Genetic variants associated with the risk of NAFLD and NASH exhibit pleiotropic effects. 1 , 2 Specifically, the non‐synonymous rs738409 C/G variant in PNPLA3 (patatin‐like phospholipase domain‐containing protein 3), which encodes I148 M protein isoforms with a significant effect on the severity of several liver‐related outcomes, 3 presents pleiotropic effects on cells that mediate the immune response. 1 , 2 Figure 1 discloses the laboratory measurements and/or haematological traits associated with variants in PNPLA3 extracted from different data sources.
FIGURE 1.
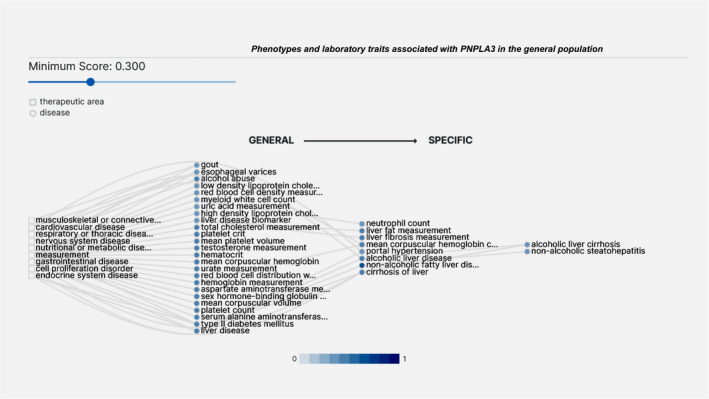
Association analysis of variants in PNPLA3 and diverse and systemic traits in the general population. The graph was performed using the open‐source, publicly available Open Targets Platform, available at https://platform.opentargets.org/, which integrates publicly available human genome‐wide association (GWAS). Graph discloses association scores higher than 0.3 (score ranges from 0 to 1; the higher the score, the stronger the association)
The COVID‐19 pandemic revealed that NAFLD and NASH, ‐either explained by the presence of overlapping co‐morbidities such as obesity and type 2 diabetes or by the presence of the systemic inflammation associated with the chronic liver condition‐, appear to be a risk factor for severe SARS‐CoV‐2 infection. 4 Hence, it was postulated that variants that predispose to NAFLD may also indirectly condition the severity of the COVID‐19 infection. This hypothesis motivated interesting candidate gene association studies. For example the UKBB (UK Biobank) dataset was initially used to develop a genetic risk score for NAFLD based on the weighted effect of variants involved in the accumulation of fat in the liver (PNPLA3‐TM6SF2‐MBOAT7‐GCKR). 5 Thus, leveraging this information, Valenti et al examined the impact of that NAFLD‐genetic risk score on the risk of COVID‐19, 5 and detected a trend of protection against COVID‐19 conferred by rs738409. 5 In addition, the authors observed a trend towards differential gene expression levels of SARS‐CoV‐2 receptors (ACE2) in a cohort of 125 obese subjects. 5
More recently, two studies from Europe highlighted the potential involvement of the rs738409 in modifying the severity of COVID‐19, including hospitalization and death rate. 6 , 7 Grimaudo et al found a direct association between the rs738409 G‐allele and severe COVID‐19 outcomes among patients aged 65 years or less. 6
On the other side, Innes et al reported a striking inverse association analysis between the rs738409 and COVID‐19 severity outcomes in 1585 UKBB participants. 7 The authors found that the rs738409‐G allele was independently associated with a reduced risk of COVID‐19 hospitalization and mortality; the protective effect remained significant after adjusting for major demographic and disease risk factors, including underlying metabolic and liver co‐morbidities. 7 In addition, Innes et al 7 performed a meta‐analysis of three independent data sources that explored the potential association between rs738409 and COVID‐19. This study included data from the FinnGen study (a population‐based biobank comprising 83 patients with COVID‐19 hospital admission and 274 SARS‐CoV‐2–positive patients without hospital admission), the Geisinger Health System (n = 854 subjects of European Ancestry, comprising 165 individuals COVID‐19 hospitalization vs 689 SARS‐CoV‐2–positive patients without hospital admission), and the study of Grimaudo et al (a total of 383 COVID‐19 patients). 6 The pooled analysis of the above‐mentioned data sources showed that the presumed protective effect of the G‐ ‘NASH‐risk allele' on the morbidity and mortality of COVID‐19 could not be further validated. 7 However, it appears that there was a trend for association with a reduced risk of COVID‐19 hospitalization/severe disease that did not reach statistical significance. 7
Likewise, Bianco et al found that the rs738409 G allele has a tendency not only to be associated with protection against COVID‐19 but also it was associated with lower C‐reactive protein levels despite higher ALT and lower albumin in severe COVID‐19 patients of European ancestry. 8 Table 1 shows a summary of current evidence of the putative association between rs738409 and COVID‐19 outcomes.
TABLE 1.
Summary of current evidence of the putative role of PNPLA3‐rs738409 in protecting against severe COVID‐19 outcomes
| Study [ref] | Study design/ population features | Sample size | Effect: OR (95% CI) | P value |
|---|---|---|---|---|
| Valenti et al 5 | Population‐based UKBB/ restricted to British ancestry (case‐control study) | 1,460 (positive for SARS‐CoV‐2 n = 526; negative n = 934) | NR a | .06 |
| 0.86 (0.71‐1.04) b | .12 | |||
| Grimaudo et al 6 | Hospital‐based Sicilian patients with laboratory‐confirmed SARS‐CoV‐2 infection | 383 (hospitalization n = 148; death n = 32) | 4.69 (1.01‐22.04) adjusted for sex and only among patients >65 years old | .035 |
| Innes et al 7 | Population‐based UKBB (only COVID‐19 patients) | 1585 patients (hospital admission n = 759; admission with pneumonia n = 450; admission requiring advanced respiratory support n = 76). | 0.79 (0.64‐0.97) hospitalization risk per G allele. c | .027 |
| 0.75 (0.57‐0.98) COVID‐19 death per G allele. c | .037 | |||
| Innes et. al. 7 | Meta‐analysis of 3 population‐based datasets (all positive COVID‐19 patients) | FinnGen study (hospital admission n = 83; without hospital admission n = 274). Geisinger Health System (hospitalization n = 165; without hospitalization n = 689) | 0.83 (0.66‐1.05) | .12 |
| Bianco et al 8 | Hospital‐based Fondazione IRCCS Ca' Granda cohort (case‐control study). | Severe COVID‐19 outcomes n = 508; healthy controls n = 889 | 0.88 (0.70‐1.10) | 0.27 |
Non‐reported, unadjusted model
Adjusted for age, sex, BMI, PC1‐10 (ethnicity), assessment centre, array batch.
Values adjusted for potential non‐specified confounding factors. All studies reported subjects of European ancestry.
These remarkable findings deserve several reflections before raising unequivocal conclusions of the putative role of rs738409 on the course of COVID‐19, one concerning the sample size of the studies and another concerning the biological plausibility of the reported association.
Specifically, a note of caution must be added to the initial enthusiasm about the role of a PNPLA3‐variant in protecting against severe COVID‐19 because it might trigger unexpected clinical consequences, including changes in the course of clinical trials and disease management. The analysis of the presumed protective effect of the variant on COVID‐19 mortality and morbidity was done on a sample without enough power to detect such association/s. 7 To detect reproducible causal variants associated with complex diseases, a sample size with statistical power is critical to the robustness of the conclusion/s of the study. For instance the study of Innes et al involved a sample of 267 patients who died due to COVID‐19 and 1318 who survived. 7 Unfortunately, the statistical power for the sample size used in the additive model to infer the presumed protective effect is ~0.52 (considering the informed frequency of the rs738409 G‐allele (20%), the COVID‐19 death prevalence of ~3%, and the explained effect of ~25% lower odds of COVID‐19 death). 7 Thereby, the results of this meta‐analysis need to be interpreted with caution until the presumed association be consistently reproduced in larger cohorts.
Yet, the reported association is not only clinically relevant, but it seems to be also plausible.
For that reason, we would like to highlight putative explanations based on the presence of broad and systemic biological effect/s of variants located in PNPLA3.
The analysis of genotype‐phenotype associations shows that PNPLA3 presents several variants that modulate epigenetic modifications, including changes in DNA methylation and histones in human immune cell types. Specifically, the intron rs2294915 C/G/T (chr22:44340904) variant that is not only in high linkage disequilibrium with rs738409 (r2 0.85) 9 but also modifies the expression of PNPLA3 10 and SAMM50, is associated with the histone modification H3K4me1 in CD14+ monocytes (beta: −0.8994, P = 3.00e‐14) from blood donors of the United Kingdom population. 11 In fact, Donati et al showed that the PNPLA3‐rs2294915 variant influences the phenotypic expression of the rs7383409 in a bimodal fashion by acting on the PNPLA3 mRNA stability probably through a cis‐expression quantitative trait locus. 10
Finally, Phenome‐wide association (PheWAS) analysis on data of the UK Biobank denotes not only beta positive ('risk') effects of rs738409 on liver‐related traits but also beta negative ('protective') effects on many non‐liver traits, including respiratory diseases and haematological‐related traits, such as neutrophil and platelet count. 2 Of note, platelet count was associated with over fivefold enhanced risk of severe COVID‐19 (OR, 5.1). 12 In addition, a higher neutrophil/monocytes ratio is another predictor of COVID19 worse outcome.
However, it is difficult to interpret how exactly the PNPLA3‐rs738409 variant may differentially affect these traits in healthy individuals with and without COVID‐19 infection. It is thus clear from the above discussion that there is a great opportunity for further genetic association studies. Epigenetic investigations based on the rs738409 can also be of help at identification of factors that may affect COVID‐19 outcomes, including not only changes in cells of the immune system and platelets but also interactions with host lipid metabolism and SARS‐CoV‐2 life cycle.
Collectively, these observations might have several implications beyond the potential presumed protective effect of rs738409 on the course of COVID‐19. For example the large and widely reproducible effect of rs738409 on the biology of NASH and fibrosis 13 has positioned PNPLA3 as an attractive target for pharmacological intervention. The first in vivo experimental study examining the effect of antisense oligonucleotide (ASO)‐mediated silencing of Pnpla3 in a knock‐in mouse model in which the human PNPLA3‐I148M mutation was introduced showed that Pnpla3 ASO therapy can improve all features of NAFLD, including liver fibrosis. 14 Following advancements in humans were done as well. For example AZD2693 is a ligand‐conjugated antisense therapy designed to inhibit the production of PNPLA3 protein. A clinical trial (ClinicalTrials.gov Identifier: NCT04483947) is recruiting volunteers to assess the safety and tolerability, pharmacokinetics and pharmacodynamics of AZD2693 in patients with NASH fibrosis and homozygous for the rs738409‐G risk allele, in whom investigators should closely monitor COVID19 susceptibility.
Hence, the COVID‐19 pandemic has brought attention to the importance of having a complete understanding not only of the PNPLA3 gene‐attributed associations with liver‐related traits but also a thorough understanding of the protein interactions, the active protein ligands, and most importantly, an accurate and comprehensive assessment of the impact of the variant pleiotropic effects.
CONFLICT OF INTERESTS
None.
ACKNOWLEDGMENT
This study was partially supported by grants PICT 2018‐889, and PICT 2016‐0135 (Agencia Nacional de Promoción Científica y Tecnológica, FONCyT), CONICET Proyectos Unidades Ejecutoras 2017, PUE 0055.
Pirola CJ, Sookoian S. PNPLA3 and COVID‐19 outcomes: Thinking outside the box might explain the biology behind pleiotropic effects of rs738409 on the immune system. Liver Int. 2021;41:2801–2804. 10.1111/liv.15043
Co‐corresponding authorship: Carlos J Pirola, Silvia Sookoian
SS and CJP should be considered joint senior authors
Handling Editor: Luca Valenti
Contributor Information
Carlos J. Pirola, Email: pirola.carlos@conicet.gov.ar.
Silvia Sookoian, Email: ssookoian@intramed.net.
REFERENCES
Author names in bold designate shared co‐first authorship.
- 1. Diogo D, Tian C, Franklin CS, et al. Phenome‐wide association studies across large population cohorts support drug target validation. Nat Commun. 2018;9:4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pirola CJ, Salatino A, Sookoian S. Pleiotropy within gene variants associated with nonalcoholic fatty liver disease and traits of the hematopoietic system. World J Gastroenterol. 2021;27:305‐320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sookoian S, Pirola CJ, Valenti L, et al. Genetic pathways in nonalcoholic fatty liver disease: insights from systems biology. Hepatol. 2020;72:330‐346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marjot T, Webb GJ, Barritt AS, et al. COVID‐19 and liver disease: mechanistic and clinical perspectives. Nat Rev Gastroenterol Hepatol. 2021;18:348‐364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Valenti L, Jamialahmadi O, Romeo S. Lack of genetic evidence that fatty liver disease predisposes to COVID‐19. J Hepatol. 2020;73:709‐711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grimaudo S, Amodio E, Pipitone RM, et al. PNPLA3 and TLL‐1 polymorphisms as potential predictors of disease severity in patients with COVID‐19. Front Cell Dev Biol. 2021;9:627914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Innes H, Buch S, Barnes E, et al. The rs738409 G Allele in PNPLA3 is associated with a reduced risk of COVID‐19 mortality and hospitalization. Gastroenterol. 2021;160:2599‐2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bianco C, Baselli G, Malvestiti F, et al. Genetic insight into COVID‐19‐related liver injury. Liver Int. 2021;41:227‐229. [DOI] [PubMed] [Google Scholar]
- 9. Sookoian S, Pirola CJ. PNPLA3, the triacylglycerol synthesis/hydrolysis/storage dilemma, and nonalcoholic fatty liver disease. World J Gastroenterol. 2012;18:6018‐6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Donati B, Motta BM, Pingitore P, et al. The rs2294918 E434K variant modulates patatin‐like phospholipase domain‐containing 3 expression and liver damage. Hepatol. 2016;63:787‐798. [DOI] [PubMed] [Google Scholar]
- 11. Chen LU, Ge B, Casale FP, et al. Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell. 2016;167:1398‐1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lippi G, Plebani M, Henry BM. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID‐19) infections: a meta‐analysis. Clin Chim Acta. 2020;506:145‐148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sookoian S, Pirola CJ. Meta‐analysis of the influence of I148M variant of patatin‐like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatol. 2011;53:1883‐1894. [DOI] [PubMed] [Google Scholar]
- 14. Lindén D, Ahnmark A, Pingitore P, et al. Pnpla3 silencing with antisense oligonucleotides ameliorates nonalcoholic steatohepatitis and fibrosis in Pnpla3 I148M knock‐in mice. Mol Metab. 2019;22:49‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]


