Abstract
We aimed to create the prediction model of in‐hospital mortality using machine learning methods for patients with coronavirus disease 2019 (COVID‐19) treated with steroid and remdesivir. We reviewed 1571 hospitalized patients with laboratory confirmed COVID‐19 from the Mount Sinai Health System treated with both steroids and remdesivir. The important variables associated with in‐hospital mortality were identified using LASSO (least absolute shrinkage and selection operator) and SHAP (SHapley Additive exPlanations) through the light gradient boosting model (GBM). The data before February 17th, 2021 (N = 769) was randomly split into training and testing datasets; 80% versus 20%, respectively. Light GBM models were created with train data and area under the curves (AUCs) were calculated. Additionally, we calculated AUC with the data between February 17th, 2021 and March 30th, 2021 (N = 802). Of the 1571 patients admitted due to COVID‐19, 331 (21.1%) died during hospitalization. Through LASSO and SHAP, we selected six important variables; age, hypertension, oxygen saturation, blood urea nitrogen, intensive care unit admission, and endotracheal intubation. AUCs using training and testing datasets derived from the data before February 17th, 2021 were 0.871/0.911. Additionally, the light GBM model has high predictability for the latest data (AUC: 0.881) (https://risk-model.herokuapp.com/covid). A high‐value prediction model was created to estimate in‐hospital mortality for COVID‐19 patients treated with steroid and remdesivir.
Keywords: COVID‐19, machine learning, mortality, New York, remdesivir, steroid
HIGHLIGHTS
-
•
Through LASSO and SHAP techniques, we selected six important variables associated with death of patients infected with COVID‐19: age, hypertension, oxygen saturation, blood urea nitrogen, intensive care unit admission, and endotracheal intubation.
-
•
Area under the curves (AUCs) using training and testing datasets derived from the data before February 17th, 2021 were 0.871/0.911.
-
•
Additionally, the light gradient boosting model model has high predictability for the latest data (AUC: 0.881).
WHAT IS NEW?
As steroids and remdesivir are the standard treatment of moderate or severe COVID patients as of April 17th 2021, a prediction model among patients treated with steroid and remdesivir is warranted.
WHAT DOES THIS ADD TO WHAT IS ALREADY KNOWN?
High‐value prediction model was created to estimate in‐hospital mortality for COVID‐19 patients treated with steroid and remdesivir.
WHAT IS THE IMPLICATION; WHAT SHOULD CHANGE NOW?
If patients are going to be treated with steroid and remdesivir, our prediction model would be useful.
1. INTRODUCTION
Coronavirus disease 2019 (COVID‐19) caused by a novel coronavirus, severe acute respiratory syndrome coronavirus 2, has spread all around the world since the first reported case in December 2019. 1 The World Health Organization declared COVID‐19 to be a pandemic on March 11, 2020 and as of April 22nd, New York City has become the epicenter. 2 , 3 , 4 On April 17th, 2021, the number of deaths due to the COVID‐19 pandemic has almost exceeded 3.0 million and the number of COVID‐19 cases reached 140 millions globally 2 ; 31 millions of which are from the United States alone.
There are several prediction models to estimate the risk of in‐hospital death for patients with COVID‐19, however, prediction models with light gradient boosting model (GBM) are scarce. 5 , 6 , 7 , 8 , 9 Light GBM is considered to reduce calculation time and it might be suitable for creation of a prediction calculator on the website. It also allows missing values for prediction, which is more advantageous than the conventional logistic regression model. Additionally, as steroids and remdesivir are the standard treatments of moderate or severe COVID patients as of April 17th 2021, 10 , 11 , 12 a prediction model among patients treated with both steroid and remdesivir is warranted. Moreover, the racial difference in death due to COVID‐19 remains uncertain although racial disparities were observed in infection rates, 13 , 14 , 15 , 16 it should be investigated whether including it into the risk model predicting mortality.
We aimed to build the prediction model for in‐hospital mortality among patients infected with COVID‐19 treated and treated with both steroid and Remdesivir in a diverse population of New York City. We also aimed to create the calculator on the website so that frontline providers can use this prediction model to identify high risk hospitalized COVID‐19 patients treated with steroid and remdesivir.
2. METHODS
This retrospective study was conducted by review of the medical records of 9565 hospitalized patients between March 1st, 2020 and March 31st, 2021 with laboratory confirmed COVID‐19 in the Mount Sinai Health system. 17 , 18 , 19 , 20 , 21 , 22 , 23 Identification of COVID‐19 required a nasopharyngeal swab, which was tested using a polymerase chain reaction.
Patients' electronic medical records were reviewed and demographics, comorbidities, vital signs at admission, laboratory data at admission, and clinical outcomes were extracted. Among 9565 patients, 1571 patients treated with both remdesivir and steroid were selected. Additionally, patients were stratified into groups, those who were discharged by February 17th, 2021 (N = 769) and those who were discharged between February 18th, 2021 and March 31th, 2021 (N = 802). Steroids were used only for moderate or severe COVID‐19 patients. 12 , 18 Only patients treated with systemic steroids (betamethasone, dexamethasone, hydrocortisone, prednisone, prednisolone, and methylprednisolone) regardless of dosage, were included.
Differences in baseline characteristics between both study periods were evaluated using the χ 2 test for categorical variables. Continuous variables are presented as means ± SD or medians [interquartile range] depending on what is appropriate for the data distribution, and categorical variables were expressed as percentages. All vital signs were recorded at time of admission. The primary outcome of interest was in‐hospital mortality. Acute kidney injury was defined as any increase of creatinine by more than 0.3 mg/dl or to more than 1.5 times baseline. 24
Two approaches were used to predict in‐hospital death for patients infected with COVID‐19: machine learning model and logistic regression model. With the machine learning model, we initially identified the important variables associated with in‐hospital death using LASSO (least absolute shrinkage and selection operator). LASSO selects variables by shrinking the coefficients of less‐important variables from logistic regression to zero. 25 Age, race, sex, asthma, chronic obstructive pulmonary disease, hypertension, obstructive sleep apnea, obesity, diabetes mellitus, chronic kidney disease, human deficiency virus, cancer, atrial fibrillation, heart failure, coronary artery disease, chronic viral hepatitis, alcoholic/nonalcoholic liver disease, peripheral vascular disease, vitals at admission, C‐reactive protein, d‐dimer, white blood cell count, hemoglobin, creatinine, blood urea nitrogen, estimated glomerular filtration rate (eGFR), intensive care unit (ICU) admission, and endotracheal intubation were included into the LASSO model. 4 , 26 The Modification of Diet in Renal Disease equation was used to estimate eGFR. 27 In addition, we constructed the SHAP (SHapley Additive exPlanations) approach to select the important variables with the light GBM using the variables selected by LASSO. This approach explains the models at the level of individual patients based on the sum of the numeric computed credit (SHAP) values of each feature. 28 , 29
After selection of important variables, the data before February 17th, 2021 (N = 769) was randomly split into training and testing datasets; 80% and 20%, respectively. Then, light GBM and a logistic regression model using the stratified K‐fold cross‐validation method were applied to the train data (K = 5). In comparison to the logistic regression model, Light GBM used “NaN” to represent missing values and were dealt separately than zero, as missing values were interpreted as containing information. 28 The hyper‐parameter optimization was performed using an implementation called “Optuna” for light GBM. For logistic regression, we used a grid search strategy to identify the best tuning hyperparameters. 30 We also used Standard Scaler to improve predictability. 31 We also performed an imputation for missing data using the library of IterativeImputer in Python for a logistic regression model. We used area under the curve (AUC) to evaluate the different models. Furthermore, we validated the model into the data between February 18th, 2021 and March 30th, 2021 (N = 802). Finally, we created a web‐based calculator to predict in‐hospital mortality due to COVID‐19.
All statistical calculations and analyses were performed on R (version 3.6.2, R Foundation for Statistical Computing, Vienna, Austria) and Python 3.7 (Python Software Foundation Delaware, USA). All p values <0.05 considered statistically significant.
This study was approved by the institutional review boards (#2000495) and conducted in accordance with the principles of the Declaration of Helsinki. The waiver of patients' informed consent was also approved by the institutional review boards.
3. RESULTS
Of the 1571 patients admitted due to COVID‐19, 331 (21.1%) died during hospitalization. Baseline characteristics across two study periods are reported in Table 1, demonstrating mostly comparable patients' characteristics except sex and race.
Table 1.
Baseline characteristics of patients admitted with COVID‐19 and treated with steroid and remdesivir stratified by discharge date
| Patients who were discharged before February 17th, 2021, N = 769 | Patients who were discharged between February 18th, 2021 and March 30th, 2021, N = 802 | p value | |
|---|---|---|---|
| Age, (mean, SD), year | 66.3 (15.6) | 65.8 (16.0) | 0.57 |
| Male, n (%) | 462 (60.1) | 431 (53.7) | 0.013 |
| Race, n (%) | 324 (42.1) | 212 (26.4) | <0.001 |
| White | 101 (13.1) | 157 (19.6) | |
| African American | 138 (17.9) | 177 (22.1) | |
| Hispanic | 50 (6.5) | 85 (10.6) | |
| Asian | 156 (20.3) | 171 (21.3) | |
| Other | |||
| Comorbidities | |||
| Asthma, n (%) | 31 (4.0) | 50 (6.2) | 0.063 |
| COPD, n (%) | 38 (4.9) | 35 (4.4) | 0.67 |
| Hypertension, n (%) | 239 (31.1) | 273 (34.0) | 0.23 |
| Diabetes mellitus, n (%) | 153 (19.9) | 181 (22.6) | 0.22 |
| Chronic kidney disease, n (%) | 28 (3.6) | 39 (4.9) | 0.28 |
| Obstructive sleep apnea, n (%) | 28 (3.6) | 13 (1.6) | 0.019 |
| Obesity, n (%) | 60 (7.8) | 84 (10.5) | 0.081 |
| HIV, n (%) | 28 (3.6) | 13 (1.6) | 0.019 |
| Cancer, n (%) | 69 (9.0) | 61 (7.6) | 0.37 |
| Atrial fibrillation, n (%) | 44 (5.7) | 63 (7.9) | 0.12 |
| Heart failure, n (%) | 36 (4.7) | 43 (5.4) | 0.62 |
| Coronary artery disease, n (%) | 88 (11.4) | 91 (11.3) | 1.00 |
| Peripheral vascular disease, n (%) | 30 (3.9) | 33 (4.1) | 0.93 |
| Alcoholic/nonalcoholic liver disease, n (%) | 13 (1.7) | 13 (1.6) | 1.00 |
| Vitals | |||
| Temperature (mean, SD) | 38.1 [37.4, 39.0] | 37.8 [37.3, 38.7] | <0.001 |
| Heart rate | 93.0 [82.0, 106.0] | 95.0 [84.0, 107.0] | 0.14 |
| (mean, SD) | |||
| Respiratory rate (mean, SD) | 20.0 [18.0, 22.0] | 20.0 [18.0, 22.0] | 0.009 |
| Systolic blood pressure (mean, SD) | 131.0 [118.0, 146.0] | 129.0 [116.0, 145.0] | 0.035 |
| Diastolic blood | 74.0 [66.0, 84.0] | 75.0 [66.0, 83.8] | 0.90 |
| Pressure (mean, SD) | |||
| O2 saturation (mean, SD) | 88.0 [81.0, 91.0] | 88.0 [80.0, 91.0] | 0.92 |
| Laboratory data | |||
| White blood cell, K/μl (mean, SD) | 7.0 [5.2, 9.7] | 6.3 [4.8, 8.5] | <0.001 |
| Hemoglobin, g/dl (mean, SD) | 13.4 [12.2, 14.6] | 13.6 [12.3, 14.7] | 0.180 |
| Blood urea nitrogen, mg/dl (median [IQR]) | 17.0 [12.0, 24.0] | 17.0 [12.0, 25.0] | 0.82 |
| Creatinine, mg/dl (median [IQR]) | 0.90 [0.74, 1.19] | 0.95 [0.76, 1.23] | 0.074 |
| Lactate dehydrogenase, U/L (median [IQR]) | 384.5 [293.0, 499.0] | 392.5 [299.5, 530.0] | 0.23 |
| C‐reactive protein, mg/L (median [IQR]) | 88.2 [50.2, 148.7] | 85.1 [46.4, 142.2] | 0.30 |
| D‐Dimer, μg/ml (median [IQR]) | 1.02 [0.65,1.81] | 1.13 [0.67, 1.97] | 0.029 |
Abbreviations: APTT, activated partial thromboplastin time; COPD, chronic obstructive pulmonary disease; COVID‐19, coronavirus disease 2019; HIV, human immunodeficiency virus; IQR, interquartile range.
Treatments and outcomes are shown in Table 2. Although the rates of therapeutic versus prophylactic anticoagulation, Tocilizumab, convalescent plasma were significantly different between the study periods. ICU admission, endotracheal intubation, acute kidney injury and in‐hospital mortality were not significantly different (Table 2).
Table 2.
In‐hospital treatment and outcomes
| Patients who was discharged before February 17th, 2021, N = 769 | Patients who was discharged between February 18th, 2021 and March 30th, 2021, N = 802 | p value | |
|---|---|---|---|
| Therapeutic anticoagulation, n (%) | 329 (42.8) | 177 (22.1) | <0.001 |
| Prophylactic anticoagulation, n (%) | 436 (56.7) | 616 (76.8) | <0.001 |
| Use of Tocilizumab, n (%) | 22 (2.9) | 51 (6.4) | 0.002 |
| Convalescent plasma, n (%) | 407 (52.9) | 116 (14.5) | <0.001 |
| ICU admission, n (%) | 229 (29.8) | 211 (26.3) | 0.14 |
| Endotracheal intubation, n (%) | 126 (16.4) | 119 (14.8) | 0.44 |
| Acute kidney injury, n (%) | 152 (19.8) | 140 (17.5) | 0.27 |
| In‐hospital mortality, n (%) | 156 (20.3) | 175 (21.8) | 0.49 |
Abbreviation: ICU, intensive care unit.
LASSO method showed the following 17 variables as important features to predict in‐hospital mortality; age, race, hypertension, coronary artery disease, heart rate, respiratory rate, systolic blood pressure, diastolic blood pressure, oxygen saturation, C‐reactive protein, d‐dimer, white blood cell count, hemoglobin, blood urea nitrogen, eGFR, ICU admission and endotracheal intubation. Then, SHAP showed six important variables; age, hypertension, oxygen saturation, blood urea nitrogen, ICU admission and endotracheal intubation (Figure 1). We created the final model with six variables. AUCs using training and testing datasets derived from the data before February 17th, 2021 were 0.871/0.911 with light GBM, and 0.952/0.918 with the logistic regression model.
Figure 1.
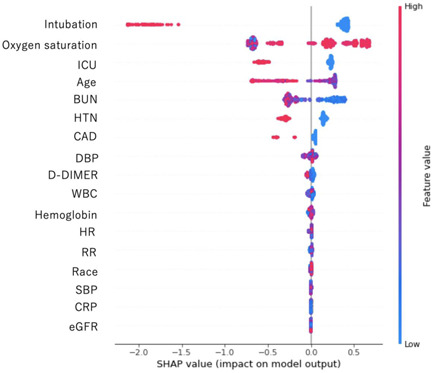
SHAP model to estimate important variables with the light gradient boosting model using the 17 variables selected by LASSO. The features are sorted in descending order by Shapley values. BUN, blood urea nitrogen; CAD, coronary artery disease; CRP, C‐reactive protein; DBP, diastolic blood pressure; eGFR, estimated glomerular filtration rate; HR, heart rate; HTN, hypertension; ICU, intensive care unit; LASSO, least absolute shrinkage and selection operator; RR, respiratory rate; SBP, systolic blood pressure; SHAP, SHapley Additive exPlanations; WBC, white blood cell count
Additionally, the light GBM model has high predictability for the data derived from February 18th, 2021 and March 30th, 2021 as well as the logistic regression model (AUC; light GBM: 0.873, logistic regression: 0.882, respectively). Figure 2 shows the calibration plots with the light GBM using the six variables.
Figure 2.
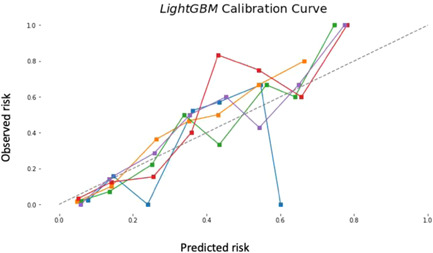
Calibration plot using the light GBM with six variables; age, hypertension, oxygen saturation, blood urea nitrogen, ICU admission and endotracheal intubation. ICU, intensive care unit; Light GBM, light gradient boosting model
The web calculator was created using light GBM as it allows missing values and both light GBM and the logistic regression models are comparable with high prediction. It can be used to calculate the risk of in‐hospital death for patients hospitalized with COVID‐19 (https://risk-model.herokuapp.com/covid). Two examples of using this calculator are shown in Figure 3A,B. Using this calculator, we could estimate the risk of death.
Figure 3.
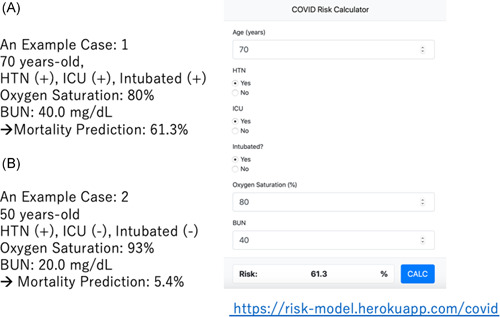
Examples of mortality prediction for patients with COVID‐19. COVID‐19, coronavirus disease 2019
4. DISCUSSION
The salient of our findings are the followings: (1) light GBM showed high AUC to predict in‐hospital mortality, which was comparable to the logistic regression model; (2) Calculator on the website using a light GBM model which allows missing values is useful to predict in‐hospital mortality.
As of April 17th, 2021, steroids and remdesivir are the standard treatment of COVID patients 10 , 11 for patients with moderate or severe COVID‐19 (oxygen saturation level <94%). As the prediction model among patients treated with steroid and remdesivir is needed and we created the risk model among those patients. Using LASSO method, age, race, hypertension, coronary artery disease, heart rate, respiratory rate, systolic blood pressure, diastolic blood pressure, oxygen saturation, C‐reactive protein, d‐dimer, white blood cell count, hemoglobin, blood urea nitrogen, eGFR, ICU admission and endotracheal intubation were selected as important features which is compatible with the previous studies. 16 , 26 , 32 , 33 , 34 Additionally, we adjusted the number of variables with SHAP to enhance convenience of the risk model, with six variables of age, hypertension, blood urea nitrogen, oxygen saturation, ICU admission, and endotracheal intubation. Our risk model is valuable to predict the risk of death for moderate or severe COVID‐19 patients treated with steroid and remdesivir. We demonstrated blood urea nitrogen as important variables rather than C‐reactive protein, d‐dimer using SHAP. 26 , 35
Another strength of this study is the website calculator, which will enable frontline providers to identify high‐risk patients immediately at the time of admission for patients requiring steroid and remdesivir. We consider risk prediction model is really useful especially when frontline providers can utilize it. It is also valuable as we could calculate the risk of death even with missing values since light GBM allows missing values to construct a model.
Racial difference in death due to COVID‐19 remains uncertain although racial disparities were observed in infection rates. 13 , 14 , 15 , 16 LASSO using our data showed that race is an important feature, however, SHAP did not reveal that we could predict in‐hospital mortality without the information of race. Ase COVID‐19 occurred among diverse patients population in New York City, 3 , 4 , 36 , 37 our model would be useful globally as COVID‐19 affects all over the world, however, more extensive validation using international data is necessary.
Moreover, gender or comorbidities were less prominent in our model, especially selected by SHAP. Although gender or comorbidities were important variables that affect mortality, 38 , 39 these variables were less important compared to six variables to predict in‐hospital mortality in our model.
Our study is not without limitations. This is a retrospective observational study and not the study to collect all variables prospectively. Although we created the risk model from our diverse cohort, our risk model needs to be validated in other populations. However, our risk model could apply to the latest data. Moreover, we did not have the information on admission data. We only have the data of admission date before February 17th, 2021 or after. However, the idea behind selecting patients with steroids and remdesivir were to select moderate or severe COVID‐19 patients and to exclude relatively the early phase of a pandemic. The mortality rate of the initial phase was relatively high compare to the second phase of COVID‐19. 40 In addition, selecting patients with steroids and remdesivir allowed us to investigate patients who were moderate or severe in any timing of hospitalization, as we have the data of vital signs at the time of admission only.
In addition, our data could be applied to only COVID‐19 patients who received steroids and remdesivir, basically for moderate or severe patients. Finally, the time between death and ICU admission or endotracheal intubation might affect the prediction model, however, we do not have that information.
In conclusion, a high‐performance prediction model was created with light GBM to estimate in‐hospital mortality for COVID‐19 patients treated with both steroid and remdesivir. Our model is useful in estimating patients' predictive mortality.
AUTHOR CONTRIBUTIONS
Toshiki Kuno, Mai Takahashi, and Natalia N. Egorova: data curation, and had full access to all the data in the study and take responsibility for the integrity of the data and accuracy of the data analysis. Toshiki Kuno: study concept and design, drafting of the manuscript. Toshiki Kuno and Mai Takahashi: statistical analysis. Yuki Sahashi, Shinpei Kawahito, Masao Iwagami, and Natalia N. Egorova: administrative, technical, or material support. Natalia N. Egorova: study supervision. All authors: acquisition, analysis, or interpretation of data; critical revision of the manuscript for important intellectual content.
Kuno T, Sahashi Y, Kawahito S, Takahashi M, Iwagami M, Egorova NN. Prediction of in‐hospital mortality with machine learning for COVID‐19 patients treated with steroid and remdesivir. J Med Virol. 2022;94:958‐964. 10.1002/jmv.27393
DATA AVAILABILITY STATEMENT
Research data are not shared.
REFERENCES
- 1. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Coronavirus COVID‐19 Global Cases by the Center for Systems Science and Enginerring at Johns Hopkins University. https://coronavirus.jhu.edu/map.html
- 3. Maeda T, Obata R, Rizk D, Kuno T. Cardiac injury and outcomes of patients with COVID‐19 in New York City. Heart Lung Circ. 2020;30:848‐853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maeda T, Obata R, Rizk DD, Kuno T. The association of interleukin‐6 value, interleukin inhibitors, and outcomes of patients with COVID‐19 in New York City. J Med Virol. 2020;93:463‐471. [DOI] [PubMed] [Google Scholar]
- 5. Vaid A, Somani S, Russak AJ, et al. Machine learning to predict mortality and critical events in a cohort of patients with COVID‐19 in New York City: model development and validation. J Med Internet Res. 2020;22(11):e24018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yadaw AS, Li YC, Bose S, Iyengar R, Bunyavanich S, Pandey G. Clinical features of COVID‐19 mortality: development and validation of a clinical prediction model. Lancet Digit Health. 2020;2(10):e516‐e525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Abdulaal A, Patel A, Charani E, Denny S, Mughal N, Moore L. Prognostic modeling of COVID‐19 using artificial intelligence in the United Kingdom: model development and validation. J Med Internet Res. 2020;22(8):e20259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. An C, Lim H, Kim DW, Chang JH, Choi YJ, Kim SW. Machine learning prediction for mortality of patients diagnosed with COVID‐19: a nationwide Korean cohort study. Sci Rep. 2020;10(1):18716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu C, Liu Z, Jiang Y, et al. Early prediction of mortality risk among patients with severe COVID‐19, using machine learning. Int J Epidemiol. 2021;49(6):1918‐1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yokoyama Y, Briasoulis A, Takagi H, Kuno T. Effect of remdesivir on patients with COVID‐19: a network meta‐analysis of randomized control trials. Virus Res. 2020;288:198137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. WHO Rapid Evidence Appraisal for COVID‐ Therapies (REACT) Working G, Sterne J, Murthy S, et al, Group WHOREAfC‐TW . Association between administration of systemic corticosteroids and mortality among critically Ill patients with COVID‐19: a meta‐analysis. JAMA. 2020;324(13):1330‐1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Group RC, Horby P, Lim WS, et al. Dexamethasone in hospitalized patients with Covid‐19. N Engl J Med. 2021;384(8):693‐704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Perkin MR, Heap S, Crerar‐Gilbert A, et al. Deaths in people from Black, Asian and minority ethnic communities from both COVID‐19 and non‐COVID causes in the first weeks of the pandemic in London: a hospital case note review. BMJ Open. 2020;10(10):e040638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vahidy FS, Nicolas JC, Meeks JR, et al. Racial and ethnic disparities in SARS‐CoV‐2 pandemic: analysis of a COVID‐19 observational registry for a diverse US metropolitan population. BMJ Open. 2020;10(8):e039849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ogedegbe G, Ravenell J, Adhikari S, et al. Assessment of racial/ethnic disparities in hospitalization and mortality in patients with COVID‐19 in New York City. JAMA Netw Open. 2020;3(12):e2026881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rodriguez F, Solomon N, de Lemos JA, et al. Racial and ethnic differences in presentation and outcomes for patients hospitalized with COVID‐19: findings from the american heart association's COVID‐19 cardiovascular disease registry. Circulation. 2020;143:2332‐2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Takahashi M, Egorova NN, Kuno T. COVID‐19 and influenza testing in New York City. J Med Virol. 2021;93(2):698‐701. [DOI] [PubMed] [Google Scholar]
- 18. Kuno T, Miyamoto Y, Iwagami M, Ishimaru M, Takahashi M, Egorova NN. The association of remdesivir and in‐hospital outcomes for COVID‐19 patients treated with steroids. J Antimicrob Chemother. 2021;76:2690‐2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kuno T, So M, Miyamoto Y, Iwagami M, Takahashi M, Egorova NN. The association of COVID‐19 antibody with in‐hospital outcomes in COVID‐19 infected patients. J Med Virol. 2021;93(12):6841–6844. 10.1002/jmv.27260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kuno T, So M, Takahashi M, Egorova NN. U shape association of hemoglobin level with in‐hospital mortality for COVID‐19 patients. J Thromb Thrombolysis. 2021;1–5. 10.1007/s11239-021-02516-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. So M, Kabata H, Takahashi M, Egorova NN, Kuno T. The association of inhaled corticosteroid before admission and survival of patients with COVID‐19. J Aerosol Med Pulm Drug Deliv. 2021;34(4):265‐267. [DOI] [PubMed] [Google Scholar]
- 22. Kuno T, Takahashi M, Egorova NN. The association between convalescent plasma treatment and survival of patients with COVID‐19. J Gen Intern Med. 2021;36(8):2528‐2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. So M, Steiger DJ, Takahashi M, Egorova NN, Kuno T. The characteristics and outcomes of critically Ill patients with COVID‐19 who received systemic thrombolysis for presumed pulmonary embolism: an observational study. J Thromb Thrombolysis. 2021;1–7. 10.1007/s11239-021-02477-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chandiramani R, Cao D, Nicolas J, Mehran R. Contrast‐induced acute kidney injury. Cardiovasc Interv Ther. 2020;35(3):209‐17. [DOI] [PubMed] [Google Scholar]
- 25. Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med. 1997;16(4):385‐395. [DOI] [PubMed] [Google Scholar]
- 26. Petrilli CM, Jones SA, Yang J, et al. Factors associated with hospital admission and critical illness among 5279 people with coronavirus disease 2019 in New York City: prospective cohort study. BMJ. 2020;369:m1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Acosta‐Ochoa I, Bustamante‐Munguira J, Mendiluce‐Herrero A, Bustamante‐Bustamante J, Coca‐Rojo A. Impact on outcomes across KDIGO‐2012 AKI criteria according to baseline renal function. J Clin Med. 2019;8:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zein JG, Wu CP, Attaway AH, Zhang P, Nazha A. Novel machine learning can predict acute asthma exacerbation. Chest. 2021;159:1747‐1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lundberg SM, Erion G, Chen H, et al. From local explanations to global understanding with explainable AI for trees. Nat Mach Intell. 2020;2(1):56‐67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Osawa I, Goto T, Yamamoto Y, Tsugawa Y. Machine‐learning‐based prediction models for high‐need high‐cost patients using nationwide clinical and claims data. NPJ Digit Med. 2020;3(1):148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cao Y, Bass GA, Ahl R, et al. The statistical importance of P‐POSSUM scores for predicting mortality after emergency laparotomy in geriatric patients. BMC Med Inform Decis Mak. 2020;20(1):86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang L, Jin J, Luo W, Gan Y, Chen B, Li W. Risk factors for predicting mortality of COVID‐19 patients: a systematic review and meta‐analysis. PLoS One. 2020;15(11):e0243124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mesas AE, Cavero‐Redondo I, Álvarez‐Bueno C, et al. Predictors of in‐hospital COVID‐19 mortality: a comprehensive systematic review and meta‐analysis exploring differences by age, sex and health conditions. PLoS One. 2020;15(11):e0241742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Takagi H, Kuno T, Yokoyama Y, et al. Ethnicity/race and economics in COVID‐19: meta‐regression of data from counties in the New York metropolitan area. J Epidemiol Community Health. 2020;75:205‐206. [DOI] [PubMed] [Google Scholar]
- 35. Liang M, He M, Tang J, et al. Novel risk scoring system for predicting acute respiratory distress syndrome among hospitalized patients with coronavirus disease 2019 in Wuhan, China. BMC Infect Dis. 2020;20(1):960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Obata R, Maeda T, Do DR, Kuno T. Increased secondary infection in COVID‐19 patients treated with steroids in New York City. Jpn J Infect Dis. 2020;74:307‐315. [DOI] [PubMed] [Google Scholar]
- 37. Obata R, Maeda T, Rizk D, Kuno T. Palliative care team involvement in patients with COVID‐19 in New York City. Am J Hosp Palliat Care. 2020;37(10):869‐872. [DOI] [PubMed] [Google Scholar]
- 38. Ueyama H, Kuno T, Takagi H, et al. Gender difference is associated with severity of coronavirus disease 2019 infection: an insight from a meta‐analysis. Crit Care Explor. 2020;2(6):e0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kuno T, Takahashi M, Obata R, Maeda T. Cardiovascular comorbidities, cardiac injury, and prognosis of COVID‐19 in New York City. Am Heart J. 2020;226:24‐25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dorrucci M, Minelli G, Boros S, et al. Excess mortality in Italy during the COVID‐19 pandemic: assessing the differences between the first and the second wave, Year 2020. Front Public Health. 2021;9:669209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Research data are not shared.


