Abstract
The SARS‐CoV‐2 virus was detected for the first time in December 2019 in Wuhan, China. Currently, this virus has spread around the world, and new variants have emerged. This new pandemic virus provoked the rapid development of diagnostic tools, therapies and vaccines to control this new disease called COVID‐19. Antibody detection by ELISA has been broadly used to recognize the number of persons infected with this virus or to evaluate the response of vaccinated individuals. As the pandemic spread, new questions arose, such as the prevalence of antibodies after natural infection and the response induced by the different vaccines. In Mexico, as in other countries, mRNA and viral‐vectored vaccines have been widely used among the population. In this work, we developed an indirect ELISA test to evaluate S1 antibodies in convalescent and vaccinated individuals. By using this test, we showed that IgG antibodies against the S1 protein of SARS‐CoV‐2 were detected up to 42 weeks after the onset of the symptoms, in contrast to IgA and IgM, which decreased 14 weeks after the onset of symptoms. The evaluation of the antibody response in individuals vaccinated with Pfizer‐BioNTech and CanSinoBio vaccines showed no differences 2 weeks after vaccination. However, after completing the two doses of Pfizer‐BioNTech and the one dose of CanSinoBio, a significantly higher response of IgG antibodies was observed in persons vaccinated with Pfizer‐BioNTech than in those vaccinated with CanSinoBio. In conclusion, these results confirm that after natural infection with SARS‐CoV‐2, it is possible to detect antibodies for up to 10 months. Additionally, our results showed that one dose of the CanSinoBio vaccine induces a lower response of IgG antibodies than that induced by the complete scheme of the Pfizer‐BioNTech vaccine.
Keywords: antibodies, CanSinoBio, COVID‐19, ELISA, Pfizer‐BioNTech, S1, SARS‐CoV‐2, vaccine
1. INTRODUCTION
In December 2019, the city of Wuhan, China, reported an outbreak of pneumonia. Later in that month, the World Health Organization declared that a novel virus was the cause of this problem, and it was initially called new coronavirus 2019 (nCoV‐2019) (WHO, 12 January, 2020). Subsequently, the International Committee on Virus Taxonomy named the virus SARS‐CoV‐2, as currently known ("The species Severe acute respiratory syndrome‐related coronavirus: classifying 2019‐nCoV and naming it SARS‐CoV‐2," 2020). The global impact of this virus is undeniable. On 11 January 2020, almost 2 weeks after the first report of this pathogen, the first death was documented. On 20 January, multiple cases were reported in Japan, South Korea, and Thailand (EWHO, 2020). The first case in the United States was declared one day later (Ghinai et al., 2020). Since then, the SARS‐CoV‐2 virus has been reported all across the globe. In Mexico, the first case was reported in February 2020, and since then it has continued to spread among the population. At the time this report was written (10 July 2021), approximately 2,764,852 cases were documented, including 246,910 deaths (SS‐México, 2021).
Reverse transcription polymerase chain reaction (RT‐PCR) is an essential assay for diagnosing coronavirus disease 19 (COVID‐19), especially during the active virus‐shedding phase. However, 2 weeks after the onset of the symptoms, the viral loads decrease (Zhang et al., 2020). Antibody assays have shown significant sensitivity. Zhao et al. (2020) showed that the median times for IgM and IgG seroconversion were 12 and 14 days, respectively. In contrast, Long, Tang, et al. (2020) demonstrated that IgM and IgG antibody seroconversion occurred on day 6. Interestingly, the analysis of seroconversion of these isotypes did not show significant differences between critical and noncritical patients (Zhao et al., 2020). However, Long, Liu, et al. (2020) showed that at 2 weeks post‐symptom onset, IgG was significantly higher in patients with severe disease than in those with non‐severe disease. The analysis of humoral response in symptomatic and asymptomatic patients showed a significant reduction in the levels of IgG in the asymptomatic group (Long, Tang, et al., 2020). This study also revealed that an important percentage of asymptomatic patients were IgG negative (40%) compared with symptomatic (12%) patients who were IgG negative (Long, Tang, et al., 2020). Antibody responses have been evaluated against the N and S proteins (Sun et al., 2020), including receptor binding domain (Roy et al., 2020) and S1 (Krähling et al., 2021). Currently, several commercial kits for the detection of antibodies against SARS‐CoV‐2 are available. Some studies have evaluated different commercial serological assays, and positivity rates vary among assays, probably due to the low antibody response of asymptomatic patients. In general, the conclusions were that antibody detection is suitable for 10 days after the onset of symptoms (Herroelen et al., 2020; Trabaud et al., 2020).
Today, one crucial question in the immune response against SARS‐CoV‐2 is the persistence of antibodies. The first reports suggested that the persistence of IgG antibodies is up to 6 months after the onset of the symptoms (Zhang et al., 2020). Similar results were observed by Liu et al. (2021). In that study, IgG antibodies were detected for 6 months. In contrast, IgM antibodies were reduced in approximately 80% of the patients. Recently, a study evaluated a cohort of 254 samples with diverse symptoms and observed that IgG titres remained detectable for 6–8 months after symptom onset (Dan et al., 2021). However, a recent report suggests that IgG antibodies can persist for 10 months (Dudreuilh et al., 2021). In that report, 2 of 84 patients showed IgG antibodies after 10 months.
Currently, several vaccines have been approved in different countries (Forni & Mantovani, 2021). For COVID‐19, there are mRNA‐based vaccines available that have shown the highest levels of protection, followed by viral vector, protein subunit, and whole‐inactivated viruses (Kim et al., 2021). In Mexico, mRNA and viral vectors are vaccines being used to immunize the population (Vacunacovid.Gob.Mx, 2021). Several reports agree the efficacy of the mRNA vaccines, particularly Pfizer‐BioNTech. Other studies have demonstrated their efficacy against the new variants of SARS‐CoV‐2 (Stankov et al., 2021). In contrast, the available information for CanSinoBio, a viral‐vector vaccine, its limited (Zhu et al., 2020) and indicates an efficacy of 65%, which places it as one of the vaccines with the lowest efficacy (Forni & Mantovani, 2021). In Mexico, as in other countries, many questions have arisen about this vaccine. Our study established an indirect enzyme‐linked immunosorbent assay (ELISA) to detect IgM‐, IgA‐, and IgG‐specific antibodies against the S1 protein of SARS‐CoV‐2 in convalescent patients. In addition to measuring the persistence of antibodies, we evaluated the response of individuals with a history of COVID‐19 or naïve individuals to COVID‐19 vaccinated with either Pfizer‐BioNTech or CanSinoBio.
2. MATERIALS AND METHODS
2.1. Samples and patients
For this study, 145 negative control serum samples were included. Sixty‐five samples were collected between 2017 and 2019 from a previous breast cancer project, and they were kindly provided by Dr. Graciela Caire‐Juvera (CIAD, A.C.). Eighty samples were collected between 2015 and 2018 from a project on seroprevalence against Cryptosporidium spp., and they were kindly provided by Dr. Olivia Valenzuela (Universidad de Sonora). These serum samples were included because no SARS‐CoV‐2 was circulating at that time.
Serum samples from RT‐PCR‐positive adult patients were included. One hundred forty‐two samples were from convalescent and non‐hospitalized recovered adult patients who were volunteer donors to collect convalescent plasma for treatment in the TERAPLASCOV‐2 trial (median age of 34; interquartile range [IQR]: 29–42; 12% women). Sixty samples were from hospitalized patients (46.5; IQR: 42.25–59; 25% women) subject to treatment with convalescent plasma in the TERAPLASCOV‐2 trial registered in ClinicalTrials.gov Identifier: NCT04356482. In this case, the samples used in this work were those obtained from previous plasma treatments.
Three consecutive samples were taken from another group of 28 patients (median age of 37; IQR: 30–58; 50% women) to evaluate the persistence of antibodies against SARS‐CoV‐2. In these cases, all patients were RT‐PCR positive and had mild to severe symptoms of COVID‐19. In this group, the first sample was taken between 1 and 5 weeks post‐symptom onset. The second and third samples were taken at approximately 12–14 and 38–40 weeks post‐symptom onset.
To evaluate the antibody response to the vaccines, we included 62 individuals (median age of 50; IQR: 31–55; 66.1 women). Twenty‐five patients were vaccinated with Pfizer‐BioNTech, and 37 were vaccinated with CanSinoBio. In the Pfizer‐BioNTech‐vaccinated group, 40% had a history of COVID‐19, and 60% were naïve to COVID‐19. In the CanSinoBio‐vaccinated group, 25% had a history of COVID‐19, and 75% were naïve to COVID‐19. In the Pfizer‐BioNTech group (two doses), three samples were collected from the patients: before the first dose of the vaccine, 10–14 days after the first dose, and 14 days after the second dose. Unfortunately, it was not possible to collect all three samples from all of the patients. In the CanSinoBio group (single dose), three samples were collected from each patient: before the vaccine, and then 2 weeks and 4 weeks after vaccination.
This work was conducted in agreement with general ethical principles, and all of the participants provided written informed consent. The protocol was approved by the Ethics Committees from the Centro de Investigación en Alimentación y Desarrollo, A.C., the Hospital General del Estado de Sonora and Hospital Central Norte de PEMEX.
2.2. Gene design and expression of the S1 protein
The S1 domain of SARS‐CoV‐2 includes amino acids 1–633 from the full Spike protein. The expression gene construct was designed with the deduced S1 sequence, preceded by a signal peptide and a Hist‐tag (6xHist) terminal carboxyl domain (Supplementary Figure S1A). The gene was synthesized and cloned into a pcDNA3.1(‐) vector by GenScript (GenScript, Piscataway, New Jersey, USA), producing the expression plasmid pcDNA3.1(‐)/SARS‐CoV‐2 S1 for a mammalian expression system.
The expression of the recombinant SARS‐CoV‐2 S1 protein was performed in the Expi293 Expression System following the manufacturers’ instructions (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Briefly, Expi293 cells were grown in Expi293 expression medium. Then, the transfection complex was prepared by mixing 30 μg of pcDNA3.1(‐)/SARS‐CoV‐2 S1 and 81 μg of Expifectamine (both diluted in OptiMEM‐I) and incubated for 20 min at room temperature. The complex was added to a flask with 75 × 106 Expi293 cells in 25.5 mL of Expi293 expression medium and incubated at 37°C and 125 rpm with 8% CO2. After 20 h, enhancers 1 and 2 were added to the medium and the cells were incubated for 4 more days until harvesting.
2.3. Recombinant S1 purification and characterization
After 4 days of transfection, the culture supernatant was harvested and clarified by centrifugation at 1600 rpm for 10 min. Then, the clarified supernatant was filtered through a 0.22‐μm filter followed by purification on immobilized metal affinity chromatography (IMAC) using a HisTrap HP column (Cytiva, Shrewsbury, Massachusetts, USA). The elution was carried out in the chromatograph ÄKTA prime plus (GE Healthcare Life Sciences, Marlborough, Massachusetts, USA) by a linear gradient from 0 to 100% elution buffer (500 mM imidazole), where the recombinant S1 protein was eluted in 26% elution buffer (approximately 150 mM imidazole), collecting 1 mL fractions. The total S1 protein purified was quantified by a Bradford assay, concentrated, and desalted with an Amicon Ultra15 (10 kDa cutoff) cartridge (Millipore, Burlington, Massachusetts, USA) followed by dilution in phosphate buffered saline (PBS) pH 7.4 to a final concentration of approximately 1 mg/mL.
For characterization of the purified recombinant S1 protein, SDS‐PAGE and western blotting were performed. Briefly, 3 μg of purified S1 was heat‐denatured and electrophoresed in a 10% polyacrylamide gel under reducing conditions followed by Coomassie blue staining. The gel was transferred to a PVDF membrane and blocked overnight with 5% nonfat milk in PBS with 0.05% of Tween 20. Afterwards, the blocked membrane was incubated with alkaline phosphatase‐conjugated mouse anti‐polyhistidine (Sigma‐Aldrich, St. Louis, Missouri, USA) for 1 h at 37°C, and BCIP®/NBT alkaline phosphatase substrate (Sigma‐Aldrich, St. Louis, Missouri, USA) was added for detection.
2.4. ELISA
SARS‐CoV‐2 S1 protein (2 μg/mL) was used to coat Maxisorp ELISA microwell plates (Thermo Fisher Scientific, Waltham, Massachusetts, USA) using 100 nM carbonate‐bicarbonate buffer, pH 9.5. ELISA plates were incubated overnight (18–19 h) at 4°C and washed once with PBS and blocked, or the supernatant was discharged and kept frozen until the blocking process. The ELISA plates were blocked with a blocking buffer containing 2% bovine serum albumin (Sigma‐Aldrich, St. Louis, Missouri, USA), 3% glucose (Fagalab, Sinaloa, México) and 0.025% of sodium azide (Sigma‐Aldrich, St. Louis, Missouri, USA). The blocking was carried out for 1 h at room temperature and washed three times with PBS/0.1% Tween 20 (PBST). Human serum samples were diluted 1:100 in PBS with 0.05% Tween 20 and 1% nonfat milk (American Bio, Canton, Massachusetts, USA) and incubated for 1 h at room temperature with agitation. Then, the wells were washed five times with PBST, and anti‐human IgG‐HRP (Polyclonal; Cat. No. A0170; Sigma‐Aldrich, St. Louis, Missouri, USA), anti‐human IgM‐HRP (Polyclonal; Cat. No. A0420; Sigma‐Aldrich, St. Louis, Missouri, USA), anti‐human IgA‐HRP (polyclonal; Cat. No. ab97220; Abcam, Cambridge, UK), anti‐human IgG1‐HRP (clone: HP6069; Cat. No. A10648; Invitrogen, Waltham, Massachusetts, USA), anti‐human IgG2‐HRP (clone: HP6014: Cat. No. 050520; Cat Bo. Life Technologies) anti‐human IgG3‐HRP (Clone: HP6047; Cat. No. 053620; Invitrogen, Waltham, Massachusetts, USA) or anti‐human IgG4 (clone: HP6025; Cat. No. A10654; Invitrogen, Waltham, Massachusetts, USA) were added to the plate and incubated for 30 min at room temperature with agitation. Anti‐IgG, anti‐IgA and anti‐IgM conjugates were diluted with antigen‐down HRP conjugate stabilizer 5× and stored at 4°C (Immunochemistry, Bloomington, Minnesota, USA). The wells were washed five times with PBST, and 50 μL of 3,3′,5,5′‐tetramethylbenzidine (Immunochemistry, Bloomington, Minnesota USA) was added for 5 min. The reaction was stopped with 50 μL of 1 M H2SO4, and the optical density (O.D.) was read at 450 nm using an automated spectrophotometer (Thermo Scientific Multiskan FC microplate photometer) during the next 5 min. Each plate included three controls: one positive control (pooled from SARS‐CoV‐2‐infected people), one negative control (pooled from SARS‐CoV‐2‐negative people) by duplicate, and blanks (n = 4). The mean of the blanks was subtracted from the absorbance of the samples, and the results were expressed as the relative INDEX (Index = absorbance/cutoff).
2.5. Statistical analysis
Statistical comparisons between negative samples and positive convalescent samples were performed using the Mann–Whitney U‐test for continuous variables. The analysis of antibody persistence and the response to the vaccine was evaluated with the Kruskal–Wallis test and Dunn's multiple comparisons test. p < .05 was considered statistically significant. Prism v8 (GraphPad, La Jolla, California, USA) was used for statistical analysis.
3. RESULTS
3.1. Production of the recombinant S1 protein of SARS‐CoV‐2
The recombinant S1 protein produced in this study (633 aa) was successfully expressed in Expi293 cells. Sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS‐PAGE) Coomassie blue staining allowed visualization of the S1 protein with an approximate weight of 130 kDa and a purity over 90% (Supplementary S1B). Western blot analysis confirmed the purification of the recombinant S1 protein by identifying the carboxyl‐terminal domain His‐tag (Supplementary S1C). It is notable that even though the estimated molecular weight from the amino acid sequence is approximately 74 kDa, the purified protein presented a higher weight. After expression, purification and desalting, the yield of recombinant SARS‐CoV‐2 S1 protein expressed in Expi293 cells was 40 mg/L of cell culture harvested at day 4 post‐transfection.
3.2. Development of the SARS‐CoV‐2 S1 ELISA
This study developed three indirect ELISA types to detect IgG, IgA and IgM antibodies against SARS‐CoV‐2 using the recombinant S1 protein as the target. First, the optimal antigen concentration for coating was evaluated and set at a concentration of 2 μg/mL (50 μL/well = 100 ng/well). Serum dilution was set at 1:100 and diluted with w/0.1% low‐free milk PBST. Serum and conjugate incubations were performed for 30 min at room temperature with slight agitation. In some experiments, the coated plates were frozen immediately at −20°C until further use or blocked for 1 h at room temperature and kept at 4°C for 2 weeks. No significant differences were observed when plates were either frozen or blocked and kept at 4°C (data not shown). After optimization, the cutoff was determined. For IgG, 145 samples with no history of SARS‐CoV‐2 were used as a negative control (Figure 1a). In this case, the cutoff value represents the mean ± 2 SD and it was set at 0.150 (mean = 0.071, 2 SD = 0.079). For IgA and IgM, 88 samples with no history of SARS‐CoV‐2 were used as negative controls (Figure 1b,c). The cutoff for IgA (mean ± 2 SD) was set at 0.180 (mean = 0.074, 2 SD = 0.105). For IgM, the cutoff (mean ± 2 SD) was set at 0.130 (mean = 0.048, 2 SD = 0.080). Few samples showed a value over the cutoff, suggesting high specificity.
FIGURE 1.
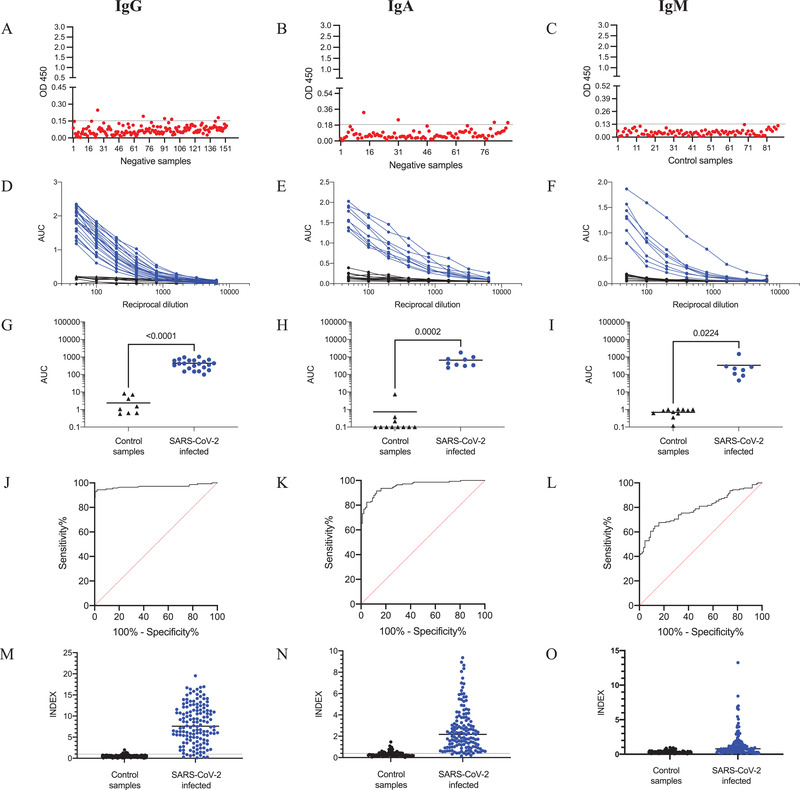
ELISA validation. (a–c): 145 human serum samples negative for SARS‐CoV‐2 were evaluated at a dilution of 1:100, and the cutoff for IgG (0.150), IgA (0.180) and IgM (0.130) was set with the mean value plus two standard deviations. (d–f): Sera from SARS‐CoV‐2‐infected individuals and negative control samples were used to evaluate the reactivity of IgG, IgA and IgM against the S1 protein of SARS‐CoV‐2. (g–i): Data from (d–f) were analyzed as the area under the curve (AUC), and the data were analyzed by unpaired Student´s test in GraphPad prism. The horizontal line represents the mean, and significant differences in each case are indicated. (j–o): Receiver operator characteristics (ROC) curves. The AUC was calculated to discriminate 145 SARS‐CoV‐2‐negative from 142 convalescent SARS‐CoV‐2‐positive samples based on INDEX (positive > 1) levels for IgG (m), IgA (n) and IgM (o)
The specificity calculated for IgG was 97.22% (confidence interval [CI], 93.04%–99.24%), and four of the 145 negative control samples were positive. For IgA, the specificity was calculated to be 97.73% (CI, 92.02%–99.72%), and two of 88 negative control samples were positive. For IgM, the specificity was 100.00% (95.89% – 100%).
To evaluate the assay performance, a serial dilution of positive samples was performed for the IgG, IgA and IgM antibodies. Initially, 22 COVID‐19 convalescent samples and 10 negative samples were used for the IgG antibodies. Similarly, COVID‐19 convalescent samples and negative samples (n = 12) were used for IgA (n = 9) and IgM (n = 8) (Figure 1d–i). These results showed that the ELISA test for IgG, IgA and IgM could significantly distinguish COVID‐19 convalescent serum samples from negative samples. Then, the diagnostic power of each ELISA was evaluated with receiver operating characteristic curves (ROCs) (Figure 1j–o). A panel of 142 convalescent sera and 145 negative samples was used. The results showed good accuracy for IgG and IgA, with an area under the ROC curve (AUC) of 0.97 ± 0.012 (95% CI [0.9468, 0.9940]) and 0.95 ± 0.012 (95% CI [0.9327, 0.9798]), respectively. In the case of the IgM, the AUC was 0.78 ± 0.029 (95% CI [0.7315, 0.8454]). To determine whether the low accuracy was because the IgM antibodies had disappeared from most convalescent samples, 60 positive samples from patients with < 15 days since the first symptoms were collected and analyzed. Supplementary S2‐A shows that the AUC for IgM using these sera was 0.90 ± 0.026 (95% CI [0.8561, 0.9594]). In further analysis, we evaluated the sera without IgG and only with IgM (Supplementary S2‐B). In this case, the AUC was 0.97 ± 0.013 (95% CI [0.9474, 1.00]), confirming the high accuracy of our assay when samples < 15 days since symptom onset were analyzed.
Finally, using a relative INDEX value (INDEX = cutoff/sample absorbance), we evaluated the IgG, IgA and IgM levels in the convalescent samples (n = 142). The results showed a different response of the isotype antibodies against SARS‐CoV‐2 in the samples. Interestingly, eight samples were negative for IgG antibodies, and only one of them had IgA (Figure 2). It is important to note that all of the patients had a confirmatory diagnosis of the virus by RT‐PCR. Unfortunately, we were not able to collect the clinical history of all of the patients to correlate the antibody response with the symptoms. Ma et al. (2020) described a different antibody response according to the severity of the disease. Patients with mild disease had low or negative antibody responses of IgG, IgA and IgM (Ma et al., 2020).
FIGURE 2.
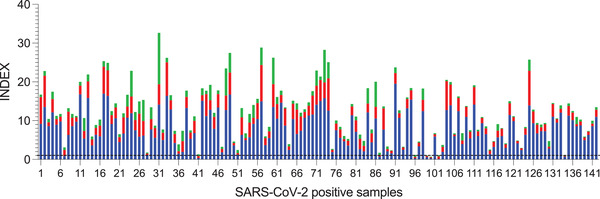
Antibody response in convalescent SARS‐CoV‐2‐positive samples. Bars represent the values of IgG (blue), IgA (red), and IgM (green) of 145 samples positive for SARS‐CoV‐2. INDEX values > 1 are positive
Overall, the sensitivity calculated for IgG was 94.37% (89.20–97.54%); 8 of 142 positive samples were negative. For IgA, the sensitivity was estimated to be 76.06% (68.18–82.82%) and 34 of 142 positive samples were negative. For IgM, the sensitivity was 59.15% (50.60–67.32), and 84 of 142 positive samples were negative. When the analysis was performed with sera from patients with < 15 days since symptom onset and without IgG, the sensitivity increased to 78.12% (CI, 60.03% – 90.72%]).
Additionally, the subtypes of IgG were evaluated in a group of 88 of 142 patients. The results (Figure 3) showed that most patients produced the IgG1 subtype, and few produced IgG3. Subtypes IgG2 and IgG4 were not detected in this cohort.
FIGURE 3.
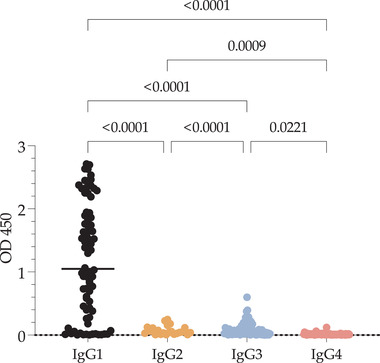
Subtypes of antibodies from SARS‐CoV‐2‐positive samples against the S1 protein. The subtype reactivity against S1 from 88 SARS‐CoV‐2‐positive samples was evaluated. Values are expressed as the OD(450), and significant differences are shown. Data were analyzed by the Kruskal–Wallis test, and multiple comparisons were performed with Dunn's test
3.3. Tracking the production of antibodies against SARS‐CoV‐2
To track the kinetics of IgG, IgA and IgM antibodies against SARS‐CoV‐2, 28 samples were collected at different time points: (1) between 1 and 5 weeks, (2) 12–14 weeks, and (3) 38–42 weeks after symptom onset. The results are presented in Figure 4. At the first time point, the median IgG antibody level was 3.75 (IQR, 2.52–4.96); IgG levels decreased at the second and third points (median 2.15, IQR, 1.45–3.75 and 1.5, IQR, 0.81–4.89, respectively). However, the differences between the first and the second and third time points were not statistically significant. No differences were observed between the second and third time points (p = .999). Five patients received the first dose of vaccine just a few days before the third sampling; this increased the median IgG and reduced the differences. The median IgA antibody level at the first time point was 5.13 (IQR, 2.55–7.85). In contrast to IgG, IgA decreased sharply (p < .0026) at the second time point (median 1.82, IQR 1.41–2.69). This tendency remained at the third time point (median 0.88, IQR 0.660–1.73), except for the four vaccinated patients, in which IgA presumably increased due to the vaccine. The median IgM antibody level at the first time point was 3.17 (IQR, 1.97–4.215). The IgM antibodies showed similar behaviour to IgA at the second time point (median 1.68, IQR 1.28–2.83), and at the third time point, IgM antibodies were not detected (median 0.58, IQR 0.25–0.58). In this case, IgM did not increase in response to the vaccine.
FIGURE 4.
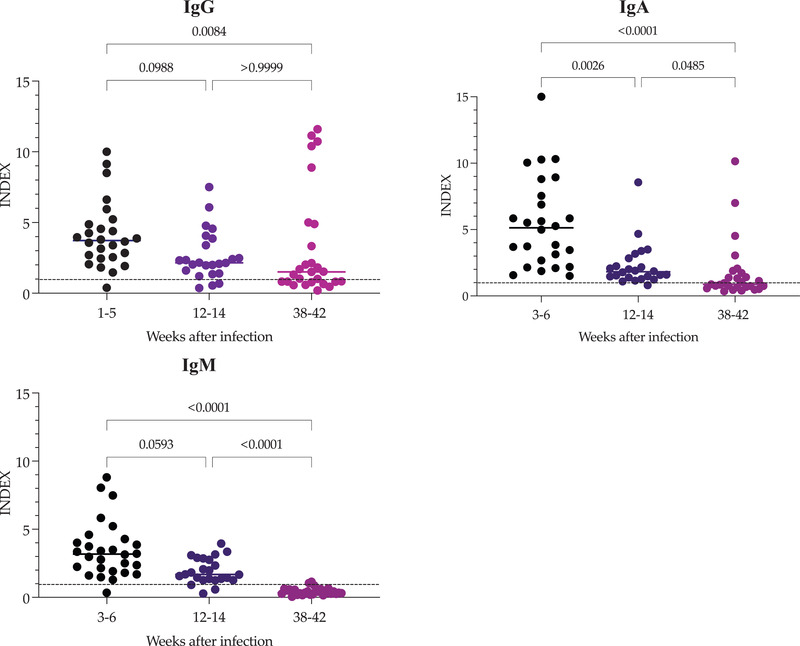
Development of antibodies in convalescent COVID‐19 individuals. Serum samples were taken after symptom onset and again at 12–14 and 38–42 weeks. Data represent the index values of the IgG, IgA and IgM antibodies against the SARS‐CoV‐2 S1 protein. The horizontal line represents the median and p value in each case. INDEX values > 1 are positive
3.4. Evaluation of the response against SARS‐CoV‐2 vaccine
In addition to the persistence of the antibodies, this work evaluated the IgG, IgA and IgM antibody response in persons with a history of COVID‐19 (the convalescent‐COVID‐19 group) and without a history of COVID‐19 (naïve‐COVID‐19 group). One group was vaccinated with Pfizer‐BioNTech and the other with CanSinoBio. Figure 5 shows the IgG, IgA and IgM responses before the vaccine, 2 weeks after the first dose, and 2 weeks after the second dose of Pfizer‐BioNTech, or 2 and 4 weeks after the first dose of CanSinoBio.
FIGURE 5.
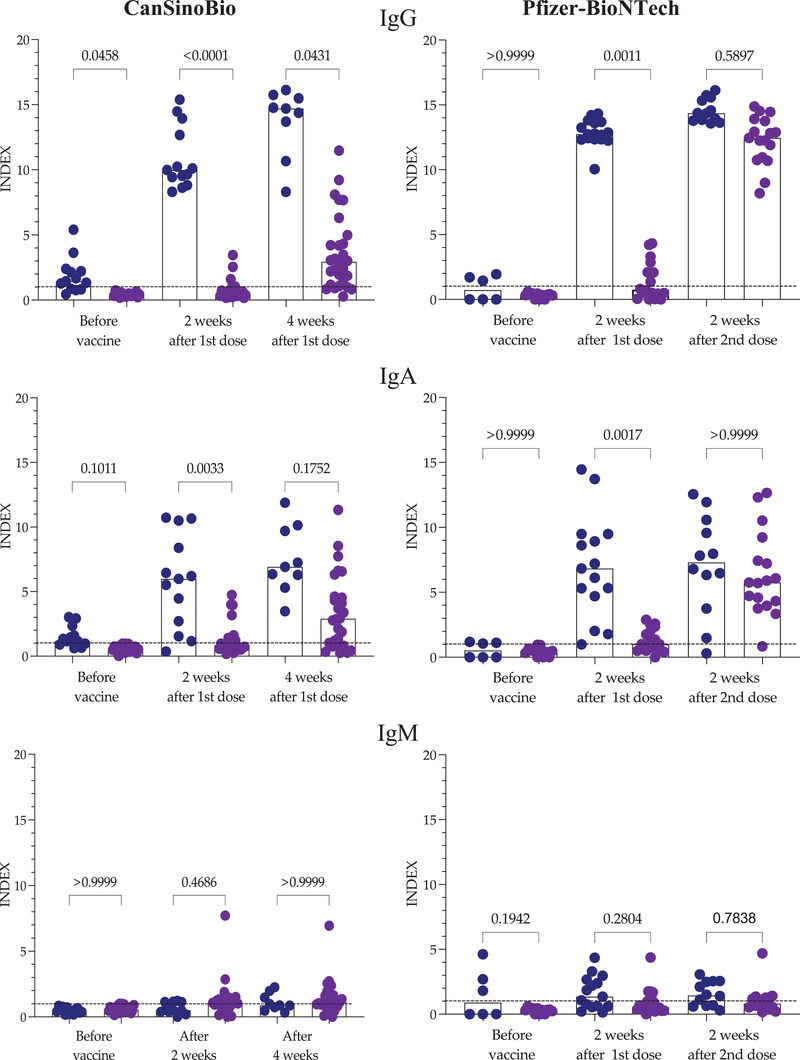
Antibodies in response to CanSinoBio or Pfizer‐BioNTech vaccine. IgG, IgA and IgM were evaluated in a group of individuals vaccinated with CanSinoBio and another with Pfizer‐BioNTech. In each case, persons with a history of COVID‐19 (blue) naïve to COVID‐19 (purple) were included. Bars represent the median, and each circle represents an individual. Differences were analyzed with the Kruskal–Wallis test and Dunn's multiple comparisons test. p values indicate a comparison of the response of individuals with a history of COVID‐19 versus naïve to COVID‐19. INDEX values > 1 are positive
The analysis of IgG antibodies showed that before vaccination, some of the patients in the COVID‐19 convalescent group had antibodies against the virus, in contrast to the COVID‐19‐naïve group. Two weeks after the first dose, all of the patients in the convalescent group developed a robust IgG response against SARS‐CoV‐2, regardless of the type of vaccine administered. In contrast, in naïve COVID‐19 patients, only 25% and 33% of individuals vaccinated with either CanSinoBio or Pfizer‐BioNTech, respectively, produced IgG, a rather poor response. Four weeks after the first dose of CanSinoBio or 2 weeks after the second dose of Pfizer‐BioNTech, the IgG response in the convalescent COVID‐19 group remained robust, with a median of 14 in both groups. However, in the naïve COVID‐19 group, differences were observed. In the group vaccinated with CanSinoBio, 17.85% (five of 28) did not present IgG, and the response in the positive individuals had a median of 2.940 (IQR: 1.28–4.823). These results were significantly different from those of the convalescent COVID‐19 (p = .0001) group. In contrast, 2 weeks after the second dose of the Pfizer‐BioNTech vaccine, all individuals in the naïve COVID‐19 group had IgG with a median of 12.45; IQR 10.85–13.83. These results did not differ significantly from those of the convalescent COVID‐19 group (p = .334).
The IgA response was quite similar to the IgG response. Two weeks after the first dose, a moderate IgA response was observed in most of the convalescent COVID‐19 group in contrast to the naïve COVID‐19 group. The median IgA levels in the convalescent COVID‐19 group vaccinated with CanSinoBio were 5.983 (IQR 2.122–9.447) and 0.855 (IQR 0.579–1.389) in the naïve COVID‐19 group. Similarly, the convalescent COVID‐19 group vaccinated with Pfizer‐BioNTech had a median IgA of 6.84 (IQR 4.700–9.490) and 1.005 (IQR 0.500–2.138) in the naïve COVID‐19 group. After 4 weeks, the IgA response did not change in the convalescent COVID‐19 group vaccinated with CanSinoBio. However, in the naïve COVID‐19 group, more individuals were IgA positive (75%), with a median of 2.900 (IQR 0.035–4.590). Two weeks after the second dose of Pfizer‐BioNTech, the IgA in the convalescent COVID‐19 group did not change. In contrast, an increase in IgA was observed in the naïve COVID‐19 group (median 5.760; IQR 4.150–8.335). Only one person remained IgA negative. Interestingly, the IgM antibody response was low in all groups regardless of the vaccine administered.
Finally, a comparison between the naïve COVID‐19 group vaccinated with CanSinoBio and Pfizer‐BioNTech is shown in Figure 6. Two weeks after vaccination, no differences were observed in the IgG or IgA response. However, after completing the vaccination scheme for both vaccines, IgG and IgA were significantly higher (p = .0001 and p = .0004, respectively) in individuals vaccinated with Pfizer‐BioNTech than in those vaccinated with CanSinoBio.
FIGURE 6.
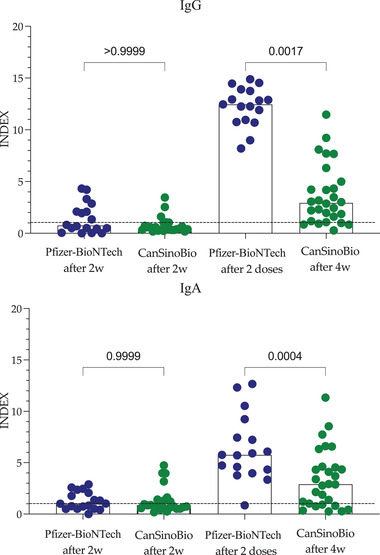
IgG and IgA antibodies of individuals naïve to COVID‐19 in response to CanSinoBio or Pfizer‐BioNTech vaccine. Bars represent the median, and each circle represents an individual. Differences were analyzed with the Kruskal–Wallis test and Dunn's multiple comparisons test. p values indicate the response of individuals with a history of COVID‐19 versus naïve to COVID‐19. INDEX values > 1 are positive
4. DISCUSSION
In this work, we standardized an indirect ELISA to detect IgM, IgG and IgA in sera from convalescent donors and evaluated the antibody persistence and the response of a group of patients with a history of COVID‐19 or those naïve to COVID‐19 vaccinated with either Pfizer‐BioNTech or CanSinoBio. Previous results have shown that most B‐cell epitopes identified have been found in the S1 domain. Only one of them recognized the amino acids missing from our recombinant S1 (657–664) (Dawood et al., 2020). In addition, approximately 90% of neutralizing antibodies are induced by the receptor‐binding domain (amino acids 306–527) (Piccoli et al., 2020). In contrast, the S2 protein is more conserved within other coronaviruses. These results confirm that the S1 protein is an excellent option for antibody detection. In terms of yield, we obtained a higher performance than the efficiency reported for other SARS‐CoV‐2 antigens, such as the whole spike protein (5 mg/L) produced in the same expression system (Amanat et al., 2020).
The specificity and sensitivity of this S1 ELISA were evaluated with 145 samples collected before December 2019 and with 142 samples from convalescent patients. The specificity for IgG and IgA was similar at 97.22% (CI, 93.04% – 99.24%) and 97.73% (CI, 92.02% – 99.72%), respectively. For IgM, the specificity was 100.00% (95.89% – 100%). These results are similar to those observed in other studies (Krähling et al., 2021). However, one limitation was that we were unable to evaluate the cross‐reactivity against other coronaviruses. The sensitivity for IgG was 94.37% (CI, 89.20% – 97.54%), and eight of 142 positive samples were negative. Interestingly, one of these negative samples was positive for IgA. The remaining seven samples were negative for IgG, IgA and IgM. All positive samples were selected from donors of convalescent plasma, and all were PCR‐confirmed for COVID‐19. The lack of antibodies in SARS‐CoV‐2‐infected patients has been reported previously (Marklund et al., 2020) and occurs in patients with mild clinical manifestations of COVID‐19. Unfortunately, we did not have access to the clinical history of these patients to confirm this observation. In IgA, the sensitivity was calculated to be 76.06% (68.18–82.82%), and 34 of 142 positive samples were negative. In contrast to IgG, the IgA antibody response in sera is inconsistent, explaining the high number of negative results observed in the positive samples. Sterlin et al. (2021) showed that IgA antibodies appear before IgG and that IgA antibodies disappear after 28 days. For IgM, the sensitivity was 59.15% (50.60–67.32) because 84 of 142 positive samples were negative. This was most likely because the patients had 3‐6 weeks elapse since the onset of the symptoms, and it has been reported that IgM decrease after the first 7 weeks (Padoan et al., 2020; Zhou et al., 2021). An additional analysis was performed with sera from patients with < 15 days since symptom onset and without IgG to explore this hypothesis further. In this case, the sensitivity increased to 78.12% (CI, 60.03% – 90.72%). These values are similar to other reports (Okba et al., 2020; Suhandynata et al., 2020). In conclusion, these results can support the accuracy of the S1 ELISA.
The dynamics of the antibody response after SARS‐CoV‐2 infection have been explored in several studies. Many of them have evaluated antibodies, especially IgG, in periods ranging from 6 to 8 months. In most, the results confirmed the presence of IgG. A small number of studies have evaluated the dynamics at 10 months (Dan et al., 2021; Liu et al., 2021). Our study evaluated IgG, IgA and IgM at three different time points. The longest was at approximately 10 months and confirmed that IgG could be detected at this time point in some patients. IgG has been used as a marker for past infection. These results confirm that in patients without vaccines, IgG can be detected until the 10th month after symptom onset. Currently, with the massive use of vaccines, the dynamics of IgG could be different. Interestingly, five of our patients received the vaccine just before the third blood sampling, and an increase in IgG antibodies was obvious. Interestingly, IgA and IgM were not detected at longer periods, in agreement with previous reports (Padoan et al., 2020).
Today, several reports support the efficacy of Pfizer‐BioNTech, in contrast to CanSinoBio. However, the available information is limited (Zhu et al., 2020). In this study, we confirmed that hybrid immunity is induced by different vaccines, including CanSinoBio. Our results showed that vaccination of convalescent COVID‐19 individuals triggered a robust response of IgG 2 weeks after vaccination. This response was observed in the groups vaccinated with Pfizer‐BioNTech and with CanSinoBio. We did not observe significant differences between the vaccines in this situation. These results demonstrated that one dose of CanSinoBio can refresh and reinforce the immunity induced by the infection, similar to other vaccines, such as Pfizer‐BioNTech (Saadat et al., 2021) . However, in naïve COVID‐19 persons, one dose of CanSinoBio induced a significantly lower response to IgG and IgA antibodies than vaccination with two doses of Pfizer‐BioNTech. Another important observation was that at 4 weeks after a single dose of CanSinoBio, 18% of the individuals remained antibody negative. We decided to evaluate the response at 4 weeks after vaccination because previous reports showed that at that point the vaccine induced a robust IgG response (Zhu et al., 2020). Unfortunately, we were not able to evaluate the presence of neutralizing antibodies in this study. However, several studies agreed that there is a correlation between the levels of IgG and neutralizing antibodies. Even with this limitation, it is possible to state that one dose of CanSinoBio in naïve COVID‐19 individuals induces moderate immunity, which is associated with the observed moderate (65%) efficacy. It is probable that a second dose of CanSinoBio could increase immunity, as well as its efficacy. These considerations are important because the new variants of the virus represent a challenge to our immunity.
In this study, we verified that in individuals with previous SARS‐CoV‐2 infection, a single dose of vaccine induces a response that can be immunologically equivalent to a full vaccine schedule in naïve individuals. Several authors have confirmed that one dose of vaccine in previously infected patients induces a strong antibody response and that naïve patients require a second dose of vaccine to induce a robust response to antibodies, in contrast to recovered individuals (Callegaro et al., 2021; Ciccone et al., 2021; Ma et al., 2020). The results obtained in our report are in agreement with these observations. In previous reports, Wang et al. (2021) showed that IgA antibody responses were low compared with IgG and that IgM was lower in patients vaccinated with RNA vaccines (Wang et al., 2021). In agreement with these results, we observed that the IgM response was low for both vaccines. This is different from the IgM response in naturally infected patients, where IgM is produced at higher levels. These results could suggest that proteins other than S are involved in the induction of IgM and that viral infection activates an additional immunological mechanism that produces a high response of IgM, in contrast to vaccines.
In conclusion, this study developed an indirect ELISA to detect IgG, IgA and IgM against SARS‐CoV‐2 spike protein S1. With this assay, it was possible to demonstrate that the persistence of antibodies continues for up to 10 months for IgG antibodies, in contrast to IgA and IgM. Additionally, we demonstrated that in individuals with a history of COVID‐19, the CanSinoBio vaccine induced a robust immune response to IgG, similar to that reported for Pfizer‐BioNTech. We also showed that in persons without a previous infection of COVID‐19, CanSinoBio induced a moderate response to antibodies after 4 weeks. These results highlight the importance of more studies to improve immunity in those vaccinated with CanSinoBio who had no history of COVID‐19 infection.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
Conceptualization, J.H.; methodology, E.A.M.G., D.H.T., M.R., S.H.V.; validation, M.G., and M.B.‐P.; formal analysis, J.H., E.A.M.G., D.H.T.; resources, A.A., O.V., E.V., A.S.‐G., F.P.‐J., L.V.; data curation, J.H., E.A.M.G., D.H.T., S.H.‐V.; writing—original draft preparation, J.H.; writing—review and editing, E.A.M.G., V.M.‐H.,; supervision, J.H.; project administration, J.H.; funding acquisition, J.H. All authors have read and agreed to the published version of the manuscript.
ETHICS STATEMENT
The authors confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to. The study was approved by the Ethics Committee of CIAD (Decision number: CONBIOÉTICA‐26‐CEI‐001‐20200122).
Supporting information
Supporting Information
Supporting Information
ACKNOWLEDGEMENTS
The authors are grateful to Nilza Cordova and Julia Real for their valuable technical assistance. This research was funded by Consejo Nacional de Ciencia y Tecnología (CONACyT), grant number 314320.
Melgoza‐González, E. A. , Hinojosa‐Trujillo, D. , Reséndiz‐Sandoval, M. , Mata‐Haro, V. , Hernández‐Valenzuela, S. , García‐Vega, M. , Bravo‐Parra, M. , Arvizu‐Flores, A. A. , Valenzuela, O. , Velázquez, E. , Soto‐Gaxiola, A. , Gómez‐Meza, M. B. , Pérez‐Jacobo, F. , Villela, L. , & Hernández, J. (2022). Analysis of IgG, IgA, and IgM antibodies against SARS‐CoV‐2 spike protein S1 in convalescent and vaccinated patients with the Pfizer‐BioNTech and CanSinoBio vaccines. Transboundary and Emerging Diseases, 69, e734–e745. 10.1111/tbed.14344
DATA AVAILABILITY STATEMENT
Raw data in this study are available from the corresponding authors on request. All requests for raw and analyzed data and materials will be reviewed by the corresponding authors to verify whether the request is subject to any intellectual property or confidentiality obligations.
REFERENCES
- Amanat, F. , Stadlbauer, D. , Strohmeier, S. , Nguyen, T. H. O. , Chromikova, V. , McMahon, M. , Jiang, K. , Arunkumar, G. A. , Jurczyszak, D. , Polanco, J. , Bermudez‐Gonzalez, M. , Kleiner, G. , Aydillo, T. , Miorin, L. , Fierer, D. S. , Lugo, L. A. , Kojic, E. M. , Stoever, J. , Liu, S. T. H. , … Krammer, F. (2020). A serological assay to detect SARS‐CoV‐2 seroconversion in humans. Nature Medicine, 26(7), 1033‐1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callegaro, A. , Borleri, D. , Farina, C. , Napolitano, G. , Valenti, D. , Rizzi, M. , & Maggiolo, F. (2021). Antibody response to SARS‐CoV‐2 vaccination is extremely vivacious in subjects with previous SARS‐CoV‐2 infection. Journal of Medical Virology, 93, 4612–4615. 10.1002/jmv.26982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccone, E. J. , Zhu, D. R. , Ajeen, R. , Lodge, E. K. , Shook‐Sa, B. E. , Boyce, R. M. , & Aiello, A. E . (2021). SARS‐CoV‐2 seropositivity after infection and antibody response to mRNA‐based vaccination (Preprint). medRxiv, 10.1101/2021.02.09.21251319 [DOI] [Google Scholar]
- Dan, J. M. , Mateus, J. , Kato, Y. , Hastie, K. M. , Yu, E. D. , Faliti, C. E. , Grifoni, A. , Ramirez, S. I. , Haupt, S. , Frazier, A. , Nakao, C. , Rayaprolu, V. , Rawlings, S. A. , Peters, B. , Krammer, F. , Simon, V. , Saphire, E. O. , Smith, D. M. , Weiskopf, D. , … Crotty, S. (2021). Immunological memory to SARS‐CoV‐2 assessed for up to 8 months after infection. Science, 371(6529), 10.1126/science.abf4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawood, R. M. , El‐Meguid, M. A. , Salum, G. M. , El‐Wakeel, K. , Shemis, M. , & Awady, E. l. , & M, K. (2020). Bioinformatics prediction of B and T cell epitopes within the spike and nucleocapsid proteins of SARS‐CoV2. Journal of Infection and Public Health, 2, 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudreuilh, C. , Roper, T. , Breen, C. , Chowdhury, P. , Douthwaite, S. , Kumar, N. , & Moutzouris, D. A . (2021). IgG SARS‐CoV‐2 antibodies persist at least for 10 months in patients on hemodialysis. Kidney International Reports, 6(7): 1961–1964. 10.1016/j.ekir.2021.03.900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EWHO (2020). Novel Coronavirus – Republic of Korea (ex‐China). https://www.who.int/docs/default‐source/coronaviruse/situation‐reports/20200121‐sitrep‐1‐2019‐ncov.pdf
- Forni, G. , & Mantovani, A. (2021). COVID‐19 vaccines: Where we stand and challenges ahead. Cell Death and Differentiation, 28(2), 626‐639. 10.1038/s41418-020-00720-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghinai, I. , McPherson, T. D. , Hunter, J. C. , Kirking, H. L. , Christiansen, D. , Joshi, K. , Rubin, R. , Morales‐Estrada, S. , Black, S. R. , Pacilli, M. , Fricchione, M. J. , Chugh, R. K. , Walblay, K. A. , Ahmed, N. S. , Stoecker, W. C. , Hasan, N. F. , Burdsall, D. P. , Reese, H. E. , Wallace, M. , … Layden, J. E. (2020). First known person‐to‐person transmission of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) in the USA. Lancet, 395, 1137–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herroelen, P. H. , Martens, G. A. , De Smet, D. , Swaerts, K. , & Decavele, A. S . (2020). Humoral immune response to SARS‐CoV‐2. American Journal of Clinical Pathology, 154(5), 610‐619. 10.1093/ajcp/aqaa140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. H. , Marks, F. , & Clemens, J. D . (2021). Looking beyond COVID‐19 vaccine phase 3 trials. Nature Medicine, 27(2), 205‐211. 10.1038/s41591-021-01230-y [DOI] [PubMed] [Google Scholar]
- Krähling, V. , Halwe, S. , Rohde, C. , Becker, D. , Berghöfer, S. , Dahlke, C. , Eickmann, M. , Ercanoglu, M. S. , Gieselmann, L. , Herwig, A. , Kupke, A. , Müller, H. , Neubauer‐Rädel, P. , Klein, F. , Keller, C. , & Becker, S. (2021). Development and characterization of an indirect ELISA to detect SARS‐CoV‐2 spike protein‐specific antibodies. Journal of Immunological Methods, 490, 112958. 10.1016/j.jim.2021.112958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C. , Yu, X. , Gao, C. , Zhang, L. , Zhai, H. , Hu, Y. , Liu, E. , Wang, Q. , Gao, Y. , Wei, D. , Zhang, D. , Han, Y. , & Zhang, X. (2021). Characterization of antibody responses to SARS‐CoV‐2 in convalescent COVID‐19 patients. Journal of Medical Virology, 93(4), 2227‐2233. 10.1002/jmv.26646 [DOI] [PubMed] [Google Scholar]
- Long, Q. X. , Liu, B. Z. , Deng, H. J. , Wu, G. C. , Deng, K. , Chen, Y. K. , Liao, P. , Qiu, J. F. , Lin, Y. , Cai, X. F. , Wang, D. Q. , Hu, Y. , Ren, J. H. , Tang, N. , Xu, Y. Y. , Yu, L. H. , Mo, Z. , Gong, F. , Zhang, X. L. , … Huang, A. L. (2020). Antibody responses to SARS‐CoV‐2 in patients with COVID‐19. Nature Medicine, 26(6), 845‐848. 10.1038/s41591-020-0897-1 [DOI] [PubMed] [Google Scholar]
- Long, Q. X. , Tang, X. J. , Shi, Q. L. , Li, Q. , Deng, H. J. , Yuan, J. , Hu, J. L. , Xu, W. , Zhang, Y. , Lv, F. J. , Su, K. , Zhang, F. , Gong, J. , Wu, B. , Liu, X. M. , Li, J. J. , Qiu, J. F. , Chen, J. , & Huang, A., L. (2020). Clinical and immunological assessment of asymptomatic SARS‐CoV‐2 infections. Nature Medicine, 26(8), 1200‐1204. 10.1038/s41591-020-0965-6 [DOI] [PubMed] [Google Scholar]
- Ma, H. , Zeng, W. , He, H. , Zhao, D. , Jiang, D. , Zhou, P. , Cheng, L. , Li, Y. , Ma, X. , & Jin, T. (2020). Serum IgA, IgM, and IgG responses in COVID‐19. Cellular & Molecular Immunology, 17(7), 773‐775. 10.1038/s41423-020-0474-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marklund, E. , Leach, S. , Axelsson, H. , Nyström, K. , Norder, H. , Bemark, M. , Angeletti, D. , Lundgren, A. , Nilsson, S. , Andersson, L. M. , Yilmaz, A. , Lindh, M. , Liljeqvist, J. Å. , & Gisslén, M. (2020). Serum‐IgG responses to SARS‐CoV‐2 after mild and severe COVID‐19 infection and analysis of IgG non‐responders. PLoS ONE, 15(10), e0241104. 10.1371/journal.pone.0241104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okba, N. M. A. , Müller, M. A. , Li, W. , Wang, C. , GeurtsvanKessel, C. H. , Corman, V. M. , Lamers, M. M. , Sikkema, R. S. , de Bruin, E. , Chandler, F. D. , Yazdanpanah, Y. , Le Hingrat, Q. , Descamps, D. , Houhou‐Fidouh, N. , Reusken, C. B. E. M. , Bosch, B. J. , Drosten, C. , Koopmans, M. P. G. , & Haagmans, B. L. (2020). Severe acute respiratory syndrome coronavirus 2‐specific antibody responses in coronavirus disease patients. Emerging Infectious Diseases, 26(7), 1478‐1488. 10.3201/eid2607.200841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padoan, A. , Sciacovelli, L. , Basso, D. , Negrini, D. , Zuin, S. , Cosma, C. , Faggian, D. , Matricardi, P. , & Plebani, M. (2020). IgA‐Ab response to spike glycoprotein of SARS‐CoV‐2 in patients with COVID‐19: A longitudinal study. Clinica Chimica Acta, 507, 164‐166. 10.1016/j.cca.2020.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli, L. , Park, Y. ‐ J. , Tortorici, M. A. , Czudnochowski, N. , Walls, A. C. , Beltramello, M. , Silacci‐Fregni, C. , Pinto, D. , Rosen, L. E. , Bowen, J. E. , Acton, O. J. , Jaconi, S. , Guarino, B. , Minola, A. , Zatta, F. , Sprugasci, N. , Bassi, J. , Peter, A. , De Marco, A. , … Bowen, J. E. (2020). Mapping neutralizing and immunodominant sites on the SARS‐CoV‐2 spike receptor‐binding domain by structure‐guided high‐resolution serology. Cell, 183(4), 1024‐1042. e1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, V. , Fischinger, S. , Atyeo, C. , Slein, M. , Loos, C. , Balazs, A. , Astudillo, M. G. , Yang, D. , Wesemann, D. R. , Charles, R. , Lafrate, A. J. , Feldman, J. , Hauser, B. , Caradonna, T. , Miller, T. E. , Murali, M. R. , Baden, L. , Nilles, E. , Ryan, E. , … Alter, G. (2020). SARS‐CoV‐2‐specific ELISA development. Journal of Immunological Methods, 484‐485, 112832. 10.1016/j.jim.2020.112832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadat, S. , Tehrani, Z. R. , Logue, J. , Newman, M. , Frieman, M. B. , Harris, A. D. , & Sajadi, M. M. (2021). Binding and neutralization antibody titers after a single vaccine dose in health care workers previously infected with SARS‐CoV‐2. JAMA, 325(14), 1467–1469. 10.1001/jama.2021.3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses . (2020). The species severe acute respiratory syndrome‐related coronavirus: Classifying 2019‐nCoV and naming it SARS‐CoV‐2. Nature Microbiology, 5(4), 536‐544. 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- SS‐México . (2021). Covid‐19 México . https://datos.covid‐19.conacyt.mx
- Stankov, M. V. , Cossmann, A. , Bonifacius, A. , Dopfer‐Jablonka, A. , Ramos, G. M. , Gödecke, N. , Scharff, A. Z. , Happle, C. , Boeck, A. ‐ L. , Tran, A. T. , Pink, I. , Hoeper, M. M. , Blasczyk, R. , Winkler, M. S. , Nehlmeier, I. , Kempf, A. , Hofmann‐Winkler, H. , Hoffmann, M. , Eiz‐Vesper, B. , … Behrens, G. M. N. (2021). Humoral and cellular immune responses against SARS‐CoV‐2 variants and human coronaviruses after single BNT162b2 vaccination. Clinical Infectious Diseases. Advance online publication. 10.1093/cid/ciab555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterlin, D. , Mathian, A. , Miyara, M. , Mohr, A. , Anna, F. , Claër, L. , Quentric, P. , Fadlallah, J. , Devilliers, H. , Ghillani, P. , Gunn, C. , Hockett, R. , Mudumba, S. , Guihot, A. , Luyt, C. E. , Mayaux, J. , Beurton, A. , Fourati, S. , Bruel, T. , … Gorochov, G. (2021). IgA dominates the early neutralizing antibody response to SARS‐CoV‐2. Science Translational Medicine, 13(577), eabd2223. 10.1126/scitranslmed.abd2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhandynata, R. T. , Hoffman, M. A. , Kelner, M. J. , McLawhon, R. W. , Reed, S. L. , & Fitzgerald, R. L . (2020). Longitudinal monitoring of SARS‐CoV‐2 IgM and IgG seropositivity to detect COVID‐19. Journal of Applied Laboratory Medicine, 5(5), 908‐920. 10.1093/jalm/jfaa079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, B. , Feng, Y. , Mo, X. , Zheng, P. , Wang, Q. , Li, P. , Peng, P. , Liu, X. , Chen, Z. , Huang, H. , Zhang, F. , Luo, W. , Niu, X. , Hu, P. , Wang, L. , Peng, H. , Huang, Z. , Feng, L. , Li, F. , … Chen, L. (2020). Kinetics of SARS‐CoV‐2 specific IgM and IgG responses in COVID‐19 patients. Emerging Microbes Infections, 9(1), 940‐948. 10.1080/22221751.2020.1762515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabaud, M. A. , Icard, V. , Milon, M. P. , Bal, A. , Lina, B. , & Escuret, V. (2020). Comparison of eight commercial, high‐throughput, automated or ELISA assays detecting SARS‐CoV‐2 IgG or total antibody. Journal of Clinical Virology, 132, 104613. 10.1016/j.jcv.2020.104613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacunacovid.Gob.Mx . (2021). Vacunación en México . http://vacunacovid.gob.mx/wordpress/informacion‐de‐la‐vacuna/
- Wang, Z. , Schmidt, F. , Weisblum, Y. , Muecksch, F. , Barnes, C. O. , Finkin, S. , Schaefer‐Babajew, D. , Cipolla, M. , Gaebler, C. , Lieberman, J. A. , Oliveira, T. Y. , Yang, Z. , Abernathy, M. E. , Huey‐Tubman, K. E. , Hurley, A. , Turroja, M. , West, K. A. , Gordon, K. , Millard, K. G. , & Nussenzweig, M. C. (2021). mRNA vaccine‐elicited antibodies to SARS‐CoV‐2 and circulating variants. Nature, 592(7855), 616‐622. 10.1038/s41586-021-03324-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . (2020). Novel coronavirus – China . https://www.who.int/csr/don/12‐january‐2020‐novel‐coronavirus‐china/en/
- Zhang, X. , Lu, S. , Li, H. , Wang, Y. , Lu, Z. , Liu, Z. , Lai, Q. , Ji, Y. , Huang, X. , Li, Y. , Sun, J. , Wu, Y. , Xu, X. , & Hou, J. (2020). Viral and antibody kinetics of COVID‐19 patients with different disease severities in acute and convalescent phases: A 6‐month follow‐up study. Virologica Sinica, 35(6), 820‐829. 10.1007/s12250-020-00329-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , Yuan, Q. , Wang, H. , Liu, W. , Liao, X. , Su, Y. , Wang, X. , Yuan, J. , Li, T. , Li, J. , Qian, S. , Hong, C. , Wang, F. , Liu, Y. , Wang, Z. , He, Q. , Li, Z. , He, B. , Zhang, T. , … Zhang, Z. (2020). Antibody responses to SARS‐CoV‐2 in patients of novel coronavirus disease 2019. Clinical Infectious Diseases, 71(16), 2027–2034. 10.1093/cid/ciaa344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, W. , Xu, X. , Chang, Z. , Wang, H. , Zhong, X. , Tong, X. , Liu, T. , & Li, Y. (2021). The dynamic changes of serum IgM and IgG against SARS‐CoV‐2 in patients with COVID‐19. Journal of Medical Virology, 93(2), 924‐933. 10.1002/jmv.26353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, F. C. , Guan, X. H. , Li, Y. H. , Huang, J. Y. , Jiang, T. , Hou, L. H. , Li, J. X. , Yang, B. F. , Wang, L. , Wang, W. J. , Wu, S. P. , Wang, Z. , Wu, X. H. , Xu, J. J. , Zhang, Z. , Jia, S. Y. , Wang, B. S. , Hu, Y. , Liu, J. J. , … Chen, W. (2020). Immunogenicity and safety of a recombinant adenovirus type‐5‐vectored COVID‐19 vaccine in healthy adults aged 18 years or older: A randomised, double‐blind, placebo‐controlled, phase 2 trial. Lancet, 396(10249), 479‐488. 10.1016/s0140-6736(20)31605-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Data Availability Statement
Raw data in this study are available from the corresponding authors on request. All requests for raw and analyzed data and materials will be reviewed by the corresponding authors to verify whether the request is subject to any intellectual property or confidentiality obligations.


