Abstract
Introduction
Numerous efforts in natural product drug development are reported for the treatment of Coronavirus. Based on the literature, among these natural plants Artemisia annua L. shows some promise for the treatment of SARS‐CoV‐2.
Objective
The main objective of our study was to determine artemisinin content by liquid chromatography electrospray ionisation tandem mass spectrometry (LC‐ESI‐MS/MS), to investigate the in vitro biological activity of artemisinin from the A. annua plants grown in Turkey with various extracted methods, to elaborate in silico activity against SARS‐CoV‐2 using molecular modelling.
Methodology
Twenty‐one different extractions were applied. Direct and sequential extractions studies were compared with ultrasonic assisted maceration, Soxhlet, and ultra‐rapid determined artemisinin active molecules by LC‐ESI‐MS/MS methods. The inhibition of spike protein and main protease (3CL) enzyme activity of SARS‐CoV‐2 virus was assessed by time resolved fluorescence energy transfer (TR‐FRET) assay.
Results
Artemisinin content in the range 0.062–0.066%. Artemisinin showed significant inhibition of 3CL protease activity but not Spike/ACE‐2 binding. The 50% effective concentration (EC50) of artemisinin against SARS‐CoV‐2 Spike pseudovirus was found greater than 50 μM (EC45) in HEK293T cell line whereas the cell viability was 94% of the control (P < 0.01). The immunosuppressive effects of artemisinin on TNF‐α production on both pseudovirus and lipopolysaccharide (LPS)‐induced THP‐1 cells were found significant in a dose dependent manner.
Conclusion
Further studies of these extracts for COVID‐19 treatment will shed light to seek alternative treatment options. Moreover, these natural extracts can be used as an additional treatment option with medicines, as well as prophylactic use can be very beneficial for patients.
Keywords: antiviral activity, Artemisia annua SARS‐CoV‐2, LC‐ESI‐MS/MS, natural products for COVID‐19
Short abstract
In this study, from the locally grown natural product 21 different extracts of Artemisia annua L. are illustrated. Robust analysis and determination method for Artemisia annua extracts using tandem LC‐MS/MS instrument was established and optimized. Promising reasonably good antiviral activity was revealed using SARS‐CoV‐2 pseudo‐neutralization assay. Additionally, comparative in silico molecular modelling studies of artemisinin with three (3) known antiviral agents (manidipine, lercaidipine, and efonidipine) was elaborated in detail.
1. INTRODUCTION
SARS‐CoV‐2 virus, which emerged in China in late 2019 and spread all over the world, caused a global pandemic (COVID‐19). 1 Until now, the total number of cases in the world is 203 million while the number of deaths is 4.3 million. There are unfortunately no specific antiviral drugs for the treatment of the COVID‐19 pandemic that affects the whole world. Although there is more than one vaccine with emergency use authorisation in the world, most of these vaccines seem to be not very effective for the variants. In addition, the right of access to the existing vaccines is not equal in every country, and there are delays in production due to the demand. Moreover, a certain majority reject the vaccination. Considering all these reasons, studies with natural products for the tr
eatment of COVID‐19 are very important.
Owing to the natural and widespread distribution of Artemisia annua L., also known as Sweet Wormwood, in wide and various geographies, and usage patterns of numerous cultures different health problems are revealed. 2 Artemisia annua has antimalarial properties and has been used frequently in traditional Chinese medicine in the treatment of malaria and in high fever since ancient times. In extended ethno‐botanical studies and archaeological studies, it was revealed that A. annua was used as fresh plant water. Artemisinin, the active ingredient of the A. annua plant, has passed World Health Organisation’s (WHO's) recommendations as the first drug of choice in malaria resistant to chloroquine and other treatments or showing cerebral involvement. 3
Studies show that A. annua has a strong antiviral effect. Li et al. tested the ethanolic extract of A. annua and determined that it was effective against coronavirus associated with SARS at a 50% effective concentration (EC50) dose of 34.5 ± 2.6 μg/mL and reported that it could be developed as an antiviral agent in the treatment of coronavirus. 4 In addition, Karamoddini et al. reported that A. annua has the most intense anti‐herpetic effect among the methanolic extracts homogenised of various Artemisia species in vitro. 5
Inflammatory response is extremely important in SARS‐CoV‐2 infection as in many diseases. It has been suggested that a severe increase in inflammatory response is observed in patients with SARS‐CoV‐2 infection due to viral replication in the lungs, and the resulting cytokine storm may be closely related to the severity of the disease. 6 , 7 To reduce the inflammatory response shaped in SARS‐CoV‐2 infection, it is recommended in different treatments besides classical anti‐inflammatory drugs. 8 It is emphasised that artemisinin and its analogues have antiviral, antifungal, anticancer and anti‐inflammatory properties in addition to their antimalarial activities. 9 Macrophages play an extremely important role in initiating and regulating the immune response. They control various cytokines they produce in the inflammatory response process through NF‐κB (nuclear factor kappa‐light‐chain‐enhancer of activated B cells). 10 Artemisinin, by regulating transcriptional signalling pathways in macrophages, consequently, reduce cytokine release by macrophages. 11 It has been reported that artemisinin in human monocytes suppresses matrix metallopeptidase 9 (MMP‐9), tumour necrosis factor alpha (TNF‐α) and interleukin‐1 beta (IL‐1β) release by regulating NF‐κB release. 12 , 13 In addition, artemisinin has been shown to have an anti‐inflammatory effect in phorbol 12‐myristate 13‐acetate (PMA)‐treated human THP‐1 cells. 13
Based on the overwhelmingly studies of A. annua extracts in the literature and the recent pandemic conditions we have been inspired to study this plant extract further. To investigate its potential antiviral properties since it is commonly grown in our region and the plant is used as a medicinal tea, especially for treating malaria. Nair et al., researched antiviral activity of dried leaf extracts of seven cultivars of A. annua against SARS‐CoV‐2. They used hot‐water leaf extracts based on artemisinin. Artemisinin has alone an antimalarial effect with an estimated 50% inhibitory concentration (IC50) of approximately 70 μM, while the derivatives of artemisinin, artesunate, artemether and dihydro artemisinin, are inactive or cytotoxic at high micromolar concentrations. 14 Many extraction methods have been investigated in the literature to obtain the artemisinin‐rich fractions. However, the plant used in these studies is A. annua, collected from different places. One of the crucial parameters that affect the active substance (secondary metabolite) content, is the location where the plant is collected. Therefore, it was more meaningful to compare the efficiency of the extraction methods in the literature. Hence, in our study we selected the most accurate and efficient extraction method which is aimed to be applicable to the industrial scale. This research was later supported by in silico molecular modelling as well as in vitro biological activity studies. Our aim and objective were to identify the best extraction method for artemisinin and subsequently investigate its antiviral properties with pseudoneutralisation assay and compare artemisinin binding motifs with known antivirals which have not appeared in the recent literature.
The novelty and advantage of our research from the literature is: (i) the location of where the plants are collected; (ii) the 21 different extraction methods with the comparison of previous studies in the literature; (iii) ultra‐rapid determination of the artemisinin from various A. annua L. extracts by liquid chromatography electrospray ionisation tandem mass spectrometry (LC‐ESI‐MS/MS), (iv) in vitro analysis of four artemisinin rich extracts with the comparison of water extracts where the artemisinin amount is not detected; (v) detailed in vitro SARS‐CoV‐2 pseudovirus neutralisation studies of artemisinin; (vi) comparative in silico molecular modelling studies of artemisinin with three known antiviral agents (manidipine, lercaidipine, and efonidipine).
2. MATERIAL AND METHODS
2.1. Chemicals and standard
The chemicals used in the experiments listed as follows: isopropanol, ethanol, methanol, n‐hexane, chloroform, acetone, dichloroethane, dichloromethane, propylene glycol methyl ether and chloroform were supplied from Merck (Darmstadt, Germany). All chemicals used in this study were of analytical reagent grade. HPLC grade acetonitrile was purchased from Sigma‐Aldrich Chemie GmbH (Steinheim, Germany). Ultra‐pure water was obtained using Direct‐Q UV3 (Millipore, Bedford, MA, USA) system. Artemisinin (C15H22O5 – CAS 63968‐64‐9, 98%) was supplied by Sigma‐Aldrich (St Louis, MO, USA).
2.2. Plant material and extraction
2.2.1. Plant material
The aerial parts of Artemisia annua L. were collected from the coordinates of Bursa‐Uludag (40.178915, 29.141146). Artemisia annua was determined by Prof. Dr. Hulusi Malyer in the herbarium of Bursa Uludag University, with the herbarium code BULU‐28872 (Supporting Information Figure S1). Necessary permissions have been obtained from Bursa Regional Directorate of Forestry, Non‐Wood Products and Services Branch Office for the supply of raw materials to be used in our studies.
2.2.2. Preparation of extracts
Soxhlet extraction, ultrasonic assisted maceration and infusion methods were used to obtain the extract with the highest artemisinin content. Initially, the shade dried and powdered aerial parts of the A. annua were kept in an ultrasonic assisted maceration for 15 minutes and then macerated at room temperature for 24 hours (Table 1). Then, extracts (AA‐2, AA‐3, AA‐5, AA‐6, AA‐7, AA‐8 AA‐11, AA‐12 and AA‐13) were concentrated in vacuo at 40°C. The highest artemisinin content was obtained by ultrasonic assisted maceration using hexane (AA‐5), ethanol (AA‐7) and dichloromethane (AA‐12) solvents. Therefore, extraction was performed by a Soxhlet apparatus using hexane (AA‐4), dichloromethane (AA‐16) and ethanol (AA‐17). In addition to this, considering the artemisinin content in AA‐4 after Soxhlet extraction, ultrasonic assisted maceration was performed with different solvents to evaluate the amount of the artemisinin in the remaining pulp. The extracts (AA‐1, AA‐9, AA‐10, AA−14 and AA‐15) were concentrated in vacuo at 40°C. Water extracts were also obtained (AA‐18) because A. annua leaves are consumed as tea. Hot water at 80°C was added to the powdered aerial parts of the A. annua and then once the sample had reached room temperature it was filtered through cheesecloth. Also, water extracts, AA‐20 and AA‐21 were obtained using the remaining pulp after Soxhlet with dichloromethane and ethanol. All water extracts were lyophilised.
TABLE 1.
Extraction processing of Artemisia annua
| Code of extracts | Solvent | Extraction condition |
|---|---|---|
| AA‐1 | 80% Methanol | Maceration with residual pulp after Soxhlet extraction using petroleum ether |
| AA‐2 | Isopropanol | Ultrasonic assisted maceration (15 minutes) + maceration for 24 hours 15 |
| AA‐3 | 95% Ethanol | Ultrasonic assisted maceration (15 minutes) + maceration for 24 hours 16 |
| AA‐4 | Hexane | Soxhlet 17 |
| AA‐5 | Hexane | Ultrasonic assisted maceration (15 minutes) + maceration for 24 hours 18 |
| AA‐6 | Methanol | Ultrasonic assisted maceration (15 minutes) + maceration for 24 hours |
| AA‐7 | Ethanol | Ultrasonic assisted maceration (15 minutes) + maceration for 24 hours |
| AA‐8 | 80% MeOH | Ultrasonic assisted maceration (15 minutes) + maceration for 24 hours |
| AA‐9 | Ethanol | Maceration with residual pulp after Soxhlet extraction using petroleum ether |
| AA‐10 | Methanol | Maceration with residual pulp after Soxhlet extraction using petroleum ether |
| AA‐11 | Dichloroethane | Ultrasonic assisted maceration (15 minutes) + maceration for 24 hours |
| AA−12 | Dichloromethane | Ultrasonic assisted maceration (15 minutes) + maceration for 24 hours |
| AA‐13 | Propylene glycol methyl ether (PGME) | Ultrasonic assisted maceration (15 minutes) + maceration for 24 hours 19 |
| AA‐14 | Dichloromethane (DCM) | Maceration with residual pulp after Soxhlet extraction using petroleum ether |
| AA−15 | Acetone | Maceration with residual pulp after Soxhlet extraction using petroleum ether |
| AA‐16 | Dichloromethane | Soxhlet |
| AA‐17 | Ethanol | Soxhlet |
| AA‐18 | Water (80°C) | Infusion |
| AA‐19 | Water (80°C) | Maceration with residual pulp after Soxhlet extraction using petroleum ether |
| AA‐20 | Water (80°C) | Maceration with residual pulp after Soxhlet extraction using dichloromethane |
| AA‐21 | Water (80°C) | Maceration with residual pulp after Soxhlet extraction using ethanol |
The solvent plant ratio was chosen as 1:10 and 10 g of dry plant was used in all extraction methods. Considering the different extraction studies in the literature, 21 different extracts were obtained.
2.3. Quantitative analysis of artemisinin using LC‐ESI‐MS/MS
An artemisinin standard was prepared by dissolving in methanol for the analysis of the obtained extracts to determine the amount of artemisinin. The artemisinin standard was used for the determination of individual artemisinin. Initially, 8 mg of artemisinin was dissolved in 2 mL of methanol and 4000 ppm (mg/L) stock was prepared and diluted in the ratio of 1:1 (v/v) (4000/2000/1000/500/250/125/62.50/31.25/15.62/7.81/3.90) range 3.9–62 ng/mL for injected in the LC‐MSMS (Shimadzu brand LC/MS‐8045 Triple Quadrupole, Kyoto, Japan) and a calibration curve was calculated and defined. 20
The regression equation of the calibration curve having R 2 = 0.9998605 was linear from the artemisinin standard concentration range 3.9–62 ng/mL (y = 53952.0x + 21331.4). Limit of detection (LOD) and limit of quantification (LOQ) were defined according to three injections made from each analytical portion. The analysis method has been validated according to a single laboratory validation approach. 21 The LOD, LOQ, and recovery for artemisinin were 1.3 ng/mL and 4.2 ng/mL.
2.4. LC‐ESI‐MS/MS analysis
Artemisia annua extracts with different methods and different solvent systems were prepared as 400 ppb. Artemisia annua extracts were prepared by dissolving in methanol for the analysis of the obtained extracts to determine the amount of artemisinin. Artemisia annua extracts were prepared by dissolving 4 mg pieces in 100 mL of methanol and 40 ppm (mg/L) stock solution was prepared and diluted in the ratio of 1:100 (v/v). Next, 1 mL of stock solution was diluted with 100 mL of methanol. The final concentration of the A. annua extracts was 400 ppb (μg/L) and was injected in the LC‐ESI‐MS/MS (Triple Quadrupole, Shimadzu) and artemisinin content was calculated according to the artemisinin calibration curve. LC‐ESI‐MS/MS data analysis was made with LabSolutions LCMS version 5.91 (Shimadzu) software.
Then the diluted extract was filtered through 0.22 μm syringe filters before quantitative analysis using LC‐ESI‐MS/MS (Triple Quadrupole) with biphenyl column in LC system (2.1 mm × 100 mm inner diameter, 2.7 μm). The LC‐ESI‐MS/MS running condition parameters are: injection volume, 2 μL; furnace temperature, 25°C; total elution time, 5 min; heat block temperature, 400°C; DL temperature, 250°C; ESI voltage, 4.0 kV. Mobile phase: 60% acetonitrile + 40% water (0.1% formic acid).
Mobile phase A consisted of 0.1% (v/v) formic acid in ultra‐pure water. Mobile phase B consisted of acetonitrile. The flow rate was 0.5 mL/min, spraying gas flow rate, 3 mL/min. LC‐MS/MS (Triple Quadrupole) mass spectrophotometer operating in positive ESI‐multiple reaction monitoring (MRM) mode was connected to the chromatographic system. Artemisinin peaks were identified by using the standards.
2.5. Screening for main protease (3CL) enzyme inhibition
Inhibitory molecules against main protease enzyme were screened with 3CL Protease (SARS‐CoV‐2), MBP‐tagged Assay Kit (#79955–2, BPS Bioscience, San Diego, CA, USA). Artemisinin and its derivatives were solubilised to 100 mM stock solutions by using dimethyl sulphoxide (DMSO). Then, 100 nM to 100 μM final concentrations were diluted with assay buffer. Maximum DMSO concentration was 1%. Assay was designed as 384 well plate and 150 ng of 3CL protease enzyme added to each well. GC376 is a well‐known 3CL protease inhibitor, and it was used as an inhibitor control and 50 μg of GC376 was used to compare with artemisinin. Artemisinin concentrations ranging from 100 nm to 100 μM, and an assay buffer was added to each well. Final concentration of 50 μM 3CL protease substrate was added to the mixture and incubated for 4 hours at room temperature. HIDEX Sense Microplate reader was used for fluorescence at 360 nm excitation and 460 nm emission wavelength. Percentage inhibitory activity was assessed as follows: Fluorescence values were subtracted from GC376 inhibitor control fluorescence value and was set as zero percentage activity and the fluorescence value from no inhibitor control was set as 100% activity. The data were evaluated with GraphPad Prism 8.0 (GraphPad, San Diego, CA, USA).
2.6. Screening for Spike/ACE‐2 binding inhibition
Spike and ACE2 binding inhibition were assessed by using SARS‐CoV‐2 Spike: ACE2 Inhibitor Screening Assay (BPS Bioscience, #79931). Artemisinin concentrations were prepared described above for screening by using same stock solutions and working concentrations. 96‐well plate was coated with SARS‐CoV‐2 Spike (S) protein and the plate was incubated overnight at 4°C. After washing the plate with assay buffer and blocking with blocking buffer, the artemisinin compounds were added to Spike protein coated well plate with different concentration and then incubated at room temperature with slow shaking for 3 hours. As final step, chemiluminescence was produced by adding Anti His‐HRP and then HRP substrate and measured by using HIDEX Sense Microplate reader.
2.7. Cell viability and SARS‐CoV‐2 pseudovirus neutralisation assay
2.7.1. Cell viability assay
2.7.1.1. Growth conditions of the cell cultures
Cercopithecus aethiops kidney (Vero E6, CRL1586), human kidney (HEK293T, CRL‐11268) and human lung adenocarcinoma (Calu‐3, HTB‐55 and A549, CCL‐185), and human normal bronchus (Beas‐2B, CRL‐9609), and human colon adenocarcinoma (Caco‐2, HBT‐37 and H1299, CRL‐5803), and mouse subcutaneous connective tissue (L929, CCL‐1) cell lines were received from American Type Culture Collection (ATCC, Manassas, VA, USA). The selected cell lines were grown in Dulbecco's Modified Eagle Medium (DMEM, p# 41965 Gibco) supplemented with 1% antibiotic‐antimycotic and 10% feotal bovine serum (FBS, p# 10500 Gibco) and incubated at 37°C and 5% carbon dioxide (CO2). The cells were harvested and used at 70–80% confluency for the following assays.
2.7.1.2. Cell viability assay
Vero E6, HEK293T, Calu‐3, A549, Beas‐2B, Caco‐2, H1299, and L929 cell lines were seeded (1 × 104 cells/well) and the plates were incubated at 37°C and 5% CO2 for 24 hours. On the day of the experiment, artemisinin in DMEM at 3.15, 6.25, 12.5, 25, 50, 100, and 200 μM concentrations was added and the cells were exposed for 24, 48, and 72 hours. The colorimetric agent 3‐[4,5‐dimethylthiazol‐2‐yl]‐2,5‐diphenyltetrazolium bromide (MTT, p# M5655, Sigma‐Aldrich) in DMEM (5 μg/mL) was placed on the cells for 4 hours. Thereafter, DMSO (p# D 8418, Sigma‐Aldrich) solution in 100 μL was added on the cells which were incubated to dissolve the formazan crystals for 2 hours. The absorbance of each plate was measured at 570 nm and 630 nm used as a reference wavelength using microplate reader (ELX800 UV, Bio‐Tek Instruments Inc., Winooski, VT, USA).
2.7.2. Pseudovirus production and infection
2.7.2.1. Transfection
HEK293T is widely used for retroviral production and highly transfectable cell lines. The pseudovirus involving SARS‐CoV‐2 S was prepared as described by the previous study. 22 Briefly, HEK293T cells were cultured (5 × 105 cells/well) in the six‐well plates. When their confluency reached to 40–60%, HEK293T cells were used for transfection. Fugene‐6 (p# E2691, Promega, Madison, WI, USA) in 10 μL as a transfection agent was added into the basal medium in 100 μL not including FBS and Pen/Strep and incubated for 5 minutes at room temperature. The reporter plasmid (lenti RRL_GFP, 450 ng), the packaging plasmid (psPAX2, 400 ng) (Addgene plasmid p# 12260), and Spike‐18aa truncated (150 ng) (Addgene plasmid p# 149541) plasmid were mixed in another tube and this plasmid was added into the fugene‐6 tube then allowed to incubate at room temperature for 25 to 30 minutes. The transfection mix was added over the cells, which was incubated for 14 to 16 hours. The fresh growth medium, DMEM including 1% Pen/Strep and 10% FBS was replaced with the old medium and the cells were allowed to transfection in the incubator for 48 hours. The pseudoviruses produced by the HEK293T cells were collected and filtered with the syringe filters (0.45 μm) then stored at −80°C until they were used for the infection assay.
2.7.2.2. Infection
For the co‐transfection of the cells, the plasmids of ACE‐2 (Addgene plasmid p#141185) and TMPRSS2 (Addgene plasmid p#145843) using 1250 ng from each plasmid were added on HEK293T cells (5 × 105 cells/well) on the six‐well plate and incubated to render the infection of the cells by pseudoviruses for 48 hours. HEK293T/ACE‐2 cells were subcultured and seeded (2 × 104 cells/well) into the black 96‐well plates and then incubated at 37°C and 5% CO2 for 24 hours. For the infection of the cells, the prepared pseudoviruses (50 μL) and artemisinin (50 μL) from each concentration (12.5, 25, and 50 μM) were added and the tubes were incubated for 60 minutes at room temperature then directly added on the HEK293T/ACE‐2 cells. To determine the infection rate of HEK293T cells with pseudoviruses, the fluorescence intensity of the infected cells due to green fluorescent protein (GFP) reporter plasmids was measured at the excitation and emission wavelengths (485–530 nm) with a fluorescence microplate reader. The neutralisation efficiency of artemisinin at the tested concentrations was determined as the relative fluorescence to the mock‐transfected cells. The cell viability of the control and the treated cells was determined by using 10% of WST‐1 calorimetric agent (p# 11644807001, Roche, Basel, Switzerland) and the absorbance was measured at 450 nm using a microplate reader.
2.7.3. TNF‐α, IL‐8, and IL‐6 enzyme‐linked immunosorbent assay (ELISA)
Human peripheral blood monocyte (THP‐1, ATCC® TIB‐202) and mouse macrophage (J774a.1, ATCC® TIB‐67) cell lines purchased from ATCC. THP‐1 monocyte cells were induced to differentiate into macrophages by 10 nM of PMA (# P8139, Sigma‐Aldrich) for 24 hours. To evaluate immunomodulatory effects of artemisinin on lipopolysaccharide (LPS)/pseudovirus induced human TNF‐α and IL‐8 production of THP‐1, and mouse IL‐6 production of J774a.1 macrophage, they were pretreated with artemisinin at concentrations of 12.5, 25, and 50 μM for 4 hours, and then the cells was treated with 0.2 or 1 mg/mL LPS (# L4391, Sigma‐Aldrich) or pseudovirus and incubated at 37°C and 5% CO2 for 20 hours. Thereafter these treatments, the cell culture supernatants were collected and analysed to determine the cytokines production using enzyme‐linked immunosorbent assay (ELISA) kit as described by the manufacturer's instructions. The level of the each examined cytokine in each treatment was quantified using a standard curve generated with the given standards in each ELISA kit.
2.8. Molecular modelling studies
2.8.1. Preparation of target proteins and the ligand
Before the molecular docking calculations, both ligand structure (artemisinin) and used SARS‐CoV‐2 target protein structures were prepared. The artemisinin three‐dimensional (3D) structure was downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov/), PubChem ID: 68827, and LigPrep was used in ligand preparation with OPLS3 force field. Epik was used in the identification of the protonation states at neutral pH. The 7CWB PDB‐coded dimer structure is used for the main protease which is solved in near physiological temperature. Corresponding target structure for Spike/ACE‐2 interface was 6M0J PDB‐coded protein. For the IL‐6 and IL‐8 targets, 1ALU and 3IL8 PDB‐coded structures were used, respectively. Crucial residues for ligand binding were used in grid‐box generation. All proteins were prepared with Protein preparation tool of Maestro at physiological pH. Missing side chains were fixed with Prime; bond orders are assigned, and hydrogen atoms were added. PROPKA was used in the protonation states of the residues. OPLS3 force field was used in the restrain minimisation with heavy atom convergence of 0.3 Å.
2.8.2. Molecular docking
A grid‐based docking approach (Glide) with standard precision (SP) is used at the docking calculations. Active site of the main protease is determined by centring the grid lattices at the centroid of a set of important residues at the binding pocket (His41, Cys145, and Glu166). Since previous studies highlighted the crucial interactions between Lys417 of Spike RBD and Asp30 of ACE‐2, this region is used at the grid generation before docking. In ligand docking, enhanced conformational sampling for the ligand is conducted and at the selection of initial positions expanding the sampling is also considered.
2.8.3. Molecular mechanics generalised born surface area (MM/GBSA) calculations
Molecular mechanics generalized born surface area (MM/GBSA) binding free energies of compounds were calculated using Prime. VSGB 2.0 solvation model at Prime module of Maestro was utilised for the top‐docking position of compounds. Atoms which are within the 3 Å from ligand were used in optimisation with OPLS3 force field.
2.9. Statistical analysis
The statistical analysis of all assays was evaluated by using GraphPad Prism 7.00 (GraphPad Software, Inc.). For the neutralisation and cell viability assays of HEK293T/ACE‐2 cell line, two‐way analysis of variance (ANOVA) and Dunnett's multiple comparisons tests were used to evaluate statistical significance within each treatment compared with its control group. The significant differences for the cytokine production within each treatment was tested by two‐way ANOVA and Tukey's multiple comparison test. The results for each assay obtained from the three independent experiments were presented as means ± standard deviation (*P < 0.01; **P < 0.001).
3. RESULTS AND DISCUSSION
3.1. Percentage extract yield of artemisinin (gram extract/gram plant)
The effects of different types of solvents and extraction method were investigated to determine the presence of artemisinin contents and yield of extraction. According to the results, highest extract yield was found to be comparatively high in water, a polar solvent, than that in non‐polar solvents which are obtained after Soxhlet extraction with petroleum ether (AA‐19), dichloromethane (AA‐20) and ethanol (AA‐21), the residual pulp was used for infusions with water, at 80°C. The extraction yields expressed relative to the weight of the initial sample (dry by product) ranged from 3.7% to 32.4% and depended on the solvent and type of extraction (Table 2).
TABLE 2.
Percentage artemisinin content of Artemisia annua extracts
| % Extract yield (g extract/g plant) | % Artemisinin | |
|---|---|---|
| AA‐1 | 4.1 | 0.003 ± 0.000 |
| AA‐2 | 14.6 | 0.057 ± 0.002 |
| AA‐3 | 9.1 | 0.065 ± 0.002 |
| AA‐4 | 8.6 | 0.066 ± 0.000 |
| AA‐5 | 9.9 | 0.058 ± 0.001 |
| AA‐6 | 10.6 | 0.047 ± 0.000 |
| AA‐7 | 7.7 | 0.063 ± 0.001 |
| AA‐8 | 5.6 | 0.005 ± 0.000 |
| AA‐9 | 6.5 | 0.041 ± 0.000 |
| AA‐10 | 6.8 | 0.008 ± 0,000 |
| AA‐11 | 10.3 | 0.028 ± 0,001 |
| AA‐12 | 9.4 | 0.061 ± 0.001 |
| AA‐13 | 16.4 | 0.038 ± 0.001 |
| AA−14 | 3.7 | ND |
| AA‐15 | 5.6 | ND |
| AA‐16 | 15.8 | 0.017 ± 0.000 |
| AA‐17 | 13.3 | 0.037 ± 0.001 |
| AA‐18 | 29.0 | 0.001 ± 0.001 |
| AA‐19 | 29.2 | ND |
| AA‐20 | 32.4 | ND |
| AA‐21 | 27.2 | ND |
ND, not determined.
Qiu et al., in their study in 2018, developed and validated an accurate and fast high‐performance liquid chromatography tandem mass spectrometry (HPLC‐MS/MS) test for the simultaneous detection of artemisinin and six synergistic components in A. annua. The run time to analyse a sample was 6.0 minutes. 20 Depending on our method and the parameters of our instrument, the analysis run time is only 5.0 minutes and the retention time of artemisinin is 1.331 minutes. Thus, according to the literature, the ultra‐rapid determination of the artemisinin from various A. annua extracts by LC‐ESI‐MS/MS method was developed.
In the literature, artemisinin has been conventionally isolated from A. annua with organic solvents such as toluene, 23 , 24 n‐hexane, 18 , 25 , 26 petroleum ether, 27 , 28 chloroform, 29 95% ethanol, 16 dichloromethane, 15 , 22 propylene glycol methyl ether, 19 isopropanol, 15 1‐butanol, 30 and water. 14 , 31 , 32
Since the amount of artemisinin found in A. annua is extremely low, different combined systems for increasing the extraction efficiency of this compound, e.g. Soxhlet, ultrasound, microwave assisted extraction and supercritical fluid extraction (SFE) (usually supercritical CO2 extraction) have been used. In recent years, ultrasound‐assisted extraction has been used to predict the best extraction parameters among extraction methods using various computational tools and mathematical modelling. 30
Ultrasonic extraction, known as a non‐thermal extraction method, shortens the processing time, therefore, provides a higher purity product, reduces energy consumption and this extraction method is an environmentally friendly technology with less solvent usage. Most of the studies have been conducted with ultrasonic probe extraction. Some researchers claim that heat treatment, although commonly used, has some negative effects on the nutrient content. Ultrasound‐assisted extraction uses ultrasound or ultrasonic agitation to increase the extraction efficiency from a solid matrix using a solvent or solvent mixture. There are studies using ultrasound‐assisted artemisinin isolation. 16 , 19 , 24 , 30 , 33
Zhang et al. extracted artemisinin from A. annua using a series of mono ether‐based solvents with an ultrasound‐assisted extraction system. Propylene glycol methyl ether (PGME) was found to be the most suitable as it is safe and of low toxicity. 19
3.2. Percentage yield of artemisinin
The regression equation of the calibration curve was y = 53952.0x + 21331.4, with R 2 = 0.9998605 (Figure S2). Slope (S) and standard deviation (δ) of the response were determined for the calibration curve. LOQ and LOD were calculated. 3 injections were made from each analytical portion. The analysis method has been validated according to a single laboratory validation approach. 21 The LOD, LOQ, and recovery for artemisinin were 1.3 ng/mL and 4.2 ng/mL (Tables 3 and 4).
TABLE 3.
Liquid chromatography electrospray ionisation tandem mass spectrometry (LC‐ESI‐MS/MS) parameters of artemisinin
| Component | Parent ion (m/z) | Precursor ion (m/z) | Product ions for identification (m/z) | Q1 (V) | Collision energy (CE) | Q3 (V) | Ionisation type |
|---|---|---|---|---|---|---|---|
| Artemisinin | 283.15 | 247.15 (CE: −9) | 209.2 | ‐14 | −12 | ‐15 | + |
| 201.2 | ‐14 | ‐12 | −22 | + | |||
| 173.1 | −14 | −16 | −18 | + | |||
| 151.3 | −19 | −18 | −29 | + | |||
| 95.0 | −30 | −28 | −19 | + |
TABLE 4.
Linear ranges, limits of detection (LODs) and limits of quantification (LOQs) of artemisinin in Artemisia annua determined by liquid chromatography electrospray ionisation tandem mass spectrometry (LC‐ESI‐MS/MS)
| Calibration curves | Correlation coefficient (R) | Range (ng/mL) | LOD (ng/mL) | LOQ (ng/mL) | |
|---|---|---|---|---|---|
| Artemisinin | y = 53952.0x + 21331.4 | 0.9998605 | 3.9–62.5 | 1.3 | 4.2 |
The extraction solvent is a very important factor in the recovery of artemisinin from A. annua so extraction is performed in solvents of different polarity as no single solvent may be reliable. Hexanes, 95% ethanol and isopropanol were identified as the most effective solvents for the extraction, resulting in the highest artemisinin content in the range 0.062–0.066% (Table 1).
Although water as the extraction solvent provided the highest extraction yield, artemisinin content was not determined. In a previous report, water used as solvent in the extraction of A. annua plant did not extract artemisinin, confirming the low solubility of artemisinin in water. 31 , 32 While artemisinin could not be determined by LC‐MS/MS in water extracts, it was reported that these extracts were effective against SARS‐CoV‐2. This result shows that the antiviral effect is probably not due to artemisinin alone. Based on this research nine artemisinin‐related compounds were investigated for their potential anti‐SARS‐CoV‐2 activities. Among these compounds were artesunate, arteannuin B, and Lumefantrine which demonstrated reasonably good antiviral activities against SARS‐CoV‐2. 34
Discovery of alternative sources of artemisinin in seven Artemisia species from Tajikistan, hexane was used with ultrasound‐assisted extraction after content of artemisinin was determined by HPLC based on dry mass of Artemisia species samples. Artemisia annua were sonicated in an ultrasonic bath at a frequency of 35 kHz for 15 minutes at room temperature after mixtures were allowed to stand for 12 hours at room temperature. Content of artemisinin were observed in different species, ranged between 0.07% and 0.45% (% dry weight plant). 18
During a vegetation period in Vietnam, where this species is indigenous, researchers followed the development of the artemisinin content of A. annua plants. Leaf, bud, flower or post‐flowering flower and fruit samples were taken at different times: vegetative (5, 6 and 8 months old), collective bud formation (9 months), in full bloom (10 months) and post‐flowering (10 months). Leaves of 5‐month‐old plants had the highest artemisinin content (0.86% dry weight) and leaf yield, then the content of artemisinin gradually decreased. 29
In the literature artemisinin is found in the range 0.01–1.4% in various parts of A. annua, including leaves, flowers, and buds. 35 Artemisia annua contains 0.06–0.50% artemisinin in natural habitats, but there are species in which the amount of artemisinin increases up to 2.0%. In another study, Peng et al. stated that the content of artemisinin of 16 lines produced by the seed of the A. annua variety varied from 0.2% to 0.9% by both gas chromatography flame ionisation detector (GC‐FID) and HPLC‐evaporative light scattering detector (ELSD). 25 The study of ElSohly et al. stated that artemisinin (0.138%, 0.140 and 0.153%) reached its maximum levels before the flowering period in A. annua cultivars analysed by HPLC. 26
Another reason why the content of artemisinin is different from the literature is the plant location. In the study conducted by Avula et al. in 2009, the artemisinin contents of A. annua samples collected from two different regions of Leszek Vincent province in South Africa were found to be 0.0687 ± 0133 and 0.015 ± 0.197 (%, w/w). 36
In another study, 11.6 mg of artemisinin/g of the feed had been obtained from 16.25% ethyl alcohol with supercritical carbon dioxide (SC‐CO2) extraction in 2 hours. More and pure artemisinin was obtained with 2 hours of ethanol modified SC‐CO2 extraction compared to 16 hours of Soxhlet n‐hexane extraction. 37
There are studies conducted with Artemisia species grown in our country such as Çoşge Senkal et al. In their study, essential oils were obtained by the water distillation method from A. annua. The chemical composition was determined using GC and GC‐MS. The essential oil contents were 0.80%, 0.96%, 1.22% and 1.38% in the harvested before flowering (BF), 50% of flowering (50%F), full flowering (FF), and after flowering (AF) stages, respectively. It is shown that the different harvest stages had a significant effect on the percentage, chemical composition, and antioxidant capacity of the essential oil from A. annua. While the essential oil was found to be low in the CD stage (0.80%), it increased in the later harvest stages (0.96%, 1.22 and 1.38% for the 50% W, LO, and LW stages, respectively). 38
In the study of Erdemoğlu et al., A. annua is as an alternative to ten different types of collected Artemisia species from different localities throughout Turkey (A. santonic L., A. taurica Willd., A. spiciger K. Koch, A. herba‐alba Asso, A. haussknechtii Boiss., A. campestris L., A. araratica Krasch., A. armeniaca Lam., A. austriaca Jacq., and A. abrotanum L.). Artemisinin was quantified using reverse phase HPLC analysis technique. However, the study showed that artemisinin could not be detected in all Artemisia species. 39
3.3. Artemisinin inhibited the 3CL activity but not Spike/ACE‐2 binding
The inhibitory effect of each compound at 100 nM, 1 μM, 10 μM and 100 μM concentrations was tested. Among the tested samples, extracts named AA‐16 and AA‐17 showed significant inhibition of 3CL protease activity reducing the activity of enzyme by 47% and 50% at 100 μM, respectively (Figure 1). The other extracts did not show significant reduction in enzyme activity. However, at higher concentrations the fluorescence obtained from the compounds were much higher than the blanks due to compounds auto fluorescence.
FIGURE 1.
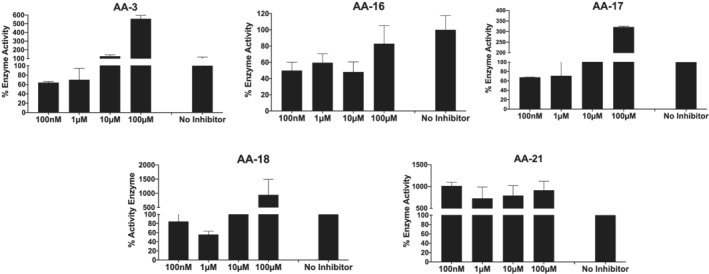
Main protease activity of different Artemisia annua extracts at different concentration. “No Inhibitor” indicates protease activity without any inhibitor molecules as positive control. Each bar represents the SARS‐CoV‐2 3CL protease activity at the different concentration of molecules
Spike inhibition of artemisinin extracts were also examined at different concentrations. Binding activity of SARS‐CoV‐2 S protein to receptor ACE‐2 was evaluated with Spike protein treated with His‐labelled ACE‐2. The binding activity was evaluated as 100% (No Inhibitor) to compare the inhibitors of interaction. AA‐3 and AA‐17 extracts seem to show decrease in percentage binding affinity on dose dependent manner; however, no inhibitors exhibited the inhibitor activity between Spike/ACE‐2 interaction at 10 μM concentration with reference to No Inhibitor control (Figure 2).
FIGURE 2.
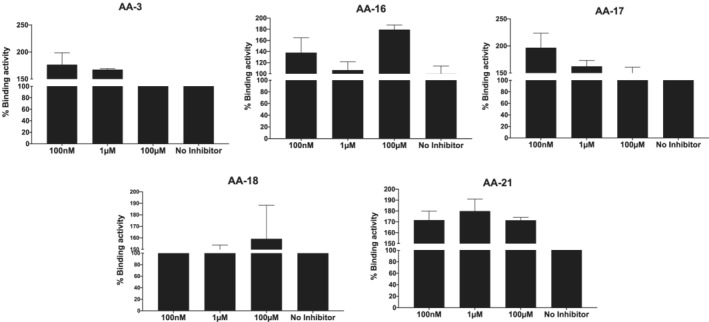
Spike and ACE‐2 binding activity with different Artemisia annua extracts treated at different concentration “No Inhibitor” indicates the binding activity without any inhibitor molecules as positive control
3.4. Cell viability and SARS‐CoV‐2 pseudovirus neutralisation assay
3.4.1. Cell viability
Cytotoxicity assays were carried out before the antiviral activity to determine the non‐cytotoxic ranges for further assays. The viability of Vero E6, HEK293T, Calu‐3, A549, Beas‐2B, Caco‐2, H1299, and L929 cell lines was measured upon 24, 48, and 72 hour exposure to the tested artemisinin concentrations by MTT calorimetric assay. As shown in Figure 3, HEK293T cells showed the most sensitive response upon artemisinin treatment, while the A549, H1299 and L929 cell lines had IC50 values of 114.87 μM, 125.83 μM, 135,56 μM and 197.87 μM, respectively. The viability of Vero E6, Calu‐3, Beas‐2B, and Caco‐2 cell lines was decreased to 77.15–82.96% at the highest concentration of artemisinin (200 μM) after the 72 hour exposure period. The IC50 values of artemisinin against the used cell lines were calculated and presented in Table 5.
FIGURE 3.
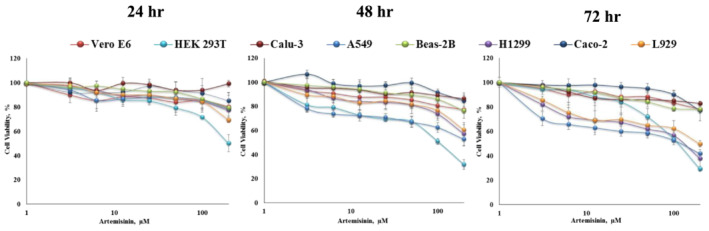
Cell viability of Vero E6, HEK293T, Calu‐3, A549, Beas‐2B, H1299, Caco‐2, and L929 cell lines upon exposure to artemisinin at the concentration range of 1 to 200 μM for 24, 48, and 72 hours. Results were measured by MTT colorimetric assay. Each dose was tested in triplicate and error bars indicate standard error of the mean of triplicates
TABLE 5.
The 50% inhibitory concentration (IC50) values of artemisinin toward the used Vero E6, HEK293T, Calu‐3, A549, Beas‐2, H1299, Caco‐2, and L929 cells lines obtained from dose–response curves. Mean ± standard deviation values were calculated from three independent experiments carried out in triplicate
| Cell lines | IC50 values of artemisinin (μM) | ||
|---|---|---|---|
| 24 hours | 48 hours | 72 hours | |
| Vero E6 | > 200 | > 200 | > 200 |
| HEK293T | > 200 | 104.23 ± 3.20 | 114.87 ± 6.42 |
| Calu‐3 | > 200 | > 200 | > 200 |
| A549 | > 200 | > 200 | 125.83 ± 9.12 |
| Beas‐2 | > 200 | > 200 | > 200 |
| H1299 | > 200 | > 200 | 135.56 ± 10.61 |
| Caco‐2 | > 200 | > 200 | > 200 |
| L929 | > 200 | > 200 | 197.87 ± 12.56 |
The IC50 value of artemisinin using MCF‐231 cell line was reported as 177 μM after 72 hour exposure. 40 As demonstrated in Figure 3, the cytotoxic effects of artemisinin on the examined cell lines were increased with the increasing dose and exposure manner. Cao et al. reported similar result for IC50 values of artemisinin for Vero E6 greater than 200 μM for 48 hour. 30 In another study, the hot water extract of A. annua containing 500 μg/mL artemisinin did not substantially decrease the cell viability of Vero E6 and Calu‐3 cell lines upon 24 hours post‐treatment. 14 The number of cells has been used to investigate antiviral, anticarcinogenic, antimalarial, and cytotoxic effects of artemisinin. A wide range of IC50 values of artemisinin in human cell lines had been reported as 160 mM for hepatoma, HepG2, 41 57.1 μM for cervix cancer (HeLa) and 1602 μM for leukemia, 42 167 μM and 178 μM for osteosarcoma cell lines, MG63 and 48B, 43 respectively. These studies demonstrate that artemisinin caused differential cytotoxic effects depending not only on the concentration and time of exposure but also on the specific target cells.
3.4.2. Pseudovirus neutralisation
Various assays have been implemented to evaluate the neutralisation efficacy of the novel drugs and vaccines on the infected cells with either the native SARS‐CoV‐2 or a pseudo type reporter virus carrying SARS‐CoV‐2 S protein. The ACE‐2 transfected cell culture was treated with therapeutic vaccines/drugs after pre‐incubation period with the virus and the potential neutralisation was measured quantifying either the viral cytopathic effects or the number of infected cells (the production of viral RNA or infectious virus). 44 For safety and versatility, pseudoviruses are very promising tools because of the increased virus infection risks and its influence on the health and economic concerns. The main factors such as cell numbers, cell types, selection of plasmids, virus inoculum in the development stage of the pseudoviruses need to be optimised to increase the efficacy of transfection and the potential infection rate. These factors of the pseudovirus neutralisation assay were optimised to evaluate inhibition potential of the examined artemisinin on the main protease (Mpro) of SARS‐CoV‐2. 45
The selected cell lines, Vero E6, HEK293T, A549, Calu‐3, and Caco‐2 have been widely used for the transduction of pseudovirus studies due to their high level of ACE‐2 expression and their availability for the high level of SARS‐CoV‐2 replication. 40 , 46 , 47 To investigate the inhibition of pseudovirus entry via artemisinin treatment, HEK293T/hACE‐2 cells were pretreated with both artemisinin at 12.5, 25, and 50 μM concentrations and with the pseudovirion of SARS‐CoV‐2. The cell viability was measured by WST‐1 calorimetric assay after 72 hours post‐transduction (Figure 4A). The significant reduction on the cell viability was only observed at the 50 μM concentration of artemisinin compared with the control (P < 0.01). As shown in Figure 4(B), the representative cytopathic effects due to the inhibition of infections at the used concentrations were not observed during neutralisation period. The neutralisation rate of the artemisinin for SARS‐CoV‐2 S pseudoviruses was calculated relative to the control (without any treatment) taken as 100%. The EC50 of artemisinin against SARS‐CoV‐2 S pseudovirus was found greater than 50 μM (EC45) in HEK293T cell line whereas EC50 for hydroxychloroquine sulphate used as a positive control was 25 μM. The cell viability was 94% at the same condition as seen in Figure 4(A). This result demonstrated that artemisinin has almost half of the antiviral efficacy of hydroxychloroquine sulphate. The higher doses of artemisinin (> 50 μM) were not used in this study due to its increased cytotoxicity. As mentioned, Cao et al. also evaluated the antiviral potential of artemisinin against SARS‐CoV‐2 in Vero E6 cell line, which was infected with SARS‐CoV‐2 virus but not with its spike pseudovirus. 30 Thus, EC50 value was reported as 64.45 μM. In another study, Vero E6 was infected with SARS‐CoV‐2 variants and its pseudovirus and EC50 values of artemisinin were reported as 0.1–8.7 μM and 70 μM, 14 respectively. Gilmore et al. tested antiviral activity of extract of A. annua containing artemisinin and EC50 value was reported as 534.4 μM. 48 The differences of EC50 values obtained from these studies and our study could be due to applied different extraction method, SARS‐CoV‐2 variants/pseudovirus, different cell sources, and neutralisation periods.
FIGURE 4.
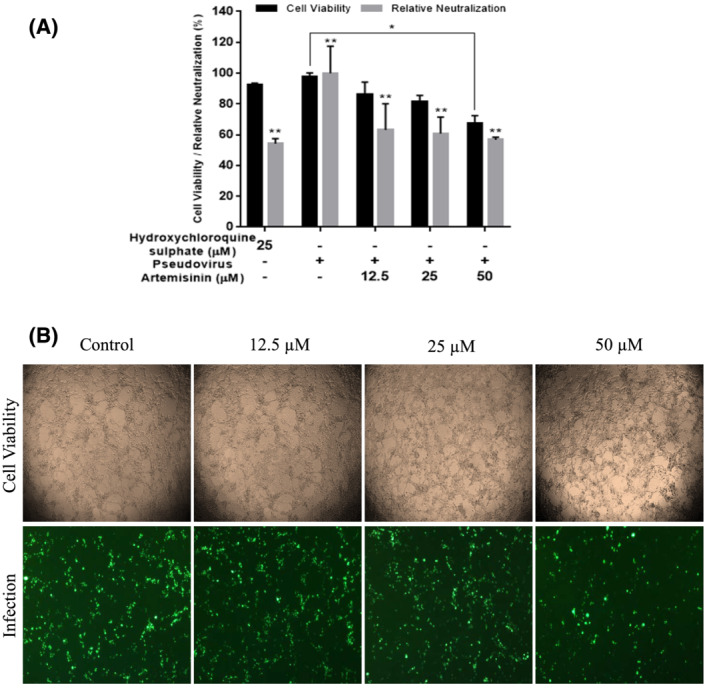
Pseudovirus neutralisation on HEK293T/hACE‐2 by artemisinin. Inhibition of entry of HEK293T/hACE‐2 cells were pretreated at 12.5, 25, and 50 μM concentrations of artemisinin and transduced with SARS‐CoV‐2 S pseudovirion. (A) The cell viability was measured using WST‐1 after 72 hours post‐transduction and the percentage relative neutralisation was calculated as the ratio of fluorescence intensity normalised to the control which is the entry efficiency of SARS‐CoV‐2 pseudoviruses without any treatment was taken as 100%. Hydroxychloroquine sulphate at 25 μM was used as a positive control the results from the three independent experiments were presented as means ± standard deviation (*P < 0.01; **P < 0.001). (B) The cell viability images of HEK293T/hACE‐2 treated with artemisinin and the fluorescence images of the cells transduced with SARS‐CoV‐2 S pseudovirion transfected with GFP
3.4.3. Immunomodulatory effects of artemisinin
As reported in the literature, large amounts of variety of pro‐inflammatory cytokines and chemokines were produced upon the SARS‐CoV‐2 infection by the infected monocytes and macrophages. 49 They stimulate the local tissue inflammation and an adverse systemic inflammatory response known as cytokine storm. Artemisinin might have potential therapeutic interventions to attenuate macrophage‐related inflammatory reactions in possible approaches for COVID‐19 treatment. In this study, the immunomodulatory effect of artemisinin on LPS or SARS‐CoV‐2 pseudovirus induced production of cytokines, TNF‐α, IL‐8, and IL‐6 in the macrophages, THP‐1 and J774a.1 was investigated. First, the cytotoxic effect of artemisinin on LPS or pseudovirus induced THP‐1 and J774a.1 cell line was examined. The cell viability of THP‐1 and J774a.1 cell was not decreased upon any of the treatments as compared with the control within each group (Figure 5A). The immunosuppressive effects of artemisinin at 25 μM and 50 μM doses on TNF‐α production of THP‐1 macrophages on both pseudovirus and LPS‐induced (positive control) was found significant in a dose dependent manner (P < 0.05) (Figure 5B). However, the immunosuppressive effects of artemisinin on the IL‐8 production of THP‐1 macrophages were only seen in the pseudovirus induced treatment (P < 0.05) as compared with the positive control (LPS‐induced) of this group (Figure 5B). IL‐6 production of J774a.1 macrophage was only significantly suppressed in the high level of LPS (1 μg/mL) induced treatment (positive control) (P < 0.05) (Figure 5B). The immunosuppressive effects of artemisinin on TNF‐α, IL‐8, and IL‐6 cytokines production of monocytes and macrophages was confirmed with other studies. Park et al. studied the immunosuppressive effects of artemisinin on LPS‐induced production of TNF‐α which was reduced only at high doses of artemisinin (100 μM). 50 In our study, the immunosuppressive effect of artemisinin on production of TNF‐α on THP‐1 macrophages was significantly observed in both SARS‐CoV‐2 S pseudovirus‐ and LPS‐induced production at 25–50 μM doses of artemisinin. Wang et al. also demonstrated the inhibition of TNF‐α, IL‐1ß, and IL‐6 at the messenger RNA (mRNA) levels in THP‐1 cells. 51 In our study, the immunosuppressive effects of artemisinin on IL‐1ß production of J774a.1 macrophages were examined but there was no significant inhibition observed (data not shown here). The reason for the differences in the results from these studies could be due to the applied different macrophage cell lines. The elevated levels of IL‐1β, IL‐6, IL‐8, and TNF‐α have been detected in COVID‐19 patients during the pandemic. 49 Therefore, the use of immunosuppressive drugs against cytokine and chemokine storm has been very crucial for the treatment of COVID‐19. Artemisinin has been reported being used in a combinational approach along with other antiviral drugs designed to evaluate the neutralisation efficacy of SARS‐CoV‐2. Artemisinin is one of the currently considered therapeutic agents as potential candidate/employed for ongoing trials and the treatment of COVID‐19 disease due to its anti‐inflammatory and immunomodulatory actions. 49
FIGURE 5.
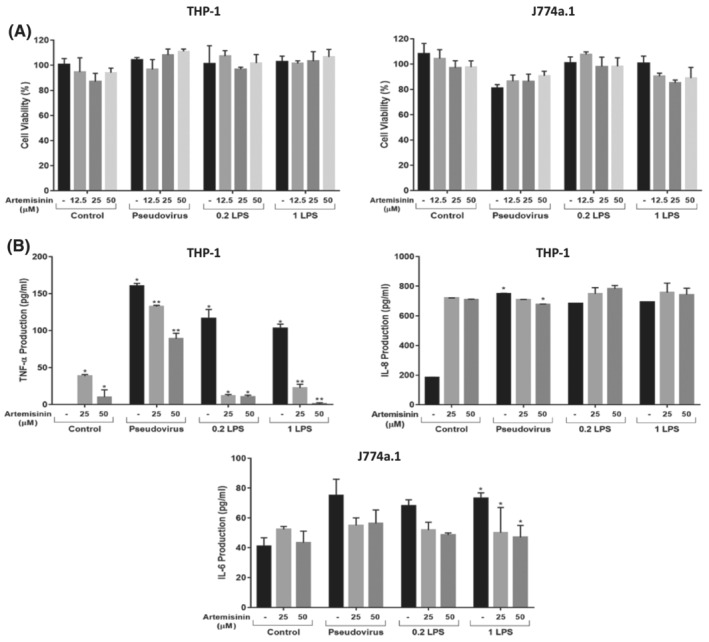
Immunomodulatory effects of artemisinin on proinflammatory cytokine production on LPS or SARS‐CoV‐2 S pseudovirion‐stimulated THP‐1 and J774a.1 macrophage cells. Human THP‐1 differentiated into the macrophages and mouse J774a.1 cells were seeded into the 96 well plates in triplicates (2 × 104/well) and incubated for 24 hours. Later, they were treated with artemisinin at the indicated concentrations for 4 hours, then LPS (μg/ml) or SARS‐CoV‐2 S pseudovirion was added. (A) After 20 hours, WST‐1 calorimetric agent was added and incubated at 37°C and 5% CO2 for 2 hours and the cell viability was measured at 450 nm with a microplate reader. (B) The cell culture supernatant was collected from each treatment and each cytokine level was determined by ELISA. The results from one representative experiment of three independent experiments were presented as means ± standard deviation (*P < 0.01; **P < 0.001)
3.5. Molecular modelling studies
Expanding sampling of conformational search is conducted throughout the molecular docking. Top‐docking scores of artemisinin at the binding pocket of SARS‐CoV‐2 main protease and Spike/ACE‐2 interface were found as −5.21 and −5.06 kcal/mol, respectively. Top‐docking positions were used in MM/GBSA calculations and binding free energies of artemisinin were calculated as −25.84 kcal/mol and −30.61 kcal/mol at main protease and Spike/ACE‐2 interface, respectively. In order to compare the docking scores of artemisinin at the SARS‐CoV‐2 main protease and Spike/ACE‐2 interface, we used reference ligands which are known inhibitors from the literature for these targets. In the work of Ghahremanpour et al. it is stated that calcium channel blockers manidipine, lercanidipine, and efonidipine inhibit activity of main protease enzyme with IC50 values of 4.8 uM, 16.2 uM, and 38.5 uM, respectively. 52 Thus, we docked these known main protease inhibitors with the same docking protocol used in the docking of artemisinin. Top‐docking scores were found as −4.88 kcal/mol, −5.72 kcal/mol, and −5.03 kcal/mol for manidipine, lercanidipine, and efonidipine, respectively. Docking scores of known main protease inhibitors and artemisinin have similar values. Figure 6 shows two‐dimensional (2D) ligand interaction diagrams of these three main protease inhibitors. Top‐docking positions of artemisinin shows that Asn142 forms hydrogen bonds with the ligand at the binding pocket of the main protease. Other residues in contact with the ligand within 3 Å were Thr25, His41, Met49, His163, Glu 166, and Gln189 (Figure 7). Corresponding residues at the Spike/ACE‐2 were Glu23, Lys26, Thr27, and Asp30 from ACE‐2 region and Lys417, Tyr421, Phe456, Arg457, and Tyr473 from Spike region (Figure 8). In order to compare the docking score of artemisinin at the Spike/ACE‐2 interface, its docking score is also compared with the known inhibitor. In the report of Bojadzic et al., it is stated that a drug‐like compound DRI‐C23041 inhibits the interaction of hACE‐2 with Spike protein. 53 Thus, DRI‐C23041 is docked with the same protocol of artemisinin at the Spike/ACE‐2 interface. Its docking score was found as −3.69 kcal/mol which is lower than the corresponding docking score of artemisinin. Figure 9 shows a 2D ligand interaction diagram of DRI‐C23041 at the Spike/ACE‐2 interface. Since experimental studies show promising results for IL‐6 and IL‐8 targets, we also docked artemisinin to these targets and docking scores were found as −3.63 and −4.75 kcal/mol, respectively.
FIGURE 6.
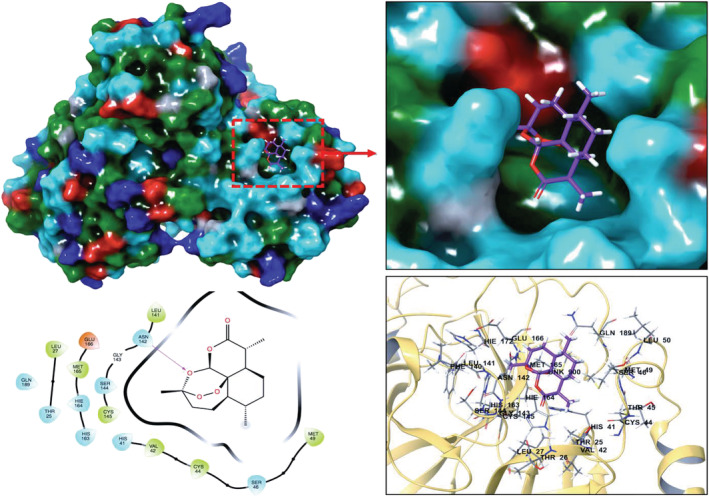
Top‐docking position of artemisinin at the binding pocket of SARS‐CoV‐2 main protease. Two‐dimensional (2D) and three‐dimensional (3D) ligand interaction diagrams are detailed in the figure
FIGURE 7.
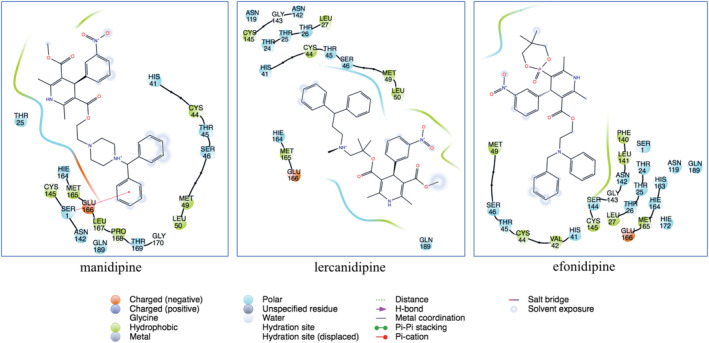
Two‐dimensional (2D) ligand interaction diagrams of known main protease inhibitors (manidipine, lercaidipine, and efonidipine) at the binding pocket of SARS‐CoV‐2 main protease. Top‐docking positions were used
FIGURE 8.
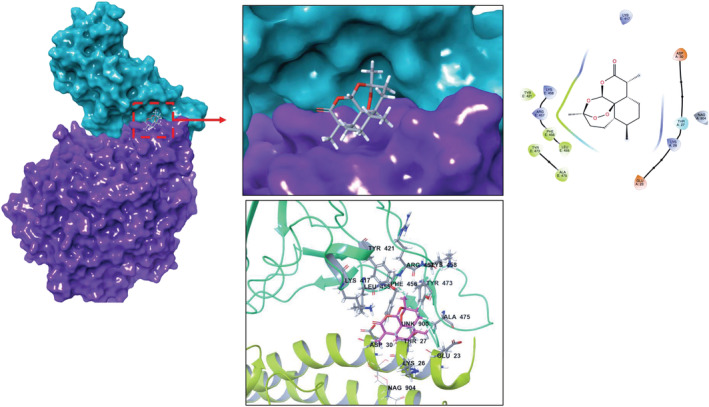
Top‐docking positions of artemisinin at the binding pocket of SARS‐CoV‐2 Spike/ACE‐2 region. Two‐dimensional (2D) and three‐dimensional (3D) ligand interaction diagrams are detailed in the figure
FIGURE 9.
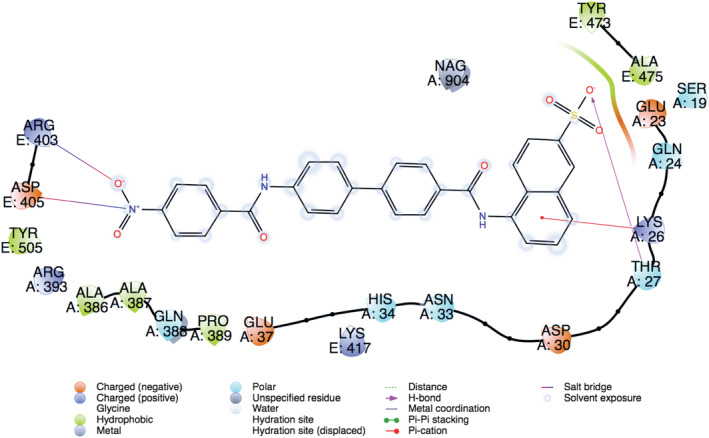
Two‐dimensional (2D) ligand interaction diagram of DRI‐C23041 (a known Spike/ACE‐2 small molecule inhibitor). Top‐docking position is used
There have been tremendous efforts worldwide to find possible cures for COVID‐19. Despite huge worldwide efforts to produce a vaccination to stop it, this deadly pandemic is still a threat to mankind. Thus, the use of prophylactically potential herbal medicines is another approach to prevent this virus spreading. In the future, effective herbal medicines will be very attractive sources as food supplements for healthy lifestyle and free of virus infections. Artemisinin can be a good candidate to develop common immunity since it can grow in most countries in the world. Moreover, this herbal extract can provide extra antiviral properties once used with common antiviral agents as an additional synergetic natural product. As a result of this study, we have demonstrated that local A. annua extracts can be considered a potential antiviral. Further studies need to be conducted especially in vivo SARS‐CoV‐2 challenge experiments will be the following goals to carry out this research on a higher level, which will be reported in due course.
Supporting information
Figure S1. Artemisia annua registered in the herbarium of Bursa Uludag University (Herbarium code: BULU‐28872)
Figure S2. Calibration curve of artemisinin standard and LS‐ESI‐MS/MS mass spectrum
ACKNOWLEDGEMENTS
This study was funded by the Scientific and Technological Research Council of Turkey (TÜBİTAK), within the programme number 18AG003. The authors would like to thank Assoc. Prof. Fatih Tornuk and Assoc. Prof. Murat Ertas for their valuable and helpful discussions as well as providing extracts. The authors also would like to thank to Atabay Pharmaceutical Fine Chemicals Inc. for use of their laboratory and LC‐ESI‐MS/MS. The authors would like to dedicate this research in the honour of deceased Prof. Hulusi Malyer who pioneered A. annua research in Turkey, unfortunately he died in December 2020 from COVID‐19.
Dogan K, Erol E, Didem Orhan M, et al. Instant determination of the artemisinin from various Artemisia annua L. extracts by LC‐ESI‐MS/MS and their in‐silico modelling and in vitro antiviral activity studies against SARS‐CoV‐2. Phytochemical Analysis. 2022;33(2):303-319. doi: 10.1002/pca.3088
Funding information Scientific and Technological Research Council of Turkey, Grant/Award Number: 18AG003
REFERENCES
- 1. Gralinski LE, Menachery VD. Return of the Coronavirus: 2019‐nCoV. Viruses. 2020;12(2):135‐143. 10.3390/v12020135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Udaykumar P. Discovery of artemisinin: The Chinese wonder drug. Muller J Med Sci Res. 2014;5(2):191‐192. 10.4103/0975-9727.135780 [DOI] [Google Scholar]
- 3. Brown GD, Liang GY, Sy LK. Terpenoids from the seeds of Artemisia annua . Phytochemistry. 2003;64(1):303‐323. 10.1016/S0031-9422(03)00294-2 [DOI] [PubMed] [Google Scholar]
- 4. Li SY, Chen C, Zhang HQ, et al. Identification of natural compounds with antiviral activities against SARS‐associated coronavirus. Antiviral Res. 2005;67(1):18‐23. 10.1016/j.antiviral.2005.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Karamoddini MK, Emami SA, Ghannad MS, Sani EA, Sahebkar A. Antiviral activities of aerial subsets of Artemisia species against Herpes Simplex virus type 1 (HSV1) in vitro. Asian Biomedicine. 2011;5(1):63‐68. 10.5372/1905-7415.0501.007 [DOI] [Google Scholar]
- 6. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395(10223):497‐506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wong CK, Lam CWK, Wu AKL, et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004;136(1):95‐103. 10.1111/j.1365-2249.2004.02415.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fu Y, Cheng Y, Wu Y. Understanding SARS‐CoV‐2‐Mediated Inflammatory Responses: From Mechanisms to Potential Therapeutic Tools. Virol Sin. 2020;35(3):266‐271. 10.1007/s12250-020-00207-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yao W, Wang F, Wang H. Immunomodulation of artemisinin and its derivatives. Science Bulletin. 2016;61(18):1399‐1406. 10.1007/s11434-016-1,105-z [DOI] [Google Scholar]
- 10. Fujiwara N, Kobayashi K. Macrophages in inflammation. Curr Drug Targets Inflamm Allergy. 2005;4(3):281‐286. 10.2174/1568010054022024 [DOI] [PubMed] [Google Scholar]
- 11. Pahl HL. Activators and target genes of Rel/NF‐$κ$B transcription factors. Oncogene. 1999;18(49):6853‐6866. 10.1038/sj.onc.1203239 [DOI] [PubMed] [Google Scholar]
- 12. Shakir L, Hussain M, Javeed A, Ashraf M, Riaz A. Artemisinins and immune system. Eur J Pharmacol. 2011;668(1–2):6‐14. 10.1016/j.ejphar.2011.06.044 [DOI] [PubMed] [Google Scholar]
- 13. Prato M, Gallo V, Giribaldi G, Aldieri E, Arese P. Role of the NF‐$κ$B transcription pathway in the haemozoin‐ and 15‐HETE‐mediated activation of matrix metalloproteinase‐9 in human adherent monocytes. Cell Microbiol. 2010;12(12):1780‐1791. 10.1111/j.1462-5822.2010.01508.x [DOI] [PubMed] [Google Scholar]
- 14. Nair MS, Huang Y, Fidock DA, et al. Artemisia annua L. extracts inhibit the in vitro replication of SARS‐CoV‐2 and two of its variants. J Ethnopharmacol. 2021;274:114016. 10.1016/j.jep.2021.114016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Laboukhi‐Khorsi S, Daoud K, Chemat S. Efficient Solvent Selection Approach for High Solubility of Active Phytochemicals: Application for the Extraction of an Antimalarial Compound from Medicinal Plants. ACS Sustainable Chem Eng. 2017;5(5):4332‐4339. 10.1021/acssuschemeng.7b00384 [DOI] [Google Scholar]
- 16. de Silva EO, Borges LL, da Conceição EC, Bara MTF. Box–Behnken experimental design for extraction of artemisinin from Artemisia annua and validation of the assay method. Rev Bras Farm. 2017;27(4):519‐524. 10.1016/j.bjp.2017.03.002 [DOI] [Google Scholar]
- 17. Klayman DL, Lin AJ, Acton N, et al. Isolation of Artemisinin (Qinghaosu) from Artemisia annua Growing in the United States. J Nat Prod. 1984;47(4):715‐717. 10.1021/np50034a027 [DOI] [PubMed] [Google Scholar]
- 18. Numonov S, Sharopov F, Salimov A, et al. Assessment of Artemisinin Contents in Selected Artemisia Species from Tajikistan (Central Asia). Medicines. 2019;6(1):23. 10.3390/medicines6010023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Y, Prawang P, Li C, et al. Ultrasonic assisted extraction of artemisinin from Artemisia annua L. using monoether‐based solvents. Green Chem. 2018;20(3):713‐723. 10.1039/C7GC03191B [DOI] [Google Scholar]
- 20. Qiu F, Wu S, Lu X, et al. Quality evaluation of the artemisinin‐producing plant Artemisia annua L. based on simultaneous quantification of artemisinin and six synergistic components and hierarchical cluster analysis. Ind Crop Prod. 2018;118(March):131‐141. 10.1016/j.indcrop.2018.03.043 [DOI] [Google Scholar]
- 21. Thompson M, Ellison LRS, Wood R. Harmonized guidelines for single‐laboratory validation of methods of analysis. In: Resulting from the Symposium on Harmonization of Quality Assurance Systems for AnalyticalLaboratories; 1999. [Google Scholar]
- 22. Qu H, Christensen KB, Fretté XC, Tian F, Rantanen J, Christensen LP. Chromatography‐Crystallization Hybrid Process for Artemisinin Purification from Artemisia annua . Chem Eng Technol. 2010;33(5):791‐796. 10.1002/ceat.200900575 [DOI] [Google Scholar]
- 23. Kohler M, Haerdi W, Christen P, Veuthey J‐L. Supercritical fluid extraction and chromatography of artemisinin and artemisinic acid. An improved method for the analysis Ofartemisia annua samples. Phytochem Anal. 1997;8(5):223‐227. [DOI] [Google Scholar]
- 24. Mannan A, Ahmed I, Arshad W, et al. Survey of artemisinin production by diverse Artemisia species in northern Pakistan. Malar J. 2010;9(1):310‐319. 10.1186/1475-2,875-9-310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peng CA, Ferreira JFS, Wood AJ. Direct analysis of artemisinin from Artemisia annua L. using high‐performance liquid chromatography with evaporative light scattering detector, and gas chromatography with flame ionization detector. J Chromatogr a. 2006;1133(1–2):254‐258. 10.1016/j.chroma.2006.08.043 [DOI] [PubMed] [Google Scholar]
- 26. ElSohly HN, Croom EM, El‐Feraly FS, El‐Sherei MM. A Large‐Scale Extraction Technique of Artemisinin from Artemisia annua . J Nat Prod. 1990;53(6):1560‐1,564. 10.1021/np50072a026 [DOI] [Google Scholar]
- 27. Qian G, Yang Y, Ren Q. Determination of Artemisinin in Artemisia annua L. by Reversed Phase HPLC. J Liq Chromatogr Relat Technol. 2005;28(5):705‐712. 10.1081/JLC-200048890 [DOI] [Google Scholar]
- 28. Klayman D. Qinghaosu (artemisinin): an antimalarial drug from China. Science. 1985;228(4703):1049‐1,055. 10.1126/science.3887571 [DOI] [PubMed] [Google Scholar]
- 29. Woerdenbag HJ, Pras N, Chan NG, et al. Artemisinin, Related Sesquiterpenes, and Essential Oil in Artemisia annua During a Vegetation Period in Vietnam. Planta Med. 1994;60(3):272‐275. 10.1055/s-2006-959,474 [DOI] [PubMed] [Google Scholar]
- 30. Cao J, Yang M, Cao F, Wang J, Su E. Well‐Designed Hydrophobic Deep Eutectic Solvents As Green and Efficient Media for the Extraction of Artemisinin from Artemisia annua Leaves. ACS Sustain Chem Eng. 2017;5(4):3270‐3,278. 10.1021/acssuschemeng.6b03092 [DOI] [Google Scholar]
- 31. Martinez‐Correa HA, Bitencourt RG, Kayano ACAV, Magalhães PM, Costa FTM, Cabral FA. Integrated extraction process to obtain bioactive extracts of Artemisia annua L. leaves using supercritical CO2, ethanol and water. Ind Crop Prod. 2017;95:535‐542. 10.1016/j.indcrop.2016.11.007 [DOI] [Google Scholar]
- 32. Lapkin AA, Walker A, Sullivan N, Khambay B, Mlambo B, Chemat S. Development of HPLC analytical protocols for quantification of artemisinin in biomass and extracts. J Pharm Biomed Anal. 2009;49(4):908‐915. 10.1016/j.jpba.2009.01.025 [DOI] [PubMed] [Google Scholar]
- 33. Appalasamy S, Lo KY, Ch'ng SJ, Nornadia K, Othman AS, Chan L‐K. Antimicrobial Activity of Artemisinin and Precursor Derived from In Vitro Plantlets of Artemisia annua L. Biomed Res Int. 2014;2014:1‐6. 10.1155/2014/215872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cao R, Hu H, Li Y, et al. Anti‐SARS‐CoV‐2 Potential of Artemisinins In Vitro. ACS Infect Dis. 2020;6(9):2524‐2,531. 10.1021/acsinfecdis.0c00522 [DOI] [PubMed] [Google Scholar]
- 35. Briars R, Paniwnyk L. Effect of ultrasound on the extraction of artemisinin from Artemisia annua . Ind Crop Prod. 2013;42:595‐600. 10.1016/j.indcrop.2012.06.043 [DOI] [Google Scholar]
- 36. Avula B, Wang Y‐H, Smillie TJ, et al. Comparison of LC–UV, LC–ELSD and LC–MS Methods for the Determination of Sesquiterpenoids in Various Species of Artemisia. Chromatographia. 2009;70(5–6):797‐803. 10.1365/s10337-009-1,237-2 [DOI] [Google Scholar]
- 37. Tzeng T, Lin Y, Jong T, Chang C. Ethanol modified supercritical fluids extraction of scopoletin and artemisinin from Artemisia annua L. Sep Purif Technol. 2007;56(1):18‐24. 10.1016/j.seppur.2007.01.010 [DOI] [Google Scholar]
- 38. Çoşge Şenkal B, Kiralan M, Yaman C. The effect of different harvest stages on chemical composition and antioxidant capacity of essential oil from Artemisia annua L. Tarım Bilimleri Dergisi. 2014;21(1):71‐78. 10.15832/tbd.19830 [DOI] [Google Scholar]
- 39. Erdemoğlu N, Orhan İ, Kartal M, Adıgüzel N, Bani B. Determination of artemisinin in selected Artemisia L. species of Turkey by reversed phase HPLC. Records Nat Prod. 2007;20072–3 July–October:36‐43. [Google Scholar]
- 40. Chu H, Chan JF‐W, Yuen TT‐T, et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS‐CoV‐2 and SARS‐CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID‐19: an observational study. The Lancet Microbe. 2020;1(1):e14‐e23. 10.1016/S2666-5247(20)30004-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Romero MR, Efferth T, Serrano MA, et al. Effect of artemisinin/artesunate as inhibitors of hepatitis B virus production in an “in vitro” replicative system. Antiviral Res. 2005;68(2):75‐83. 10.1016/j.antiviral.2005.07.005 [DOI] [PubMed] [Google Scholar]
- 42. Efferth T, Herrmann F, Tahrani A, Wink M. Cytotoxic activity of secondary metabolites derived from Artemisia annua L. towards cancer cells in comparison to its designated active constituent artemisinin. Phytomedicine. 2011;18(11):959‐969. 10.1016/j.phymed.2011.06.008 [DOI] [PubMed] [Google Scholar]
- 43. Jirangkul P, Srisawat P, Punyaratabandhu T, Songpattanaslip T, Mungthin M. Cytotoxic effect of artemisinin and its derivatives on human osteosarcoma cell lines. J Med Assoc Thail = Chotmaihet Thangphaet. 2014;97(Suppl 2):S215‐S221. http://www.ncbi.nlm.nih.gov/pubmed/25518197 [PubMed] [Google Scholar]
- 44. Tan CW, Chia WN, Qin X, et al. A SARS‐CoV‐2 surrogate virus neutralization test based on antibody‐mediated blockage of ACE2–spike protein–protein interaction. Nat Biotechnol. 2020;38(9):1073‐1078. 10.1038/s41587-020-0631-z [DOI] [PubMed] [Google Scholar]
- 45. Yang L, Pei R, Li H, et al. Identification of SARS‐CoV‐2 entry inhibitors among already approved drugs. Acta Pharmacol SinPublished online. 2020;42(8):1347‐1353. 10.1038/s41401-020-00556-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271‐280.e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jennifer H, Azaibi T, Xiaoyan L, et al. Isolation and characterization of SARS‐CoV‐2 from the first US COVID‐19 patient. BioRxiv. 2020:1‐18. 10.1101/2020.03.02.972935 [DOI] [Google Scholar]
- 48. Gilmore K, Zhou Y, Ramirez S, et al. In vitro efficacy of Artemisinin‐based treatments against SARS‐CoV‐2. BioRxiv. 2020:1‐23. 10.1101/2020.10.05.326637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jafarzadeh A, Chauhan P, Saha B, Jafarzadeh S, Nemati M. Contribution of monocytes and macrophages to the local tissue inflammation and cytokine storm in COVID‐19: Lessons from SARS and MERS, and potential therapeutic interventions. Life Sci. 2020;257:118102. 10.1016/j.lfs.2020.118102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Park KH, Yoon YD, Han S‐B, et al. Artemisinin inhibits lipopolysaccharide‐induced interferon‐β production in RAW 264.7 cells: implications on signal transducer and activator of transcription‐1 signaling and nitric oxide production. Int Immunopharmacol. 2012;14(4):580‐584. 10.1016/j.intimp.2012.09.012 [DOI] [PubMed] [Google Scholar]
- 51. Wang Y, Huang Z, Wang L, et al. The anti‐malarial artemisinin inhibits pro‐inflammatory cytokines via the NF‐κB canonical signaling pathway in PMA‐induced THP‐1 monocytes. Int J Mol Med. 2011;27(2):233‐241. 10.3892/ijmm.2010.580 [DOI] [PubMed] [Google Scholar]
- 52. Ghahremanpour MM, Tirado‐Rives J & Deshmukh M et al. Identification of 14 Known Drugs as Inhibitors of the Main Protease of SARS‐CoV‐2. bioRxiv: the preprint server for biology. Published online 2020. Doi: 10.1101/2020.08.28.271957 [DOI] [PMC free article] [PubMed]
- 53. Bojadzic D, Alcazar O, Chen J, et al. Small‐Molecule Inhibitors of the Coronavirus Spike: ACE2 Protein–Protein Interaction as Blockers of Viral Attachment and Entry for SARS‐CoV‐2. ACS Infect Dis. 2021;7(6):1519‐1534. 10.1021/acsinfecdis.1c00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Artemisia annua registered in the herbarium of Bursa Uludag University (Herbarium code: BULU‐28872)
Figure S2. Calibration curve of artemisinin standard and LS‐ESI‐MS/MS mass spectrum


