Abstract
Background
COVID‐19 convalescent plasma (CCP) ideally contains high titers of (neutralizing) anti‐SARS‐CoV‐2 antibodies. Several scalable immunoassays for CCP selection have been developed. We designed an enzyme‐linked immunosorbent assay (ELISA) that measures neutralizing antibodies (of all isotypes) in plasma by determining the level of competition between CCP and a mouse neutralizing antibody for binding to the receptor binding domain (RBD) of SARS‐CoV‐2.
Methods
Plasma was collected from 72 convalescent individuals and inhibition of viral infection was determined by plaque reduction neutralization (PRNT50). The level of neutralizing antibodies was measured in the novel competition ELISA and in a commercially available ELISA that measures inhibition of recombinant ACE2 binding to immobilized RBD. These results were compared with a high throughput chemiluminescent microparticle immunoassay (CMIA).
Results
The results from both ELISAs were correlating, in particular for high titer CCP (PRNT50 ≥ 1:160) (Spearman r = .73, p < .001). Moderate correlation was found between the competition ELISA and CMIA (r = .57 for high titer and r = .62 for low titer CCP, p < .001). Receiver operator characteristic analysis showed that the competition ELISA selected CCP with a sensitivity and specificity of 61% and 100%, respectively. However, discrimination between low and high titer CCP had a lower resolution (sensitivity: 34% and specificity: 89%).
Conclusion
The competition ELISA screens for neutralizing antibodies in CCP by competition for just a single epitope. It exerts a sensitivity of 61% with no false identifications. These ELISA designs can be used for epitope mapping or for selection of CCP.
Keywords: CCP, convalescent plasma, competition ELISA, COVID‐19, neutralizing antibody, SARS‐CoV‐2
1. INTRODUCTION
The coronavirus disease (COVID‐19) has caused over 3.9 million deaths worldwide. 1 Several therapies, either prophylactic or therapeutic, are under investigation or in (emergency) use. Transfusion of COVID‐19 convalescent plasma (CCP), defined as plasma collected from individuals that have recovered from COVID‐19, has been proposed as a potential therapeutic. Major advantages of CCP therapy are the availability early on in an epidemic, (relatively) low cost, and the well‐known safety profile of plasma transfusion. However, as all substances of human origin, plasma is inherently variable including antibody content directed at (inhibiting) SARS‐CoV‐2.
Studies of CCP transfusion in small cohorts of immunocompromised patients suggest that it is highly efficacious and safe. 2 , 3 , 4 Nonetheless, results from trials with a large sample size of immunocompetent COVID‐19 patients suggest that it decreases mortality or severe illness only when a “high titer” of neutralizing antibodies is present in plasma and when administered early in the course of the disease. 5 , 6 , 7
The selective recruitment of “high titer” donors has been challenging, because these high titers of antibodies are observed in just a subset of patients. 8 , 9 , 10 The plaque reduction neutralization test (PRNT) is considered the gold standard for detecting and measuring neutralizing antibodies that mediate viral inhibition. However, it is time‐consuming, expensive, requires live virus handling, and thus biosafety level 3 facilities with expert staff and is subjected to biological variability (e.g., cell lines and viral strains). It therefore may become rate limiting for plasma availability, especially during the peak of an outbreak. Immunoassays using recombinant SARS‐CoV‐2 antigen(s) and multiwell designs allow higher throughput and can be performed in laboratories of lower biosafety level. 11 Certain serology assays that determine the relative quantity of anti‐SARS‐CoV‐2 antibodies in plasma have been shown to correlate with PRNT. 12 However, immunoassays that directly measure inhibitory capacity of CCP may probably be more selective for the presence of (high titer) neutralizing antibodies. Such assays are based on the inhibition of the interaction between the recombinant SARS‐CoV‐2 receptor binding domain (RBD) and the recombinant human angiotensin converting enzyme‐2 (ACE2) soluble receptor fragment. 13 , 14
Here, we describe a different enzyme‐linked immunosorbent assay (ELISA) that measures neutralizing antibodies in CCP based on the competition between neutralizing antibodies (of all isotypes) in plasma of convalescent individuals and a single commercially available neutralizing mouse monoclonal antibody (mAb), for binding to RBD. We have assessed whether there is a correlation between the results obtained by this ELISA and those obtained by (a) a commercially available ELISA that measures inhibition of recombinant ACE2 binding to immobilized RBD and (b) a high throughput serology test that screens for the presence of anti‐RBD IgG antibodies.
2. MATERIALS AND METHODS
2.1. Convalescent and non‐immune plasma
Plasma samples from 74 separate donations were collected from 72 donors who had recovered from a SARS‐CoV‐2 infection. Previous infection needed to be confirmed by either RT‐PCR, chest computed tomography, or serology assays. Donors were scheduled for testing at least 14 days after resolution of symptoms. Test samples were transferred to the laboratory performing PRNT (Rega Institute of Medical Research, KU Leuven, Belgium). In addition, plasma samples collected from 24 different individuals before the SARS‐CoV‐2 pandemic (i.e. 2018–2019) were used as naïve, non‐immune controls. Signed, informed consent was obtained from each donor.
2.2. Competition ELISA
Microtiterplates in 96‐well format (Cat # 655092, Greiner Bio‐one, Kremsmunster, Austria) were coated overnight with 100 μl of purified SARS‐CoV‐2 (2019‐nCoV) Spike RBD‐His Recombinant Protein (1 μg/ml) (YP_009724390.1) (Arg319‐Phe541) (Cat # 40592‐V08H, Sino Biological, Beijing, China) in phosphate buffered saline (PBS) pH 7.4 at 4°C. During blocking with assay buffer (PBS with 1% [wt/vol] bovine serum albumin [BSA]), a 7‐step, 2‐fold dilution series of donor plasma was prepared in a separate blocked low‐binding microplate (Cat # 655101, Greiner Bio‐one, Kremsmunster, Austria). Each plasma dilution was mixed with the mouse anti‐RBD neutralizing mAb solution in assay buffer (Cat # 40592‐MM57, Sino Biological, Beijing, China) at a 9:1 volume ratio, yielding a final mAb concentration of 0.55 nM. These sample series were transferred to the RBD‐coated microtiterplate and incubated for 1 hour at room temperature. For all samples, technical triplicates were performed, unless otherwise indicated. Finally, the wells were incubated with a rabbit anti‐mouse IgG secondary antibody conjugated with horseradish peroxidase (Cat # 315‐035‐008, Jackson Immunoresearch, West Grove, PA, US) at a 1:60,000 dilution (1.33 ng/well) in assay buffer for 1 h at room temperature. Chromogenic development was with 3,3′,5,5′‐tetramethylbenzidine substrate solution (Cat # T444, Sigma‐Aldrich, St. Louis, MO, USA) for 10 min at room temperature and quenched with 1M sulfuric acid. Optical density was measured in a spectrophotometer (Plate Reader Infinite F200 PRO, TECAN, Männedorf, Switzerland) at a wavelength of 450 nm (OD450nm). Coating efficiency and precision of the blank samples (i.e. control with no plasma added) were assessed (Figures S1 and S2).
2.3. Inhibition ELISA
The inhibition ELISA from ACROBiosystems (Newark, DE, US) (Cat # EP‐105) was performed according to the manufacturer's instructions except for following modifications: (a) the recommended coating buffer (15.0 mM Na2CO3, 35.0 mM NaHCO3, 7.7 mM NaN3, pH 9.6) was changed to PBS (pH 7.4) and (b) the blocking and dilution buffer (PBS with 0.05% [v/v] Tween‐20 [pH 7.4] and 2.0% [wt/vol] or 0.5% [wt/vol] BSA, respectively) by assay buffer. (c) The lowest dilution of donor plasma was adjusted to 90% (vol/vol), instead of 50% (vol/vol) in assay buffer. These adjustments were made to allow a paired experimental design for our competition versus the commercial inhibition ELISA.
2.4. Chemiluminescent microparticle immunoassay (CMIA)
Serology testing specific for IgG binding to RBD was assessed using the i2000SR Architect robot (Abbott Diagnostics, Lake County, Illinois) equipped with the CE‐IVD labeled SARS‐CoV‐2 IgG II Quant assay (Cat # 6S60). Samples were processed according to the manufacturer's recommendations. In brief, plasma sample and RBD‐coated paramagnetic microparticles were mixed with assay diluent and incubated. Wells were incubated with anti‐human IgG acridinium‐labeled conjugate and bound antibody was quantified by the addition of “pre‐trigger” and “trigger” solutions to initiate the chemiluminescent reaction. Results were generated by plotting the measured values against the assay‐specific calibration curve, generated by a four‐parameter logistic fit. An assay‐specific positive and negative quality control as well as two non‐assay‐specific positive controls were included. A plasma sample is considered positive for anti‐RBD IgG if signal‐to‐cut‐off (S/CO) is equal or above the threshold value of 50.0 AU/ml.
2.5. Plaque reduction neutralization test (PRNT50)
This assay was conducted with a Belgian clinical isolate (SARS‐2‐CoV/Belgium/GHB‐03021/2020, GISAID accession number EPI_ISL_407976, passage 5) as previously described. 2 , 15 In brief, plasma sample was heat inactivated for 30 min at 56°C to minimize complement activity. Following incubation of a 6‐step serial plasma dilution with 400 plaque forming units (pfu) of SARS‐CoV‐2 in 96‐well plates seeded with Vero E6 cells (1 h, 37°C, humidified 5% CO2 atmosphere), a 1% (wt/vol) agarose (SeaKem LE agarose, Lonza, Belgium) overlay was added (4 days, 37°C). Following overlay with 1% (wt/vol) neutral red/1% (wt/vol) agarose (24 h, 37°C), plaques were counted. Technical duplicates were performed at all times. Virus neutralization titers were reported as 50% reduction (PRNT50) in the number of plaques in comparison to a non‐neutralizing antibody control. Convalescent plasma samples of known PRNT50 titer are presented as CCP20 if the titer was 1:20, CCP40 if the titer was 1:40, etc.
2.6. Data analysis
For the competition and the inhibition ELISA, background OD450nm values obtained from uncoated control wells were subtracted from raw OD450nm data before analysis. These specific OD450nm values were normalized to the signal obtained in the absence of plasma, that is, maximal binding and no competition or inhibition. These normalized data are referred to as “relative OD450nm.” For both ELISAs, data were plotted as log transformed relative OD450nm as a function of log transformed plasma volume fraction (%) (Figure S3). The Area Under the Curve (AUC), a measure for the rate of competition or inhibition, 13 was calculated using Prism version 9 (GraphPad Software Inc., San Diego, CA, USA).
Correlation outcomes were obtained by calculating the Spearman's rank correlation r value. The intra‐ and inter‐assay coefficients of variation (CV) were calculated across experiments using CCP samples containing varying PRNT50 titers from 1:40 to 1:640, to cover a broad range in signal. Inter‐assay CV was determined by testing each sample in duplicate on five different microtiter plates, whereas intra‐assay CV was determined by testing the CCP samples in quintuplicate within the same microtiter plate.
The discriminating power of the competition ELISA was calculated by receiver operator characteristic (ROC) analysis: first, using 98 plasma samples consisting of non‐immune (n = 24) and CCP samples with PRNT50 ≥ 1:20 (n = 74), and second, using the 74 CCP samples consisting of PRNT50 < 1:160 (n = 36) and PRNT50 ≥ 1:160 CCP samples (n = 38). The cut‐off value, the diagnostic sensitivity, and specificity were determined by ROC analysis using Prism.
3. RESULTS
3.1. Assay design
Humoral immunity to SARS‐CoV‐2 is defined by the presence of antibodies in plasma of (former) COVID‐19 patients. Some of these antibodies can neutralize the virus–host interaction which is biochemically based on molecular binding of the viral Spike protein to the host ACE2 receptor (Figure 1A). The aim of this study was to test a simple, fast, and low‐cost ELISA to detect neutralizing antibodies in plasma.
FIGURE 1.
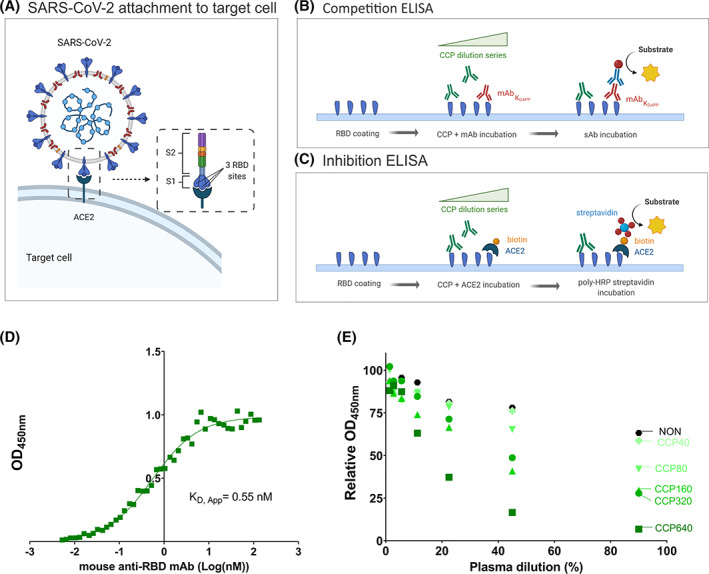
ELISA design to measure neutralizing antibodies in CCP. (A) The adhesion of SARS‐CoV‐2 to the target cell is mediated by the interaction between Spike protein (S1 and S2) of SARS‐CoV‐2 (composed of 3 RBD sites) with the ACE2 host receptor. (B) Competition ELISA measuring the amount of neutralizing murine anti‐RBD mAb (red) binding to immobilized RBD in the presence of varying concentrations of CCP. Detection of bound mAb is with peroxidase labeled secondary anti‐mouse antibody (blue). (C) Inhibition ELISA measuring the amount of biotinylated (orange) ACE2 binding to immobilized RBD in the presence of varying concentrations of CCP (green). Detection of bound ACE2 is with peroxidase labeled streptavidin. (D) For selection of the optimal mAb concentration, binding of mAb to immobilized RBD was investigated. Absorbance (OD450nm) as a function of increasing amounts of mAb (log transformed concentration in nM) indicates saturable binding with a KD,App of 0.55 nM. (E) Results of the competition ELISA using a selected series of CCP of known PRNT50 titer as indicated by color coding. The black dots assigned “NON” were from a pool of non‐immune plasma (n = 15) as a negative control. All 7‐step titrations were conducted in triplicate and mean values are given [Color figure can be viewed at wileyonlinelibrary.com]
We designed a competition ELISA (Figure 1B) and compared it to an inhibition ELISA (Figure 1C). The former detects binding of a murine neutralizing anti‐RBD mAb, while the latter detects binding of biotinylated recombinant ACE2 to RBD. The presence of inhibitory antibodies in plasma will either compete (competition ELISA) or inhibit (inhibition ELISA) the respective interaction. To maximize ELISA sensitivity, half‐maximal binding of the anti‐RBD mAb is required. Therefore, its apparent dissociation constant (KD,app) was determined by a series of titration experiments (Figure 1D). The KD,app was 0.55 nM. Any antibody in plasma at sufficiently high concentration and binding to the same epitope will lower the signal.
Results from five examples of CCP samples with varying PRNT50 values and a pool (n = 15) of non‐immune control plasma samples are presented in Figure 1E. At low dilution of non‐immune control plasma circa 20% apparent competition was found, indicating a matrix effect of naïve plasma. The five CCP examples in Figure 1E effectively competed to varying degrees with the mouse mAb for binding to RBD. A linearity study has been conducted on these five serially diluted CCP samples (Figure S4). A linear relationship between titer and dilution was observed for low to high PRNT50‐predefined titers (R2 > 0.8). The area under the curve (AUC) will be used as an analytical outcome parameter to describe the rate of inhibition in a given CCP relative to non‐immune control plasma (Figure S3).
3.2. Assay validation
A cohort of 74 CCP samples of varying PRNT50 titer was analyzed pairwise in the competition and inhibition ELISA (Figure 2). The signal of the non‐immune control was used to set a statistical threshold AUC above which CCP was deemed “positive” (upper dotted lines). This value corresponded to the mean (lower dotted lines) plus two standard deviations (2SD) of the non‐immune control and was 0.256 for the competition ELISA and 0.581 for the inhibition ELISA. Using this criterion, just 28 of 74 CCP samples or 38% were considered as positive by the competition ELISA (Figure 2A,C) and 56 of 74 CCP samples or 76% were positive by inhibition ELISA (Figure 2B,D). The inhibition ELISA displayed a higher dynamic range compared with the competition ELISA. Despite this apparent difference in sensitivity, for every single sample, signal trends were comparable between assays. This was most prominent for high titer CCP (Figure 2A,B) and less prominent for low titer CCP (Figure 2C,D). The highest PRNT50 titers of 1:640 did not stand out as the highest signals in either ELISA compared with medium CCP320 and CCP160 titers. In addition, large variability between different donors within each PRNT50 bin was observed in both ELISAs.
FIGURE 2.
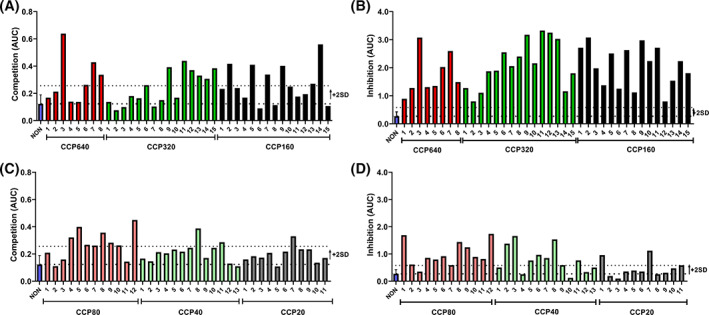
Comparison between competition and inhibition ELISAs. (A) Bar graph representing the AUC of individual high titer (PRNT50 ≥ 1:160) CCP screened in the competition ELISA. (B) Bar graph representing the AUC of the same individual high titer (PRNT50 ≥ 1:160) CCP samples as in panel A screened by the inhibition ELISA. (C) Bar graph representing the AUC of individual low titer (PRNT50 ≤ 1:80) CCP screened in the competition ELISA. (D) Bar graph representing the AUC of the same individual low titer (PRNT50 ≤ 1:80) CCP samples as in panel C screened by the inhibition ELISA. The lower dotted line represents the mean of a non‐immune plasma pool (n = 15) of 23 technical replicates, each tested in triplicate (blue bar). The upper dotted line represents the threshold value at 2SD. All samples were tested in triplicate and mean values given. Error bars represent SD [Color figure can be viewed at wileyonlinelibrary.com]
Assay specificity was determined for both ELISAs using 24 different non‐immune samples from before the pandemic (Figure S5). No false positives were found in the competition ELISA and three samples were found borderline false positive in the inhibition ELISA. The inter‐assay CV of the competition ELISA was ≤31% for low titer CCP (≤1:40) and ≤14% for medium to high titer CCP (≥1:80) samples. The intra‐assay CV was <9% over the entire CCP working range.
3.3. Correlation between assays
Correlation between both ELISAs was stronger for high titer (≥1:160) CCP (r = .73, p < .0001) (Figure 3A) compared with low titer (≤1:80) CCP (r = .46, p < .005) (Figure 3B). The correlation between the competition ELISA and the high‐throughput CMIA was moderate for both high titer (r = .57, p < .002, Figure 3C) and low titer (r = .62, p < .0001, Figure 3D) CCP. In Figure 3, PRNT50 titers are color coded. No apparent clustering of titers was observed in all three assays though low titer CCP generally appear in the lower dynamic range of the inhibition ELISA (AUC of maximally 2.0) and the CMIA (S/CO of maximally 4000).
FIGURE 3.
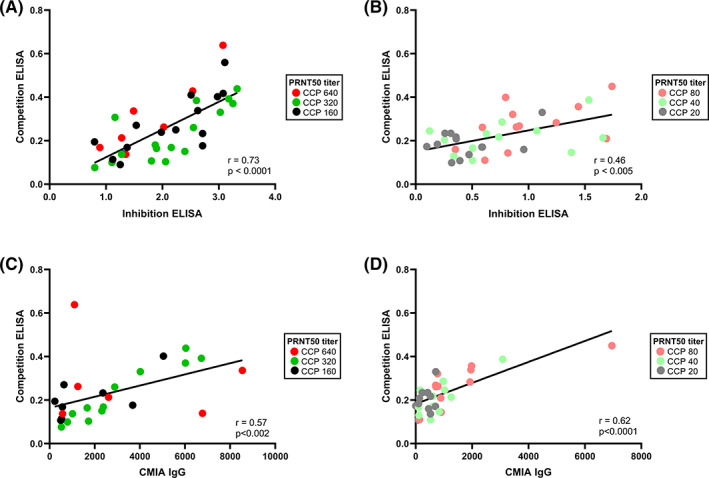
Correlation between the competition and inhibition ELISAs and between the inhibition ELISA and CMIA. (A) Correlation between the competition and inhibition ELISA for high titer CCP (PRNT50 ≥ 1:160) (n = 38). (B) Correlation between the competition and inhibition ELISA for low titer CCP (PRNT50 ≤ 1:80) (n = 36). (C) Correlation between the competition ELISA and CMIA for high titer CCP (n = 27). (D) Correlation between the competition ELISA and CMIA for low titer CCP (n = 33). In all panels, color coding is used to indicate the PRNT50 bin to which each individual CCP belongs (see legend in frame). Statistical outcomes are given in the bottom right corner [Color figure can be viewed at wileyonlinelibrary.com]
3.4. CCP selection by ELISA
Receiver operator characteristic analysis was performed using 24 non‐immune and 74 CCP samples of varying PRNT50 titer. Four arbitrary threshold values were investigated using data from the competition ELISA yielding diagnostic sensitivities and specificities (Table 1A). At a threshold of 0.189, specificity was 100% and sensitivity was 61% (Figure 4A). For the inhibition ELISA, specificity was 100% and sensitivity 81% at a threshold value of 0.568 (Figure 4B). The discrepancy between both ELISAs is larger when assessing discrimination between low titer (≤1:80) CCP and high titer (≥1:160) CCP (Figure 4C,D). In the CCP cohort (n = 74), 36 were low titer (≤1:80) and 38 were high titer (≥1:160). For any ELISA to have a specificity of 100%, sensitivity decreases to 5% and 63% for the competition (Table 1B) and inhibition (Figure 4D) ELISA, respectively. The optimal threshold for the competition ELISA was 0.330 yielding a diagnostic specificity of 89% with just 34% true positive high titer CCP (Figure 4C). Consequently, the predictive value (both positive and negative) was acceptable for the competition ELISA only when used for distinguishing CCP from non‐immune plasma (Table 1A).
TABLE 1.
Threshold values and corresponding diagnostic sensitivity and specificity for the competition ELISA, determined by ROC analysis
| Threshold | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | PPV (%) | NPV (%) |
|---|---|---|---|---|
| (A) Non‐immune plasma versus CCP | ||||
| >0.183 | 60.8 (49.4–71.1) | 100 (86.2–100) | 100 | 0 |
| >0.159 | 77.3 (66.3–85.1) | 87.5 (69.0–95.7) | 100 | 0 |
| >0.138 | 82.4 (72.2–89.4) | 79.2 (59.5–90.8) | 100 | 0 |
| >0.106 | 94.6 (86.9–97.9) | 62.5 (42.7–78.8) | 100 | 0 |
| (B) Low titer CCP versus high titer CCP | ||||
| >0.505 | 5.26 (0.94–17.3) | 100 (90.4–100) | 100 | 76.0 |
| >0.401 | 18.4 (9.22–33.4) | 97.2 (85.8–99.9) | 68.7 | 78.1 |
| >0.330 | 34.2 (21.2–50.1) | 88.9 (74.7–95.6) | 50.7 | 80.2 |
| >0.233 | 52.6 (37.3–67.5) | 58.3 (42.2–72.9) | 29.6 | 79.7 |
Note: (A) Results for discrimination between non‐immune plasma and CCP. (B) Results for discrimination between low (≤1:80) and high (≥ 1:160) titer CCP. The positive predictive value (PPV) and negative predictive value (NPV) are given in (A) assuming a prevalence of 100%, that is, the donor provides evidence of former SARS‐CoV‐2 infection and (B) assuming a prevalence of 25% ≥ CCP160 in the population.
FIGURE 4.
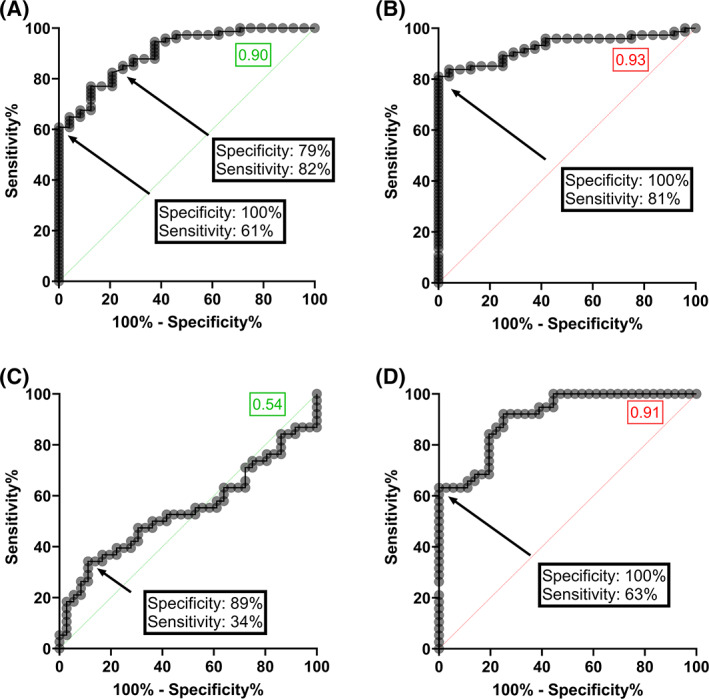
ROC curves for the competition and inhibition ELISAs. (A, B) ROC curves generated from data of the competition ELISA (panel A) or the inhibition ELISA (panel B) discriminating non‐immune (n = 24) from CCP (n = 74) at a threshold of 0.183 (panel A) or 0.568 (panel B). (C,D) ROC curves generated from data of the competition ELISA (panel C) or the inhibition ELISA (panel D) discriminating high (PRNT50 ≥ 1:160) from low (PRNT50 ≤ 1:80) CCP at a threshold of 0.330 (panel C) or 1.781 (panel D). Areas under the curve for the competition and inhibition ELISA are shown in green and red inset boxes, respectively [Color figure can be viewed at wileyonlinelibrary.com]
4. DISCUSSION
In Belgium, three clinical assessments are ongoing for CCP transfusion, two randomized clinical trials (access numbers NCT04429854 and NCT04558476) and one monitored access program. In all the three cases, CCP distribution to hospitals has been restricted to high titer CCP only (arbitrarily defined in Belgium as PRNT50 ≥ 1:160). This neutralizing antibody titer was determined by PRNT, currently considered as the gold standard for testing viral inhibition. Yet, off‐site PRNT testing has several disadvantages that may be solved by on‐site high throughput tests that do not require stringent biocontainment. The fastest available alternative is a serology immunoassay that measures the relative quantity of antibodies directed against relevant SARS‐CoV‐2 antigens, such as spike, RBD, and/or nucleocapsid protein. However, such tests do not discriminate between neutralizing and non‐neutralizing antibodies.
In the context of a European effort to better understand the laboratory tests that quantify antibody content in CCP, several collaborative goals were set. 16 One was to find the best suited immunoassay specifically detecting inhibitory antibodies in a (semi‐)quantitative way. 14 In the current study, we therefore developed a simple and cost‐effective competition ELISA for the screening of neutralizing antibody titers in CCP. We hypothesized that the polyclonal mix of neutralizing antibodies in CCP would compete with a single inhibitory mouse mAb, given that the RBD–ACE2 interaction is limited to just 17 and 20 amino acids, respectively. 17
To test this, a panel of 74 CCP samples was used and data were compared with a commercially available inhibition ELISA. Notwithstanding the differences in assay design, the signals of both ELISAs correlated well, in particular for high titer CCP. Correlation between the competition ELISA and the high throughput CMIA serology test was moderate. This may be caused by a different assay design, for instance measuring total antibody versus IgG only. Several studies were published that report acceptable correlation between serology testing and virus neutralization tests. 12 , 18 , 19 From our data, however, “correlation” between the outcome in PRNT and the results of both ELISAs was poor and variability within each PRNT50 bin was high. However, the CVs of these ELISAs are low to moderate and hence do not explain this observation. The CV of the PRNT that was used to determine virus neutralization is unknown but may be high given that this is an assay that requires live cell and virus culture. So poor correlation may be caused by inherent variation in the PRNT as well. In theory, inhibition of viral infection by CCP may be accomplished in ways beyond steric interference of RBD–ACE2 interactions, which might be detected in PRNT but not in an ELISA set‐up. It has been hypothesized, for example, that neutralizing antibodies targeting the N‐terminal domain of the viral spike might elucidate different neutralizing mechanisms such as restricting pre‐ to post‐fusion conformational changes of the S protein. 20 , 21 Most recovered COVID‐19 patients, however, bear antibodies to RBD 9 suggestive of at least a major antigenic determinant in that viral protein sequence. 22 , 23
The challenge is recruitment of adequate numbers of donors to meet demand, while still ensuring that those who are recruited have sufficiently high titers of antibodies to be effective. 11 Therefore, the discriminating power of neutralization tests is utmost important. The competition ELISA was acceptable in selecting CCP from non‐immune plasma but not good enough to discriminate low from high titer CCP. A major reason is the limited dynamic range/sensitivity of our ELISA. The threshold value calculated from the mean + 2SD of repeated non‐immune control analysis and statistically ruling out 95% of negative plasma was 0.255 for the competition ELISA. A lower threshold (>0.183) was determined by ROC analysis, which resulted in a diagnostic sensitivity of 61% with no false identifications. Further optimization of the ELISA by decreasing matrix effect/noise from non‐immune plasma or optimization of the dye‐antibody ratio would be necessary to allow discrimination between low and high titer CCP. There are some additional limitations to be highlighted. First, only a relatively small number of samples (n = 74) from convalescent plasma donors have been tested so far. Second, the proposed ELISA set‐up in this study is based on the competition between neutralizing antibodies present in CCP and a commercially available mouse anti‐RBD antibody. Consequently, if blocking of the RBD–ACE2 interaction is mediated by neutralizing antibodies targeting epitopes remote from the primary mouse mAb, this might not be detected. Finally, because plasma of convalescent donors is a polyclonal mixture of antibodies, the concentration level and affinity of neutralizing antibodies targeting this specific epitope will influence the diagnostic sensitivity of our assay.
However, the fact that the competition ELISA assay is based on the competition for just one (mAb) epitope and not the entire RBD–ACE2 interface is of particular interest, as it nonetheless discriminates CCP from non‐immune plasma with a sensitivity of 61% compared with 81% for the inhibition ELISA. This implies that the mouse mAb binds to a pivotal antigenic sequence also recognized by the humoral system of many humans exposed to SARS‐CoV‐2.
Like all viruses, evolutionary pressure causes mutations, and for RBD, at least 44 actual amino acid changes have been identified. 24 Like any other diagnostic test based on the original Wuhan strain, competition for binding to the original RBD sequence is only useful when the donors have been infected with strains (very) similar to the Wuhan sequence. Mutations in RBD that significantly alter the protein's tertiary structure will inevitably evoke an altered panel of antibodies in both humans and mice. This therefore may affect assay performance.
However, as a proof of principle, this type of assay may be of benefit for screening CCP that specifically targets mutant SARS‐CoV‐2. Monoclonal neutralizing mAbs that have an epitope specifically recognizing RBD mutations close to or in the actual ACE2 interface can be exploited for targeted CCP screening. The N501Y mutation 17 for instance adds an aromatic core to the interface and increases the interaction force with the ACE2 receptor. 25 , 26 A competition ELISA with a mAb against an epitope that harbors this sequence will specifically select for CCP containing neutralizing antibodies that will inhibit mutants carrying N501Y. The proposed ELISA design may be used for these specific cases.
In conclusion, a competition ELISA can contribute to (1) the elucidation of the potential mechanism of neutralizing SARS‐CoV‐2 antibodies by epitope mapping 27 , 28 and (2) the selection of CCPs with high titers of neutralizing antibodies targeting a mutated strain with increased viral infectivity.
CONFLICT OF INTEREST
The authors have disclosed no conflicts of interest.
Supporting information
Appendix S1. Supporting Information.
ACKNOWLEDGMENTS
We thank all donors who have voluntarily donated (convalescent) plasma. We thank Belgian Red Cross Flanders employees who have participated in recruitment, plasma collection, component manufacturing, CCP (sample) distribution, and quality assessment during the pandemic. We thank Prof. Piet Maes for performing and analyzing PRNT data. We thank Gaia Mori, Prof. Dr. Pierre Tiberghien, and Catherine Hartmann for the expert coordination of the SUPPORT‐E project. Figure 1A–C was created in BioRender.com.
Wouters E, Verbrugghe C, Devloo R, Debruyne I, De Clippel D, Van Heddegem L, et al. A novel competition ELISA for the rapid quantification of SARS‐CoV‐2 neutralizing antibodies in convalescent plasma. Transfusion. 2021;61:2981–2990. 10.1111/trf.16652
Abbreviations: ACE2, angiotensin converting enzyme‐2; CCP, COVID‐19 convalescent plasma; COVID‐19, Coronavirus disease; CMIA, chemiluminescent microparticle immunoassay; ELISA, enzyme‐linked immunosorbent assay; mAb, monoclonal antibody; PRNT50, plaque reduction neutralization test; RBD, receptor binding domain.
Funding information Foundation for Scientific Research of the Belgian Red Cross‐Flanders; Horizon 2020 Framework Programme, Grant/Award Number: 101015756
REFERENCES
- 1.Coronavirus/COVID‐19 worldwide cases live data & statistics. 2020. Available from: https://covidstatistics.org/
- 2. Betrains A, Godinas L, Woei‐A‐Jin FJSH, Rosseels W, Van Herck Y, Lorent N, et al. Convalescent plasma treatment of persistent severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) infection in patients with lymphoma with impaired humoral immunity and lack of neutralising antibodies. Br J Haematol. 2021;192:1100–5. [DOI] [PubMed] [Google Scholar]
- 3. Hueso T, Pouderoux C, Péré H, Beaumont A‐L, Raillon L‐A, Ader F, et al. Convalescent plasma therapy for B‐cell‐depleted patients with protracted COVID‐19. Blood. 2020;136:2290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. London J, Boutboul D, Lacombe K, Pirenne F, Heym B, Zeller V, et al. Severe COVID‐19 in patients with B cell alymphocytosis and response to convalescent plasma therapy. J Clin Immunol. 2021;41:356–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Joyner MJ, Carter RE, Senefeld JW, Klassen SA, Mills JR, Johnson PW, et al. Convalescent plasma antibody levels and the risk of death from Covid‐19. N Engl J Med. 2021;18:1015–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Klassen SA, Senefeld JW, Johnson PW, Carter RE, Wiggins CC, Shoham S, et al. The effect of convalescent plasma therapy on COVID‐19 patient mortality: systematic review and meta‐analysis. Mayo Clin Proc. 2021;96(5):1262–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. U.S. Food and Drug Administration . Revised EUA for convalescent plasma. 2021. Available from: https://www.fda.gov/media/141477/download
- 8. Schlickeiser S, Schwarz T, Steiner S, Wittke K, Al Besher N, Meyer O, et al. Disease severity, fever, age, and sex correlate with SARS‐CoV‐2 neutralizing antibody responses. Front Immunol. 2020;11:628971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee WT, Girardin RC, Dupuis AP, Kulas KE, Payne AF, Wong SJ, et al. Neutralizing antibody responses in COVID‐19 convalescent sera. J Infect Dis. 2021;223:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cervia C, Nilsson J, Zurbuchen Y, Valaperti A, Schreiner J, Wolfensberger A, et al. Systemic and mucosal antibody responses specific to SARS‐CoV‐2 during mild versus severe COVID‐19. J Allergy Clin Immunol. 2021;147:545–57. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wouters E, Steenhuis M, Schrezenmeier H, Tiberghien P, Harvala H, Feys HB, et al. Evaluation of SARS‐CoV‐2 antibody titers and potency for convalescent plasma donation: a brief commentary. Vox Sang. 2021;116:493–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harvala H, Robb ML, Watkins N, Ijaz S, Dicks S, Patel M, et al. Convalescent plasma therapy for the treatment of patients with COVID‐19: assessment of methods available for antibody detection and their correlation with neutralising antibody levels. Transfus Med. 2021;31:167–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abe KT, Li Z, Samson R, Samavarchi‐Tehrani P, Valcourt EJ, Wood H, et al. A simple protein‐based surrogate neutralization assay for SARS‐CoV‐2. JCI Insight. 2020;5:e142362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Steenhuis M, van Mierlo G, Derksen NI, Ooijevaar‐de Heer P, Kruithof S, Loeff FL, et al. Dynamics of antibodies to SARS‐CoV‐2 in convalescent plasma donors. Clin Transl Immunol. 2021;10:e1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sanchez‐Felipe L, Vercruysse T, Sharma S, Ma J, Lemmens V, Van Looveren D, et al. A single‐dose live‐attenuated YF17D‐vectored SARS‐CoV‐2 vaccine candidate. Nature. 2020;590:320–5. [DOI] [PubMed] [Google Scholar]
- 16. SUPPORTing high quality evaluation of COVID‐19 convalescent plasma throughout EUROPE. Available from: https://www.support-e.eu/
- 17. Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS‐CoV‐2 spike receptor‐binding domain bound to the ACE2 receptor. Nature. 2020;581:215–20. [DOI] [PubMed] [Google Scholar]
- 18. Bal A, Pozzetto B, Trabaud MA, Escuret V, Rabilloud M, Langlois‐Jacques C, et al. Evaluation of high‐throughput SARS‐CoV‐2 serological assays in a longitudinal cohort of patients with mild COVID‐19: clinical sensitivity, specificity and association with virus neutralization test. Clin Chem. 2021;29:742–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Okba NMA, Müller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, et al. Severe acute respiratory syndrome coronavirus 2‐specific antibody responses in coronavirus disease patients. Emerg Infect Dis. 2020;26:1478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chi X, Yan R, Zhang J, Zhang G, Zhang Y, Hao M, et al. A neutralizing human antibody binds to the N‐terminal domain of the Spike protein of SARS‐CoV‐2. Science. 2020;369:650–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cerutti G, Guo Y, Zhou T, Gorman J, Lee M, Rapp M, et al. Potent SARS‐CoV‐2 neutralizing antibodies directed against spike N‐terminal domain target a single supersite. Cell Host Microbe. 2021;29:819–833.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dejnirattisai W, Zhou D, Ginn HM, Duyvesteyn HME, Supasa P, Case JB, et al. The antigenic anatomy of SARS‐CoV‐2 receptor binding domain. Cell. 2021;15:2183–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goodhue Meyer E, Simmons G, Grebe E, Gannett M, Franz S, Darst O, et al. Selecting COVID‐19 convalescent plasma for neutralizing antibody potency using a high‐capacity SARS‐CoV‐2 antibody assay. Transfusion. 2021;61:1160–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guruprasad L. Human SARS CoV‐2 spike protein mutations. Proteins. 2021;89:569–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Santos JC, Passos GA. The high infectivity of SARS‐CoV‐2 B.1.1.7 is associated with increased interaction force between Spike‐ACE2 caused by the viral N501Y mutation. bioRxiv. 2021. 10.1101/2020.12.29.424708 [DOI]
- 26. Starr TN, Greaney AJ, Addetia A, Hannon WW, Choudhary MC, Dingens AS, et al. Prospective mapping of viral mutations that escape antibodies used to treat COVID‐19. Science. 2021;371:850–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li T, Han X, Wang Y, Gu C, Wang J, Hu C et al. A key linear epitope for a potent neutralizing antibody to SARS‐CoV‐2 S‐RBD. bioRxiv. 2020. 10.1101/2020.09.11.292631 [DOI]
- 28. Mor M, Werbner M, Alter J, Safra M, Chomsky E, Lee JC, et al. Multi‐clonal SARS‐CoV‐2 neutralization by antibodies isolated from severe COVID‐19 convalescent donors. PLoS Pathog. 2021;17:e1009165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supporting Information.


