Abstract
COVID‐19, caused by the SARS‐CoV‐2 virus, has become a significant global public health problem, with a wide variety of clinical manifestations and disease progression outcomes. LncRNAs are key regulators of the immune response and have been associated with COVID‐19 risk infection. Previous studies focused mainly on in‐silico analysis of lncRNA expression in the lungs or peripheral blood cells. We evaluated the expression of lncRNAs NEAT1, MALAT1, and MIR3142 in saliva and nasopharyngeal swab from SARS‐CoV‐2 positive (n = 34) and negative patients (n = 46). A higher expression of the lncRNAs NEAT1 and MALAT1 (p < 0.05) were found in positive samples. NEAT1 had a higher expression mainly in saliva samples (p < 0.001), and MALAT1 was upregulated in nasopharyngeal samples (p < 0.05). Area under the ROC curve for NEAT1 in saliva was 0.8067. This study was the first to investigate the expression of lncRNAs in saliva and nasopharyngeal samples of COVID‐19 patients, which gives new insights into the initial response to infection and infectivity and may provide new biomarkers for severity and targets for therapy.
Keywords: COVID‐19, lncRNA, MALAT1, nasopharyngeal samples, NEAT1, saliva, SARS‐CoV‐2
1. “HOT TOPICS”
LncRNAs (long noncoding RNAs) are transcripts larger than 200 nucleotides in length that do not appear to have protein‐coding potential (Cabili et al., 2011), although some of them may produce small functional peptides (Choi et al., 2019). Further to complex regulation in multiple cell processes, lncRNAs are emerging as critical regulators of immune responses (Bocchetti et al., 2021; Heward & Lindsay, 2014), and some may control innate and adaptive cell types (Chen et al., 2017). Antiviral immune responses are also influenced by lncRNAs (Liu et al., 2020; Ouyang et al., 2016; Qiu et al., 2018), but information regarding a role in COVID‐19 patients is limited.
In 2020, Vishnubalaji et al. performed a more in‐depth analysis of transcriptome data from primary normal human bronchial epithelial cells during SARS‐CoV‐2 infection and lung biopsies derived from COVID‐19 patients. They included the lncRNA transcriptional portrait in response to SARS‐CoV‐2 infection in lung cells. Among the regulated lncRNAs were NEAT1, MALAT1, and miR3142HG all of which have been previously associated with immune response and viral infection.
Considering the relevance of saliva and nasopharyngeal cells in infectivity and initial SARS‐CoV‐2 infection, we analyzed the expression of those lncRNAs in nasopharyngeal swab (NPS) and saliva samples from 41 individuals (39 NPS/saliva paired samples, 2 NPS without saliva counterpart). The National Research Ethics Committee approved the study (CAAE: 31687620.2.0000.0096), and samples were collected from healthcare workers in a tertiary public hospital in Curitiba, Brazil, after obtaining written informed consent. The samples were designated according to SARS‐CoV‐2 detection by RT‐qPCR as “negative” (n = 23 individuals and 46 samples) or “positive” (n = 18 individuals and 34 samples). All positive samples were wild type SARS‐CoV‐2, using genotyping methodology previously described (Adamoski et al., 2021). The median age of the negative group was 40 (±10.9) and 41.5 (±10.3) for the positive samples. For NEAT1, MALAT1, and miR3142HG lncRNAs expression, RNA was extracted from 200 μL of sample with Trizol Reagent (Invitrogen). A total of 70 ng of total RNA was treated with DNAse I (1U), and the absence of DNA contamination was confirmed by a retrotranscription (RT) minus reaction. For RT, the Superscript III kit (Invitrogen) was used. qPCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems) and 10 nM of each primer. The relative expression was calculated by the 2−ΔΔ ct method with two internal controls (B‐actin/RNU6).
Positive samples for SARS‐CoV‐2 exhibited a significant upregulation of the lncRNAs NEAT1 and MALAT1 (p < 0.05) (Figure 1). For NEAT1, this difference was observed mainly in saliva samples (p < 0.001), when comparing only NPSs from negative and positive samples, this difference was not observed. In contrast, MALAT1 was observed with higher expression only in NPS positive samples (p < 0.05), when considering only saliva samples, no difference was observed. MIR3142HG expression was detected in less than 17.5% of the samples analyzed and it was removed from further analysis.
FIGURE 1.
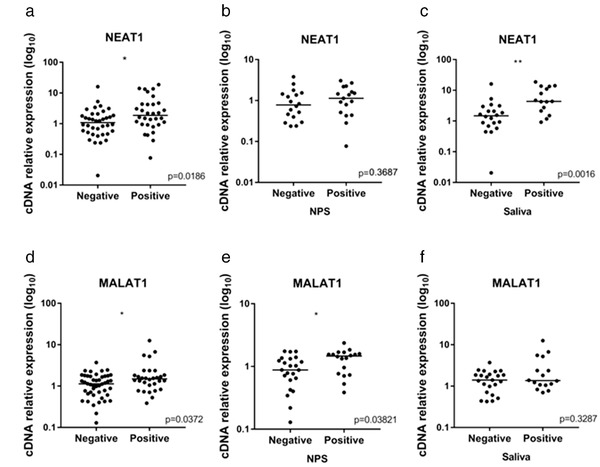
Relative expression levels of lncRNAs in negative and positive samples for SARS‐CoV‐2. (a) NEAT1 expression including nasopharyngeal swab (NPS) and saliva samples. (b) NEAT1 expression in NPS samples. (c) NEAT1 expression in saliva samples. (d) MALAT1 expression including NPS and saliva samples. (e) MALAT1 expression in NPS samples. (f) MALAT1 expression in saliva samples. *p < 0.05 and **p < 0.01, Mann–Whitney test
When analyzing paired samples, including NPS and saliva from the same patient, NEAT1 showed a higher expression in saliva samples, when comparing negative and positive samples together (p < 0.001), only negative (p < 0.05), and only positive samples (p < 0.002). The expression of MALAT1 was also higher in saliva samples, but this difference was only observed among all (negative and positive) samples (p < 0.001) and negative samples only (p < 0.001). As swab positive samples had high MALAT1 expression, differences between swab and saliva samples were not evidenced among positive individuals.
For determining the potential of both lncRNAs as possible biomarkers for the infection, receiver operating characteristic (ROC) curves were calculated. The specificity was significant for MALAT1 in NPS and NEAT1 in saliva. The AUC was 0.8067 (p < 0.001) for NEAT1, showing a high power for distinguishing positive and negative saliva samples. A significant positive correlation was also observed, by Spearman's correlation test, between NEAT1 and MALAT1 expression in both negative and positive samples (r = 0.48, p < 0.01).
In summary, this study observed a differential expression of NEAT1 and MALAT1 lncRNAs comparing infected and noninfected individuals with SARS‐CoV‐2 in saliva or NPS samples. For NEAT1, this difference was observed mainly in saliva samples. In contrast, MALAT1 high expression was prominent in positive nasopharyngeal samples.
Both lncRNAs have been associated with immune system responses. NEAT1 is an inflammatory regulator that promotes activation of inflammasomes in macrophages (Zhang et al., 2019) and induces inflammatory cytokines such as interleukin‐6 (IL‐6) and CXCL8 (Tang et al., 2020). IL‐6 and the NLRP3 inflammasome are primary immune components in responses to SARS‐CoV‐2 infection (Paniri & Akhavan‐Niaki, 2020). MALAT1 also exerts various immunological effects including the mediation of NLRP3 inflammasome activation (Menon & Hua, 2020; Yu et al., 2018). Through NF‐κB and HIF‐1α activation, MALAT1 increases production of inflammatory cytokines, such as IL‐6 and TNF‐α, to promote inflammatory cell infiltration and tissue damage (Tian et al., 2018). Furthermore, MALAT1 has been linked to M1‐like activity in macrophages, promoting pulmonary inflammation and injury (Cui et al., 2019).
Most recent studies investigated lncRNA expression in lung cells or in the peripheral blood of COVID‐19 patients, and we did not find any analyses of saliva or nasopharyngeal cells. Vishnubalaji et al., in 2020, generated lncRNAs transcriptome data from SARS‐CoV‐2 infected bronchial epithelial cells and demonstrated upregulation of NEAT1 and MALAT1. Elevated expressions of these lncRNAs were also shown in bronchoalveolar lavage fluid from COVID‐19 patients (Moazzam‐Jazi et al., 2021). Additionally, in peripheral blood mononuclear cells, increased NEAT1 expression was found in severe COVID‐19 patients compared to moderate patients and healthy individuals (Tang et al., 2020). Also, by a single‐cell transcriptomic analysis in the respiratory tract and peripheral blood, NEAT1 and MALAT1 were differentially expressed between mild and severe patients (Huang et al., 2021).
SARS‐CoV‐2′s first entry is through mouth and nose cells, and differential expression in these sites may be associated with an initial response to infection, infectivity, viral dosage, and clinical association. The present study presents NEAT1 and MALAT1 high expression in saliva and nasopharyngeal samples in SARS‐CoV‐2 positive individuals, and these results contribute to a better understanding of COVID‐19. NEAT1 and MALAT1 are important components of antivirus immune responses and may provide, in the future, new targets for severity‐related diagnostic measures or therapy.
CONFLICT OF INTEREST
The authors have stated explicitly that there are no conflicts of interest in connection with this article.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1111/omi.12351
ACKNOWLEDGMENTS
This work was supported by the: PROPLAN/Federal University of Parana, FINEP‐Funder of Studies and Projects, Ministry of Science, Technology and Innovation‐Brazil, CAPES (001‐Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) and CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico). We are grateful to the Setor de Ciências Biológicas of the Federal University of Parana and all team of volunteers at SCB‐UFPR COVID‐19 team.
Rodrigues, A. C. , Adamoski, D. , Genelhould, G. , Zhen, F. , Yamaguto, G. E. , Araujo‐Souza, P. S. , Nogueira, M. B. , Raboni, S. M. , Bonatto, A. C. , Gradia, D. F. , & Carvalho de Oliveira, J. (2021). NEAT1 and MALAT1 are highly expressed in saliva and nasopharyngeal swab samples of COVID‐19 patients. Molecular Oral Microbiology, 36, 291–294. 10.1111/omi.12351
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon request.
REFERENCES
- Adamoski D., Oliveira J. C. d., Bonatto A. C., Wassem R., Nogueira M. B., Raboni S. M., Trindade E. d. S., Souza E. M. d., SCB‐UFPR COVID‐19 team , & Gradia D. F. (2021). Large‐scale screening of asymptomatic for SARS‐CoV‐2 variants of concern and rapid P.1 takeover, Curitiba, Brazil. MedRxiv, 2021.06.18.21258649. medRxiv [Google Scholar]
- Bocchetti, M. , Scrima, M. , Melisi, F. , Luce, A. , Sperlongano, R. , Caraglia, M. , Zappavigna, S. , & Cossu, A. M. (2021). LncRNAs and immunity: Coding the immune system with noncoding oligonucleotides. International Journal of Molecular Sciences, 22(4), 1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili, M. N. , Trapnell, C. , Goff, L. , Koziol, M. , Tazon‐Vega, B. , Regev, A. , & Rinn, J. L. (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & Development, 25(18), 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. G. , Satpathy, A. T. , & Chang, H. Y. (2017). Gene regulation in the immune system by long noncoding RNAs. Nature Immunology, 18(9), 962–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, S.‐W. , Kim, H.‐W. , & Nam, J.‐W. (2019). The small peptide world in long noncoding RNAs. Briefings in Bioinformatics, 20(5), 1853–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, H. , Banerjee, S. , Guo, S. , Xie, N. , Ge, J. , Jiang, D. , Zörnig, M. , Thannickal, V. J. , & Liu, G. (2019). Long noncoding RNA Malat1 regulates differential activation of macrophages and response to lung injury. JCI Insight, 4(4), e124522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heward, J. A. , & Lindsay, M. A. (2014). Long non‐coding RNAs in the regulation of the immune response. Trends in Immunology, 35(9), 408–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, K. , Wang, C. , Vagts, C. , Raguveer, V. , Finn, P. W. , & Perkins, D. L. (2021). LncRNAs NEAT1 and MALAT1 differentiate inflammation in severe COVID‐19 patients. MedRxiv: The Preprint Server for Health Sciences, medRxiv [Google Scholar]
- Liu, W. , Wang, Z. , Liu, L. , Yang, Z. , Liu, S. , Ma, Z. , Liu, Y. , Ma, Y. , Zhang, L. , Zhang, X. , Jiang, M. , & Cao, X. (2020). LncRNA Malat1 inhibition of TDP43 cleavage suppresses IRF3‐initiated antiviral innate immunity. Proceedings of the National Academy of Sciences of the United States of America, 117(38), 23695–23706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon, M. P. , & Hua, K.‐F. (2020). The long non‐coding RNAs: Paramount regulators of the NLRP3 inflammasome. Frontiers in Immunology, 11, 569524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazzam‐Jazi, M. , Lanjanian, H. , Maleknia, S. , Hedayati, M. , & Daneshpour, M. S. (2021). Interplay between SARS‐CoV‐2 and human long non‐coding RNAs. Journal of Cellular and Molecular Medicine, 25, 5823–5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang, J. , Hu, J. , & Chen, J. (2016). LncRNAs regulate the innate immune response to viral infection. Wiley Interdisciplinary Reviews. RNA, 7(1), 129–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniri, A. , & Akhavan‐Niaki, H. (2020). Emerging role of IL‐6 and NLRP3 inflammasome as potential therapeutic targets to combat COVID‐19: Role of lncRNAs in cytokine storm modulation. Life Sciences, 257, 118114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, L. , Wang, T. , Tang, Q. , Li, G. , Wu, P. , & Chen, K. (2018). Long non‐coding RNAs: Regulators of viral infection and the interferon antiviral response. Frontiers in Microbiology, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, H. , Gao, Y. , Li, Z. , Miao, Y. , Huang, Z. , Liu, X. , Xie, L. , Li, H. , Wen, W. , Zheng, Y. , & Su, W. (2020). The noncoding and coding transcriptional landscape of the peripheral immune response in patients with COVID‐19. Clinical and Translational Medicine, 10(6), e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, H. , Wu, M. , Zhou, P. , Huang, C. , Ye, C. , & Wang, L. (2018). The long non‐coding RNA MALAT1 is increased in renal ischemia‐reperfusion injury and inhibits hypoxia‐induced inflammation. Renal Failure, 40(1), 527–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnubalaji, R. , Shaath, H. , & Alajez, N. M. (2020). Protein coding and long noncoding RNA (lncRNA) transcriptional landscape in SARS‐CoV‐2 infected bronchial epithelial cells a role for interferon and inflammatory response. Genes, 11(7), 760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, S. , Dong, B. , Tang, L. , & Zhou, S. (2018). LncRNA MALAT1 sponges miR‐133 to promote NLRP3 inflammasome expression in ischemia‐reperfusion injured heart. International Journal of Cardiology, 254, 50. [DOI] [PubMed] [Google Scholar]
- Zhang, P. , Cao, L. , Zhou, R. , Yang, X. , & Wu, M. (2019). The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nature Communications, 10(1), 1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.


