To the Editor,
1.
The emergence of severe acute respiratory syndrome 2 (SARS‐CoV‐2) variants of concern (VOC) have posed a serious threat to the control of the disease worldwide. Of note are the B.1.1.7 (alpha), B.1.351 (beta), and P.1 (gamma) that were first reported from the United Kingdom, South Africa, and Brazil, respectively. The B.1.351 contains characteristic mutations in the receptor‐binding domain of spike glycoprotein (S) protein: K417N, E484K, and N501Y that have functional significance. 1 As of May 7, 2021, 14 543 sequences of B.1.351 lineage have been reported from 85 countries around the world. 2 Since the start of the pandemic, the National Institute of Health, Pakistan has been involved in the surveillance of SARS‐CoV‐2 and although the B.1.1.7 cases have been detected, 3 the whole‐genome sequence of B.1.351 has not been reported in Pakistan. We hereby report the genomic diversity of the first two sequences of the B.1.351 variant detected from Pakistan.
On April 24 and 26, 2021, two oropharyngeal samples were received at the Department of Virology, National Institute of Health for the detection of SARS‐CoV‐2 as part of routine surveillance. Briefly, viral RNA was extracted using the MagMAX™ Viral/Pathogen Nucleic Acid Isolation Kit on the KingFisher Flex™ Purification System (Thermo Fisher Scientific). The TaqPath™ COVID‐19 CE‐IVD RT‐PCR kit (Thermo Fisher Scientific) was used for the detection of SARS‐CoV‐2. These two samples were selected for whole‐genome sequencing due to amplification of spike gene on TaqPath™ assay and having low C t value (<22 for S gene).
For whole‐genome sequencing, the cDNA synthesis and amplification were performed according to the Primal‐Seq Nextera XT protocol (version 2) using SuperScript™ IV VILO™ Master Mix (Invitrogen) and Q5® High‐Fidelity 2X Master Mix (New England BioLabs) respectively, 4 with the ARTIC nCoV‐2019 Panel (Integrated DNA Technologies, Inc.). Illumina DNA Prep Kit (Illumina, Inc.) was used for library preparation and subjected to sequencing on Illumina iSeq 100 (Illumina, Inc.). The read quality of sequenced files was assessed using the FastQC tool (v0.11.9). 5 The data were processed and analyzed as per recommended guidelines of the Centers for Diseases Control and Prevention. 6
On real‐time PCR, the sample NIH‐S6 (EPI_ISL_1969999) showed a C t value of 24 for N‐gene, 23 for ORF1ab, and 21 for S‐gene. The patient was a 28‐year‐old female who attended a funeral ceremony in her neighborhood on April 20, 2021, before infection. The patient had mild symptoms of having body aches, loss of taste and smell, and had no travel history. Similarly, the C t values of sample NIH‐S12 (EPI_ISL_1969995) were 14 for N‐gene, 16 for ORF1ab, and 14 for S‐gene. The patient was a 30‐year‐old male with low‐grade fever and body aches and had no travel history. Whole‐genome sequencing results showed the detection of B.1.351 in both cases having characteristic mutations in the spike: D80A, D215G, D614G, E484K, K417N, N501Y, and A701V (Table 1). Based on phylogenetic analysis, Pakistani sequences were closely related to the B.1.351 (Figure 1).
Table 1.
Genome‐wide amino acid mutations
| S. No. | GISAID ID | PANGO lineage | ORF1a | ORF1b | ORF3a | N | S | E | ORF9b |
|---|---|---|---|---|---|---|---|---|---|
| 1. | EPI_ISL_1969995 | B.1.351 | T265I, S1612L, K1655N, K3353R, L3829F | P314L, S936L | Q57H, S171L | G30R, T205I | D80A, D215G, K417N, E484K, N501Y, D614G, A701V | P71L | M26I |
| 2. | EPI_ISL_1969999 | B.1.351 | T265I, S1612L, K1655N, K3353R | P314L | Q57H, S171L | G30R, T205I | D80A, D215G, K417N, E484K, N501Y, D614G, A701V | P71L | M26I |
Figure 1.
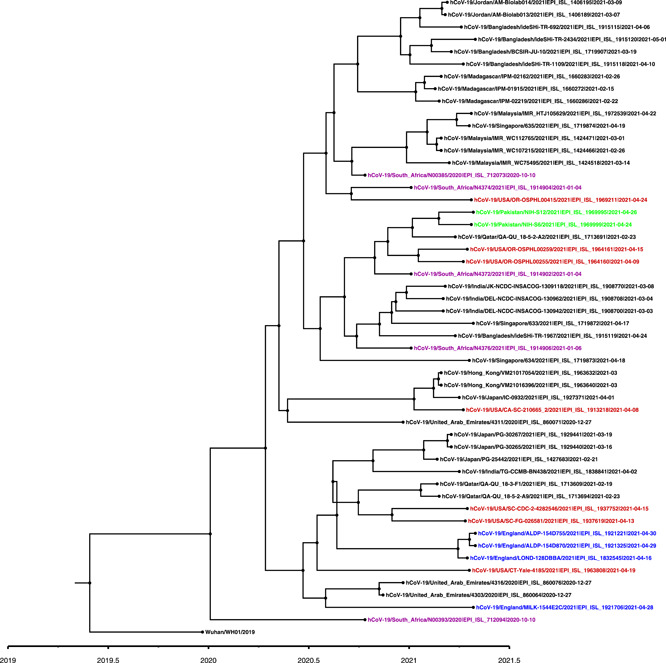
A time‐scaled maximum likelihood phylogenetic tree of B.1.351 variant viruses. For phylogenetic analysis, Wuhan reference SARS‐CoV‐2 (EPI_ISL_529213) sequence was used and closely related sequences to the study strains were downloaded from GISAID using BLAST (https://www.gisaid.org/). In addition, representative B.1.351 sequences from neighboring countries were also included in the analysis followed by multiple sequence alignment using MAFTT. The phylogenetic tree was constructed using BEAST.v1.10.4 to estimate divergence times. The Pakistani sequences are highlighted in green color, USA in Red, England in blue, and South Africa in magenta. A timescale for the evolution of strain is shown at the bottom of the tree
As Pakistan fights against the COVID‐19 pandemic, a total of 864 557 cases and 19 106 deaths have been reported until May 10, 2021, with a high number of cases (n = 283 192) seen during the third wave (https://covid.gov.pk/stats/pakistan). Based on our laboratory data (spike gene target failure cases using COVID‐19 TaqPath™ kit; Thermo Fisher Scientific), the recent surge in cases correlates with the detection of the B.1.1.7 variant in the country. Conversely, no data is available on the prevalence of other VOCs including B.1.351, which is primarily attributed to the limited genomic surveillance carried out at the national level (<0.1% genomic sequencing). The two B.1.351 cases reported in our study belonged to the capital city Islamabad where an average of 370 cases per day (total n = 7773) was reported in the last three weeks (April 20 to May 10, 2021). The fact that both cases had no travel history highlights the local circulation of B.1.351 in the indigenous population and warrants large‐scale genomic surveillance to track the spread of this variant.
A major challenge in the control of COVID‐19 in Pakistan is low vaccination coverage underlined by the administration of only 3 629 065 doses, whereby <1% of the population has been fully vaccinated until May 10, 2021. Pakistan's COVID‐19 vaccination program has mainly relied on Sinopharm and CanSino vaccines in addition to the limited doses of the Oxford AstraZeneca vaccine. The efficacy of Sinopahrm and CanSino vaccines against the B.1.351 variant is unknown albeit the former has shown a weakened effect. 7 Moreover, the Oxford AstraZeneca vaccine has been shown to provide limited protection against mild to moderate COVID‐19 caused by B.1.351, however, data on its efficacy against severe disease is pending. 8 In contrast, the Pfizer BioNTech mRNA vaccine was found to be effective against infection and disease caused by the B.1.351 in a recent study. 9 The emergence of the B.1.351 variant and its probable spread in Pakistan can pose a threat to the control of the disease in the country as witnessed in Bangladesh where the variant has been associated with a resurgence in COVID‐19 cases and reinfections. 10 Therefore, continued surveillance and gathering data on the efficacy of the vaccines against COVID‐19 caused by B.1.351 and other VOCs are needed for informed policy decisions in countries like Pakistan that have limited options for vaccine access.
AUTHOR CONTRIBUTIONS
Conceptualization: Massab Umair, Aamer Ikram, and Muhammad Salman. Methodology: Massab Umair, Rana M. Suleman, Nazish Badar, Muhammad Ammar, Qasim Ali, AH. Formal analysis: Massab Umair, Syed A. Haider, Zaira Rehman, AH, and Rana M. Suleman. Resources: Massab Umair, Aamer Ikram, and Muhammad Salman. Writing—original draft preparation: Massab Umair, Zaira Rehman, and Syed A. Haider. Writing—review and editing: Massab Umair, Aamer Ikram, Muhammad Salman, Rana M. Suleman, and Nazish Badar. All authors have read and agreed to the published version of the manuscript.
REFERENCES
- 1. Wibmer CK, Ayres F, Hermanus T, et al. SARS‐CoV‐2 501Y.V2 escapes neutralization by South African COVID‐19 donor plasma. Nat Med. 2021;27(4):622‐625. [DOI] [PubMed] [Google Scholar]
- 2. Latif AA, Mullen JL, Alkuzweny M, et al. B.1.351 Lineage Report. Accessed May 10, 2021. https://outbreak.info/situation-reports?pango=B.1.351%26selected=ZAF%26loc=ZAF%26loc=USA%26loc=USA_US-CA
- 3. Umair M, Ikram A, Salman M, et al. Importation of SARS‐CoV‐2 variant B.1.1.7 in Pakistan. J Med Virol. 2021;93:2623‐2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Quick J, Grubaugh ND, Pullan ST, et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat Protoc. 2017;12(6):1261‐1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Andrews S. FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics. Babraham Institute: Cambridge, UK; 2010.
- 6. Paden CR, Tao Y, Queen K, et al. Rapid, sensitive, full‐genome sequencing of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26(10):2401‐2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sinopharm's COVID‐19 vaccine remained active against S.Africa variant, effect reduced—lab study . Reuters. Accessed February 3, 2021. https://www.reuters.com/article/us-health-coronavirus-china-vaccineidUSKBN2A30DT
- 8. Madhi SA, Baillie V, Cutland CL, et al. Efficacy of the ChAdOx1 nCoV‐19 Covid‐19 vaccine against the B.1.351 variant. N Engl J Med. 2021;384:1885‐1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Abu‐Raddad LJ, Chemaitelly H, Butt AA. Effectiveness of the BNT162b2 Covid‐19 vaccine against the B.1.1.7 and B.1.351 Variants. N Engl J Med. 2021;385:187‐189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Saha S, Tanmoy AM, Hooda Y, et al. COVID‐19 rise in Bangladesh correlates with increasing detection of B.1.351 variant. BMJ Glob Health. 2021;6:e006012. [DOI] [PMC free article] [PubMed] [Google Scholar]


