Abstract
We report a corona virus disease (COVID‐19) case with unprecedented viral complexity. In the first severe episode, two different severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) strains (superinfection) were identified within a week. Three months after discharge, the patient was readmitted and was infected in a nosocomial outbreak with a different strain, suffering a second milder COVID‐19 episode.
Keywords: COVID‐19, genomics, reinfection, SARS‐CoV‐2, superinfection
1. FIRST COVID‐19 EPISODE (E1)
A sixty‐five‐year‐old male with past medical history of hypertension was admitted in the emergency department on 6 February 2020 (Day 0–E1). He was diagnosed of hypertensive cerebellar haemorrhage with intraventricular and subarachnoid haemorrhage required decompressive craniectomy. The patient was moved postoperatively to the neurosurgery ward (Day 4). The condition of the patient deteriorated on Day 50: diminished oxygen saturation (93%), no consolidation on chest X‐ray, with lymphopenia of 800, IL‐6 of 4.6, CRP of 1.1, and D‐dimer of 800 ng/ml. On the same day, the patient´s severe acute respiratory syndrome coronavirus 2 (SARS‐COV2) reverse transcription‐polymerase chain reaction (RT‐PCR) (Ct value not available, Specimen 1) result was positive; he was moved to a corona virus disease (COVID‐19) ward in a different building and given lopinavir/ritonavir and hydroxychloroquine for 10 days. On Day 69, a second SARS‐CoV‐2 RT‐PCR assay was done with positive result (Ct 19; Specimen 2). The patient was returned to the neurosurgery ward, which had been turned into a COVID‐19 area. On Day 74, radiological worsening was noted in the left lung, coinciding with an inflammatory process: lymphocytopenia (0.7 × 103/μl), low platelet count (125 × 103/μl), increased DD (989 ng/ml), CRP of 4.4 mg/dl, ferritin of 778 μg/L, and IL6 of 13.6 pg/ml. Treatment with methylprednisolone was initiated, and a SARS‐CoV‐2 RT‐PCR (Specimen 3) was performed on the same day (Day 74) with positive result (Ct 25). SARS‐CoV‐2 RT‐PCR assay performed on Day 87 was negative, as well as subsequent tests on Days 108, 117, 147, 195, and 206. The patient was discharged on Day 207 to a long‐term facility due to persistent generalized muscle weakness.
2. SECOND COVID‐19 EPISODE (E2)
On December 6 (3 months after the previous discharge) (Day 304), the patient was admitted due to a fortuitous fall from a seated position. He was diagnosed of head trauma and subdural hematoma over left frontoparietal lobe. At admission, severe malnutrition and muscular hypotrophy were found. On Day 350, he developed fever, and SARS‐COV2 RT‐PCR result was positive (Ct 16; Specimen 4); no SARS‐COV2 IgG antibodies were detected (SARS‐CoV‐2 IgG II Quant Reagent Kit; Abbott, Chicago, USA). There were no signs of consolidations on chest X‐rays; the patient presented mild hypoxemia and completed a 5‐day course of dexamethasone (6 mg/day). By Day 364, respiratory syndrome was controlled and positive serology for SARS‐COV‐2 determined (IgG of > 40,000 UA/ml).
3. GENOMIC ANALYSIS
Two positive SARS‐CoV‐2 RT‐PCR from E1 (Specimens 2 and 3) and one positive specimen (Specimen 4) from E2 were available for whole genome sequencing (WGS). Whole genome amplification was done with an Artic_nCov‐2019_V3 panel of primers (Integrated DNA Technologies, Inc., Coralville, Iowa, USA) (artic.network/ncov‐2019). Libraries were prepared using the Nextera DNA Flex Library Preparation Kit (Illumina lnc., California, USA) and sequenced in pools on the Miseq system (Illumina Inc, California, USA). Sequences were deposited at GISAID (Specimen 1: EPI_ISL_1547368, Specimen 3: EPI_ISL_1547363, Specimen 2 not deposited due to heterozygous calls) and raw sequences for the three specimens in ENA (Project reference: PRJEB47864). An in‐house analysis pipeline was applied on the sequencing reads (https://github.com/pedroscampoy/covid_multianalysis). Briefly, the pipeline involves (1) removal of human reads with Kraken; (2) pre‐processing and quality assessment of FASTQ files using fastp v0.20.1 (arguments: –cut tail, –cut‐window‐size, –cut‐mean‐quality, –max_len1, –max_len2) and fastQC v0.11.9; (3) mapping with BWA v0.7.17 and variant calling using IVAR v1.2.3, using the Wuhan‐1 sequence (NC_045512.2) as reference; and (4) calibration of occasional low coverage positions using joint variant calling.
A B.1.258 lineage strain (Strain 1, Figure 1) was identified in E1 from the Specimen 2. When analyzing the data from the Specimen 3, many heterozygous calls were obtained (Table S1 and Figure S1), suggesting the presence of two SARS‐CoV‐2 strains. Cross‐contamination with a positive specimen from another patient was ruled out by confirming identical human genetic content (short tandem repeat human DNA profile analysis, Figure 2) in Specimens 2 and 3. Thus, the heterozygous calls indicated superinfection with a different strain (Strain 2; lineage B.1). Frequency determination of alternative alleles in the heterozygous calls allowed us to determine that superinfecting Strain 2 was over‐represented (59%–79% frequency, Table S1). Strains 1 and 2 sequences were analyzed along with 2249 SARS‐CoV‐2 sequences from specimens collected from COVID‐19 cases among the Madrid population throughout the pandemic. A tree with the results was created, and Strains 1 and 2 are presented on the branch corresponding to the strains circulating in the first COVID‐19 wave (before July 2020, Figure 1).
FIGURE 1.
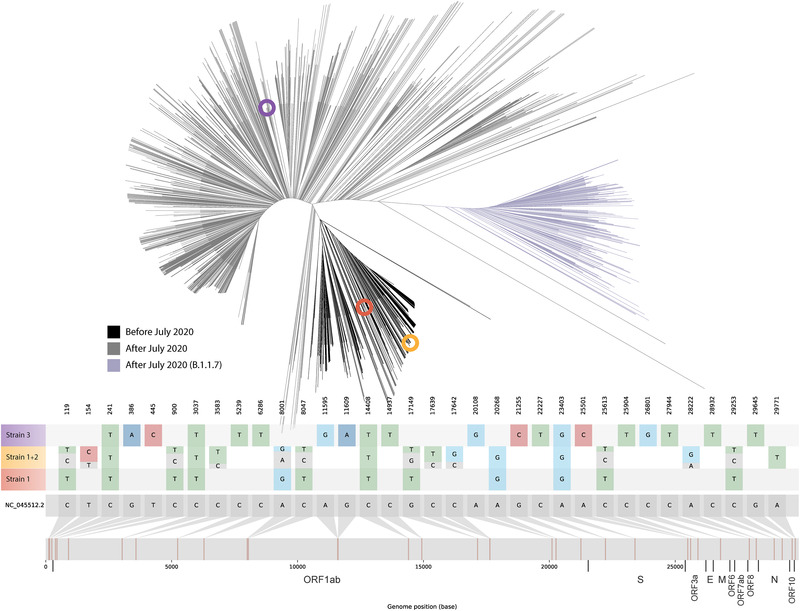
Upper panel: maximum likelihood unrooted tree with genetic distances of the three identified strains (highlighted as coloured circles within the phylogenetic tree; Strain 1 in orange, Strains 1 + 2 in ochre, and Strain 3 in purple) based on sequence alignment of 2249 positive severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) samples from cases among the Madrid population throughout the pandemic. Coloured branches are used for SARS‐CoV‐2 sequences from specimens collected during the first COVID‐19 wave (black) and after July 2020 (). The B.1.1.7 variant is shown in purple. Lower panel: location of single nucleotide polymorphisms along the SARS‐CoV‐2 reference genome for the three identified strains. Relative allele frequency of each strain (Strains 1 and 2; see also Table S1) in the superinfection event is indicated for the positions with heterozygous calls. Fasta files were deposited in GISAID (accession numbers strain 1 ‐ EPI_ISL_1547368 strain 2 ‐ EPI_ISL_1547369 and strain 3 ‐ EPI_ISL_1547363). FASTQ files were deposited at the European Nucleotide Archive (https://www.ebi.ac.uk; project reference: PRJEB47864)
FIGURE 2.
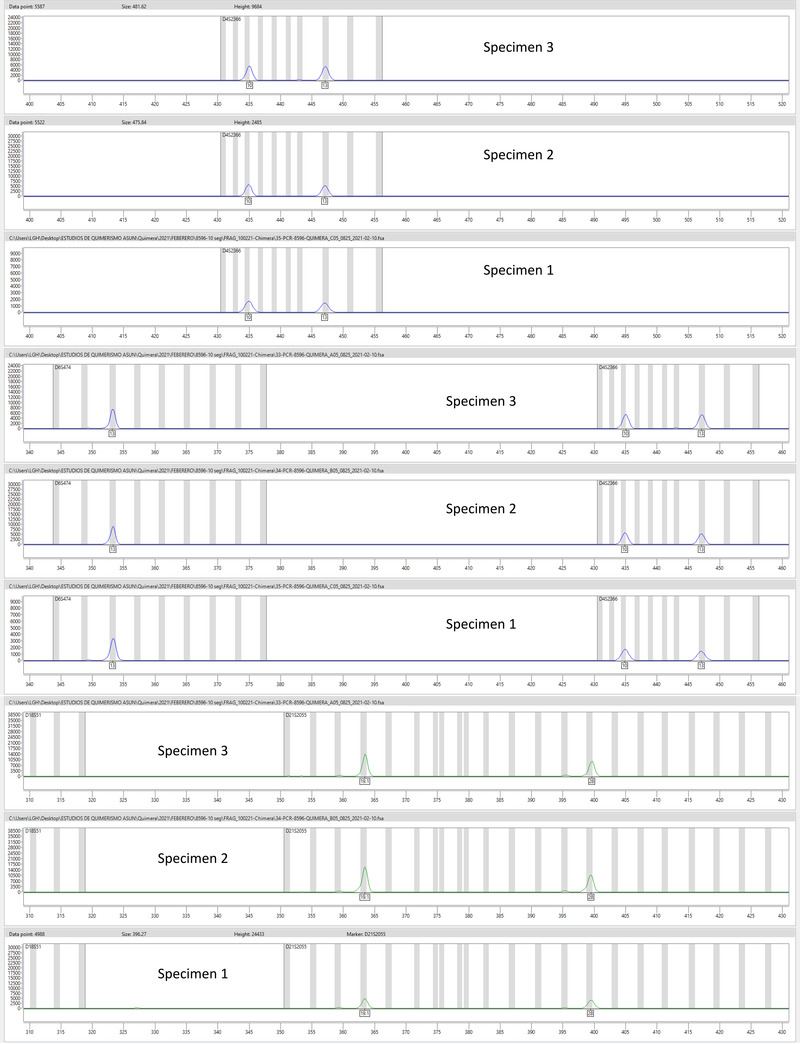
Short tandem repeat‐PCR results on the specimens used for the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) reverse transcription‐polymerase chain reaction (RT‐PCR) (also sequenced). Twelve non‐coding STR loci and the gender‐specific locus amelogenin were examined
It could be argued that our findings of two simultaneous SARS‐CoV‐2 strains in our patient might correspond to evolution of a pre‐existing strain. Different facts make this option highly unlikely. First, Strain 2 was detected in our patient just 5 days after the identification of Strain 1, which is a time lapse too short to allow the acquisition of such diversity (13 differential single nucleotide polymorphisms (SNPs) between them; Table 1). Moreover, if an evolutionary path existed between Strains 1 and 2, the evolved strain should conserve the SNPs already fixed in the parental strain and subsequently acquire new SNPs absent in the parental strain. This does not correspond to the SNPs distribution between Strains 1 and 2 (Strain 1 harbours seven SNPs not present in Strain 2 and another six SNPs are found in Strain 2 but not in Strain 1, Table 1). All these aspects together reinforce the superinfection with an unrelated strain as the most likely explanation and allow us to rule out an alternative evolutionary explanation.
A new strain (Strain 3; lineage B.1.177) was identified in E2 from Specimen 4, with 16 SNPs not shared with Strains 1 or 2, and without the 14 SNPs identified in Strains 1 and 2 (Figure 1 and Table 1). This indicated that COVID‐19 E2 was a reinfection. Strain 3 sequence was positioned in the phylogenetic tree among the sequences from strains circulating after July 2020, ruling out its circulation during the first episode of our case. Short tandem repeat human DNA analysis confirmed that specimens from the two sequential COVID‐19 episodes were from the same individual (Figure 2).
TABLE 1.
Differential single nucleotide polymorphisms (SNPs) between the three strains in the study
| SNP | Strain 1 | Strain 2 | Strain 3 |
|---|---|---|---|
| C119T | 1 | 0 | 0 |
| T154C | 0 | 1 | 0 |
| C241T | 1 | 1 | 1 |
| G386A | 0 | 0 | 1 |
| T445C | 0 | 0 | 1 |
| C900T | 1 | 0 | 0 |
| C3037T | 1 | 1 | 1 |
| C3583T | 0 | 1 | 0 |
| C5239T | 0 | 0 | 1 |
| C6286T | 0 | 0 | 1 |
| A8001G | 1 | 0 | 0 |
| C8047T | 1 | 0 | 0 |
| A11595G | 0 | 0 | 1 |
| G11609A | 0 | 0 | 1 |
| C14408T | 1 | 1 | 1 |
| C14937T | 0 | 0 | 1 |
| G17149T | 1 | 0 | 0 |
| C17639T | 0 | 1 | 0 |
| C17642G | 0 | 1 | 0 |
| A20108G | 0 | 0 | 1 |
| A20268G | 1 | 1 | 0 |
| G21255C | 0 | 0 | 1 |
| C22227T | 0 | 0 | 1 |
| A23403G | 1 | 1 | 1 |
| A25501C | 0 | 0 | 1 |
| C25613T | 1 | 0 | 0 |
| C25904T | 0 | 0 | 1 |
| C26801G | 0 | 0 | 1 |
| C27944T | 0 | 0 | 1 |
| A28222G | 0 | 1 | 0 |
| C28932T | 0 | 0 | 1 |
| C29253T | 1 | 0 | 0 |
| G29645T | 0 | 0 | 1 |
| A29771T | 0 | 1 | 0 |
E1 SARS‐CoV‐2 superinfection occurred during the first COVID‐19 wave, when prevalence of SARS‐COV‐2 among our population was very high (1182 cases/100,000 inhabitants) and most hospitalized cases were COVID‐19 patients. Moreover, our patient was highly dependent due to neurological damage and remained hospitalized in two different buildings. Thus, the patient was probably exposed to different nosocomial circulating strains that might have caused E1 superinfection. To the best of our knowledge, only another likely SARS‐CoV‐2 superinfection has been reported (Tarhini et al., 2021), in an immunosuppressed patient, for whom two SARS‐CoV‐2 strains were identified on Day 56. There is higher risk of prolonged viral shedding in immunosuppressed SARS‐CoV‐2 positive cases (Choi et al., 2020), which may explain the superinfection. Our patient was not immunosuppressed and superinfection was detected only 1 week after the identification of a single‐strain infection in a previous specimen. Two co‐existing SARS‐CoV‐2 strains have been also reported in a COVID‐19 case who was reinfected 26 days after the first infection (Lee, 2020).
E1 was severe and identification of two coinfecting strains coincided with patient´s clinical deterioration, suggesting some clinical impacts of superinfection on the severity of E1. On the other hand, E2, associated with a third different strain, was milder, similar to other reinfection reports (Van Elslande et al., 2020), with no consolidation on X‐ray and mild hypoxemia. E2 infection resulted from a nosocomial exposure, as Strain 3 was responsible for a hospital outbreak involving at least 11 cases (zero SNPs among them) from three wards. An epidemiological survey confirmed that the health care worker who attended the patient and had shared a room with our case was also infected with the same strain (zero SNPs), suggesting a potential role in the reinfection.
Summarizing, we describe a COVID‐19 case with unprecedented viral complexity in SARS‐CoV‐2 infection. Initially, the patient was superinfected by two different strains within a short period, followed months later by a COVID‐19 reinfection by a third distinct strain.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
This study was approved by the ethical research committee of Gregorio Marañón Hospital (REF: MICRO.HGUGM.2020‐042). Informed consent was obtained from the patient for the publication of this case report.
AUTHOR CONTRIBUTIONS
Conception, design, analysis, and manuscript revision: Laura Pérez Lago. Data compilation, clinical data, and epidemiological research: Martha Kestler. Bioinformatic analysis: Pedro J. Sola‐Campoy. Methodology, investigation, and data analysis: Cristina Rodriguez‐Grande. Epidemiological research and data analysis: Rubén Francisco Flores‐García. Bioinformatic analysis: Sergio Buenestado‐Serrano. Analysis and investigation: Marta Herranz. Data compilation and data analysis: Luis Alcala. Data analysis and investigation: Carolina Martínez‐Laperche. Data analysis and sequencing: Julia Suárez‐González. Data analysis, methodology, and investigation: Pilar Catalán. Resources and manuscript revision: Patricia Muñoz. Conception, design, analysis, and manuscript writing: Darío García de Viedma.
GREGORIO MARAÑÓN MICROBIOLOGY‐ID COVID‐19 STUDY GROUP
Adán‐Jiménez (Javier), Alcalá (Luis), Aldámiz (Teresa), Alonso (Roberto), Álvarez (Beatriz), Álvarez‐Uría (Ana), Arias (Alexi), Arroyo (Luis Antonio), Berenguer (Juan), Bermúdez (Elena), Bouza (Emilio), Buenestado‐Serrano (Sergio), Burillo (Almudena), Candela (Ana), Carrillo (Raquel), Catalán (Pilar), Cercenado (Emilia), Cobos (Alejandro), de la Cueva (Victor Manuel), Díez (Cristina), Escribano (Pilar), Estévez (Agustín), Fanciulli (Chiara), Galar (Alicia), García (Mª Dolores), García de Viedma (Darío), Gijón (Paloma), González (Adolfo), Guillén (Helmuth) Guinea (Jesús), Haces (Laura Vanessa), Herranz (Marta), Kestler (Martha), López (Juan Carlos), Losada (Carmen Narcisa), Machado (Marina), Marín (Mercedes), Martín (Pablo), Montilla (Pedro), Muñoz (Patricia), Olmedo (María), Padilla (Belén), Palomo (María), Parras (Francisco), Pérez‐Granda (María Jesús), Pérez‐Lago (Laura), Pérez (Leire), Pescador (Paula), Reigadas (Elena), Rico‐Luna (Carla Margarita), Rincón (Cristina), Rodríguez (Belén), Rodríguez (Sara), Rodríguez‐Grande (Cristina), Rojas (Adriana), Ruiz‐Serrano (María Jesús), Sánchez (Carlos), Sánchez (Mar), Serrano (Julia), Sola Campoy (Pedro J), Tejerina (Francisco), Valerio (Maricela), Veintimilla (Mª Cristina), Vesperinas (Lara), Vicente (Teresa), de la Villa (Sofía).
Supporting information
SUPPORTING INFORMATION
ACKNOWLEDGEMENT
We are grateful to Dainora Jaloveckas (cienciatraducida.com) for editing and proofreading assistance.
Pérez‐Lago, L. , Kestler, M. , Sola‐Campoy, P. J. , Rodriguez‐Grande, C. , Flores‐García, R. F. , Buenestado‐Serrano, S. , Herranz, M. , Alcalá, L. , Martínez‐Laperche, C. , Suárez‐González, J. , Catalán, P. , Muñoz, P. , & García de Viedma, D. , Gregorio Marañón Microbiology‐ID COVID 19 Study Group . (2021). SARS‐CoV‐2 superinfection and reinfection with three different strains. Transboundary and Emerging Diseases, 1–6. 10.1111/tbed.14352
DATA AVAILABILITY STATEMENT
The data that support the findings of this study (FastA files) are openly available in GISAID at https://www.gisaid.org/. Reference numbers (Strain 1 ‐ EPI_ISL_1547368, Strain 2 ‐ EPI_ISL_1547369, and Strain 3 ‐ EPI_ISL_1547363).
REFERENCES
- Choi, B. , Choudhary, M. C. , Regan, J. , Sparks, J. A. , Padera, R. F. , Qiu, X. , Solomon, I. H. , Kuo, H. H. , Boucau, J. , Bowman, K. , Adhikari, U. D. , Winkler, M. L. , Mueller, A. A. , Hsu, T. Y. T. , Desjardins, M. , Baden, L. R. , Chan, B. T. , Walker, B. D. , Lichterfeld, M. , …, Li, J. Z. (2020). Persistence and Evolution of SARS‐CoV‐2 in an Immunocompromised Host. The New England Journal of Medicine, 383(23), 2291–2293. 10.1056/NEJMc2031364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. S. , Kim, S. Y. , Kim, T. S. , Hong, K. H. , Ryoo, N. H. , Lee, J. , Park, J. H. , Cho, S. I. , Kim, M. J. , Kim, Y. G. , Kim, B. , Shin, H. S. , Oh, H. S. , Seo, M. S. , Gwon, T. R. , Kim, Y. , Park, J. S. , Chin, B. S. , Park, W. B. , … Seong, M. W. (2020). Evidence of severe acute respiratory syndrome coronavirus 2 reinfection after recovery from mild coronavirus disease 2019. Clinical Infectious Diseases. 10.1093/cid/ciaa1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarhini, H. , Recoing, A. , Bridier‐Nahmias, A. , Rahi, M. , Lambert, C. , Martres, P. , Lucet, J. C. , Rioux, C. , Bouzid, D. , Lebourgeois, S. , Descamps, D. , Yazdanpanah, Y. , Le Hingrat, Q. , Lescure, F. X. , & Visseaux, B. (2021). Long term SARS‐CoV‐2 infectiousness among three immunocompromised patients: From prolonged viral shedding to SARS‐CoV‐2 superinfection. The Journal of infectious diseases, 223(9), 1522–1527. 10.1093/infdis/jiab075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Elslande, J. , Vermeersch, P. , Vandervoort, K. , Wawina‐Bokalanga, T. , Vanmechelen, B. , Wollants, E. , Laenen, L. , André, E. , Ranst, M. V. , Lagrou, K. , & Maes, P. (2020). Symptomatic SARS‐CoV‐2 reinfection by a phylogenetically distinct strain. Clinical Infectious Diseases, 73(2), 354–356. 10.1093/cid/ciaa1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPORTING INFORMATION
Data Availability Statement
The data that support the findings of this study (FastA files) are openly available in GISAID at https://www.gisaid.org/. Reference numbers (Strain 1 ‐ EPI_ISL_1547368, Strain 2 ‐ EPI_ISL_1547369, and Strain 3 ‐ EPI_ISL_1547363).


