed using TreeTime and visualized in Auspice
4. DISCUSSION
The COVID‐19 pandemic has affected every continent, resulting in a global health crisis that has been aggravated by emergence of different variants of the virus. Amongst the SARS‐CoV‐2 variants of concern, B.1.1.7 (Alpha) became the predominant lineage worldwide after the initial detection from UK in September, 2020. The first case of alpha variant from Pakistan was reported in December, 2020 26 followed by a rapid spread of this infectious variant in indigenous population 27 that triggered a spike of cases in March, 2021 leading to the third wave. Although the prevalence and sustained circulation of alpha variant in Pakistan is well established, data on the presence and genomic diversity of other lineages and VOCs is limited mainly due to the lack of genomic surveillance. Therefore, the current study aimed to detect and explore the genomic diversity of SARS‐CoV‐2 variants (other than B.1.1.7) prevalent in Pakistan during the third wave. Our results showed a high percentage of delta (B.1.617.2) (n = 26) and beta (B.1.351) (n = 27) variants among the samples selected for whole‐genome sequencing from May 01 to June 09, 2021 and serve as the first report on the detection and exploration of genomic diversity of delta variant from Pakistan. The delta variant, which was first identified in India during September, 2020, caused a ferocious second wave in country with 414 188 cases reported on May 06, 2021. As of July 31, 2021, the delta variant has been linked to the recent surge in COVID‐19 cases in regional countries such as Nepal, Bangladesh, Russia, and Indonesia with the reported prevalence of 81%, 64%, 56%, and 32%, respectively. 28 Similarly, UK has witnessed a rise in cases due to delta variant during the first 3 weeks of June, 2021 with health authorities fearing a possible third wave. The first case of delta variant from Pakistan (detected in the current study) was a 39‐year‐old male from Islamabad whose sample was collected on May 16, 2021 and sent to NIH for laboratory testing. Additionally, 15 cases of delta variant were detected from Islamabad and one each from Azad Kashmir and Rawalpindi highlighting the need of large‐scale genomic surveillance. Additionally, the beta variant was discovered for the first time in October, 2020 in Nelson Mandela Bay metropolitan area of South Africa's Eastern Cape Province. As of July 30, 2021, there were reports of beta resurgence in France and Spain, with 14.2% of Spanish and 1.9% of French submissions to GISAID in the last 4 weeks. 29 Recently, the beta variant was responsible for causing a second wave in Bangladesh with highest number of cases (7626) reported on April 7, 2021 and 112 deaths on April 19, 2021. The beta variant is also linked to recent surges in countries such as Botswana (49%), Philippines (41% prevalence), Qatar (24% prevalence), Malaysia (22% prevalence), and Turkey (14% prevalence) according to GISAID data as of July 31, 2021. In the current study, for the first time, we had found high number of beta variant cases mainly from Islamabad and Karachi. These findings highlight the hotspot locations and suggests possible upsurge of beta cases along with delta ones.
In the current study, we have identified 11 cases of inbound travelers from Afghanistan, UAE, Oman, Saudi Arabia, Bahrain, and Italy with delta, beta, and gamma variants of SARS‐CoV‐2 despite the entry restrictions from Bangladesh, India, Brazil, South Africa, and Iran. The National Command and Operation Center (NCOC) has been regularly updating the list of countries with travel restrictions. On April 12, 2021 the NCOC, on the basis of epidemiological risk and emergence of mutants, released the list of 23 countries with restricted entry in Pakistan and later on June 12, 2021 with an update of 26 countries (including, Iran, Indonesia, Iraq, and Sri Lanka). Keeping in view of importations of new SARS‐CoV‐2 variants through travelers, the health ministry has released guidelines for the travelers with mandatory rapid antigen testing for all the passengers entering in Pakistan. Those with the positive antigen test will have to undergo PCR testing. Pakistan not only have air travelers but also frequent “on ground border movement” through Afghanistan and Iran borders. The presence of delta variant in travelers from Afghanistan gives an indication that viral movement is taking place and can potentially be one of the major source of influx of VOCs and VOIs of SARS‐CoV‐2. Rapid antigen test is now mandatory for all the pedestrian inbound travelers as per the new policy made by the NCOC on May 5, 2021. Despite the testing facilities at all the entry points, new variants of SARS‐CoV‐2 are finding their way in entry and circulation in Pakistani population. This demands for effective implementation of vigilant antigen testing protocols at all port of entries across the country. Keeping in view the genomic surveillance is critical in mapping the transmission portfolio across borders, the samples from positive SARS‐CoV‐2 travelers must undergone whole genome sequencing to identify the VOIs and VOCs. This will ultimately help to devise strategies of viral containment.
For the first time in Pakistan, we have also found one significant mutation E484Q in a 52‐year‐old male patient infected with delta variant (B.1.617.2), traveling back from Bahrain. The E484Q mutation has a prevalence of only 0.1% in B.1.617.2 as of July 31, 2021. The viruses with the mutations, L452R and E484Q, are more resistant to monoclonal antibodies, including bamlanivimab, and convalescent plasma, 30 and also have a role in increased transmissibility due to enhanced ACE2 binding based on the structural impact of these mutations in the furin cleavage site. 14 Moreover, in one female patient, a less prevalent spike mutation (about 0.5%), L5F was observed. This mutation in combination with D614G had demonstrated increased infectivity and enhanced transmissibility. 31 In case of beta variant, one of the patient is having a travel history of the Middle East carried two rare non‐synonymous spike mutations, A27S and G181V, having a global prevalence of 5.7% and 0.1%, respectively according to GISAID data (July 31, 2021). Furthermore, in two patients a rare spike mutation (about 1%), A879S, was reported. This mutation along with D614G had been found to decrease the sensitivity of convalescent sera and thus more likely to evade immune responses. 31 In one of the sample, K444R mutation was observed that exhibit increased binding affinity to the human ACE2 receptor based on an in silico study. 32 These findings emphasize the critical nature of continuous monitoring of amino acid changes in the spike region for vaccine development and therapeutic antibodies.
It is also critical to contain the variant spread and build national immunity through the vaccine rollout. Pakistan is currently lagging behind other countries in the global campaign for mass vaccination against COVID‐19. As of July 31, 2021, only 6.31 million people are fully vaccinated that comprises 2.9% of the total population and 23.3 million people are partially vaccinated that comprises about 10.8% of the total population. 33 Currently, in Pakistan, mostly Chinese vaccines, that is, Sinopharm, Cansino Bio, and Sinovac are being administered. The efficacy of these vaccines against beta and delta variants has not been reported so far. In many countries, however, there is a growing concern regarding their efficacy against delta variants owing to recent worldwide outbreaks and vaccine breakthrough cases in fully vaccinated people. Indonesia recently reported the death of 131 health care workers who were mostly inoculated with Sinovac. 34 This raises the serious question of whether the vaccines used in the nationwide campaign can contain any future upsurge of COVID‐19 cases due to the emergence of immune‐evading variants. To address this concern, several countries have announced the use of other vaccines for booster doses. Though conclusive evidence supporting the need for so‐called “booster” shots has yet to emerge, the health officials from Thailand to Bahrain and the United Arab Emirates have announced that they will offer the additional doses to some people already immunized with vaccines manufactured by Chinese manufacturers and AstraZeneca. 35 Such booster doses will also be a challenge for developing countries like Pakistan where vaccination rate is already very low and the priority is to vaccinate the vulnerable/susceptible population first.
Pakistan has confronted a 4‐month‐long (March–June 2021) third wave of COVID‐19 with the highest positivity rate reaching 11.6% in April, then declined to 2.35% in June and had recently started to rise again to 7.73% as of July 31, 2021, showing early signs of possible fourth wave. 36 In conclusion, the high prevalence of beta and delta variants reported in our study coupled with limited sequencing capacity (less than 1% of the total reported cases), lower vaccination rates against COVID‐19, less efficient screening and quarantine protocols of inbound travelers, and ease in lockdown restrictions, that is, the opening of restaurants, tourism sector, educational institutes, outdoor marriage ceremonies starting from May 24, 2021, 37 suggests a possible increase in the number of cases in the near future and provides an early warning to national health authorities to take timely decisions and devise suitable interventions to contain the possible fourth wave.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Conceptualization: Massab Umair, Aamer Ikram, and Muhammad Salman; Methodology: Massab Umair, Muhammad Suleman Rana, Nazish Badar, Muhammad Ammar, Qasim Ali. Formal analysis: Massab Umair, Syed Adnan Haider, Zaira Rehman, and RMuhammad Salman. Resources: Massab Umair, Aamer Ikram, and Muhammad Salman; Writing—original draft preparation: Massab Umair, Zaira Rehman, and Syed Adnan Haider. Writing—review and editing: Massab Umair, Aamer Ikram, Muhammad Salman, RMuhammad Salman, Nazish Badar. All authors have read and agreed to the published version of the manuscript.
Umair M, Ikram A, Salman M, et al. Genomic surveillance reveals the detection of SARS‐CoV‐2 delta, beta, and gamma VOCs during the third wave in Pakistan. J Med Virol. 2022;94:1115‐1129. 10.1002/jmv.27429
DATA AVAILABILITY STATEMENT
All the sequences generated in the current study are submitted to the GISAID under the accession numbers: EPI_ISL_2894980, EPI_ISL_2438599, EPI_ISL_2438598, EPI_ISL_2313082, EPI_ISL_2314809, EPI_ISL_2894979, EPI_ISL_2313098, EPI_ISL_2313099, EPI_ISL_2313112, EPI_ISL_2313114, EPI_ISL_2314185, EPI_ISL_2313081, EPI_ISL_2313084, EPI_ISL_2894975, EPI_ISL_2894976, EPI_ISL_2894978, EPI_ISL_2438665, EPI_ISL_2438683, EPI_ISL_2434247, EPI_ISL_2757747, EPI_ISL_2757753, EPI_ISL_2757748, EPI_ISL_2757749, EPI_ISL_2757750, EPI_ISL_2757751, EPI_ISL_2757752, EPI_ISL_2757754, EPI_ISL_2757745, EPI_ISL_2757746, EPI_ISL_2757735, EPI_ISL_2757740, EPI_ISL_2757755, EPI_ISL_2313086, EPI_ISL_2434976, EPI_ISL_2434982, EPI_ISL_2894974, EPI_ISL_2894977, EPI_ISL_2434174, EPI_ISL_2894982, EPI_ISL_2438666, EPI_ISL_2894983, EPI_ISL_2438547, EPI_ISL_2434781, EPI_ISL_2757743, EPI_ISL_2757744, EPI_ISL_2757734, EPI_ISL_2757736, EPI_ISL_2757737, EPI_ISL_2757738, EPI_ISL_2757739, EPI_ISL_2757741, EPI_ISL_2757757, EPI_ISL_2757758, EPI_ISL_2757759, EPI_ISL_2757760, EPI_ISL_2757761, EPI_ISL_2757762, EPI_ISL_275775
REFERENCES
- 1. van Oosterhout C, Hall N, Ly H, Tyler KM. COVID‐19 Evolution During the Pandemic–Implications of New SARS‐CoV‐2 Variants on Disease Control and Public Health Policies. Taylor & Francis; 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rahimi A, Mirzazadeh A, Tavakolpour S. Genetics and genomics of SARS‐CoV‐2: A review of the literature with the special focus on genetic diversity and SARS‐CoV‐2 genome detection. Genomics. 2020;113:1221‐1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. Tracking SARS‐CoV‐2 variants. Accessed July 31, 2021. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
- 4. Nelson G, Buzko O, Spilman P, et al. Molecular dynamic simulation reveals E484K mutation enhances spike RBD‐ACE2 affinity and the combination of E484K, K417N and N501Y mutations (501Y. V2 variant) induces conformational change greater than N501Y mutant alone, potentially resulting in an escape mutant. BioRxiv. 2021. 10.1101/2021.01.13.426558 [DOI] [Google Scholar]
- 5. Liu Y, Liu J, Plante KS, et al. The N501Y spike substitution enhances SARS‐CoV‐2 transmission. BioRxiv. 2021. 10.1101/2021.03.08.434499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Starr TN, Greaney AJ, Hilton SK, et al. Deep mutational scanning of SARS‐CoV‐2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182(5):1295‐1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gu H, Chen Q, Yang G, et al. Adaptation of SARS‐CoV‐2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369(6511):1603‐1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wibmer CK, Ayres F, Hermanus T, et al. SARS‐CoV‐2 501Y. V2 escapes neutralization by South African COVID‐19 donor plasma. Nature Med. 2021;27(4):622‐625. [DOI] [PubMed] [Google Scholar]
- 9. Greaney AJ, Starr TN, Gilchuk P, et al. Complete mapping of mutations to the SARS‐CoV‐2 spike receptor‐binding domain that escape antibody recognition. Cell Host Microbe. 2021;29(1):44‐57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Greaney AJ, Loes AN, Crawford KHD, et al. Comprehensive mapping of mutations to the SARS‐CoV‐2 receptor‐binding domain that affect recognition by polyclonal human serum antibodies. bioRxiv. 2021;31:425021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu K, Werner AP, Moliva JI, et al. mRNA‐1273 vaccine induces neutralizing antibodies against spike mutants from global SARS‐CoV‐2 variants. BioRxiv. 2021. 10.1101/2021.01.25.427948 [DOI] [Google Scholar]
- 12. Starr TN, Greaney AJ, Dingens AS, Bloom JD. Complete map of SARS‐CoV‐2 RBD mutations that escape the monoclonal antibody LY‐CoV555 and its cocktail with LY‐CoV016. Cell Rep Med. 2021;2(4):100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zahradnik J, Marciano S, Shemesh M, et al. SARS‐CoV‐2 RBD in vitro evolution follows contagious mutation spread, yet generates an able infection inhibitor. BioRxiv. 2021. 10.1101/2021.01.06.425392 [DOI] [Google Scholar]
- 14. Cherian S, Potdar V, Jadhav S, et al. Convergent evolution of SARS‐CoV‐2 spike mutations, L452R, E484Q and P681R, in the second wave of COVID‐19 in Maharashtra, India. BioRxiv. 2021. 10.1101/2021.04.22.440932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.COVID‐19 Pakistan Cases. Government of Pakistan. Accessed July 31, 2021. https://covid.gov.pk/stats/pakistan
- 16. Quick J. nCoV‐2019 sequencing protocol v2 (GunIt) V. 2. Protocols. io; 2020. Accessed February 3, 2021.
- 17. Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010; 2017.
- 18. Li H, Durbin R. Fast and accurate long‐read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26(5):589‐595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Institute B . Picard tools; 2016, Broad Institute, GitHub repository.
- 20. Paden CR, Tao Y, Queen K, et al. Rapid, sensitive, full‐genome sequencing of severe acute respiratory syndrome coronavirus 2. Emerging Infect Dis. 2020;26(10):2401‐2405. https://wwwnc.cdc.gov/eid/article/26/10/20-1800_article [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. O'Toole A, Scher E, Underwood A, et al. Pangolin: lineage assignment in an emerging pandemic as an epidemiological tool; 2020.
- 22. Huddleston J, Hadfield J, Sibley T, et al. Augur: a bioinformatics toolkit for phylogenetic analyses of human pathogens. J Open Source Softw. 2021;6(57):2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059‐3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nguyen L‐T, Schmidt HA, von Haeseler A, Minh BQ. IQ‐TREE: a fast and effective stochastic algorithm for estimating maximum‐likelihood phylogenies. Mol Biol Evol. 2015;32(1):268‐274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sagulenko P, Puller V, Neher RA. TreeTime: maximum‐likelihood phylodynamic analysis. Virus Evol. 2018;4(1):vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Umair M, Ikram A, Salman M, et al. Importation of SARS‐CoV‐2 variant B. 1.1. 7 in Pakistan. J Med Virol. 2021;93:2623‐2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Umair M, Salman M, Rehman Z, et al. Proliferation of SARS‐CoV‐2 B. 1.1. 7 variant in Pakistan—a short surveillance account. Front Public Health. 2021;9:702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Latif AA, Mullen JL, Alkuzweny M, et al. and the Center for Viral Systems Biology. outbreak.info; 2021. Accessed 22 July 2021. https://outbreak.info/situation-reports?pango=B.1.617.2%26loc=NPL%26loc=BGD%26loc=RUS
- 29.COVID: the beta variant is surging in mainland Europe—should the UK be worried?; 2021. Accessed July 30, 2021. https://theconversation.com/covid-the-beta-variant-is-surging-in-mainland-europe-should-the-uk-be-worried-164815
- 30. Gottlieb RL, Nirula A, Chen P, et al. Effect of bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID‐19: a randomized clinical trial. JAMA. 2021;325(7):632‐644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li Q, Wu J, Nie J, et al. The impact of mutations in SARS‐CoV‐2 spike on viral infectivity and antigenicity. Cell. 2020;182(5):1284‐1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ortega JT, Pujol FH, Jastrzebska B, Rangel HR. Mutations in the SARS‐CoV‐2 spike protein modulate the virus affinity to the human ACE2 receptor, an in silico analysis. EXCLI J. 2021;20:585‐600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.National Command and Operation Center (NCOC), G.o.P. Accessed July 31, 21. https://ncoc.gov.pk/covid-vaccination-en.php
- 34.COVID infections imperil Indonesia's vaccinated health workers, and hospitals. REUTERS. https://www.reuters.com/world/asia-pacific/covid-infections-imperil-indonesias-vaccinated-health-workers-hospitals-2021-07-07/, Accessed July 23, 2021.
- 35.COVID vaccine plans tweaked around the world in attempt to boost effectiveness against Delta variant. Accessed July 23, 2021. https://www.abc.net.au/news/2021-07-22/countries-change-vaccine-advice-to-combat-delta-sinovac-pfizer/100307864
- 36.COVID19 Dashboard for Pakistan. Ministry of Health Services & Regulation. https://covid.gov.pk/
- 37.NCOC allows staggered reopening of educational institutes, outdoor dining from May 24, DAWN. https://www.dawn.com/news/1624532
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the sequences generated in the current study are submitted to the GISAID under the accession numbers: EPI_ISL_2894980, EPI_ISL_2438599, EPI_ISL_2438598, EPI_ISL_2313082, EPI_ISL_2314809, EPI_ISL_2894979, EPI_ISL_2313098, EPI_ISL_2313099, EPI_ISL_2313112, EPI_ISL_2313114, EPI_ISL_2314185, EPI_ISL_2313081, EPI_ISL_2313084, EPI_ISL_2894975, EPI_ISL_2894976, EPI_ISL_2894978, EPI_ISL_2438665, EPI_ISL_2438683, EPI_ISL_2434247, EPI_ISL_2757747, EPI_ISL_2757753, EPI_ISL_2757748, EPI_ISL_2757749, EPI_ISL_2757750, EPI_ISL_2757751, EPI_ISL_2757752, EPI_ISL_2757754, EPI_ISL_2757745, EPI_ISL_2757746, EPI_ISL_2757735, EPI_ISL_2757740, EPI_ISL_2757755, EPI_ISL_2313086, EPI_ISL_2434976, EPI_ISL_2434982, EPI_ISL_2894974, EPI_ISL_2894977, EPI_ISL_2434174, EPI_ISL_2894982, EPI_ISL_2438666, EPI_ISL_2894983, EPI_ISL_2438547, EPI_ISL_2434781, EPI_ISL_2757743, EPI_ISL_2757744, EPI_ISL_2757734, EPI_ISL_2757736, EPI_ISL_2757737, EPI_ISL_2757738, EPI_ISL_2757739, EPI_ISL_2757741, EPI_ISL_2757757, EPI_ISL_2757758, EPI_ISL_2757759, EPI_ISL_2757760, EPI_ISL_2757761, EPI_ISL_2757762, EPI_ISL_275775


