Abstract
Background
Emerging data support detectable immune responses for months after severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection and vaccination, but it is not yet established to what degree and for how long protection against reinfection lasts.
Methods
We investigated SARS‐CoV‐2‐specific humoral and cellular immune responses more than 8 months post‐asymptomatic, mild and severe infection in a cohort of 1884 healthcare workers (HCW) and 51 hospitalized COVID‐19 patients. Possible protection against SARS‐CoV‐2 reinfection was analyzed by a weekly 3‐month polymerase chain reaction (PCR) screening of 252 HCW that had seroconverted 7 months prior to start of screening and 48 HCW that had remained seronegative at multiple time points.
Results
All COVID‐19 patients and 96% (355/370) of HCW who were anti‐spike IgG positive at inclusion remained anti‐spike IgG positive at the 8‐month follow‐up. Circulating SARS‐CoV‐2‐specific memory T cell responses were detected in 88% (45/51) of COVID‐19 patients and in 63% (233/370) of seropositive HCW. The cumulative incidence of PCR‐confirmed SARS‐CoV‐2 infection was 1% (3/252) among anti‐spike IgG positive HCW (0.13 cases per 100 weeks at risk) compared to 23% (11/48) among anti‐spike IgG negative HCW (2.78 cases per 100 weeks at risk), resulting in a protective effect of 95.2% (95% CI 81.9%–99.1%).
Conclusions
The vast majority of anti‐spike IgG positive individuals remain anti‐spike IgG positive for at least 8 months regardless of initial COVID‐19 disease severity. The presence of anti‐spike IgG antibodies is associated with a substantially reduced risk of reinfection up to 9 months following asymptomatic to mild COVID‐19.
Keywords: COVID‐19, humoral response, long‐term immunity, reinfection, SARS‐CoV‐2

Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has infected millions of people worldwide and has caused a global crisis. As new waves of the pandemic cause significant morbidity and mortality around the globe, the extent to which previously infected individuals are protected from reinfection becomes increasingly pivotal.
Long‐term adaptive immunity likely relies on both humoral and cellular contributions. It is now well‐established that the majority of COVID‐19 cases seroconvert [1, 2], but immunoglobulin G (IgG) antibody kinetics clearly depend on the antigen used in the assays which has resulted in discrepancies between studies [2, 3, 4, 5]. The spike glycoprotein and the nucleocapsid protein are the most commonly used antigens in currently available serology assays. The spike glycoprotein contains the receptor‐binding domain, rendering it the main target for neutralizing antibodies and vaccine development. Anti‐spike IgG antibodies have been shown to remain relatively stable for at least 5 months [4] and correlate well with neutralization of authentic SARS‐CoV‐2 [4].
Although T cells do not prevent infections on their own, CD4+ T cells, and in particular T follicular helper (Tfh) cells, are crucial for the generation of neutralizing antibodies while antigen‐specific memory CD8+ T cells are key players in clearing virus‐infected cells upon infection and reinfection. SARS‐CoV‐2‐specific CD4+ and CD8+ T cells have been identified in the peripheral blood of approximately 100% and 70% of COVID‐19 patients shortly after recovery, respectively [6]. Further, a recent longitudinal study investigating both humoral and cellular immune memory found that the majority of 36 COVID‐19 cases followed up for 6 months remained positive for anti‐spike IgG as well as circulating CD4+ T cells, including Tfh cells, whereas the percentage of cases with measurable circulating memory CD8 T cells declined over time [5]. These findings, although based on relatively small cohorts, support a robust immunological memory lasting for at least 6 months. Analyses of cellular immune responses are, however, more complicated than antibody analyses and therefore less investigated. Assessments of SARS‐CoV‐2‐specific T cell memory responses are further hampered by the presence of cross‐reactive memory T cells stemming from prior encounters with endemic human coronaviruses, and SARS‐CoV‐2 cross‐reactive memory T cells are detectable in up to 50% of individuals not exposed to SARS‐CoV‐2 [6, 7, 8, 9]. The functional role, if any, of cross‐reactive T cells in COVID‐19 disease progression and herd immunity, however, remains under debate [10].
The COMMUNITY study is an ongoing longitudinal cohort study investigating long‐term immunological responses after SARS‐CoV‐2 infection in 2149 healthcare workers (HCW) and 118 hospitalized COVID‐19 patients [11, 12]. Blood samples were first obtained at study inclusion in April‐June 2020 and are from then on collected prospectively every 4 months. Here, we investigated SARS‐CoV‐2‐specific humoral and cellular immune responses at least 8 months post‐infection. In addition, we assessed the extent to which previously infected individuals are protected from reinfection through a 3‐month weekly polymerase chain reaction (PCR) screening of HCW that had seroconverted 7 months prior to the start of the 3‐month screening with seronegative HCW serving as controls.
Results
All 51 convalescent COVID‐19 patients and 96% of the 370 HCW who had been anti‐spike‐IgG positive at study inclusion remained anti‐spike IgG positive at the 3‐month follow‐up. Anti‐spike IgG levels were two‐fold higher in convalescent COVID‐19 patients (n = 51) than in HCW ≥ 8 months post‐infection (n = 370) (medians [IQR] 0.90 [0.76–0.98] and 0.48 [0.28–0.72], respectively; p = 5×10–16), in line with a disease severity‐dependent humoral response. The levels of anti‐spike IgG were also significantly higher in the group of HCW ≤ 4 months post‐infection (n = 259) than in HCW ≥ 8 months post‐infection (n = 370) (medians [IQR] 0.58 [0.37–0.76] and 0.48 [0.28–0.72], respectively; p = 0.002), although the differences were modest, implying relatively stable levels over time after asymptomatic to mild disease. The levels were similar in HCW 5–8 months post‐infection (n = 116) and HCW ≥ 8 months post‐infection (n = 370) (medians [IQR] 0.57 [0.30–0.74] and 0.48 [0.28–0.72], respectively; p = 0.4), and in HCW ≤ 4 months post‐infection (n = 259) and HCW 5–8 months post‐infection (n = 116) (medians [IQR] 0.58 [0.37–0.76] and 0.57 [0.30–0.74], respectively; p = 0.2) Fig. 1a.
Fig. 1.
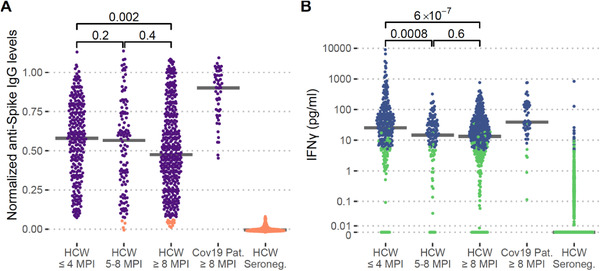
Long‐term humoral and cellular immune responses in healthcare workers (HCW) and COVID‐19 patients. Normalized anti‐spike IgG levels (a) and concentration of background‐adjusted interferon‐gamma (IFN‐γ) levels after SARS‐CoV‐2‐specific peptide stimulation of whole blood (b) in HCW less than or equal to 4 months post‐infection (HCW ≤ 4 MPI), n = 259, HCW 5–8 months post‐infection (HCW 5–8 MPI), n = 116, HCW at least 8 months post‐infection (HCW ≥ 8 MPI), n = 370, hospitalized COVID‐19 patients at least 8 months post‐infection (Cov19 Pat ≥ 8 MPI), n = 51, and anti‐spike IgG negative HCW at all sampling time points, n = 1076. Purple and orange: Anti‐Spike IgG seropositive and seronegative, respectively. Blue and green: Positive and negative IFN‐γ response to the SARS‐CoV‐2‐specific peptide pool, respectively. p‐values are shown with brackets
The presence and magnitude of SARS‐CoV‐2 T cell memory responses were assessed in all study participants based on IFN‐γ levels after stimulation of fresh whole blood using two different SARS‐CoV‐2 peptide pools. To assess a specific SARS‐CoV‐2 T cell memory response, we generated a SARS‐CoV‐2‐specific peptide pool with no more than five amino acid stretches aligning with endemic coronaviruses (Table S1) and an estimated HLA‐coverage of 97%. Samples were stimulated with this peptide pool alongside a commercially available SARS‐CoV‐2 peptide pool with an immunodominant peptide region present in the majority of endemic coronaviruses [13] (own unpublished data). Based on the set binary scoring, T cell responses (IFN‐γ+) were detected in response to the SARS‐CoV‐2‐specific peptide pool in 88% (45/51) of convalescent COVID‐19 patients and in 63% (234/370) of HCW ≥ 8 months post‐infection. The levels of IFN‐γ‐responses were three‐fold higher in COVID‐19 patients (n = 51) than in HCW ≥ 8 months post‐infection (n = 370) (medians [IQR] 39 [24–130] and 13 [3.2–38] pg/ml, respectively; p = 3*10–8), indicating a disease severity‐dependent immune memory. The levels were also significantly higher in HCW ≤ 4 months post‐infection (n = 258) than in HCW 5–8 months post‐infection (n = 116), (medians [IQR] 25 [7–72] and 15 [4.2–34] pg/ml; p = 8×10–4) and in HCW ≥ 8 months post‐infection (n = 370) (median [IQR] 13 [3.2–38] pg/ml, p = 6×10–7). No significant difference was, however, found between HCW 5–8 months post‐infection and HCW ≥ 8 months post‐infection (n = 370) (p = 0.6), suggesting that the cellular immune memory wanes over the first few months after infection and then stabilizes (Fig. 1b). Notably, only 1.5% (16/1076) of seronegative HCW responded to the SARS‐CoV‐2‐specific peptide pool, arguing against the generation of a SARS‐CoV‐2‐specific T cell immunity in the absence of seroconversion. All (n = 51) convalescent patients and 96% (357/370) of HCW ≥ 8 months post‐infection responded to the broader SARS‐CoV‐2 peptide pool, harboring an immunodominant epitope overlapping with endemic coronaviruses. However, also 43% (462/1076) of HCW who had been seronegative at all sample time points responded to this peptide pool (Fig. S1), possibly due to previous exposure to endemic coronaviruses.
There were no significant differences in anti‐spike IgG levels or in specific SARS‐CoV‐2 T cell memory responses ≥ 8 months post‐infection between HCW with and without COVID‐19 patient contact during the study period (median [IQR] 0.50 [0.29–0.72] and 0.46 [0.30–0.68]; p = 0.7 and 14.0 [3.0–41] and 9.0 [2.3–20] pg/ml; p = 0.2, respectively).
There were no differences in age (median [IQR] age 43 [33–52] in HCW ≤ 4 months post‐infection, 42.5 [32–50] in HCW 5–8 months post‐infection, and 44 [34–53] in HCW ≥ 8 months post‐infection [p = 0.6, 0.8 and 0.5]) or gender (odds ratios [95% confidence intervals] 0.84 [0.65–1.6] for HCW ≤ 4 months post‐infection vs. HCW 5–8 months post‐infection [p = 0.7], 0.97 [0.62–1.5] for HCW ≤ 4 months post‐infection vs. HCW ≥ 8 months post‐infection [p = 0.9] and 1.1 [0.63–2.2] for HCW 5–8 months post‐infection vs. ≥ HCW 8 months post‐infection [p = 0.8]) between the seropositive HCW groups.
To address the capacity of post‐infection immune responses to protect against reinfection, a weekly SARS‐CoV‐2 qPCR screening was performed on 252 HCW who had seroconverted at study inclusion (7 months prior to initiation of the qPCR screening) and 48 SARS‐CoV‐2 seronegative HCW. Adherence to screening test was high with a sampling median at 11 of 12 weeks in both groups. The cumulative incidence of qPCR‐confirmed SARS‐CoV‐2 infection was 1% (3/252) among anti‐spike IgG positive HCW (0.13 per 100 weeks at risk) compared to 23% (11/48) among anti‐spike IgG negative HCW (2.78 per 100 weeks at risk) (Fig. 2); rendering an incident rate ratio of 0.05 (95% CI 0.01–0.18) and a protective effect of 95.2% (95% CI 81.9%–99.1%) for HCWs that had seroconverted. These findings imply a substantially reduced risk of SARS‐CoV‐2 reinfection for up to 9 months following mild COVID‐19. Notably, this period was marked by high risk for viral exposure as shown by the high rate of SARS‐CoV‐2 infection in the SARS‐CoV‐2 seronegative HCW group working in the same environment.
Fig. 2.

Incidence of three‐month weekly qPCR screening. The cumulative incidence of qPCR‐confirmed SARS‐CoV‐2 infection in healthcare workers who had seroconverted (anti‐spike IgG; red line) 7 months prior to initiation of the qPCR screening, and in anti‐spike IgG negative healthcare workers (blue line)
Discussion
Our findings support a robust immune memory for at least 8 months following asymptomatic to mild COVID‐19. Although the magnitudes of both humoral and cellular immune responses were found to be dependent on disease severity, our results show that asymptomatic to mild COVID‐19 is associated with a substantially reduced risk of reinfection for up to at least 9 months.
Neutralizing antibodies have earlier been shown to persist for up to 5 months following mild COVID‐19 [4]. Here, we measured both detectable anti‐spike IgG antibodies and T cell memory at least 8 months post‐infection as well as the risk of reinfection in seropositive participants up to 9 months following asymptomatic to mild COVID‐19. Although reports of reinfection have been relatively few considering the global spread of the virus, the magnitude of immune responses required to confer protection against reinfection is not well known. A recent study investigated the relationship between anti‐spike IgG antibodies and the risk of SARS‐CoV‐2 reinfection in a large cohort of 1265 anti‐spike IgG positive HCW and found substantially lower risk of reinfection among HCW with anti‐spike IgG antibodies, including titers below the positive threshold [14]. Test frequency was, however, relatively low, on average once every 10–13 weeks. Another large retrospective cohort study using deidentified longitudinally linked commercial laboratory data found that the presence of SARS‐CoV‐2 antibodies was associated with a reduced risk of having a positive diagnostic nucleic acid amplification test [15]. Both these large studies, although providing important insights into post‐infection immunity, are however hampered by low rates of testing [14] as well as selection bias [15], and asymptomatic infections may thus have gone undetected. The high compliance with weekly test frequency and the notably high risk for viral exposure as shown by the high rate of SARS‐CoV‐2 infection in seronegative HCW working in the same environment in our study strengthen these prior findings and extend the data to up to 9 months post‐infection. We found that the presence of anti‐spike IgG not only protected against symptomatic COVID‐19, but also asymptomatic infection, which is crucial considering that asymptomatic cases likely act as silent drivers of the pandemic [16]. Notably, protection level in the group of anti‐spike IgG positive HCW up to 9 months following detected seroconversion was similar to demonstrated vaccine efficacy of mRNA immunization [17, 18]. These findings may contribute to the scientific ground in vaccine prioritization, such as directing vaccines towards seronegative individuals in countries with vaccine shortage and low vaccine coverage [19].
Although T cells do not protect against infection on their own, robust adaptive long‐term immunity relies on the interplay between humoral and cellular immune memory compartments. Assessments of SARS‐CoV‐2‐specific T cell memory responses are challenging due to the presence of cross‐reactive memory T cells stemming from prior encounters with endemic human coronaviruses. All convalescent COVID‐19 patients and the vast majority of anti‐spike IgG positive HCW (96%) were found to retain a T cell memory response towards the SARS‐CoV‐2 peptide pool entailing an immunodominant epitope overlapping with endemic coronaviruses. Notably, however, a large portion of seronegative HCW (43%, 462/1076) also responded to this peptide pool, likely due to previous exposure to endemic coronaviruses. In contrast, only 1.5% (16/1076) of the seronegative HCW responded to the in‐house generated SARS‐CoV‐2‐specific peptide pool with a predicted 97% HLA class I and II combined coverage. The in‐house peptide pool encompasses peptides generated by the genomic variants of the P13L as well as the D614G mutation. Using this peptide pool, the majority, albeit a smaller proportion, of convalescent COVID‐19 patients (88%, 45/51) and anti‐spike IgG positive HCW (63%, 234/370) still responded. Whether a portion of SARS‐CoV‐2 infected individuals mount a cellular immune response in the absence of seroconversion, and whether a pre‐existing cross‐reactive T cell immunity stemming from prior exposure to other coronaviruses may influence the trajectory of the pandemic has been under extensive debate [7, 10, 20]. Although the exclusion of immunodominant epitopes overlapping with endemic coronaviruses may underestimate the SARS‐CoV‐2 cellular immune response, our results argue against a significant portion of COVID‐19 cases in whom the infection is cleared solely by a T cell mediated immune response without seroconversion. This is further supported by the large cohort studies from Iceland [2] and the United States [1] showing that the vast majority of qPCR‐confirmed COVID‐19 cases undergo seroconversion. These findings support that anti‐spike IgG seroprevalence surveys reflect the majority of COVID‐19 convalescents. The functional role of a pre‐existing immune memory towards endemic coronaviruses in COVID‐19 disease progression, however, remains to be investigated [10].
This study is strengthened by the large sample size, frequent blood, nasal/oropharyngeal swabs and saliva samplings along with the high rate of follow‐up. Limitations include the fact that the cohort of individuals with asymptomatic to mild COVID‐19 is entirely composed of HCW. Considering the high viral exposure and transmission in hospital settings [11], the durable immune memory seen in this cohort may thus in part reflect repeated viral encounters boosting the immune memory. Importantly, the magnitude of immune response did not, however, differ between HCW with and without COVID‐19 patient contact, implying that the cohort is representative of an age‐matched community population. The HCW cohort is furthermore composed of a majority of women, and possible gender‐driven differences are not encompassed in these analyses. The frequencies of a measurable circulating T cell response over time will depend on the individual peptide pool to capture the cytotoxic as well as the helper T cell response. The in‐house‐generated peptide pool in this study may be slightly skewed to a class I epitope coverage (88%) compared to a class II coverage (77%), possibly favoring a CD8 T cell response readout, which has been reported to decline over time in circulation more rapidly than CD4 responses [5]. The study was further conducted during a time period without known prevalence of emerging variants of SARS‐CoV‐2, and the risk of reinfection caused by these is not encompassed in these analyses.
Taken together, our findings support a broad immune memory for at least 8 months following asymptomatic to mild COVID‐19. We furthermore show that the presence of anti‐spike IgG is associated with a substantially reduced risk of reinfection up to 9 months following asymptomatic to mild COVID‐19. In an era of limited vaccine supplies, taking serostatus into consideration in COVID‐19 vaccine prioritization strategies may extend the benefits of vaccination.
Methods
Study population and study design
The COMMUNITY study enrolled 2149 HCW and 118 hospitalized COVID‐19 patients at Danderyd Hospital, Stockholm, Sweden, between 9 April and 8 June 2020. Blood samples were first obtained at study inclusion and are from then on collected prospectively approximately every 4 months (mean [SD] 125 [8] and 121 [15] days until first follow‐up of HCW and patients respectively, and mean [SD] 263 [8] and 260 [15] days until second follow‐up of HCW and patients, respectively). Detailed symptomatology is obtained through a smartphone app system using standardized questionnaires prior to each blood sampling. Clinical, demographic and serological data at study inclusion have been presented elsewhere [11, 12]. At the 8‐month follow‐up, a total of 1884 HCW and 51 COVID‐19 patients remained in the study, rendering an 88% follow‐up rate of HCW and 43% follow‐up for patients (Fig. 3). COVID‐19 patients who did not come for the follow‐ups were either diseased (n = 14) or did not answer repeated invitations (n = 53). HCW who did not come for follow‐up did not answer on repeated invitations (n = 265). The majority of HCW were women (85%, 1606/1884), and the median age was 46 (IQR 35–54) years. A total of 1669/1884 HCW (89%) worked with patients, of which 1344 of 1669 HCW (81%) had COVID‐19 patient contact. HCW were stratified into four groups depending on serostatus; seropositive at study inclusion (i.e., ≥ 8 months post‐infection, n = 370), seroconversion between study inclusion and the 4‐month follow‐up (i.e., 5–8 months post‐infection, n = 116), seroconversion between the 4‐month follow‐up and the 8‐month follow‐up (i.e., ≤ 4 months post‐infection, n = 259), and seronegative at all timepoints (n = 1076). HCW who were seronegative at study inclusion and did not attend the 4‐month follow‐up (n = 63) were excluded from the current analyses. Among the 370 HCW who were seropositive at study inclusion, 9% (33/370) reported to have had no symptoms, 78% (286/370) reported to have had mild symptoms and 13% (47/370) reported to have had moderate symptoms. The COVID‐19 patient group was predominantly male (67%, 34/51), and the median age was 60 (IQR 50–66) years. The study was approved by the Swedish Ethical Review Authority (dnr 2020‐01653), and informed consent was obtained from all study participants.
Fig. 3.
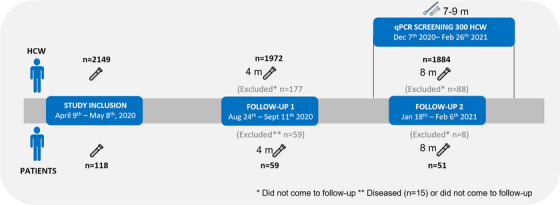
Study timeline. Flow chart depicting number of healthcare workers (HCW) and COVID‐19 patients at each follow‐up and qPCR screening sub‐study. Patients indicate COVID‐19 patients
Serological assays
The serological assay was performed utilizing a bead‐based high‐throughput multiplex assay based on the FlexMap3D (Luminex Corp.) platform [11, 21]. Anti‐Spike IgG was measured using beads with immobilized in‐house produced spike trimers containing the prefusion‐stabilized glycoprotein ectodomain [22]. To account for inter‐assay variability, raw serological data (median fluorescence intensity, arbitrary units) were normalized to the mean of 12 negative and four positive controls included in every assay. Seropositivity was defined as six times the standard deviation above the mean of the 12 negative control samples that were included for inter‐assay normalization. The 12 negative controls were thoroughly selected from 2090 pre‐pandemic samples to represent the general distribution of background signals. Based on a separate method validation using 331 positive control samples collected at least 17 days after symptom onset or positive qPCR‐test and 2090 negative control samples collected before 2020, the sensitivity and specificity were determined to be 99.7% (330 of 331 positive, 98.3–100.0, 95% CI) and 98.1% (2050 of 2090 negative, 97.4–98.6, 95% CI), respectively, for the anti‐Spike IgG classification [21]. All samples were analyzed at a dilution of 1:50, utilizing a broad dynamic range of 104 arbitrary units.
Whole blood interferon‐gamma (IFN‐γ) release assay (IGRA)
The detection of SARS‐CoV‐2 T cell responses was performed using two SARS‐CoV‐2 peptide pools. A SARS‐CoV‐2‐specific peptide pool was generated comprising a selection of 16 SARS‐CoV‐2‐specific peptides covering the SARS‐CoV‐2 spike, nucleocapsid protein, membrane protein, and open reading frame 3 and 7 with peptide stretches containing no more than 5‐mer length overlap with endemic coronaviruses (Table S1). The peptides were synthesized and delivered with a purity of >95% and with an estimated HLA‐coverage of 97% (class I and II combined). A second SARS‐CoV‐2 peptide pool (Mabtech, Sweden) containing 47 peptides covering multiple SARS‐CoV‐2 antigens with a mean purity of 80% (60%–99%) was used in parallel on all samples. This peptide pool contains a reported immunodominant epitope in the nucleocapsid protein region overlapping with endemic coronaviruses within the sequence LSPRWYFYYLGTGPEAGL [13]. Peripheral blood was collected in lithium heparin tubes and 500 μl was added to tubes containing glucose (2 mg/ml whole blood) and 0.9% NaCl with and without the stimulant (in‐house generated peptide pool 1 μg/ml whole blood, Mabtech SARS‐CoV‐2 peptide pool 0.5 μg/ml whole blood or Mabtech monoclonal anti‐CD3 clone CD3‐2 0.1 μg/ml whole blood) within 6 h. Samples were subsequently incubated for 20 h at 37°C with 5% CO2, after which plasma was collected and IFN‐γ, and IL‐2 levels were analyzed using Mesoscale Discovery V‐plex kit (Meso Scale Diagnostics, Maryland, USA). A positive control stimulation (anti‐CD3) was included for 59 seropositive and 88 seronegative samples to assess for cell viability and assay robustness, showing significantly elevated IFN‐γ and IL‐2 responses to the two peptide pools and anti‐CD3 stimulation compared to the negative control (unstimulated) in seropositive samples (Fig. S2). To further confirm that the IFN‐γ secretion in response to the SARS‐CoV‐2 peptide mix stems from T cells, a peptide stimulation in the presence of brefeldin was performed followed by intracellular flow cytometry analysis. We noted a clear IFN‐γ response in CD3+ cells, while the CD3‐gated cells showed no IFN‐γ secretion (n = 2, data not shown). The whole cohort was run with a negative control sample (unstimulated) and the peptide pool stimulations. Results are plotted with background sample deducted (Figs. 1B and S2). For a binary T cell response scoring, a sample was scored positive if the IFN‐γ level was above the mean average of vehicle stimulated blood of the whole cohort (HCW and patient cohort respectively) and if ≥ two‐fold above its own vehicle stimulated level. IFN‐γ levels are displayed on a pseudo‐logarithmic scale.
Three‐month weekly qPCR screening
Weekly SARS‐CoV‐2 qPCR screening was conducted in self‐collected nasal and oropharyngeal swabs and saliva on a sub‐cohort of 300 HCW. Two hundred and fifty‐two HCW had seroconverted at study inclusion 7 months prior to initiation of the qPCR screening, and 48 HCW were anti‐spike IgG negative at sampling 3 months prior and served as control group. qPCR screening continued for 12 consecutive weeks between 7 December and 26 February 2021. Participants were considered to be at risk for infection to the end of the 12‐week screening period or until a positive qPCR test result or until COVID‐19 vaccination whichever occurred first. SARS‐CoV‐2 RT‐PCR was performed on all collected samples. Participants collected nasal and oropharyngeal swabs and saliva in a 1‐ml tube with storage buffer. Samples were mixed with Trizol; RNA was extracted using PSS magLEAD 12gC, after which RT‐qPCR was performed. Primers (CATGTGTGGCGGTTCACTATATGT and TGTTAAARACACTATTAGCATAWGCAGT) and probe (FAM‐CAGGTGGAACCTCATCAGGAGATGC‐QSY) targeting the RNA dependent RNA polymerase region were used.
Statistical analyses
Group comparisons were performed using the Wilcoxon rank‐sum test. T cell activation levels are displayed on a pseudo‐logarithmic scale (R package scales version 1.1.1). Group comparisons on repeated measures data were performed using the Friedman test with post‐hoc pairwise Wilcoxon signed‐rank tests (R package rstatix version 0.6.0). The post‐hoc tests were FDR corrected using the Benjamini‐Hochberg method. Data visualization and statistical analyses were performed in R [23]. Incidence‐rate comparison was made in STATA version 16.1 (StataCorp, LP) using the STATA command iri.
Funding information
Region Stockholm; Knut and Alice Wallenberg foundation; Jonas & Christina af Jochnick foundation; Lundblad family foundation; Science for Life Laboratory (SciLifeLab); Erling‐Persson family foundation; Svenska Sällskapet för Medicinsk Forskning; Swedish Research Council; Centrum for Innovativ Forskning (CIMED).
Conflict of interest
The authors declare no competing interests.
Supporting information
Supporting information
Acknowledgements
The authors are grateful to Carola Jonsson, Camilla Redhevon, Sofia Englund, Lisa Törnlöf, Sofie Lundin, Nelly Romero, Carin Erlandsson, Maria Johansson, Monica Hoffman, Erica Åhlander, Anna Weimar, Malgorzata Wolski, Frehiywot Tesfatsioni and Helene Andersson at Danderyd Hospital for assisting in administration and blood sampling. We would like to thank Richard Scholvin for technical support with the smartphone app and assisting with data information and Carina Rudberg and Christina Einarsson for assisting with data collection. The Protein Factory at KTH is acknowledged for protein production and purification and Sofia Bergström, Shaghayegh Bayati and Sara Mravinacova at KTH and SciLifeLab for technical assistance. The authors would like to acknowledge support of Clinical Biomarkers Facility at SciLifeLab Uppsala Sweden for providing assistance in cytokine analyses.
Havervall S, Ng H, Falk AJ, Greilert‐Norin N, Månberg A, Marking U, et al. Robust humoral and cellular immune responses and low risk for reinfection at least 8 months following asymptomatic to mild COVID‐19. J Intern Med. 2022;291:72–80.
Data availability statement
The anonymized datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- 1. Wajnberg A, Mansour M, Leven E, Bouvier NM, Patel G, Firpo‐Betancourt A, et al. Humoral response and PCR positivity in patients with COVID‐19 in the New York City region, USA: an observational study. Lancet Microbe. 2020;1(7):e283–e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gudbjartsson DF, Norddahl GL, Melsted P, Gunnarsdottir K, Holm H, Eythorsson E, et al. Humoral immune response to SARS‐CoV‐2 in Iceland. N Engl J Med. 2020;383(18):1724–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bonifacius A, Tischer‐Zimmermann S, Dragon AC, Gussarow D, Vogel A, Krettek U, et al. COVID‐19 immune signatures reveal stable antiviral T cell function despite declining humoral responses. Immunity. 2021;54(2):340–54. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wajnberg A, Amanat F, Firpo A, Altman DR, Bailey MJ, Mansour M, et al. Robust neutralizing antibodies to SARS‐CoV‐2 infection persist for months. Science (New York, NY). 2020;370(6521):1227–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dan JM, Mateus J, Kato Y, Hastie KM, Yu ED, Faliti CE, et al. Immunological memory to SARS‐CoV‐2 assessed for up to 8 months after infection. Science. 2021;371(6529):eabf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T cell responses to SARS‐CoV‐2 coronavirus in humans with COVID‐19 disease and unexposed individuals. Cell. 2020;181(7):1489–501. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sekine T, Perez‐Potti A, Rivera‐Ballesteros O, Stralin K, Gorin JB, Olsson A, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID‐19. Cell. 2020;183(1):158–68. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Braun J, Loyal L, Frentsch M, Wendisch D, Georg P, Kurth F, et al. SARS‐CoV‐2‐reactive T cells in healthy donors and patients with COVID‐19. Nature. 2020;587(7833):270–4. [DOI] [PubMed] [Google Scholar]
- 9. Mateus J, Grifoni A, Tarke A, Sidney J, Ramirez SI, Dan JM, et al. Selective and cross‐reactive SARS‐CoV‐2 T cell epitopes in unexposed humans. Science (New York, NY). 2020;370(6512):89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lipsitch M, Grad YH, Sette A, Crotty S. Cross‐reactive memory T cells and herd immunity to SARS‐CoV‐2. Nat Rev Immunol. 2020;20(11):709–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rudberg AS, Havervall S, Manberg A, Jernbom Falk A, Aguilera K, Ng H, et al. SARS‐CoV‐2 exposure, symptoms and seroprevalence in healthcare workers in Sweden. Nat Commun. 2020;11(1):5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Havervall S, Rosell A, Phillipson M, Mangsbo SM, Nilsson P, Hober S, et al. Symptoms and functional impairment assessed 8 months after mild COVID‐19 among health care workers. JAMA. 2021;325(19):2015–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, et al. SARS‐CoV‐2‐specific T cell immunity in cases of COVID‐19 and SARS, and uninfected controls. Nature. 2020;584(7821):457–62. [DOI] [PubMed] [Google Scholar]
- 14. Lumley SF, O'Donnell D, Stoesser NE, Matthews PC, Howarth A, Hatch SB, et al. Antibody status and incidence of SARSCoV‐2 infection in health care workers. N Engl J Med. 2021;384(6):533–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harvey RA, Rassen JA, Kabelac CA, Turenne W, Leonard S, Klesh R, et al. Association of SARS‐CoV‐2 seropositive antibody test with risk of future infection. JAMA Intern Med. 2021;181(5):672–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Buitrago‐Garcia D, Egli‐Gany D, Counotte MJ, Hossmann S, Imeri H, Ipekci AM, et al. Occurrence and transmission potential of asymptomatic and presymptomatic SARS‐CoV‐2 infections: a living systematic review and meta‐analysis. PLoS Med. 2020;17(9):e1003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, et al. Efficacy and safety of the mRNA‐1273 SARS‐CoV‐2 vaccine. N Engl J Med. 2021;384(5):403–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and efficacy of the BNT162b2 mRNA covid‐19 vaccine. N Engl J Med. 2020;383(27):2603–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bubar KM, Reinholt K, Kissler SM, Lipsitch M, Cobey S, Grad YH, et al. Model‐informed COVID‐19 vaccine prioritization strategies by age and serostatus. medRxiv. 2021. 10.1101/2020.09.08.20190629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sette A, Crotty S. Pre‐existing immunity to SARS‐CoV‐2: the knowns and unknowns. Nat Rev Immunol. 2020;20(8):457–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hober S, Olofsson J, Andersson E, Bergstrom S, Falk AJ, Mravinacova S, et al. Systematic evaluation of SARS‐CoV‐2 antigens enables a highly specific and sensitive multiplex serological COVID‐19 assay. Clin. Transl. Immunol. 2021;10(7):e1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, et al. Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Core Team R. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Data Availability Statement
The anonymized datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.


