Abstract
The Wnt/Wg signaling pathway functions during development to regulate cell fate determination and patterning in various organisms. Two pathways are reported to lie downstream of Wnt signaling in vertebrates. The canonical pathway relies on the activation of target genes through the β-catenin–Lef/TCF complex, while the noncanonical pathway employs the activation of protein kinase C (PKC) and increases in intracellular calcium to induce target gene expression. cDNA subtractive hybridization between a cell line that overexpresses Wnt-1 (C57MG/Wnt-1) and the parental cell line (C57MG) was performed to identify downstream target genes of Wnt-1 signaling. Among the putative Wnt-1 target genes, we have identified a mouse homolog of the gene encoding human transcription factor basic transcription element binding protein 2 (mBTEB2). The mBTEB2 transcript is found at high levels in mammary tissue taken from a transgenic mouse overexpressing Wnt-1 (both tissue prior to active proliferation and tumor tissue) but is barely detectable in wild-type mouse mammary glands. The regulation of mBTEB2 by Wnt-1 signaling in tissue culture occurs through a β-catenin–Lef/TCF-independent mechanism, as it is instead partially regulated by PKC. The Wnt-1-induced, PKC-dependent activation of mouse BTEB2 in C57MG cells, as well as the ability of Wnt-1 to stabilize β-catenin in these cells, is consistent with the hypothesis that both the noncanonical and canonical Wnt pathways are activated concomitantly in the same cell. These results suggest that mBTEB2 is a biologically relevant target of Wnt-1 signaling that is activated through a β-catenin-independent, PKC-sensitive pathway in response to Wnt-1.
The Wnt/Wg signal transduction pathway is an evolutionarily conserved pathway that plays an important role in the developmental program of various organisms (for reviews, see references 1, 3, 12, and 23). Genetic epistasis tests in Drosophila, in combination with biochemical experiments performed in Xenopus and in tissue culture cell lines, have established a model for Wnt/Wg signaling. In the absence of Wnt/Wg signaling, the cell undertakes active measures to maintain low cytoplasmic levels of the oncoprotein β-catenin or Armadillo, its Drosophila homolog. β-Catenin is a multifunctional molecule that acts at the plasma membrane in adherens junctions, can be found in the cytoplasm, and is also detected in the nucleus as part of a transcription factor complex. Upon phosphorylation at its amino terminus by glycogen synthetase kinase 3β (GSK-3β), β-catenin is targeted for ubiquitin-mediated degradation by a complex of proteins consisting of GSK-3β, the tumor suppressor protein adenomatous polyposis coli (APC), axin, and a member of the SCF ubiquitin ligase complex, β-TrCP/Slimb (28).
Under these conditions, the downstream target genes of Wnt-1 and/or β-catenin signaling presumably remain untranscribed due to the inability of β-catenin to form a functional transcription factor complex with one of its protein binding partners, the Lef/TCF family of transcription factors (2, 30). Furthermore, the activity of a corepressor protein, groucho, in conjunction with Lef/TCF proteins and CREB binding protein, keeps these target genes repressed in the absence of Wnt/Wg signaling (6). Binding of Wnt to its receptor Frizzled, however, initiates a cascade of signaling events that results in β-catenin stabilization. β-Catenin levels now rise in the cytoplasm, reaching a critical amount that enables it to bind to a Lef/TCF protein. This complex then translocates to the nucleus, where it serves as a transcription factor to activate target gene expression.
Besides playing a role in differentiation and development, the Wnt/Wg signaling pathway has also been implicated in tumorigenesis. The Wnt-1 oncogene was originally characterized as int-1, a gene whose activation upon insertion of the mouse mammary tumor virus results in the formation of mouse mammary tumors (33, 34). Although mutations in Wnt-1 have never been implicated in human cancer, mutations in several components of the Wnt signaling pathway, such as APC and β-catenin, have been linked to tumorigenesis. APC is a tumor suppressor gene that is mutated in up to 80% of human colon carcinomas (29). Mutations of APC result in the development of a form of inherited colon carcinoma called familial adenomatous polyposis. Individuals with this condition develop multiple colonic polyps throughout their life, predisposing them to colon cancer. The majority of APC mutations delete the sites in the protein that bind β-catenin and foster its degradation. Hence, elevated levels of β-catenin can also contribute to tumorigenesis. High levels of β-catenin are associated with several human cancers, including colon carcinomas, melanomas, pilomatricomas, and hepatocellular carcinomas, due either to a nonfunctioning APC protein or to mutations that eliminate the phosphorylation sites within β-catenin (7, 9, 28, 32, 37).
The diverse roles of Wnts in both development and tumorigenesis have fostered the search for Wnt target genes in these processes. Wg signaling in Drosophila is known to transcriptionally activate the expression of engrailed and Ultrabithorax through the Armadillo-Drosophila dTCF complex (36, 45). Besides these Drosophila homeobox genes, Wnt signaling through β-catenin results in the transcriptional induction of two additional homeobox genes in Xenopus, siamois (5) and twin (22). Other target genes of Wnt/Wg that are transcriptionally activated through a β-catenin–Lef/TCF transcription factor complex include the oncogenes cyclin D1 (39, 43), c-myc (14), Wnt-1-induced secreted protein 1 (WISP-1) (35, 47), and c-jun and fra-1 (2, 26), as well as the Xenopus fibronectin gene (11), connexin43 (44), and matrilysin (8).
The canonical Wnt signaling pathway relies on the activation of target gene expression by a β-catenin–Lef/TCF transcription factor complex in response to the binding of Wnt to its receptor, Frizzled. There is evidence, however, that Wnts can signal through a β-catenin–Lef/TCF-independent mechanism to activate downstream gene expression. This noncanonical pathway relies on the phosphatidylinositol (PI) pathway to activate protein kinase C (PKC) and raise levels of intracellular calcium Ca2+ in order to regulate target gene expression (the Wnt/Ca2+ pathway) (28). Xenopus Wnt-5a, (XWnt-5a), a Wnt that neither induces ectopic axis formation nor stabilizes β-catenin, employs this pathway to exert its effects (10). The activation of the PI pathway occurs for only specific Wnts and Frizzled receptors and is an event that is independent of β-catenin stabilization. Thus, it is apparent that not all Wnts initiate the transcriptional activation of their target genes through the β-catenin–Lef/TCF complex, suggesting that β-catenin stabilization is not the sole result of all Wnt signaling and that other pathways can be stimulated upon binding of Wnt to the Frizzled receptor.
The current list of Wnt/Wg target genes contains primarily those genes whose expression is activated through the β-catenin–Lef/TCF complex. In an attempt to identify additional downstream target genes of the Wnt-1 signaling pathway that are relevant to transformation and tumorigenesis, we have performed cDNA subtractive hybridization between a Wnt-1-expressing mouse mammary epithelial cell line (C57MG/Wnt-1) and the parental cell line (C57MG) (35). Among the putative Wnt-1 target genes, we have identified a mouse homolog of the gene encoding the human transcription factor basic transcription element binding protein 2 (mBTEB2) as a Wnt-1-responsive gene. The mBTEB2 transcript is found at high levels in tissues taken from Wnt-1 transgenic mouse (both normal tissue [tissue prior to active proliferation] and tumor tissue) but is barely detectable in wild-type mouse mammary glands. Furthermore, transcriptional activation of this gene by Wnt-1 signaling in tissue culture occurs through a mechanism that is independent of β-catenin–Lef/TCF-mediated transcription. Interestingly, the response of the mBTEB2 promoter to Wnt-1 signaling is dependent on the activation of PKC, a known transducer of the XWnt-5a signal which also does not rely on β-catenin-mediated transcription (40, 41). These results suggest that mBTEB2 is a biologically relevant target of Wnt-1 signaling that is activated through a β-catenin-independent, PKC-sensitive pathway in response to Wnt-1.
MATERIALS AND METHODS
Cell culture and reagents.
C57MG and 293 cell lines were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum and penicillin-streptomycin (GIBCO/BRL). The quail fibrosarcoma cell lines QT6 and QT6Wnt-1, as well as the Wnt-1 transgenic mouse mammary tumor and wild-type mammary gland specimens, were obtained from Harold Varmus. C57MG cells were a gift from Anthony Brown. QT6Wnt-1 was maintained in DMEM supplemented with Geneticin (400 μg/ml; GIBCO/BRL). All transfections were performed using the Lipofectamine reagent as directed by the manufacturer (GIBCO/BRL). Stable C57MG cell lines were generated by cotransfection of a hygromycin resistance marker along with the mBTEB2 promoter deletion construct of interest. After 48 h, cells were divided into plates containing 25, 75, or 150 U of hygromycin per ml. Stable pools were collected and subsequently passaged in the presence of 150 U of hygromycin per ml. Phorbol 12-myristate 13-acetate (PMA) was acquired from Sigma. The mitogen-activated protein kinase (MAPK) kinase (MEK) inhibitor U0126 and the myristoylated PKC inhibitor were both obtained from Promega. The Egr-1 monoclonal antibody was provided by Santa Cruz Biotechnology, and the β-catenin monoclonal antibody (2E1) was kindly provided by Linda Bullions. The phosphothreonine monoclonal antibody was purchased from Calbiochem. Protein A-Sepharose beads were obtained from Sigma. The secondary antibodies anti-rabbit immunoglobulin G-horseradish peroxidase and protein A-peroxidase were provided by Santa Cruz Biotechnology and Boehringer Mannheim, respectively.
Retrovirus infections.
The retrovirus expression vector pBabePuro containing a Wnt-1 or Wnt-4 cDNA insert was used to infect C57MG cells. Briefly, retroviruses were prepared by calcium phosphate transfection of the packaging cell line BOSC 23. The retrovirus-containing supernatant was isolated 48 h posttransfection and divided into 3-ml aliquots. These virus stocks were used to infect C57MG cells at approximately 50% confluence in the presence of Polybrene (8 μg/ml) and 1 ml of fetal bovine serum. After 48 h, cells were divided among plates containing puromycin (final concentration, 2.5 μg/ml). Approximately 85% of the cells demonstrated puromycin resistance after 48 h, after which time 100% of the cells grew in puromycin. Cells were passaged in the presence of puromycin to maintain expression of Wnt-1 or Wnt-4.
Coculture assays.
In the QT6Wnt-1 cell line, Wnt-1 expression is driven by the mouse mammary tumor virus promoter. In the coculture assay, QT6 and QT6Wnt-1 cells are plated in 10-cm-diameter tissue culture dishes and allowed to grow for 48 h at 37°C, after which time they are approximately 30% confluent. C57MG cells are trypsinized and added to the tissue culture dishes containing QT6 or QT6Wnt-1 cells. Cells are cocultured for various time periods at 37°C in 5% CO2. The Wnt-1 extracellular matrix (ECM) and control ECM used in the coculture assays were prepared by growing QT6 and QT6Wnt-1 cells to approximately 90% confluency and scraping the quail cells off of the tissue culture dishes. C57MG cells were then added to the Wnt-1 or control ECM, and coculture assays were performed as described previously. This technique leaves behind Wnt-1 protein in the ECM, and an increase in β-catenin protein in C57MG cells grown on a Wnt-1 ECM can be observed, suggesting that enough Wnt-1 protein is trapped in the ECM to stabilize β-catenin.
DNA transfections.
Transient and stable transfections were performed with Opti-MEM (GIBCO/BRL) and Lipofectamine (GIBCO/BRL), using protocols provided by the manufacturer. For luciferase assays, cells were transiently transfected with the appropriate reporter along with a β-galactosidase plasmid in order to normalize for transfection efficiency. For coculture luciferase assays, luciferase activity was normalized to protein concentration used in the assay.
Luciferase assays.
All mBTEB2 promoter fragments were subcloned into the pGL2Basic luciferase reporter plasmid (Promega). Luciferase assays were performed using a Dual-Light kit (Tropix, Inc.) according to the manufacturer's directions. Luciferase activity was measured using a TR717 microplate luminometer (Tropix). All luciferase measurements were made from triplicate transfections and averaged after normalization to β-galactosidase activity or to protein concentration. The TopFlash luciferase reporter and dn TCF constructs were kindly provided by Bert Vogelstein. Richard Goodman kindly supplied the wild-type CREB and dn CREB (KCREB) constructs. Myc epitope-tagged wild-type Xenopus β-catenin in the pCS2/MT expression vector and the empty pCS2/MT expression vector were generously provided by Barry Gumbiner. Subsequent β-catenin constructs, generated through site-directed mutagenesis by Lifeng Xu, included the following: 4145β-catenin (β-catenin mutant having a threonine and serine at positions 41 and 45, respectively, mutated to alanine; stable mutant with longer half-life), wt β-catenin (wild-type β-catenin), NLS (4145β-catenin mutant containing the simian virus 40 large T antigen nuclear localization signal sequence; localized to nucleus [immunofluorescence]), and 4145ΔC (4145β-catenin mutant having a deletion of the carboxy terminus [putative transcriptional activation domain]).
Isolation of mBTEB2 promoter fragments. (i) PCR.
A Mouse GenomeWalker kit (Clontech) was used to isolate three overlapping fragments of the mBTEB2 promoter (1.1, 1.3, and 1.5 kb). This kit contains five libraries of uncloned, adapter-ligated genomic DNA fragments and relies on nested PCR in order to amplify a region of interest. The 5′ primers (AP1 and AP2) are based on the adapter sequence and supplied by the kit. The 3′ primers used were designed based on the mBTEB2 sequence supplied by Genentech. The sequence of the first 3′ primer (5′-GGCCTGCCATAGAAACATTAAGGGT-3′) lies within the putative coding region of the mouse homolog and encompasses amino acids 13 to 21. The sequence of the second 3′ primer (5′-TTTGTAAACTGGGCATGTCTAGATA-3′) lies further 5′ to the sequence of the first 3′ primer and encompasses the putative start site of translation of the mouse homolog. All PCR products were subjected to sequence analysis and identified by the BLAST program. PCR was then used to introduce a 3′ XhoI site on the 1.3- and 1.5-kb fragments to facilitate cloning. These mBTEB2 promoter fragments were then digested with MluI (a site in the 5′ adapter sequence) and XhoI and were cloned into the pGL2Basic luciferase reporter plasmid (Promega). 5′ MluI digestion followed by 3′ blunt-end ligation was used to clone the 1.1-kb fragment into this reporter plasmid.
(ii) Genomic library screening.
The 129 SVJ mouse genomic library, lambda FIX II vector (Stratagene), was successfully screened with the 1.5-kb mBTEB2 promoter fragment obtained from the GenomeWalker PCR assay. Approximately 2 × 107 plaques were initially screened, and after three rounds of screening a positive clone was identified by sequence analysis. The isolated clone was only slightly larger (approximately 1.6 kb) than the original fragment used to screen the library. This mBTEB2 promoter fragment was also cloned into the pGL2Basic luciferase reporter plasmid (Promega).
Generation of mBTEB2 promoter deletion constructs.
A QuikChange site-directed mutagenesis kit (Stratagene) was used to generate a series of 3′ deletion constructs using the 1.6-kb promoter fragment as a template. PCR primers were designed such that the constructs shared a common 5′ primer and had a different 3′ primer. The 5′ primer and 3′ primer for each deletion construct contain a SacI site and an XhoI site, respectively, to allow for cloning into the pGL2Basic luciferase reporter plasmid (Promega). The following PCR primers were used to create the various deletion constructs indicated below: 5′ primer, 5′-TAACCCGGGAGGTACCGAGCTCTTA-3′ (SacI site underlined); TATA, 5′-CCGCTCGAGCGGGCTTCTGTGTGTG-3′ (XhoI site underlined); Lef, 5′-CCGCTCGAGCGGCCTGTGCAAATCT-3′ (XhoI site underlined); CREB1, 5′-CCGCTCGAGCGGCTACGACATGTCT-3′ (XhoI site underlined); and CREB2, 5′-CCGCTCGAGCGGCAGTTCTCAGGTG-3′ (XhoI site underlined).
RNA isolation and Northern blot analysis.
Total RNA was isolated from cultured cells using an RNeasy Mini kit (Qiagen) or from tissues using the Trizol reagent (GIBCO/BRL) as directed by the manufacturer. Northern blot analysis was performed using at least 5 μg of total RNA. RNA was electrophoresed overnight on a 1.2% agarose gel containing 6% formaldehyde in circulating 1× E buffer (18 mM Na2HPO4, 2 mM NaH2PO4). RNA was transferred to a Nytran membrane using the Turboblotter transfer system (Schleicher & Schuell). Membranes were either baked under vacuum at 80°C for 2 h or UV cross-linked (Stratagene Stratalinker) prior to 2 h of prehybridization in 5 ml of Church buffer (0.2 M NaH2PO4, 0.3 M Na2HPO4, 1% bovine serum albumin, 7% sodium dodecyl sulfate [SDS], 1 mM EDTA) at 65°C; 250 ng of the appropriate DNA was then labeled for a probe by random priming in the presence of [α-32P]dCTP using a PrimeIt RmT kit (Stratagene). Blots were hybridized with the labeled probe overnight in Church buffer at 65°C. Blots were then rinsed twice in 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS and washed twice in this solution for 30 min at 65°C. The final two washes were performed in 0.2× SSC–0.1% SDS and 0.1× SSC–0.1% SDS, respectively, for 30 min at 65°C. Blots were wrapped in Saran Wrap and exposed to X-Omat film for at least 8 h. The signals were quantitated using a PhosphorImager. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control.
In situ hybridization.
Nonradioactive in situ hybridizations were performed as described previously (17). An 800-bp human BTEB2 coding region was isolated by PCR from human colorectal carcinoma cell line HT29 genomic DNA (prepared using a Qiagen QIAmp blood kit) using a 5′ primer containing a BamHI site (underlined; 5′-CGGGATCCCGCATGAACGTCTTCCT-3′) and a 3′ primer containing an EcoRI site (underlined; 5′-CGGAATTCCGCCTTCTATTGTATCT-3′). This PCR product was subjected to sequencing, and the BLAST program was used to assess its identity. The BTEB2 product was cloned into the pBluescript II KS(−) phagemid (Stratagene) such that the T7 RNA polymerase could be used to transcribe a sense probe and T3 RNA polymerase could be used to transcribe an antisense probe. The pGEM-4 vector (Promega) containing the Wnt-1 cDNA was used to generate Wnt-1 antisense and sense riboprobes using T7 and SP6 RNA polymerases, respectively. All riboprobes were created using a Riboprobe in vitro transcription system kit (Promega), in which a digoxigenin-labeled UTP is provided in the ribonucleotide mix, according to the manufacturer's instructions.
Three- to four-micrometer serial mammary tumor and mammary gland tissue sections taken from a Wnt-1 transgenic mouse, as well as wild-type mammary gland tissue sections, were obtained from Genentech and used for in situ hybridization. Hybridizations were performed in a humidified chamber overnight at 65°C. Sections were washed in successive solutions and then blocked for 1 h at room temperature in 2% goat serum–bovine serum albumin (2 mg/ml) in 1× TBST (10× stock is 8 g of NaCl, 0.2 g of KCl, 25 ml of 1 M Tris [pH 7.5], and 1 ml of Tween 20). An antibody to digoxigenin conjugated to alkaline phosphatase (Boehringer Mannheim) was diluted in blocking buffer (1:500) and incubated overnight at 4°C. Sections were subsequently rinsed in 1× TBST for 3 h and then washed in NTMT (100 mM Tris [pH 9.5], 50 mM MgCl2, 100 mM NaCl, 0.1% Tween 20) for 30 min. Staining was revealed in the dark using NTMT containing 20 μl each of 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium (Boehringer Mannheim). Pictures were taken using a bright-field microscope. Serial sections were also stained with hematoxylin to visualize the tissue sections.
Labeled immunoprecipitation.
The parental C57MG cell line and C57MG cells infected with the Wnt-1, Wnt-4, and empty vector retroviruses were grown in 10-cm-diameter tissue culture dishes. Cells were prestarved for 30 min in 4 ml of DMEM minus methionine plus 2% dialyzed fetal calf serum. Cells were labeled for 18 h in the presence of a mixture of 35S-labeled methionine and cysteine 75 μCi/ml; (Expre35S35S; NEN). Cells were lysed in 500 μl of lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.5% Nonidet P-40, 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 mM pepstatin, 1 mM E-64), and 107 trichloro-acetic acid-precipitable cpm was immunoprecipitated using a monoclonal antibody to β-catenin for 4 h in the presence of protein A-Sepharose beads (Sigma). The immunoprecipitates were washed three times with 500 μl of SNNTE (50 mM Tris [pH 7.4], 5 mM EDTA, 5% sucrose, 1% Nonidet P-40, 0.5 M NaCl) and one time with 1 ml of radioimmunoprecipitation assay buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% [wt/vol] sodium deoxycholate). The proteins were eluted and separated on an SDS–8% polyacrylamide gel. The gel was fixed in 30% methanol–10% acetic acid for 20 min and then soaked in a 1 M sodium salicylate–1.5% glycerol solution for 20 min. The gel was wrapped in Saran Wrap and dried on a gel dryer for 1 h, after which time it was exposed to X-Omat film for 24 h.
Nucleotide sequence accession number.
The GenBank accession number for the mBTEB2 promoter sequence is AF285184.
RESULTS
Identification of downstream target genes of the Wnt-1 signaling pathway.
To identify putative downstream target genes of the Wnt signaling pathway, cDNA subtraction analysis (13) was performed between a cell line overexpressing Wnt-1 and the parental cell line. C57MG cells, a mouse mammary epithelial cell line, were chosen because they provide a good in vitro model system for Wnt-1 signaling, and it is within this type of cell in the mouse mammary gland that aberrant activation of Wnt-1 results in mammary hyperplasia (for example, see references 4 and 18). To effect high levels of Wnt-1 expression, the C57MG cells were infected with a Wnt-1 retrovirus (see Materials and Methods) that contains the mouse Wnt-1 cDNA under control of the viral long terminal repeat promoter and a puromycin resistance selectable marker. Control infections for this experiment included C57MG cells infected with either an empty vector retrovirus or a Wnt-4 retrovirus. Wnt-4 is normally expressed in the developing mouse mammary gland and during pregnancy, and it has never been demonstrated to possess any oncogenic activities (31). These cell lines will be referred to as C57MG/Wnt-1, C57MG/Wnt-4, and C57MG/Vector to distinguish among the Wnt-1-, Wnt-4-, and vector-infected cells, respectively.
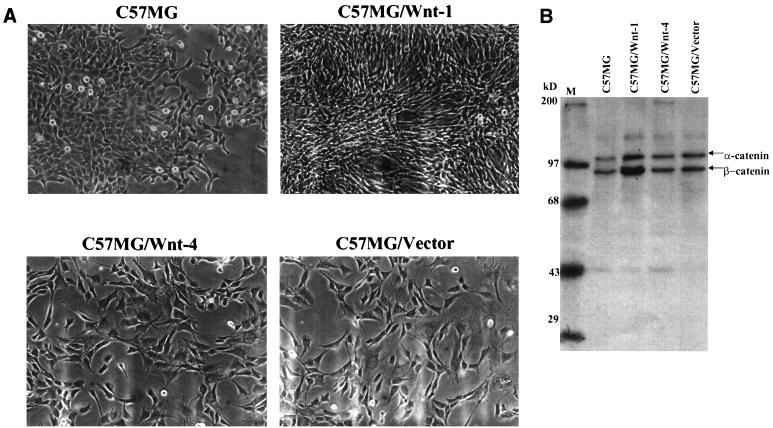
Prior to performing the cDNA subtractive hybridization, we assessed the effects of Wnt-1 on C57MG cell morphology and on β-catenin levels. One of the hallmarks of Wnt-1 overexpression in C57MG cells is the drastic morphology change exhibited by these cells (33). C57MG cells normally form monolayers that are relatively cuboidal. Overexpression of Wnt-1 in these cells through the pBabePuro retrovirus, however, causes the cells to become very refractile and elongated (Fig. 1A). The morphology of C57MG/Wnt-1 cells is in striking contrast to that seen for C57MG/Wnt-4 and C57MG/Vector cells, which both appear to be morphologically indistinguishable from the parental C57MG cell line. Thus, a Wnt-1 retrovirus infection of C57MG cells appears to at least morphologically mimic that elsewhere reported (34). To assess the levels of β-catenin in the retrovirus-infected cell lines, each cell line (parental C57MG, C57MG/Wnt-1, C57MG/Wnt-4, and C57MG/Vector) was metabolically labeled and immunoprecipitated using a monoclonal antibody to β-catenin (Fig. 1B). Approximately fivefold more total β-catenin was immunoprecipitated from the C57MG/Wnt-1 cells than from the other three cell lines. Thus, the Wnt-1 signaling pathway functions to stabilize β-catenin in the Wnt-1-infected cells, the expected outcome for Wnt-1 overexpression.
FIG. 1.
(A) C57MG cells are morphologically transformed by infection with a Wnt-1 retrovirus. C57MG cells were infected with either a Wnt-1, Wnt-4, or empty vector retrovirus (see Materials and Methods for details) and selected in puromycin after 48 h. (B) Labeled immunoprecipitation for β-catenin protein. C57MG cells were infected with either the Wnt-1 (C57MG/Wnt-1), Wnt-4 (C57MG/Wnt-4), or empty vector (C57MG/Vector) retrovirus. The parental cell line (C57MG), along with the other three retrovirus-infected cell lines, were labeled for 18 h with Expre35S35S (NEN). Cell lysates were prepared, and 107 cpm was immunoprecipitated with a β-catenin monoclonal antibody. Bound proteins were loaded on an SDS–8% polyacrylamide gel and visualized by autoradiography. M, markers.
To identify those transcripts that are up-regulated in response to Wnt-1 signaling, we performed PCR-select cDNA subtractive hybridization between C57MG/Wnt-1 cells and the parental C57MG cell line (35). C57MG cells which had been stably expressing Wnt-1 for 3 weeks after retrovirus infection were chosen for this analysis, as it would allow for the identification of both direct and indirect targets of Wnt-1 signaling. The majority of the genes having a known function were up-regulated at least twofold at the mRNA level in response to Wnt-1 signaling. Only a minority were found to be false positives, as determined by Northern blot analysis (data not shown). Furthermore, two of the genes identified in the screen, mouse connexin43 and mouse WISP-1, have recently been shown to be targets of the Wnt-1 signaling pathway (35, 44). The identification of at least two targets of the Wnt-1 signaling pathway, one of which was isolated independently of this screen, suggests that the screen is a useful method by which to identify novel targets of this signaling pathway. For one of these putative Wnt-1 targets, mBTEB2, expression at the mRNA level was approximately fivefold higher in C57MG/Wnt-1 cells than in the parental or C57MG/Vector cells; therefore, mBTEB2 was chosen for further analysis.
mBTEB2 is a Wnt-1-responsive gene in vitro.
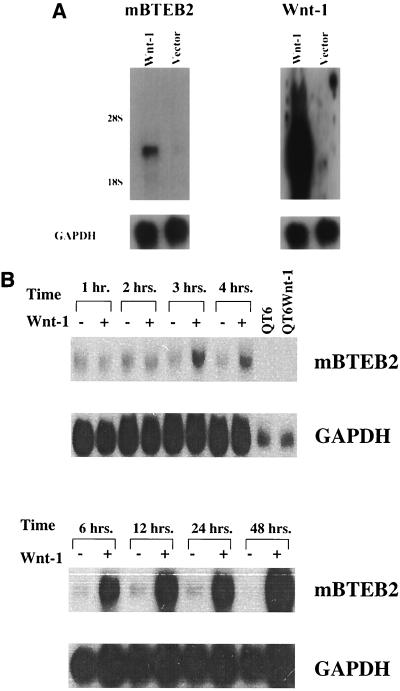
BTEB2 is a member of the Sp1 family of transcription factors and has recently been reported to function in vivo to regulate the expression of the smooth muscle myosin heavy chain B gene (46). To confirm that mBTEB2 is indeed a true transcriptional target of the Wnt-1 signaling pathway, three independent experiments were performed in vitro. In an independent experiment, C57MG cells were infected with the Wnt-1 and empty vector retrovirus, and RNA was isolated from cells 3 weeks after infection in order to mimic the conditions under which the original screen was conducted. As shown by Northern blot analysis, the mBTEB2 transcript is reproducibly up-regulated approximately fivefold in the C57MG/Wnt-1 cells compared to the C57MG/Vector control cells (Fig. 2A). In addition, semiquantitative reverse transcription-PCR indicates that the mBTEB2 mRNA is induced in the Wnt-1-infected cells compared to the parental control cell line (data not shown).
FIG. 2.
(A) mBTEB2 induction in Wnt-1-expressing C57MG cells. C57MG cells were infected with a Wnt-1 and empty vector retrovirus (as a control) and passaged in the presence of puromycin (2.5 μg/ml) for 3 weeks. Northern blot analysis on 5 μg of total RNA was performed to assess mBTEB2 and Wnt-1 mRNA expression levels in each cell type (Wnt-1 and Vector). Signals were initially visualized by autoradiography and then quantitated with a PhosphorImager. Blots were later probed for GAPDH as a loading control. (B) Time course of induction of mBTEB2 in response to Wnt-1 signaling. C57MG cells were cocultured with either the parental QT6 cell line (derived from a quail fibrosarcoma) or the QT6Wnt-1 cell line, which overexpresses and secretes Wnt-1. Total RNA was isolated from the cocultures at the indicated times, and Northern blot analysis was performed on 10 μg of total RNA using an mBTEB2 probe. Signals were initially visualized by autoradiography and quantified by PhosphorImager analysis. Blots were probed for GAPDH as a loading control. QT6 and QT6Wnt-1 lanes, containing RNA derived from QT6 and QT6Wnt-1 cells, respectively, show that the mBTEB2 probe does not cross-hybridize with quail RNAs.
To assess the temporal regulation of mBTEB2 by Wnt-1 signaling, coculture of a Wnt-1-expressing cell line (QT6Wnt-1) with C57MG cells was performed for various time periods. This method of coculture establishes a paracrine signaling such that the Wnt-1 secreted by QT6Wnt-1 cells can elicit a response from C57MG cells (the activation of target gene expression) (18). In the coculture assay, Wnt-1 transcriptionally activates the mBTEB2 promoter approximately fourfold after 3 h of coculture, and this transcriptional induction persists through a 48-h coculture experiment (Fig. 2B). Furthermore, regulation of the mBTEB2 promoter by Wnt-1 signaling is in fact due to transcription, as coculture experiments performed in the presence of actinomycin D, an inhibitor of RNA polymerase II, prevent the transcriptional activation of mBTEB2 by Wnt-1 signaling (data not shown). These data suggest that mBTEB2 is a transcriptional target of the Wnt-1 signaling pathway in vitro.
mBTEB2 is a Wnt-1-responsive gene in vivo.
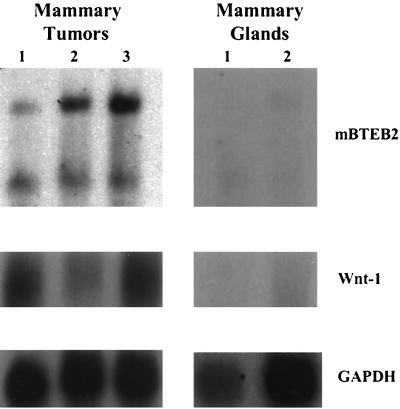
To assess the biological relevance of mBTEB2 as a target gene for Wnt-1 signaling, mBTEB2 transcript levels were measured in Wnt-1 transgenic mouse mammary tumors and normal wild-type mouse mammary glands. Northern blot analysis of these tissues demonstrates that the mBTEB2 transcript is found at higher levels in three independent Wnt-1 transgenic mouse mammary tumors and at significantly lower levels in the wild-type mouse mammary glands (Fig. 3). The presence of two transcripts for mBTEB2 in the mammary tumor and mammary gland samples is not unexpected, given the identification of two mRNA species for human BTEB2 in the testis (42). From this experiment, it is apparent that the absolute level of Wnt-1 mRNA expression does not necessarily correlate with mBTEB2 transcript levels. Rather, the expression of Wnt-1 itself is enough to transcriptionally induce mBTEB2 in the mouse mammary tumor samples.
FIG. 3.
mBTEB2 mRNA in Wnt-1 transgenic mouse mammary tumors and in normal, wild-type mouse mammary glands. Total RNA was isolated from three independent Wnt-1 transgenic mouse mammary tumors and from two different wild-type mouse mammary glands. Northern blot analysis was performed on 10 μg of total RNA to assess expression levels of Wnt-1 and mBTEB2. Signals were visualized by autoradiography, and blots were reprobed for GAPDH as a loading control.
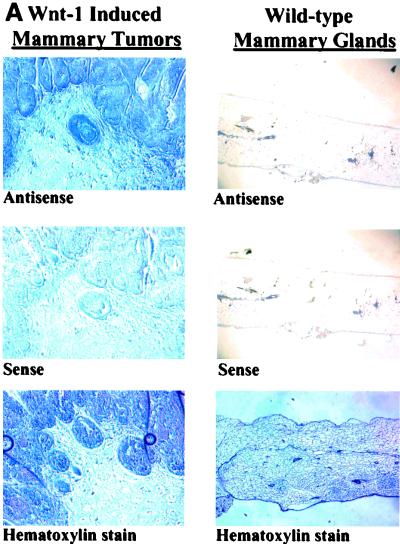
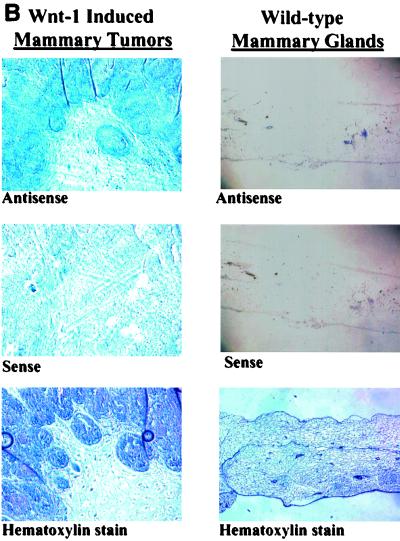
These data are further confirmed by in situ hybridizations performed with serial mammary tumor and mammary gland sections obtained from Wnt-1 transgenic mice and wild-type mice (Fig. 4). As expected, the Wnt-1 antisense probe recognizes the Wnt-1 transcript in tissues derived from the Wnt-1 transgenic mouse (both tumor and nontumor), but no Wnt-1 mRNA is detectable in the wild-type mouse mammary gland. Analysis of Wnt-1 transgenic mouse mammary tumor sections using the mBTEB2 antisense probe shows that mBTEB2 mRNA is found at high levels in areas where Wnt-1 is expressed. Very little, if any, mBTEB2 mRNA can be detected in the normal, wild-type mouse mammary gland tissue sections. Furthermore, the nontumor mammary tissue taken from the Wnt-1 transgenic mouse (which still expresses high levels of Wnt-1 mRNA) has an elevated level of mBTEB2 mRNA which in some areas of the tissue section overlaps but is not entirely coincident with Wnt-1 expression. Perhaps this is due to the fact that Wnt-1 is a secreted protein and that not every cell type in the mouse mammary gland expresses the appropriate Frizzled receptor to permit the transduction of the Wnt-1 signal (and the subsequent activation of mBTEB2). These results establish a clear correlation between overexpression of Wnt-1 and increased levels of the mBTEB2 transcript in vivo.
FIG. 4.
In situ hybridization of mBTEB2 mRNA in Wnt-1 transgenic mouse mammary tissues and adjacent mammary gland tissue and in wild-type mouse mammary glands. Nonradioactive in situ hybridization was performed using either a human BTEB2 antisense or sense probe, or a Wnt-1 antisense or sense probe, on serial sections obtained from a Wnt-1 transgenic mouse mammary gland tumor or adjacent normal tissue or from a wild-type mouse (see Materials and Methods for details). (A) Wnt-1 mRNA expression in a Wnt-1-induced mammary tumor and in a wild-type mammary gland; (B) mBTEB2 mRNA expression in a Wnt-1-induced mammary tumor and in a wild-type mammary gland; (C) Wnt-1 and mBTEB2 mRNA expression in Wnt-1 transgenic mouse mammary gland tissue. Sections were also stained with hematoxylin to visualize the tissue.
The mBTEB2 promoter is regulated by a Wnt-1-dependent, β-catenin-independent, mechanism. (i) Isolation of mBTEB2 promoter fragments.
The mBTEB2 promoter/enhancer region was isolated to determine the mechanism of its regulation by Wnt-1 signaling. Overlapping fragments of the mBTEB2 promoter were obtained through a combination of mouse genomic library screening and PCR (see Materials and Methods for details). Several mBTEB2 promoter fragments were identified, including a minimal 1.1-kb promoter fragment that, after cloning into a luciferase expression plasmid, activates luciferase expression in 293 cells to fivefold-higher levels in the absence of Wnt-1 than the control plasmid. All of the mBTEB2 promoter fragments were cloned into the pGL2Basic luciferase reporter plasmid. Sequence analysis of the mBTEB2 promoter fragments revealed the presence of four putative CREB binding sites, four putative GC boxes, a putative TATA box, and two imperfect Lef/TCF binding sites.
Wnt-1 regulation of and identification of negative regulatory elements in the mBTEB2 promoter.
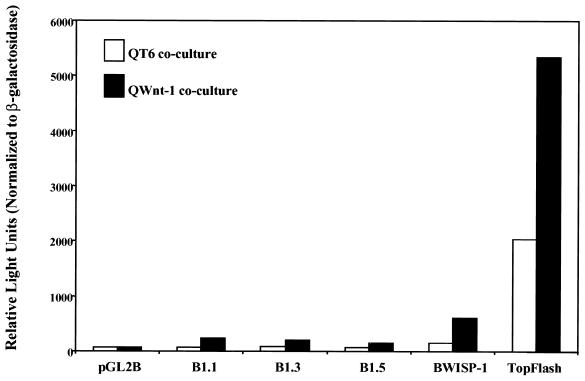
Transient transfection of the mBTEB2 promoter constructs into C57MG cells, followed by the coculture of the transfected cells with Wnt-1-expressing cells, confirmed that these constructs are responsive to Wnt-1 (Fig. 5). This response, however, did not completely recapitulate the response of the endogenous promoter to Wnt-1, as the luciferase activity was only about threefold higher in the presence of Wnt-1 using the 1.1-kb promoter. The two positive controls used in this assay were TopFlash, an artificial luciferase reporter which contains four tandem repeats of the Lef/TCF binding site cloned upstream of a minimal c-fos promoter, and WISP-1, a known Wnt-1- and β-catenin-responsive gene (47). The WISP-1 construct contains the WISP-1 promoter (approximately 5 kb) cloned into the pGL2Basic luciferase reporter plasmid (BWISP-1). The luciferase read-out from the TopFlash coculture experiment indicates that this promoter is activated approximately 2.5-fold by Wnt-1 signaling due to the Wnt-1-induced stabilization of β-catenin. BWISP-1 is also activated to high levels by Wnt-1 signaling in this coculture assay. These results indicate that the endogenous mBTEB2 promoter contains additional regulatory elements which modulate its response beyond the threefold level observed in these assays. Interestingly, the coculture of cells containing the largest promoter fragment (1.5 kb) with Wnt-1-expressing cells actually shows a diminished response to Wnt-1 compared to the smaller promoter fragments. Similar results were obtained upon generation of stable C57MG cell lines containing these mBTEB2 promoter reporters and the coculture of these cells with Wnt-1-expressing cells (data not shown).
FIG. 5.
mBTEB2 promoter fragments are transcriptionally induced in response to Wnt-1 signaling in coculture assay. C57MG cells were transiently transfected with the pGL2Basic luciferase reporter plasmid, or various pGL2Basic luciferase reporters containing mBTEB2 promoter fragments of different sizes, and a β-galactosidase plasmid. The two positive controls used in this assay include WISP-1, a Wnt-1 and β-catenin target gene (47), and TopFlash. Forty-eight hours after transfection, the cells were cocultured with QT6 and QT6Wnt-1 cells for 24 h. Luciferase activity was measured and normalized to β-galactosidase activity. The results shown are averages of three independent experiments.
We constructed a series of 3′ deletion mutants of the 1.5-kb mBTEB2 promoter fragment to examine how different sequence elements within the mBTEB2 promoter respond to Wnt-1 signaling. Coculture assays using these deletion constructs revealed that the promoter fragment having the strongest response to Wnt-1 signaling is approximately 1.1 kb and consists of primarily the 5′ promoter region and approximately 200 bp downstream of the putative TATA box (data not shown). This construct (the Lef construct) terminates after the first putative Lef/TCF binding site, and it does not contain any CREB binding sites but does retain three of the four GC box sequences. These results corroborate the previous coculture results with the larger mBTEB2 promoter fragments, suggesting that the additional 5′ and 3′ sequences found in the larger mBTEB2 promoter fragments contain negative regulatory elements that diminish the luciferase response of this promoter fragment to Wnt-1 signaling.
mBTEB2 transcriptional activation does not require high levels of β-catenin.
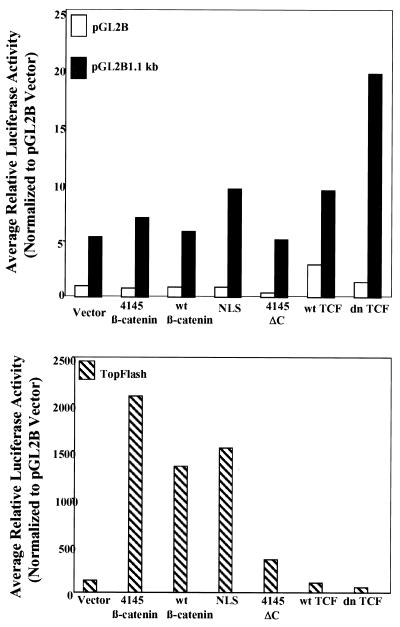
Given the presence of two imperfect Lef/TCF binding sites within the mBTEB2 promoter fragments and the finding that many Wnt target genes rely on the β-catenin–Lef/TCF transcription factor complex for transcriptional activation, it was important to assess the ability of these promoter fragments to respond to various β-catenin and TCF mutants. The mBTEB2 promoter fragments were cloned into the pGL2Basic luciferase reporter vector and tested in transient transfection assays in both 293 cells and C57MG cells, in combination with various β-catenin mutant plasmids and TCF constructs (see Materials and Methods for further details). All of the β-catenin mutant plasmids express proteins at similar levels relative to one another and have a half-life of longer than 6 h (in comparison, wild-type β-catenin has a half-life of only 30 min) (47). Cotransfection of the 4145β-catenin, NLS, or wtβ-catenin construct enhanced the luciferase activity of the 1.1-kb mBTEB2 promoter construct less than twofold in both 293 cells (averages of two independent triplicate experiments are shown in Fig. 6) and C57MG cells (data not shown). These β-catenin plasmids readily transcriptionally activate TopFlash, the luciferase reporter used as the positive control. Furthermore, cotransfection of a wild-type TCF plasmid (wt TCF) does not alter the luciferase activity of the mBTEB2 promoter construct. Cotransfection of the dn TCF plasmid, which can bind to DNA but cannot bind to β-catenin, appears to slightly enhance the luciferase response of the 1.1-kb mBTEB2 promoter construct, in striking contrast to the decreased luciferase response observed for TopFlash. Similar results were obtained from transient cotransfections using the 1.5-kb mBTEB2 promoter reporter (data not shown). These results suggest that the β-catenin–Lef/TCF transcription factor complex does not regulate the transcriptional activity of this promoter in response to Wnt-1 signaling.
FIG. 6.
The mBTEB2 promoter is not responsive to elevated levels of β-catenin or TCF protein. Transient transfections were performed in 293 cells and C57MG cells (data not shown), using 2.0 μg of total plasmid DNA (0.5 μg of reporter plasmid, 1.0 μg of β-catenin or TCF plasmid, and 0.5 μg of vector). The reporter constructs used included the pGL2Basic luciferase reporter vector (pGL2B) as a negative control, TopFlash as a positive control, and one of the mBTEB2 promoter reporters (pGL2B 1.1, 1.3, 1.5 kb). The results are shown for the 1.1-kb promoter construct. Luciferase activity of the transfected cells was measured after 48 h using a luminometer. Cells were also cotransfected with a β-galactosidase reporter to normalize for transfection efficiency such that the reported luciferase values are normalized to β-galactosidase activity. The results are representative of three independent experiments, each done in triplicate.
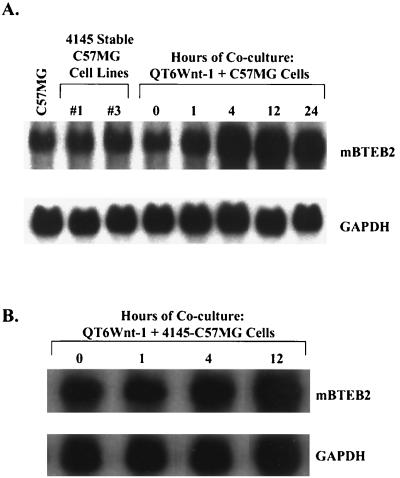
The effect of β-catenin on the endogenous mBTEB2 promoter in C57MG cells was also investigated. Two independent stable C57MG cell lines which overexpress the nondegradable, stable form of β-catenin (4145β-catenin) were generated, and the transcript levels of mBTEB2 were compared to the levels of mBTEB2 mRNA in C57MG cells cocultured with Wnt-1-expressing cells. The half-life of the β-catenin protein produced in these stable cell lines is longer than 6 h (47). Expression of β-catenin at high levels does not transcriptionally induce the mBTEB2 promoter, whereas Wnt-1 signaling has the opposite effect, suggesting that the transcriptional response of the mBTEB2 promoter to Wnt-1 occurs through a β-catenin-independent mechanism (Fig. 7A). In addition, coculture of these C57MG cell lines overexpressing 4145β-catenin with Wnt-1-expressing cells demonstrates an increase in the mBTEB2 mRNA to comparable levels observed in the coculture of Wnt-1-expressing cells with C57MG cells (Fig. 7B). These results corroborate the previous endogenous promoter results and further suggest that mBTEB2 is regulated independently of high levels of β-catenin.
FIG. 7.
Wnt-1 signaling transcriptionally activates the mBTEB2 promoter through a β-catenin-independent mechanism in both the parental C57MG cell line and the C57MG cell line overexpressing 4145β-catenin. (A) Two C57MG cell lines that stably express 4145β-catenin were generated, and cocultured with QT6Wnt-1 cells for the indicated times. mBTEB2 mRNA levels were determined by Northern blot analysis using 10 μg of total RNA isolated from the parental C57MG cell line, the two stable 4145β-catenin-expressing cell lines, and the QT6Wnt-1-plus-C57MG cocultures. Blots were reprobed for GAPDH as a loading control. Signals were visualized by autoradiography and quantitated using a PhosphorImager. (B) The sample designated #1 was cocultured with QT6Wnt-1 cells for various times, and Northern blot analysis was carried out as for panel A.
Role of MAPK and PMA/PKC in regulation of the mBTEB2 promoter by Wnt-1.
Given that the mBTEB2 promoter is transcriptionally regulated by Wnt-1 signaling in a β-catenin-independent manner, other models for its regulation were postulated to explain how Wnt-1 signaling activates this promoter. The human gene is expressed at high levels in smooth muscle cells, and it is transcriptionally regulated by the early growth response gene 1 (Egr-1) protein product in a phorbol ester (PMA)- and MAPK-dependent manner (19). The presence of several GC boxes (sequences to which Egr-1 can bind) within the mBTEB2 promoter suggested that perhaps the mouse promoter is regulated in a similar fashion. Therefore, MAPK and a PMA-sensitive isoform of PKC (PMA/PKC) were tested to determine if they regulate the mBTEB2 promoter and what role, if any, these molecules play in the response of this promoter to Wnt-1 signaling.
Role of MAPK.
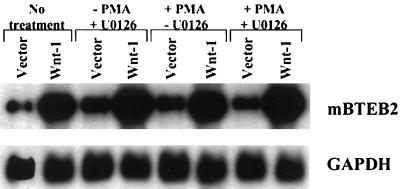
The human BTEB2 promoter has been reported to be regulated through a PMA-sensitive, MAPK-dependent mechanism (19). We therefore set out to determine whether the mouse promoter alone, or in the presence of Wnt-1 signaling, is regulated in a similar fashion. C57MG/Wnt-1 and C57MG/Vector cells were pretreated for 1 h with 10 μM MEK inhibitor U0126 and then treated with 10 nM PMA for 4 h; Northern blot analysis was then performed to assess the mBTEB2 transcript levels. The results indicate that the MAPK pathway may negatively regulate the Wnt-1 signaling pathway, as there is a reproducible increase in the mBTEB2 transcript levels in C57MG/Wnt-1 cells treated with U0126 (Fig. 8). Treatment with only PMA causes an increase in mBTEB2 mRNA in C57MG/Vector cells but no further increase in C57MG/Wnt-1 cells treated with PMA. These results suggest that regulation of the mBTEB2 promoter is sensitive to PMA and that the MAPK pathway negatively regulates the response of the promoter to Wnt-1 signaling. It is not yet clear whether transcriptional activation of the mBTEB2 promoter by Wnt-1 signaling occurs through a PMA-dependent pathway or if two different pathways (one sensitive to PMA and one activated by Wnt-1 signaling) act on the mBTEB2 promoter.
FIG. 8.
Regulation of the mBTEB2 promoter by Wnt-1 signaling does not depend on the MAPK pathway. C57MG cells stably infected with either the Wnt-1 (Wnt-1) or empty vector (Vector) retrovirus were pretreated with 10 μM U0126 (an inhibitor of MEK) or an equal volume of water for 1 h. PMA was added to a final concentration of 10 nM (or the equivalent volume of water), and the cells were incubated for 4 h at 37°C. Total RNA was isolated from each sample, and Northern blot analysis was performed on 5 μg of total RNA to determine the effects of PMA and U0126 treatments on mBTEB2 transcript levels. The blot was also probed for GAPDH to normalize for differences in loading. Signals were visualized by autoradiography and quantitated using a PhosphorImager.
Role of PMA/PKC.
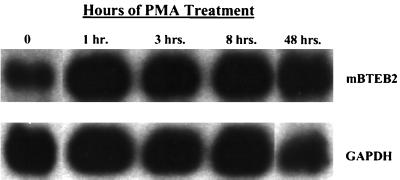
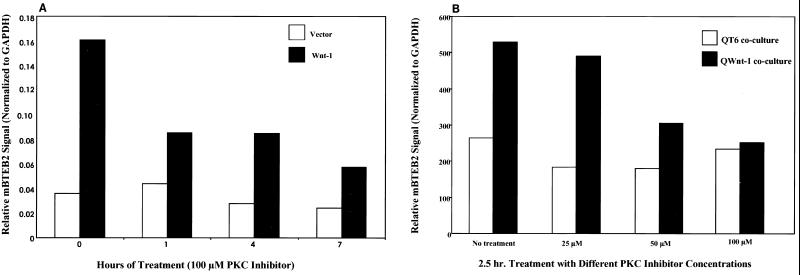
The mBTEB2 promoter is transcriptionally activated by low concentrations (10 nM) of the phorbol ester PMA (Fig. 9),, and high concentrations (100 μM) result in cell death (data not shown). These results are in close agreement with the ability of PMA to transcriptionally activate the human promoter (19). Experiments were designed to investigate whether PKC activation was necessary for the activation of the mBTEB2 promoter in response to Wnt-1 signaling. This was an attractive idea given the role of other Wnts which modulate gene expression through the activation of PKC and Ca2+ fluxes (the so-called Wnt/Ca2+ pathway) (reference 28 and references therein). Northern blot analysis was performed on C57MG/Wnt-1 and C57MG/Vector cells that had been treated for various times in the presence or absence of 100 μM myristoylated PKC inhibitor. These results suggest that the inhibition of PKC in the Wnt-1-infected cells decreases the transcriptional response of mBTEB2 to Wnt-1 signaling by approximately 50% (Fig. 10A). No change is apparent upon treatment of the empty vector-infected cells with the PKC inhibitor. This inhibition is also observed in C57MG cells cocultured with Wnt-1-expressing cells (data not shown). Furthermore, the Wnt-1-dependent transcriptional activation of mBTEB2 is sensitive to inhibition of PKC in a dose-dependent manner, in both the coculture assay and in retrovirus-infected cells (shown for cocultured cells in Fig. 10B). These results suggest that at least half of the response of mBTEB2 to Wnt-1 signaling depends on the activation of PKC, and that further downstream of the Wnt-1 signal there are other pathways that can contribute to the transcriptional activation of this promoter in response to Wnt-1 signaling.
FIG. 9.
Transcription of the mBTEB2 promoter is induced in response to low concentrations (10 nM) of the phorbol ester PMA. C57MG cells were incubated with 10 nM PMA for the indicated times, and total RNA was isolated. Northern blot analysis was performed using 5 μg of total RNA to assess mBTEB2 mRNA levels. The blot was probed for GAPDH to control for differences in loading. Signals were visualized by autoradiography and quantitated using a PhosphorImager.
FIG. 10.
Inhibition of PKC activity decreases the Wnt-1-dependent transcriptional activation of mBTEB2 in a dose-dependent manner. (A) C57MG cells infected with the Wnt-1 (C57MG/Wnt-1) or empty vector (C57MG/Vector) retrovirus were treated in the presence or absence of 100 μM myristoylated PKC inhibitor for the indicated times. Northern blot analysis was performed on 5 μg of total RNA in order to assess the expression of mBTEB2 mRNA. The blot was probed for GAPDH as a loading control. Signals were quantitated using a PhosphorImager, and the data are presented in graphical form. (B) C57MG cells were cocultured with QT6 or QT6Wnt-1 cells for 2.5 h in the presence or absence of various concentrations of a myristoylated PKC inhibitor. Northern blot analysis, signal quantitation, and graphical analysis were carried out as described for panel A.
DISCUSSION
Using cDNA subtraction between a Wnt-1-transformed cell line (C57MG/Wnt-1) and the parental cell line (C57MG), we have identified a novel target of the Wnt-1 signaling pathway, a mouse homolog of the human transcription factor BTEB2. In independent retrovirus infections of C57MG cells, as well as in a coculture assay with Wnt-1-expressing cells, mBTEB2 is transcriptionally up-regulated in response to Wnt-1 signaling. The mBTEB2 promoter is activated by 3 h after exposure to Wnt-1, and this activation persists at high levels throughout a 48-h coculture assay and up to 3 weeks in a Wnt-1 retrovirus infection of C57MG cells. To determine if mBTEB2 is up-regulated by the Wnt-1 signaling pathway in the absence of new protein synthesis, coculture experiments were performed in the presence of cycloheximide. Treatment with cycloheximide in the absence of coculture, however, induces mBTEB2 mRNA expression in C57MG cells (data not shown), eliminating this approach. That the mBTEB2 transcript is strongly and rapidly up-regulated by Wnt-1 signaling suggests that Wnt-1 regulates the mBTEB2 promoter through the activation of another transcription factor. In addition, mBTEB2 is a biologically relevant target of Wnt-1 signaling in vivo. Wnt-1 transgenic mouse mammary tumors and mouse mammary gland tissue prior to tumor formation express high levels of mBTEB2 mRNA, establishing a clear correlation between high Wnt-1 expression levels and the transcriptional up-regulation of mBTEB2. Low levels of mBTEB2 mRNA are detected in wild-type mouse mammary glands.
Surprisingly, while the mBTEB2 gene is consistently responsive to Wnt-1 signaling, it is not regulated through the β-catenin–Lef/TCF pathway. Transient transfection experiments using various mBTEB2 promoter constructs in combination with stable β-catenin mutants, as well as studies on the endogenous mBTEB2 promoter, indicate that the response of the promoter to Wnt-1 is not mediated by β-catenin–Lef/TCF. Rather, mBTEB2 appears to be responsive to Wnt-1 signaling in a manner that depends on the activation of PKC. This is a novel finding, given that most of the reported Wnt-1/Wg target genes respond to the growth factor through the binding of the β-catenin–Lef/TCF complex within their promoters. In the transfection experiments performed in 293 cells, the mBTEB2 promoter appears to be transcriptionally regulated by the presence of high levels of dominant negative TCF, as promoter activity is twofold higher than for the control (Fig. 6). One possible explanation for this increase is that the mutated TCF construct is now unable to interact with another factor that, when in a complex with TCF, normally negatively regulate transcription from this promoter. The twofold increase, however, was not robust enough to warrant further investigation. The activation of Wnt target gene expression through the use of a PKC/Ca2+-dependent pathway is a mechanism used by other Wnts, such as XWnt-5a, which do not signal through the β-catenin–Lef/TCF complex. Thus, mBTEB2 is a transcriptional target of Wnt-1 signaling that does not rely on the β-catenin–Lef/TCF complex but is instead activated at least in part (50%) through a PKC-dependent pathway.
Human BTEB2 was originally cloned from a human placenta cDNA library using the BTEB cDNA as a probe (38). The BTEB2 gene encodes a 219-amino-acid protein with an amino-terminal proline-rich transcriptional activation region, a basic region, and three zinc fingers of the C2H2 type that mediate DNA binding (16). A biological function for BTEB2 was recently demonstrated in the rabbit aorta, in which BTEB2 binds to a region (SE1) in the promoter of the rabbit embryonic smooth muscle myosin heavy chain B gene (SMemb/NMHC-B) and regulates its expression in a rabbit smooth muscle cell line (46). Furthermore, using the model system of vascular smooth muscle cells, the transcriptional regulation of human BTEB2 has been shown to be dependent on the binding of Egr-1 within its promoter in a PMA- and MAPK-dependent manner (19). Like the human BTEB2 promoter, the mouse promoter is regulated through a PMA-sensitive pathway. The mouse promoter, however, is not modulated by the MAPK pathway in the absence of Wnt-1 signaling. In striking contrast to the regulation of the human promoter, the Wnt-1 signaling pathway activates a MAPK pathway that negatively regulates the expression of mBTEB2.
The canonical Wnt signaling pathway employs the β-catenin–Lef/TCF complex to transcriptionally activate target genes. Recent evidence suggests that some Wnts can signal through a β-catenin–Lef/TCF-independent pathway to activate target gene expression. This pathway, known as the Wnt/Ca2+ pathway, uses the PI pathway, and its activation of the second messengers PKC and calcium, to regulate the expression of downstream genes (28). Calcium is a well-characterized second messenger molecule, and it appears that increases in intracellular calcium levels are the end result of XWnt-5A signaling. This effect is specific to Xwnt-5A, a Wnt that does not normally induce ectopic axis formation in embryos but instead perturbs morphogenetic movements upon overexpression (10). Injection of Xwnt-5A RNA into zebrafish embryos elicits increases in intracellular calcium, whereas injection of Xwnt-8 RNA, representing a Wnt that normally induces ectopic axis formation, does not (41). In addition, injection of either Xwnt-5A or rat Frizzled 2 (Rfz-2) RNA into zebrafish embryos has been shown to increase intracellular calcium, and coinjection of these RNAs into zebrafish embryos leads to a synergistic effect on calcium release (40).
Moreover, injection of Xenopus embryos with RNAs for Xwnt-5A and Rfz-2 results in the translocation of PKC to the plasma membrane in a manner that is dependent on G-protein signaling, while injection of Rfz-1 (which functions to stabilize β-catenin in response to Wnt signaling) has no effect on PKC (38). Xwnt-5a and Rfz-2 can also stimulate PKC activity in an in vitro kinase assay. In addition, other Frizzled RNAs have been tested for the ability to induce PKC movement to the membrane as well as induce expression of two β-catenin target genes, Xnr-3 and siamois. Interestingly, Frizzled receptors which induce translocation of PKC to the membrane (mouse Frizzled 3 [Mfz3], Mfz4, and Mfz6) cannot activate the expression of Xnr-3 and siamois, whereas expression of Frizzled receptors that do not cause a change in PKC location (Mfz7 and Mfz8) do activate β-catenin-mediated gene expression (38).
A recent report, however, argues that Wnt signaling through one receptor can activate both the canonical (β-catenin stabilization) and noncanonical (calcium and PKC) Wnt signaling pathways. Xenopus frizzled 7 (Xfz7) encodes a protein that can activate both the canonical and noncanonical downstream pathways in response to different Wnts (27). Injection of Xfz7 RNA into the ventral side of Xenopus embryos does not cause secondary axis formation, but it does inhibit cell movement and decreases the adhesive properties of the cells expressing the receptor. This phenotype for a Frizzled receptor is consistent with it having a role in the noncanonical pathway, and overexpression of Xfz7 RNA does indeed lead to translocation of PKC to the plasma membrane. The Xfz7 receptor, however, can also activate the canonical Wnt signaling pathway upon binding of the Xwnt-8b ligand, as demonstrated by the transcriptional activation of siamois and Xnr-3 (genes activated by the β-catenin–Lef/TCF complex). Thus, this study shows that a Frizzled receptor can discriminate among different Wnt ligands and select which downstream pathway(s) becomes activated upon binding of the ligand to the receptor.
The parental C57MG cell line and the C57MG/Wnt-1 cell line express high levels of Mfz6 RNA (which encodes a protein that can induce PKC translocation but not elevate β-catenin levels) and lower levels of Mfz7 and Mfz8 RNA (which encode proteins that can stabilize β-catenin levels but not induce PKC translocation) (D. Pennica, personal communication). Although low levels of Mfz2 RNA are also detected in these cell lines, mammary gland and mammary tumor tissue taken from the Wnt-1 transgenic mouse exhibit high levels of Mfz2 RNA (Pennica, personal communication). Presumably, this mouse homolog of Rfz2 functions to regulate PKC, and the presence of high levels of Mfz2 RNA in these particular tissues is in good agreement with the up-regulation of mBTEB2.
The presence of different Frizzled proteins on the surface of C57MG and C57MG/Wnt-1 cells suggests that binding of Wnt-1 to one or more Frizzled proteins may permit both the translocation of PKC to the plasma membrane (and its subsequent activation) and the stabilization of β-catenin concomitantly in the same cell. In light of the recent evidence reported for Xfz7, it is highly probable that one (or more) of the Frizzled receptors on the surface of C57MG cells are capable of binding Wnt-1 and eliciting both the canonical and noncanonical downstream pathways. The presence of Mfz2 and Mfz6 in these cells suggests a mechanism by which Wnt-1 signaling may activate PKC, for these Frizzleds have been reported to foster PKC activation (21). In our experiments, it appears that both pathways are activated in Wnt-1-treated C57MG cells because the transcriptional activation of mBTEB2 relies, in part, on the activation of PKC. Furthermore, β-catenin levels are also stabilized, presumably due to the ability of Wnt-1 to interact with those Frizzled receptors that permit the stabilization of β-catenin (Mfz7 and Mfz8). These results are also in agreement with the previous finding that another Wnt family member, Xwnt-5a, can bind to both the Hfz5 and Rfz2 receptors and elicit the canonical and noncanonical downstream pathways, respectively (15).
In summary, we have identified a novel target gene, mBTEB2, whose expression is correlated with Wnt-1 signaling. Surprisingly, mBTEB2 is transcriptionally activated through a β-catenin-independent mechanism. The regulation of this gene by Wnt-1 appears to occur, in part, through a PKC-dependent pathway, although this particular pathway is not the only means by which Wnt-1 signaling activates this promoter. The results presented here suggest a model in which Wnt-1 binding to one or more Frizzled receptors on the surface of C57MG cells results in both the activation of PKC and the concurrent stabilization of β-catenin. The activation of two downstream pathways could occur through the binding of Wnt-1 to two different Frizzled receptors, each eliciting a separate downstream pathway, or binding of Wnt-1 to one receptor, which in turn activates both pathways. Given the recent observation that those Wnts which activate PKC can also elicit downstream responses through the activation of calcium/calmodulin-dependent protein kinase II (21), it is also possible that Wnt-1 signaling can regulate the mBTEB2 promoter through the activation of this kinase and its subsequent downstream effectors. The transcript levels of mBTEB2 remain high throughout Wnt-1 signaling, possibly due to its ability to positively autoregulate its own promoter, a phenomenon that occurs for other GC box binding factors such as mouse gut-enriched Krüppel-like factor (24).
Although the human BTEB2 promoter is transcriptionally regulated by the Egr-1 protein, no change in Egr-1 protein levels were detected in the presence of Wnt-1 signaling. Furthermore, experiments performed to assess changes in the phosphorylation status of threonine residues in Egr-1 in response to Wnt-1 signaling were not definitive and appeared negative. Thus, we can conclude that the mouse and human BTEB2 promoters are regulated differently with respect to the role of Egr-1. This is not surprising, given the sequence difference in their promoter/enhancer regions. Comparison of these two sequences reveals some homology in the 5′ untranslated region but little homology further 5′ within the promoter/enhancer region (see reference 19 for the sequence of the human BTEB2 promoter/enhancer region and GenBank accession number AF285184 for the partial mBTEB2 promoter/enhancer sequence).
The 50% decrease in the Wnt-1-induced activation of the mBTEB2 promoter in the presence of the PKC inhibitor implies that other pathways that regulate the mBTEB2 promoter are also activated by Wnt-1 binding to Frizzled. Experiments performed using the MEK inhibitor U0126 suggest that Wnt-1 signaling in C57MG cells activates an ERK (extracellular signal-regulated kinase)-type MAPK pathway that negatively regulates the mBTEB2 promoter. Although PKC is a known activator of the ERK-based MAPK cascade (20, 26), the PKC-dependent and MAPK-dependent pathways activated by Wnt-1 in C57MG cells are mutually exclusive because treatment with the PKC inhibitor results in a down-regulation of mBTEB2 transcript levels. Thus, regulation of the mBTEB2 promoter in response to Wnt-1 signaling involves the activation of several downstream pathways, two of which include a MAPK/ERK-based signaling pathway and a PKC-dependent pathway.
It will be of interest to determine what other intracellular pathways are activated in response to Wnt-1 and in turn impinge upon the mBTEB2 promoter. In particular, it is not yet known whether the Egr-1 protein regulates the mBTEB2 promoter in the same manner as it regulates the human BTEB2 promoter. Furthermore, it will be of interest to determine how changes in intracellular calcium levels affect the response of the mBTEB2 promoter to Wnt-1 signaling and whether this effect depends on the coupling of G proteins to the Frizzled receptor. The identification of mBTEB2 as a Wnt-1-responsive gene will aid in the search for new target genes of Wnt-1 signaling whose expression is modulated by mBTEB2.
ACKNOWLEDGMENTS
This work was supported by NIH cancer training grant T32 CA-09528. L. T. Ziemer was supported in part by a fellowship from the New Jersey Commission on Cancer Research (98-2001-CCR-00).
We thank Y. Ahmed for critical reading of the manuscript.
REFERENCES
- 1.Arias A M, Brown A M, Brennan K. Wnt signalling: pathway or network? Curr Opin Genet Dev. 1999;9:447–454. doi: 10.1016/s0959-437x(99)80068-9. [DOI] [PubMed] [Google Scholar]
- 2.Behrens J, von Kries J P, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 3.Bejsovec A. Wnt signalling shows its versatility. Curr Biol. 1999;9:R684–R687. doi: 10.1016/s0960-9822(99)80439-4. [DOI] [PubMed] [Google Scholar]
- 4.Bradley R S, Brown A M C. A soluble form of Wnt-1 protein with mitogenic activity on mammary epithelial cells. Mol Cell Biol. 1995;15:4616–4622. doi: 10.1128/mcb.15.8.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brannon M, Gomperts M, Sumoy L, Moon R T, Kimelman D. A β-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev. 1997;11:2359–2370. doi: 10.1101/gad.11.18.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavallo R A, Cox R T, Moline M M, Roose J, Polevoy G A, Clevers H, Peifer M, Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- 7.Chan E F, Gat U, McNiff J M, Fuchs E. A common human skin tumour is caused by activating mutations in β-catenin. Nat Genet. 1999;21:410–413. doi: 10.1038/7747. [DOI] [PubMed] [Google Scholar]
- 8.Crawford H C, Fingleton B M, Rudolph-Owen L A, Goss K J, Rubinfeld B, Polakis P, Matrisian L M. The metalloproteinase matrilysin is a target of beta-catenin transactivation in intestinal tumors. Oncogene. 1999;18:2883–2891. doi: 10.1038/sj.onc.1202627. [DOI] [PubMed] [Google Scholar]
- 9.de La Coste A, Romagnolo B, Billuart P, Renard C A, Buendia M A, Soubrane O, Fabre M, Chelly J, Beldjord C, Kahn A, Perret C. Somatic mutations of the β-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc Natl Acad Sci USA. 1998;95:8847–8851. doi: 10.1073/pnas.95.15.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du S, Purcell S, Christian J, McGrew L, Moon R. Identification of distinct classes and functional domains of Wnts through expression of wild-type and chimeric proteins in Xenopus embryos. Mol Cell Biol. 1995;15:2625–2634. doi: 10.1128/mcb.15.5.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gradl D, Kuhl M, Wedlich D. The Wnt/Wg signal transducer beta-catenin controls fibronectin expression. Mol Cell Biol. 1999;19:5576–5587. doi: 10.1128/mcb.19.8.5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gumbiner B M. Signal transduction of β-catenin. Curr Opin Cell Biol. 1995;7:634–640. doi: 10.1016/0955-0674(95)80104-9. [DOI] [PubMed] [Google Scholar]
- 13.Gurskaya N G, Diatchenko L, Chenchik A, Siebert P D, Khaspekov G L, Lukyanov K A, Vagner L L, Ermolaeva O D, Lukyanov S A, Sverdlov E D. Equalizing cDNA subtraction based on selective suppression of polymerase chain reaction: cloning of Jurkat cell transcripts induced by phytohemagglutinin and phorbol 12-myristate 13-acetate. Anal Biochem. 1996;240:90–97. doi: 10.1006/abio.1996.0334. [DOI] [PubMed] [Google Scholar]
- 14.He T C, Sparks A B, Rago C, Hermeking H, Zawel L, da Costa L T, Morin P J, Vogelstein B, Kinzler K W. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 15.He X, Saint-Jeannet J P, Wang Y, Nathans J, Dawid I, Varmus H. A member of the Frizzled protein family mediating axis induction by Wnt-5a. Science. 1997;275:1652–1654. doi: 10.1126/science.275.5306.1652. [DOI] [PubMed] [Google Scholar]
- 16.Imataka H, Sogawa K, Yasumoto K, Kikuchi Y, Sasano K, Kobayashi A, Hayami M, Fujii-Kuriyama Y. Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-4501A1 gene. EMBO J. 1992;11:3663–3671. doi: 10.1002/j.1460-2075.1992.tb05451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnston S H, Rauskolb C, Wilson R, Prabhakaran B, Irvine K D, Vogt T F. A family of mammalian Fringe genes implicated in boundary determination and in the Notch pathway. Development. 1997;124:2245–2254. doi: 10.1242/dev.124.11.2245. [DOI] [PubMed] [Google Scholar]
- 18.Jue S F, Bradley R S, Rudnicki J A, Varmus H E, Brown A M C. The mouse Wnt-1 gene can act via a paracrine mechanism in transformation of mammary epithelial cells. Mol Cell Biol. 1992;12:321–328. doi: 10.1128/mcb.12.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawai-Kowase K, Kurabayashi M, Hoshino Y, Ohyama Y, Nagai R. Transcriptional activation of the zinc finger transcription factor BTEB2 gene by Egr-1 through mitogen-activated protein kinase pathways in vascular smooth muscle cells. Circ Res. 1999;85:787–795. doi: 10.1161/01.res.85.9.787. [DOI] [PubMed] [Google Scholar]
- 20.Kazlauskas A, Cooper J A. Protein kinase C mediates platelet-derived growth factor-induced tyrosine phosphorylation of p42. J Cell Biol. 1988;106:1395–1402. doi: 10.1083/jcb.106.4.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kühl M, Sheldahl L C, Malbon C C, Moon R T. Ca2+/calmodulin-dependent protein kinase II is stimulated by Wnt and Frizzled homologs and promotes ventral cell fates in Xenopus. J Biol Chem. 2000;275:12701–12711. doi: 10.1074/jbc.275.17.12701. [DOI] [PubMed] [Google Scholar]
- 22.Laurent M N, Blitz I L, Hashimoto C, Rothbacher U, Cho K W. The Xenopus homeobox gene twin mediates Wnt induction of goosecoid in establishment of Spemann's organizer. Development. 1997;124:4905–4916. doi: 10.1242/dev.124.23.4905. [DOI] [PubMed] [Google Scholar]
- 23.Magee A I. Cell adhesion molecules and intracellular signalling: from fly to man. Cell Signal. 1995;7:165–170. doi: 10.1016/0898-6568(94)00090-x. [DOI] [PubMed] [Google Scholar]
- 24.Mahatan C S, Kaestner K H, Geiman D E, Yang V W. Characterization of the structure and regulation of the murine gene encoding gut-enriched Krüppel-like factor (Krüppel-like factor 4) Nucleic Acids Res. 1999;27:4562–4569. doi: 10.1093/nar/27.23.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mann B, Gelos M, Siedow A, Hanski M L, Gratchev A, Ilyas M, Bodmer W F, Moyer M P, Riecken E O, Buhr H J, Hanski C. Target genes of β-catenin-T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proc Natl Acad Sci USA. 1999;96:1603–1608. doi: 10.1073/pnas.96.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marquardt B, Frith D, Stabel S. Signalling from TPA to MAP kinase requires protein kinase C, raf and MEK: reconstitution of the signalling pathway in vitro. Oncogene. 1994;9:3213–3218. [PubMed] [Google Scholar]
- 27.Medina A, Reintsch W, Steinbeisser H. Xenopus frizzled 7 can act in canonical and non-canonical Wnt signaling pathways: implications on early patterning and morphogenesis. Mech Dev. 2000;92:227–237. doi: 10.1016/s0925-4773(00)00240-9. [DOI] [PubMed] [Google Scholar]
- 28.Miller J R, Hocking A M, Brown J D, Moon R T. Mechanism and function of signal transduction by the Wnt/β-catenin and Wnt/Ca2+ pathways. Oncogene. 1999;18:7860–7872. doi: 10.1038/sj.onc.1203245. [DOI] [PubMed] [Google Scholar]
- 29.Miyoshi Y, Hagase H, Ando H, Horii A, Ichii S, Nakatsuru S, Aoki T, Miki Y, Mori T, Nakamura Y. Somatic mutations of the APC gene in colorectal tumors: mutation cluster region in the APC gene. Hum Mol Genet. 1992;1:229–233. doi: 10.1093/hmg/1.4.229. [DOI] [PubMed] [Google Scholar]
- 30.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. XTcf-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 31.Moon R T, Brown J D, Torres M. WNTs modulate cell fate and behavior during vertebrate development. Trends Genet. 1997;13:157–162. doi: 10.1016/s0168-9525(97)01093-7. [DOI] [PubMed] [Google Scholar]
- 32.Morin P J, Sparks A B, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K W. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 33.Nusse R. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982;31:99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 34.Nusse R, Varmus H E. Wnt genes. Cell. 1992;69:1073–1087. doi: 10.1016/0092-8674(92)90630-u. [DOI] [PubMed] [Google Scholar]
- 35.Pennica D, Swanson T A, Welsh J W, Roy M A, Lawrence D A, Lee J, Brush J, Taneyhill L A, Deuel B, Lew M, Watanabe C, Cohen R L, Melhem M F, Finley G G, Quirke P, Goddard A D, Hillan K J, Gurney A L, Botstein D, Levine A J. WISP genes are members of the connective tissue growth factor family that are up-regulated in Wnt-1-transformed cells and aberrantly expressed in human colon tumors. Proc Natl Acad Sci USA. 1998;95:14717–14722. doi: 10.1073/pnas.95.25.14717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riese J, Yu X, Munnerlyn A, Eresh S, Hsu S-C, Grosschedl R, Bienz M. LEF-1, a nuclear factor coordinating signaling inputs from wingless and decapentaplegic. Cell. 1997;88:777–787. doi: 10.1016/s0092-8674(00)81924-8. [DOI] [PubMed] [Google Scholar]
- 37.Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Stabilization of β-catenin by genetic defects in melanoma cell lines. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 38.Sheldahl L C, Park M, Malbon C C, Moon R T. Protein kinase C is differentially stimulated by Wnt and Frizzled homologs in a G-protein-dependent manner. Curr Biol. 1999;9:695–698. doi: 10.1016/s0960-9822(99)80310-8. [DOI] [PubMed] [Google Scholar]
- 39.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. The cyclin D1 gene is a target of the β-catenin/LEF-1 pathway. Proc Natl Acad Sci USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slusarski D C, Corces V G, Moon R T. Interaction of Wnt and a Frizzled homologue triggers G-protein-linked phosphatidylinositol signalling. Nature. 1997;390:410–413. doi: 10.1038/37138. [DOI] [PubMed] [Google Scholar]
- 41.Slusarski D C, Yang-Snyder J, Busa W B, Moon R T. Modulation of embryonic intracellular Ca2+ signaling by Wnt-5A. Dev Biol. 1997;182:114–120. doi: 10.1006/dbio.1996.8463. [DOI] [PubMed] [Google Scholar]
- 42.Sogawa K, Imataka H, Yamasaki Y, Kusume H, Abe H, Fujii-Kuriyama Y. cDNA cloning and transcriptional properties of a novel GC box-binding protein, BTEB2. Nucleic Acids Res. 1993;21:1527–1532. doi: 10.1093/nar/21.7.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tetsu O, McCormick F. β-Catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 44.van der Heyden M A, Rook M B, Hermans M M, Rijksen G, Boonstra J, Defize L H, Destree O H. Identification of connexin43 as a functional target for Wnt signalling. J Cell Sci. 1998;111:1741–1749. doi: 10.1242/jcs.111.12.1741. [DOI] [PubMed] [Google Scholar]
- 45.van der Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J. Armadillo co-activates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 46.Watanabe N, Kurabayashi M, Shimomura Y, Kawai-Kowase K, Hoshino Y, Manabe I, Watanabe M, Aikawa M, Kuro-o M, Suzuki T, Yazaki Y, Nagai R. BTEB2, a Kruppel-like transcription factor, regulates expression of the SMemb/Nonmuscle myosin heavy chain B (SMemb/NMHC-B) gene. Circ Res. 1999;85:182–191. doi: 10.1161/01.res.85.2.182. [DOI] [PubMed] [Google Scholar]
- 47.Xu L, Corcoran R B, Welsch J W, Pennica D, Levine A J. WISP-1 is a Wnt-1- and β-catenin-responsive oncogene. Genes Dev. 2000;14:585–595. [PMC free article] [PubMed] [Google Scholar]