Like most vaccines, the effectiveness of COVID‐19 vaccines developed so far is not 100% and a small percentage of fully vaccinated individuals still develop symptomatic or asymptomatic SARS‐CoV‐2 infections. 1 In addition, the emerging of novel mutations is likely to help the virus evade vaccines especially in regions with low vaccination coverage. 2
In this study, we analyzed SARS‐CoV‐2 infections in two healthcare workers (HCWs) fully vaccinated with the Pfizer‐BioNTech COVID‐19 vaccine (BNT162b2) and subsequently diagnosed with postvaccination SARS‐CoV‐2 on a background of IgG antibodies.
Epidemiological and clinical data including, symptoms initial analysis and observations were obtained from the National electronic system for COVID‐19 (Table 1).
Table 1.
SARS‐CoV‐2 Alpha (B.1.1.7) postvaccination infections among two healthcare workers, March 08–18, 2021
| Patients (SARS‐CoV‐2 sequence GISAID accession IDs) | Case 1 (Sequence ID 1805746) | Case 2 (Sequence ID 1805747) |
|---|---|---|
| Age | 43 | 44 |
| Sex | F | F |
| Hospital name | UOGHa | UH‐ISULb |
| Position in the hospital | Midwife | Doctor |
| Healthcare ward | COVID‐19 | Endocrinology and metabolic diseases |
| Date of the first vaccine | Dec 13, 2020 | Jan 20, 2021 |
| Date of the second vaccine | Jan 07, 2021 (25 days after the first vaccine) | Feb 10, 2021 (21 days after the first vaccine) |
| Test method for detecting anti ‐ SARS‐CoV‐2 antibodies | BIOMERIEUX, The VIDAS® SARS‐COV‐2 IgG ELFA (Enzyme‐Linked Fluorescent Assay) | Abbott, SARS‐CoV‐2 II Quant Abbott Architect, for the qualitative and quantitative determination of IgG antibodies to SARS‐CoV‐2 |
| Date of test for anti ‐ SARS‐CoV‐2 antibodies | Mar 18, 2021 (70 days after the second vaccine) | Mar 5, 2021 (23 days after the second vaccine) |
| SARS‐CoV‐2 antibodies after the second vaccination. Test values and limit values | 24, limit >1 | 22 495 AU/ml, limit >50 |
| Date of the first positive real‐time PCR test for SARS‐CoV‐2 after the second vaccination | Mar 18, 2021 (70 days after second vaccination) | Mar 8, 2021 (26 days after second vaccination) |
| Real‐time PCR test for SARS‐CoV‐2 results | Positive (gen E ‐ 25,20; gen N ‐ 25,73; RdRp ‐ 24,92) | Positive (ORF 1ab ‐ 25,39; gen E ‐ 24,55; gen N ‐ 23,79) |
| Indication for testing | Symptoms | Contact with positive patients |
| Presumed exposure source | Family member | Patients |
| Date of obtaining symptoms after the second vaccination | Mar 18, 2021–Apr 04, 2021 (70 days after the second vaccine) | No symptoms |
| Clinical symptoms | Severe persistent cough, runny nose, fatigue, headache, fever 37.5, nausea | No symptoms |
| Mutations in Gene: | None‐synonymous substitutionsc | |
| ORF1a | T1001I, A1708D, I2230T, S3675_F3677del (TCTGGTTTT) | T1001I, A1708D, I2230T, S3675_F3677del (TCTGGTTTT) |
| Gene: ORF1b | P314L, V1092F | P314L, K1383R, I2166L |
| Gene: S | H69_V70del (ACATGT), Y144del (TAT), N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | H69_V70del (ACATGT), Y144del (TAT), N501Y, A570D, D614G, P681H, T716I, S982A, D1118H |
| Gene: ORF3a | A54S | Y145F |
| Gene: ORF7a | T111I | |
| Gene: ORF8 | Q27* (CAA27TAA), R52I, Y73C | Q27*(CAA27TAA), R52I, Y73C |
| Gene: N | D3L, R203K, G204R, S235F | D3L, R203K, G204R, S235F |
| Gene: ORF14 | G50N | G50N |
University Obstetrics and Gynecology Hospital “Mother's Home”, Sofia, Bulgaria.
University Hospital “Tsaritsa Yoanna” ‐ ISUL, Sofia, Bulgaria.
Bolded nonsynonymous substitutions indicate notable mutations and deletions within the S gene‐specific to SARS‐CoV‐2 alpha (B.1.1.7) lineage.
Blood samples of the HCWs were tested for anti‐SARS‐CoV‐2 IgG antibodies. The analysis was performed using two different serological tests: The VIDAS® SARS‐COV‐2 IgG Enzyme‐Linked Fluorescent Assay (Biomerieux) and SARS ‐CoV‐2 II Quant Abbott Architect (Abbott).
Viral RNA was extracted from nasal swabs using an ExiPrep 48 Viral DNA/RNA Kit (Bioneer) following the manufacturer's instructions. Real‐time PCR was performed using GeneFinder™ COVID‐19 Plus RealAmp Kit (OSANG Healthcare Co., Ltd.).
Whole‐genome next‐generation sequencing (NGS) of SARS‐CoV‐2 was performed by using a modified ARTIC v3 tailed amplicon method and Illumina MiSeq v2 reagent kit with 500 cycles (Illumina).
Pangolin COVID‐19 Lineage Assigner Tool v3.1.7. was used to define the variant classification. 3
The dataset for the phylogenetic analysis contained sequences from both samples under investigation together with 112 other randomly selected SARS‐CoV‐2 sequences isolated in Bulgaria and the reference sequence employed by GISAID (EPI_ISL_402124).
Sequence alignments were performed using MAFFT version 7. 4 All Bulgarian SARS‐CoV‐2 sequences were deposited in GISAID databases, the dataset and sequence accession IDs are available upon request.
The potential phylogenetic relationship of the SARS‐CoV‐2 S gene clades was evaluated by approximate maximum‐likelihood phylogenies using the GTR nucleotide substitution model in FastTree v2.1.10. 5
Nonsynonymous mutations were defined by the Internet available Genome Detective Coronavirus Typing Tool. 6
Case 1 is a 43‐year‐old woman, working in the COVID‐19 ward during the second pandemic wave in the country, and Case 2 is a 44‐year‐old woman, who works in the Endocrinology and metabolic diseases ward, both with no evidence of chronic illness, see Table 1.
Vaccination in each individual was performed according to the manufacturer's recommendations, none of them was hospitalized and both have since fully recovered.
Alpha (B.1.1.7) variant was identified in both cases which is consistent with the widespread circulation (over 99%) of this lineage in Bulgaria at the time of patient identification.
The phylogenetic analysis revealed that the sequences isolated from the postvaccination SARS‐CoV‐2 infection were phylogenetically similar and fall into the branch formed by the largest number of sequences, see Figure 1.
Figure 1.
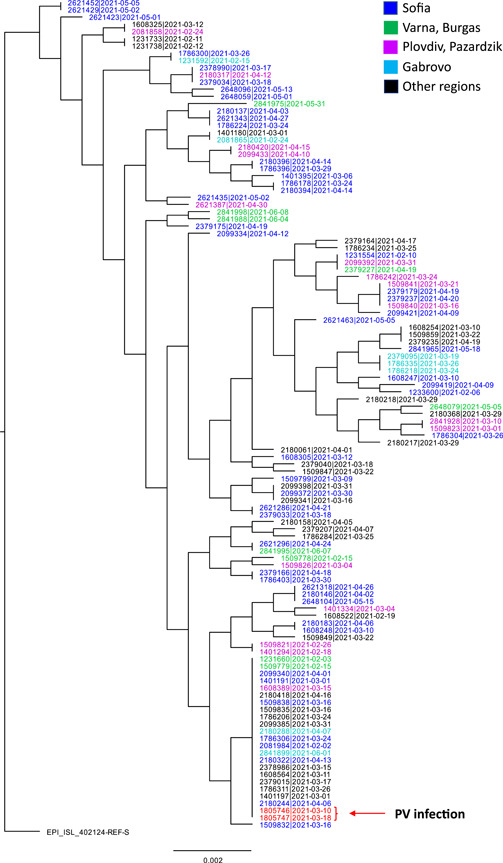
Approximate maximum likelihood (ML) phylogenies reconstructed with 115S gene sequences isolated in Bulgarian and the reference sequence employed by GISAID (EPI_ISL_402124) using the GTR nucleotide substitution model implemented in FastTree v2.1.10. The red arrow marked “PV infection”, indicates the positions of the two postvaccination sequences marked in red. The different regions of the country (Sofia, Varna and Burgas, Plovdiv, Gabrovo, and other regions) from which the sequences were isolated are marked with different colors
Both postvaccination isolates exhibited 7 nonsynonymous mutations in the spike protein, D614G and 6 substitutions which are the defining SNPs of the alpha linage, including N501Y, A570D, P681H, T716I, S982A, D1118H, see Table 1. In addition, two deletions were found in the spike protein: H69_V70del (ACATGT) which has been reported to be associated with this variant, and Y144del (TAT), both located in the N‐terminal domain.
Notably, both individuals were found positive for IgG antibodies after vaccination before infection, and yet infection was not prevented. None of them were hospitalized; Case 1 with mild symptoms and Case 2 asymptomatic, which is in line with the known high effectiveness of the vaccine against severe COVID‐19 see Table 1. 7 Although the course of the infection was not life‐threatening, it raises questions related to asymptomatic carriers and the possibility of transmitting the infection to other nonimmune individuals especially in the healthcare setting. 8 In addition, the question of whether vaccinated individuals can transmit the infection remains open.
Our analysis indicated that the postvaccination infections were caused by the SARS‐CoV‐2 Alpha variant which was also widely circulating with over 99% of the isolates during the spring 2021 COVID‐19 wave in Bulgaria.
It is interesting to note that the rapid ubiquitous dissemination of alpha lineage across the country is also in parallel with the results of the phylogenetic analysis which indicates that sequences isolated from various geographical locations were dispersed throughout the topology of the entire phylogenetic tree across different branches.
Vaccination is a reliable and effective tool for disease control, however, newly emerging mutations can compromise vaccines and accelerate the spread of the disease. Our findings indicate the need for careful monitoring of the viral genome of the regions targeted by the vaccines and analysis of the postvaccination immunity. These issues draw attention to further research in favor of public health for the rapid response to the COVID‐19 pandemic.
ACKNOWLEDGMENTS
The study was supported by a grant from the Ministry of Education and Science, Bulgaria (contract: KП‐06‐H43/1 27.11.2020); by the European Regional Development Fund through Operational Program Science and Education for Smart Growth 2014–2020, Grant BG05M2OP001‐1.002‐0001‐C04 “Fundamental Translational and Clinical Investigations on Infections and Immunity”.
CONFLICT OF INTERESTS
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Ivailo Alexiev and Ivan Ivanov conceived and designed the study. Ivailo Alexiev and Ivan Ivanov analyzed the data. Ivailo Alexiev and Ivan Ivanov prepared the draft manuscript. Nelly Korsun, Ivan Stoikov, Reneta Dimitrova, Lubomira Grigorova, Ivelina Trifonova, Veselin Dobrinov, Iliana Grigorova, Alexey Savov, Boryana Asenova, Massimo Ciccozzi, and Todor Kantardjiev reviewed the draft and all authors contributed to the final version, which was approved by all authors for submission.
ETHICS STATEMENT
This study was approved by the Ethical Committee at the National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (NCIPD IRB 00006384).
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ivailo Alexiev and Ivan Ivanov equally contributed to the study.
REFERENCES
- 1. Amit S, Beni A, Biber A, Grinberg A, Leshem E, Regev‐Yochay G. Postvaccination COVID‐19 among Healthcare Workers, Israel. Emerg Infect Dis. 2021;27:1220‐1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weisblum Y, Schmidt F, Zhang F, et al. Escape from neutralizing antibodies by SARS‐CoV‐2 spike protein variants. eLife. 2020;9:e61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rambaut A, Holmes EC, O′Toole Á, et al. A dynamic nomenclature proposal for SARS‐CoV‐2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5(11):1403‐1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20:1160‐1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Price MN, Dehal PS, Arkin AP. FastTree 2‐approximately maximum‐likelihood trees for large alignments. PLoS One. 2010;5:e9490. 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cleemput S, Dumon W, Fonseca V, et al. Genome Detective Coronavirus Typing Tool for rapid identification and characterization of novel coronavirus genomes. Bioinform. 2020;36(11):3552‐3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid‐19 vaccine. NEJM. 2020;383:2603‐2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. European Centre for Disease Prevention and Control . Rapid Risk Assessment “Risk related to the spread of new SARS‐CoV‐2 variants of concern in the EU/EEA – first update”, Accessed January 21, 2021. https://www.ecdc.europa.eu/sites/default/files/documents/COVID-19-risk-related-to-spread-of-new-SARS-CoV-2-variants-EU-EEA-first-update.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


