Abstract
Fully automated immunoassays for detecting severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) antibodies that are strongly correlated with neutralization antibodies (nAbs) are clinically important because they enable the assessment of humoral immunity after infection and vaccination. Access SARS‐CoV‐2 immunoglobulin M (IgM) and immunoglobulin G (IgG) II antibody tests are semi‐quantitative, fully automated immunoassays that detect anti‐receptor‐binding domain (RBD) antibodies and might reflect nAb levels in coronavirus disease 2019 (COVID‐19). However, no studies have investigated the clinical utility of these tests in association with nAbs to date.
To evaluate the clinical utility of Access SARS‐CoV‐2 IgM and IgG II antibody tests and their correlation with the SARS‐CoV‐2 surrogate virus neutralization test (sVNT) that measures nAbs in patients with COVID‐19, we analyzed 54 convalescent serum samples from COVID‐19 patients and 89 serum samples from non‐COVID‐19 patients. The presence of anti‐RBD antibodies was detected using Access SARS‐CoV‐2 IgM and IgG II antibody tests, while nAbs were measured by sVNT.
The sensitivity and specificity of sVNT were 94.4% and 98.9%, respectively. There were strong positive correlations between the inhibition values of sVNT and the results of the Access SARS‐CoV‐2 IgM (R = 0.95, R 2 = 0.90, p < 0.001) and IgG II antibody tests (R = 0.96, R2 = 0.92, p < 0.001). In terms of the presence of nAbs, the sensitivity and specificity were 98.1% and 98.9% in the IgM assay and 100.0% and 100.0% in the IgG II assay, respectively.
The Access SARS‐CoV‐2 IgM and IgG II antibody tests showed high sensitivity and specificity for the detection of nAbs in COVID‐19 patients and might be alternatives for measuring nAbs.
Keywords: COVID‐19, IgG, IgM, neutralization antibody, SARS‐CoV‐2
1. INTRODUCTION
Coronavirus disease 2019 (COVID‐19) is an emerging disease caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). The spike (S) and nucleocapsid (N) structural proteins of SARS‐CoV‐2 are currently the main targets for antibody testing. The S protein contains two subunits (S1 and S2), and the receptor‐binding domain (RBD) in S1 is responsible for the recognition of angiotensin‐converting enzyme‐2, a human cell surface receptor. 1 Because antibodies specific for the RBD in S1 can neutralize SARS‐CoV‐2 by inhibiting its entry into host cells, anti‐RBD antibodies work as protective immune and neutralizing antibodies (nAbs). 2 , 3 Several effective vaccines have been developed using the S protein and vaccination programs have started worldwide. 4 , 5 , 6 , 7 Serological assays for detecting anti‐RBD antibodies and nAbs are becoming increasingly important for the evaluation of humoral immunity against SARS‐CoV‐2 not only after vaccination but also in past infections.
Currently, there is widespread use of fully automated immunoassays for the detection of anti‐SARS‐CoV‐2 antibodies with high sensitivity and specificity. Conventional neutralization assays require trained staff, specific equipment, and time‐consuming methods. The association between the results of fully automated immunoassays for anti‐SARS‐CoV‐2 antibodies and the presence of nAbs has mainly been explored because those assays are used as substitutes for neutralization assays. Although several assays are indeed useful to assess protective immunity against SARS‐CoV‐2, there is wide variability in their correlation with nAbs. 8 , 9 , 10 , 12 , 13 , 14 Semi‐quantitative chemiluminescent enzyme immunoassays using the RBD protein have been launched by Beckman Coulter, Inc. and include the Access SARS‐CoV‐2 immunoglobulin M (IgM) antibody test and the Access SARS‐CoV‐2 immunoglobulin M (IgG) II antibody test, which are new versions of the Access SARS‐CoV‐2 IgG antibody test previously granted Emergency Use Authorization by the US Food and Drug Administration. 15 However, no study has evaluated the association between the results of these antibody tests and the titers of nAbs to date.
Here, we evaluated the reliability of the thresholds of the Access SARS‐CoV‐2 IgM and IgG II antibody tests for predicting nAb levels using convalescent sera from COVID‐19 patients and sera from non‐COVID‐19 patients.
2. METHODS
2.1. Patients with COVID‐19, clinical data, and serum samples
Patient information was collected retrospectively from hospital electronic medical records. The study included a total of 54 serum samples collected from 45 COVID‐19 patients who were hospitalized at Saitama Medical University Hospital in Japan from February 11, 2020 to December 31, 2020. SARS‐CoV‐2 RNA was detected in all the patients by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) according to the Japanese Infectious Disease Law. 16 Briefly, the patients ranged in age from 33 to 94 years (median: 69 years; interquartile range [IQR]: 53–77 years); there were 32 men (71.1%) and 13 women (28.9%). Among the 45 patients, 35 (77.8%) required oxygen therapy during hospitalization. All serum samples used in this study were collected at ≥14 days after disease onset (median: 17 days; IQR: 16–20) and stored at −80°C until use.
2.2. Non‐COVID‐19 patients and serum samples
We also analyzed 89 serum samples collected from 89 patients with fever at Saitama Medical University Hospital in Japan, from February 11, 2020 to December 31, 2020. All patients were negative for SARS‐CoV‐2 at least once by RT‐qPCR and were then clinically diagnosed by their doctor as having a disease other than COVID‐19. Briefly, the patients ranged in age from 20 to 93 years (median: 73 years; IQR: 48–82 years); there were 49 men (55.1%) and 40 women (44.9%). All serum samples were stored at −80°C until use.
2.3. Detection of IgM and IgG antibodies for the RBD protein of SARS‐CoV‐2
IgM and IgG antibody assays against the SARS‐CoV‐2 RBD protein were performed using the Access SARS‐CoV‐2 IgM and IgG II Antibody Tests (Beckman Coulter, Inc.) according to the manufacturer's instructions. The results were interpreted as positive (signal for test sample/signal at cut‐off value [S/CO] ≥1.0) and negative (S/CO <1.0) in accordance with the manufacturer's instructions for the Access SARS‐CoV‐2 IgM Antibody Test, and as positive (≥10 arbitrary units [AU]/ml) and negative (<10 AU/ml) for the Access SARS‐CoV‐2 IgG II Antibody Test. All serum samples were evaluated in duplicate, and the average value for these measurements was defined as the test result.
2.4. Detection of nAbs against SARS‐CoV‐2
To measure nAbs against RBD of SARS‐CoV‐2, the SARS‐CoV‐2 surrogate virus neutralization test (sVNT) (GenScript) was used. For this assay, each serum sample was diluted 1/10 in sample dilution buffer, and an equal volume of horseradish peroxidase‐conjugated RBD solution was added to each diluted sample according to the manufacturer's instructions; each sample was tested in duplicate. To perform accurate correlation analysis, serum samples must be diluted appropriately to capture the entire linear range, but in this study, the sample dilution was fixed to 1/10 in accordance with the provided instructions. After incubation at 37°C for 30 min, 100 µl of each sample was added to the wells of a 96‐well plate and incubated at 37°C for 30 min. The plates were washed four times with wash solution. The enzymatic reaction was developed with 100 µl/well TMB solution at 25°C for 15 min. Thereafter, the reaction was stopped using a 50 µl/well stop solution, and the plates were read at 450 nm using an enzyme‐linked immunosorbent assay reader. The inhibition rate (%) was determined using the following formula: (1 − sample OD450 value/negative control OD450 value) × 100, according to the manufacturer's instructions.
2.5. Determination of the cut‐off value for nAbs against SARS‐CoV‐2
According to the manufacturer's instructions, the cut‐off value for sVNT for the presence of SARS‐CoV‐2 nAbs is inhibition ≥20%. However, sVNT has not been validated in the Japanese population. Therefore, we determined the optimal cut‐off value for our patient population as recommended by the manufacturer (https://www.genscript.com/). The optimal cut‐off value for sVNT was determined by maximizing Youden's index based on the receiver operating characteristic (ROC) curves using the COVID‐19‐positive (n = 54) and ‐negative (n = 89) samples.
2.6. Statistical analysis
Continuous variables are expressed as the mean and SD or median and IQR and were compared using the t test and the Wilcoxon rank‐sum test for parametric and nonparametric data, respectively. Linear regression analysis was used to assess the relationship between each continuous variable. Rho values were analyzed by the Spearman rank‐order correlation coefficient. A two‐sided p value <0.05 was considered statistically significant. All statistical analyses were conducted using R (v4.0.2; R Foundation for Statistical Computing, Vienna, Austria; http://www.R-project.org/).
3. RESULTS
3.1. Sensitivity and specificity of the access IgM and IgG antibody assays for COVID‐19
Distributions of S/CO and AU/ml for the Access IgM and IgG II assays for COVID‐19 and non‐COVID‐19 sera are shown in Figure 1A,B, respectively. In the IgM assay, the median S/CO value of the COVID‐19 samples was 16.8 (IQR: 7.0–37.8), while that of the non‐COVID‐19 samples was 0.2 (IQR: 0.2–0.2) (p < 0.001). In the IgG II assay, the median S/CO value of the COVID‐19 samples was 198.7 (IQR: 137.2–427.3), while that of the non‐COVID‐19 samples was 0.2 (IQR: 0.1–0.5) (p < 0.001). The area under the curve of the IgM assay was 0.99 and that of the IgG II assay was 0.99 (Figure 2A). The cut‐off value of the IgM assay determined from ROC curve analysis to maximize Youden's index was S/CO ≥1.00 and that of the IgG II assay was 10 AU/ml, and these values were concordant with the manufacturer's instructions. The overall sensitivity was 96.3% (95% confidence interval [CI]: 87.3%–99.5%) and specificity was 100% (95% CI: 95.9%–100%) in the IgM assay and 94.4% (95% CI: 84.6%–98.8%) and 98.9% (95% CI: 93.9%–99.9%) in the IgG II assay, respectively (Table 1).
Figure 1.
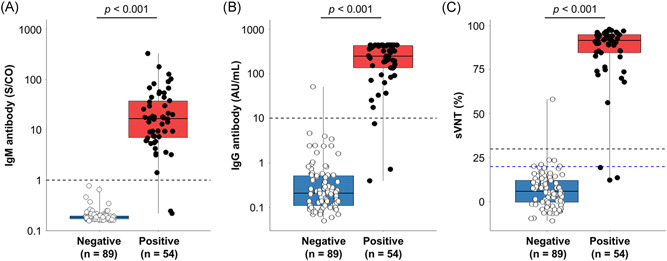
(A) Distribution of IgM and (B) IgG antibody levels for receptor‐binding domain protein and (C) inhibition of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) using the surrogate virus neutralization test (sVNT). Plot shows the distribution of the test results for each method. Black plots, coronavirus disease 2019 (COVID‐19) cases; white plots, non‐COVID‐19 cases; black horizontal dashed line, optimal cut‐off value based on the test results; and blue horizontal dashed line, cut‐off value provided by the manufacturer. IgG, immunoglobulin G; IgM, immunoglobulin M
Figure 2.
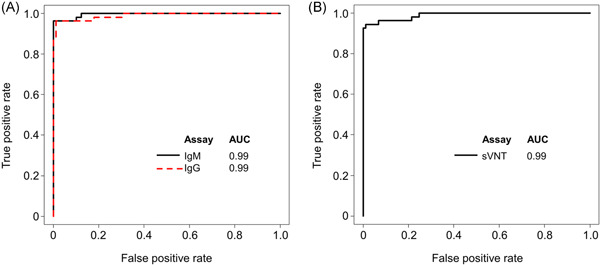
(A) Receiver operating characteristic curves of the IgM and IgG II antibody assays for receptor‐binding domain protein and of the (B) surrogate virus neutralization test (sVNT) for SARS‐CoV‐2. (A) Black line, IgM antibody assay; red dashed line, IgG antibody assay. (B) Black line, sVNT. AUC, area under the curve; IgG, immunoglobulin G; IgM, immunoglobulin M; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
Table 1.
Sensitivity, specificity, and cut‐off values of the IgM and IgG II antibody assays and nAbs for COVID‐19
| Specificity | Sensitivity | |||||||
|---|---|---|---|---|---|---|---|---|
| Non‐COVID‐19 samples | Detected | Not detected | Specificity (95% CI) | COVID‐19 samples | Detected | Not detected | Sensitivity (95% CI) | |
| IgM antibody assay | ||||||||
| Cut‐off S/CO ≥1.0 | 89 | 0 | 89 | 100% | 54 | 52 | 2 | 96.3% |
| (95.9–100) | (87.3–99.5) | |||||||
| IgG II antibody assay | ||||||||
| Cut‐off IgG ≥10 AU/ml | 89 | 1 | 88 | 98.9% | 54 | 51 | 3 | 94.4% |
| (93.9–99.9) | (84.6–98.8) | |||||||
| sVNT | ||||||||
| Cut‐off inhibition ≥20% | 89 | 6 | 83 | 93.3% | 54 | 51 | 3 | 94.4% |
| (85.9–97.5) | (84.6–98.8) | |||||||
| Cut‐off inhibition ≥30% | 89 | 1 | 88 | 98.9% | 54 | 51 | 3 | 94.4% |
| (93.9–99.9) | (84.6–98.8) | |||||||
Abbreviations: AU, arbitrary units; CI, confidence interval; COVID‐19, coronavirus disease 2019; IgG, immunoglobulin G; IgM, immunoglobulin M; nAb, neutralization antibody; S/CO, signal for the test sample/signal at cut‐off value; sVNT, surrogate virus neutralization test.
3.2. Alternative cut‐off point, sensitivity, and specificity for sVNT
The alternative cut‐off value for sVNT was determined using serum samples from COVID‐19 patients (n = 54) and non‐COVID‐19 patients (n = 89). Median inhibition was 91.4% (IQR: 84.6%–94.8%) for COVID‐19 and 5.9% (IQR: 0%–12.1%) for non‐COVID‐19 in sVNT (p < 0.001) (Figure 1C). In the ROC curve analysis, the area under the curve of sVNT was 0.99, and the alternative cut‐off value was determined to be ≥30% inhibition (Figure 2B). Sensitivity was 94.4% (95% CI: 84.6%–98.8%) and specificity was 98.9% (95% CI: 93.9%–99.9%) when using the alternative cut‐off value of ≥30% inhibition, and 94.4% (95% CI: 84.6%–98.8%) and 93.3% (95% CI: 85.9%–97.5%), respectively, when using the ≥20% inhibition cut‐off value provided by the manufacturer (Table 1). One non‐COVID‐19 patient was confirmed positive by both the sVNT and the IgG II antibody assay. Finally, 52 and 91 serum samples were determined to be positive and negative by sVNT, respectively.
3.3. Correlations of the IgM and IgG II antibody assays with sVNT
Regression analysis was conducted to evaluate the association between the results of sVNT and the results of the IgM and IgG II antibody assays (Figure 3A,B). Among the 143 serum samples collected from COVID‐19 and non‐COVID‐19 patients, there were strong positive correlations between inhibition values of sVNT and the values of the IgM (R = 0.95, R 2 = 0.90, p < 0.001) and IgG II (R = 0.96, R 2 = 0.92, p < 0.001) antibody assays. When using the alternative cut‐off value of sVNT (inhibition ≥30%), the sensitivity and specificity results of sVNT were 98.1% (95% CI: 89.7%–99.9%) and 98.9% (95% CI: 95.9%–100%) in the IgM assay, and 100% (95% CI: 93.2%–100%) and 100.0% (95% CI: 95.9%–100%) in the IgG II assay, respectively (Table 2).
Figure 3.
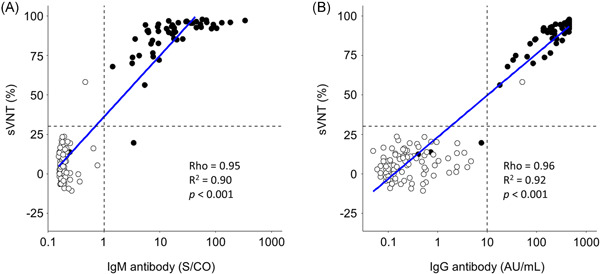
(A) Regression analyses between the inhibition of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) in the surrogate virus neutralization test (sVNT) and the values from the IgM and (B) IgG II antibody assays for receptor‐binding domain protein. Regression analyses were conducted by linear regression. Rho (R) values were analyzed by the Spearman rank‐order correlation coefficient. Black plots, coronavirus disease 2019 (COVID‐19) cases; white plots, non‐COVID‐19 cases; and blue line, fitting curve. AU, arbitrary units; COVID‐19, coronavirus disease 2019; IgG, immunoglobulin G; IgM, immunoglobulin M; S/CO, signal for test sample/signal at the cut‐off value
Table 2.
Sensitivity and specificity of the Access IgM and IgG II antibody assays for the presence of nAbs
| Specificity | Sensitivity | |||||||
|---|---|---|---|---|---|---|---|---|
| Negative sVNT samples | Detected | Not detected | Specificity (95% CI) | Positive sVNT samples | Detected | Not detected | Sensitivity (95% CI) | |
| IgM antibody assay | ||||||||
| Cut‐off S/CO ≥1.0 | 91 | 1 | 90 | 98.9% | 52 | 51 | 1 | 98.1% |
| (94.0–99.9) | (89.7–99.9) | |||||||
| IgG antibody assay | ||||||||
| Cut‐off IgG ≥10 (AU/ml) | 91 | 0 | 91 | 100% | 52 | 52 | 0 | 100% |
| (93.2–100) | ||||||||
| (96.0–100) | ||||||||
Abbreviations: AU, arbitrary units; CI, confidence interval; COVID‐19, coronavirus disease 2019; IgG, immunoglobulin G; IgM, immunoglobulin M; nAb, neutralization antibody; S/CO, signal for test sample/signal at cut‐off value; sVNT, surrogate virus neutralization test.
4. DISCUSSION
In this study, we showed that the results of the Access SARS‐CoV‐2 IgM and IgG II antibody tests were strongly correlated with those of sVNT among patients with COVID‐19 in the convalescence period (R 2 of IgM: 0.90; R 2 of IgG: 0.92).
nAbs against SARS‐CoV‐2 in COVID‐19 patients were assessed using sVNT, and the results have been shown to correlate strongly with those of conventional pseudovirus‐based VNTs. 3 , 17 We determined an alternative cut‐off value for sVNT by using sera collected from Japanese COVID‐19 and non‐COVID‐19 patients in our target population because the sensitivity, specificity, and cut‐off value of sVNT can vary in a population‐based manner. The clinical specificity of sVNT improved from 93.3% (95% CI: 85.9%–97.5%) using the manufacturer's cut‐off value (≥20% inhibition) to 98.9% (95% CI: 93.9%–99.9%) using the alternative cut‐off value (≥30% inhibition), but the sensitivity remained unchanged at 94.4% (95% CI: 84.6%–98.8%). Fafi‐Kremer et al. 18 showed that 2% of patients with COVID‐19 do not produce nAbs, even in the convalescence period. In our study, serum samples collected from three COVID‐19 cases tested negative by sVNT. Our findings also provide support for the hypothesis that a subset of patients has insufficient humoral immunity after SARS‐CoV‐2 infection and suggest the importance of evaluating humoral immunity by an immunological assay.
Our study showed that the IgM and IgG II antibody tests for anti‐RBD antibodies had high diagnostic sensitivity (IgM: 96.3% [95% CI: 87.3%–99.5%]; IgG: 94.4% [95% CI: 84.6%–98.8%]) and specificity (IgM: 100% [95% CI: 95.9%–100%]; IgG: 98.9% [95% CI: 93.9%–99.9%]). Under the widespread rollout of vaccines targeting the S and RBD proteins, fully automated immunoassays for measuring antibodies against the N protein might be the most relevant for COVID‐19 surveillance. 19 On the other hand, immunoassays for measuring anti‐S and anti‐RBD antibodies might be relevant for evaluating humoral immunity against SARS‐CoV‐2. Indeed, the results of the IgG II antibody test were consistent with those of the sVNT using the alternative cut‐off value (sensitivity: 100% [95% CI: 93.2%–100%]; specificity: 100.0% [95% CI: 95.9%–100%]). Although the IgM antibody test showed high clinical performance for predicting the presence of nAbs (sensitivity: 98.1% [95% CI: 89.7%–99.9%]; specificity: 98.9% [95% CI: 95.9%–100%]), its sensitivity and specificity were inferior to those of the IgG II antibody assay. Generally, IgG antibodies for viruses have a longer half‐life than IgM antibodies 20 ; thus, an IgG II test is desirable when considering a substitute for an nAb assay in the convalescence period.
Our study had several limitations. First, serum samples collected from patients with a negative RT‐qPCR result for SARS‐CoV‐2 in 2020 were used as negative controls, although sera from febrile patients collected before SARS‐CoV‐2 began spreading in 2019 should have ideally been used. Cohort studies have shown false‐negative RT‐qPCR results for SARS‐CoV‐2 in patients with COVID‐19. 21 , 22 Therefore, it was not possible to determine whether the positive results of the two antibody tests in patients classified as non‐COVID‐19 were false positives for the antibody tests or false negatives for the RT‐qPCR tests. Second, serum samples were collected from only a small number of patients who were hospitalized and confirmed to have COVID‐19. Previous reports demonstrated that nAb titers correlated strongly with disease severity and then declined rapidly after recovery compared with anti‐spike IgG levels. 23 Therefore, large‐scale multi‐national investigations are still required to determine whether the Access SARS‐CoV‐2 IgG II antibody test might be a suitable substitute for the nAb assay.
5. CONCLUSION
The Access SARS‐CoV‐2 IgM and IgG II antibody tests showed high sensitivity and specificity for the detection of anti‐SARS‐CoV‐2 antibodies in COVID‐19 and also showed strong positive correlations with the results of sVNT. In particular, the Access SARS‐CoV‐2 IgG II antibody test might be an alternative to an nAb assay as a surveillance tool.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Takuya Maeda: study conception and design; Kazuo Imai and Takuya Maeda: clinical data collection and data analysis; Yutaro Kitagawa, Masaru Matsuoka, Katsumi Kubota, Ai Fukada, and Sakiko Noguchi: experiment and data analysis; Yutaro Kitagawa and Kazuo Imai: manuscript drafting and editing; Momoko Sato, Tomohito Takada, and Takuya Maeda: manuscript revision; Norihito Tarumoto, Shigefumi Maesaki, and Shinichi Takeuchi: study supervision. All authors have read and approved the final manuscript.
ETHICS STATEMENT
This study was reviewed and approved by the Institutional Review Board of Saitama Medical University Hospital (Approval Number: 20148.01).
ACKNOWLEDGMENTS
We thank the clinical laboratory technicians and all members of the COVID‐19 Task Force at Saitama Medical University Hospital.
Kitagawa Y, Imai K, Matsuoka M, et al. Evaluation of the correlation between the access SARS‐CoV‐2 IgM and IgG II antibody tests with the SARS‐CoV‐2 surrogate virus neutralization test. J Med Virol. 2021;94:335‐341. 10.1002/jmv.27338
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor‐binding domain complexed with receptor. Science. 2005;309:1864‐1868. [DOI] [PubMed] [Google Scholar]
- 2. Ju B, Zhang Q, Ge J, et al. Human neutralizing antibodies elicited by SARS‐CoV‐2 infection. Nature. 2020;584:115‐119. [DOI] [PubMed] [Google Scholar]
- 3. Tan CW, Chia WN, Qin X, et al. A SARS‐CoV‐2 surrogate virus neutralization test based on antibody‐mediated blockage of ACE2‐spike protein‐protein interaction. Nat Biotechnol. 2020;38:1073‐1078. [DOI] [PubMed] [Google Scholar]
- 4. Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid‐19 vaccine. N Engl J Med. 2020;383:2603‐2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baden LR, El Sahly HM, Essink B, et al. Efficacy and safety of the mRNA‐1273 SARS‐CoV‐2 vaccine. N Engl J Med. 2020;384:403‐416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Voysey M, Clemens SAC, Madhi SA, et al. Safety and efficacy of the ChAdOx1 nCoV‐19 vaccine (AZD1222) against SARS‐CoV‐2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99‐111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Logunov DY, Dolzhikova IV, Shcheblyakov DV, et al. Safety and efficacy of an rAd26 and rAd5 vector‐based heterologous prime‐boost COVID‐19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet. 2021;397:671‐681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Padoan A, Bonfante F, Pagliari M, et al. Analytical and clinical performances of five immunoassays for the detection of SARS‐CoV‐2 antibodies in comparison with neutralization activity. EBioMedicine. 2020;62:103101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Muecksch F, Wise H, Batchelor B, et al. Longitudinal serological analysis and neutralizing antibody levels in coronavirus disease 2019 convalescent patients. J Infect Dis. 2021;223:389‐398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bal A, Pozzetto B, Trabaud MA, et al. Evaluation of high‐throughput SARS‐CoV‐2 serological assays in a longitudinal cohort of patients with mild COVID‐19: clinical sensitivity, specificity and association with virus neutralization test. Clin Chem. 2021;67(5):742‐752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tang MS, Case JB, Franks CE, et al. Association between SARS‐CoV‐2 neutralizing antibodies and commercial serological assays. Clin Chem. 2020;66:1538‐1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bonelli F, Sarasini A, Zierold C, et al. Clinical and analytical performance of an automated serological test that identifies S1/S2‐neutralizing IgG in COVID‐19 patients semiquantitatively. J Clin Microbiol. 2020;58‐e01224‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gniadek TJ, Thiede JM, Matchett WE, et al. SARS‐CoV‐2 neutralization and serology testing of COVID‐19 convalescent plasma from donors with nonsevere disease. Transfusion. 2021;61:17‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Therrien C, Serhir B, Bélanger‐Collard M, et al. Multicenter evaluation of the clinical performance and the neutralizing antibody activity prediction properties of ten high throughput serological assays used in clinical laboratories. J Clin Microbiol. 2021;59(3):e02511‐e02520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. U.S. Food and Drug Administration . https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/eua-authorized-serology-test-performance. [Latest accessed 13/June/2021].
- 16. Shirato K, Nao N, Katano H, et al. Development of genetic diagnostic methods for detection for novel coronavirus 2019(nCoV‐2019) in Japan. Jpn J Infect Dis. 2020;73:304‐307. [DOI] [PubMed] [Google Scholar]
- 17. Valcourt EJ, Manguiat K, Robinson A, et al. Evaluation of a commercially‐available surrogate virus neutralization test for severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2). Diagn Microbiol Infect Dis. 2021;99:115294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fafi‐Kremer S, Bruel T, Madec Y, et al. Serologic responses to SARS‐CoV‐2 infection among hospital staff with mild disease in eastern France. EBioMedicine. 2020;59:102915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. National SARS‐CoV‐2 Serology Assay Evaluation Group . Performance characteristics of five immunoassays for SARS‐CoV‐2: a head‐to‐head benchmark comparison. Lancet Infect Dis. 2020;20:1390‐1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gudbjartsson DF, Norddahl GL, Melsted P, et al. Humoral Immune Response to SARS‐CoV‐2 in Iceland. N Engl J Med. 2020;383:1724‐1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Z, Bi Q, Fang S, et al. Insight into the practical performance of RT‐PCR testing for SARS‐CoV‐2 using serological data: a cohort study. Lancet Microbe. 2021;2:e79‐e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baron RC, Risch L, Weber M, et al. Frequency of serological non‐responders and false‐negative RT‐PCR results in SARS‐CoV‐2 testing: a population‐based study. Clin Chem Lab Med. 2020;58:2131‐2140. [DOI] [PubMed] [Google Scholar]
- 23. Legros V, Denolly S, Vogrig M, et al. A longitudinal study of SARS‐CoV‐2‐infected patients reveals a high correlation between neutralizing antibodies and COVID‐19 severity. Cell Mol Immunol. 2021;18(2):318‐x327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


