Abstract
During the COVID‐19 pandemic, genetic variants of SARS‐CoV‐2 have been emerging and spreading around the world. Several SARS‐CoV‐2 endemic variants were found in United Kingdom, South Africa, Japan, and India between 2020 and April 2021. Studies have shown that many SARS‐CoV‐2 variants are more infectious than early wild strain and produce immune escape. These SARS‐CoV‐2 variants have brought new challenges to the prevention and control of COVID‐19. This review summarizes and analyzes the biological characteristics of different amino acid mutations and the epidemic characteristics and immune escape of different SARS‐CoV‐2 variants. We hope to provide scientific reference for the monitoring, prevention, and control measures of new SARS‐CoV‐2 variants and the development strategy of the second‐generation vaccine.
Keywords: COVID‐19, immune escape, mutations, SARS‐CoV‐2 variants
1. INTRODUCTION
Over the last two decades, SARS‐CoV‐2 is the third coronavirus known to cause severe acute respiratory disease in humans, following SARS‐CoV in 2003 and MERS‐CoV in 2012. 1 , 2 , 3 Compared to MERS and SARS limited to relatively more minor populations, coronavirus disease 2019 (COVID‐19) caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has affected the whole world, wreaking havoc on healthcare systems and costing millions of lives. 4 , 5 , 6 Critical illness includes acute respiratory distress syndrome, coagulopathies, septic shock and multiple organ injuries, including heart injury, 7 kidney injury, 8 liver injury, 9 and gastrointestinal symptoms. 10 As of September 22, 2021, COVID‐19 spread fast to more than 200 countries, there have been 229 373 963 confirmed cases of COVID‐19, including 4 705 111 deaths (https://www.who.Int/).
SARS‐CoV‐2 has ~30 kb genome and encodes four structural proteins including Spike (S), Envelope (E), Membrane (M), and Nucleocapsid (N) proteins, six accessory proteins open reading frame (ORF) (ORF3a, ORF6, ORF7a, ORF7b, ORF8, and ORF10) and 16 nonstructural proteins (NSP1−NSP16). 11 , 12 The SARS‐CoV‐2 spike protein is cleaved by furin into S1 subunit and S2 subunit. S1 subunit consists of an N‐terminal domain (NTD) and the receptor‐binding domain (RBD), and is responsible for binding to the host‐cell ACE2 receptor. Whereas, the S2 subunit includes the trimeric core of the protein and is responsible for membrane fusion. 13 , 14 The structural proteins constitute the mature virion, whereas the nonstructural proteins of CoVs are indispensable for viral replication and transcription. 15 The substitution, deletion and insertion of amino acid sites, which occurred in spike protein and the ORF of SARS‐CoV‐2, led to many virus variants. Furthermore, these mutations may alter the virus biological characteristics, including increasing transmissibility and generating immune escape from innate or acquired immune responses. 16 , 17
At the end of January 2020, the D614G mutant, which turns Aspartic acid (Asp) into Glycine (Gly) at site 614 in the amino acid sequence of spike, was first discovered in the UK and quickly became the significant epidemic strain in the world and attracted widespread attention. 18 , 19 During the COVID‐19 pandemic, genetic variants of SARS‐COV‐2 have been emerging and spreading around the world. 20 , 21 Therefore, it is significant to understand the biological characteristics of amino acid mutations, the epidemiological characteristics, and vaccine reactivity of new SARS‐CoV‐2 variants for the surveillance, prevention, and control of COVID‐19.
1.1. The epidemiological characteristics of “variant of concern (VOCs)” and their crucial amino acid mutations
The established nomenclature systems for naming and tracking SARS‐CoV‐2 genetic lineages by GISAID, Nextstrain and Pango are currently and will remain in use by scientists (https://covlineages.org/resources/pangolin.html). The SARS‐CoV‐2 variants were classified as VOCs and “Variant of Interest, VOI” by WHO. At present, WHO described four VOCs, namely, Alpha B.1.1.7 (known as 20I/501Y.V1, VOC 202012/01), 22 Beta B.1.351 (known as 501Y.V2), 23 Gamma P.1 (known as 501Y.V3), 24 and Delta B.1.617.2 (known as 478 K.V1). 25
1.1.1. Key mutations related to enhancing the infectivity of VOC
1.1.1.1. D614G
D614G mutation is present in all VOC. In vivo and ex vivo studies found that in the early stages of virus infection, the D614G viruses exhibited significantly faster droplet transmission between hamsters, and the virus with D614G mutation resulted in a 0.5 to twofold higher gene expression than wild strain. 26 The infection efficiency of D614G pseudovirus was reported to be 8–10 fold higher than wild strain. D614G is hypothesized to “shift the RBD to an 'up' conformation, promoting binding with the ACE2 receptor, leading to enhanced virion infectivity.” 27 Moreover, clinical samples infected with D614G mutant had a high titer of SARS‐CoV‐2 RNA. 28 Therefore, both clinical infection studies and animal experiments of pseudovirus infection suggested that the virus with D614G mutation have higher infectivity than wild strain.
1.1.1.2. N501Y
N501Y mutation located in the RBD region and appeared in B.1.17, B.1.351, P1. Structural modeling data showed that RBD with N501Y mutation could form a potential aromatic ring–ring interaction and an additional hydrogen bond with ACE2, and these interactions made the binding tightness of N501Y‐RBD to ACE2 was 10‐fold than wild strain. 29 In addition, N501Y mutation can decrease the polarity of critical residues in RBD, increasing the affinity between RBD and cellular surface ACE2. 30 , 31 Interestingly, the binding affinity of N501Y‐RBD to ACE2 was much higher than K417N/T‐E484K‐N501Y‐RBD. 32 Therefore, N501Y mutation can enhance the binding affinity and tightness of RBD to ACE2, increasing the chance of the virus infecting host cells.
1.1.1.3. L452R
L452R mutation located in the receptor‐binding motif (RBM) region and appeared in B.1.617 lineages and B.1.427/B.1.429. Starr et al. used quantitative deep mutation scanning found that L452R mutation could increase the expression of S protein by 0. 32 times and enhanced the infectivity of the virus. 30 In addition, B.1.167.1 and B.1.167.3 lacks E484K‐N501Y mutation but shows a unique L452R‐E484Q double mutation in RBM of S protein. Using combined structural modeling and biophysical approach, researchers revealed that B.1.167 variants with L452R‐E484Q double mutations possess a stronger binding affinity to the host‐cell receptor ACE2, and has a ability to evade humoral immunity. 33
1.1.1.4. HV69⁃70del
There were multiple amino acid mutations in the NTD of VOC. By analyzing the S gene sequence from December 1, 2019, to October 24, 2020 in the GISAID database, 90% deletions were found in the NTD region in 1108 sequences of the S gene with deletion mutation. 34 HV69‐70del is present in B.1.1.7 and B.1.258, and it had often emerged after some mutations known to increase binding affinity of S protein to the ACE2 receptor or confer immune escape, such as N501Y, N439K, Y453F. 35 Through the pseudovirus model found that B.1.1.7 containing N501Y without HV69‐70del mutation significantly reduced its infectivity. Structural modeling indicated that the B.1.1.7 with HV69‐70del induces more rapid cell–cell fusion and the formation of multi‐nucleated cells. Whereas, repairing these two amino acids can lead to reduced the infectivity of B.1.1.7 and reduced cell‐cell fusion kinetics back to wild strain level. 36
Furthermore, an experiment in vitro indicated that D796H mutation produces immune escape but reduces infectivity. When D796H and HV69‐70del appeared in SARS‐CoV‐2 variants simultaneously, HV69‐70del can compensate for the reduction of infectivity caused by D796H mutation. 37 The above study demonstrated that HV69⁃70del is responsible for increasing the infectivity of B.1.1.7 by increasing the rate of S2/S2 cleavage and cell–cell membrane fusion, and can compensate for the reduction of infectivity caused by immune escape mutations.
1.1.1.5. T478K
Compared to B.1.617.1 and B.1.617.3, B.1.617.2 lacks the E484Q mutation and has a unique T478K mutation in the S protein. An in silico molecular dynamics study on the S protein structure has predicted that the T478K mutation may increase the electrostatic and steric hindrance of the S protein and increase the binding affinity of RBD to cellular surface ACE2. 38
1.1.1.6. P681H/R
The furin protease cleavage sites located between S1 and S2 subunits of S protein of SARS‐CoV‐2, which contain amino acid site: 681–685. The cleavage of this region is key to the entry of the virus into human host cells. P681H is present in the Alpha VOC and the Theta VOI, and P681R is found in the Delta VOC and the Kappa VOI, respectively. Previous studies have noted that P681H and P681R may increase S1/S2 cleavage by furin‐like proteases and enhance virus‐host cell membrane fusion. 39
1.1.2. Ability to spread more quickly
1.1.2.1. B.1.1.7
B.1.1.7 was first detected in New York in November 2020. 22 B.1.1.7 variant had 10 key amino acid mutations accumulated in Spike (S) protein (HV69‐70del, Y144del, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H). 40 Three mutations of B.1.1.7 with the greatest potential to affect the virus's transmissibility are H69‐V70del, N501Y, and P681H. Epidemiological studies and dynamic modeling methods suggested that the transmissibility of the B.1.1.7 in Britain was increased by 43%–90% and became the dominant strain in the United Kingdom. B.1.1.7 was reported in the United States in December 2020, and the transmissibility of B.1.1.7 was 59%–74% higher than wild strain in Denmark, Switzerland, United States. 41 Moreover, the viral loads was higher in B.1.1.7 samples than in non‐B. 1.1.7 samples, with cycle threshold value (Ct) (mean Ct 28.8 vs. 32.0 for B.1.1.7 vs. non‐B.1.1.7, p = 0.0085) and genomic read depth (1280 vs. 831 for B.1.1.7 vs. non‐B.1.1.7, p = 0.0011). 42 (lower Ct values are correlated with larger amounts of virus in the sample).
1.1.2.2. B.1.351
B.1.351 was first detected in Nelson Mandela Bay, South Africa, in early October 2020. 23 One month later, it replaced the circulating viruses and became the dominant strain in South Africa. B.1.351 has several biologically significant mutations in S protein, including D80A, D215G, LLA241‐243del, K417N, E484K, N501Y, D614G, A701V. 40 In Zambia, the number of COVID‐19 patients infected with B1.351 from 44 cases to 700 from December 1–10 to January 1–10, 2021. Another study in South Africa showed that B.1.351 was 50% more transmissible than wild strain and had a higher viral loads in samples infected with B.1.351 variant (Ct < 30). 43 , 44
1.1.2.3. P.1
P.1, a strain of B.1.1.28, was initially detected during routine screening of foreign passengers from Brazil at Haneda Airport in Tokyo, Japan, on January 10, 2021. 24 P.1 has 17 unique gene mutations, and its S protein accumulated 12 amino acid mutations, including L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F. 40 A two‐category dynamical model that integrates genomic and mortality data estimated that P.1 was 1.7–2.4‐times more transmissible than wild strain, with Ct values of E gene and N gene decreased by 1.43 [95% confidence interval [CI]: 0.17–2.60, p = 0.029] and 1.91 (95% CI: 0.49–3.23, p = 0.01), respectively. 24
1.1.2.4. B.1.617.2
Recently, Delta (B.1.617.2) was earliest reported in India but has spread globally. 25 Except for D614G, B.1.617.2 accumulated eight amino acid mutations in the S protein, including T19R, G142D, FR156‐157del, R158G, L452R, T478K, P681R, D950N. 40 Delta has been linked to the second outbreak of COVID‐19 in Nepal, southeast Asia, and South Africa. In the United Kingdom, Delta seems to be around 60% more transmissible than B.1.1.7. In France, the number of COVID‐19 cases infected with Delta increases by 50–150 cases per day, accounting for 2%–4% of the total new cases of COVID‐19. In addition, Delta is rising fast in the United States, particularly in the Midwest and southeast. Using a rapid genotyping test, the genomics company Helix in San Mateo, California has found that the number of COVID‐19 cases caused by B.1.1.7 fell from more than 70% of cases in the end of April to 42% as of mid‐June 2021 in the United States, with the rise of Delta variant driving much of the shift. 45
Delta was first identified in Guangzhou, Guangdong, China, on May 21, 2021. A preprint 46 reported that the time interval from exposure to the first polymerase chain reaction (PCR) positive was 4 days (interquartile range [IQR]: 3.00–5.00) in the Delta epidemic 2021 and 6 days (IQR: 5.00–8.00) during the 2020 epidemic. The relative viral loads of cases infected with the Delta variant (n = 62, Ct = 24.00 for the ORF1ab gene, IQR: 19.00–29.00) were 1260 times higher than wild strain (n = 63, Ct = 34.31 for ORF1ab gene, IQR: 31.00–36.00) when SARS‐CoV‐2 was first detected by PCR. Moreover, 80.65% of samples infected with the Delta variant contained >6 × 105 copies/ml in oropharyngeal swabs when the viruses were first detected, compared to 19.05% of samples infected with wild strain contained >6 × 105 copies/ml. Epidemiological investigation showed that typical clinical symptoms were observed 2–3 days after infection with Delta. The fifth generation of cases emerged just ten days after the first case infected with Delta. Moreover, the basic transmission (Basic reproduction number, R0) was 4.04–5.0 was higher than the wild strain (R0: 2.2–3.77). 47 (The basic transmission value [R0] means that the infected person can transmit the pathogen to several other people). The above results indicated that Delta could be more transmissible during the early stage of the infection and has higher viral loads.
Altogether, these results indicated that multiple amino acid mutations (D614G, N501Y, L452R, P681H/R, T478K, HV69‐70del) which can enhance the infectivity of the virus appeared in the RBD and NTD of S protein of VOC. By increasing the expression of Spike and the interaction forces (aromatic ring–ring interaction, hydrogen bond) bond between spike and host cell ACE2 or increasing the rate of S1/S2 cleavage, these mutations can increase the binding affinity and binding tightness of SARS‐CoV‐2 spike to hACE2 receptor, and leading to enhance the infectivity of VOC variants, as shown in Figures 1 and 2 and Table 1. Compared with the wild strain, the transmission speed and infection rate of VOC variants were increased. In addition, the viral loads in samples infected with VOC variants was higher than wild strain, and the Ct value was less than 30. It is worth noting that Delta may be more transmissible than other VOC variants (B.1.1.7, B.1.351, and P.1), spreading to 54 countries and rapidly replacing the Alpha variant in the United Kingdom 47 , 48 and the United States. 49
Figure 1.
The biological characteristics of key amino acid mutations of Spike in B.1.1.7 and B.1.617.2 variant. Mutations (HV69‐70del, N501Y, D614G, P681H/R, L452R, T478K) could increase the binding affinity and binding tightness of SARS‐CoV‐2 spike to hACE2 receptor, or increase cell‐cell membrane fusion, result in increasing the infectivity of B.1.1.7 and B.1.617.2 variant. 144del and L452R mutations generated resistance to the neutralization activity of mAbs, convalescent plasma, and post‐vaccination serum against B.1.1.7 and B.1.617.2 variant, respectively. del, deletion; FP, fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; IC, intracellular domain; NTD, N‐terminal domain; RBD, receptor‐binding domain; RBM, receptor binding motif; SD1, subdomain 1; SD2, subdomain 2; TD, transmembrane domain
Figure 2.
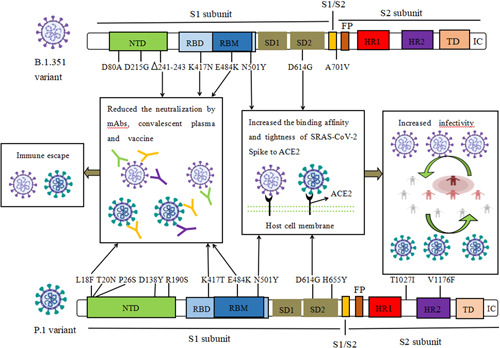
The biological characteristics of key amino acid mutations of Spike in B.1.351 and P.1 variant. Mutations including N501Y, D614G could increase the binding affinity and binding tightness of SARS‐CoV‐2 spike to hACE2 receptor, and result in increasing the infectivity of B.1.1.7 and B.1.617.2 variant. 241‐243del, L18F, K417N/T, E484K mutations generated resistance to the neutralization activity of mAbs, convalescent plasma, and postvaccination serum against B.1.351 and P.1 variant. del, deletion; FP, fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; IC, intracellular domain; NTD, N‐terminal domain; RBD, receptor‐binding domain; RBM, receptor binding motif; SD1, subdomain 1; SD2, subdomain 2; TD, transmembrane domain
Table 1.
The epidemiological characteristics of SARS‐CoV‐2 variants and their immune escape
| WHO label | Alpha | Beta | Gamma | Delta | Lambda |
|---|---|---|---|---|---|
| Pango lineage | B.1.1.7 | B.1.351 | P.1 | B.1.617.2 | C.37 |
| Next strain | S:501Y.V1 | S:501Y.V2 | S:501Y.V3 | S:478K | S:452Q |
| GISAID clade | GR/501Y.V2 | GH/501Y.V2 | GR/501Y.V3 | G/478K.V1 | GR/452Q.V1 |
| Amino acid mutations in the spike protein | HV69‐70del, Y144del, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | D80A, D215G, LLA241‐243del, K417N, E484K, N501Y, D614G, A701V | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F | T19R, G142D, FR156‐157del, R158G, L452R, T478K, D614G, P681R, D950N | G75V, T76I, 247‐252del, L452Q, F490S, D614G, T859N |
| Increased the rate of infection than that of wild strain (References) | Increased by 43%–90% in UK 35 Increased by 59%–74% in Denmark, USA 36 | Increased by 50% in South Africa 39 | Increased by 1.7–2.4‐fold in Brazil 20 | Increased by 60% than B.1.1.7 variant in India 40 | 81% new cases in Peru 89 |
| Increased the rate of hospitalization and mortality than that of wild strain (References) | Increased by 11% for hospitalization, 53 1.4% for ICU 53 and 35% for mortality 51 | Increased by 19.3% for hospitalization 53 and 2.3% for ICU 53 | Increased by 20% for hospitalization 53 and 2.1% for ICU 53 | Increased by 120% for hospitalization,287% for ICU admission and 137% for death 66 | To be confirmed |
| Immune escape | + | ++++ | ++ | ++ | To be confirmed |
Abbreviations: AA, amino acid mutation; del, deletion.
Resistance to partial monoclonal antibodies (mAbs)
Resistance to partial mAbs, convalescent plasma and partial vaccine.
Resistance even immune escape to many mAbs, convalescent plasma and many vaccine.
1.1.3. Non‐spike mutations
Several studies have confirmed that coronavirus accessory proteins play a role in virus‐host interactions, pathogenesis, and virulence. 50 Among the accessory proteins, ORF3a is the largest one containing 274 amino acids in SARS‐CoV. 51 Cell surface localization of ORF3a in SARS‐CoV potentiates viral entry within the host, and ORF3a is also implicated in ion channel formation and modulates the release of virus from the host cell. 51 , 52
1.1.3.1. ORF1ab and ORF3a mutations
Analysis of SARS‐CoV‐2 gene polymorphism found that several independent recurrent mutations in NSP6, NSP7, NSP12, NSP13 encoded by ORF1ab are identified as mutational hotspot are closely associated with inter‐species transmission and virulence. 21 , 53 Several mutations in ORF3a appear in B.1.1.7, B.1.351, P.1, and B.1.167.2. Parinita Majumdar et al. analyzed two group COVID‐19 positive cases with different infection rates and mortality found that ORF3a mutations of SARS‐CoV‐2 are associated with a higher infection and mortality rate. Thirteen different amino acid mutations in ORF3a (P25L, Q57H, K67E, V90F, Y109C, R126T, D142N, W149L, D155Y, Y156N, T176I, T217I, G251V) were deleterious, and these mutations resulted in the loss of predicted motifs and B‐cell epitope as found in wild‐type (WT) ORF3a protein. 53
1.1.4. Higher risk of hospitalization and mortality in patients infected with VOC than wild strain
An initial matched case‐control study from London reported no significant difference in the risk of hospitalization or mortality of cases infected with B.1.1.7 compared to other existing variants. 42 However, several studies have subsequently reported that the risk ratios of hospitalization were from 1.15 to 1.43 for patients infected with B.1.1.7 compared with the non‐B.1.1.7 group. 54 , 55 , 56 Retrospective observational studies performed in the UK reported that the mortality hazard ratio of patients infected with B.1.1.7 was 1.64 (95% CI: 1.32–2.04, p < 0.0001) compared with individuals with non‐B.1.1.7 variant 57 and estimated 35% (12%–64%) increased risk of death associated with B.1.1.7 variant. 58
Both the matched and unmatched multi‐variable analysis found that a more significant proportion of VOC cases were admitted to hospital (B.1.1.7 11.0%; B.1.351 19.3%, and P.1 20.0%; p < 0.001) and ICU admission (B.1.1.7 1.4%, B.1.351 2.3% and P.1 2.1%, p < 0.005) compared with non‐VOC cases (7.5% for hospitalization and 0.6% for ICU admission). 59
For Delta, preliminary evidence from England and Scotland suggested that people infected with Delta are about twice as likely to hospitalization as those infected with B.1.1.7. 45 Patients infected with Delta have an increased risk of hospitalization: hazard ratio (HR) 1.85 (95% CI: 1.39–2.47) compared to B.1.1.7 or ancestral strains in Scotland. 60 Similarly, a retrospective cohort study from Ontario, Canada showed that compared to non‐VOC variants, the adjusted elevation in risk associated with N501Y‐positive VOC variants (B1.1.17, B.1.351, and P.1) was 59% (49%–69%) for hospitalization, 105% (82%–134%) for ICU admission and 61% (40%–87%) for death. In addition, the risk of patients infected with Delta was 120% (93%–153%) for hospitalization, 287% (198%–399%) for ICU admission and 137% (50%–230%) for death. 61
Based on these data, patients infected with VOC variants were more likely to be admitted to hospitals and ICU than non‐VOC cases. Delta may cause severe diseases than N501Y‐positive VOC variants (B1.1.17, B.1.351, and P.1), and cases infected with Delta had a higher risk of hospitalization and mortality, as shown in Table 1.
1.2. Immune escape of VOC
1.2.1. Key mutations related to enhancing immune escape of VOC
1.2.1.1. E484K
The Spike protein is the dominant neutralization target of monoclonal antibodies (mAbs), convalescent plasma and vaccines. The virus would likely need to accumulate multiple mutations in the Spike to evade immunity induced by vaccines or by natural infection. 62 E484K mutation located in RBM and is present in B.1.351, p.1 VOC variants and in the VOIs Eta (B.1.525), Iota (B.1.526), Theta (P.3), and Zeta (p.2). 40 Many studies indicated that the E484K mutation generated apparent resistance to mAbs and convalescent plasma. 63 , 64 , 65 Collier Da et al. found that E484K mutation reduced the neutralization activity of the BNT162B2 vaccine‐induced antibody and 61% (19 of 31) mAbs against the virus. 66
1.2.1.2. L452R
L452R is present in the Delta, Kappa (B.1.617.1), and Epsilon (B.1.427/9). L452R mutation could not only increase the transmissibility of the virus but also cause immune escape. Studies found that the pseudovirus carrying L452R mutation could escape the neutralization activity of mAbs and convalescent plasma 67 and was significantly resistant to mAbs X593 and P2B‐2F6. 68
1.2.1.3. K417N/T
K417N and K417T are present in B.1.351 and P.1, respectively. Using pseudovirus models found that the neutralization activity of 29.4% (5/17) mRNA vaccine‐induced neutralizing antibodies against pseudoviruses carrying the K417N mutation was at least 10% lower than wild strain. 69 Data reported in another study showed that the neutralization activity of 35% mAbs (6 of 17) was decreased by 4‐fold against pseudoviruses carrying K417N or K417N‐N501Y mutations. 70
1.2.1.4. The deletion in the NTD
Despite the RBD is the dominant neutralization targeted by mAbs. Evidence indicated that the NTD of the SARS‐CoV‐2 Spike has a substantial role in antigenicity. 71 Andreano et al. reported that convalescent plasma thoroughly neutralized the live virus at the initial stage of coculture, but after 45 days of cultivation, the deletion of F140 in the NTD N3 loop led to partial resistant to the neutralization activity of convalescent plasma. After coculture 80 days, an insertion in the NTD N5 loop containing a new glycan sequon generated a complete resistance to the neutralization activity of convalescent plasma. In line with, computational modeling predicted that the NTD loops also have some key mutations, including the F140 deletion in loop N3 and the 11‐amino‐acid insertion in loop N5 that introduces a novel N‐glycan sequon at position N248d. These mutations remodel this critical antigenic region and have the potential to obstruct or hide the binding to neutralizing epitopes, and could effectively eliminate the neutralization of some antibodies. 72 In addition, it is reported that Y141‐144del can enable the B.1.1.7 pseudovirus to escape the neutralization of mAbs (4A8, S2X28, S2M28, and S2X333). 73 Another study showed that the B.1.351 variant carrying LAL242–244del mutations in the NTD was most resistant to current mAbs and convalescent plasma, followed by the P.1 variant and the B.1.1.7 variant. 74
1.2.2. Escape from the neutralization activity of mAbs and convalescent plasma
Studies showed that the neutralization activity of some mAbs and convalescent plasma against B.1.1.7, B.1.351, P.1 pseudovirus was reduced. 75 One study demonstrated that convalescent plasma shows a significant reduction in neutralization activity against B.1.351 pseudovirus (9.4‐fold) and shows a modest reduction against P.1 pseudovirus (2.4‐fold) and no significant reduction against the B.1.1.7 pseudovirus. 76 Wibmer et al. found that 48% (21/44) of the neutralizing antibodies isolated from convalescent plasma lost neutralization against B.1.351. 77 Similarly, it is reported that the neutralization susceptibility of 45% of convalescent plasma against Delta variant was reduced by approximately 3‐fold to 10‐fold and 5% of convalescent plasma against Delta variant was reduced by >10‐fold, respectively. 33 , 75 In addition, the neutralization activity of 30% (6/20) mAbs against the Delta variant was reduced more than 5‐fold. Interestingly, the neutralization curves assay found that the neutralizing activity of plasma from individuals infected with B.1.351 and P.1 showed loss entirely against Delta variant, suggesting that individuals infected with B.1.351 and P.1 may be at risk of reinfection with Delta variant. 75
1.2.3. Resistance to antibody‐mediated immunity elicited by vaccines
Live‐virus and pseudovirus neutralization assays indicated that the neutralizing activity of sera elicited by some vaccine against B.1.351variant showed a significant reduction: BNT162b2 (10.4‐fold), 78 Moderna mRNA (6.4–27.7‐fold), 79 , 80 , 81 Novavax subunit vaccine (14.5‐fold), 81 respectively. By contrast, postvaccination serum elicited by some vaccines exhibited a modest reduction in the neutralizing activity against B.1.1.7: BNT162b2 (less than 3‐fold), 78 Moderna mRNA vaccine (2–2.3‐fold), 79 Novavax subunit vaccine (2‐fold). 79 In addition, the P.1 variant also showed more significant decreases with both BNT162b2 (6.7‐fold 78 ) and Moderna mRNA vaccine (4.5–4.8‐fold) 82 postvaccination serum, respectively. Furthermore, it is reported that approximately 15% displayed 3‐fold to the 10‐fold reduced neutralizing activity of sera elicited by the BNT162b vaccine against the Delta variant. 75 , 78
Similarly, the neutralization activity of sera elicited by the inactivated‐virus vaccines‐BBIBP‐CorV (Sinopharm) and CoronaVac (Sinovac) against B.1.1.7 pseudovirus and B.1.351 pseudovirus was decreased by 2.0‐fold, 2.5–3.3‐fold, respectively. 83 A randomized controlled trial showed that the neutralization activity of sera elicited by ChAdOx1 adenovirus vector vaccine against B.1.1.7 and B.1.351 live‐virus was reduced by 2.5–8.9‐fold and 4.1–31.5‐fold, respectively. 84
Altogether, mutations in RBD region (K417N/T, S477N, E484K, F490s) and NTD region (F140del, Y144del, LLA242‐244del) may lead to the transfer of the antigen spectrum of S protein of SARS‐CoV‐2. These mutations occur in VOC variants and generate resistance to mAbs, convalescent plasma, and vaccine, as shown in Figures 1 and 2, and Table 1. Molecular dynamic simulations have pointed out “K417N‐E484K‐N501Y” triple mutations induces S protein conformational change was greater than N501Y or E484K alone, allowing the virus‐carrying “K417N‐E484K‐N501Y” triple mutations to be a more effective escape from the neutralization activity. 74 All these results indicated that B.1.351 variant was most resistant to the neutralization activity of most mAbs, convalescent plasma, and postvaccination serum, followed by P.1 variant and B.1.1.7 variant, primarily due to triple mutations” K417N‐E484K‐N501Y” occur in B.1.351 variant.
1.3. The epidemiological characteristics of “variants of interest (VOIs)” and their key amino acid mutations
July 1, 2021, the WHO Epidemiological update described seven VOIs, namely B.1.427/B.1.429 (Epsilon), Zeta (P.2), Eota (B.1.525), P.3, Iota (B.1.526), Kappa (B.1.617.1), Lambda (C.37) (WHO (2021a) (https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/).
1.3.1. N439K
N439K was located in RBM and present in B.1.258 variant. 85 It was found that the RBD region of Spike carrying N439K mutation could form a strong noncovalent salt bridge with ACE2, which could enhance the binding affinity of spike to ACE2. 35 Compared with the D614G epidemic strain, the neutralization activity of sera elicited by BNT162B2 vaccine against the variants with N439K mutation was not significantly changed. 80
1.3.2. S477N
S477N mutation existed in some B.1.526 strains, but it did not coexist with E484K mutation simultaneously. Singh et al. found that S477N mutation could enhance the binding affinity of spike to ACE2 and can attenuate the neutralization of some mAbs and convalescent plasma. 86
1.3.3. L452Q, del247‐252, F490S
Recently, Lambda (C.37) was first reported in South America on June 16, 2021. 87 , 88 Lambda showed an several mutation in the spike protein, including G75V, T76I, del247‐252, L452Q, F490S, D614G, T859N. 89 A preprint from the University of Tokyo revealed that T76I and L452Q mutations contributed to the higher infectivity. The RSYLTPGD246‐66 253 N mutation, a unique 7‐amino‐acid deletion mutation in the NTD and F490S, is responsible for evasion from neutralizing antibodies. 90
1.3.4. Epsilon (B.1.427/B.1.429)
It was reported that in California, the incidence of B.1.427/B.1.429 variants increased from 0% to >50% of sequenced cases from September 1, 2020, to January 29, 2021, exhibiting an 18.6%–24% increase in transmissibility relative to wild strains. 91 In throat swab samples, the viral load of B.1.427/B.1.429 was approximately twice that of the nonmutant virus. 92 The neutralization activity of antibody assays showed B.1.427/B.1.429 variants have 4.0–6.7‐fold and 2.0‐fold reduction in neutralizing titers from convalescent patients and vaccine recipients, respectively. 91 The neutralization activity of the sera elicited by the Moderna mRNA vaccine and Novavax subunit vaccine against B.1.427/B.1.429 variant was decreased by 2.0‐fold 80 and 2.5‐fold, 81 respectively.
1.3.5. Lambda (C.37)
As of July 6, 2021, Lambda (C.37) variant accounted for about 81% of new cases and is responsible for the second peak of COVID‐19 in Peru. It was reported that about a third of new cases were infected with the Lambda variant in Chile, and this variant accounted for 37% total of COVID‐19 cases between April 2 and May 19 in Argentina. 89 As of July 6, GISAID data showed the Lambda variant had been reported in 31 countries (https://www.gisaid.org; as of July 6, 2021).
One study from the University of Chile also found that the neutralization activity of the CoronaVac vaccine against Lambda variant was reduced by 67% than wild strain. 93 Another preprint paper from the University of Tokyo revealed that the Lambda variant has two abilities associate with the massive infection: increased infectivity and increased immune escape. 90 Although, the Lambda variant was classified as VOI by WHO. Based on all above results, Lambda variants have a strong potential to cause new massive pandemics in the future. However, there are few studies about the epidemiological characteristics of Lambda variant. It is significant to understand the biological characteristics of amino acid mutations, the epidemiological characteristics, and the vaccine efficacy of the Lambda variant.
1.3.6. Other VOIs
In contrast to the Delta VOC, the Kappa (B.1.617.1) has not demonstrated increased transmissibility. However, the Kappa variant has a greater ability to evade humoral immunity than the Delta variant. Several studies demonstrated that about 40% had 3‐fold to 10‐fold and 15% had >10‐fold reduced neutralizing activity of convalescent plasma against the Kappa variant. Similarly, approximately 55% displayed 3‐fold to the 10‐fold and 5% had >10‐fold reduced neutralizing activity of sera elicited by an mRNA vaccine against the Kappa variant. 33 , 75 , 94 In addition, the P.2 variant is circulating in Rio de Janeiro, Brazil, and has resulted in two separate clinical cases of secondary infection. 95 , 96 Geometric mean neutralization titers against B.1.617.1 were reduced 2.7‐fold (p < 0.0001) for the Pfizer‐BioNTech vaccine serum and 2.6‐fold (p < 0.0001) for the Oxford‐AstraZeneca vaccine than wild strain. 75
2. CONCLUSIONS
In this review, we described the characteristics of several amino acid mutations in RBD, NTD, furin protease cleavage sites, and ORF, including D614G, N501Y, L452R, N439K, S477N, HV69‐70del, E484K/Q, K417N/T, S477N, Y144Del, LLA242‐244Del, P681H/R, ORF3a mutations, N439K, S477N, L452Q, T76I, del247‐252, F490S. These mutations alter VOC and VOI variants' biological behavior, including more transmissibility, higher risk of hospitalization and mortality, and increased immune escape than wild strain. What is more, the Delta variant may be more transmissible and has a higher risk of hospitalization and mortality than other VOC variants (B.1.1.7, B.1.351, and P.1 variant).
CONFLICT OF INTERESTS
All authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Dandan Tian conceived and wrote the manuscript and prepared figures. Yanhong Sun and Jianming Zhou contributed to the data collection and prepared the table. Qing Ye contributed to the modification and revision of the manuscript. All authors contributed to this article and approved the submitted versions.
Tian D, Sun Y, Zhou J, Ye Q. The global epidemic of SARS‐CoV‐2 variants and their mutational immune escape. J Med Virol. 2022;94:847‐857. 10.1002/jmv.27376
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Graham RL, Baric RS. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross‐species transmission. J Virol. 2010;84(7):3134‐3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Su S, Wong G, Shi W, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490‐502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee N, Hui D, Wu A, et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348(20):1986‐1994. [DOI] [PubMed] [Google Scholar]
- 4. Tsang KW, Ho PL, Ooi GC, et al. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348(20):1977‐1985. [DOI] [PubMed] [Google Scholar]
- 5. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814‐1820. [DOI] [PubMed] [Google Scholar]
- 6. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ye Q, Lu D, Shang S, et al. Crosstalk between coronavirus disease 2019 and cardiovascular disease and its treatment. ESC Heart Fail. 2020;7(6):3464‐3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Han X, Ye Q. Kidney involvement in COVID‐19 and its treatments. J Med Virol. 2021;93(3):1387‐1395. [DOI] [PubMed] [Google Scholar]
- 9. Tian D, Ye Q. Hepatic complications of COVID‐19 and its treatment. J Med Virol. 2020;92(10):1818‐1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ye Q, Wang B. The mechanism and treatment of gastrointestinal symptoms in patients With COVID‐19. Am J Physiol Gastrointestinal liver Physiol. 2020;319(2):G245‐G252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pachetti M, Marini B, Benedetti F, et al. Emerging SARS‐CoV‐2 mutation hot spots include a novel RNA‐dependent‐RNA polymerase variant. J Transl Med. 2020;18(1):179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271‐280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lan J, Ge J, Yu J, et al. Structure of the SARS‐CoV‐2 spike receptor‐binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215‐220. [DOI] [PubMed] [Google Scholar]
- 15. Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Volz E, Hill V, McCrone JT, et al. Evaluating the effects of SARS‐CoV‐2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184(1):64‐75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Giovanetti M, Benedetti F, Campisi G, et al. Evolution patterns of SARS‐CoV‐2: snapshot on its genome variants. Biochem Biophys Res Commun. 2021;538:88‐91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Korber B, Fischer WM, Gnanakaran S, et al. Tracking changes in SARS‐CoV‐2 spike: evidence that D614G increases infectivity of the COVID‐19 virus. Cell. 2020;182(4):812‐827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Groves DC, Rowland‐Jones SL, Angyal A. The D614G mutations in the SARS‐CoV‐2 spike protein: implications for viral infectivity, disease severity and vaccine design. Biochem Biophys Res Commun. 2021;538:104‐107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sheikh JA, Singh J, Singh H, et al. Emerging genetic diversity among clinical isolates of SARS‐CoV‐2: lessons for today. Infect Genet Evol. 2020;84:104330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Van Dorp L, Acman M, Richard D, et al. Emergence of genomic diversity and recurrent mutations in SARS‐CoV‐2. Infect Genet Evol. 2020;83:104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leung K, Shum MH, Leung GM, Lam TT, Wu JT. Early transmissibility assessment of the N501Y mutant strains of SARS‐CoV‐2 in the United Kingdom, October to November 2020. Euro Surveill. 2021;26(1):2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Makoni M. South Africa responds to new SARS‐CoV‐2 variant. Lancet. 2021;397(10271):267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Faria NR, Mellan TA, Whittaker C, et al. Genomics and epidemiology of the P.1 SARS‐CoV‐2 lineage in Manaus, Brazil. Science. 2021;372(6544):815‐821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Singh J, Rahman SA, Ehtesham NZ, Hira S, Hasnain SE. SARS‐CoV‐2 variants of concern are emerging in India. Nat Med. 2021;27(7):1131‐1133. [DOI] [PubMed] [Google Scholar]
- 26. Hou YJ, Chiba S, Halfmann P, et al. SARS‐CoV‐2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370(6523):1464‐1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou B, Thao T, Hoffmann D, et al. SARS‐CoV‐2 spike D614G variant confers enhanced replication and transmissibility. bioRxiv. 2020. 10.1101/2020.10.27.357558 [DOI] [Google Scholar]
- 28. Daniloski Z, Jordan TX, Ilmain JK, et al. The Spike D614G mutation increases SARS‐CoV‐2 infection of multiple human cell types. eLife. 2021;10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu H, Zhang Q, Wei P, et al. The basis of a more contagious 501Y.V1 variant of SARS‐CoV‐2. Cell Res. 2021;31(6):720‐722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Starr TN, Greaney AJ, Hilton SK, et al. Deep mutational scanning of SARS‐CoV‐2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182(5):1295‐1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Laffeber C, de Koning K, Kanaar R, Lebbink J. Experimental evidence for enhanced receptor binding by rapidly spreading SARS‐CoV‐2 variants. J Mol Biol. 2021;433(15):167058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Planas D, Bruel T, Grzelak L, et al. Sensitivity of infectious SARS‐CoV‐2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med. 2021;27(5):917‐924. [DOI] [PubMed] [Google Scholar]
- 33. Edara VV, Lai L & Sahoo MK et al. Infection and vaccine‐induced neutralizing antibody responses to the SARS‐CoV‐2 B.1.617.1 variant. bioRxiv. 2021. 10.1101/2021.05.09.443299 [DOI] [PMC free article] [PubMed]
- 34. McCarthy KR, Rennick LJ, Nambulli S, et al. Recurrent deletions in the SARS‐CoV‐2 spike glycoprotein drive antibody escape. Science. 2021;371(6534):1139‐1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thomson EC, Rosen LE, Shepherd JG, et al. Circulating SARS‐CoV‐2 Spike N439K variants maintain fitness while evading antibody‐mediated immunity. Cell. 2021;184(5):1171‐1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meng B, Kemp SA, Papa G, et al. Recurrent emergence of SARS‐CoV‐2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep. 2021;35(13):109292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kemp SA, Collier DA, Datir RP, et al. SARS‐CoV‐2 evolution during treatment of chronic infection. Nature. 2021;592(7853):277‐282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pascarella S, Ciccozzi M, Zella D, et al. SARS‐CoV‐2 B.1.617 Indian variants: are electrostatic potential changes responsible for a higher transmission rate? J Med Virol. 2021. 10.1002/jmv.27210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Garcia‐Beltran WF, Lam EC, St. DK, et al. Multiple SARS‐CoV‐2 variants escape neutralization by vaccine‐induced humoral immunity. Cell. 2021;184(9):2372‐2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Harvey WT, Carabelli AM, Jackson B, et al. SARS‐CoV‐2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409‐424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Davies NG, Abbott S, Barnard RC, et al. Estimated transmissibility and impact of SARS‐CoV‐2 lineage B.1.1.7 in England. Science. 2021;372:6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Frampton D, Rampling T, Cross A, et al. Genomic characteristics and clinical effect of the emergent SARS‐CoV‐2 B.1.1.7 lineage in London, UK: a whole‐genome sequencing and hospital‐based cohort study. Lancet Infect Dis. 2021;21(9):1246‐1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Golubchik T, Katrina AL, Matthew H, et al. Early analysis of a potential link between viral load and the N501Y mutation in the SARS‐COV‐2 spike protein. medRxiv. 2021. 10.1101/2020.10.27.357558 [DOI]
- 44. Pearson CAB, Russell TW, Davies NG, et al. Estimates of severity and transmissibility of novel South Africa SARS‐CoV‐2 variant. 501Y.V2. Retrieved January 15, 2021, from https://cmmid.github.io/topics/covid19/reports/sa-novel-variant/2021_01_11_Transmissibility_and_severity_of_501Y_V2_in_SA.pdf
- 45. Callaway E. Delta coronavirus variant: scientists brace for impact. Nature. 2021;595(7865):17‐18. [DOI] [PubMed] [Google Scholar]
- 46. Li B, Deng A, Li K, et al. Viral infection and transmission in a large, well‐traced outbreak caused by the SARS‐CoV‐2 Delta variant. medRxiv. 2021. 10.1101/2021.07.07.21260122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Challen R, Dyson L, Overton CE, et al. Early epidemiological signatures of novel SARS‐CoV‐2 variants: establishment of B.1.617.2 in England. medRxiv. 2021. 10.1101/2021.06.05.21258365 [DOI] [Google Scholar]
- 48. Riley S, Wang H, Eales O, et al. REACT‐1 round 12 report: resurgence of SARS‐CoV‐2 infections in England associated with increased frequency of the Delta variant. MedRxiv. 2021. 10.1101/2021.06.17.21259103 [DOI] [Google Scholar]
- 49. Bolze A, Cirulli ET, Luo S, et al. Rapid displacement of SARS‐CoV‐2 variant B.1.1.7 by B.1.617.2 and P.1 in the United States. MedRxiv. 2021. 10.1101/2021.06.20.21259195 [DOI] [Google Scholar]
- 50. Issa E, Merhi G, Panossian B, Salloum T, Tokajian S. SARS‐CoV‐2 and ORF3a: nonsynonymous mutations, functional domains, and viral pathogenesis. mSystems. 2020;5(3):e00266‐20. 10.1128/mSystems.0026620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu DX, Fung TS, Chong KK, Shukla A, Hilgenfeld R. Accessory proteins of SARS‐CoV and other coronaviruses. Antiviral Res. 2014;109:97‐109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhong NS, Zheng BJ, Li YM, et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. 2003;362(9393):1353‐1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Majumdar P, Niyogi S. ORF3a mutation associated with higher mortality rate in SARS‐CoV‐2 infection. Epidemiol Infect. 2020;148:e262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bager P, Wohlfahrt J, Fonager J, et al. Risk of hospitalisation associated with infection with SARS‐CoV‐2 lineage B.1.1.7 in Denmark: an observational cohort study. Lancet Infect Dis. 2021. 10.1016/S1473-3099(21)00290-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dabrera G, Allen H & Zaidi A et al. Assessment of mortality and hospital admissions associated with confirmed infection with SARS‐CoV‐2 Variant of Concern VOC‐202012/01 (B.1.1.7) a matched cohort and time‐to‐event analysis. SSRN; 2021. https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3802578 [DOI] [PMC free article] [PubMed]
- 56. Nyberg T, Twohig KA, Harris RJ, et al. risk of hospital admission for patients with SARS‐CoV‐2 variant B.1.1.7: cohort analysis. BMJ. 2021;373:n1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Grint DJ, Wing K, Williamson E, et al. Case fatality risk of the SARS‐CoV‐2 variant of concern B.1.1.7 in England, November 16 to February 5. Euro Surveill. 2021;26(11):2100256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Davies NG, Jarvis CI, CMMID COVID‐ Working G, et al. Increased mortality in community‐tested cases of SARS‐CoV‐2 lineage B.1.1.7. medRxiv. 2021. 10.1101/2021.02.01.21250959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Funk T, Pharris A, Spiteri G, et al. Characteristics of SARS‐CoV‐2 variants of concern B.1.1.7, B.1.351 or P.1: data from seven EU/EEA countries, weeks 38/2020 to 10/2021. Euro Surveill. 2021;26(16):2100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sheikh A, McMenamin J, Taylor B, Robertson C Public Health Scotland and the EAVE II C. SARS‐CoV‐2 Delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet. 2021;397(10293):2461‐2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fisman DN, Ashleigh RT. Progressive increase in virulence of novel SARS‐CoV‐2 variants in Ontario, Canada. bioRxiv. 2021. 10.1101/2021.07.05.21260050 [DOI] [Google Scholar]
- 62. Chi X, Yan R, Zhang J, et al. A neutralizing human antibody binds to the N‐terminal domain of the spike protein of SARS‐CoV‐2. Science. 2020;369(6504):650‐655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Weisblum Y, Schmidt F, Zhang F, et al. escape from neutralizing antibodies by SARS‐CoV‐2 spike protein variants. eLife. 2020;9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Baum A, Fulton BO, Wloga E, et al. Antibody cocktail to SARS‐CoV‐2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369(6506):1014‐1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liu Z, VanBlargan LA, Bloyet LM, et al. Identification of SARS‐CoV‐2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29(3):477‐488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Collier DA, De Marco A, Ferreira I, et al. Sensitivity of SARS‐CoV‐2 B.1.1.7 to mRNA vaccine‐elicited antibodies. Nature. 2021;593(7857):136‐141. [DOI] [PubMed] [Google Scholar]
- 67. Liu Z, VanBlargan LA, Bloyet LM, et al. Landscape analysis of escape variants identifies SARS‐CoV‐2 spike mutations that attenuate monoclonal and serum antibody neutralization. bioRxiv. 2020. 10.1101/2020.11.06.372037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li Q, Wu J, Nie J, et al. The impact of mutations in SARS‐CoV‐2 spike on viral infectivity and antigenicity. Cell. 2020;182(5):1284‐1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang Z, Schmidt F, Weisblum Y, et al. mRNA vaccine‐elicited antibodies to SARS‐CoV‐2 and circulating variants. Nature. 2021;592(7855):616‐622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li Q, Nie J, Wu J, et al. SARS‐CoV‐2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. 2021;184(9):2362‐2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Suryadevara N, Shrihari S, Gilchuk P, et al. Neutralizing and protective human monoclonal antibodies recognizing the N‐terminal domain of the SARS‐CoV‐2 spike protein. Cell. 2021;184(9):2316‐2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Andreano E, Piccini G, Licastro D, et al. SARS‐CoV‐2 escape in vitro from a highly neutralizing COVID‐19 convalescent plasma. bioRxiv. 2020. 10.1101/2020.12.28.424451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. McCallum M, De Marco A, Lempp FA, et al. N‐terminal domain antigenic mapping reveals a site of vulnerability for SARS‐CoV‐2. Cell. 2021;184(9):2332‐2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang R, Zhang Q, Ge J, et al. Analysis of SARS‐CoV‐2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species. Immunity. 2021;54(7):1611‐1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liu C, Ginn HM, Dejnirattisai W, et al. Reduced neutralization of SARS‐CoV‐2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184(16):4220‐4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Liu Y, Liu J, Xia H, et al. Neutralizing activity of BNT162b2‐elicited serum. N Engl J Med. 2021;384(15):1466‐1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wibmer CK, Ayres F, Hermanus T, et al. SARS‐CoV‐2 501Y.V2 escapes neutralization by South African COVID‐19 donor plasma. Nat Med. 2021;27(4):622‐625. [DOI] [PubMed] [Google Scholar]
- 78. Lustig Y, Zuckerman N, Nemet I, et al. Neutralising capacity against Delta (B.1.617.2) and other variants of concern following Comirnaty (BNT162b2, BioNTech/Pfizer) vaccination in health care workers, Israel. Euro Surveill. 2021;26(26):2100557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Shen X, Tang H, McDanal C, et al. SARS‐CoV‐2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe. 2021;29(4):529‐539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wu K, Werner AP, Koch M, et al. Serum neutralizing activity elicited by mRNA‐1273 vaccine. N Engl J Med. 2021;384(15):1468‐1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Shen X, Tang H, Pajon R, et al. Neutralization of SARS‐CoV‐2 variants B.1.429 and B.1.351. N Engl J Med. 2021;384(24):2352‐2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wang P, Casner RG, Nair MS, et al. Increased resistance of SARS‐CoV‐2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021;29(5):747‐751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wang GL, Wang ZY, Duan LJ, et al. Susceptibility of circulating SARS‐CoV‐2 variants to neutralization. N Engl J Med. 2021;384(24):2354‐2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhang Y, Hu YS. tolerability, and immunogenicity of an inactivated SARS‐CoV‐2 vaccine in healthy adults aged 18‐59 years: a randomised, double‐blind, placebo‐controlled, phase 1/2 clinical trial. Lancet Infect Dis. 2021;21:181‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Broňa B, Kristína B, Viktória H. A SARS‐CoV‐2 mutant from B.28 lineage with ∆H69/∆V70 deletion in the spike protein circulating in Central Europe in the fall 2020. Virus Genes. 2021;27:1‐5. 10.1007/s11262-021-01866-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Singh A, Steinkellner G, Köchl K, Gruber K, Gruber CC. Serine 477 plays a crucial role in the interaction of the SARS‐CoV‐2 spike protein with the human receptor ACE2. Sci Rep. 2021;11(1):4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. COVID‐19 Weekly Epidemiological Update Global overview . https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---15-june-2021 [DOI] [PMC free article] [PubMed]
- 88.Nextstrain/community/quipupe/C37_lineage. https://nextstrain.org/community/quipupe/C37_lineage
- 89.Novel sublineage within B.1.1.1 currently expanding in Peru and Chile, with a convergent deletion in the ORF1a gene (Δ3675‐3677) and a novel deletion in the Spike gene (Δ246‐252, G75V, T76I, L452Q, F490S, T859N)—SARS‐CoV‐2 coronavirus/nCoV‐2019 Genomic Epidemiology—Virological. https://virological.org/t/novel-sublineage-within-b-1-1-1-currently-expanding-in-peru-and-chile-with-a-convergent-deletion-in-the-orf1a-gene-3675-3677-and-a-novel-deletion-in-the-spike-gene-246-252-g75v-t76i-l452q-f490s-t859n/685
- 90. Izumi K, Ryosuke N, Wu J, et al. SARS‐CoV‐2 Lambda variant exhibits higher infectivity and immune resistance. bioRxiv. 2021. 10.1101/2021.07.28.454085 [DOI] [Google Scholar]
- 91. Deng X, Garcia‐Knight MA, Khalid MM, et al. Transmission, infectivity, and antibody neutralization of an emerging SARS‐CoV‐2 variant in California carrying a L452R spike protein mutation. medRxiv. 2021. 10.1101/2021.03.07.21252647 [DOI] [Google Scholar]
- 92. Drew RJ, O'Donnell S, LeBlanc D, McMahon M, Natin D. The importance of cycle threshold values in interpreting molecular tests for SARS‐CoV‐2. Diagn Microbiol Infect Dis. 2020;98(3):115130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Acevedo ML, Palomares LA, Bustamante A, et al. Infectivity and immune escape of the new SARS‐CoV‐2 variant of interest Lambda. MedRxiv. 2021. 10.1101/2021.06.28.21259673 [DOI] [Google Scholar]
- 94. Hoffmann M, Hofmann‐Winkler H, Krüger N, et al. SARS‐CoV‐2 variant B.1.617 is resistant to bamlanivimab and evades antibodies induced by infection and vaccination. Cell Rep. 2021;36(3):109415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Nonaka C, Franco MM, Graf T, et al. Genomic evidence of SARS‐CoV‐2 reinfection involving E484K spike mutation, Brazil. Emerg Infect Dis. 2021;27(5):1522‐1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Resende PC, Bezerra JF, Teixeira Vasconcelos RH, et al. Severe acute respiratory syndrome coronavirus 2 P.2 lineage associated with reinfection case, Brazil, June‐October 2020. Emerg Infect Dis. 2021;27(7):1789‐1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


