Abstract
Numerous studies have demonstrated that overexpression or aberrant expression of the HMGI(Y) family of architectural transcription factors is frequently associated with both neoplastic transformation of cells and metastatic tumor progression. Little is known, however, about the molecular roles played by the HMGI(Y) proteins in these events. Here we report that human breast epithelial cells harboring tetracycline-regulated HMGI(Y) transgenes acquire the ability to form both primary and metastatic tumors in nude mice only when the transgenes are actively expressed. Unexpectedly, the HMG-Y, rather than the HMG-I, isoform of these proteins is the most effective elicitor of both neoplastic transformation and metastatic progression in vivo. Furthermore, expression of either antisense or dominant-negative HMGI(Y) constructs inhibits both the rate of proliferation of tumor cells and their ability to grow anchorage independently in soft agar. Array analysis of transcription profiles demonstrates that the HMG-I and HMG-Y isoform proteins each modulate the expression of distinctive constellations of genes known to be involved in signal transduction, cell proliferation, tumor initiation, invasion, migration, induction of angiogenesis, and colonization. Immunohistochemical analyses of tumors formed in nude mice indicate that many have undergone an epithelial-mesenchymal transition in vivo. Together, these findings demonstrate that overexpression of the HMGI(Y) proteins, more specifically, the HMG-Y isoform protein, is causally associated with both neoplastic transformation and metastatic progression and suggest that induction of integrins and their signaling pathways may play significant molecular roles in these biological events.
The mammalian HMG-I, HMG-Y, and HMGI-C proteins are members of the HMGI(Y) family of nonhistone chromatin proteins that have been implicated in both positive and negative regulation of gene transcription in vivo (reviewed in references 14 and 96). The human HMG-I (11.5-kDa) and HMG-Y (10.4-kDa) proteins are produced by translation of alternatively spliced mRNAs coded for by the Hmgiy gene at chromosomal locus 6p21 (39), while the related, but distinct, HMGI-C protein (12 kDa) is coded for by the Hmgi-c gene at chromosomal locus 12q15 (15). Members of the HMGI(Y) family are often referred to as architectural transcription factors because of their ability to regulate gene activity through the recognition and alteration of the structure of DNA and chromatin substrates (14, 100). The HMGI(Y) proteins not only bind with high specificity to the narrow minor groove of A · T-rich, random sequence B-form DNA (97, 107) but also have the ability to bind with high affinity to bent or distorted DNA structures such as synthetic four-way junction DNAs (55, 56), supercoiled plasmid substrates (87), and nucleosome core particles (98, 100). The ability of the HMGI(Y) proteins to bind to a variety of DNA substrates is a consequence of the high degree of intrinsic flexibility of the proteins, particularly within their A · T hook DNA-binding motifs (97) whose structure in a cocomplex with a synthetic substrate has recently been determined (61). In addition to recognizing structure rather than sequence, the HMGI(Y) proteins also have the ability to bend, straighten, unwind, and supercoil DNA substrates (14, 33, 75, 87). It is this unusual combination of substrate-binding characteristics, combined with the ability to specifically interact with other transcription factors, that allows the HMGI(Y) proteins to function as gene-regulatory factors in vivo by participating in the formation of stereospecific multiprotein enhanceosome complexes (66, 122). Among the known transcription factors that interact with HMGI(Y) in vitro or in vivo are NF-κB, ATF-2/c-Jun, Elf-1, Oct-2, Oct-6, SRF, NF-Y, PU-1, RAR, Sp1, NFAT, and others (14, 19, 24, 57, 58, 66, 86, 121). Their many potential protein partners provide the HMGI(Y) proteins with considerable flexibility in regulating the transcriptional activity of a large number of different genes in vivo (14), among which some, such as the intercellular adhesion molecule E-selectin (77), often exhibit aberrant expression patterns in human malignancies (6) whereas others, such as tumor necrosis factor beta (35) and beta interferon (IFN-β) (83), are associated with angiogenesis, a necessary component of solid-tumor formation.
The level of expression of HMGI(Y) mRNAs and proteins is low (68, 69, 82) or undetectable (40, 44) in most differentiated or nonproliferating normal cells but is rapidly induced in response to growth-stimulatory factors such as serum (67, 69), transforming growth factor α (TGF-α) (95), epidermal growth factor (EGF) (59), platelet-derived growth factor, and fibroblast growth factor (FGF) (74), as well as phorbol esters (22, 39), beta 1 interferon (IFN-β1), and endotoxin (91). These observations, combined with the fact that deletion of the Hmgi-c gene results in the diminutive pygmy phenotype in mice (7), implicate the HMGI(Y) proteins in the control of cell proliferation. Consistent with this suggestion, in neoplastically transformed cells, as well as in embryonic cells that have not yet undergone overt differentiation, the constitutive level of HMGI(Y) gene products is often exceptionally high, with increasing concentrations being correlated with increasing degrees of malignancy or metastatic potential (14, 40, 44, 116). This correlation is so consistent and widespread in tumors that it has been suggested that elevated concentrations of HMGI(Y) proteins are diagnostic markers of both neoplastic transformation (43, 44) and increased metastatic potential (13, 117). For example, HMGI(Y) overexpression has been suggested to be a diagnostic indicator for human prostate tumors (118), thyroid neoplasia (18), uterine cervix cancers (4), and colorectal cancers (1), among others (reviewed in reference 116).
Of particular interest for the present study is the fact that increased expression of HMGI(Y) proteins has been correlated with the increased metastatic potential of both mouse (94, 95) and human (59, 79) mammary epithelial cancers. Importantly, HMGI(Y) overexpression has also recently been detected in primary human breast cancers by a combination of serial analysis of gene expression and array technologies (85, 92). Overexpression of full-length HMGI(Y) proteins induces anchorage-independent growth of both mouse breast epithelial cells (94) and Rat-1 fibroblast cells (132) in soft agarose, as does overexpression of either truncated or chimeric forms of the HMGI-C protein in murine fibroblasts (37). Furthermore, antisense-mediated suppression of HMGI(Y) protein synthesis suppresses retrovirus-induced neoplastic transformation of rat thryoid cells (8), as well as inducing apoptotic death in anaplastic human thyroid carcinoma cell lines but not in normal thyroid cells (102). Of additional interest is the fact that chromosomal rearrangements in which the A · T hook DNA-binding domains of the HMGI(Y) proteins are fused to ectopic peptide sequences to form chimeric oncogenic proteins are among the most common lesions in human cancer, being observed in a high percentage of benign mesenchymal neoplasias, including lipomas, leiomyomas, fibroadenomas, aggressive myxomas, pulmonary hamartomas, and endometrial polyps (reviewed in references 54 and 116). These previous studies, when considered together, present a compelling case for involvement of the HMGI(Y) proteins, and their A · T hook DNA-binding motifs, in both neoplastic transformation and metastatic tumor progression. Nevertheless, they did not directly and unambiguously demonstrate a causal in vivo relationship between the HMGI(Y) proteins and cancer in intact organisms. These previous studies also did not elucidate the molecular events underlying the biological consequences that overexpression or aberrant expression of HMGI(Y) proteins might have on processes such as oncogenic transformation, increased tumor metastatic potential, and aggressive neoplastic malignancy.
In the present study, we demonstrated that nontumorigenic human breast epithelial cells containing either a tetracycline-regulated HMG-I or HMG-Y transgene acquire the abilities to both grow anchorage independently in soft agar and form metastatic tumors when injected into nude mice only when the HMGI(Y) proteins are overexpressed. While overexpression of either the HMG-Y or HMG-I protein alone readily induced neoplastic transformation, as evidenced by the acquired ability of cells to grow in soft agar, somewhat surprisingly, only the HMG-Y isoform elicited efficient tumor formation and metastasis in vivo when expressing cells were injected into nude mice. Furthermore, expression of either antisense or dominant-negative HMGI(Y) constructs in tumor cells inhibited both their rate of proliferation and their ability to grow anchorage independently in soft agar. Analysis of the transcription profiles of cells employing cDNA arrays demonstrates that overexpression of HMGI(Y) proteins modulates the expression of a complex set of genes that are known to be involved in cell signaling, proliferation, tumor initiation, invasion, migration, induction of angiogenesis, and colonization. Importantly, immunohistochemical analyses of tumors formed in nude mice by cells overexpressing HMGI(Y) suggest that they have undergone an epithelial-mesenchymal transition (EMT) in vivo, an event consistent with the results obtained from gene transcription array analyses. These data represent direct experimental support for the hypothesis that overexpression of the HMGI(Y) proteins is causally associated with neoplastic transformation, metastatic progression, and mesenchymal transition of human breast epithelial cells in the context of an intact living organism.
MATERIALS AND METHODS
Cell culture and selection of transgenic clones.
The human breast epithelial cell line MCF-7 (catalog no. HTB-22) was obtained from the American Type Culture Collection (ATCC; Manassas, Va.) and was grown in Dulbecoco's modified Eagle's medium (DMEM) supplemented with 2 mM l-glutamine, 10 mM HEPES, 10% fetal calf serum (FCS), penicillin G sodium at 100 U/ml, and streptomycin sulfate at 100 μg/ml and maintained as recommended by the supplier. MCF-7/PKCα cells (a generous gift of D. K. Ways, East Carolina University School of Medicine [128] were grown in the same medium as MCF-7 cells but supplemented with G418 at 100 μg/ml. The human epithelial cell lines Hs578Bst (ATCC catalog no. HTB-125) and Hs578T (ATCC catalog no. HTB-126), as well as the human HeLa cervical carcinoma cell line (ATCC catalog no. CCL-2), were also obtained from the ATCC and maintained as recommended by the supplier. The tetracycline-regulated M/tet, M/tet/Vec, M/tet/HA-I, and M/tet/HA-Y human breast epithelial cells were obtained as follows. The parental M/tet (MCF-7/Tet-off) cell line was purchased from Clontech, Palo Alto, Calif. (catalog no. C30071, lot no. 7080502). The M/Tet cell line was derived by stable transfection of MCF-7 cells with a tetracycline-controlled transgene coding for the tetracycline transactivator protein and exhibits normal slow-growth characteristics, is contact inhibited, and does not form tumors when injected into nude mice. The cell lines designated M/tet/HA-I or M/tet/HA-Y (e.g., M/tet/HA-IC7, M/tet/HA-YC21, etc.) are clonal derivatives of M/tet cells that have been stably transfected with a plasmid vector (Clontech) containing the tetracycline response element (pTRE) driving the expression of either a hemagglutinin (HA)-tagged HMG-I or HA-tagged HMG-Y cDNA transgene (69). The control M/tet/Vect cells are clonal derivatives of M/tet cells that have been stably transfected with the empty pTRE vector without any insert. M/tet cells were cultured in DMEM containing 10% FCS (tetracycline free), 2 mM l-glutamine, G418 at 100 μg/ml (for maintenance of selection of the gene for the tetracycline transactivator protein), penicillin G sodium at 100 U/ml, and streptomycin sulfate at 100 μg/ml. Selection and maintenance of the M/tet/Vect, M/tet/HA-I, and M/tet/HA-Y cell clones were done with the same medium used for the M/tet cells, except that it contained either hygromycin at 100 μg/ml for M/tet/Vect and M/tet/HA-I cells or zeocin at 50 μg/ml for M/tet/HA-Y cells. In addition, the M/tet/HA-I and M/tet/HA-Y clones were routinely maintained in tetracycline at 2 to 10 μg/ml (depending on the individual clone) to keep expression of the HA-I and HA-Y transgenes turned off.
Construction and use of HMGI(Y) plasmid expression vectors.
Wild-type HMG-I and wild-type HMG-Y plasmid expression vectors were created by subcloning the PCR products of the coding regions of human HMG-I and HMG-Y cDNAs (69) into the pcDNA3.1/Zeo+ plasmid expression vector (Invitrogen) between the HindIII and BamHI sites of the polylinker. The construction and use of both the antisense expression vector pRcCMVIGMH (57, 99) and the dominant-negative expression vector HMG-I (cytomegalovirus-HMGImII,mIII) (58) have been described previously. HA-tagged HMG-I and HA-tagged HMG-Y expression vectors were created by fusing a synthetic DNA oligonucleotide encoding the peptide MYPYDVPDYASL from the influenza virus HA protein (i.e., HA tag) in frame to the N-terminal end of the HMG-I and HMG-Y cDNAs by standard PCR protocols (3). The PCR fragments containing either HA-tagged HMG-I or HA-tagged HMG-Y were then subcloned into the pTRE expression vector, and the sequences of the resulting expression constructs were confirmed by automated DNA sequencing prior to use.
Cell transfection and clone selection.
Plasmid expression vectors were introduced into cells by employing either the DMRIE-C, the Lipofectamine, or the Lipofectamine Plus transfection reagent following the manufacturer's (Gibco BRL, Gaithersburg, Md.) instructions. Transfection efficiencies in transient-transfection experiments using antisense and dominant-negative HMGI(Y) expression vector constructs were routinely monitored by cotransfection of a cytomegalovirus-based plasmid vector expressing green fluorescent protein (GFP) at a DNA ratio of 1:1. The total viable cells and viable GFP-expressing cells were counted using a UV microscope. The percentage of GFP-expressing cells was then obtained and served as an estimate of the transfection efficiency. In many experiments, transfection efficiencies were also monitored by cotransfection of cells with a luciferase reporter plasmid and determination of luminescence activity in cell lysates using a luciferase assay kit (catalog no. E4030; Promega, Madison, Wis.) and a Top Count Luminometer (Packard Instruments).
Cell proliferation assays.
Cells were trypsinized and suspended in DMEM with 10% FCS and counted using either a Coulter Counter (Coulter Corp., Hialeah, Fla.) or a hemacytometer. Cell viability was assessed using trypan blue exclusion (to obtain the percentage of viable cells). For growth curve determinations, 2 × 104 or 4 × 104 viable cells were initially seeded in 2 ml of medium per 35-mm diameter dish. In all experiment, cells in four or more separate culture dishes were counted on each day of a time course study in order to ensure statistical accuracy of the growth curves.
Acid extraction and Western blot analysis of total cellular HMGI(Y) proteins.
Acid extraction of HMG proteins from cells with 5% perchloric acid was performed as previously described (99). The acid-soluble proteins, which include not only HMGI(Y) but the other HMG proteins and histone H1 as well, were electrophoretically separated on either a 4 to 20% gradient or a sodium dodecyl sulfate (SDS)–15 or 18% polyacrylamide gel electrophoresis (PAGE) minigel and then transferred to an Immobilon-P membrane (Millipore) by electrotransfer overnight. Immunodetection of the HMG-I (Y) proteins on the membrane was performed using a rabbit polyclonal anti-HMG-I antibody (MR-19) whose production and use have been described previously (99) and an enhanced-chemiluminescence detection procedure using the SuperSignal substrate supplied by Pierce Chemical Co., Rockford, Ill.
Immunoprecipitation and Western blot assays of HA-tagged proteins.
After washing with phosphate-buffered saline, cells were directly lysed in their culture dishes (about 5 × 106 cells/dish) by addition of 1 ml of radioimmunoprecipitation assay buffer (1× phosphate-buffered saline, 1% Nonidet P-40, 00.5% sodium deoxycholate, 0.1% SDS, phenylmethylsulfonyl fluoride and aprotinin [Sigma catalog no. A6279] at 30 μg/ml each, and sodium orthovanadate at 10 μl/ml). The lysate was passed through a 21-gauge needle to shear the DNA and incubated on ice for 60 min. Particulates were removed from the lysate by centrifugtion at 15,000 × g for 20 min at 4°C, and the supernatant containing the total soluble cellular proteins was collected. Approximately 50 μl of anti-HA tag mouse monoclonal antibody (12CA5; a generous gift of J. J. Chen [16]) was added to 100 μl of lysate, and the mixture was incubated on ice for 1 h. Protein A-agarose beads (50 μl; Sigma) were then added, and the mixture was incubated overnight at 4°C while rotating. The beads were collected by centrifugation, and the bound proteins were removed by washing the beads three times with radioimmunoprecipitation assay buffer. An equal volume of SDS-PAGE sample buffer was added to the eluate, and the mixture was boiled for 2 min. The proteins in the eluate were separated by electrophoresis on an SDS–15% PAGE minigel and then transferred to a nitrocellulose membrane using a semidry electrophoretic transfer cell (Trans-Blot SD; Bio-Rad). The enhanced-chemiluminescence detection procedure described above was then used to identify the membrane-bound, HA-tagged proteins.
Soft agar anchorage-independent growth assays.
Seven milliliters of molten 0.6% Difco agar (44°C) in DMEM with 10% FCS and the desired antibiotic (such as G418, tetracycline, hygromycin, or zeocin, etc.) was poured into 60-mm diameter petri dishes and allowed to solidify for 30 min. The desired numbers of cells and the desired antibiotics were then layered over the hardened agar in 1.5 ml of molten (44°C) 0.33% low melting temperature Difco agar containing DMEM and 10% FCS, and the dishes were allowed to sit for 10 min before transfer to an incubator. Cultures were incubated at 37°C in 5% CO2 with over 90% humidity for about 30 days. The colonies were stained using a 0.25-mg/ml solution of [2-(p-iodophenyl)-3-γ(p-nitropheyl)-5-phenyl tetrazolium chloride (Sigma) for 6 to 24 h at 37°C in a 5% CO2 incubator. Colonies containing more than 50 cells were visually counted.
Analysis of gene expression profiles using cDNA arrays and quantitative RT-PCR.
The commercial Atlas Human Cancer cDNA Expression Array kit (catalog no. 7742-1) supplied by Clontech was used to establish gene expression array profiles. The isolation of total cellular RNA, the preparation of cDNA probes, and the hybridization of the cDNA array membranes followed the protocols supplied by the manufacturer (Clontech manual PT3140-1, version PR89832). Following hybridization, the membranes were exposed to a PhosphorImager screen, the screen was developed, and the resulting hybridization images were analyzed and quantitatively assessed using ImageQuant software (Molecular Dynamics, Inc.).
Standard protocols for quantitative reverse transcriptase PCR (RT-PCR; 63) were used to evaluate the levels of specific gene transcripts in cells. Total cellular RNA was isolated using Trizol reagent (Life Technologies, GIBCO; catalog no. 15596) following the instructions of the manufacturer. A mixture of 1 μg of RNA and 20 μM primer poly(T25) oligonucleotides (Collaborative Research) was reverse transcribed with 10 U of Moloney murine leukemia virus reverse transcriptase (Promega Corp.) in a PCR thermal cycler at 42°C for 60 min to produce cDNA, and the mixture was then heat denatured and cooled. Diluted samples of this cDNA were then mixed with two different PCR primer pairs, one that is specific for the gene transcript of interest and a second that is specific for a constitutively synthesized message that acts as an internal reference standard (e.g., hypoxanthine-guanine phosphoribosyltransferase [HPRT]) for the amplification reaction. The mixture was then PCR amplified in a thermal cycler for 25 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 1 min. The resulting RT-PCR products were separated by either electrophoresis on a native agarose gel or non-denaturing PAGE and detected and quantified by PhosphorImager analysis.
Nude mouse tumor formation assays.
Female BALB/c (nu/nu) mice 4 to 6 weeks old were purchased from The Jackson Laboratory, Bar Harbor, Maine. For tumor formation assays, approximately 5 × 106 M/tet/Vect cells, HA–HMG-I-expressing cells, or HA–HMG-Y-expressing cells in serum-free medium were injected into either the mammary fat pad or the subcutaneous space under the skin of the nude mice. The mice were maintained in a germfree facility, and tumor incidence and size were observed at regular intervals and documented.
Histology and immunohistochemistry.
Tumor nodules were fixed in 10% formalin and embedded in paraffin, and 4- to 6-μm sections were mounted onto slides and stained with hematoxylin and eosin by the histochemistry core facility of the Center for Reproductive Biology, Washington State University. Immunostaining of tissue sections that had been preserved in Histochoice fixative (Sigma) followed standard protocols. Antigen retrieval for vimentin detection was done by microwave heating in citrate buffer. Before adding horseradish peroxidase-conjugated reagents, endogenous peroxidase activity was blocked by a methanol-H2O2 solution. For immunolocalization studies, tissue sections were reacted with a primary antibody (either a monoclonal mouse antibody or a polyclonal rabbit antibody), washed, and then reacted with either biotinylated goat anti-mouse immunoglobulin G (IgG; Vector Laboratories; 1:200) or biotinylated goat anti-rabbit IgG (Bio-Rad; 1:1,000). Biotinylated antibodies were visualized by reaction with horseradish peroxidase-streptavidin reagent (Vector Laboratories) and diaminobenzidine staining, followed by counter staining of nuclei with Meyer's hematoxylin. The antibodies and dilutions used for tumor immunostaining were anti-HA tag mouse monoclonal antibody 12CA5 (1:50 dilution), anti-pan-cytokeratin (C11) mouse monoclonal antibody (1:50 dilution; Santa Cruz Biotechnology), and anti-vimentin (V9) mouse monoclonal antibody (1:50 dilution; Santa Cruz Biotechnology). Both the mouse monoclonal anti-human procollagen alpha type I antibody M-38 (1:50 dilution; developed by J. A. McDonald) and the mouse monoclonal anti-procollagen alpha type III antibody SP1.D8 (1:50 dilution; developed by H. Furthmayr) were obtained from the Developmental Studies Hybridoma Bank, a contract facility supported by the National Institute of Child Health and Human Development and maintained by the Department of Biological Sciences, University of Iowa, Iowa City. Rabbit polyclonal antibody 8691 (1:40 dilution), a general anti-human alpha 1 (XI) collagen antibody, and rabbit polyclonal antibody 1703 (1:40 dilution), specific for the v2 and vlav2 isoforms of human alpha 1 (XI) collagen associated with mesenchymal and not cartilaginous tissues, were both generous gifts from Julia Oxford, Oregon Health Sciences University.
RESULTS
Endogenous HMGI(Y) protein levels correlate with the tumorigenic potential of human mammary epithelial cells.
In preliminary experiments, a number of different normal and malignant human breast epithelial cell types were examined for both the levels of endogenous HMGI(Y) proteins and the ability to respond to artificial manipulation of these levels by the introduction of antisense or dominant-negative HMGI(Y) expression vectors. Among those tested were two matched pairs of human mammary epithelial cell lines (lines Hs578Bst and Hs578T and lines MCF-7 and MCF-7/PKCα). Each pair was originally derived from the same parent, but the individually lines exhibit markedly different tumorigenic phenotypes. The aneuploid Hs578T cell line originated from a human mammary epithelial carcinoma (46) and grows aggressively in soft agar, exhibits invasive properties in in vitro matrigel outgrowth assays, and causes tumors that extensively metastasize in immunodeficient nude mice (111). In contrast, the diploid Hs578Bst cell line is a nontumorigenic myoepithelial cell line established from normal breast tissue of the same patient (46) but, unlike Hs578T, does not grow in soft agar, is not invasive in matrigel assays, and does not cause tumors in nude mice. The human MCF-7 breast epithelial cell line is estrogen receptor positive and retains many of the biochemical and phenotypic characteristics of normal mammary epithelial cells (76). It has only a very limited ability to grow in soft agar (128), is noninvasive in matrigel assays, and does not form tumors in nude mice (111, 128). On the other hand, the MCF-7/PKCα cell line is a derivative of MCF-7 cells that has been stably transformed with a transgenic cDNA that codes for the transforming oncogene protein kinase Cα (PKCα). This PKCα-over expressing cell line readily growns in soft agar in vitro, is invasive in matrigel assays, and readily produces aggressive tumors that metastasize when injected into nude mice (128).
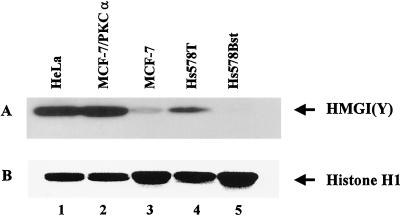
Figure 1A shows a Western blot in which the endogenous HMGI(Y) proteins (which migrate together under these gel conditions) present in MCF-7 (lane 3), MCF-7/PKCα (lane 2), Hs578Bst (lane 5), and Hs578T (lane 4) cells were detected by specific anti-HMGI(Y) polyclonal antibody MR-19 (99). Also shown as a positive control on this blot are the amounts of endogenous HMGI(Y) proteins found in the malignant HeLa cervical adenocarcinoma cell line (lane 1), which is known to contain high levels of these proteins (81). Figure 1B shows a Commassie blue-stained gel of the H1 proteins present in the same protein samples used for the immunoblot in panel A that served as an internal standard for total protein loading on the gels. From Fig. 1, it is evident that the amount of endogenous HMGI(Y) proteins present in these mammary epithelial cell lines closely correlates with their degree of in vivo tumorigenicity, a finding consistent with many previous studies (14, 116). Specifically, the nontumorigenic Hs578 and MCF-7 cell lines contain low or nondetectable levels of endogenous HMGI(Y) proteins whereas these proteins are exceedingly abundant in the highly malignant Hs578T, MCF-7/PKCα, and HeLa cell lines.
FIG. 1.
Expression of endogenous HMGI(Y) proteins correlates with tumor phenotype. Equal amounts of total cellular protein (∼5 μg) extracted from cells with different tumorigenic potentials were separated by electrophoresis on a 4 to 20% gradient polyacrylamide gel, and the proteins were transferred onto a nitrocellulose membrane. (A) Western blot obtained with rabbit MR19 polyclonal antibody against the HMGI(Y) proteins. Under the electrophoretic conditions employed, the HMG-I and HMG-Y isoforms of the proteins comigrate as a single band. Lanes: 1, HeLa cells; 2, MCF-7/PKCα cells; 3, MCF-7 cells; 4, Hs578T cells; 5, Hs578Bst cells. (B) Histone H1 proteins on the same membrane stained with Coomassie brilliant blue. The H1 histones served as an internal standard for equal protein loading on the gel.
Antisense HMGI(Y) expression inhibits cell proliferation and anchorage-independent growth.
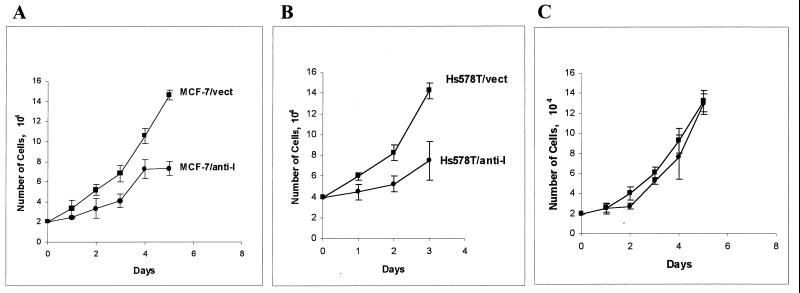
To further explore the role of HMGI(Y) in tumorigenesis, transfection experiments were performed in order to modulate the amounts of these proteins in various epithelial cell lines in order to determine whether there is a mechanistic connection between the endogenous level of HMGI(Y) proteins and cell proliferation rates or other phenotypic characteristics. Previous studies have demonstrated that the endogenous levels of HMGI(Y) proteins can be dramatically reduced in cells by the introduction of antisense expression vectors (8, 102) and that both antisense and dominant-negative expression vectors can significantly inhibit the in vivo transcription of genes regulated by the HMGI(Y) proteins (57, 58, 121). Figure 2 shows the results of experiments in which either the antisense pRcCMVIGMH (57) vector or, as a control, the empty parental pRcCMV vector without any insert was individually transfected into MCF-7, Hs578T, and HeLa cells. The transfectants were selected with G418 for a week, and pools of the G418-resistant cells from each cell line were then subcultured and their relative growth rates were determined by cell counting. As is evident from Fig. 2, expression of antisense HMGI(Y) inhibits the growth of MCF-7 cells (panel A) and Hs578T cells (panel B) but not HeLa cells (panel C). Control experiments with cotransfected reporter genes as internal reference standards demonstrated that this difference in the degree of growth inhibition in the different cell lines as a result of antisense HMGI(Y) expression is not due to the differences in transfection efficiency between the different cell lines (data not shown). The question therefore arises of why growth of the MCF-7 and Hs578T cells is significantly inhibited by antisense HMGI(Y) expression but growth of HeLa cells is not. A likely answer lies in the relative levels of endogenous HMGI(Y) proteins found in these different cell lines (Fig. 1). As shown in Fig. 2D, when the levels of HMGI(Y) proteins in the transfected cells were determined by Western blot analysis, it was seen that the antisense vector reduced the amount of proteins remaining in the MCF-7 and Hs578T cells (lanes 3 and 5, respectively) to a far greater extent than it did in the HeLa cells (lane 7). Thus, it is reasonable to propose that the reason why the growth rate of HeLa cells is unaffected by the antisense expression vector is that it is unable to reduce the very high concentrations of endogenous HMGI(Y) proteins found in HeLa cells to a level that would slow cell proliferation. Nevertheless, these transfection results clearly indicate that antisense HMGI(Y) expression impacts the growth rate of cells with low-to-moderate levels of endogenous HMGI(Y) proteins. Additional transfection experiments also demonstrated that antisense expression inhibited the anchorage-independent growth of some lines of mammary epithelial tumor cells in soft agar (data not shown). Taken together, the results from these two independent lines of antisense experiments indicate that reduction of endogenous HMG-I(Y) levels in cells can lead to a decrease in their proliferation rates and a reduction of the ability of some tumor cells to grow anchorage independently.
FIG. 2.
Expression of antisense HMG-inhibits tumor cell proliferation. MCF-7 (A), Hs578T (B), and HeLa (C) cells were transfected with 6 μg of either the empty pRcCMV expression vector (■) or the pRcCMV-IGMH (antisense HMG-I) vector (●). The transfectants were selected with G418 at 150 μg/ml in culture medium for 7 days, and then the viable cells were subjected to a cell growth rate assay. Data were collected from three independent experiments with four dishes for each time point per experiment. (D) Western blots of HMGI(Y) proteins in MCF-7 cells (lanes 2 and 3), Hs578T cells (lanes 4 and 5), and HeLa cells (lanes 6 and 7) transfected with either the empty control vector (lanes 2, 4, and 6) or the antisense HMG-I vector (lanes 3, 5, and 7). Approximately equal amounts of total acid-soluble proteins were loaded into all of the lanes, and the Western blots were probed with a rabbit polyclonal antibody (MR19) against the HMGI(Y) proteins. On these gels, the HMG-I and HMG-Y proteins comigrated as a single band.
Overexpression of exogenous HMGI(Y) proteins promotes anchorage-independent growth in soft agar.
A series of preliminary transfection experiments demonstrated that transfection of MCF-7 cells with a wild-type HMG-I or HMG-Y vector whose transcriptional expression was driven by a highly efficient constitutive promoter resulted in a marked increase in the total amount of HMGI(Y) proteins in the recipient cells and also resulted in a significant increase in their ability to grow anchorage independently in soft agar (data not shown). There is, however, an intrinsic problem that interferes with an unambiguous interpretation of these and other, similar studies (37, 94, 132). Namely, owing to the fact that HMGI(Y) proteins can influence the transcriptional activity of the host cell's endogenous Hmgiy gene promoter (unpublished observations), it is difficult, if not impossible, to distinguish between the amount of HMGI(Y) proteins in the transfected cells contributed by expression of the introduced transgene and the amount of these proteins contributed by expression of the endogenous host cell gene.
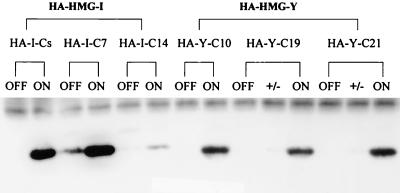
To circumvent the problem of protein identification, plasmid expression vectors were constructed that produced either HMG-I or HMG-Y proteins containing a nine-amino-acid peptide derived from the HA protein of influenza virus fused to their N-terminal ends. Furthermore, these transgenes were under the transcriptional control of a tetracycline-regulated promoter element (30) and were transfected into a genetically engineered MCF-7 cell line that synthesizes the Tet regulatory protein, whose activity was controlled by the presence or absence of tetracycline in the cell culture medium. Specifically, in the Tet-off expression system employed, genetically engineered MCF-7 cells that were stably transfected with either the HA-tagged HMG-I or HMG-Y gene produced large amounts of the corresponding HA-tagged protein in the absence of tetracycline in the culture medium. In contrast, when tetracycline was added to the cell culture medium, production of the HA-tagged transgenic protein was shut off. Thus, by using this tetracycline-regulated gene transcription system, the expression of either the HA–HMG-I or HA–HMG-Y transgenic protein in MCF-7 cells could be reversibly controlled at will and the resulting levels of protein expression could be quantitatively assessed. A major advantage of using HA-tagged proteins is that the exogenous HMG-I(Y) proteins can be easily distinguished from the endogenous proteins in transgenic cells by means of a monoclonal antibody (12CA5) directed specifically against the HA tag peptide that was used for immunoprecipitation of HA-tagged proteins during screening for clones expressing high levels of exogenous HMGI(Y) proteins (30). As shown in Fig. 3, by using immunoprecipitation and Western blotting analysis, several independently derived transgenic MCF-7 clones that express high levels of either HA–HMG-I or HA–HMG-Y protein were identified. In these experiments, the HA–HMGI(Y) proteins were first immunoprecipitated from the transgenic clones by using the 12CA5 antibody and then the HMG-I and HMG-Y proteins in the immunoprecipitates were identified by Western blotting using the MR-18 antibody directed against wild-type HMGI(Y) proteins. As is evident in Fig. 3, all of the clonal MCF-7 cell lines containing the HA-tagged HMG-I (i.e., HA–I-Cs, HA–I-C7, and HA–I-C14) or HMG-Y gene produced high concentrations of the respective HA-tagged transgenic protein when the gene was transcriptionally on (i.e., in the absence of tetracycline). In contrast, expression of either the HA–HMG-I or HA–HMG-Y transgenic protein could be completely inhibited (i.e., turned off) in these transgenic cells by the addition of 2 to 10 μg of tetracycline (depending on the clone) to the cell culture medium, with the exception of clone of HA–I-C7, which required higher levels of the drug to be completely repressed (data not shown). Importantly, control Western blot experiments in which total acid-soluble proteins isolated from the on cells were run under electrophoretic conditions that separated the HMG-I from the HMG-Y isoform protein also demonstrated high-level expression of the HA-tagged transgenic proteins in the on transfected cells when an anti-HMGI(Y) antibody was used as a probe (data not shown).
FIG. 3.
Identification of HA–HMG-I- and HA–HMG-Y-expressing clones using immunoprecipitation and Western blots. Total cellular proteins were isolated from 5 × 106 cells from individual transgenic MCF7 clones treated without (ON) or with (OFF) tetracycline at 10 μg/ml, and the extracts were immunoprecipitated using the 12CA5 monoclonal antibody directed against the HA tag peptide of the transgenic proteins. The precipitated proteins were separated by SDS–15% PAGE and then transferred onto a nitrocellulose membrane. Western blotting of the membrane was performed using the MR19 antibody against the HMGI(Y) proteins. Under these electrophoresis conditions, the HMG-I and HMG-Y isoform proteins comigate on the gel. Individual transgenic clones expressing either the HMG-I (HA–I-Cs, HA–I-7C, and HA–IC14) or the HMG-Y (HA–Y-C10, HA–Y-C19, and HA–Y-C21) isoform protein are indicated. The +/− designation indicates that tetracycline was added (+) to expressing ON cells (−) several days prior to protein extraction in order to inhibit transgene expression.
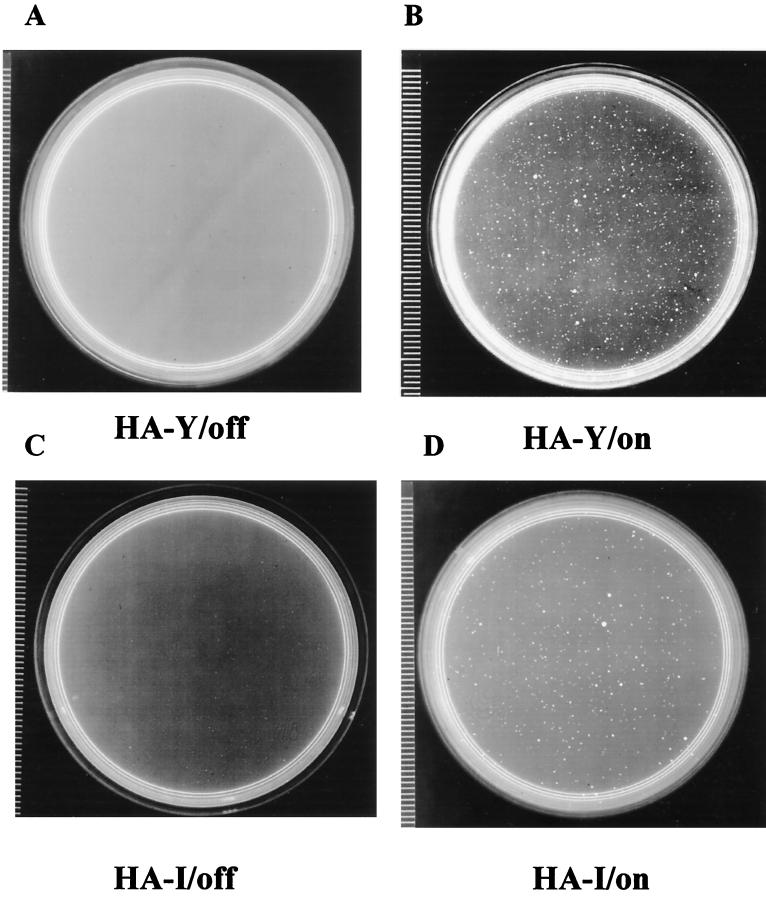
To test the effects of expression of the exogenous HMG-I or HMG-Y protein, MCF-7 clones producing these transgenic proteins were tested in the soft agar assay for the ability to exhibit anchorage-independent growth. Figure 4 shows a typical example of cells containing the exogenous HMG-I or HMG-Y-encoding gene growing in soft agar with (expression off) or without (expression on) tetracycline treatment. As summarized in Table 1, the HMG-Y-expressing clones HA–Y-C10, HA–Y-C19, and HA–Y-C21 produced, depending on the clone, 13- to 39-fold more colonies in soft agar when the gene is on than when it is off. Similarly, HMG-I-expressing clones HA–I-C14 and HA–I-Cs formed about 10- to 20-fold more colonies. We therefore conclude from these experiments that overexpression of either the HMG-I or HMG-Y isoform protein in MCF-7 cells induces reproducible changes in the phenotype of these cells, including an ability to exhibit anchorage-independent growth in soft agar.
FIG. 4.
Anchorage-independent growth of MCF-7/HA-I and MCF-7/HA-Y clones. Assays for growth in soft agar of MCF-7/HA-Y cells (A and B) and MCF-7/HA-I cells (C and D) were performed as described in Materials and Methods. The agar medium in the plates shown in panels A and C contained tetracycline at 10 μg/ml (i.e., transgene expression was off), whereas those in panels B and D were without tetracycline (i.e., transgene expression was on).
TABLE 1.
Overexpression of HA–HMG-I(Y) proteins in MCF-7 cells leads to anchorage-independent growth in soft agar
| Clone | Avga no. of colonies ± SD
|
Fold difference | |
|---|---|---|---|
| On, without tet | Off, with tet | ||
| HA–Y-C10 | 245.50 ± 21.20 | 6.25 ± 1.70 | 39.3 |
| HA–Y-C19 | 151.75 ± 19.92 | 11.25 ± 3.09 | 13.5 |
| HA–Y-C21 | 143.75 ± 10.37 | 5.75 ± 1.70 | 25.0 |
| HA–I-C14 | 78.00 ± 12.19 | 8.00 ± 2.16 | 9.8 |
| HA–I-Cs | 204.25 ± 25.32 | 10.25 ± 2.87 | 20.0 |
| M7CC21 | 28.25 ± 2.45 | 5.00 ± 2.16 | 5.7 |
| M/Tet/Vect | 17.00 ± 2.94 | 4.50 ± 2.08 | 3.7 |
Average values based on eight independent dishes in two experiments.
Array and RT-PCR analyses of gene transcription profiles in cells overexpressing HMG-I and HMG-Y.
To identify the tumor-promoting genes that are altered by HMG-I and HMG-Y overexpression in vivo, Atlas Human Cancer cDNA Expression Arrays (Clontech Inc.) were used to generate tumor gene expression profiles. The array was a carefully selected collection of cDNA fragments arranged on nylon membranes for rapid assessment of the differential gene expression of 588 tumor-associated genes. These well-characterized genes broadly represented many crucial cellular signaling pathways and other complex biological functions that have been associated at the molecular level with changes underlying tumor progression and metastasis.
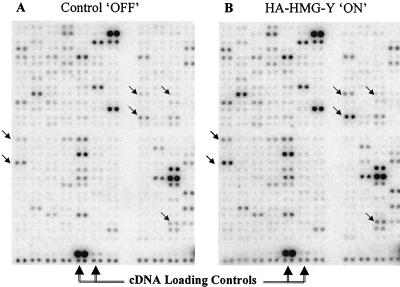
Using these arrays, gene transcription expression profiles were compared in two clonal transgenic cell lines, HA–Y-C21 and HA–I-C7, which, as shown in Fig. 3, express high levels of HA-tagged HMG-Y and HA-tagged HMG-I proteins, respectively, when on. For each of these experiments, total RNA was isolated from both control (i.e., off) cells not expressing transgenic proteins and from cells expressing high levels of HA-tagged protein (i.e., on) and reverse transcribed into cDNA with 32P labeling. The two cDNA probe populations were then separately hybridized with two identical Atlas Array nylon membranes on which the 588 genes were spotted in duplicate. The membranes were then developed by exposure to a phosphorimaging screen, and the expression profiles of the two RNA populations were compared side by side, as shown in the representative result for HA–HMG-Y–OFF and HA–HMG-Y–ON profiles in Fig. 5. Similar hybridization profiles were generated for HA–HMG-I–OFF and HA–HMG-I–ON clones (data not shown).
FIG. 5.
Array analysis of gene expression in transgenic cells over- expressing HA–HMG-Y. Total RNA was isolated from control off M/tet/Vect cells not expressing HMG-Y and on HA–Y-C21 cells expressing high levels of HA–HMG-Y protein and reverse transcribed into cDNA with 32P labeling. The two cDNA probes were separately hybridized with identical Atlas Array nylon membranes (Clontech) on which 588 genes associated with human cancer were spotted in duplicate. The membranes were then developed by exposure to a phosphorimaging screen, and the profiles of the two RNA populations were quantitatively compared. (A) One small section of the large array membrane hybridized with a probe from control off cells. (B) A corresponding region of a duplicate membrane hybridized with a probe from HA–HMG-Y on cells. The small diagonal arrows indicate corresponding positions on the two membrnaes where significant differences in spot intensities can be observed.
The intensity of each of the hybridizing spots on the membrane was quantified by PhosphorImager analysis using six housekeeping genes placed on each array membrane in order to normalize the hybridization signals. These housekeeping genes (which generated nearly equal-intensity hybridization signals for all of the samples being compared) were those for HPRT, ubiquitin, ribosomal protein S9, 23-kDa highly basic protein, beta-actin, and phospholipase A2. In addition, the hybridization signals of cDNAs surrounding a specific target cDNA were used both as references for normalization and for confirmation of relative intensities.
The criterion we chose for identifying the most important differentially regulated genes is that their expression had to be altered fourfold or more compared to other populations of mRNAs. A 4-fold difference as a standard is quiet stringent considering that many other studies using gene array analyses have considered 1.4- to 3-fold differences in expression to be biologically significant (2, 34, 64, 92). Furthermore, this degree of difference in transcriptional expression can be easily detected and independently confirmed by quantitative RT-PCR techniques (see below). Table 2 lists some of the proteins encoded by genes that are induced or repressed in the HA–Y-C21 cell line that is overexpressing the transgenic HMG-Y protein. Of the 588 genes in the array, 33 (5.6%) showed a 10-fold or greater difference in their level of expression and 52 (8.8%) were differentially expressed in the range of 9- to 4-fold. Thus, a total of 85 genes (∼14%) had significantly altered levels of expression whereas 503 genes (∼86%) exhibited changes in expression of less than fourfold. Of the 85 genes with significantly altered expression, 72 (∼12%) were upregulated and 13 (∼2%) were downregulated, resulting in an up-to-down ratio of 6, indicating that significantly more genes are positively regulated by overexpression of the transgenic HMG-Y protein. Similar analyses of gene expression profiles in cells overexpressing the HMG-I isoform protein were performed yielding generally comparable results (data not shown).
TABLE 2.
Proteins encoded by genes in transgenic human MCF-7 breast epithelial cells whose transcriptional expression is modulated by overexpression of HMG-Y protein as determined by cDNA array analyses
| Categorya | Fold increase | Fold decrease | |
|---|---|---|---|
| Cell cycle and growth regulators | |||
| cdc25A | 98.6 | ||
| cdk5 activator isoform P391 precursor | 35.8 | ||
| NEDD5 protein homolog | 35.2 | ||
| TPKII regulatory subunit | 17.2 | ||
| CLK-1 | 14.6 | ||
| stk1 | 9.3 | ||
| cdc25C | 9.1 | ||
| Jun N-terminal kinase 2 | 7.4 | ||
| p38 MAPK | 5.7 | ||
| CDC10 protein homolog | 5.2 | ||
| Cyclin C | 5.1 | ||
| CDK inhibitor p19INK4d | 4.4 | ||
| N-myc | 4 | ||
| Cyclin A | 3.6 | ||
| raf | 3.3 | ||
| Cyclin D | 3.1 | ||
| Intermediate filament markers | |||
| Cytokeratin 10, type 1 | 9.3 | ||
| Vimentin | 5.6 | ||
| Cytokeratin 12, type 1 | 3.1 | ||
| Apoptosis regulators | |||
| TRAR 15 | 6.9 | ||
| Pig 11 | 3.5 | ||
| Oncogenes and tumor suppressors | |||
| MET | 4.9 | ||
| RBQ1 | 3.9 | ||
| DNA damage response, repair, and recombination genes | |||
| DNase X | 7.5 | ||
| DNA methyltransferase | 6 | ||
| DNA ligase III | 4.8 | ||
| ATM | 3.9 | ||
| Cell fate and development regulators | |||
| Frizzled-5 | 161.1 | ||
| Wnt-13 | 14.6 | ||
| Jagged-1 | 7.5 | ||
| Notch-4 | 4.1 | ||
| WNT-10B | 4.3 | ||
| Frizzled homolog | 3.9 | ||
| Receptors | |||
| GARP | 27.9 | ||
| FGF2b | 29.3 | ||
| FGFR1 | 24.7 | ||
| Thrombopoietin receptor MPL | 13.2 | ||
| Her-4; ERBB4 | 10.5 | ||
| NGFR | 6.4 | ||
| Erythropoietin receptor | 5.2 | ||
| HGF activator like | 5 | ||
| TGFβ receptor III | 4.6 | ||
| 5T4 oncofetal antigen | 4.3 | ||
| ERBB3 | 3.9 | ||
| EGF receptor | 3.5 | ||
| IGFBP-6 | 3.4 | ||
| Cell adhesion, motility, and invasion genes | |||
| Collagen type I | 85.9 | ||
| Integrin alpha-6 | 60.6 | ||
| Laminin B1 | 24.7 | ||
| Collagen type VI alpha-1 | 24.4 | ||
| Collagen type XI pro-alpha-1 | 18.7 | ||
| Integrin beta-8 | 15.9 | ||
| Collagen type III pro-alpha-1 | 15.7 | ||
| Collagen type XVI alpha-1 | 11.4 | ||
| Collagen type XVIII alpha | 10.7 | ||
| Integrin beta-1 | 10.7 | ||
| Integrin alpha-E | 6.5 | ||
| Thrombopoietin 1 precursor | 6.2 | ||
| LAMA-4 | 6 | ||
| Integrin alpha-8 | 5.2 | ||
| Collagen type VIII alpha-1 | 4.5 | ||
| Cell adhesion kinase beta; Pyk2 | 4.5 | ||
| Tenascin-R | 4.2 | ||
| Collagen type IV alpha-3 | 4.1 | ||
| Integrin beta-3 | 4.1 | ||
| Integrin alpha-1 | 4.1 | ||
| Collagen type VI alpha-2 | 3.9 | ||
| Angiogensis regulator: FGFR2 precursor | 29.3 | ||
| Invasion regulators | |||
| PAI-3 | 74.2 | ||
| MMP-16 | 13.5 | ||
| MDC9 | 5.8 | ||
| PAI-1 | 5.4 | ||
| MMP-13 | 4.7 | ||
| MMP-8 | 4.3 | ||
| TIMP-3 | 4.6 | ||
| Rho family small GTPases and their regulators | |||
| RhoC | 3.9 | ||
| Putative Rho-Rac G exchange factor | 3.8 | ||
| RhoG | 3.7 | ||
| RhoGDI gamma | 3.5 | ||
| Cell cell interaction genes | |||
| Cadherin 12 | 294.4 | ||
| H-cadherin | 20.7 | ||
| Desmoglein 2 | 18.4 | ||
| Desmoglein type 1 | 11 | ||
| HCK | 5.1 | ||
| LERK-2, LERK-L | 4.7 | ||
| M-cadherin | 3.8 | ||
| Growth factors and cytokines | |||
| Interleukin-11 | 56.9 | ||
| FGF-6 | 14.57 | ||
| SF, alternative transcript | 14.5 | ||
| GDNF | 12.2 | ||
| Osteogenic protein 2 | 9.4 | ||
| LIF | 7.4 | ||
| Leukocyte IFN-inducible peptide | 6.5 | ||
| FHF-1 | 6.2 | ||
| Interleukin-14 | 5.7 | ||
| Interleukin-10 | 5.3 | ||
| Interleukin-12p35 | 5.2 | ||
| IFN-α | 4.6 | ||
| Interleukin-17 | 4.5 | ||
| FGF-7 | 4.1 | ||
| BMP-6 | 4.1 | ||
| IFN-bata-1 | 4 | ||
| Interleukin-13 | 3.9 | ||
| IFN-γ | 3.8 | ||
| Interleukin-12 p40 | 3.8 | ||
| FGF-9 | 3.8 |
The genes are listed in various categories related to human cancer as described by the manufacturer of the cDNA arrays (Clontech).
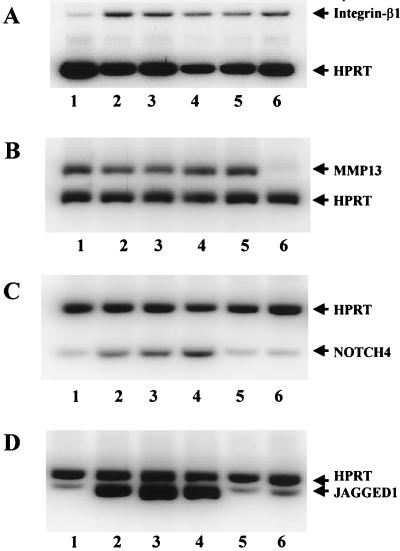
To independently verify the reliability of the cDNA array hybridization data, quantitative RT-PCR analyses were also performed on selected genes for three independently derived HMG-Y-expressing clones and two independently derived HMG-I-expressing clones. As shown in Fig. 6, among the representative genes analyzed by RT-PCR were those coding for integrin-β1 (panel A), matrix metalloprotease-13 (MMP13) (panel B), Notch-4 (panel C), and Jagged-1 (panel D). In each RT-PCR, amplification of HPRT transcripts served as an internal control. Quantitative assessment of these and other RT-PCR results indicated that the differential expression patterns and the relative expression levels of these genes are similar to those observed with the cDNA arrays, confirming the reliability of the array expression profile data. Interestedly, and perhaps significantly, these detection methods confirm that HMG-Y and HMG-I upregulate integrin-β1 and MMP13 to about the same extent, but only HMG-Y upregulates Notch-4 and Jagged-1. This is the first experimental data demonstrating differential regulation of gene expression by the HMG-Y and HMG-I isoform proteins. Northern blot analysis of representative mRNAs isolated from on and off cell clones likewise confirmed both the array and RT-PCR results (unpublished data).
FIG. 6.
RT-PCR analysis of gene expression profiles regulated by HMG-I and HMG-Y indicates that the two protein isoforms do not have identical functions in vivo. The DNA-free RNAs from two HMG-Y-expressing clones (M8AC10 and M8AC19), two HMG-I-expressing clones (M7CC7 and M7CCs), and control M/tet/Vect cells were subjected to semiquantitative RT-PCRs as described in Materials and Methods. The PCR products were electrophoretically separated by SDS–15% nondenaturing PAGE. The results of this analysis demonstrate that while both HMG-I and HMG-Y upregulate the expression of integrin β1 and MMP13, HMG-Y, but not HMG-I, upregulates Notch-4 and Jagged-1. (A) Co-RT-PCR of integrin β1 and HPRT messages from different transgenic cell clones. Lanes: 1, M/Tet/Vec; 2, M8AC10; 3, M8AC19; 4, M8AC21; 5, M7CC7; 6, M7CCs. (B) Co-RT-PCR of MMP13 and HPRT messages from different transgenic cell clones. Lanes: 1, M7CCs; 2, M7CC7; 3, M8AC21; 4, M8AC19; 5, M8AC10; 6, M/Tet/Vec. (C) Co-RT-PCR of Notch-4 and HPRT messages from different transgenic cell clones. Lanes: 1, M/Tet/Vec; 2, M8AC10; 3, M8AC19; 4, M8AC21; 5, M7CC7; 6, M7CCs. (D) Co-RT-PCR of Jagged-1 and HPRT messages from different transgenic cell clones. Lanes: 1, M/Tet/Vec; 2, M8AC10; 3, M8AC19; 4, M8AC21; 5, M7CC7; 6, M7CCs.
Genes regulated by HMG-Y overexpression.
The 588 cDNAs present on the membrane array represent 13 different categories of genes that have been reported to play key roles in different biological processes of tumor progression and metastasis. These categories include the following: A, cell cycle and growth regulators (79 genes); B, intermediate filament markers (19 genes); C, apoptosis regulators (69 genes); D, oncogenes and tumor suppressors (29 genes); E, DNA damage response, repair, and recombination genes (33 genes); F, cell fate and development regulators (23 genes); G, receptors (42 genes); H, cell adhesion, motility, and invasion genes (82 genes); I, angiogenesis regulators (16 genes); J, invasion regulators (39 genes); K, Rho family small GTPases and their regulators (21 genes); L, cell-cell interaction genes (38 genes); M, growth factors and cytokines (98 genes).
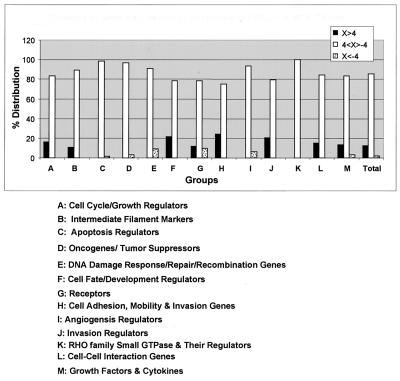
The 85 genes whose transcriptional expression was significantly altered in response to overexpression of HMG-Y protein were distributed in all 13 categories. However, as shown in Fig. 7, the patterns of alteration of gene expression were found to be quite different within the various individual categories. For example, in category H (i.e., cell adhesion, motility, and invasion genes) and category J (i.e., invasion regulators), expression of ∼24% (20 of 82) and ∼21% (8 of 39) of the genes, respectively, were significantly altered and all were upregulated. In marked contrast, in category E (i.e., DNA damage response, repair, and recombination genes), alteration of the expression of only ∼9% (3 of 33) of the genes was observed and all of these were downregulated. It may be argued that the grouping and classification of the cDNA clones of the Atlas Array (as suggested by the manufacturer) are in some ways artificial and/or arbitrary; nevertheless, the observed expression pattern variations provide strong evidence that overexpression of the HMGI(Y) proteins differentially regulates, either directly or indirectly, the transcriptional activity of various categories of genes in vivo.
FIG. 7.
Diversity of genes that are regulated by overexpressed HMG-Y proteins. Transcription profiles determined by cDNA array analyses indicate that overexpression of HMG-Y protein either up- or downregulates a broad range of genes thought to be involved with various aspects of neoplastic transformation, tumor progression, and metastasis in human cells. The graph depicts the percentages of genes in various groups or categories of cancer-related genes present on the cDNA array membrane. Conservatively, we consider only those genes whose expression is either up- or downregulated by a factor of 4 or more to be the most important ones modulated in vivo by HMG-Y overexpression. At the bottom is a list of the various categories of genes represented on the cDNA array membrane. For example, in group G, HMG-Y upregulates ∼12% and downregulates ∼10% of the receptor genes present on the membrane.
Overexpression of HMG-Y promotes tumor formation and metastasis in nude mice.
The tumorigenicity and metastatic potential of MCF-7 cells overexpressing tetracycline-regulated exogenous HMG-I and HMG-Y were evaluated by injection of cells expressing these HA-tagged proteins from a number of different clones into immunodeficient BALB/c (nu/nu) nude mice. When 5 × 106 cells were injected into either the mammary fat pad or the subcutaneous space, the HMG-Y-expressing clones (HA–Y-C19 and HA–Y-C21) formed tumors. No mice injected with HA–I-C7 clones expressing the HMG-I protein or control clones expressing only the empty M/tet parental vector developed tumors within a 3-month observation period, as summarized in Table 3. When inoculated subcutaneously into the backs of mice, HA–Y-C19 cells formed solid primary tumor nodules with local invasion of the surrounding tissues (data not shown). Morphological examination indicated that these tumors were well circumscribed and bordered by what appeared to be a network of thin, fibrous tissue. Histological examination of the HA–HMG-Y-induced primary tumors revealed that they were composed of solid masses of tightly packed carcinoma cells with a highly necrotic center. No metastases of cells to other areas of the body were observed when cells were injected subcutaneously (unpublished observations).
TABLE 3.
Overexpression of HMG-Y proteins leads to tumor growth in immunodeficient nu−/nu− nude mice
| Clone | No. of mice with tumors/total no. of mice used | Avg latency (no. of days) ± SD |
|---|---|---|
| HA–Y-C21 | 3/5 | 50 ± 20 |
| HA–Y-C19 | 1/2 | 35 |
| HA–I-C7 | 0/5 | |
| M/tet | 0/4 | |
| MCF-7/PKCα | 1/2 | 15 |
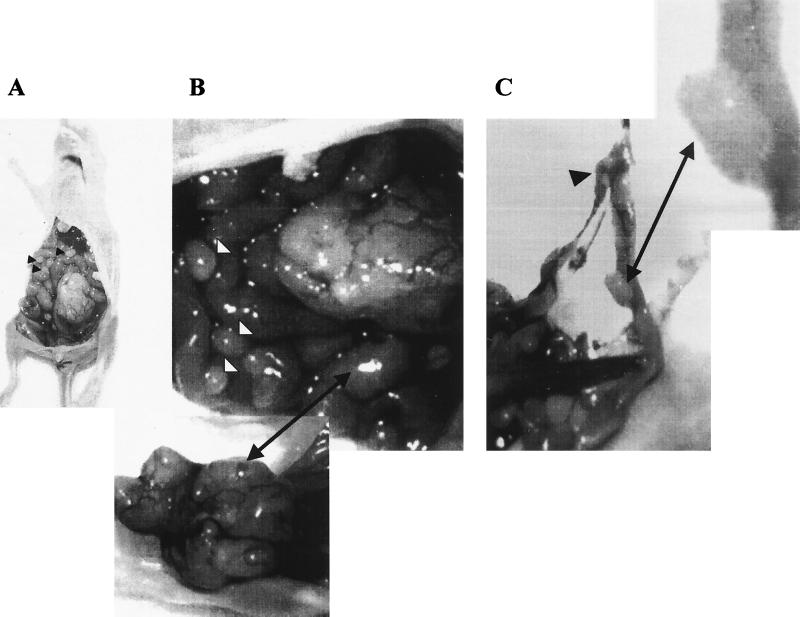
In marked contrast, when HA–HMG-Y-expressing cells were inoculated directly into mammary fat pats, not only were primary tumors formed at the site of injection but numerous secondary tumors were found to have metastasized into most of the mesenchymal tissues of the abdominal cavity, as well as into the connective tissues surrounding and supporting various organs (Fig. 8A to C). The sites of metastasis included the mesentery supporting the large and small intestines and the colon, the greater omentum, the mesentery lining of the diaphragm, and the fibrous capsule around the kidneys. Histological examination revealed that these secondary metastatic tumors were often collagen-rich carcinomas that showed a complex, pleiomorphic cytology with different regions of the tumors exhibiting markedly different morphological characteristics (Fig. 9). Some areas of the metastatic carcinomas exhibited a compact, more-or-less epithelial cell-like, cobblestone morphology (Fig. 9A and B), whereas other areas were more loosely organized and often surrounded by, or infiltrated with, collagen fibers (Fig. 9C). In some cases, the metastatic tumors were not confined to the mesentery and fibrous connective tissues but exhibited considerable anaplasia with invasion of underlying organ stroma structures, such as the striated muscle tissue of the diaphragm (Fig. 9B). Importantly, the presence of blood vessels within or near masses of tumor tissue was commonly observed (e.g., Fig. 9A and B), suggesting that the tumor cells had induced angiogenesis and/or that they had extravasated from nearby blood vessel during their migration and metastasis.
FIG. 8.
MCF-7 cells overexpressing HMG-Y protein form tumors in nude mice. Metastatic tumors developed in the mesentery membranes and in the lining of the peritoneal cavity when the mammary fat pads of nude mice were injected with approximately 5 × 106 cells (in this case, clone HA–Y-21) expressing the HA–HMG-Y protein. Many of the metastatic tumor nodules were seen in mesenteric membranes within and lining the peritoneal cavity (A), as well as in the greater omentum and other mesenteries supporting the small intestines (B) and in the mesenteries supporting and lining the colon (C).
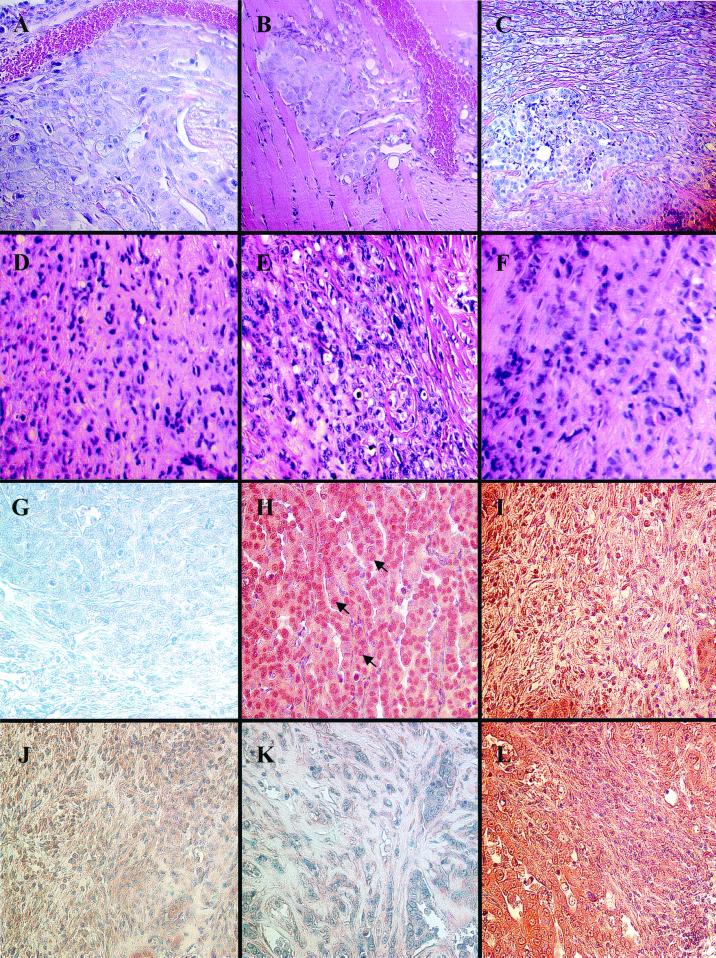
FIG. 9.
Histological analysis of sections of metastatic tumors formed in nude mice overexpressing HA–HMG-Y protein. (A to F) Hematoxylin-and-eosin staining. (G to L) Immunohistochemical analysis with specific antibodies and counterstaining with Meyer's hematoxylin. (A) Paraffin section of a secondary metastatic carcinoma with a cobblestone-like epithelial morphology (stained blue) formed by HA–Y-C21 cells located near a blood capillary shown in the upper part of the picture. Magnification, ×100. (B) Section of a secondary metastatic carcinoma formed by clone HA–Y-C21 cells showing that the tumor (blue) has invaded the striated muscle of the diaphragm (pink with striations) in close proximity to blood capillaries filled with red cells. Magnification, 100×. (C) Region of a metastatic tumor showing the coexistence of collage-rich and non-collagen-rich areas within the tumor mass. In this section, an island of cobblestone-like carcinoma cells containing little extracellular collagen (lower left) is surrounded by areas of more elongated, spindle-like cells with a dense extracellular collagen matrix (stained pink). Magnification, 40×. (D to F) Regions of carcinosarcoma-like cells found in three independently derived HA–Y-C21-induced tumors. Magnification, 60×. (G) Carcinoma region reacted with a control nonimmune mouse IgG antibody. Magnification, 100×. (H) Carcinoma region reacted with mouse monoclonal IgG (12CA5) specifically directed against the HA tag peptide. Magnification, 100×. Note the prominent nuclear staining (arrows), although cytoplasmic staining is also evident. (I) Carcinosarcoma-like region reacted with anti-HA tag mouse monoclonal IgG. Magnification, 40×. (J) Carcinosarcoma-like region reacted with mouse monoclonal IgG (M-38) directed against human fibroblast procollagen type I (which has no cross-reactivity with mouse collagens). Magnification, 40×. (K) Disorganized region of tumor reacted with a polyclonal rabbit antibody (1703) directed against the v2 and vlav2 isoforms of human alpha 1 (XI) collagen associated with mesenchymal, but not cartilagenous, tissues. Magnification, ×60. (L) Mixed carcinoma and carcinosarcoma-like regions reacted with mouse monoclonal IgG (V9) directed against human vimentin (no reaction with vimentin of mouse origin or other intermediate filaments. Magnification, ×40.
Another commonly observed feature in many of the HMG-Y-induced metastatic tumors was the presence of areas of less organized, more elongated cells with variant morphologies that contained appreciable concentrations of extracellular collagen. For example, as shown in Fig. 9C, islands of epithelioid carcinoma cells with little apparent collagen were often observed adjacent to nearby areas of more elongated and spindle-shaped tumor cells surrounded by a dense, collagen-rich matrix. Nevertheless, as illustrated in Fig. 9D to F, perhaps the most interesting and potentially significant feature observed in the metastatic tumors was the presence of localized areas of less differentiated cells that seemed to possess an unorganized, carcinosarcoma-like morphology (38). These results suggest that some of the metastatic tumor cells have undergone a phenotypic change from their original epithelial cell-like phenotype to a more mesenchymal cell-like phenotype as a consequence of overexpression of the HMG-Y protein. Such phenotypic changes require extensive alterations in gene expression patterns and are called EMTs (51). The phenomenon of EMT is a common feature of both embryonic development (123) and advanced epithelial tumors where epithelial cells have dedifferentiated to a more fibroblast-like state and regained the ability to invade, migrate, and/or proliferate in an uncontrolled fashion (9). The EMT of epithelial tumors is characterized by a number of phenotypic changes, including loss of cell-to-cell interactions; the acquisition of a more fibroblast-like cellular morphology, the downregulation of epithelial cell products such as cytokeratins, the upregulation of mesenchymal cell marker proteins such as vimentin, a marked increase in the production of cytoplasmic procollagen and extracellular collagen fibers, and the acquisition of a motility machinery that allows cells to interact in three dimensions with the extracellular matrix (ECM) (9, 10).
HMG-Y overexpression induces EMT in tumor cells.
Immunohistochemical staining of tumor sections was performed using a number of monoclonal and polyclonal antibodies directed against specific proteins to confirm that the tumors formed in nude mice had, indeed, originated from the transgenic MCF-7 cells and to further investigate the possibility that these cells had undergone an EMT. The antibodies used for these studies were selected on the basis of the gene products indicated by cDNA array analysis to be produced in the transgenic cells (Table 2) and on the basis of the histological appearance of the tumors themselves (Fig. 9A to F). For example, reaction of tumor sections with a monoclonal antibody directed against the HA tag peptide show that the transgenic human HMG-Y protein was localized primarily in the nuclei (but with detectable cytoplasmic staining) of cells in both regions of carcinoma morphology (Fig. 9H) and regions of carcinosarcoma morphology (Fig. 9I). These results unambiguously demonstrate that the tumors originated from the injected human MCF-7 cells overexpressing the HMG-Y protein, a fact also confirmed by the ability to reisolate tetracycline-regulated transgenic cell lines from the tumors by growth in selective medium (data not shown).
The list of genes differentially regulated by HMG-Y shown in Table 2 indicates that a number of types of collagen proteins, especially pro-alpha I-type collagens, are dramatically upregulated in the HMG-Y-overexpressing transgenic cells. This finding is confirmed by the immunoreactive staining of the cytoplasm of tumor cells with a peptide monoclonal antibody that is specific for human fibroblast pro-alpha I-type collagen and does not cross-react with mouse proteins (Fig. 9J). Likewise, a polyclonal rabbit antibody directed against the isoforms of human alpha I (XI) collagen that are associated with mesenchymal and noncartilaginous tissues stains the extensive extracellular collagen accumulations found in tumor tissues, indicating that the tumor cells are synthesizing and secreting these proteins (Fig. 9K). These findings are significant because excess collagen production is a characteristic feature of EMT. High levels of type I and type III procollagen proteins are also observed in the most aggressive human breast cancers, with the level of collagen expression being positively correlated with the degree of malignancy of the tumor (12, 71). The cDNA array analyses also indicated that expression of the gene coding for the classical mesenchymal cell marker protein vimentin, a diagnostic protein for EMT, is significantly upregulated (∼5.6-fold) in transgenic cells. This expression is confirmed by the reaction of tumor tissue with a monoclonal antibody that is specific for human vimentin and does not cross-react with other human intermediate filament proteins, with mouse vimentin, or with other murine proteins (Fig. 9L). Finally, a pancytokeratin monoclonal antibody against human proteins that does not cross-react with mouse cytokeratins also stains the cytoplasm of tumor cells, consistent with the array analyses (Table 2) that indicated that cytokeratins 10 and 12 are upregulated in the transgenic cells (data not shown). Although cytokeratins are usually thought of as epithelial cell differentiation markers, the most aggressive human breast cancers that have undergone EMT coexpress both vimentin and cytokeratins (28, 109), as do some malignant mouse mammary epithelial tumors that have undergone in EMT (113). Together, these immunostaining results not only demonstrate the transgenic human cell origin of the tumors formed in nude mice but are also entirely consistent with previous results indicating that some of the HMG-Y-overexpressing cells having undergone an EMT.
DISCUSSION
HMGI(Y) proteins are causal agents in tumor progression.
As recounted in the introduction, published reports of human clinical studies, as well as numerous studies of animal and cell culture model systems, have firmly established that aberrant expression or overexpression of members of the HMGI(Y) gene family positively correlates with neoplastic transformation and metastatic progression of a wide variety of tumors. Nevertheless, the results reported here represent the first direct demonstration that overexpression of the HMGI(Y) proteins is causally involved in modulating the expression of genes involved in all stages of tumor progression from transformation to anchorage-independent growth in vitro, to the formation of primary and metastatic tumors in nude mice, and the induction of in EMT in vivo. The current data unambiguously confirm the long-held suspicion the Hmgiy is a bone fide proto-oncogene and also provide insights into probable molecular events underlying its in vivo mode of action.
The results of gene array analyses revealed that induced overexpression of HMGI(Y) proteins in human mammary epithelial cells modulated, either upward or downward, the transcriptional expression of constellations of genes associated with regulation of cell signaling, cell proliferation, cell migration, tissue invasion, induction of angiogenesis, and metastatic colonization (Table 2; Fig. 7). Whether the HMGI(Y) proteins function directly as transcription factors by regulating the promoters of these genes or whether their effect is indirect and is mediated through other intermediary genes or proteins is unknown. Nevertheless, it is quite remarkable how closely the cDNA expression array profiles induced by HMGI(Y) overexpression correspond to the phenotypic patterns of the metastatic tumors formed by transgenic cells injected into nude mice. Examination of the pathology in host mice clearly showed that the HMG-Y transgene-containing cells injected into the mammary fat pad had metastasized to many distant sites in the mesenteries lining the body cavity and various organs and had also invaded muscle tissues, such as the diaphragm (Fig. 8 A to C). These biological results could only be achieved if many of the tumor progression gene products detected by the cDNA array analyses were indeed functional in vivo, a conclusion also supported by the histochemical and immunolocalization results shown in Fig. 9.
In vivo, the HMG-I and HMG-Y isoform proteins are not functionally equivalent.
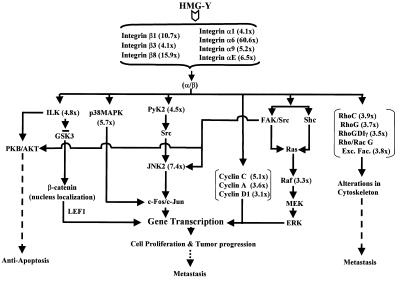
One of the most unexpected findings in this study was that transgenic cells overexpressing the HMG-Y isoform protein were much more effective in inducing both primary and metastatic tumors when injected into nude mice than were transgenic cells overexpressing the HMG-I isoform protein (Table 3). In less than 2 months following injection, over half (four of seven) of the nude mice injected with cells overexpressing the HMG-Y protein had developed tumors whereas during this same time period none of the five mice injected with cells overexpressing HMG-I developed tumors. Only much later (after about 4 months) did one of the mice injected with cells overexpressing the HMG-I protein develop small primary tumors (data not shown). Thus, HMG-Y is far more efficient in promoting tumor progression in vivo than HMG-I even though these isoform proteins differ only by an internal deletion of 11 amino acids in the former. These results were initially puzzling, since expression of both isoform proteins efficiently induced anchorage-independent growth in soft agar (Fig. 4; Table 1). Nevertheless, they were consistent with an earlier report demonstating that the tumor promoter 12-O-tetradecanoylphorbol acetate preferentially induces HMG-Y protein expression in transformation-sensitive, but not in transformation-resistant, mouse JB6 epithelial cells in culture (22). The cumulative data therefore strongly imply that, in addition to sharing many common biological functions, the HMG-I and HMG-Y proteins also exhibit important differences in their in vivo functions. This view is further supported by the finding that in vivo, the HMG-Y protein contains many more secondary biochemical modifications than the HMG-I isoform and that these modifications differentially affect the substrate-binding properties of the two proteins (5). A potential difference in function between the two isoforms is likewise supported by the results of the cDNA expression profiles and RT-PCR experiments reported earlier (Fig. 5 and 6; Table 2). The data from these experiments demonstrate that the HMG-I and HMG-Y proteins do, indeed, regulate the transcriptional expression of a large constellation of common genes but also indicate that, in addition, each protein can regulate its own independent set of genes. For example, both proteins upregulate the expression of genes such as those which encode the p38 mitogen-activated protein kinase (MAPK), collagen type XVIII, integrin β1, MMP13, and TIMP (tissue inhibitor of MMP3), but on the other hand, only HMG-Y upregulates genes such as those for Notch-4, Jagged-1, and Wnt-10B (Fig. 6) and only HMG-I downregulates the genes for Wnt-10B and Wnt-8B (data not shown). It is reasonable to suspect that the constellation of genes regulated by both the HMG-I and HMG-Y proteins is responsible for the anchorage-independent growth in soft agar of cells overexpressing these proteins. On the other hand, it is likely that some combination of the individual genes that are either up-or downregulated by the HMG-Y or HMG-I isoform protein is responsible for the much greater in vivo tumorigenic and metastatic potential of the HMG-Y protein. It should be stressed that the cDNA array analyses reported here sampled the transcriptional activity of only a very limited number of cellular genes. To fully address the issue of the extent to which the HMG-I and HMG-Y isoform proteins have distinct biological functions in tumor progression, a much more comprehensive transcriptional expression analysis employing high-density oligonucleotide array technology is necessary.
Genes modulated by HMGI(Y) overexpression.
The list of genes differentially regulated by HMG-Y overexpression in Table 2 demonstrates a broad range of functional activity of the HMG-Y architectural transcription factor. During mouse development, very high expression of HMGI(Y) proteins has been detected in all embryonic tissues up to 8.5 days of gestation, a period during which the most critical events of organogenesis start to take place, and its expression continues at lower levels to the end of gestation (17, 78). Based on these patterns of embryonic expression, the suggestion has been made that the HMGI(Y) proteins are not only involved with cell proliferation but also are likely to be involved with the establishment of various cell types during organogenesis. This suggestion is supported by the present data, which demonstrate that overexpression of HMG-Y significantly upregulates genes known to be involved in development, such as those for Frizzled 5 (161-fold), Wnt-13 (14.6-fold), Wnt-10B (4.1-fold), as well as Jagged-1 and its receptor, Notch-4 (7.5- and 4.1-fold, respectively). HMG-Y also upregulates a number of genes involved in both cell cycle regulation and signal transduction. These include the genes for CLK-1 (14.6-fold), cdc25A (98.6-fold), cdc25B (9.1-fold), cyclin C (5.1-fold), JNK2 (7.4-fold), and p38 MAPK (5.7-fold) and others. Each of these genes has been demonstrated not only to be involved in either the control of cell cycle or signaling processes but also to participate in neoplastic transformation and/or tumor progression in various systems. For example, the genes coding for the protein phosphatase enzymes cdc25A and cdc25B are well known as G1/S transition and G2/M transition regulators, respectively. In addition, they are all proto-oncogenes whose overexpression has been implicated in many types of tumors, including breast cancers (60, 135).
HMG-Y modulates expression of EMT genes.
Although studies analyzing the molecular characteristics of both mesenchymal and epithelial cells are still at an early stage, a group of genes has already been identified that is both characteristic of these two different cell types and influential in EMT (reviewed in reference 9). Analysis of the cDNA profile of epithelial cells overexpressing HMG-Y protein indicates that transcriptional expression of several EMT marker genes is modulated in HMG-Y overexpressing cells (Table 2). Among the group of upregulated genes are type I, III, and IV collagens, vimentin, certain cytokeratins, a number of matrix metalloproteases, basic FGF receptor 1, FGF7, and SF (scatter factor-hepatocyte growth factor) protein. Amoung the group of downregulated genes are those coding for the EGF receptor and TGF-β receptor III, as well as those coding for both the Met and FGFR2b proteins.
As noted previously, excess collagen production is a characteristic feature of EMT and clinical studies have correlated high levels of procollagen proteins with the most aggressive human breast cancers and those with the worst prognosis (12, 71). The cells of many of these highly malignant breast tumors also coexpress elevated levels of both vimentin and cytokeratins (29, 108, 113).
Extracellular and membrane-bound MMPs are a family of enzymes involved in much of the degradation of ECM molecules that occurs in developmental and pathological processes, including tumor invasion and metastasis and EMTs (84, 113). Overexpression of HMG-Y in human epithelial cells induces the expression of MMP16 (a potent activator of gelatinase MMP2) 13.5-fold, that of MMP13 (collagenase 3) 4.7-fold, that of MMP8 (collagenase 2 or neutrophil collagenase) 4.3-fold, and that of MDC9 (a membrane-anchored metalloprotease-disintegrin) 5.8-fold. Investigations have implicated the expression of all of these MMPs in the in vivo spread of tumor cells. For example, increased expression of MMP13 has been observed in many different human cancers, including breast carcinoma (53). Interestingly, in these breast carcinomas, MMP13 is expressed by tumor stromal cells or fibroblast-like tumor cells (also referred to as mesenchymal cells) (53). In addition to upregulating metalloproteases, overexpression of HMG-Y also increases the metalloprotease inhibitor TIMP-3 and plaminogen activator inhibitors PAI-1 and PAI-3 4.6-, 5.4-, and 74.2-fold, respectively. Overexpression of both TIMP-3 (125) and PAI-1 (101) has been detected in breast carcinomas, and high levels of PAI-1 are associated with a higher incidence of lymph node involvement in breast cancer and poor response to tamoxifen therapy (65). The role of MMP-inhibitory proteins such as TIMP-3 and PAI in tumor invasion and metastasis is unresolved and controversial, but many investigator now lean toward the view that there is a delicate interplay between these inhibitory proteins and the MMPs that regulate the invasiveness of tumor cells by controlling cellular adhesion to, and release from, the ECM (27).
SF and its specific receptor, c-Met, have been implicated in EMTs during both development (110) and tumor invasion and metastasis (9). SF is a potent pleiotrophic cytokine with multiple biological effects on various epithelial cells, including morphogenesis, proliferation, migration, and differentiation. It promotes development, regeneration, and reconstitution of normal organ architecture. The normal physiological roles played by SF and c-Met have been established in embryological studies. In the majority of tissues, SF is expressed exclusively on mesenchymal cells and c-Met, a receptor tyrosine kinase, is expressed on epithelial cells (110). The exchange of signals between the mesenchymal and epithelial cells has long been recognized as a major driving force in embryogenesis, and SF and c-Met constitute one such major paracrine signaling system (114). In the development of epithelium lined organs, for example, SF-expressing mesenchymal cells play an essential role in inducing epithelial cells to proliferate, migrate, and differentiate in a coordinated fashion. The c-Met receptor on the surface of the epithelial cells transduces the SF signal to induce motogenic, mitogenic, and morphogenetic and differentiation activities. The effects of SF and c-Met on the EMT and the generation of motile cells in normal developmental processes appear to be indistinguishable from the generation of motile carcinoma cells during tumor metastasis in vivo. Clinical studies have shown that the level of expression of SF by breast carcinoma cells correlates with the state of tumor progression (134). Interestingly, overexpression of HMG-Y in human breast epithelial cells upregulates SF 14.5-fold and downregulates c-Met 4.9-fold (Table 2). This strongly suggests that, in some cases, overexpression of HMG-Y promotes an EMT by coordinately upregulating the expression of mesenchymal cells genes and downregulating the expression of epithelial cell genes.
FGF7, which is produced by fibroblastic cells, and its receptor, FGFR2b, which is found on the surface of epithelial cells, represent another example of a paracrine system that actively mediates communication between mesenchymal and epithelial cells during embryonic development (130). In this case, too, overexpression of HMG-Y results in 4.1-fold upregulation of FGF7 and 29.3-fold down-regulate of FGFR2b in transgenic epithelial cells, again reinforcing the idea that overexpression induces EMT by coordinated up-and downregulation of genes.
HMGI(Y) upregulates integrins and their signaling pathways.
Integrins are a large family of transmembrane heterodimeric proteins that are the principal receptors on mammalian cells for the binding of ECM proteins. They are also the primary mediators of cell-cell interactions. At least 20 different integrin heterodimers, comprised of 14 types of α subunits and 8 types of β subunits, are known (62). Many of the different α and β subunits come in alternatively spliced forms which are expressed in a tissue-specific manner with different cell types producing multiple, but distinctive, sets of heterodimers. Although expressed on many different cell types, integrins are expressed predominantly in the mesenchyme during embryonic development (36).
Integrins functions as transmembrane linkers (or integrators) and mediate the interactions between the cytoskeleton, cytoplasmic kinases, and the ECM. Owing to this property, integrins enable the inside of cells and the ECM to communicate with each other across the plasma membrane in a bidirectional fashion (20, 41). That is, the extracellular binding activity of integrins is regulated from the inside of the cell (inside-out signaling) while binding of the ECM elicits signals that are transmitted into the cell (outside-in signaling). As one of the key signaling transducers, integrins influence cell shape, movement, proliferation, development, differentiation, and death (62). These cellular effects involve changes in the expression of many genes, including those coding for integrins themselves. Transcription factors are often the final executors of signals emanating from integrins and mediate the changes observed in gene expression patterns.
Overexpression of HMG-Y significantly upregulates the expression of several integrin genes, including those coding for the β1, β3, β8, α1, α6, α9, and αE proteins, as well as those coding for certain of the cellular components of integrin signaling pathways, such as integrin-linked kinase (ILK), protein tyrosine kinase 2 (also known as cell adhesion kinase β), c-Jun N-terminal kinase 2, p38 MAPK, and RhoC, a member of the Rho family of small GTP-hydrolyzing proteins (Table 2; Fig. 10). Since the cDNA array used in the present expression studies included only a restricted subset of the many genes known to be involved in integrin signaling, the data set is incomplete. Nevertheless, even with the limited amount of information available, it is clear that overexpression of HMG-Y upregulates the expression of many integrins and their signaling pathways and these, in turn, are likely to be causally involved in tumor progression.
FIG. 10.
Diagram of integrins and their signaling pathways that are upregulated by overexpression of HMGI(Y) proteins in human mammary epithelial cells (see text for details). PyK2, protein tyrosine kinase 2; JNK2, c-Jun N-terminal kinase 2; FAK, focal adhesion kinase; ERK, extracellular response kinase; Rho/Rac G., Exc. Fac., Rho/Rac G exchange factor.
Of particular interest is the fact that HMG-Y overexpression upregulates transcription of the integrin β1 gene. Integrins containing the β1 subunit are normally activated through binding to components of the ECM. The cytoplasmic domain of the β1 subunit is thought to interact directly with components of the actin cytoskeleton, such as α actinin and talin, thereby forming focal adhesion plaques (49) in the cytoplasm at points of contact between integrins and the ECM. The essential nature of the integrin β1 protein in linking the ECM and the actin cytoskeleton was shown by gene knockout experiments which demonstrated that β1 integrin deficiency in mice results in inner-cell mass failure and peri-implantation lethality (112). Elevated levels of β1 integrins are correlated with high proliferation potential and also serve as markers for stem cells (70, 90, 120). Overexpression of β1 integrin has been detected in many types of cancers and associated with metastasis. In human breast cancer, the total β1 integrin level is significantly higher in malignant epithelial cells than in nontumorigenic cells (129). By using a function-blocking antibody that inhibits β1 integrin, malignant breast epithelial cells have been demonstrated to undergo a striking morphological and functional normalization; moreover, these reverted cells became growth arrested and were found to exhibit reduced tumorigenicity in nude mice (129). In ovarian carcinomas, aggregation of β1 integrins mediates tumor invasion and metastasis by stimulation of the expression of MMP2 and MT1 MMP (31). The notion of β1 integrins promoting metastasis has been supported by other studies as well. Targeted disruption of the β1 integrin gene in T-cell lymphomas greatly reduces their metastatic capacity (115). Reciprocally, re-expression of β1 integrin in β1-deficient epithelial cells led to a malignant, cell-scattering phenotype that requires activation of the Rho family of GTPases (45). Most interestingly, small-cell lung cancer cells are apparently able to utilize β1 integrin to resist chemotherapeutic agents (103). In small-cell lung cancer tumors, β1 integrin-mediated adhesion of cells to ECM proteins around the tumor enhances tumorigenicity and activates protein tyrosine kinase activity that inhibits the activation of caspase-3 and apoptosis that is normally induced by chemotherapeutic agents (103).
The α9 protein is a recently identified integrin subunit that forms only one integrin heterodimer, the α9β1 integrin (90). Expression of α9β1 has been detected on various types of cells, including human carcinomas (90, 120). α9β1 promotes cell proliferation and migration when it interacts with tenascin, one of the components of the ECM (136), the gene for which is also upregulated (4.2-fold) by HMG-Y overexpression (Table 2). The cellular effects of α9β1 are mediated by phosphorylation and activation of the protein tyrosine kinase focal adhesion kinase (FAK) and the mitogen-activated kinase Erk2 (136). Recent studies have shown that α9β1 expressed on neutrophil cells interacts with VCAM-1, which is expressed on activated endothelial cells, and mediates neutrophil passage through the endothelial cell layer (105, 120). The implication of these studies in terms of cancer progression is striking. Overexpression of α9β1 in metastatic cells may well lead to tumor cells gaining extravasation capabilities and thus allow them to colonize various sites at the end state of the metastatic process. As with α9β1, overexpression of α6β1 can also contribute to cell proliferation and migration in mammary and prostate carcinomas (23, 80). Overexpression of HMG-Y upregulates the integrin β1 subunit 10.7-fold and the integrin subunits α9, α6, and β1, 5.2-, 60.6-, and 10.7-fold, respectively (Table 2).
As diagrammed in Fig. 10, these results support a model in which HMG-Y promotes tumor cell proliferation and metastasis, at least in part, through control of the level of expression of integrins and their signaling pathways. The following findings support such a model. Engagement of integrin β1-containing heterodimers activates ILK, a focal adhesion plaque-associated serine/threonine protein kinase that is emerging as a key regulatory protein that functions at one of the early convergence points of integrin-and growth factor-stimulated signaling (25, 50). The active form of ILK phosphorylates and activates PKB (also called Akt), a potent antiapoptosis enzyme that enable many types of tumors to evade programmed cell death (26, 32). ILK also directly phosphorylates glycogen synthase kinase 3, glycogen synthesis kinase thereby inhibiting 3 activity and leading to a dramatic relocalization of β-catenin to the cell nucleus (124). In the nucleus, β-catenin pairs with lymphoid enhancer factor 1 to form a complex that acts as an active transcription factor and plays an essential role in tumor progression (reviewed in reference 60). Interestingly, overexpression of ILK in epithelial cells leads to a complete EMT that is accompanied by a loss of cell-cell adhesions and an increase in vimentin expression (88, 133). In addition, overexpression of ILK also leads to anchorage-independent cell growth, as determined by growth in soft agar, suppression of apoptosis, increased invasion of the ECM, and tumorigenicity in nude mice (50, 93).
Heterodimers containing integrin β1 subunits are also know to be involved with activation of protein tyrosine kinase 2, the p38 MAPK, the focal adhesion kinase, and the Shc signaling pathways (reviewed in references 20, 41, and 127). Overexpression of integrins and the activation of one or more of the resulting signaling pathways are often associated with both tumor progression and increased metastatic potential of cancer cells (11, 72, 73, 106, 119).
Integrins also regulate cell spreading, migration, and scattering through activation of the Rho family of proteins, GTPases that interact with multiple downstream effectors to control the actin cytoskeleton (47, 126). Recent work by Clark et al. (21) has also demonstrated that overexpression of the wild-type RhoC protein alone induces a poorly metastatic human melanoma cell line to become highly metastatic when injected into nude mice whereas a dominant-negative Rho inhibits metastasis. Analysis of the phenotype of cells expressing dominant-negative Rho or RhoC indicated that the RhoC GTPase is important in invasion by tumor cells (21). Importantly, as shown in Table 2, overexpression of HMG-Y induces the upregulation of a number of Rho family proteins, including RhoC (3.9-fold), RhoG (3.7-fold) RhoGDIγ (3.5-fold), and Rho-Rac G exchange factor (3.8-fold), a finding consistent with the proposed model the involvement of the HMGI(Y) proteins in the promotion of tumor metastasis (Fig. 10).
Implications for human cancer.
There are more than 100 distinct types of cancer in humans, and subtypes of tumors can be found in each specific organ. Available evidence suggests that cancer cells have defects in the regulatory mechanisms that govern normal cell proliferation and homeostasis. The complexity and variety of tumor types raise the question of how many distinct regulatory mechanisms within a normal cell must be disrupted in order for it to become cancerous. The current view is that tumorigenesis is a multstep process (i.e., the multihit theory) and that each step reflects a genetic alteration that drives the progressive transformation of normal human cells into highly malignant derivatives. However, all types of cancers have common hallmarks, i.e., have acquired several common capabilities (48). Therefore, the simplicity elucidated from the complexity provokes another, even more important question i.e. whether there is an essential mechanism that is “hard wired” throughout all types of cancer? The cumulative results of numerous correlative studies have shown that HMGI(Y) genes are overexpressed in such diverse human epithelial tumor types as malignant prostate, breast, thyroid, and colon cancers, as well as aberrantly expressed in benign mesenchymal tumors such as a leiomyomas, adenomas, hamartomas, and lipomas. Recent studies employing gene expression array analyses of transcription profiles have also confirmed the overexpression of HMGI(Y) in biopsies of many human breast tumors (85, 92, 104). An even more interesting finding is that these array analyses also show that many of the genes listed in Table 2 are likewise overexpressed in surgical breast tumor biopsies, with the expression of several (including Notch-4, cyclin A, Rho GDL, and others) closely paralleling the expression of HMGI(Y) in the tumors (92) (e.g., see the primary data tables at the website http://genome-www.stanford.edu/molecularportraits). Combined with the presented results, the cumulative data therefore suggest that HMGI(Y) may play the role of a master gene, perhaps one of several, whose overexpression impacts multiple steps involved in breast cancer neoplasia. If so, a second critical question arises: which initial events turn on the overexpression of the Hmgiy gene?
The promoter regions of both the human (39; M. L. Pedulla, N. Treff, L. Beckenbauer, L. M. Resar, and R. Reeves, submitted for publication:) and mouse (132) Hmgiy genes contain binding sites for the transcription factors AP-1 and c-Myc. Phorbol esters, tumor-promoting substances that activate AP-1 transcription factors, have been demonstrated to stimulate transcription of both the human (89) and mouse (22) genes. In addition, Resar and her colleagues also recently demonstrated that the c-Myc protein, along with its partner, Max, regulates transcription of the mouse Hmgiy gene in vivo and that overexpression of either c-Myc or HMGI(Y) proteins leads to anchorage-independent growth of Rat-1 cells in soft agarose (132). These findings, coupled with the fact that mutations in the adenomatous polyposis coli tumor suppressor gene leads to overexpression of c-Myc and uncontrolled cell growth (52), logically lead to a number of plausible multistep mechanisms that might explain the widespread involvement of HMGI(Y) in neoplastic transformation and metastatic progression. In one experimentally testable scenario, the initial tumor lesion is envisioned as a mutation in a tumor suppressor gene (e.g., adenomatous polyposis coli) which allows the aberrant expression or overexpression of one or more proto-oncogenes (e.g., c-myc) coding for transcription factors that can induce the promoter of the Hmgiy gene. These initial lesions might result in neoplastic transformation of the cells but not necessarily lead to permanent activation of the Hmgiy gene. Subsequent exposure of these mutation-containing cells to substances that promoter tumor progression (e.g., phorbol esters that activate AP-1 transcription factors) could then induce constitutive overexpression of the Hmgiy gene (c.f. reference 22), setting in motion a cascade of events leading to tumor progression and metastasis. Regardless of the actual sequence of molecular events involved, however, the present work demonstrates that the HMGI(Y) proteins are attractive targets for antitumor drug development and also for other, novel forms of cancer treatment.
ACKNOWLEDGMENTS
This work was supported by NIH grant RO1-GM46352 (to R.R.) and U.S. Army Breast Cancer Research Grant DAMD17-96-16249 (to Y.L.).
We sincerely thank M. Nissen and other members of both the Reeves laboratory and that of M. Griswold (Washington State University) for helpful discussions and technical assistance during the course of this work. Our thanks also go to Charles W. Leathers (Washington State University College of Veterinary Medicine) for assistance with histological analyses, Adrea Cupp (Center for Reproductive Biology, Washington State University) for advice on immunohistochemical staining, and Eric Nilsson (also of the Center for Reproductive Biology, Washington State University) for assistance with tumor dissections.
ADDENDUM
It has recently been reported (131) that Rat-1a fibroblast cells overexpressing either the HMG-I, HMG-Y, or HMGI-C protein form tumors and distant metastases when injected into nude mice.
REFERENCES
- 1.Abe N, Watanabe T, Sugiyama M, Uchimura H, Chiappetta G, Fusco A. Determination of high mobility group I(Y) expression level in colorectal neoplasias: a potential diagnostic marker. Cancer Res. 1999;59:1169–1174. [PubMed] [Google Scholar]
- 2.Alizadeh A A, Eisen M B, Davis R E, Ma C, Lossos I S, Rosenwald A, Boldrick J C, Sabet H, Tran T, Yu X, Powell J I, Yang L, Marti G E, Moore T, Hudson J J, Lu L, Lewis D B, Tibshirani R, Sherlock G, Chan W C, Greiner T C, Weisenburger D D, Armitage J O, Warnke R, Staudt L M. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 1999;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1988. [Google Scholar]
- 4.Bandiera A, Bonifacio D, Manfioletti G, Mantovani F, Rustighi A, Zanconati F, Fusco A, Di Bonito L, Giancotti V. Expression of HMGI(Y) proteins in squamous intraepithelial and invasive lesions of the uterine cervix. Cancer Res. 1998;58:426–431. [PubMed] [Google Scholar]
- 5.Banks G C, Li Y, Reeves R. Differential in vivo modifications of the HMGI(Y) nonhistone chromatin proteins modulate nucleosome and DNA interactions. Biochemistry. 2000;39:8333–8346. doi: 10.1021/bi000378+. [DOI] [PubMed] [Google Scholar]
- 6.Banks R E, Gearing A J, Hemingway I K, Norlfolk D R, Perren T J, Selby P J. Circulating intercellular adhesion molecule-1(ICAM-1), E-selectin and vascular cell adhesion molecule-1 (VCAM-1) in human malignancies. Br J Cancer. 1993;68:122–124. doi: 10.1038/bjc.1993.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson K F, Chada K. Mini-mouse phenotypic characterization of a transgenic insertional mutant allelic to “pygmy.”. Genet Res. 1995;64:27–33. doi: 10.1017/s0016672300032511. [DOI] [PubMed] [Google Scholar]
- 8.Berlingieri M T, Manfioletti G, Santoro M, Bandiera A, Visconti R, Giancotti V, Fusco A. Inhibition of HMG-IC protein synthesis suppresses retrovirally induced neoplastic transformation of rat thyroid cells. Mol Cell Biol. 1995;15:1545–1553. doi: 10.1128/mcb.15.3.1545. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Birchmeier C, Birchmeier W, Brand-Saberi B. Epithelial-mesenchymal transitions in cancer progression. Acta Anat. 1996;156:217–226. doi: 10.1159/000147848. [DOI] [PubMed] [Google Scholar]
- 10.Birchmeier W, Behrens J, Weidner K M, Hulsken J, Birchmeier C. Epithelial differentiation and the control of metastasis in carcinomas. Curr Top Microbiol Immunol. 1996;213:117–135. doi: 10.1007/978-3-642-61109-4_6. [DOI] [PubMed] [Google Scholar]
- 11.Boudreau N, Bissell M J. Extracellular matrix signaling: integration of form and function in normal and malignant cells. Curr Opin Cell Biol. 1998;10:640–646. doi: 10.1016/s0955-0674(98)80040-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown L F, Guidi A J, Schnitt S J, Van De Water L, Iruela-Arispe M L, Yeo T K, Tognazzi K, Dvorak H F. Vascular stroma formation in carcinoma in situ, invasive carcinoma, and metastatic carcinoma of the breast. Clin Cancer Res. 1999;5:1041–1056. [PubMed] [Google Scholar]
- 13.Bussemakers M J, van de Ven W J, Debruyne F M, Schalken J A. Identification of high mobility group protien I(Y) as potential progression marker for prostate cancer by differential hybridization analysis. Cancer Res. 1991;51:606–611. [PubMed] [Google Scholar]
- 14.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 15.Chau K Y, Patel U A, Lee K L, Lam H Y, Crane-Robinson C. The gene for the human architectural transcription factor HMGI-C consists of five exons each coding for a distinct functional element. Nucleic Acids Res. 1995;23:4262–4266. doi: 10.1093/nar/23.21.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Silver D P, Walpita D, Cantor S B, Gazdar A F, Tomlinson G, Couch F J, Weber B L, Ashley T, Livingston D M, Scully R. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell. 1998;2:317–328. doi: 10.1016/s1097-2765(00)80276-2. [DOI] [PubMed] [Google Scholar]
- 17.Chiappetta G, Avantaggiato V, Visconti R, Fedele M, Battista S, Trapasso F, Merciai B M, Fidanza V, Giancotti V, Santoro M, Simeone A, Fusco A. High level expression of the HMGI(Y) gene during embryonic development. Oncogene. 1996;13:2439–2446. [PubMed] [Google Scholar]
- 18.Chiappetta G, Tallini G, De Biasio M C, Manfioletti G, Martinez-Tello F J, Pentimalli F, de Nigris F, Mastro A, Botti G, Fedele M, Berger N, Santoro M, Giancotti V, Fusco A. Detection of high mobility group I HMGI(Y) protein in the diagnosis of thyroid tumors: HMGI(Y) expression represents a potential diagnostic indicator of carcinoma. Cancer Res. 1998;58:4193–4198. [PubMed] [Google Scholar]
- 19.Chin M T, Pellacani A, Wang H, Lin S S, Jain M K, Perrella M A, Lee M E. Enhancement of serum-response factor-dependent transcription and DNA binding by the architectural transcription factor HMG-I(Y) J Biol Chem. 1998;273:9755–9760. doi: 10.1074/jbc.273.16.9755. [DOI] [PubMed] [Google Scholar]
- 20.Clark E A, Brugge J S. Integrins and signal transduction pathways: the road taken. Science. 1995;268:233–239. doi: 10.1126/science.7716514. [DOI] [PubMed] [Google Scholar]
- 21.Clark E A, Golub T R, Lander E S, Hynes R O. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406:532–535. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 22.Cmarik J L, Li Y, Ogram S A, Min H, Reeves R, Colburn N H. Tumor promoter induces high mobility group HMG-Y protein expression in transformation-sensitive but not -resistant cells. Oncogene. 1998;16:3387–3396. doi: 10.1038/sj.onc.1201888. [DOI] [PubMed] [Google Scholar]
- 23.Cress A E, Rabinovitz I, Zhu W, Nagle R B. The alpha 6 beta 1 and alpha 6 beta 4 integrins in human prostate cancer progression. Cancer Metastasis Rev. 1995;14:219–228. doi: 10.1007/BF00690293. [DOI] [PubMed] [Google Scholar]
- 24.Currie R A. Functional interaction between the DNA binding domain of NF-Y and the high mobility group protein HMG-I(Y) J Biol Chem. 1997;272:30880–30888. doi: 10.1074/jbc.272.49.30880. [DOI] [PubMed] [Google Scholar]
- 25.Dedhar S, Williams B, Hannigan G. Integrin-linked kinase (ILK): a regulator of integrin and growth-factor signalling. Trends Cell Biol. 1999;9:319–323. doi: 10.1016/s0962-8924(99)01612-8. [DOI] [PubMed] [Google Scholar]
- 26.Delcommenne M, Tan C, Gray V, Rue L, Woodgett J, Dedhar S. Phosphoinositide-3-OH kinase-dependent regulation of glycogen synthase kinase 3 and protein kinase B/AKT by the integrin-linked kinase. Proc Natl Acad Sci USA. 1998;95:11211–11216. doi: 10.1073/pnas.95.19.11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng G, Curriden S A, Wang S, Rosenberg S, Loskutoff D J. Is plasminogen activator inhibitor-1 the molecular switch that governs urokinase receptor-mediated cell adhesion and release? J Cell Biol. 1996;134:1563–1571. doi: 10.1083/jcb.134.6.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Domagala W, Lasota J, Bartkowiak J, Weber K, Osborn M. Vimentin is preferentially expressed in human breast carcinomas with low estrogen receptor and high Ki-67 growth fraction. Am J Pathol. 1990;136:219–227. [PMC free article] [PubMed] [Google Scholar]
- 29.Domagala W, Wozniak L, Lasota J, Weber K, Osborn M. Vimentin is preferentially expressed in high-grade ductal and medullary, but not in lobular breast carcinomas. Am J Pathol. 1990;137:1059–1064. [PMC free article] [PubMed] [Google Scholar]
- 30.Eldredge E R, Chiao P J, Lu K P. Use of tetracycline operator system to regulate oncogene expression in mammalian cells. Methods Enzymol. 1995;254:481–491. doi: 10.1016/0076-6879(95)54033-4. [DOI] [PubMed] [Google Scholar]
- 31.Ellerbroek S M, Stack M S. Membrane associated matrix metalloproteinases in metastasis. Bioessays. 1999;21:940–949. doi: 10.1002/(SICI)1521-1878(199911)21:11<940::AID-BIES6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 32.Evan G, Littlewood T. A matter of life and cell death. Science. 1998;281:1317–1322. doi: 10.1126/science.281.5381.1317. [DOI] [PubMed] [Google Scholar]
- 33.Falvo J V, Thanos D, Maniatis T. Reversal of intrinsic DNA bends in the IFN beta gene enhancer by transcription factors and the architectural protein HMG I(Y) Cell. 1995;83:1101–1111. doi: 10.1016/0092-8674(95)90137-x. [DOI] [PubMed] [Google Scholar]
- 34.Fambrugh D, McClure K, Kazlauskas A, Lander E S. Diverse signaling pathways activated by growth factor receptors induce broadly overlapping, rather than independent, sets of genes. Cell. 1999;97:727–741. doi: 10.1016/s0092-8674(00)80785-0. [DOI] [PubMed] [Google Scholar]
- 35.Fashena S J, Reeves R, Ruddle N H. A poly(dA-dT) upstream activating sequence binds high-mobility group I protein and contributes to lymphotoxin (tumor necrosis factor-β) gene regulation. Mol Cell Biol. 1992;12:894–903. doi: 10.1128/mcb.12.2.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fassler R, Georges-Labouesse E, Hirsch E. Genetic analyses of integrin function in mice. Curr Opin Cell Biol. 1996;8:641–646. doi: 10.1016/s0955-0674(96)80105-0. [DOI] [PubMed] [Google Scholar]
- 37.Fedele M, Berlingieri M T, Scala S, Chiariotti L, Viglietto G, Rippel V, Bullerdiek J, Santoro M, Fusco A. Truncated and chimeric HMGI-C genes induce neoplastic transformation of NIH3T3 murine fibroblasts. Oncogene. 1998;17:413–418. doi: 10.1038/sj.onc.1201952. [DOI] [PubMed] [Google Scholar]
- 38.Firth C H, Ward J M. Color atlas of neoplastic and non-neoplastic lesions in aging mice. Amsterdam, The Netherlands: Elsevier Press; 2000. [Google Scholar]
- 39.Friedmann M, Holth L T, Zoghbi H Y, Reeves R. Organization, inducible-expression and chromosome localization of the human HMG-I(Y) nonhistone protein gene. Nucleic Acids Res. 1993;21:4259–4267. doi: 10.1093/nar/21.18.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giancotti B V, Berlingieri M T, DiFiore P P, Fusco A, Vecchio G, Crane-Robinson C. Changes in nuclear proteins on transformation of rat epithelial thyroid cells by murine sarcoma retrovirus. Cancer Res. 1985;45:6051–6057. [PubMed] [Google Scholar]
- 41.Giancotti F G, Ruoslahti E. Integrin signaling. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 42.Giancotti V, Bandiera A, Fusco A, Marzari R, Coles B, Goodwin G H. Comparison of multiple forms of high mobility group proteins in rodent and human cells. Identification of the human high mobility group I-C protein. Eur J Biochem. 1991;198:211–216. doi: 10.1111/j.1432-1033.1991.tb16003.x. [DOI] [PubMed] [Google Scholar]
- 43.Giancotti V, Buratti E, Perissin L, Zorzet S, Balmain A, Portella G, Fusco A, Goodwin G H. Analysis of the HMGI nuclear proteins in mouse neoplastic cells induced by different procedures. Exp Cell Res. 1989;184:538–545. doi: 10.1016/0014-4827(89)90352-2. [DOI] [PubMed] [Google Scholar]
- 44.Giancotti V, Pani B, D'Andrea P, Berlingieri M T, DiFiore P P, Fusco A, Vecchio G, Philip R, Crane-Robinson C, Nicolas R H, Wright C A, Goodwin G H. Elevated levels of a specific class of nuclear phosphoproteins in cells transformed with v-ras and v-mos oncogenes and by co-transfection with c-myc and polyoma middle T antigens. EMBO J. 1987;6:1981–1987. doi: 10.1002/j.1460-2075.1987.tb02461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gimond C, van Der F, van Delft S, Brakebusch C, Kuikman I, Collard J G, Fassler R, Sonnenberg A. Induction of cell scattering by expression of beta1 integrins in beta1-deficient epithelial cells requires activation of members of the rho family of GTPases and downregulation of cadherin and catenin function. J Cell Biol. 1999;147:1325–1340. doi: 10.1083/jcb.147.6.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hackett A J, Smith H S, Springer E L, Owens R B, Nelson-Rees W A, Riggs J L, Gardnee M B. Two syngenic cell lines from human breast tissue: the aneuploid mammary epithelial (Hs578T) and the diploid myoepithelial (Hs578Bst) cell lines. J Natl Cancer Inst. 1977;58:1795–1800. doi: 10.1093/jnci/58.6.1795. [DOI] [PubMed] [Google Scholar]
- 47.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 48.Hanahan D, Weinberg R A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 49.Hannigan G, Dedhar S. Protein kinase mediators of integrin signal transduction. J Mol Med. 1997;75:35–44. doi: 10.1007/s001090050084. [DOI] [PubMed] [Google Scholar]
- 50.Hannigan G E, Leung-Hagesteijn C, Fitz-Gibbon L, Coppolino M G, Radeva G, Filmus J, Bell J C, Dedhar S. Regulation of cell adhesion and anchorage-dependent growth by a new beta 1-integrin-linked protein kinase. Nature. 1996;379:91–96. doi: 10.1038/379091a0. [DOI] [PubMed] [Google Scholar]
- 51.Hay E D. An overview of epithelio-mesenchymal transformation. Acta Anat. 1995;154:8–20. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- 52.He T C, Sparks A B, Rago C, Hermeking H, Zawel L, da Costa L T, Morin P J, Vogelstein B, Kinzler K W. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 53.Heppner K J, Matrisian L M, Jensen R A, Rodgers W H. Expression of most matrix metalloproteinase family members in breast cancer represents a tumor-induced host response. Am J Pathol. 1996;149:273–282. [PMC free article] [PubMed] [Google Scholar]
- 54.Hess J L. Chromosomal translocations in benign tumors. Am J Clin Pathol. 1998;109:251–261. doi: 10.1093/ajcp/109.3.251. [DOI] [PubMed] [Google Scholar]
- 55.Hill D A, Pedulla M L, Reeves R. Directional binding of HMG-I(Y) on four-way junction DNA and the molecular basis for competitive binding with HMG-1 and histone H1. Nucleic Acids Res. 1999;27:2135–2144. doi: 10.1093/nar/27.10.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hill D A, Reeves R. Competition between HMG-I(Y), HMG-1 and histone H1 on four-way junction DNA. Nucleic Acids Res. 1997;25:3523–3531. doi: 10.1093/nar/25.17.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Himes S R, Coles L S, Reeves R, Shannon M F. High mobility group protein I(Y) is required for function and for c-Rel binding to CD28 response elements within the GM-CSF and IL-2 promoters. Immunity. 1996;5:479–489. doi: 10.1016/s1074-7613(00)80503-8. [DOI] [PubMed] [Google Scholar]
- 58.Himes S R, Reeves R, Attema J, Nissen M, Li Y, Shannon M F. The role of high-mobility group I(Y) proteins in expression of IL-2 and T cell proliferation. J Immunol. 2000;164:3157–3168. doi: 10.4049/jimmunol.164.6.3157. [DOI] [PubMed] [Google Scholar]
- 59.Holth L T, Thorlacius A E, Reeves R. Effects of epidermal growth factor and estrogen on the regulation of the HMG-I/Y gene in human mammary epithelial cell lines. DNA Cell Biol. 1997;16:1299–1309. doi: 10.1089/dna.1997.16.1299. [DOI] [PubMed] [Google Scholar]
- 60.Hunter T. Oncoprotein networks. Cell. 1997;88:333–346. doi: 10.1016/s0092-8674(00)81872-3. [DOI] [PubMed] [Google Scholar]
- 61.Huth J R, Bewley C A, Nissen M S, Evans J N, Reeves R, Gronenborn A M, Clore G M. The solution structure of an HMG-(Y)-DNA complex defines a new architectural minor groove binding motif. Nat Struct Biol. 1997;4:657–665. doi: 10.1038/nsb0897-657. [DOI] [PubMed] [Google Scholar]
- 62.Hynes R O. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 63.Innis M A, Gelfand D H, Sninsky J J, White T K. PCR protocols: a guide to applications. New York, N.Y: Academic Press, Inc.; 1990. [Google Scholar]
- 64.Iyer V R, Eisen M B, Ross D T, Schuler G, Moore T, Lee J F, Trent J M, Staudt L M, Hudson J J, Boguski M S, Lashkari D, Shalon D, Botstein D, Brown P O. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 65.Janicke F, Schmitt M, Pache L, Ulm K, Harbeck N, Hofler H, Graeff H. Urokinase (uPA) and its inhibitor PAI-1 are strong and independent prognostic factors in node-negative breast cancer. Breast Cancer Res Treat. 1993;24:195–208. doi: 10.1007/BF01833260. [DOI] [PubMed] [Google Scholar]
- 66.John S, Reeves R B, Lin J-X, Child R, Leiden J M, Thompson C B, Leonard W J. Regulation of cell-type-specific interleukin-2 receptor α-chain gene expression: potential role of physical interactions between Elf-1, HMG-I(Y), and NF-κB family proteins. Mol Cell Biol. 1995;15:1786–1796. doi: 10.1128/mcb.15.3.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johnson K R, Disney J E, Wyatt C R, Reeves R. Expression of mRNAs encoding mammalian chromosomal proteins HMG-I and HMG-Y during cellular proliferation. Exp Cell Res. 1990;187:69–76. doi: 10.1016/0014-4827(90)90118-t. [DOI] [PubMed] [Google Scholar]
- 68.Johnson K R, Lehn D A, Elton T S, Barr P J, Reeves R. Complete murine cDNA sequence, genomic structure, and tissue expression of the high mobility group protein HMG-I(Y) J Biol Chem. 1988;263:18338–18342. [PubMed] [Google Scholar]
- 69.Johnson K R, Lehn D A, Reeves R. Alternative processing of mRNAs encoding mammalian chromosomal high-mobility-group proteins HMG-I and HMG-Y. Mol Cell Biol. 1989;9:2114–2123. doi: 10.1128/mcb.9.5.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jones P H, Watt F M. Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell. 1993;73:713–724. doi: 10.1016/0092-8674(93)90251-k. [DOI] [PubMed] [Google Scholar]
- 71.Jukkola A, Tahtela R, Tholix E, Vuorinen K, Blanco G, Risteli L, Risteli J. Aggressive breast cancer leads to discrepant serum levels of the type I procollagen propeptides PINP and PICP. Cancer Res. 1997;57:5517–5520. [PubMed] [Google Scholar]
- 72.Kornberg L J. Focal adhesion kinase and its potential involvement in tumor invasion and metastasis. Head Neck. 1998;20:745–752. doi: 10.1002/(sici)1097-0347(199812)20:8<745::aid-hed14>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 73.Kumar C C. Signaling by integrin receptors. Oncogene. 1998;17:1365–1373. doi: 10.1038/sj.onc.1202172. [DOI] [PubMed] [Google Scholar]
- 74.Lanahan A, Williams J B, Sanders L K, Nathans D. Growth factor-induced delayed early response genes. Mol Cell Biol. 1992;12:3919–3929. doi: 10.1128/mcb.12.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lehn D A, Elton T S, Johnson K R, Reeves R. A conformational study of the sequence specific binding of HMG-I(Y) with the bovine interleukin-2 cDNA. Biochem Int. 1988;16:963–971. [PubMed] [Google Scholar]
- 76.Levenson A S, Jordan V C. MCF-7: the first hormone-responsive breast cancer cell line. Cancer Res. 1997;57:3071–3078. [PubMed] [Google Scholar]
- 77.Lewis H, Kaszubska W, DeLamarter J F, Whelan J. Cooperativity between two NF-kB complexes, mediated by high-mobility-group protein I(Y), is essential for cytokine-induced expression of the E-selectin promoter. Mol Cell Biol. 1994;14:5701–5709. doi: 10.1128/mcb.14.9.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu J, Schiltz J F, Shah P C, Benson K F, Chada K K. Genomic structure and expression of the murine Hmgi(y) gene. Gene. 2000;246:197–207. doi: 10.1016/s0378-1119(00)00073-1. [DOI] [PubMed] [Google Scholar]
- 79.Liu W M, Guerra-Vladusic F K, Kurakata S, Lupu R, Kohwi-Shigematsu T. HMG-I(Y) recognizes base-unpaired regions of matrix attachment sequences and its increased expression is directly linked to metastatic breast cancer phenotype. Cancer Res. 1999;59:5695–5703. [PubMed] [Google Scholar]
- 80.Lochter A, Navre M, Werb Z, Bissell M J. alpha1 and alpha2 integrins mediate invasive activity of mouse mammary carcinoma cells through regulation of stromelysin-1 expression. Mol Biol Cell. 1999;10:271–282. doi: 10.1091/mbc.10.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lund T, Holtlund J, Fredriksen M, Laland S G. On the presence of two new high mobility group-like proteins in HeLa S3 cells. 1983. FEBS Lett. 1983;152:163–167. doi: 10.1016/0014-5793(83)80370-6. [DOI] [PubMed] [Google Scholar]
- 82.Lundberg K, Karlson J R, Ingebrigtsen K, Holtlund J, Lund T, Laland S G. On the presence of the chromosomal proteins HMG-I and HMG-Y in rat organs. Biochim Biophys Acta. 1989;1009:277–279. doi: 10.1016/0167-4781(89)90113-9. [DOI] [PubMed] [Google Scholar]
- 83.Maniatis T, Whittemore L A, Du W, Fan C M, Keller A D, Palombella V J, Thanos D N. Positive and negative control of human interferon-beta gene expression. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. pp. 1193–1220. [Google Scholar]
- 84.Matrisian L M. The matrix-degrading metalloproteinases. Bioessays. 1992;14:455–463. doi: 10.1002/bies.950140705. [DOI] [PubMed] [Google Scholar]
- 85.Nacht M, Ferguson A T, Zhang W, Petroziello J M, Cook B P, Gao Y H, Maguire S, Riley D, Coppola G, Landes G M, Madden S L, Sukumar S. Combining serial analysis of gene expression and array technologies to identify genes differentially expressed in breast cancer. Cancer Res. 1999;59:5464–5470. [PubMed] [Google Scholar]
- 86.Nagpal S, Ghosn C, DiSepio D, Molina Y, Sutter M, Klein E S, Chandraratna R A. Retinoid-dependent recruitment of a histone H1 displacement activity by retinoic acid receptor. J Biol Chem. 1999;274:22563–22568. doi: 10.1074/jbc.274.32.22563. [DOI] [PubMed] [Google Scholar]
- 87.Nissen M S, Reeves R. Changes in superhelicity are introduced into closed circular DNA by binding of high mobility group protein I/Y. J Biol Chem. 1995;270:4355–4360. doi: 10.1074/jbc.270.9.4355. [DOI] [PubMed] [Google Scholar]
- 88.Novak A, Hsu S C, Leung-Hagesteijn C, Radeva G, Papkoff J, Montesano R, Roskelley C, Grosschedl R, Dedhar S. Cell adhesion and the integrin-linked kinase regulate the LEF-1 and beta-catenin signaling pathways. Proc Natl Acad Sci USA. 1998;95:4374–4379. doi: 10.1073/pnas.95.8.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ogram S A, Reeves R. Differential regulation of a multipromoter gene: selective 12-O-tetradecanoylphrobol-13-acetate induction of a single transcription start site in the HMG-I/Y gene. J Biol Chem. 1995;270:14235–14242. doi: 10.1074/jbc.270.23.14235. [DOI] [PubMed] [Google Scholar]
- 90.Palmer E L, Ruegg C, Ferrando R, Pytela R, Sheppard D. Sequence and tissue distribution of the integrin alpha 9 subunit, a novel partner of beta 1 that is widely distributed in epithelia and muscle. J Cell Biol. 1993;123:1289–1297. doi: 10.1083/jcb.123.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pellacani A, Chin M T, Wiesel P, Ibanez M, Patel A, Yet S F, Hsieh C M, Paulauskis J D, Reeves R, Lee M E, Perrella M A. Induction of high mobility group-I(Y) protein by endotoxin and interleukin-1beta in vascular smooth muscle cells. Role in activation of inducible nitric oxide synthase. J Biol Chem. 1999;274:1525–1532. doi: 10.1074/jbc.274.3.1525. [DOI] [PubMed] [Google Scholar]
- 92.Perou C M, Sorlie T, Eisen M B, van de Rijn M, Jeffrey S S, Rees C A, Pollack J R, Ross D T, Johnsen H, Akslen L A, Fluge O, Pergamenschikov A, Williams C, Zhu S X, Lonning P E, Borresen-Dale A L, Brown P O, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 93.Radeva G, Petrocelli T, Behrend E, Leung-Hagesteijn C, Filmus J, Slingerland J, Dedhar S. Overexpression of the integrin-linked kinase promotes anchorage-independent cell cycle progression. J Biol Chem. 1997;272:13937–13944. doi: 10.1074/jbc.272.21.13937. [DOI] [PubMed] [Google Scholar]
- 94.Ram T, Reeves R, Hosick H. Genetics and molecular biology of breast cancer; meeting abstracts. N.Y: Cold Spring Harbor Laboratory Press, Cold Spring Laboratory; 1992. Expression of the chromatin regulatory protein HMG-I is an accurate diagnostic marker for neoplastic progression of mammary epithelial cells; p. 65. [Google Scholar]
- 95.Ram T G, Reeves R, Hosick H L. Elevated high mobility group-I(Y) gene expression is associated with progressive transformation of mouse mammary epithelial cells. Cancer Res. 1993;53:2655–2660. [PubMed] [Google Scholar]
- 96.Reeves R. Structure and function of the HMGI(Y) family of architectural transcription factors. Environ Health Perspect. 2000;108(Suppl. 5):803–809. doi: 10.1289/ehp.00108s5803. [DOI] [PubMed] [Google Scholar]
- 97.Reeves R, Nissen M S. The A · T-DNA-binding domain of mammalian high mobility group I chromosomal proteins: a novel peptide motif for recognizing DNA structure. J Biol Chem. 1990;265:8573–8582. [PubMed] [Google Scholar]
- 98.Reeves R, Nissen M S. Interaction of high mobility group-I(Y) nonhistone proteins with nucleosome core particles. J Biol Chem. 1993;268:21137–21146. [PubMed] [Google Scholar]
- 99.Reeves R, Nissen M S. Purification and assays for high mobility group HMG-I(Y) protein function. Methods Enzymol. 1999;304:155–187. doi: 10.1016/s0076-6879(99)04011-2. [DOI] [PubMed] [Google Scholar]
- 100.Reeves R, Wolffe A P. Substrate structure influences binding of the non-histone protein HMG-I(Y) to free and nucleosomal DNA. Biochemistry. 1996;35:5063–5074. doi: 10.1021/bi952424p. [DOI] [PubMed] [Google Scholar]
- 101.Reilly D, Christensen L, Duch M, Nolan N, Duffy M J, Andreasen P A. Type-1 plasminogen activator inhibitor in human breast carcinomas. Int J Cancer. 1992;50:208–214. doi: 10.1002/ijc.2910500209. [DOI] [PubMed] [Google Scholar]
- 102.Scala S, Portella G, Fedele M, Chiappetta G, Fusco A. Adenovirus-mediated suppression of HMGI(Y) protein synthesis as potential therapy of human malignant neoplasias. Proc Natl Acad Sci USA. 2000;97:4256–4261. doi: 10.1073/pnas.070029997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sethi T, Rintoul R C, Moore S M, MacKinnon A C, Salter D, Choo C, Chilvers E R, Dransfield I, Donnelly S C, Strieter R, Haslett C. Extracellular matrix proteins protect small cell lung cancer cells against apoptosis: a mechanism for small cell lung cancer growth and drug resistance in vivo. Nat Med. 1999;5:662–668. doi: 10.1038/9511. [DOI] [PubMed] [Google Scholar]
- 104.Sgroi D C, Teng S, Robinson G, LeVangie R, Hudson J R J, Elkahloun A G. In vivo gene expression profile analysis of human breast cancer progression. Cancer Res. 1999;59:5656–5661. [PubMed] [Google Scholar]
- 105.Shang T, Yednock T, Issekutz A C. Alpha-9 beta-1 integrin is expressed on human neutrophils and contributes to neutrophil migration through human lung and synovial fibroblast barriers. J Leukoc Biol. 1999;66:809–816. doi: 10.1002/jlb.66.5.809. [DOI] [PubMed] [Google Scholar]
- 106.Shaw L M. Integrin function in breast carcinoma progression. J Mammary Gland Biol Neoplasia. 1999;4:367–376. doi: 10.1023/a:1018766317055. [DOI] [PubMed] [Google Scholar]
- 107.Solomon M, Strauss F, Varshavsky A. A mammalian high mobility group protein recognizes any stretch of six A-T base pairs in duplex DNA. Proc Natl Acad Sci USA. 1986;83:1276–1280. doi: 10.1073/pnas.83.5.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sommers C L, Byers S W, Thompson E W, Torri J A, Gelmann E P. Differentiation state and invasiveness of human breast cancer cell lines. Breast Cancer Res Treat. 1994;31:325–335. doi: 10.1007/BF00666165. [DOI] [PubMed] [Google Scholar]
- 109.Sommers C L, Skerker J M, Chrysogelos S A, Bosseler M, Gelmann E P. Regulation of vimentin gene transcription in human breast cancer cell lines. Cell Growth Differ. 1994;5:839–846. [PubMed] [Google Scholar]
- 110.Sonnenberg E, Meyer D, Weidner K M, Birchmeier C. Scatter factor/hepatocyte growth factor and its receptor, the c-Met tyrosine kinase, can mediate a signal exchange between mesenchyme and epithelia during mouse development. J Cell Biol. 1993;123:223–235. doi: 10.1083/jcb.123.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Staebler A, Sommers C, Mueller S C, Byers S, Thompson E W, Lupu R. Modulation of breast cancer progression and differentiation by the gp30/heregulin. Breast Cancer Res Treat. 1994;31:175–182. doi: 10.1007/BF00666151. [DOI] [PubMed] [Google Scholar]
- 112.Stephens L E, Sutherland A E, Klimanskaya I V, Andrieux A, Meneses J, Pedersen R A, Damsky C H. Deletion of beta 1 integrins in mice results in inner cell mass failure and peri-implantation lethality. Genes Dev. 1995;9:1883–1895. doi: 10.1101/gad.9.15.1883. [DOI] [PubMed] [Google Scholar]
- 113.Sternlicht M D, Lochter A, Sympson C J, Huey B, Rougier J P, Gray J W, Pinkel D, Bissell M J, Werb Z. The stromal proteinase MMP3/stromelysin-1 promotes mammary carcinogenesis. Cell. 1999;98:137–146. doi: 10.1016/s0092-8674(00)81009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stoker M, Gherardi E, Perryman M, Gray J. Scatter factor is a fibroblast-derived modulator of epithelial cell mobility. Nature. 1987;327:239–242. doi: 10.1038/327239a0. [DOI] [PubMed] [Google Scholar]
- 115.Stroeken P J, van Rijthoven E A, van der Valk M A, Roos E. Targeted disruption of the beta1 integrin gene in a lymphoma cell line greatly reduces metastatic capacity. Cancer Res. 1998;58:1569–1577. [PubMed] [Google Scholar]
- 116.Tallini G, Dal Cin P. HMGI(Y) and HMGI-C dysregulation: a common occurrence in human tumors. Adv Anat Pathol. 1999;6:237–246. [PubMed] [Google Scholar]
- 117.Tamimi Y, van der Poel H G, Denyn M, Umbas R, Karthaus H F, Debruyne F M, Schalken J A. Increased expression of high mobility group protein I(Y) in high grade prostate cancer determined by in situ hybridization. Cancer Res. 1993;53:5512–5516. [PubMed] [Google Scholar]
- 118.Tamimi Y, van der Poel H G, Karthaus H F, Debruyne F M, Schalken J A. A retrospective study of high mobility group protein I(Y) as a progression marker for prostate cancer determined by in situ hybridization. Br J Cancer. 1996;74:573–578. doi: 10.1038/bjc.1996.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tamura M, Gu J, Tran H, Yamada K M. PTEN gene and integrin signaling in cancer. J Natl Cancer Inst. 1999;91:1820–1828. doi: 10.1093/jnci/91.21.1820. [DOI] [PubMed] [Google Scholar]
- 120.Taooka Y, Chen J, Yednock T, Sheppard D. The integrin alpha-9 beta-1 mediates adhesion to activated endothelial cells and transendothelial neutrophil migration through interaction with vascular cell adhesion molecule-1. J Cell Biol. 1999;145:413–420. doi: 10.1083/jcb.145.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Thanos D, Maniatis T. The high mobility group protein HMG-I(Y) is reguired for NF-κB-dependent virus induction of the human interferon-beta gene. Cell. 1992;71:777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- 122.Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 123.Thiery J P, Chopin D. Epithelial cell plasticity in development and tumor progression. Cancer Metastasis Rev. 1999;18:31–42. doi: 10.1023/a:1006256219004. [DOI] [PubMed] [Google Scholar]
- 124.Troussard A A, Tan C, Yoganathan T N, Dedhar S. Cell-extracellular matrix interactions stimulate the AP-1 transcription factor in an integrin-linked kinase- and glycogen synthase kinase 3-dependent manner. Mol Cell Biol. 1999;19:7420–7427. doi: 10.1128/mcb.19.11.7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Uria J A, Stahle-Backdahl M, Seiki M, Fueyo A, Lopez-Otin C. Regulation of collagenase-3 expression in human breast carcinomas is mediated by stromal-epithelial cell interactions. Cancer Res. 1997;57:4882–4888. [PubMed] [Google Scholar]
- 126.Van Aelst L, D'Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 127.Wary K K, Mainiero F, Isakoff S J, Marcantonio E E, Giancotti F G. The adaptor protein Shc couples a class of integrins to the control of cell cycle progression. Cell. 1996;87:733–743. doi: 10.1016/s0092-8674(00)81392-6. [DOI] [PubMed] [Google Scholar]
- 128.Ways D K, Kukoly C A, deVente J, Hooker J L, Bryant W O, Posekany K J, Fletcher D J, Cook P P, Parker P J. MCF-7 breast cancer cells transfected with protein kinase C-alpha exhibit altered expression of other protein kinase C isoforms and display a more aggressive neoplastic phenotype. J Clin Investig. 1995;95:1906–1915. doi: 10.1172/JCI117872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Weaver V M, Petersen O W, Wang F, Larabell C A, Briand P, Damsky C, Bissell M J. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Weinberg S, Liao X, Peters K G, Blessing M, Yuspa S H, Weiner R A W L. Targeted expression of a dominant-negative FGF receptor mutant in the epidermis of transgenic mice reveals a role for FGF in keratinocyte organization and differentiation. EMBO J. 1993;12:2635–2643. doi: 10.1002/j.1460-2075.1993.tb05924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wood L J, Maher J F, Bunton T E, Resai L M. The oncogenic properties of the HMG-I gene family. Cancer Res. 2000;60:4256–4261. [PubMed] [Google Scholar]
- 132.Wood L J, Mukherjee M, Dolde C E, Xu Y, Maher J F, Bunton T E, Williams J B, Resar L M S. HMG-I/Y, a new c-Myc target gene and potential oncogene. Mol Cell Biol. 2000;20:5490–5502. doi: 10.1128/mcb.20.15.5490-5502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wu C, Keightley S Y, Leung-Hagesteijn C, Radeva G, Coppolino M, Goicoechea S, McDonald J A, Dedhar S. Integrin-linked protein kinase regulates fibronectin matrix assembly, E-cadherin expression, and tumorigenicity. J Biol Chem. 1998;273:528–536. doi: 10.1074/jbc.273.1.528. [DOI] [PubMed] [Google Scholar]
- 134.Yamashita J, Ogawa M, Yamashita S, Nomura K, Kuramoto M, Saishoji T, Shin S. Immunoreactive hepatocyte growth factor is a strong and independent predictor of recurrence and survival in human breast cancer. Cancer Res. 1994;54:1630–1633. [PubMed] [Google Scholar]
- 135.Yao Y, Slosberg E D, Wang L, Hibshoosh H, Zhang Y J, Xing W Q, Santella R M, Weinstein I B. Increased susceptibility to carcinogen-induced mammary tumors in MMTV-Cdc25B transgenic mice. Oncogene. 1999;18:5159–5166. doi: 10.1038/sj.onc.1202908. [DOI] [PubMed] [Google Scholar]
- 136.Yokosaki Y, Monis H, Chen J, Sheppard D. Differential effects of the integrins alpha-9 beta-1, alpha-v beta-3, and alpha-v beta-6 on cell proliferative responses to tenascin. Roles of the beta subunit extracellular and cytoplasmic domains. J Biol Chem. 1996;271:24144–24150. doi: 10.1074/jbc.271.39.24144. [DOI] [PubMed] [Google Scholar]