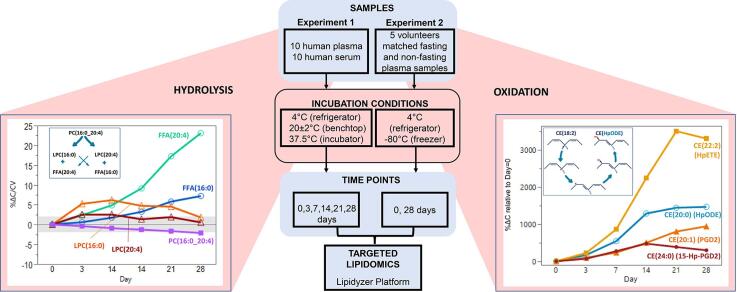
Graphical abstract
Abbreviations: FFA, Free Fatty Acid; DAG, Diacylglycerol; TAG, Triacylglycerol; CE, Cholesteryl ester; PC, Phosphatidylcholine; LPC, Lysophosphatidylcholine; PE, Phosphatidylethanolamine; LPE, Lysophosphatidylethanolamine; SM, Sphingomyelin; CER, Ceramide; PL, Phospholipid; LysoPL, Lysophospholipid; HpODE, hydroperoxyoctadecadienoic acid; HpETE, hydroperoxyeicosatetraenoic acid; PGD2, prostaglandin D2; 15-Hp-PGD2, 15-hydroperoxy-prostaglandin D2; PLA1, phospholipase A1; PLA2, phospholipase A2
Keywords: Lipidomics, Stability, Degradation, Oxidation, Hydrolysis, Cholesteryl Ester, Triglycerides, Fatty Acids
Highlights
-
•
Lipid degradation was induced by storage at typical sample preparation temperatures.
-
•
Identification of low abundance lipids that increased several-fold during incubation.
-
•
Enzymatic hydrolysis and oxidation were identified as likely causes of degradation.
-
•
Most high abundance lipids that degrade do so within method variability.
Abstract
Large epidemiological studies often require sample transportation and storage, presenting unique considerations when applying advanced lipidomics techniques. The goal of this study was to acquire lipidomics data on plasma and serum samples stored at potential preanalytical conditions (e.g., thawing, extracting, evaporating), systematically monitoring lipid species for a period of one month. Split aliquots of 10 plasma samples and 10 serum samples from healthy individuals were kept in three temperature-related environments: refrigerator, laboratory benchtop, or heated incubator. Samples were analyzed at six different time points over 28 days using a Bligh & Dyer lipid extraction protocol followed by direct infusion into a lipidomics platform using differential mobility with tandem mass spectrometry. The observed concentration changes over time were evaluated relative to method and inter-individual biological variability. In addition, to evaluate the effect of lipase enzyme levels on concentration changes during storage, we compared corresponding fasting and post-prandial plasma samples collected from 5 individuals. Based on our data, a series of low abundance free fatty acid (FFA), diacylglycerol (DAG), and cholesteryl ester (CE) species were identified as potential analytical markers for degradation. These FFA and DAG species are typically produced by endogenous lipases from numerous triacylglycerols (TAGs), and certain high abundance phosphatidylcholines (PCs). The low concentration CEs, which appeared to increase several fold, were likely mass-isobars from oxidation of other high concentration CEs. Although the concentration changes of the high abundant TAG, PC, and CE precursors remained within method variability, the concentration trends of FFA, DAG, and oxidized CE products should be systematically monitored over time to inform analysts about possible pre-analytical biases due to degradation in the study sample sets.
Introduction
Lipidomics in clinical Research
The lipidome represents the collection of thousands of distinct lipid molecular species over a wide range of abundances that are unique to the physiology of each organism, circulating biofluid, cell type, and cellular compartment. Analysis of the lipidome (lipidomics) has become a new biochemical discipline through the use of highly multiplexed mass spectrometry (MS) platforms. As lipidomics continues to increase in use as a clinical analysis tool, a thorough understanding of the stability of lipid molecules in biological samples during the preanalytical phase and during sample preparation prior to MS detection is essential [1]. Systematically collected data that would aid the development of guidelines is sparse.
The primary reason for the lack of sample stability is the dynamic nature of the lipidome in vivo. The concentration and relative abundance of individual lipid species is regulated by the concerted action of hundreds of enzymes and transfer proteins. Several enzymes that circulate in the blood remain active in vitro. The activity of these enzymes is observable through concentration changes of lipid classes and lipid species, as differentiated by polar head group or backbone, and covalently attached fatty acyl groups, respectively. A single freeze–thaw cycle can have measurable effects on absolute concentrations of some lipid species [2]. Even at temperatures as low as −80 °C, long term storage can introduce slow, but detectable, changes [3]. For example, lipases hydrolyze fatty acyl (FA) chains on glycerol and phosphatidylglycerol backbones. FA group hydrolysis of highly abundant lipids yields otherwise lower abundant lyso-species and free fatty acids (FFA). The possible enzymatic precursors of FFAs include triacylglycerol (TAG), diacylglycerol (DAG), phosphatidylcholine (PC), phosphatidylethanolamine (PE). Additionally, lysophospholipid species, such as lyso-PC (LPC), and lyso-PE (LPE) can be hydrolyzed and release a second FFA. Like glycerolipids, but with a synthetically and structurally different head group, sphingolipids, including sphingomyelin (SM) and ceramide (CER) species, also contain FA chains linked by an amide bond to a sphingosine backbone, and FFA release requires a different set of enzymatic pathways. Similarly, FFA release from cholesteryl ester (CE) species requires specialized enzymes.
Following sample collection, the chemical integrity of lipid molecules is also vulnerable to oxidation due to the reactivity of double bonds on long carbon chains. Polyunsaturated fatty acyl (PUFA) chains are especially vulnerable [4]. Additionally, oxidation can produce derivatives with MS signatures that overlap those of other naturally occurring intact lipids (mass isobars), especially in unit mass resolution mode tandem MS/MS applications. For example, FA(18:2) can be oxidized into FA(HpODE) (hydroperoxyoctadecadienoic acid), which has the same unit mass as FA(20:0). Thus, oxidation of the naturally high abundant CE(18:2) by a fraction of one percent to CE(HpODE) leads to a false increase in the detection of CE(20:0) by several fold due to its relatively low natural abundance. Theoretically, numerous other mass isobars could be formed due to oxidation.
Current literature contains insufficient experimental data on the effect of storage conditions on fluctuations in the levels of specific lipid species, especially in the context of relative abundance, method variability, and inter-individual differences. Such data is crucial to properly evaluate scientific conclusions drawn from lipidomics data. Systematic collection of such experimental data is time consuming, but also important, as the lipidomics field moves toward developing guidelines for interlaboratory comparison studies, standard operating procedures, and quality control.
The main goal of this study was to evaluate the effect of various temperature conditions on lipidomics analysis results over a period of four weeks using plasma and serum samples from 20 different individual donors, and plasma samples freshly collected from 5 individuals after fasting and 2 h after the consumption of a fatty drink. Our data provide evidence of temperature dependent lipolysis and oxidative degradation. The observed changes in lipid species over time were evaluated relative to method variability, inter-individual variability, and differences between corresponding fasting and post-prandial sample collections.
Materials and methods
Chemicals and reagents
HPLC grade methanol (MeOH), dichloromethane (DCM), 1-propanol, 2-propanol, and water were purchased from Fisher Scientific (Waltham, MA, U.S.A.). Ammonium acetate (NH4AcO) was obtained from MilliporeSigma (St. Louis, MO, U.S.A.). The internal standards kit for the Lipidyzer platform was purchased from Sciex (Framingham, MA, U.S.A). Human plasma for quality control (QC) was purchased from BioIVT, Inc. (Westbury, NY, U.S.A.).
Human Serum and Plasma samples
Ten human plasma and ten serum samples from healthy individuals were purchased from BioIVT, Inc. (Westbury, NY, U.S.A.). Sample collection was conducted in accordance with an IRB approved protocol. Samples were kept frozen at −80 °C until the start of the experiment.
In addition, ten fresh plasma samples from five volunteers were received from the Clinical Nutrition Research Laboratory of Emory University (Atlanta, U.S.A). Five healthy volunteers consumed a fatty drink containing 100 g of a long chain triglyceride emulsion, in the form of a commercial liquid drink (Calogen, from Nutricia, Inc.). Each volunteer provided one plasma sample after at least eight hours of fasting, and another sample two hours after the fatty drink consumption. The collected samples were transported on ice and stored at 4 °C for no>18–20 h before analysis.
Ethics statement
All samples were collected and handled anonymously. The project was approved as research not involving identifiable human subjects under U.S. Health and Human Services Department Policy for Protection of Human Research Subjects codified of Federal Regulations at 45 CFR part 46.
Static Incubation experiment at different temperatures
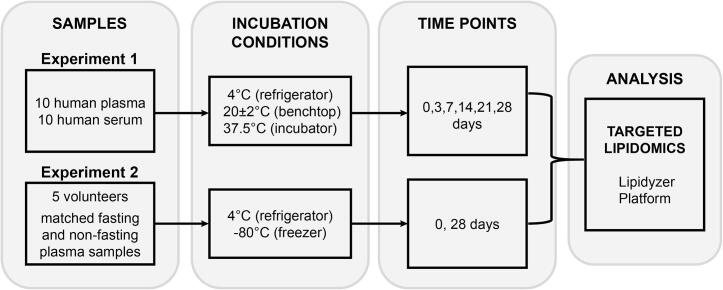
Ten serum and ten plasma samples collected from 20 individuals were shipped on dry ice and stored at −80 °C until first thaw. For monitoring sample stability over time (Experiment 1), after thawing at 4 °C, samples were distributed into three sets of three 200 µL aliquots. One set was stored at 4 °C (“refrigerator”), a second set at 20 ± 2 °C (room temperature, “benchtop”) and the third set at 37.5 °C (“incubator”). Experiment 1 was performed without repeats. Experiment 2 was designed to compare sample stability in freshly drawn fasting and postprandial (2 h) plasma. The fresh, never frozen, samples were transported on ice on the same day, immediately distributed into sets of two 200 µL aliquots per sample, and stored at 4 °C and −80 °C. In Experiment 2, analyses were performed in duplicate. From each stored aliquot for both experiments, 25 µL was used at different time points (0, 3, 7, 14, 21, and 28 days) for targeted lipidomics analysis (Fig. 1).
Fig. 1.
Workflow for the static incubation experiment at different temperatures.
Lipid extraction
Lipids were extracted using a modified Bligh and Dyer extraction protocol [5], that included 2 mL methanol, 1 mL dichloromethane (DCM), 1 mL water, and 25 µL internal standard mixture (AB Sciex, P\N 5040156) added to 25 µL plasma or serum, and vortex mixed. The resulting monophasic mixture was incubated at room temperature (20 ± 2 °C) for 30 min followed by a 10-minute centrifugation at 1200 RPM. The lower layer containing lipids was transferred to a separate tube. The lipid extraction was repeated a second time, the lower phases were merged and evaporated under nitrogen to dryness, and reconstituted with 250 µLof buffer containing 50:50 (v:v) DCM:MeOH and 10 mM NH4AcO.
LC-DMS-MS/MS Analysis
The targeted lipidomics analysis was performed using a Lipidyzer platform (AB Sciex, USA). A 50 µL aliquot of the extracts was injected into a constant 50 µL/min flow of 50:50 (v:v) DCM:MeOH and 10 mM NH4AcO buffer and directly infused into the triple quadrupole Sciex QTRAP 5500 mass spectrometer. The infusion was repeated using two different mass spectrometry methods, each run in positive and in negative modes. The first method used the SelexION Differential Mobility Spectrometry (DMS) with 1-propanol as a gas modifier to analyze PC, PE, SM, LPC, and LPE species. The second method was run without DMS to analyze TAG, DAG, CER, CE, and FFA species. For instrument tuning and suitability testing, the vendor provided tuning mix (P\N 5040141) and a system suitability kit (P\N 5040407) that was analyzed monthly. The overall method reproducibility was established using quality control (QC) material prepared from pooled human plasma samples. All MS data processing was performed with the Lipidomics Workflow Manager software, including lipid species identification and concentration calculation in nmol/mL based on isotope dilution principles, multiplying signal area ratios with the nmol amount of corresponding isotope-labeled internal standard in the infused extracts, and dividing by the volume of plasma or serum extracted.
Quality Control Procedures
Samples were measured immediately at each of the time points (0, 3, 7, 14, 21, and 28 days), in two batches based on sample type (plasma or serum). Therefore, each batch included 10 individual unknowns stored at 3 temperature conditions (for a total of 30 unknowns), along with 2–4 QCs and 2 solvent blanks (at the beginning and the end of each batch). An overall mean, standard deviation, and CV of the method were determined previously by characterization runs (n = 100) over one year and used for determination of tolerance limits for total lipid class concentrations. The quality control charts by lipid class were evaluated using Westgard rules, to accept or reject batches for that day. Overall method CV by species was also determined based on the method characterization runs. Lipid species that were < 1% of the lipid class on average in the QC samples were excluded from the list of species of interest and not used in statistical analysis. Applying the < 1% species abundance cutoff also meant the exclusion of species with missing values. Extraction blanks consisting of PBS and internal standard were analyzed to ascertain average background concentration levels for each lipid species (CBlank). After applying the < 1% species abundance cutoff, the 95th percentile of the blank concentrations were compared to the 5th percentile of the unknowns. There was only substantial overlap for DAGs and FFAs. This bias was minimized by subtracting the mean of the extraction blanks from all species values in the unknowns. Correction for inter-day bias (batch effects) was determined based on the deviation of average daily batch QC (n = 2–4) from the QC mean during the study from all batches (n = 18 for Experiment 1, n = 7 for Experiment 2). Batch corrections were applied by species in all unknown samples. Outliers in the data were identified by visual examination, resulting exclusion of one sample on one day in Experiment 1. PCA plots were generated with samples and QCs to examine the effect of batch correction. To visually observe the potential for batch effects, principal component analysis was performed and component 1 was plotted against component 2. PCA plots for QC samples are shown in supplemental Figure S1.
Data analysis
The reported lipid species concentrations were imported into the statistical analysis software, JMP (SAS Institute, U.S.A). The species abundance in each sample was calculated by dividing the lipid species concentrations by the sum of all species concentrations within the same lipid class. Only species with > 1% average abundance by class were evaluated, otherwise denoted.
Lipids in the plasma or serum samples were measured on the day of first thaw (Experiment 1), within 24 h of collection (Experiment 2). These concentrations were used as a Day = 0 reference point (C0). The change in absolute concentration (ΔC) was calculated by subtracting C0 from the concentrations of aliquots kept at different temperature conditions for a varying number of days (Ci), ΔC = Ci-C0. Because day-to-day bias of the absolute ΔC values for high concentration species could be greater due to pipetting errors and variation in extraction efficacy, ΔC was normalized to C0, giving relative concentration changes, %ΔC = ((Ci – C0)/C0))*100. Because %ΔC could have proportionately greater noise interference for low concentration species, or for lipid classes with lower ionization efficiency (i.e., positive versus negative ionization modes), %ΔC was divided by the method CV of each species that was established based on the mean and standard deviation of repeated analyses of a QC pool. As mentioned above, method CV for each lipid species was determined from over 100 technical repeats during a period of one year. The %ΔC/CV values for each lipid species and lipid class were calculated by storage temperature and timepoint. Trends in %ΔC/CV values inside the ± 2 range were contributed to method variability.
Since concentration trends during Experiment 1 at refrigerator temperature (22 ± 2 °C) were less obvious, a paired t-test was performed on the mean differences between Day = 0 and Day = 28 for all lipid species separately for plasma and serum. Confidence intervals were calculated for each analysis and significance was determined if the value 0 was not between the upper and lower 95% confidence limits. Significant species for refrigerated plasma are listed in Supplemental Table S4 and for serum in Supplemental Table S5. Lack of significance indicated a wider range of inter-individual variation than the change due to degradation between Day = 0 and Day = 28.
For Experiment 2, a paired t-test was performed on the mean differences between Day = 0 and Day = 28 for all lipid species, separately for fasting and postprandial samples stored at various temperatures. Confidence intervals were calculated for each analysis and significance was determined if the value 0 was not between the upper and lower 95% confidence limits. Lack of significance indicated a wider range of inter-individual variation than the change due to different levels of lipases in the pre- and postprandial samples, or degradation between Day = 0 and Day = 28.
Results and discussion
Experimental design
We chose three conditions to monitor changes in lipid classes and lipid species, which we refer to as refrigerator (4 °C), bench (20 ± 2 °C), and incubator (37.5 °C) conditions. These temperature conditions could occur during routine sample handling, transport, extraction, evaporation, or MS analysis of plasma and serum samples, but for a relatively short time (i.e., 30 min to 2 h). To observe quantifiable trends in concentration increases or decreases of lipid species, the plasma and serum aliquots were monitored for a period of 28 days. We assume that similar changes may occur in freezers even at −20 to −80 °C after months and years of storage. We evaluated the concentration changes relative to Day = 0 to identify lipid species that are most vulnerable to potential enzymatic hydrolysis or chemical oxidation during sample storage. Monitoring sample degradation across 20 individual samples allowed for the assessment of how biased analytical results from degradation can lead to confounding scientific conclusions in larger human population studies.
Trends in total lipid class concentration changes
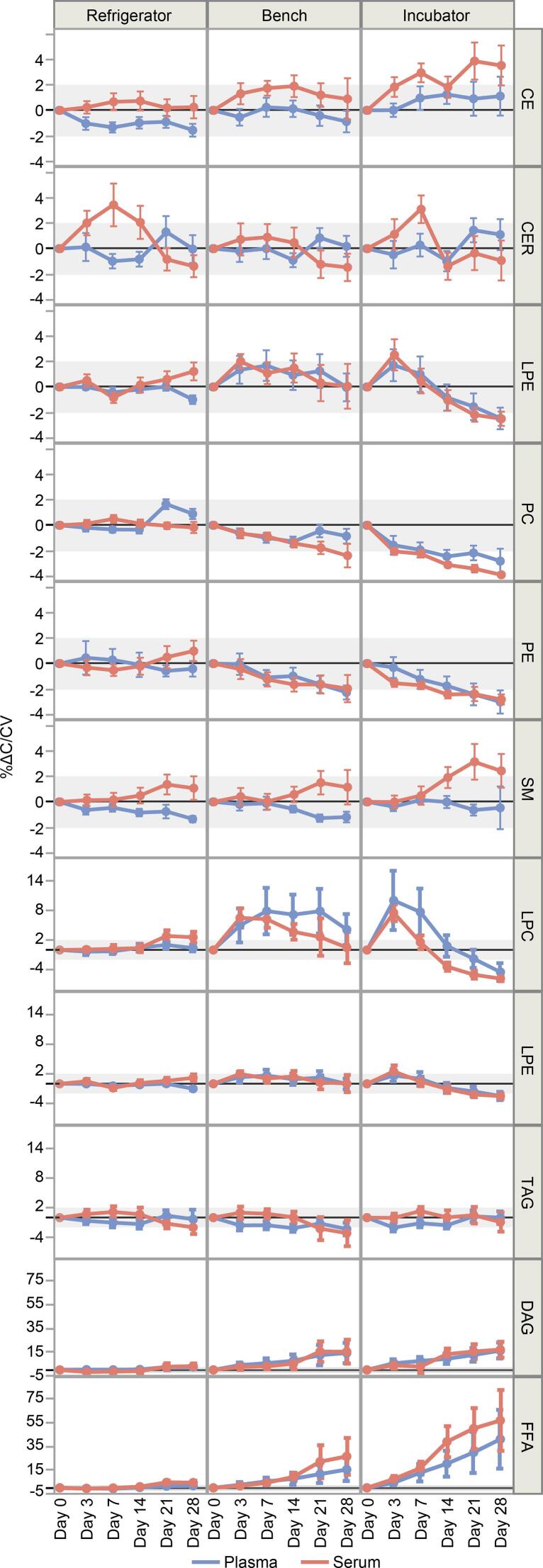
CEs, PCs and TAGs normally combine to make up ∼ 74% of total plasma lipids [6]. The concentration of these lipid classes remained within method variability after 1 month of storage at 4 °C. Surprisingly, changes in TAG levels remained within the range of method variability at all temperature conditions, even in an incubator at 37.5 °C, %ΔC/CV < 2 (Fig. 2). This is probably due to the large number of TAG species with < 1% abundance, thus relatively low signal/noise ratio, preventing the quantitative observation of concentration changes outside the range of method variability. On the contrary, other lipid classes are dominated by relatively few individual species, allowing more precise quantification of concentration changes as a sum of the lipid class, outside the range of method variability. PCs and PEs decreased over time while DAGs and FFAs increased, consistent with FA group hydrolysis in the samples. Lysophospholipids (Lyso-PLs) showed an initial increase then decrease, which is also expected because LPCs and LPEs are hydrolysis products of PCs and PEs, but can be hydrolyzed further to FFAs. In terms of stoichiometric balance, the absolute concentration decreases of the potential substrates (i.e., TAGs, PCs, and PEs) were proportionate with the concentration increases of the products (i.e., DAGs and FFAs). In other words, the number of moles of DAGs and FFAs produced was comparable to the number of moles of TAGs, PCs and PEs lost. Because of their large concentration differences, the changes in DAG, LPC, and FFA concentrations were outside the range of method variability (%ΔC/CV > 2), while the changes in TAGs and PCs were within the range of method variability (%ΔC/CV < 2).
Fig. 2.
Plasma (n = 10) and serum (n = 10) %ΔC/CV values by class totals over the course of 28 days in refrigerator, bench, and incubator. Error bars indicate standard deviation.
SM and CER species concentrations, in all temperature conditions, varied mostly within the range of %ΔC/CV of ± 2, indicating stability (Fig. 2). This is not surprising, since SMs and CERs contain a sphingosine backbone, unlike TAGs and PCs with glycerol backbones. Although hydrolysis of SM headgroups can lead to the formation of CERs, the changes in analogous SM and CER species did not correspond with each other. Furthermore, SMs and CERs typically contain saturated or monounsaturated FAs, unlike TAGs and PCs with often two or more double bonds on one of the FA groups. Thus, SMs are expected to be chemically less vulnerable to oxidative degradation [7].
Changes in lipid species concentrations by endogenous lipase activity
While the FA carbon chain lengths and number of double bonds on phospholipids can be determined by unit resolution MS/MS, the sn1 and/or sn2-positions of the FA pairs cannot. However, for the majority of phospholipids (PLs) in human samples, it can be assumed that the sn1 positions typically contain fewer double bonds than the sn2 position. Similarly, the position of the single FA on LPCs and LPEs cannot be determined. In the case of TAG species, the carbon chain lengths and number of double bonds can be determined for only one of the three FA groups, but without position identification.
Within each lipid class at Day = 0, the abundance of the species was in general agreement with literature reports for human plasma or serum [6], [8], [9]. The mean species data collected on various days from plasma and serum samples at three storage conditions is provided in supplementary information Table S1 and S2, respectively. In terms of ΔC and %ΔC/CV, the dominant species showed similar trends to their corresponding total lipid class levels. Changes in minor species with < 10% abundance changed significantly within 28 days without substantially affecting the total concentration of the corresponding lipid class.
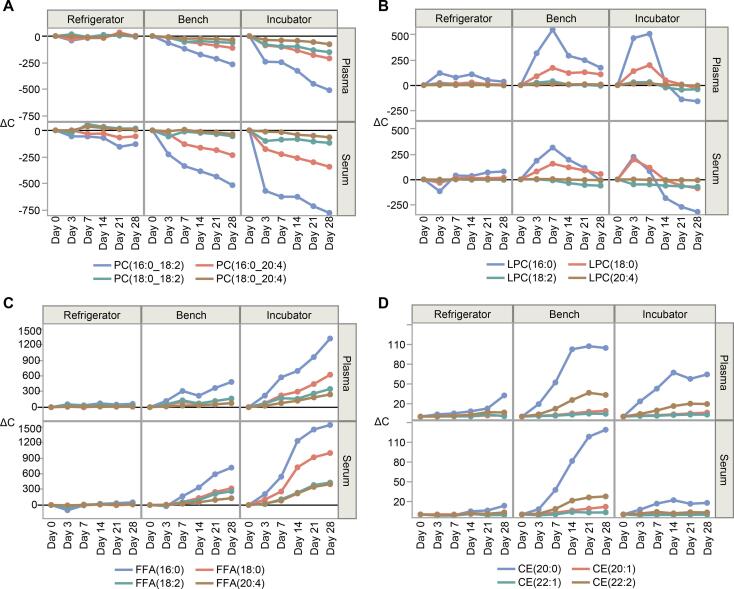
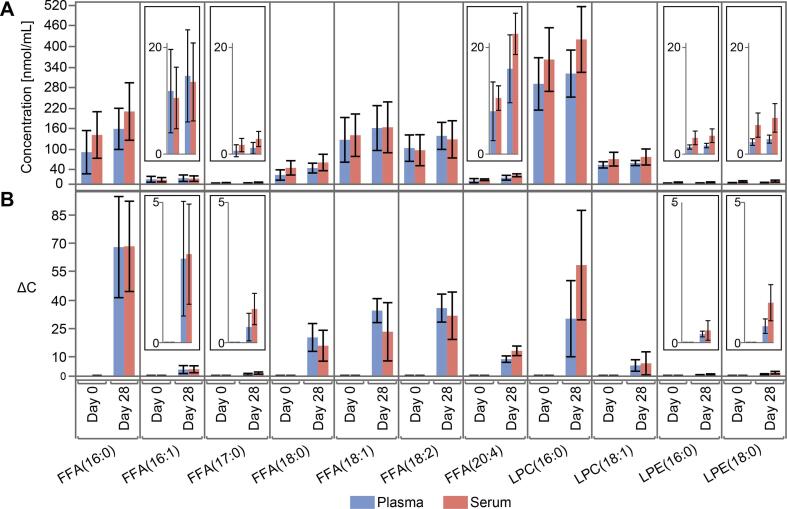
FFAs can be produced by endogenous lipase enzymes acting on many lipid species by cleaving the fatty acids from sn1, sn2, or sn3 positions of their glycerol or glycerophosphate backbones. FFA concentrations exhibited substantial increases within the 28 days in the refrigerator and on the bench (Fig. 3C and Supplement Table S2). The most substantial increases were observed for four FFAs: palmitic acid (16:0), stearic acid (18:0), linoleic acid (18:2), and arachidonic acid (20:4).
Fig. 3.
Plasma and serum absolute concentration changes (ΔC) by each day relative to Day = 0 for all storage conditions in species significantly changed in plasma and serum during Experiment 1. PCs (A) and their associated hydrolysis products LPCs (B) and FFAs (C). Selected plasma CEs (D) relative to Day = 0 that are likely isobaric oxidized CEs. For CE species we did not apply 1% abundance filter due to isobaric species having low concentrations.
DAGs are metabolically transient molecules involved in several cell-signaling pathways and tend to exist at relatively low concentrations in plasma, with a range of ∼ 0.1 – 9.7 nmol/mL (Supplemental Table S2). Unfortunately, many of the DAG species in our method have high solvent background relative to the sample levels. Some DAG species appeared to increase several-fold in the incubator (up to 20-fold relative to Day = 0, and up to 200-fold relative to the blanks), including DAG(16:0_18:1), (16:0_18:2), (18:0_18:2), (18:1_18:1) and (18:1_18:2), however, the causes of these increases are difficult to explain since DAGs can be produced in the sample by FA loss from TAGs, as well as by the loss of phosphocholine/ethanolamine head groups from PCs and PE [10].
Lyso-PLs can act as both substrates and products of endogenous lipase enzymes [11]. Enzymatic hydrolysis of a PC or PE yields one FFA and one LPC or LPE as products. For the most abundant Lyso-PLs, our data showed an initial increase in concentration, reaching a maximum by Day = 7 on the bench and in the incubator. This increase in LysoPL concentration was followed by a steady but substantial decrease. In the refrigerator, LysoPLs only increased (Fig. 3B). In other studies, LPC/PC ratio is recommended as a marker for degradation [12], however, this may be misleading considering the dynamic, time-sensitive behavior of LPC and LPE from our data.
The most noticeable trends were seen for PL species containing a 16:0 or 18:0 FA in one of the positions (likely sn1) and a PUFA in the other (likely sn2) (Fig. 3). Decreases of PC(16:0_18:2), corresponded with increases of LPC(16:0) and FFA(18:2), and likewise, PC(18:0_20:4) with LPC(18:0) and FFA(20:4). Similar corresponding patterns were found for other structurally related triads of PC, LPC, and FFA species, and of PE, LPE, and FFA.
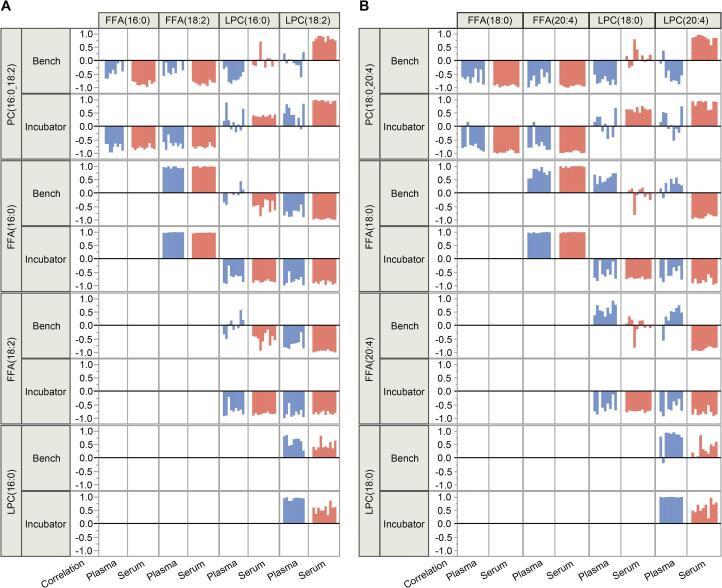
We hypothesize that degradation of PCs would be linked with changes in structurally corresponding LPCs and FFAs through either enzymatic or chemical hydrolysis. A pairwise correlation analysis was performed to assess the correlations of PC(16:0_18:2) and PC(18:0_20:4) with their corresponding FFAs and LPCs, using 10 individual samples, and 2 samples types, at 2 conditions, across several time points.In the case of PCs vs. FFAs, we observed inverse correlations between PCs and FFAs in both plasma and serum, both on the bench and incubator (Fig. 4). These correlations can be explained by substrate-product relationships through hydrolysis. In plasma vs. serum, hydrolysis was more dependent on incubation conditions, stronger correlations were seen in the incubator relative to the bench, indicating a preference for a higher temperature.
Fig. 4.
Correlation coefficients (r) for select PCs and their FFA + LPC hydrolysis products in bench and incubator conditions. (A) PC(16:0_18:2), FFA(16:0), FFA(18:2), LPC(16:0), LPC(18:2). (B) PC(18:0_20:4), FFA(18:0), FFA(20:4), LPC(18:0), LPC(20:4).
The pattern of correlations between PCs vs. LPCs, were more nuanced, both in terms of the magnitude and direction of the correlations. The correlations were also dependent on sample type (i.e., serum and plasma), temperature condition, and FA group saturation. Serum samples both on the bench and in the incubator showed positive correlation between PCs and LPCs. The positive PC-LPC correlation was more equivocal for the unsaturated LPC, in contrast to the saturated LPC product which preferred higher temperature (Fig. 4).
Plasma samples in the incubator also showed weak positive correlation between PCs and LPCs. The correlations with the unsaturated LPC were stronger than with the saturated LPC product (as with serum). However, plasma samples on the bench showed ambiguous correlation with the unsaturated LPC and negative correlations with the saturated LPC product (opposite of serum) (Fig. 4).
The explanation for these differences by serum vs. plasma, as well as by saturation of the FA groups, may be in the kinetics of the complex multistep reaction of the hydrolysis of PC to LPC and FFA. The difference between unsaturated and saturated LPC indicates the influence of enzymatic selectivity, rather than simple chemical degradation. The enzymes that are most likely involved in the PC-LPC reactions are PLA1 and PLA2. The EDTA in plasma can chelate the Ca++ (necessary for clotting), a known cofactor of PLA1 and PLA2. The reduced PLA1 and PLA2 activity changes the rate of the first reaction (PC to LPC) relative to the second reaction (LPC to FA). The kinetic balance of the two reactions becomes altered, as seen in our plasma vs. serum data.
Oxidation products
Double bonds on carbon chains of FA groups are well known to be prone to oxidation by molecular oxygen facilitated by endogenous enzymes. For example, CE(18:2) is typically the single most abundant lipid in blood, and it is a substrate of the oxidase enzyme ALOX-15 [13], [14], producing CE(HpODE), which happens to be a unit mass isobar of CE(20:0). CE(20:0) comprises only a small percentage of total cholesteryl esters ∼ 0.3% of the total CE class (Supplemental S2 and S3). However, even in the refrigerator, we saw its concentration increase with %ΔC of ∼ 40% in 14 days and ∼ 100% in 21 days. Interestingly, the increase of the CE(20:0) isobar (likely CE(HpODE)) was higher on the bench than in the incubator. This may be due to the known low temperature optimum demonstrated in mammalian 15-LOX (20 °C–35 °C) [15], therefore lower activity at > 35 °C [14].
There are other known oxidized lipids and FAs mass isobars: prostaglandin D2 (PGD2) and FA20:1; hydroperoxyeicosatetraenoic acid (HpETE) (336.23 m/z) and FA22:2 (336.30 m/z); and 15-hydroperoxy-prostaglandin D2 (15-Hp-PGD2) (368.36 m/z) as FA24:0 (368.30 m/z). We indeed found CE(20:1), CE(22:2), and CE(24:0) showing similar patterns as CE(20:0), including the observation of greater %ΔC on the bench than in the incubator (Fig. 3D).
Comparison of changes in refrigerator with inter-individual variation.
Of the conditions used in Experiment 1 (Fig. 1), refrigerator storage is the most likely occurrence in a laboratory setting. Bench and incubator storage conditions were included in this study to rapidly induce trends that occur at a much slower rates in the refrigerator. Since we monitored sample degradation in 20 individual samples (10 plasma and 10 serum), we were able to assess inter-individual variation versus degradation processes in both matrices. We performed ANOVA on the differences between Day = 0 and 28, separately for plasma and serum data from the refrigerator for all lipid species after filtering as described. The FFAs with ≥ 1% class abundance that were significantly higher after 28 days in the refrigerator (Fig. 5) included FFAs (16:0), (16:1), (17:0) (18:0), (18:1), (18:2) and (20:4). Additionally, certain Lysophospholipids showed significant increases in both plasma and serum (Fig. 5, Table S4). We believe that these lipid species should be used with caution in studies using archived plasma or serum samples. We found that the degradation changes in refrigerated samples (28 days) for most species, besides FFAs and a few Lyso-PLs, were within the range of inter-individual variation.
Fig. 5.
Significant species in plasma and serum from Experiment 1, by ANOVA analysis of mean difference between Day = 0 and Day = 28 in the Refrigerator. (A) Mean absolute concentrations (C, nmol/mL) on Day = 0 and Day = 28 with 95% confidence interval error bars. (B) Mean absolute concentration differences (ΔC) between Day = 0 and Day = 28 with 95% confidence interval error bars.
Degradation in fasting vs. postprandial plasma samples
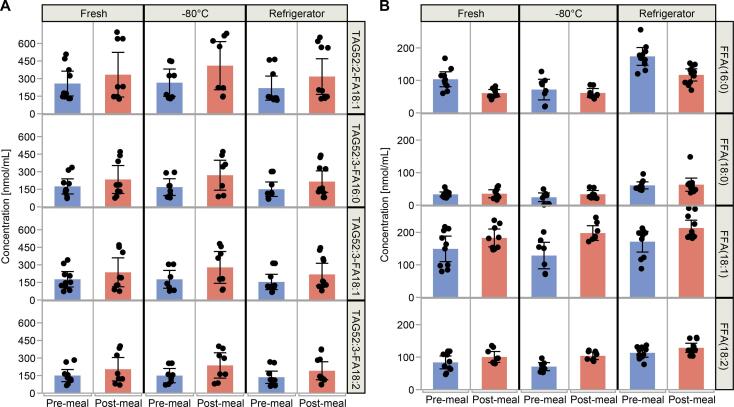
Most concentration trends that were seen in the previously frozen samples discussed above can be explained by the activity of endogenous lipases in the samples. Lipases are expected to be up-regulated after food intake. We also acquired data from paired plasma samples from 5 individuals collected before and after consumption of a fatty drink. These were fresh, never frozen aliquots analyzed within 24 h of collection and after storage for 28 days in the refrigerator and at −80 °C. ANOVA for paired differences of fasting and postprandial samples from Experiment 2 shows that for lipids with > 1% class abundance, 4 TAGs were significantly higher 2 h after the meal, compared to the fasting state. These 4 TAGs contained (16:0), (18:1) or (18:2) FA groups and can be attributed to the fat intake (Fig. 6A). Lipids with no significant effect from fat intake, but positive change during storage were among those identified in Experiment 1: FFAs (18:0), (18:2), (20:3), and (20:4); LPCs (16:0), (18:0) and (18:1); and LPE(18:0) (Table S4). FFA(16:0) was the only lipid that seemed to be significantly affected both by fat intake (levels of lipase enzymes) and storage time (regardless of fat intake), although in contrasting ways. FFA(16:0) was negatively affected by the meal test, but positively affected by storage in the refrigerator (Fig. 6B). It is likely a fatty meal may induce more complex postprandial metabolic changes than the possible upregulation of lipase enzymes. In this sense, more controlled experiments are necessary. Nonetheless, the example of FFA(16:0) demonstrates that changes during storage may have led to opposite conclusions about the effect of fat intake.
Fig. 6.
Significant species from Experiment 2, by ANOVA analysis of matched fasting (pre-meal) and postprandial (post-meal) samples, comparing freshly drawn plasma (Fresh) to aliquots stored at two different temperatures (Refrigerator, 4 °C and Freezer, −80 °C) for 28 days. (A) Significant TAGs absolute concentrations in nmol/mL. (B) Significant FFAs absolute concentrations in nmol/mL.
Limitations and Outlook
There are limitations worth noting in experimental design. Although we have implied likely isobaric overlaps between oxidized and non-oxidized fatty acid species, for example oxidized CE(18:2) and CE(20:0), the data does not provide direct proof that this is the case. In order to more precisely identify oxidized species, future experiments should employ appropriate internal standards combined with tandem high resolution mass spectrometry and liquid chromatography retention time. Another limitation is in the lack of kinetic enzyme data in our study. We discuss the action of several enzymes and their roles in lipid degradation, however we did not measure the enzymes’ concentrations or activity. MS-based absolute concentration measurements and/or kinetic activity assays for the lipases and oxidases mentioned could address this shortcoming. Additionally, our experiments extended over the course of multiple days. Additional timepoints might be of interest, including more frequent sampling earlier in the degradation process, especially for bench (22 °C) and incubator (37.5 °C), allowing for better assessment of the rates of change in other plausible laboratory work timeframes.
In this study we artificially enhanced degradation to highlight potential processes occurring during lipid analysis and storage conditions. Future studies should also address long term storage at sub-zero temperatures. Of note, our interest in degradation arose from our own experience with samples that had been purportedly stored at –80C for > 5 years. Upon lipidomic analysis, these samples showed clear evidence of degradation, particularly with the emergence of a CE(20:0) signal likely associated with the generation of oxidized CE(18:2) due to isobaric overlap at unit resolution. As lipidomic techniques continue to evolve, opening up new areas of analysis that can be applied to historical samples stored for long periods of time, the understanding of possible chemical alterations to lipids during storage is imperative.
Conclusions
While it is recognized that leaving samples in the refrigerator (or worse – on the bench, heated evaporator, etc.) for extended periods of time is poor practice, we wanted to include these conditions in the study to better understand sample degradation and to accelerate degradation pathways. These experiments shed light on analytical biases that might arise after sample storage even in −20 to −80 °C freezers, especially for months, years, and decades. We identified several DAGs, FFAs, and CEs that are usually expected to be at very low but quantifiable levels in plasma and serum samples, but the concentration of these specific species can rise several-fold due to endogenous lipase and oxidase activity in stored samples. DAG products most likely came from simultaneous degradation of numerous TAGs, and the observed FFAs were likely produced primarily from certain high abundance PCs. The CEs that showed significant increases were, in actuality, likely mass isobars of oxidation products produced from high concentration CEs, particularly CE(18:2). If these DAGs, FFAs, and CEs are found at relatively high levels, it should raise alarm about the applicability of the samples for lipidomics analysis. Ideally, during the specimen collection process, characterized aliquots should be stored along with the study sample sets. These characterized aliquots can be monitored over time for the list of the DAG, FFA, and CE species identified in our experiments. Alternatively, when such characterized aliquots are not available, concentrations of these suggested species can be expressed in terms of ratios relative to SMs, since SMs are least susceptible to enzymatic degradation and oxidation. For example, the concentration or signal ratio relative to SMs for the suggested DAGs, FFAs, and CEs may significantly correlate with collection date, collection site, etc. If these correlations do exist, then preanalytical conditions may have been poorly controlled, and, therefore, concerns should be raised about possible biases due to sample handling.
Disclaimer
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention. Use of trade names and commercial sources is for identification only and does not constitute endorsement by the U.S. Department of Health and Human Services, or the U.S. Centers for Disease Control and Prevention.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmsacl.2021.10.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Ellervik C., Vaught J. Preanalytical Variables Affecting the Integrity of Human Biospecimens in Biobanking. Clin. Chem. 2015;61(7):914–934. doi: 10.1373/clinchem.2014.228783. [DOI] [PubMed] [Google Scholar]
- 2.Zivkovic A.M., Wiest M.M., Nguyen U.T., Davis R., Watkins S.M., German J.B. Effects of sample handling and storage on quantitative lipid analysis in human serum. Metabolomics. 2009;5(4):507–516. doi: 10.1007/s11306-009-0174-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haid M., Muschet C., Wahl S., Römisch-Margl W., Prehn C., Möller G., Adamski J. Long-Term Stability of Human Plasma Metabolites during Storage at −80 °C. J. Proteome Res. 2018;17(1):203–211. doi: 10.1021/acs.jproteome.7b00518. [DOI] [PubMed] [Google Scholar]
- 4.Wagner-Golbs A., Neuber S., Kamlage B., Christiansen N., Bethan B., Rennefahrt U., Schatz P., Lind L. Effects of Long-Term Storage at −80 °C on the Human Plasma Metabolome. Metabolites. 2019;9(5):99. doi: 10.3390/metabo9050099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Canadian journal of biochemistry and physiology. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 6.Bowden J.A., Heckert A., Ulmer C.Z., Jones C.M., Koelmel J.P., Abdullah L., Ahonen L., Alnouti Y., Armando A.M., Asara J.M., Bamba T., Barr J.R., Bergquist J., Borchers C.H., Brandsma J., Breitkopf S.B., Cajka T., Cazenave-Gassiot A., Checa A., Cinel M.A., Colas R.A., Cremers S., Dennis E.A., Evans J.E., Fauland A., Fiehn O., Gardner M.S., Garrett T.J., Gotlinger K.H., Han J., Huang Y., Neo A.H., Hyötyläinen T., Izumi Y., Jiang H., Jiang H., Jiang J., Kachman M., Kiyonami R., Klavins K., Klose C., Köfeler H.C., Kolmert J., Koal T., Koster G., Kuklenyik Z., Kurland I.J., Leadley M., Lin K., Maddipati K.R., McDougall D., Meikle P.J., Mellett N.A., Monnin C., Moseley M.A., Nandakumar R., Oresic M., Patterson R., Peake D., Pierce J.S., Post M., Postle A.D., Pugh R., Qiu Y., Quehenberger O., Ramrup P., Rees J., Rembiesa B., Reynaud D., Roth M.R., Sales S., Schuhmann K., Schwartzman M.L., Serhan C.N., Shevchenko A., Somerville S.E., St John-Williams L., Surma M.A., Takeda H., Thakare R., Thompson J.W., Torta F., Triebl A., Trötzmüller M., Ubhayasekera S.J.K., Vuckovic D., Weir J.M., Welti R., Wenk M.R., Wheelock C.E., Yao L., Yuan M., Zhao X.H., Zhou S. Harmonizing lipidomics: NIST interlaboratory comparison exercise for lipidomics using SRM 1950-Metabolites in frozen human plasma. J. Lipid Res. 2017;58(12):2275–2288. doi: 10.1194/jlr.M079012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fagone P., Jackowski S. Membrane phospholipid synthesis and endoplasmic reticulum function. J. Lipid Res. 2009;50(SUPPL.):S311–S316. doi: 10.1194/jlr.R800049-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardner M.S., Kuklenyik Z., Lehtikoski A., Carter K.A., McWilliams L.G., Kusovschi J., Bierbaum K., Jones J.I., Rees J., Reis G., Pirkle J.L., Barr J.R. Development and application of a high throughput one-pot extraction protocol for quantitative LC-MS/MS analysis of phospholipids in serum and lipoprotein fractions in normolipidemic and dyslipidemic subjects. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2019;1118-1119:137–147. doi: 10.1016/j.jchromb.2019.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burla B., Arita M., Arita M., Bendt A.K., Cazenave-Gassiot A., Dennis E.A., Ekroos K., Han X., Ikeda K., Liebisch G., Lin M.K., Loh T.P., Meikle P.J., Orešič M., Quehenberger O., Shevchenko A., Torta F., Wakelam M.J.O., Wheelock C.E., Wenk M.R. MS-based lipidomics of human blood plasma: A community-initiated position paper to develop accepted guidelines. J. Lipid Res. 2018;59(10):2001–2017. doi: 10.1194/jlr.S087163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hustad S., Eussen S., Midttun Ø., Ulvik A., Van De Kant P.M., Mørkrid L., Gislefoss R., Ueland P.M. Kinetic modeling of storage effects on biomarkers related to B vitamin status and one-carbon metabolism. Clin. Chem. 2012;58(2):402–410. doi: 10.1373/clinchem.2011.174490. [DOI] [PubMed] [Google Scholar]
- 11.Law S.-H., Chan M.-L., Marathe G.K., Parveen F., Chen C.-H., Ke L.-Y. An Updated Review of Lysophosphatidylcholine Metabolism in Human Diseases. Int. J. Mol. Sci. 2019;20(5):1149. doi: 10.3390/ijms20051149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anton G., Wilson R., Yu Z.-H., Prehn C., Zukunft S., Adamski J., Heier M., Meisinger C., Römisch-Margl W., Wang-Sattler R., Hveem K., Wolfenbuttel B., Peters A., Kastenmüller G., Waldenberger M., Kim K.H. Pre-Analytical Sample Quality: Metabolite Ratios as an Intrinsic Marker for Prolonged Room Temperature Exposure of Serum Samples. PLoS ONE. 2015;10(3):e0121495. doi: 10.1371/journal.pone.0121495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kühn H., Barnett J., Grunberger D., Baecker P., Chow J., Nguyen B., Bursztyn-Pettegrew H., Chan H., Sigal E. Overexpression, purification and characterization of human recombinant 15-lipoxygenase, Biochimica et Biophysica Acta (BBA)/Lipids and Lipid. Metabolism. 1993;1169(1):80–89. doi: 10.1016/0005-2760(93)90085-n. [DOI] [PubMed] [Google Scholar]
- 14.Belkner J., Stender H., Kühn H. The rabbit 15-lipoxygenase preferentially oxygenates LDL cholesterol esters, and this reaction does not require vitamin E. J. Biol. Chem. 1998;273(36):23225–23232. doi: 10.1074/jbc.273.36.23225. [DOI] [PubMed] [Google Scholar]
- 15.Mei G., Di Venere A., Nicolai E., Angelucci C.B., Ivanov I., Sabatucci A., Dainese E., Kuhn H., Maccarrone M. Structural properties of plant and mammalian lipoxygenases, Temperature-dependent conformational alterations and membrane binding ability. Biochemistry. 2008;47(35):9234–9242. doi: 10.1021/bi800638v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.